Abstract
The accumulation of beneficial mutations on competing genetic backgrounds in rapidly adapting populations has a striking impact on evolutionary dynamics. This effect, known as clonal interference, causes erratic fluctuations in the frequencies of observed mutations, randomizes the fixation times of successful mutations, and leaves distinct signatures on patterns of genetic variation. Here, we show how this form of “genetic draft” affects the forward-time dynamics of site frequencies in rapidly adapting asexual populations. We calculate the probability that mutations at individual sites shift in frequency over a characteristic timescale, extending Gillespie’s original model of draft to the case where many strongly selected beneficial mutations segregate simultaneously. We then derive the sojourn time of mutant alleles, the expected fixation time of successful mutants, and the site frequency spectrum of beneficial and neutral mutations. Finally, we show how this form of draft affects inferences in the McDonald–Kreitman test and how it relates to recent observations that some aspects of genetic diversity are described by the Bolthausen–Sznitman coalescent in the limit of very rapid adaptation.
Keywords: genetic draft, adaptation, clonal interference
THE effects of linkage between beneficial mutations in altering evolutionary dynamics and the structures of genealogies in adapting populations has been recognized for nearly a century, particularly in the context of the evolutionary advantage of sex (Muller 1932). In both asexually reproducing organisms and in regions of low recombination in sexual organisms, the chance congregation of beneficial mutations on competing genetic backgrounds skews evolutionary dynamics. Because of this “clonal interference” effect, the success of a mutation depends not only on its fitness effect, but also on the quality of the genetic background in which it occurs and the fortune of the mutant’s progeny in amassing more beneficial mutations (Smith and Haigh 1974; Gerrish and Lenski 1998; Gillespie 2000, 2001; Kim and Orr 2005).
Recent work in experimental evolution has confirmed that clonal interference is widespread in large adapting laboratory microbial and viral populations (de Visser et al. 1999; Miralles et al. 1999; de Visser and Rozen 2006; Kao and Sherlock 2008; Lang et al. 2011, 2013). Several recent studies also suggest that classic “hard” selective sweeps may be rare in Drosophila (Sella et al. 2009; Karasov et al. 2010) and humans (Pritchard et al. 2010; Hernandez et al. 2011), implying that models that better account for linkage between sites need to be explored. As a result, in recent years there has been an influx of theoretical work describing the effects of clonal interference on the evolution of large populations (see Park et al. 2010 for a recent review).
This work has provided a good understanding of evolutionary dynamics in the regime of rare interference, where the number of strongly beneficial mutations segregating in a population is rarely more than two (Gerrish and Lenski 1998; Gillespie 2000, 2001; Kim and Stephan 2003; Park and Krug 2007). However, in large populations many beneficial mutations can segregate simultaneously and the population can maintain substantial variation in fitness. This decreases the importance of each mutation’s intrinsic fitness effect relative to the quality of the genetic background on which it occurs. Long-term evolutionary dynamics in these populations are therefore driven primarily by the stochastic introduction of mutants at the high-fitness tip of the population’s fitness distribution and the fluctuation in the lineage sizes of these super-fit mutants when rare. Several models have been introduced to study evolution in these strong selection, strong mutation regimes (Tsimring et al. 1996; Rouzine et al. 2003; Desai and Fisher 2007; Hallatschek 2011). This work has successfully described the rate of adaptation and the variation in fitness within a population (Desai and Fisher 2007; Park et al. 2010; Rouzine et al. 2008), and the fitness effects of fixed mutations (Fogle et al. 2008; Neher et al. 2010; Good et al. 2012; Fisher 2013), while ignoring the specific mutations that underlie these population-wide quantities.
In this work, we use these earlier theoretical treatments as the basis for analyzing the evolutionary dynamics of individual mutations (i.e., their frequencies over time and their eventual fates). To do so, we study the forward-time dynamics of specific mutant lineages on the backdrop of the population’s fitness distribution, concentrating specifically on quantities relevant and measurable in experiments: the trajectories and sojourn times of polymorphic sites, fixation times of successful mutants, and some basic patterns of genetic diversity.
