An enzyme of the host legume Medicago truncatula exclusively cleaves nodulation signals (Nod factors) from the microsymbiont Sinorhizobium meliloti but lacks activity toward chitin or nonmodified chitin oligosaccharides.
Abstract
Nodule formation induced by nitrogen-fixing rhizobia depends on bacterial nodulation factors (NFs), modified chitin oligosaccharides with a fatty acid moiety. Certain NFs can be cleaved and inactivated by plant chitinases. However, the most abundant NF of Sinorhizobium meliloti, an O-acetylated and sulfated tetramer, is resistant to hydrolysis by all plant chitinases tested so far. Nevertheless, this NF is rapidly degraded in the host rhizosphere. Here, we identify and characterize MtNFH1 (for Medicago truncatula Nod factor hydrolase 1), a legume enzyme structurally related to defense-related class V chitinases (glycoside hydrolase family 18). MtNFH1 lacks chitinase activity but efficiently hydrolyzes all tested NFs of S. meliloti. The enzyme shows a high cleavage preference, releasing exclusively lipodisaccharides from NFs. Substrate specificity and kinetic properties of MtNFH1 were compared with those of class V chitinases from Arabidopsis (Arabidopsis thaliana) and tobacco (Nicotiana tabacum), which cannot hydrolyze tetrameric NFs of S. meliloti. The Michaelis-Menten constants of MtNFH1 for NFs are in the micromolar concentration range, whereas nonmodified chitin oligosaccharides represent neither substrates nor inhibitors for MtNFH1. The three-dimensional structure of MtNFH1 was modeled on the basis of the known structure of class V chitinases. Docking simulation of NFs to MtNFH1 predicted a distinct binding cleft for the fatty acid moiety, which is absent in the class V chitinases. Point mutation analysis confirmed the modeled NF-MtNFH1 interaction. Silencing of MtNFH1 by RNA interference resulted in reduced NF degradation in the rhizosphere of M. truncatula. In conclusion, we have found a novel legume hydrolase that specifically inactivates NFs.
Nitrogen-fixing bacteria (rhizobia) establish a root nodule symbiosis with legume plants. Rhizobial infection and subsequent nodule formation depend on bacterial signaling, namely nodulation factors (NFs), which are perceived by specific LysM receptor kinases of the host plant (Antolín-Llovera et al., 2012). NFs are lipochitooligosaccharides (i.e. modified chitin oligosaccharides with a fatty acyl moiety replacing the N-acetyl group at the nonreducing end). In most cases, the oligosaccharide moiety of NFs consists of four or five β-1,4-linked GlcNAc residues. Depending on the rhizobial strain, NFs possess diverse additional substitutions on the oligosaccharide backbone (Perret et al., 2000). Such structural modifications of NFs influence the binding of NFs to the LysM receptor kinases. Hence, NF signaling in host plants culminating in the subsequent expression of symbiosis-specific genes and nodule formation is only triggered by specific NFs (Perret et al., 2000; Antolín-Llovera et al., 2012).
Due to structural similarities to chitin, various NFs can be hydrolyzed by plant chitinases (Staehelin et al., 1994a, 1994b; Goormachtig et al., 1998; Minic et al., 1998; Schultze et al., 1998; Ovtsyna et al., 2000). Chitinases (EC 3.2.1.14) are defined as enzymes that cleave β-1,4 glycosidic linkages in chitin (poly-GlcNAc) and chitin oligosaccharides (Collinge et al., 1993). Chitin is a major polysaccharide in fungal cell walls and exoskeletons of arthropods. Plants do not possess chitin but use chitinases as defense proteins against chitin-containing antagonists, such as fungi and arthropods. Various plant chitinases possess additional lysozyme activity (i.e. they can cleave β-1,4-linkages between N-acetylmuramic acid and GlcNAc in bacterial peptidoglycan; Collinge et al., 1993). Furthermore, certain plant chitinases appear to hydrolyze endogenous substrates such as GlcNAc-containing arabinogalactan proteins (De Jong et al., 1992; van Hengel et al., 2001). Based on their amino acid sequence, chitinolytic enzymes of plants belong either to glycoside hydrolase family 18 (class III and V chitinases) or family 19 (class I, II, and IV chitinases; Henrissat, 1991).
Class V chitinases of plants, also classified as pathogen-related protein family 11 (PR-11) proteins, form a separate clade within the phylogenetic tree of family 18 glycoside hydrolases (Karlsson and Stenlid, 2009). A phylogenetic tree of class V chitinases and closely related proteins is shown Supplemental Figure S1. In contrast to other plant chitinase classes, only a few class V proteins have been characterized. Nearly two decades ago, the first class V chitinase (NtChiV; also named Pz chitinase) was purified from tobacco mosaic virus-inoculated leaves of tobacco (Nicotiana tabacum). This defense-related enzyme efficiently cleaves chitin oligosaccharides (Heitz et al., 1994; Melchers et al., 1994; Brunner et al., 1998). Recently, enzymatic characterization and protein crystal structure analysis were performed with the NtChiV protein expressed in Escherichia coli as well as with AtChiC, a related class V chitinase from Arabidopsis (Arabidopsis thaliana; Ohnuma et al., 2011a, 2011b). Furthermore, a class V chitinase purified from the gymnosperm Cycas revoluta exhibits transglycosylation activity at high concentrations of chitin oligosaccharides (Taira et al., 2009, 2010). In addition to these enzymes, a class V chitinase-related lectin (RobpsCRA) from the legume tree Robinia pseudoacacia has been characterized. This protein binds to high-Man N-glycans but lacks catalytic activity (Van Damme et al., 2007). Similarly, the extracellular domain of the chitinase-related receptor-like kinase 1 of tobacco is related to class V chitinases (Kim et al., 2000).
In nodule symbiosis, host chitinases have the potential to inactivate NFs. When tested in root hair deformation bioassays, the purified cleavage products are at least 1,000-fold less active (Heidstra et al., 1994; Staehelin et al., 1994b). In fact, NFs produced by compatible rhizobia were found to be rapidly degraded in the rhizosphere of legumes (Heidstra et al., 1994; Staehelin et al., 1994b, 1995; Ovtsyna et al., 2000, 2005). NFs with a shorter oligosaccharide backbone and carrying additional chemical substitutions are usually more resistant against degradation by chitinases (Staehelin et al., 1994a, 1994b; Minic et al., 1998; Schultze et al., 1998; Ovtsyna et al., 2000). NodSm-IV(C16:2, Ac, S), an O-acetylated and sulfated tetrasaccharide with a C16:2 fatty acid chain (Lerouge et al., 1990), is the most abundant NF produced by Sinorhizobium meliloti (Supplemental Fig. S2). This NF is resistant to hydrolysis by all plant chitinases tested so far (Schultze et al., 1998). Nevertheless, when added to roots of the host plant alfalfa (Medicago sativa), NodSm-IV(C16:2, Ac, S) was rapidly degraded to a lipodisaccharide, and increased NF hydrolysis was observed when roots were pretreated with NFs (Staehelin et al., 1995). NFs were also found to induce an NF-cleaving activity in the rhizosphere of pea (Pisum sativum) plants, and this induction did not happen in NF signaling-deficient pea mutants (Ovtsyna et al., 2000, 2005). Similarly, chitinolytic activities of certain chitinase isoforms of legumes are induced during symbiosis with rhizobia or in response to purified NFs (Staehelin et al., 1992; Xie et al., 1999; Ovtsyna et al., 2005). Furthermore, transcript levels of the class III chitinase gene Srchi13 in the tropical legume Sesbania rostrata are strongly stimulated during symbiosis, and the encoded protein expressed in E. coli can degrade NFs of Azorhizobium caulinodans to nonidentified cleavage products (Goormachtig et al., 1998). In the model legume Medicago truncatula, expression of the class V chitinase gene MtChit5 is induced during symbiosis with S. meliloti (Salzer et al., 2004). Taken together, these findings raise the question of whether legumes possess chitinase-related enzymes that specifically cleave NFs.
In this study, we show that the MtChit5 protein of M. truncatula lacks chitinase activity but efficiently hydrolyzes NFs of S. meliloti. Therefore, we renamed MtChit5 to MtNFH1 (for M. truncatula Nod factor hydrolase1). A substrate-binding model, supported by point mutation analysis, provides a molecular explanation for the identified MtNFH1-NF interaction.