We begin by introducing our model and briefly summarizing earlier results that describe the dynamics of the population’s fitness distribution. We then demonstrate that the growth of the high-fitness “nose” of this distribution is dominated by a small number of successful, founding mutants. Since this high-fitness nose will eventually come to dominate the population, the long-term success of a given polymorphism is largely determined by its representation (or lack thereof) among this small class of stochastically fluctuating, high-fitness individuals. This allows us to model adaptation as a series of replacements of each fittest class by a new, fitter class over a typical replacement timescale. We show how this leads to a distribution of transition probabilities that describe how the frequency of each polymorphism changes in each stochastic jump from one fittest class to the next. This process bears some resemblance to several recent models of adaptation in populations with highly skewed offspring distributions (Schweinsberg 2003; Eldon and Wakeley 2006; Der et al. 2012). However, whereas in these earlier models a jump in offspring frequency is assumed to be an explicit feature of the offspring distribution, in this work these jumps emerge organically from the dynamics of the underlying model.
We next use our derived transition probabilities to calculate various diversity statistics, providing an alternative forward-time perspective that complements earlier structured coalescent approaches to these questions (Desai et al. 2013). We first calculate the site frequency spectrum of beneficial and neutral mutations, which has not yet been explicitly derived for this class of models. We then use our results to make predictions regarding the fates of mutations in experiments, particularly on the sojourn time of these mutations and the time to fixation of a successful mutant. Finally, in the Discussion we describe a decay to neutrality exhibited by mutations in these populations, comment on the relationship between our results and the Bolthausen–Sznitman coalescent, analyze the implications of our results for interpreting widely used tests for adaptation, and consider the extension of our model to evolution on more complex fitness landscapes.
Our work complements earlier analysis of related questions in facultatively sexual populations (Neher and Shraiman 2011), which neglects new mutations and focus instead on fluctuations in the frequencies of individual polymorphisms driven by recombination into higher or lower fitness backgrounds. By contrast, we focus on either asexual populations or on tightly linked genomic regions of sexual populations, where recombination can be neglected compared to selection and new mutations, and study instead the fluctuations in polymorphism frequencies driven by new mutations. Despite the different sources of draft explored in these two models, our results bear some similarities to the findings of Neher and Shraiman (2011), a parallel that we revisit in Discussion.
Model
We study the evolution of a large asexually reproducing population of constant size N, using the model introduced in Desai and Fisher (2007) (summarized below). This model assumes that beneficial mutations of a single fitness effect s occur at a constant beneficial mutation rate Ub per genome and are drawn from an effectively infinite number of possible sites. The use of a single fitness effect allows the fitness of an individual in the population to be described solely by the number of beneficial mutations k it carries, with the absolute fitness of an individual given by wk = (1 + s)k ≈ 1 + ks for s ≪ 1. More complicated effects, such as frequency-dependent selection and epistasis, are neglected in this analysis (although the model is easily modified to include some simple sign epistasis; see Discussion). Finally, we assume that the population is evolving in the strong selection, strong mutation regime. Specifically, this means that Ns ≫ 1, s/Ub ≫ 1, NUb ≫ 1, meaning that the selective forces, selective forces relative to mutations, and incoming mutations per generation are all large.
In the next few paragraphs we review the primary features of this model that are pertinent to our analysis, which are justified in detail by Desai and Fisher (2007). This model describes the population as a traveling wave in fitness space, wherein the deterministic evolution in the bulk of the wave is combined with a careful stochastic treatment of the birth and fluctuation in lineage sizes of mutants at the high-fitness nose of the distribution. Specifically, the population is characterized according to the number of individuals nk in each fitness class k, where the term fitness class refers to the class of individuals carrying k beneficial mutations. At each generation, nk changes according to the effects of genetic drift, incoming and outgoing mutations, and selection. If nk is sufficiently large, then selection trumps the effects of the other two evolutionary forces, and the rate of change of nk is
| (1) |
where t is taken in units of generations and 〈ks〉 is the (time-dependent) mean fitness of the population. A fitness class will enter this regime of deterministic growth shortly after the effect of selective forces overcomes the effect of drift, which occurs shortly after the entire fitness class reaches a population size ∼ 1/(k − 〈k〉)s, at which point we say that it is established. Given our assumption that s ≫ Ub, the probability for a fitness class that has not yet established to generate a more fit establishing lineage is extremely low. Thus, the population is well described by a deterministically growing/shrinking set of fitness classes and one stochastically fluctuating class , where are defined to be the minimum/maximum k s.t. .