RESULTS
MtNFH1 Encodes an NF-Cleaving Enzyme
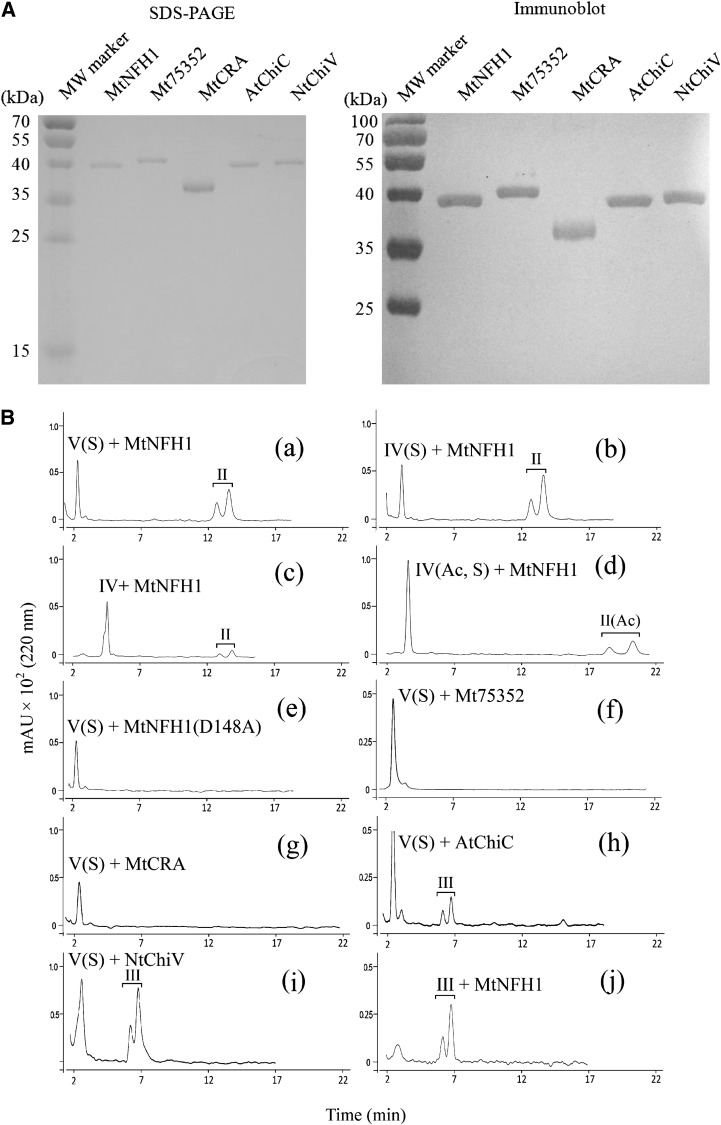
In previous work, we identified a symbiosis-related chitinase V gene, MtChit5, in M. truncatula ecotype R108-1. The expression of this gene is strongly induced in nodule symbiosis (Salzer et al., 2004). What is the catalytic activity of the enzyme encoded by this gene? To answer this question, the corresponding DNA was cloned into vector pET28b in order to express it as a His-tagged recombinant protein in E. coli BL21 (DE3). Two related genes of M. truncatula (Mt75352 and MtCRA) known to be expressed in M. truncatula (Supplemental Table S1; Supplemental Fig. S1) were also cloned and expressed in E. coli. The Mt75352 sequence encodes a putative class V chitinase, whereas MtCRA is related to the previously characterized lectin RobpsCRA of R. pseudoacacia (Van Damme et al., 2007). For comparison, the class V chitinases AtChiC of Arabidopsis (Ohnuma et al., 2011a) and NtChiV of tobacco (Ohnuma et al., 2011b) were cloned and expressed in a similar way. AtChiC and NtChiV show significant amino acid sequence homology to MtChit5 (sequence identity, 42% and 39%, respectively). The recombinant proteins were isolated by nickel affinity purification and analyzed by SDS-PAGE. Rabbit antiserum raised against the recombinant MtChit5 (renamed MtNFH1; see below) was cross-reactive with the other four proteins, indicating successful purification of the recombinant proteins (Fig. 1A). Purified proteins were then used for enzyme assays with pentameric and tetrameric NFs (Supplemental Fig. S2). These substrates had been HPLC purified from S. meliloti strain 1021 (pEK327) (Schultze et al., 1992). We found that the purified product of the MtChit5 gene rapidly hydrolyzed all the NFs (Table I) and therefore renamed it MtNFH1. Representative HPLC results with NF substrates and acylated cleavage products (lipooligosaccharides after n-butanol extraction) are shown in Figure 1B. The lipodisaccharide NodSm-II(C16:2) was formed when either pentameric NodSm-V(C16:2, S) or tetrameric NodSm-IV(C16:2, S) was used as a substrate (Fig. 1B, a and b). NodSm-IV(C16:2), a chemically desulfated NF, was also hydrolyzed by MtNFH1, albeit to a lesser extent (Fig. 1B, c). Interestingly, MtNFH1 was also able to cleave the O-acetylated tetramer NodSm-IV(C16:2, Ac, S); the more slowly migrating O-acetylated lipodisaccharide NodSm-II(C16:2, Ac) was detected by HPLC (Fig. 1B, d). Fractions containing lipodisaccharides were collected, and the chemical structures of lipodisaccharides were verified by mass spectrometry (Supplemental Fig. S3; Supplemental Table S2).
Figure 1.
Purification of recombinant proteins and activity test with NF substrates. A, SDS-PAGE analysis (left) and immunoblot detection (right) of His-tagged MtNFH1, Mt75352, MtCRA, AtChiC, and NtChiV expressed in E. coli BL21 (DE3). Purified proteins (1 μg) were analyzed on SDS-PAGE gels stained with Coomassie Brilliant Blue R-250. Immunoblot analysis was performed with a rabbit serum raised against MtNFH1 and the 3,3′-diamino-benzidine reagent. MW, Molecular mass. B, Separation of purified NFs and acylated cleavage products on a Nova Pak C18 column. The NF substrates were incubated with the indicated proteins at 37°C, extracted with n-butanol, and subjected to reverse-phase HPLC analysis. The cleavage products were separated into anomers (double peaks). Chromatograms are as follows: MtNFH1 (0.9 μg mL−1) incubated with 40 μm NodSm-V(C16:2, S) in 0.1 mL for 20 min (a), with 40 μm NodSm-IV(C16:2, S) in 0.1 mL for 40 min (b), with 20 μm desulfated NodSm-IV(C16:2) in 0.1 mL for 120 min (c), with 60 μm NodSm-IV(C16:2, Ac, S) in 0.1 mL for 40 min (d); the mutant protein MtNFH1(D148A) (0.9 μg mL−1) incubated with 20 μm NodSm-V(C16:2, S) in 0.1 mL for 4 h (e); Mt75352 (0.9 μg mL−1) incubated with 20 μm NodSm-V(C16:2, S) in 0.1 mL for 4 h (f); MtCRA (3 μg mL−1) incubated with 20 μm NodSm-V(C16:2, S) in 0.1 mL for 4 h (g); AtChiC (3 μg mL−1) incubated with 40 μm NodSm-V(C16:2, S) in 0.1 mL for 2 h (h); NtChiV (3 μg mL−1) incubated with 40 μm NodSm-V(C16:2, S) in 0.1 mL for 2 h (i); and MtNFH1 (0.9 μg mL−1) incubated with 20 μm NodSm-III(C16:2, S) in 0.1 mL for 4 h (j); II, NodSm-II(C16:2); II(Ac), NodSm-II(C16:2, Ac); III, NodSm-III(C16:2); IV, NodSm-IV(C16:2); IV(S), NodSm-IV(C16:2, S); IV(Ac, S), NodSm-IV(C16:2, Ac, S); V(S), NodSm-V(C16:2, S).
Table I. Activities of MtNFH1, AtChiC, and NtChiV with the indicated substrates.
| Substrate | Enzyme Activitya |
Assay | ||
|---|---|---|---|---|
| MtNFH1 | AtChiC | NtChiV | ||
| nkat mg−1 | ||||
| NodSm-V(C16:2, S) | 156.5 ± 8.1 | 0.37 ± 0.06 | 1.27 ± 0.23 | Reverse-phase HPLC analysis (Nova Pak C18 column)b |
| NodSm-IV(C16:2, S) | 121.8 ± 21.7 | NDc | ND | |
| NodSm-IV(C16:2, Ac, S) | 46.5 ± 4.2 | ND | ND | |
| NodSm-III(C16:2) | ND | ND | ND | |
| (GlcNAc)6 | ND | 393.1 ± 26.8 | 205.9 ± 29.8 | Reverse-phase HPLC analysis (amino column)d |
| (GlcNAc)5 | ND | 266.0 ± 19.9 | 171.1 ± 19.0 | |
| Colloidal chitin | ND | 0.020 ± 0.004 | 0.012 ± 0.001 | Ehrlich's reagent |
| Glycolchitin | ND | 13.49 ± 3.32 | 7.66 ± 0.22 | Lever assay |
| CM-chitin-RBV | ND | 0.83 ± 0.22e | 1.33 ± 0.03 | Colorimetric assay |
| M. lysodeikticus cells | ND | ND | ND | Lysozyme assay |
aData are shown for the indicated 6×His-tagged recombinant proteins. Proteolytic removal of the tag with factor Xa did not affect the enzyme activity of MtNFH1 as determined with NodSm-IV(C16:2, S). Mt75352 and MtCRA lacked enzymatic activity when tested either with NFs or chitinous substrates. Activities were determined at 37°C with a substrate concentration of 150 μm for NFs from S. meliloti or the lipotrisaccharide NodSm-III(C16:2), 3.6 mm for (GlcNAc)6, 4.5 mm for (GlcNAc)5, approximately 10 mg mL−1 for colloidal chitin, 20 mg mL−1 for glycolchitin, 0.9 mg mL−1 for CM-chitin-RBV, and 0.45 mg mL−1 for M. lysodeikticus cells. Data indicate means ± sd from at least three independently purified enzyme preparations. bFormation of the lipodisaccharide NodSm-II(C16:2) or NodSm-II(C16:2, Ac) by MtNFH1 and formation of the lipotrisaccharide NodSm-III(C16:2) by AtChiC or NtChiV. cND, Not detected. dAtChiC and NtChiV degraded (GlcNAc)6 into (GlcNAc)3 or (GlcNAc)2 and (GlcNAc)4; (GlcNAc)4 was further degraded into (GlcNAc)2; (GlcNAc)5 was cleaved into (GlcNAc)3 and (GlcNAc)2. eEnzyme activity expressed as ΔA550 mg−1 protein s−1.