Although in principle one could consider the transient dynamics by which an initially clonal population attains a steady distribution of relative fitnesses, we are instead interested in the regime where this equilibrium distribution has already been reached and is maintained over timescales that are long compared to the typical establishment time of a new fitness class. In other words, the population has been evolving long enough to attain some typical steady-state fitness profile, but not long enough to begin to deplete the supply of beneficial mutations (which validates our infinite-sites approximation). In this case, the width of the distribution is set by an equilibrium between the influx and growth of highly fit mutants (which increases the width of the distribution) and the advancement of the mean fitness (decreasing this width). The size of this width q (defined as the mean number of mutations between the mean fitness class and the largest not-yet-established class) is given to a good approximation by
| (2) |
(Desai and Fisher 2007, Equation 4), with higher-order corrections given in Desai and Fisher (2007, Equation 39). Similarly, τk, defined roughly as the random variable denoting the time between the establishment of fitness class k − 1 and k, has expectation value
| (3) |
(Desai and Fisher 2007, Equation 36), with γE ≈ 0.577 the Euler gamma constant. Note that the variable is called τq in Desai and Fisher (2007), where it contains an erroneous factor of (q − 1) instead of q within the logarithm.
A more accurate derivation of the true time between establishments, accounting primarily for the non-negligible effect of incoming mutations shortly after a class establishes, is derived in Brunet et al. (2008), whereas a more careful discussion of the correct interpretation of τk is given in Desai and Fisher (2007). However, the precise distribution of τk is not important for our analysis because throughout the rest of this article we take time in units of establishment of each new lead class.
If q is not small, the mean relative fitness of any individual class does not change too much in the course of one establishment time. In this case, the dynamics of fitness classes are excellently approximated by a staircase model, in which the fitness of every class is held constant over the course of the establishment of a new class. In the time it takes each new class to establish, the mean fitness will typically increase by s, and hence the relative fitness of all classes decreases accordingly. Thus, the number of individuals in each class grows exponentially at a rate that is well approximated as diminishing by discrete steps of s after each establishment time τk. Under these assumptions, the number of individuals in a class k with ℓ mutations more or less than the mean is approximately
| (4) |
A more detailed discussion of this formula is given in Desai and Fisher (2007, p. 1774). Note that by averaging the fitness distribution over all times we recover a Gaussian distribution with variance σ2 = v = s/〈τk〉, where v is the rate of adaptation.
We are primarily interested in the growth of fitness classes when they are still expanding near the high-fitness tip of the wave, which is where mutations destined to reach appreciable frequencies first occur. Thus, we examine the growth of the class k = kmax, i.e., the largest not-yet-established class. The number of individuals in this lead class at short times is (Desai and Fisher 2007, Equation 31)
| (5) |
Note that we have arbitrarily defined the origin of time such that t = τk corresponds to the establishment time of class k, which occurs when nk = 1/qs individuals. By a simple transformation,
| (6) |
where we have defined
| (7) |
In formulation (5), the stochasticity of the growth is encapsulated in the establishment time τk; in formulation (6), the random variable σk encodes how much the class deviates from its typical size at long times. The random variable σk is chosen to have the simple generating function [obtained by a transformation of the generating function of nk(t) given in Equation 29 of Desai and Fisher (2007)],
| (8) |
Note that σk is singularly well suited for extracting the contribution of independent lineages. To see this, we note that nk(t) denotes the growth of class k, given that it is fed by an exponentially (and deterministically) expanding class k − 1 (which we simply call the feeding class)
| (9) |
which is supplying mutants to class k of fitness qs at a rate Ub per genome per generation. To probe the contribution of particular haplotypes from the feeding class, we decompose the growth of class (k − 1) into
| (10) |
where xℓ,k−1 is the fraction of the feeding class constituted by haplotype ℓ. Note that if each haplotype is plentiful enough to expand deterministically (an assumption we discuss in more detail below), then each haplotype grows at the same rate as the class as a whole, and the xℓ,k−1 are time-independent constants. In this case, the growth of the lead class may be written as
| (11) |
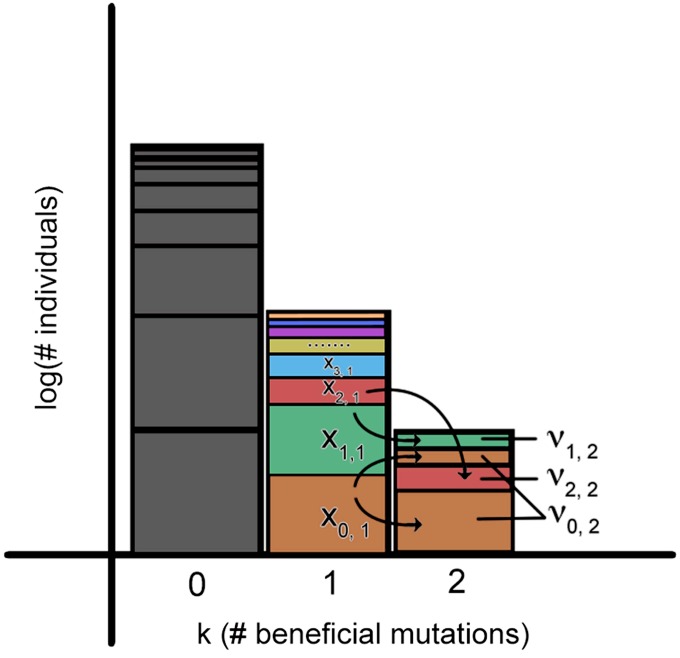
where νℓ,k is the random variable denoting the contribution from haplotype ℓ to class k from class k − 1. Note that it is not necessary that ; in fact, σk may deviate strongly from unity if a mutation occurs and establishes in the lead class anomolously early. As a result, νℓ,k is not the frequency of the lineage derived from haplotype ℓ in class k. Rather, νℓ,k is the random variable that encodes how quickly that lineage establishes and expands in class k relative to its typical growth. The frequency of the derived lineage, then, is xℓ,k = νℓ,k/σk. This setup is illustrated in Figure 1. Since each haplotype in class k − 1 expands exponentially, the lead class k is fed by m independent feeding processes, where m is the total number of haplotypes in class (k − 1). From this, we can essentially repeat the derivation for the generating function of σk done in Desai and Fisher (2007) for νℓ,k by adding appropriate factors of xℓ,k−1 in the growth of the feeding class, eventually obtaining
| (12) |
Note that the assumption of independence between each νℓ,k implicitly assumes that the size of the lead nk ≪ N. In this case, feedback effects, whereby the growth of each νℓ,k affects the advancement of the mean fitness, and thereby the growth of other νℓ,k, may safely be neglected. When class k leaves the lead and this assumption begins to break down, the frequency of each lineage ℓ derived from the corresponding haplotype in class k − 1 is already frozen in class k (an assumption justified in detail in the Appendix) and the lineage is expanding deterministically. Furthermore, because the rate of expansion of both the lineage and the class as a whole are set by the relative fitness of the class, at long times the frequencies xℓ,k do not change. The process will then begin anew, with a new set of haplotypes in class k at frequencies xℓ,k feeding a new most-fit class, k + 1. Hence, the number of individuals descended from a lineage ℓ at a time t, η(t), is given simply by .
Figure 1.
Illustration of the stochastic process by which lineages in a class k − 1 contribute to a class k through further beneficial mutations. Shown are the three fittest classes of the traveling wave. Class 1 is broken down into haplotypes {0, 1, 2, …ℓ} at frequencies {x0,1, x1,1, …, xℓ,1} within class 1, which grow deterministically. At the instant illustrated here, haplotype 0 (at frequency x0,1 within class 1) has generated two beneficial mutations. These mutations occurred at two different timepoints in the past, grew stochastically, and now cumulatively form a significant fraction of class 2. Individuals derived from haplotype 0 now form a fraction x0,2 = ν0,2/(ν0,2 + ν1,2 + ν2,2) of this class. Analogous observations can be made for the other haplotypes. Before class 2 begins generating mutants destined to establish in class 3, the frequencies of its haplotypes will be frozen up to small fluctuations, and the process repeats with a new set of frequencies {x0,2, x1,2, …, xℓ,2}.