MtNFH1(D148A), a mutant protein in which the conserved Asp residue Asp-148 of the predicted catalytic center was changed to Ala, did not show enzyme activity (Fig. 1B, e). Mt75352 and MtCRA, proteins of M. truncatula related to MtNFH1, were unable to hydrolyze the NF substrates (Fig. 1B, f and g). AtChiC of Arabidopsis hydrolyzed pentameric NodSm-V(C16:2, S), but the cleavage products were different: the more rapidly migrating lipotrisaccharide NodSm-III(C16:2) was formed (Fig. 1B, h). Similarly, the tobacco enzyme NtChiV released NodSm-III(C16:2) from the pentameric substrate (Fig. 1B, i), as reported previously (Schultze et al., 1998). The structure of the product NodSm-III(C16:2) was confirmed by mass spectrometry (Supplemental Fig. S3; Supplemental Table S2). As compared with MtNFH1, hydrolytic activities of AtChiC and NtChiV for the substrate NodSm-V(C16:2, S) were more than 100-fold lower (Table I). Tetrameric NFs were neither cleaved by AtChiC nor by NtChiV (Schultze et al., 1998). MtNFH1 was unable to hydrolyze purified NodSm-III(C16:2), providing evidence that the enzyme directly releases lipodisaccharides from the NF substrates (Fig. 1B, j).
MtNFH1 Specifically Cleaves NFs
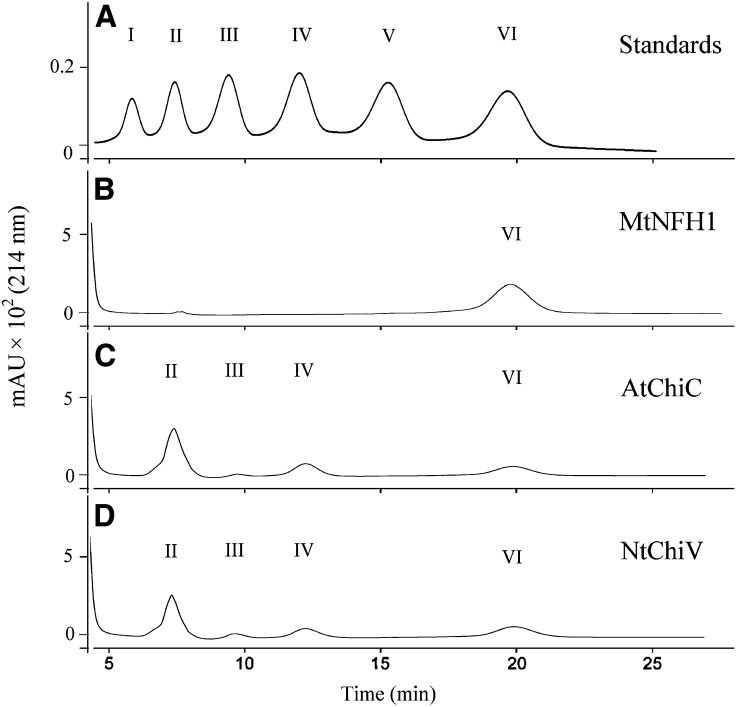
We further tested whether MtNFH1 can hydrolyze nonmodified chitin oligosaccharides (oligomers of GlcNAc), which likely represent the natural substrates for AtChiC and NtChiV. The hexamer (GlcNAc)6 and the pentamer (GlcNAc)5 were used in enzyme assays with purified proteins. After incubation, chitin oligosaccharides were separated on an amino column. Surprisingly, MtNFH1 protein samples, which had shown such a high activity with NF substrates, completely lacked activity with these nonmodified chitin oligosaccharides (Table I; Fig. 2). To test a possible inhibitory effect of chitin oligosaccharides on NF degradation, MtNFH1 was incubated with NodSm-V(C16:2, S) and a 500-fold higher concentration of (GlcNAc)5. NFs and acylated degradation products were then analyzed by HPLC. Formation of NodSm-II(C16:2) by MtNFH1 was not affected by (GlcNAc)5, as shown in Supplemental Figure S4.
Figure 2.
MtNFH1 does not hydrolyze nonmodified chitin oligosaccharides. Examples of HPLC chromatograms showing separation of the substrate (GlcNAc)6 and cleavage products are presented. After incubation (37°C) with the indicated proteins, reaction mixtures (in 25 mm sodium acetate buffer, pH 5.0; 30-μL test volume) were diluted with an equal volume of 50% acetonitrile and directly loaded onto the amino column. A, (GlcNAc)n standards (n = 1–6; 10 nmol). B, (GlcNAc)6 (5.8 mm) incubated with MtNFH1 (4.5 μg mL−1) for 3 h. C, (GlcNAc)6 (5.8 mm) incubated with AtChiC (5.0 μg mL−1) for 30 min. D, (GlcNAc)6 (5.8 mm) incubated with NtChiV (5.0 μg mL−1) for 30 min. I, GlcNAc; II, (GlcNAc)2; III, (GlcNAc)3; IV, (GlcNAc)4; V, (GlcNAc)5; VI, (GlcNAc)6; mAU, milliabsorbance units.
In contrast to MtNFH1, purified AtChiC and NtChiV proteins efficiently hydrolyzed nonmodified chitin oligosaccharides in accordance with previous reports (Brunner et al., 1998; Ohnuma et al., 2011a, 2011b). (GlcNAc)6 was cleaved into (GlcNAc)3 or into (GlcNAc)4 and (GlcNAc)2. The product (GlcNAc)4 was then further hydrolyzed into (GlcNAc)2. The substrate (GlcNAc)5 was cleaved into (GlcNAc)3 and (GlcNAc)2 (Table I; Fig. 2).
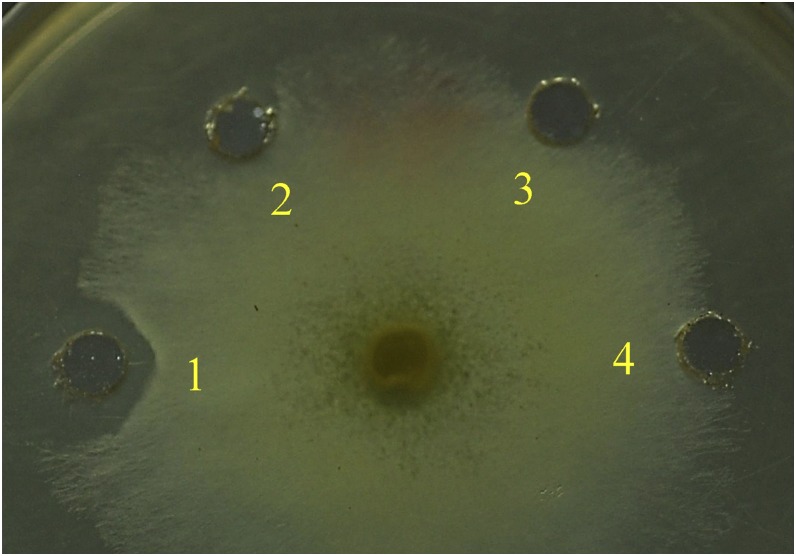
When colloidal chitin, glycolchitin, and carboxymethyl-chitin-Remazol Brilliant Violet 5R (CM-chitin-RBV) were tested as substrates, AtChiC and NtChiV clearly showed enzymatic activity, while MtNFH1 was completely inactive (Table I). The Mt75352 and MtCRA proteins were similarly inactive. MtNFH1, like AtChiC and NtChiV, lacked lysozyme activity as measured with Micrococcus lysodeikticus cells (Table I), which is in agreement with a previous report on NtChiV (Heitz et al., 1994). NtChiV inhibited hyphal growth of the fungus Trichoderma viride (Melchers et al., 1994; Ohnuma et al., 2012), and this was confirmed in a similar bioassay in this study. The MtNFH1 protein lacked such antifungal activity, however (Fig. 3).
Figure 3.
MtNFH1 does not inhibit the growth of the fungus T. viride. Purified proteins (3.2 μg in 50 μL) of NtChiV (1), AtChiC (2), or MtNFH1 (3) were pipetted into peripheral wells close to a growing mycelium of T. viride (inoculation at the center of the agar plate). The sodium acetate buffer (25 mm, pH 5.0) without enzyme was added as a control (4). The plate shown (demonstrating growth inhibition activity for NtChiV) was photographed after an incubation for 14 h at 27°C.
Kinetic Studies Indicate Efficient NF Hydrolysis at Low Substrate Concentrations
Kinetic studies with varying concentrations of NFs were performed to determine Km and catalytic rate constant (kcat) values of the purified proteins (Table II; Supplemental Fig. S5). MtNFH1 showed Km values at micromolar concentrations with all tested NFs of S. meliloti. A slightly higher Km and a lower kcat value were determined for the O-acetylated substrate NodSm-IV(C16:2, Ac, S) as compared with NFs without an O-acetyl group. Similarly, low Km values were also obtained for AtChiC and NtChiV releasing NodSm-III(C16:2) from NodSm-V(C16:2, S). As compared with MtNFH1, however, only low kcat values were determined.