Some objections to this formalism might immediately be raised. First, the number of haplotypes m in class k − 1 is increasing in time due to incoming mutations from the previous class k − 2. However, we show in the Appendix that incoming mutations typically stop contributing significantly to a class shortly after it establishes, and certainly by the time that it itself begins feeding establishing mutants to the next class. Thus, while m may be strictly increasing in time, the combined contribution of these new haplotypes affects the frequencies of all other sites only negligibly. Second, a considerable fraction of these haplotypes could be at frequencies xℓ,k−1 such that xℓ,k−1nk−1(t) ≲ 1/((q − 1)s), meaning that these haplotypes cannot be modeled deterministically. This objection becomes important when considering fluctuations in the frequencies of haplotypes that are rare in a given class. On the other hand, if a polymorphic site xℓ,k−1 is sufficiently common, its growth by the time the class begins supplying establishing mutants may be modeled deterministically. In this case, the above formalism is well suited to predict the contribution of that lineage ℓ to subsequent fitness classes.
In what follows, since we are interested only in the dynamics of one particular lineage at a time, we drop the ℓ subscript and set the initial fitness class (i.e., the fitness class in which the lineage frequency first begins to be tracked) at k = 0. The strengths and realm of validity for all of the above assumptions have been studied by ourselves and others in previous work (Desai and Fisher 2007; Brunet et al. 2008; Rouzine et al. 2008; Desai et al. 2013; Fisher 2013).
Transition Probabilities
From the considerations of the previous section, we see that evolutionary dynamics in these populations are driven by two factors: the deterministic growth and decay of existing clones—governing the short-time dynamics—and the stochastic introduction and expansion of new super-fit mutants, which govern the population’s long-term evolution. Furthermore, the fate of any particular mutation is determined by two factors: (1) the genetic background in which it occurs and (2) the success of its progeny in amassing additional beneficial mutations more quickly than competing backgrounds. When many beneficial mutations segregate simultaneously, the strictness of these two constraints requires that mutations with any nonnegligible chance of fixing (or even rising to an appreciable frequency) must have been founded near the nose of the fitness distribution. The vast majority of beneficial mutations are thus “wasted” on the bulk of the distribution, where the mutant’s lineage is doomed to eventual extinction.
We are largely concerned with those mutations that successfully rise to appreciable frequencies and thus largely consider the dynamics of those mutations that are founded on “good” genetic backgrounds. Once established, one of these new, high-fitness clones will grow, stagnate, and diminish in its class deterministically as dictated by Equation 1. At any moment in time, the population can be divided into many such expanding and contracting “bubbles,” which fully determine frequency dynamics over short timescales of (τk). However, many of the interesting long-term dynamics are determined by the stochastic origination and establishment of super-fit mutants from these deterministically expanding clones, which drive the success or failure of particular lineages, mutations, or entire evolutionary trajectories.
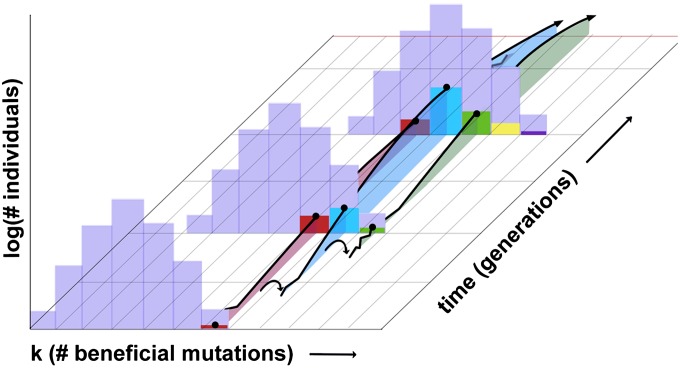
These ideas are expressed more concretely in Figure 2, which shows the distribution of fitness classes at three distinct timepoints. A clone that is about to establish in the first timepoint is growing deterministically in the second timepoint and diminishing in the third timepoint, before finally going extinct as the population evolves to higher and higher fitness. Normally, this would mean that the contribution of the clone’s lineage is also extinct; however, the lineage avoids this fate by jumping into the next fitness class through the creation of a new, super-fit mutant when the class is still small and expanding very rapidly. This new mutant establishes and expands, and because it occurs very early, comes to form a significant fraction of the next fitness class, as exemplified in the second timepoint of Figure 2. Although only one of these jumps is shown in Figure 2, a given lineage may jump many times into the next class, with each successive jump, on average, contributing a smaller and smaller fraction of individuals to that class. This new clone will then deterministically expand, contract, and go extinct in the new class, although its lineage may survive by jumping into the next fitness class sufficiently early to eventually constitute a significant fraction of that class, shown by the second jump in Figure 2. The process continues ad infinitum, until the sum of all the contributions of a given lineage to a class vanishes or constitutes the entire class, in which case the lineage is then destined to go extinct or sweep, respectively.