Table II. Km and kcat values for MtNFH1, AtChiC, and NtChiV with NFs as substrates.
| Substrate | MtNFH1 |
AtChiC |
NtChiV |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Km | kcat | kcat Km−1 | Km | kcat | kcat Km−1 | Km | kcat | kcat Km−1 | |
| μm | s−1 | mm−1 s−1 | μm | s−1 | mm−1 s−1 | μm | s−1 | mm−1 s−1 | |
| NodSm-V(C16:2, S) | 52.7 ± 2.3a | 8.4 ± 0.6 | 159.3 ± 17.6 | 80 ± 6.3 | 0.033 ± 0.001 | 0.41 ± 0.02 | 68 ± 6.4 | 0.088 ± 0.004 | 1.3 ± 0.2 |
| NodSm-IV(C16:2, S) | 60.1 ± 4.7 | 7.9 ± 0.2 | 132.5 ± 13.4 | NDb | ND | ND | ND | ND | ND |
| NodSm-IV(C16:2, Ac, S) | 97.3 ± 12.2 | 3.5 ± 0.1 | 36.4 ± 5.5 | ND | ND | ND | ND | ND | ND |
Data indicate means ± sd from enzyme tests (37°C) with three independently purified protein preparations. bND, No activity detected.
The results of kinetic studies with AtChiC and NtChiV for the substrates (GlcNAc)6 and (GlcNAc)5 are summarized in Supplemental Table S3. Relatively high Km values in the millimolar range were determined for these substrates. On the other hand, the kcat values for (GlcNAc)6 and (GlcNAc)5 were found to be more than 2 orders of magnitude higher as compared with those for NodSm-V(C16:2, S) shown in Table II.
Docking Simulation Predicts a Specific Binding Cleft for a Fatty Acid Chain
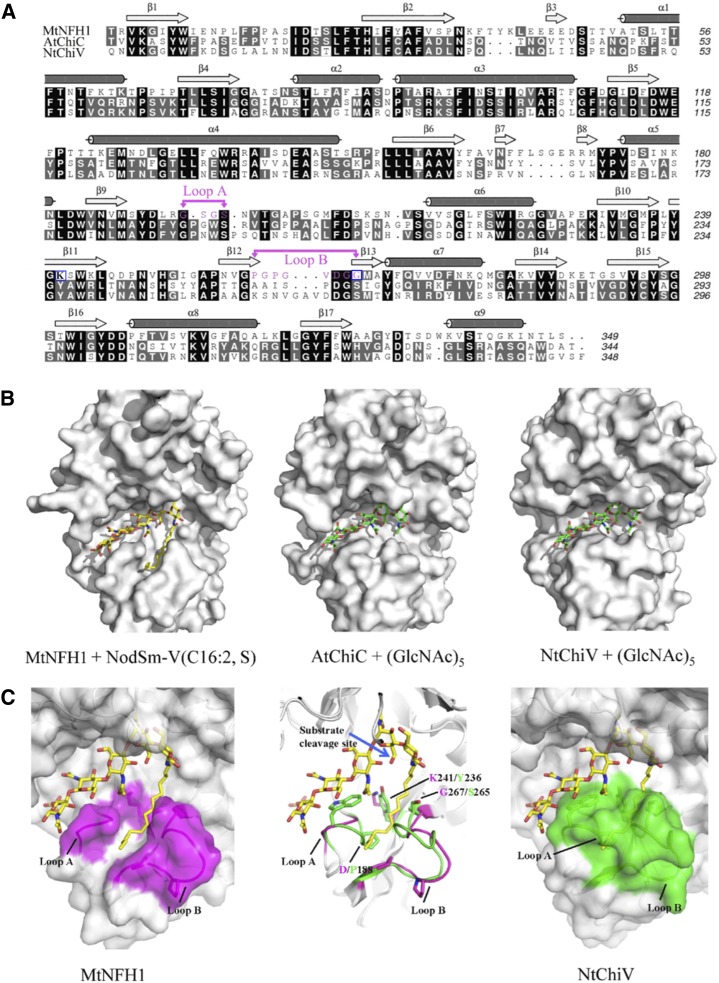
Since MtNFH1 cleaves NFs with their fatty acyl decorations, but not the simple chitin oligosaccharides, we wondered whether the fatty acid chain was required for correct substrate binding. To analyze substrate recognition on a molecular level, a three-dimensional model of MtNFH1 was constructed by homology modeling, using the crystal structures of AtChiC or NtChiV as structural templates. The structures of the substrates NodSm-V(C16:2, S) and (GlcNAC)5 were then manually docked to the active site of the proteins. Aligned primary sequences of the three proteins and the modeled substrate-protein complexes are shown in Figure 4. All three enzymes possess a predicted substrate-binding pocket for GlcNAc residues. Noteworthy, two loops in MtNFH1 (loops A and B) are predicted to contribute to the formation of the binding cleft for the C16:2 fatty acid moiety. Loop A consists of amino acid residues between strand 9 and helix 6 (GSGS motif), and loop B is formed by residues between strands 12 and 13 (GPGPGVDGG motif; Fig. 4A). In contrast to the AtChiC and NtChiV sequences, these two loops of MtNFH1 are shorter and are Gly rich, which results in a prominent cleft that can accommodate the C16:2 fatty acid moiety of NodSm-V(C16:2, S) (Fig. 4B). Based on our model, NF substrates perfectly fit to MtNFH1 in this way and are then cleaved into NodSm-II(C16:2) and a sulfated chitin oligosaccharide. Hence, MtNFH1 targets NFs at their nonreducing ends. On the other hand, there is apparently no space in AtChiC and NtChiV to accommodate the fatty acid moiety at this position (Fig. 4B), which corresponds to the observation that they cannot release NodSm-II(C16:2) from NodSm-V(C16:2, S).
Figure 4.
Homology modeling and docking simulation of NodSm-V(C16:2, S) to MtNFH1. A, Structure-based sequence alignment of MtNFH1 with AtChiC and NtChiV. Identical amino acid residues are shown on a black background, homologous residues on a gray background, and dashes indicate gaps; α-helices and β-strands are marked above the sequences. Amino acid residues of loops A and B in MtNFH1 are marked in magenta and the point-mutated residues K241 and G267 are framed in blue. B, Models of MtNFH1 with NodSm-V(C16:2, S), AtChiC with (GlcNAc)5, and NtChiV with (GlcNAc)5. C, MtNFH1 (magenta, left) has a prominent cleft between loops A and B that can bind the fatty acid moiety of NodSm-V(C16:2, S). NtChiV (green, right) and AtChiC lack such a binding cleft. The structures of MtNFH1 (magenta) and NtChiV (green) are superimposed to illustrate the expected conformational differences (middle). The cleavage site of NodSm-V(C16:2, S) hydrolyzed by MtNFH1 is indicated by a blue arrow.
A superimposed model of MtNFH1 and NtChiV indicates changes in sequence/structure that lead to the formation of the fatty acid-binding cleft (Fig. 4C). Specifically, the Tyr and Ser residues of NtChiV (Tyr-236 and Ser-265), and the corresponding residues in AtChiC are substituted in MtNFH1 by a Lys and Gly at the corresponding positions (Lys-241 and Gly-267). These two residues are located at the entrance of the cleft between loops A and B, and these sterically more bulky Tyr and Ser residues in AtChiC and NtChiV are predicted to block the formation of a cleft. Furthermore, NtChiV and AtChiC have an extra Pro residue (Pro-188 in NtChiV) that is absent in loop A of MtNFH1. The deletion of this Pro residue in MtNFH1 results in a shorter loop A and thus enlarges the predicted fatty acid-binding cleft.
Point Mutation Analysis Supports the Modeled NF-MtNFH1 Interaction
Our model predicts that Lys-241 and Gly-267 of MtNFH1, which guard the entrance of the fatty acid-binding cleft, should be important for substrate recognition. To test this hypothesis, we constructed a double mutant of MtNFH1 (K241Y/G267S) in which Lys-241 is substituted by Tyr and Gly-267 is substituted by Ser (corresponding to Tyr-236 and Ser-265 in NtChiV, respectively). When assayed with NFs as substrates, the MtNFH1(K241Y/G267S) protein showed approximately 2.5-fold increases in Km and approximately 2-fold decreases in kcat, which resulted in an approximately 5-fold reduced activity (kcat/Km; Table III). These data provide experimental support for the modeled NF-MtNFH1 interaction.
Table III. Differences of Km and kcat values of MtNFH1(K241Y/G267S) and wild-type MtNFH1.
| Protein | NodSm-V(C16:2, S) |
NodSm-IV(C16:2, S) |
||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat Km−1 | Km | kcat | kcat Km−1 | |
| % | ||||||
| MtNFH1 | 100a | 100 | 100 | 100 | 100 | 100 |
| MtNFH1(K241Y/G267S) | 271.7 ± 2.3b | 54.6 ± 0.7 | 19.9 ± 0.4 | 245 ± 1.7 | 51.6 ± 4.3 | 21 ± 1.7 |
For absolute values, see Table II. bData (relative values in relation to the wild-type protein; expressed in percentages) indicate means ± sd deduced from enzyme assays (37°C) using three independently purified protein preparations and various amounts of NodSm-V(C16:2, S) and NodSm-IV(C16:2, S), respectively.
Silencing of MtNFH1 by RNA Interference Increases NF Levels in the Rhizosphere
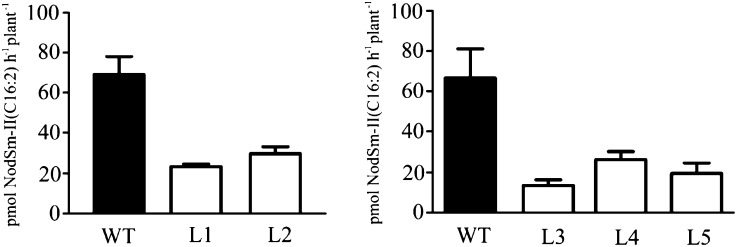
When adding NFs of S. meliloti to roots of alfalfa, they are rapidly degraded by an NF-cleaving extracellular activity (Staehelin et al., 1994b, 1995). To investigate whether MtNFH1 encodes an enzyme that contributes to NF degradation in the rhizosphere, we transformed M. truncatula ecotype R108-1 with Agrobacterium tumefaciens carrying pCAMBIA-MtNFH1i. This RNA interference (RNAi) plasmid contains a construct made of the cauliflower mosaic virus (CaMV) 35S promoter in tandem and a fragment of MtNFH1, which is inserted in both the sense and antisense directions. These sense and antisense sequences are separated by a spacer region. Furthermore, the vector carries a GUS gene under the control of the CaMV 35S promoter, which allows it to distinguish between transformed and nontransformed plants. Primary transformants were regenerated from A. tumefaciens-infiltrated leaf discs on selective medium containing hygromycin. Young seedlings from these RNAi lines (T2 generation) were first treated for 1 d with 0.1 μm NodSm-IV(C16:2, S) in order to stimulate the NF-cleaving activity in the rhizosphere. The NF degradation test was then performed by incubation in 15 μm NodSm-IV(C16:2, S). After removal of the seedlings, the NFs and the released NodSm-II(C16:2, S) were extracted from the medium by n-butanol and quantified by HPLC analysis. Finally, the seedlings were used for the determination of GUS activity, and plants lacking blue staining were excluded from the experiment. Compared with wild-type plants, an up to 5-fold reduction in NF hydrolysis was measured for plants transformed with pCAMBIA-MtNFH1i, indicating that silencing of MtNFH1 increases NF levels in the rhizosphere (Fig. 5).
Figure 5.
Hydrolysis of NodSm-IV(C16:2, S) by roots of M. truncatula seedlings transformed with A. tumefaciens carrying pCAMBIA-MtNFH1i. Roots of seedlings (T2 generation) from five independent RNAi lines (named L1–L5) and wild-type plants (WT) were individually pretreated with 0.1 μm NodSm-IV(C16:2, S) for 1 d and then incubated with 15 μm NodSm-IV(C16:2, S) for 18 h. The NF substrate and formed NodSm-II(C16:2) were extracted from the medium with n-butanol and separated by reverse-phase HPLC using a Nova Pak C18 column. Data indicate means ± sd from two representative test series (in total, 24 transgenic and seven wild-type plants). NF-cleaving activities of all five RNAi lines are significantly reduced as compared with wild-type plants (Kruskal-Wallis test; P < 0.03).
DISCUSSION
In this work, we have biochemically characterized MtNFH1, which is encoded by a symbiosis-stimulated M. truncatula gene (Salzer et al., 2004). The enzyme shows sequence similarities to class V chitinases but lacks chitinase activity. Instead, MtNFH1 cleaves NFs of the microsymbiont S. meliloti. Notably, MtNFH1 hydrolyzes NodSm-IV(C16:2, Ac, S), which is structurally rather different from nonmodified chitin oligosaccharides. Based on the capacity to release lipodisaccharides from NFs (this study) and the increase of MtNFH1 transcripts in NF-treated roots (Salzer et al., 2004), it is likely that MtNFH1 is an ortholog of the previously identified NF-cleaving enzyme of alfalfa, which could be partially purified from root exudates due to its concanavalin A-binding properties (Staehelin et al., 1995). It is likely that MtNFH1 is also a secreted N-glycosylated protein. This assumption is supported by the prediction of an N-terminal signal peptide and four potential N-glycoslation sites in MtNFH1 (Supplemental Table S1) as well as our finding that MtNFH1 RNAi-silenced plants show reduced NF-cleaving activity in the rhizosphere (Fig. 5).
After the initiation of NF signaling, MtNFH1 probably inactivates excess amounts of NFs, as has been suggested for a related hydrolase activity of alfalfa (Staehelin et al., 1994b, 1995), Srchi13 of S. rostrata (Goormachtig et al., 1998), and an unknown pea enzyme (Ovtsyna et al., 2000, 2005). Such a feedback response might control the degree of rhizobial invasion into host cells at different symbiotic stages. NFs can trigger responses on root hairs at nanomolar to picomolar concentrations. However, for developing infection threads and for initiating nodules, it has been proposed that higher NF levels that are present over a longer period of time are required (Perret et al., 2000). As the Km values of MtNFH1 for NFs are in the micromolar concentration range, we suggest that the substrate concentration for the enzyme is not saturated under natural conditions. Future work is required to examine whether NF levels in the rhizosphere have an impact on establishing symbiosis with S. meliloti. Because our RNAi lines show reduced NF-cleaving activity, we currently test them for their effects on nodule formation.
Most plants produce a large set of various chitinases, and this diversity may reflect the importance of these proteins during plant defense against chitinious pathogens, particularly fungi. However, the evolutionary driving force for this diversity remains unclear, and it has been proposed that plant chitinases are coevolving with chitinase inhibitors from pathogens (Rausher, 2001). Here, we show that MtNFH1 lacks chitinase activity and instead cleaves NFs of S. meliloti. This can be considered as a typical example of neofunctionalization (i.e. adaptation of an enzyme to a novel microbial substrate reminiscent to receptor-ligand coevolution). In fact, class V chitinases are present in ancient land plants, including the primitive gymnosperm C. revoluta (Taira et al., 2009, 2010). We suggest that MtNFH1 evolved from an ancestral defense-related chitinase and that gene duplication opened the possibility to develop a symbiotic enzyme that has the capacity to inactivate short and structurally modified NFs such as NodSm-IV(C16:2, Ac, S). In contrast to MtNFH1, recombinant MtCRA and Mt75352 were unable to hydrolyze NFs and may possess unknown nonsymbiotic functions. MtCRA is likely an ortholog of RobpsCRA, which is a nonchitinolytic lectin with binding preference for the pentasaccharide core structure of high-Man N-glycans (Van Damme et al., 2007). It remains to be seen whether MtCRA and perhaps MtNFH1 possess similar carbohydrate-binding activities.
NF receptors of legumes possess extracellular LysM domains, which strongly interact with NFs and only weakly with nonmodified chitin oligosaccharides (Broghammer et al., 2012). Similarly, the symbiotic ectoapyrase (lectin nucleotide phosphohydrolase) of the legume Dolichos biflorus shows a higher binding affinity for NFs than for chitin oligosaccharides, as determined by a competitive chitin-binding assay (Etzler et al., 1999). In this study, we found that MtNFH1 perfectly discriminates between NFs and chitin oligosaccharides. Accordingly, the Km values of MtNFH1 for NFs are in the micromolar concentration range, whereas chitin oligosaccharides are neither substrates nor inhibitors. In contrast, the class V chitinases AtChiC and NtChiV can only hydrolyze the pentameric NF, NodSm-V(C16:2, S). In comparison with MtNFH1, their activities degrading this NF are low due to low kcat values. Interestingly, as reflected by the low Km value for NodSm-V(C16:2, S), AtChiC and NtChiV show significantly higher affinities for this NF than for chitin oligosaccharides (Table II; Supplemental Table S3). These data suggest a further possible role for class V chitinases and related proteins, namely to cleave NF-like signals of mycorrhizal fungi. Roots of tobacco and M. truncatula, but not Arabidopsis, can establish symbiosis with arbuscular mycorrhizal fungi, and a set of fungal lipochitooligosaccharides have been identified recently (Maillet et al., 2011). However, MtNFH1 expression was found to be induced in S. meliloti-inoculated M. truncatula roots, whereas transcript levels remained low in mycorrhizal roots colonized by the fungus Glomus intraradices (Salzer et al., 2004).
Crystallization of AtChiC and NtChiV (Ohnuma et al., 2011a, 2011b) provided useful structural templates for homology modeling of the MtNFH1 protein structure. Docking of NFs to MtNFH1 resulted in a model in which the C16:2 fatty acid moiety fits to a specific binding cleft and apparently helps to bring the carbohydrate moiety in the catalytic pocket to a correct position. The point mutant MtNFH1(D148A) lacked enzyme activity (Fig. 1B, e), indicating that the canonical DXDXE motif of functional glycosyl hydrolase family 18 enzymes (Karlsson and Stenlid, 2009) is also critical for the hydrolysis of NFs. Furthermore, kinetic data of a MtNFH1 mutant protein, modified in the region of the predicted binding cleft for the fatty acid chain (Table III), provide support for the modeled MtNFH1-NF interaction. It is worth noting that the predicted torsion angle between the sugar moiety and the fatty acid chain in the MtNFH1-NF interaction is similar to the corresponding torsion angle in NF-receptor interaction models, in which NFs of S. meliloti were individually docked to each of the three LysM domains of a Nod factor perception receptor of M. truncatula (Mulder et al., 2006).
It is particularly intriguing that MtNFH1 completely lacks activity when incubated with nonmodified chitin oligosaccharides. As chitin oligosaccharides are smaller than NFs, they likely reach the catalytic center of MtNFH1 without steric hindrance. However, the predicted carbohydrate-binding pocket of MtNFH1 is bigger than that of AtChiC or NtChiV (Fig. 4B), and there is apparently less steric hindrance to fix the flexible chitin oligosaccharide chain into the correct orientation of the active site of MtNFH1. In other words, chitin oligosaccharides cannot adopt a correct conformation required for catalysis. In contrast, NFs with their C16:2 fatty acid moiety fully occupy the substrate-binding pocket in the correct orientation and exactly present the scissile bond to the active site residues of MtNFH1.
MATERIALS AND METHODS
Gene Cloning
DNA sequences encoding proteins without predicted signal peptides from Medicago truncatula ecotype R108-1 (MtNFH1, Mt75352, and MtCRA; Supplemental Table S1), Arabidopsis (Arabidopsis thaliana) ecotype Columbia (AtChiC; accession no. BT029539) and tobacco (Nicotiana tabacum ‘Xanthi’ [NtChiV]; accession no. X78325) were cloned into the expression vector pET28b (Novagen, Merck) and verified by sequencing. MtNFH1 without signal peptide sequence was also cloned into vector pET30 as described by the supplier of the LIC Cloning Kit (Novagen). PCR-based site-directed mutagenesis techniques were used for the introduction of point mutations into MtNFH1. Primers used and plasmids constructed are shown in Supplemental Tables S4 and S5, respectively.
To construct an RNAi plasmid for the silencing of MtNFH1 in M. truncatula, a 667-bp fragment of MtNFH1 (MtNFH1Δ1-94/Δ761-1152) was cloned into pBS-RNAi in the sense and antisense orientations (separated by a 360-bp spacer; Limpens et al., 2004). The primers and plasmid constructs are shown in Supplemental Tables S4 and S5, respectively. Finally, the entire 2,604-bp cassette [containing a double CaMV 35S promoter, the 667-bp fragment in the sense orientation, the spacer, the 667-bp fragment in the antisense orientation and a poly(A) terminator] was cloned into the binary vector pCAMBIA1305.1, which contains a GUS gene under the control of the CaMV 35S promoter (http://www.cambia.org/). The resulting RNAi plasmid, named pCAMBIA-MtNFH1i, was used for the transformation of Agrobacterium tumefaciens strain EHA105 by electroporation.
Phylogenetic Analysis
For the identification of related sequences, the amino acid sequence of MtNFH1 was used as a query sequence for a protein BLAST (BLASTP algorithm) database search (Altschul et al., 1990) at the National Center for Biotechnology Information home page (http://blast.ncbi.nlm.nih.gov/Blast.cgi). By selecting the protein database parameter “green plants,” a total of 91 related sequences were obtained. The Mt59366, Mt75352, and Mt59600 sequences were extracted from the M. truncatula Genome Project database (Mt3.5 CDS database) by using MtNFH1 as a query sequence in a nucleotide Blast (BLASTN algorithm) similarity search (http://blast.jcvi.org/er-blast/index.cgi?project=mtbe). The nucleotide sequences were then translated into predicted amino acid sequences using the splicing prediction software SplicePreditor (http://bioservices.usd.edu/splicepredictor/). The unrooted phylogenic tree was then constructed with the MEGA5 program using the neighbor-joining method (Tamura et al., 2011). The evolutionary distances were computed using the Poisson correction method.
Protein Expression and Purification
Escherichia coli BL21 (DE3) cells carrying the constructed plasmids were cultivated in Luria-Bertani medium, and recombinant protein expression was induced by 0.4 mm isopropyl-β-d-thiogalactopyranoside (18°C for 20 h). Proteins were purified by nickel affinity chromatography with nickel-nitrilotriacetic acid agarose resin beads based on the manufacturer’s protocol for protein purification under denaturing or native conditions (Qiagen). Where indicated, the His tag was removed from MtNFH1 [purified from E. coli BL21 (DE3) carrying pET30-MtNFH1] by using the protease factor Xa according to the supplier’s recommendations (New England Biolabs).
Protein Analysis and Antibodies
Purified proteins were separated by SDS-PAGE, and gels were stained with Commassie Brilliant Blue R-250. Protein contents of protein bands were then compared with bovine serum albumin standards. In addition, protein contents were photometrically measured according to the method of Bradford (1976). His-tagged MtNFH1 protein, purified under denaturing conditions according to the manufacturer’s protocol (Qiagen), was used for immunization of a New Zealand rabbit (provided by the Experimental Animal Centre of Sun Yat-sen University of Medical Science, Guangzhou, China). For immunoblot analysis, proteins were separated by SDS-PAGE and then electrophoretically transferred onto nitrocellulose membranes (Schleicher & Schuell BioScience). Membranes were incubated with the polyclonal rabbit antiserum against MtNFH1 (1:7,000 dilution) and goat anti-rabbit IgG antiserum coupled to horseradish peroxidase. Finally, blots were developed with 3,3′-diamino-benzidine according to the supplier’s recommendations (Boster).
Purification of NFs
NodSm-V(C16:2, S), NodSm-IV(C16:2, S), and NodSm-IV(C16:2, Ac, S) were purified from Sinorhizobium meliloti 1021 (pEK327) (Schultze et al., 1992). Bacteria were grown in modified glucose-Tris-sodium succinate medium (Kiss et al., 1979), and NFs were purified by HPLC according to previously described procedures (Schultze et al., 1992; Staehelin et al., 1994b, 1995). Briefly, culture supernatants (adjusted to pH 7.5 with HCl) were extracted with n-butanol and concentrated under reduced pressure. For the purification of NodSm-IV(C16:2, Ac, S), NFs were fractionated by reverse-phase HPLC (Nova Pak C18, 3.9 × 150 mm, particle size 4 μm; Waters) under isocratic conditions using 30% (v/v) acetonitrile/water containing 40 mm ammonium acetate as the mobile phase. NodSm-IV(C16:2, Ac, S), corresponding to a more slowly migrating peak, was collected and the sample dried under reduced pressure. For the purification of NodSm-V(C16:2, S) and NodSm-IV(C16:2, S), the NFs were first completely deacetylated (incubation in 50 mm Tris-HCl, pH 9.0, for 8 h) and then HPLC purified under the same conditions. Finally, NFs were desalted on a Polygosil C18 column (Staehelin et al., 2000). Desulfated NodSm-IV(C16:2) was obtained from NodSm-IV(C16:2, S) by mild acid hydrolysis in methanol-HCI followed by HPLC purification (Demont et al., 1993).
Enzyme Assays with NFs
Purified proteins and NFs at specified amounts were incubated in 25 mm sodium acetate buffer (pH 5.0) at 37°C. Nondegraded NFs and the acylated cleavage products (lipodisaccharides and lipotrisaccharides) were extracted with an equal volume of double-distilled n-butanol and dried in a Speed-Vac evaporator. Samples were taken up in 1 μL of dimethyl sulfoxide (DMSO) and analyzed by reverse-phase HPLC (Nova Pak C18; Waters) using 36% (v/v) acetonitrile/water containing 40 mm ammonium acetate as the mobile phase (Staehelin et al., 1994b). HPLC-purified cleavage products released from NFs were analyzed by mass spectrometry. For measurement of activities and determination of kinetic parameters, the incubation times varied to ensure that the product formation was less than 25% of the substrate amount. Kinetic parameters were deduced by using GraphPad Prism version 5.00 (GraphPad Software).
An NF hydrolysis assay with intact roots was performed as described previously (Staehelin et al., 1994b, 1995). Briefly, experiments were performed with young seedlings from wild-type plants of M. truncatula ecotype R108-1 and MtNFH1 RNAi-silenced lines (five lines, T2 generation, nine to 19 single plants per line). Seedlings were first incubated in 1-mL syringes filled with 400 μL of Jensen medium (Van Brussel et al., 1982) containing 0.5% (v/v) DMSO and 0.1 μm NodSm-IV(C16:2, S) at 24°C in the dark. The following day, the pretreated seedlings were transferred to new 1-mL syringes filled with 400 μL of Jensen medium containing 0.5% (v/v) DMSO and 15 μm NodSm-IV(Cl6:2, S). After incubation at 24°C for 18 h in the dark, seedlings were removed and plants from RNAi lines were stained for GUS activity. NodSm-IV(C16:2, S) and formed NodSm-II(C16:2) were extracted from the medium with an equal volume of distilled n-butanol. Dried samples were resuspended in 1 μL of DMSO and subjected to HPLC analysis as described above. Data were statistically analyzed with the nonparametric Kruskal-Wallis test, which is suitable for unequal replications.
Mass Spectrometric Analysis
HPLC fractions containing NodSm-II(C16:2), NodSm-II(C16:2, Ac), or NodSm-III(C16:2) were collected, and samples were dried and desalted on a Polygosil C18 column. Matrix-assisted laser-desorption ionization (MALDI)-time of flight (TOF) analysis was performed with an Ultraflex III MALDI-TOF/TOF mass spectrometer (Bruker Daltonics) in positive ionization mode. Samples were ionized with a smartbeam I UV laser (at a wavelength of 355 nm). Samples were then resuspended in sterilized water and mixed with the matrix (5 mg mL−1 2,5-dihydroxybenzoic acid in 30% acetonitrile). The matrix:sample mixture ratio was 1:1, and the final volume was 2 μL. The mixture was applied to an Anchorchip probe (Bruker Daltonics) and air dried. Observed ion masses corresponded to [M+H]+ and [M+K]+ due to protonation and the formation of potassium adducts.
Enzyme Assays with Chitin Oligosaccharides
(GlcNAc)2, (GlcNAc)3, (GlcNAc)4, (GlcNAc)5, and (GlcNAc)6 were purchased from Seikagaku Kogyo. Enzyme activities were tested by incubating chitin oligosaccharides with different enzymes in 25 mm sodium acetate buffer, pH 5.0, at 37°C. After incubation, reaction mixtures were diluted with an equal volume of 50% acetonitrile and then loaded onto a μBondapak NH2 HPLC column (0.8 × 10 cm; Waters) using 67% (v/v) acetonitrile/water as the mobile phase. The flow rate was 1 mL min−1, and the oligosaccharides were detected by ultraviolet A214. For measurement of enzyme activities and determination of kinetic parameters, the incubation times varied to ensure that the amount of product formation was less than 25% of the substrate amount. Kinetic parameters were deduced with the help of GraphPad Prism version 5.00 (GraphPad Software).
Enzyme Assays with Chitinous Substrates
All assays were performed at 37°C with specified amounts of purified His-tagged proteins in 25 mm sodium acetate buffer (pH 5.0) and substrate concentrations indicated in Table I. Colloidal chitin was obtained by acetylation of chitosan (Sigma-Aldrich) using acetic anhydride (Molano et al., 1977). Recombinant N-acetylglucosaminidase from M. truncatula was used to convert released oligosaccharides into GlcNAc, which was quantified with a p-dimethylaminobenzaldehyde solution (Ehrlich’s reagent; Reissig et al., 1955). Glycolchitin was obtained by acetylation of glycol chitosan (Sigma-Aldrich) with acetic anhydride (Molano et al., 1977; Trudel and Asselin, 1990). Reducing end sugars of hydrolyzed glycolchitin were measured with the Lever assay (Lever, 1972). CM-chitin-RBV was bought from Loewe Biochemica. The enzyme activity was photometrically determined based on the manufacturer’s instructions (Wirth and Wolf, 1990).
Lysozyme Assay and Fungal Growth Inhibition Test
Lyophilized Micrococcus lysodeikticus cells were purchased from Sigma-Aldrich. The lysozyme assay with purified His-tagged proteins was performed photometrically as described previously (Boller et al., 1983; Brunner et al., 1998). Effects of purified His-tagged proteins in 25 mm sodium acetate buffer (pH 5.0) on a growing mycelium of the fungus Trichoderma viride (Guangdong Culture Collection Center) were tested at 27°C according to a previously established procedure (Schlumbaum et al., 1986). Under these conditions, the MtNFH1 protein remained stable for at least 20 h as measured with the substrate NodSm-IV(C16:2, S).
Homology Modeling and Substrate-Docking Simulation
The crystal structures of class V chitinases from tobacco (NtChiV; Protein Data Bank [PDB] codes 3ALF and 3ALG) and Arabidopsis (AtChiC; PDB code 3AQU) served as structural templates for homology modeling. The sequence alignment used for modeling was obtained interactively by using the program Swiss-PdbViewer (http://spdbv.vital-it.ch/). A model for MtNFH1 was created by using the program MODELER (Fiser and Šali, 2003). The structure of substrate NodSm-V(C16:2, S) was built based on the structure of (GlcNAc)5 in complex with chitinase B from Serratia marcescens (SmChiB; PDB code 1E6N). The substrate NodSm-V(C16:2, S) was then docked manually to the active site of MtChi5, guided by the conserved interactions between the chitinase and the carbohydrate moiety of NodSm-V(C16:2, S). The docked structure of MtChit5 in complex with NodSm-V(C16:2, S) was energy minimized by the program GROMACS (Van Der Spoel et al., 2005) using the topology generated by the program PRODRG (Schüttelkopf and van Aalten, 2004).
Transformation of M. truncatula with pCAMBIA-MtNFH1i
The transformation of M. truncatula ecotype R108-1 was performed by infiltrating leaf discs with A. tumefaciens strain EHA105 carrying the RNAi plasmid pCAMBIA-MtNFH1i. The transformation and regeneration procedures were based on protocols described by Hoffmann et al. (1997) and Trinh et al. (1998). Briefly, infiltrated leaf explants were placed on petri dishes containing proembryogenic callus induction medium (SHMab) supplemented with acetosyringone. After 3 d of cocultivation with the bacteria, the explants were transferred to new plates containing SHMab medium supplemented with 500 μg mL−1 cefotaxime and 40 μg mL−1 hygromycin for selection. Healthy calli emerging from the explants were transferred to plates containing embryogenesis medium (SHM2) supplemented with hygromycin. Plates were changed every 2 weeks until somatic proembryos turned green. The embryos were then kept on SHM2 medium without hygromycin. Plantlets with shoots were transferred onto one-half-strength SHM2 plates and placed vertically to induce root development. Finally, regenerated plantlets were transferred to pots (filled with vermiculite and expanded clay; ratio of 3:1) and used for seed production in a temperature-controlled greenhouse.
GUS Staining
M. truncatula seedlings transformed with the RNAi plasmid pCAMBIA-MtNFH1i were stained for GUS activity with X-Gluc solution [0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide in 100 mm potassium phosphate buffer, pH 7, 0.1% (v/v) Triton X-100, 10 mm K3Fe(CN)6, and 10 mm K4Fe(CN)6] at 37°C. Wild-type M. truncatula plants served as a control.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KC833515 (MtNFH1), KC833513 (Mt75352), and KC833514 (MtCRA).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Unrooted phylogenic tree of MtNFH1 and 94 predicted plant proteins with related amino acid sequences.
Supplemental Figure S2. Chemical structures of NFs and acylated cleavage products used in this study.
Supplemental Figure S3. Positive-ion MALDI-TOF mass spectra of the cleavage products NodSm-II(C16:2), NodSm-II(C16:2, Ac), and NodSm-III(C16:2) purified by HPLC.
Supplemental Figure S4. The degradation of NodSm-V(C16:2, S) by MtNFH1 is not affected by a 500-fold higher concentration of (GlcNAc)5.
Supplemental Figure S5. Examples of velocity versus NF substrate concentration curves for MtNFH1.
Supplemental Table S1. MtNFH1, Mt75352, and MtCRA sequences of M. truncatula ecotype R108-1 used in this study.
Supplemental Table S2. Positive-ion MALDI-TOF analysis of the cleavage products NodSm-II(C16:2), NodSm-II(C16:2, Ac), and NodSm-III(C16:2).
Supplemental Table S3. Km and kcat for AtChiC and NtChiV with (GlcNAc)6 or (GlcNAc)5 as a substrate.
Supplemental Table S4. Primers used in this study.
Supplemental Table S5. Plasmids constructed in this study.
Acknowledgments
We thank Yongjin Liu and Shaoyun Song (Sun Yat-sen University) for performing MALDI-TOF mass spectrometric analysis; Wen-Hui Hu (Sun Yat-sen University) for cloning AtChiC into pET28b; Eva Kondorosi (CNRS) for providing S. meliloti 1021 (pEK327); and Qi Sun, Li-Ming Liang, Jing Cheng, Jin-Song Xiong, and Christian Wagner (Sun Yat-sen University) for help with various aspects of this work.
Glossary
- NF
nodulation factor
- CM-chitin-RBV
carboxymethyl-chitin-Remazol Brilliant Violet 5R
- kcat
catalytic rate constant
- RNAi
RNA interference
- CaMV
cauliflower mosaic virus
- DMSO
dimethyl sulfoxide
- MALDI
matrix-assisted laser-desorption ionization
- TOF
time of flight
- PDB
Protein Data Bank
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Antolín-Llovera M, Ried MK, Binder A, Parniske M. (2012) Receptor kinase signaling pathways in plant-microbe interactions. Annu Rev Phytopathol 50: 451–473 [DOI] [PubMed] [Google Scholar]
- Boller T, Gehri A, Mauch F, Vögeli U. (1983) Chitinase in bean leaves: induction by ethylene, purification, properties, and possible function. Planta 157: 22–31 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109: 13859–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F, Stintzi A, Fritig B, Legrand M. (1998) Substrate specificities of tobacco chitinases. Plant J 14: 225–234 [DOI] [PubMed] [Google Scholar]
- Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K. (1993) Plant chitinases. Plant J 3: 31–40 [DOI] [PubMed] [Google Scholar]
- De Jong AJ, Cordewener J, Lo Schiavo F, Terzi M, Vandekerckhove J, Van Kammen A, De Vries SC. (1992) A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 4: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demont N, Debellé F, Aurellé H, Dénarié J, Promé JC. (1993) Role of the Rhizobium meliloti nodF and nodE genes in the biosynthesis of lipo-oligosaccharidic nodulation factors. J Biol Chem 268: 20134–20142 [PubMed] [Google Scholar]
- Etzler ME, Kalsi G, Ewing NN, Roberts NJ, Day RB, Murphy JB. (1999) A nod factor binding lectin with apyrase activity from legume roots. Proc Natl Acad Sci USA 96: 5856–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Šali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374: 461–491 [DOI] [PubMed] [Google Scholar]
- Goormachtig S, Lievens S, Van de Velde W, Van Montagu M, Holsters M. (1998) Srchi13, a novel early nodulin from Sesbania rostrata, is related to acidic class III chitinases. Plant Cell 10: 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra R, Geurts R, Franssen H, Spaink HP, Van Kammen A, Bisseling T. (1994) Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol 105: 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz T, Segond S, Kauffmann S, Geoffroy P, Prasad V, Brunner F, Fritig B, Legrand M. (1994) Molecular characterization of a novel tobacco pathogenesis-related (PR) protein: a new plant chitinase/lysozyme. Mol Gen Genet 245: 246–254 [DOI] [PubMed] [Google Scholar]
- Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 280: 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Trinh TH, Leung J, Kondorosi A, Kondorosi E. (1997) A new Medicago truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Mol Plant Microbe Interact 10: 307–315 [Google Scholar]
- Karlsson M, Stenlid J. (2009) Evolution of family 18 glycoside hydrolases: diversity, domain structures and phylogenetic relationships. J Mol Microbiol Biotechnol 16: 208–223 [DOI] [PubMed] [Google Scholar]
- Kim YS, Lee JH, Yoon GM, Cho HS, Park SW, Suh MC, Choi D, Ha HJ, Liu JR, Pai HS. (2000) CHRK1, a chitinase-related receptor-like kinase in tobacco. Plant Physiol 123: 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss GB, Vincze É, Kálmán Z, Forrai T, Kondorosi Á. (1979) Genetic and biochemical analysis of mutants affected in nitrate reduction in Rhizobium meliloti. J Gen Microbiol 113: 105–118 [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. (1990) Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784 [DOI] [PubMed] [Google Scholar]
- Lever M. (1972) A new reaction for colorimetric determination of carbohydrates. Anal Biochem 47: 273–279 [DOI] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R. (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55: 983–992 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Melchers LS, Apotheker-de Groot M, van der Knaap JA, Ponstein AS, Sela-Buurlage MB, Bol JF, Cornelissen BJC, van den Elzen PJM, Linthorst HJM. (1994) A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J 5: 469–480 [DOI] [PubMed] [Google Scholar]
- Minic Z, Brown S, De Kouchkovsky Y, Schultze M, Staehelin C. (1998) Purification and characterization of a novel chitinase-lysozyme, of another chitinase, both hydrolysing Rhizobium meliloti Nod factors, and of a pathogenesis-related protein from Medicago sativa roots. Biochem J 332: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molano J, Durán A, Cabib E. (1977) A rapid and sensitive assay for chitinase using tritiated chitin. Anal Biochem 83: 648–656 [DOI] [PubMed] [Google Scholar]
- Mulder L, Lefebvre B, Cullimore J, Imberty A. (2006) LysM domains of Medicago truncatula NFP protein involved in Nod factor perception: glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology 16: 801–809 [DOI] [PubMed] [Google Scholar]
- Ohnuma T, Numata T, Osawa T, Mizuhara M, Lampela O, Juffer AH, Skriver K, Fukamizo T. (2011a) A class V chitinase from Arabidopsis thaliana: gene responses, enzymatic properties, and crystallographic analysis. Planta 234: 123–137 [DOI] [PubMed] [Google Scholar]
- Ohnuma T, Numata T, Osawa T, Mizuhara M, Vårum KM, Fukamizo T. (2011b) Crystal structure and mode of action of a class V chitinase from Nicotiana tabacum. Plant Mol Biol 75: 291–304 [DOI] [PubMed] [Google Scholar]
- Ohnuma T, Taira T, Fukamizo T. (2012) Antifungal activity of recombinant class V chitinases from Nicotiana tabacum and Arabidopsis thaliana. J Appl Glycosci 59: 47–50 [Google Scholar]
- Ovtsyna AO, Dolgikh EA, Kilanova AS, Tsyganov VE, Borisov AY, Tikhonovich IA, Staehelin C. (2005) Nod factors induce nod factor cleaving enzymes in pea roots: genetic and pharmacological approaches indicate different activation mechanisms. Plant Physiol 139: 1051–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovtsyna AO, Schultze M, Tikhonovich IA, Spaink HP, Kondorosi É, Kondorosi Á, Staehelin C. (2000) Nod factors of Rhizobium leguminosarum bv. viciae and their fucosylated derivatives stimulate a nod factor cleaving activity in pea roots and are hydrolyzed in vitro by plant chitinases at different rates. Mol Plant Microbe Interact 13: 799–807 [DOI] [PubMed] [Google Scholar]
- Perret X, Staehelin C, Broughton WJ. (2000) Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 64: 180–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausher MD. (2001) Co-evolution and plant resistance to natural enemies. Nature 411: 857–864 [DOI] [PubMed] [Google Scholar]
- Reissig JL, Storminger JL, Leloir LF. (1955) A modified colorimetric method for the estimation of N-acetylamino sugars. J Biol Chem 217: 959–966 [PubMed] [Google Scholar]
- Salzer P, Feddermann N, Wiemken A, Boller T, Staehelin C. (2004) Sinorhizobium meliloti-induced chitinase gene expression in Medicago truncatula ecotype R108-1: a comparison between symbiosis-specific class V and defence-related class IV chitinases. Planta 219: 626–638 [DOI] [PubMed] [Google Scholar]
- Schlumbaum A, Mauch F, Vögeli U, Boller T. (1986) Plant chitinases are potent inhibitors of fungal growth. Nature 324: 365–367 [Google Scholar]
- Schultze M, Quiclet-Sire B, Kondorosi E, Virelizer H, Glushka JN, Endre G, Géro SD, Kondorosi A. (1992) Rhizobium meliloti produces a family of sulfated lipooligosaccharides exhibiting different degrees of plant host specificity. Proc Natl Acad Sci USA 89: 192–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze M, Staehelin C, Brunner F, Genetet I, Legrand M, Fritig B, Kondorosi E, Kondorosi A. (1998) Plant chitinase/lysozyme isoforms show distinct substrate specificity and cleavage site preference towards lipochitooligosaccharide Nod signals. Plant J 16: 571–580 [Google Scholar]
- Schüttelkopf AW, van Aalten DMF. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60: 1355–1363 [DOI] [PubMed] [Google Scholar]
- Staehelin C, Granado J, Müller J, Wiemken A, Mellor RB, Felix G, Regenass M, Broughton WJ, Boller T. (1994a) Perception of Rhizobium nodulation factors by tomato cells and inactivation by root chitinases. Proc Natl Acad Sci USA 91: 2196–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin C, Müller J, Mellor RB, Wiemken A, Boller T. (1992) Chitinase and peroxidase in effective (fix+) and ineffective (fix−) soybean nodules. Planta 187: 295–300 [DOI] [PubMed] [Google Scholar]
- Staehelin C, Schultze M, Kondorosi E, Kondorosi A. (1995) Lipo-chitooligosaccharide nodulation signals from Rhizobium meliloti induce their rapid degradation by the host plant alfalfa. Plant Physiol 108: 1607–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin C, Schultze M, Kondorosi E, Mellor RB, Boller T, Kondorosi A. (1994b) Structural modifications in Rhizobium meliloti Nod factors influence their stability against hydrolysis by root chitinases. Plant J 5: 319–330 [Google Scholar]
- Staehelin C, Schultze M, Tokuyasu K, Poinsot V, Promé JC, Kondorosi É, Kondorosi Á. (2000) N-Deacetylation of Sinorhizobium meliloti Nod factors increases their stability in the Medicago sativa rhizosphere and decreases their biological activity. Mol Plant Microbe Interact 13: 72–79 [DOI] [PubMed] [Google Scholar]
- Taira T, Fujiwara M, Dennhart N, Hayashi H, Onaga S, Ohnuma T, Letzel T, Sakuda S, Fukamizo T. (2010) Transglycosylation reaction catalyzed by a class V chitinase from cycad, Cycas revoluta: a study involving site-directed mutagenesis, HPLC, and real-time ESI-MS. Biochim Biophys Acta 1804: 668–675 [DOI] [PubMed] [Google Scholar]
- Taira T, Hayashi H, Tajiri Y, Onaga S, Uechi G, Iwasaki H, Ohnuma T, Fukamizo T. (2009) A plant class V chitinase from a cycad (Cycas revoluta): biochemical characterization, cDNA isolation, and posttranslational modification. Glycobiology 19: 1452–1461 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh TH, Ratet P, Kondorosi E, Durand P, Kamaté K, Bauer P, Kondorosi A. (1998) Rapid and efficient transformation of diploid Medicago truncatula and Medicago sativa ssp. falcata lines improved in somatic embryogenesis. Plant Cell Rep 17: 345–355 [DOI] [PubMed] [Google Scholar]
- Trudel J, Asselin A. (1990) Detection of chitin deacetylase activity after polyacrylamide gel electrophoresis. Anal Biochem 189: 249–253 [DOI] [PubMed] [Google Scholar]
- Van Brussel AAN, Tak T, Wetselaar A, Pees E, Wijffelman CA. (1982) Small Leguminosae as test plants for nodulation of Rhizobium leguminosarum and other rhizobia and agrobacteria harbouring a leguminosarum sym-plasmid. Plant Sci Lett 27: 317–325 [Google Scholar]
- Van Damme EJ, Culerrier R, Barre A, Alvarez R, Rougé P, Peumans WJ. (2007) A novel family of lectins evolutionarily related to class V chitinases: an example of neofunctionalization in legumes. Plant Physiol 144: 662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. (2005) GROMACS: fast, flexible, and free. J Comput Chem 26: 1701–1718 [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Tadesse Z, Immerzeel P, Schols H, van Kammen A, de Vries SC. (2001) N-Acetylglucosamine and glucosamine-containing arabinogalactan proteins control somatic embryogenesis. Plant Physiol 125: 1880–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth SJ, Wolf GA. (1990) Dye-labelled substrates for the assay and detection of chitinase and lysozyme activity. J Microbiol Methods 12: 197–205 [Google Scholar]
- Xie ZP, Staehelin C, Wiemken A, Broughton WJ, Müller J, Boller T. (1999) Symbiosis-stimulated chitinase isoenzymes of soybean (Glycine Max (L.) Merr.). J Exp Bot 50: 327–333 [Google Scholar]