Figure 2.
The frequency trajectory of a successful mutation on a high-fitness background. The population’s fitness profile is shown for three timepoints, along with one haplotype present in each class. A mutation founded in the first timepoint (in red) expands and constitutes a significant fraction of its fitness class in the second timepoint. As the population adapts, the founding fitness class begins to shrink, along with the number of individuals in the lineage constituting that class. However, this lineage generates a beneficial mutant (shaded in blue) between the first and second timepoints, as shown by the first arrow. This mutant drifts randomly for a short while before establishing and eventually comprising a significant fraction of the next fitness class. In this way, the original successful mutation “jumps” into the next fitness class and is potentially able to avoid extinction. This process repeats, as the second (blue) lineage generates another successful (green) mutant that comes to populate a significant fraction of the next class, and so on.
The key distribution describing these dynamics is the jump probability ρ(xk|xk−1, …, x0), the probability of finding a lineage at frequency xk in fitness class k, given that it was at frequencies xk−1, xk−2, … x0 in the k previous fitness classes. Essentially, ρ(xk|xk−1, …, x0) gives the probability distribution of the sum of the frequencies of each clone in fitness class k that originated from a lineage at some frequency x0 in fitness class 0 and jumped through k − 1 intermediate classes. Under our particular model, the derivation of ρ(xk|xk−1, …, x0) becomes much simpler because the frequency of a lineage in fitness class k is determined only by its frequency in fitness class k − 1. This is equivalent to the statement that when a class begins feeding establishing mutants to the next class (i.e., when the jumps in Figure 2 occur), the frequencies of lineages in the feeding class are already frozen. In this case, one may consider the frequencies of lineages evolving in analogy to the entire population: the frequencies of lineages in the classes are frozen, and the frequencies of mutants in the lead are fluctuating. Thus the transition process is a Markov chain, meaning that the long-time frequency of a lineage in fitness class k, xk, is simply ρ(xk|xk−1, …, x0) = ρ(xk|xk−1). Of course this viewpoint sacrifices precision for the sake of clarity, since frequencies of lineages in the lead will continue to fluctuate after the lead establishes. It is only when a class typically begins to supply establishing mutants to the nose that the frequencies of its lineages will be frozen. Regardless, there will typically be one class with fluctuating frequencies (either the lead or next-to-lead class shortly after it establishes) and the rest of the population with lineage frequencies already frozen. This is by no means an obvious assumption and is discussed in detail in the Appendix.
In Equation 12, we gave the generating function of νk (the contribution of a particular lineage to a fitness class k, given that the frequency of the lineage in class k − 1 is xk−1). In class k, the lineage will grow as
| (13) |
where the exact proportionality constant is not relevant for our analysis. Similarly, we may denote the contribution of that lineage’s complement by —that is, the contribution to k from those individuals whose ancestors in class k − 1 were not derived from the chosen lineage. Naturally, is described by the following generating function,
| (14) |
with , and σk defined in Equation 6. Then, ρ(xk|xk−1) is derived from the two generating functions to be
| (15) |
which is corroborated by results of forward-time simulations (see Figure 3). This is readily extended to ρ(xk|x0), the distribution of a given lineage frequency k fitness class steps forward, by the simple replacement α → αk. The derivation of Equation 15 and the extension to arbitrary time steps k are given in the Appendix. Note that this transition probability was also stated, without detailed derivation, in Desai et al. (2013). Importantly, parameters such as N, s, and Ub factor into this distribution only through the parameter α = 1 − 1/q ≈ 1 − log(s/Ub)/(2 log(Ns)).
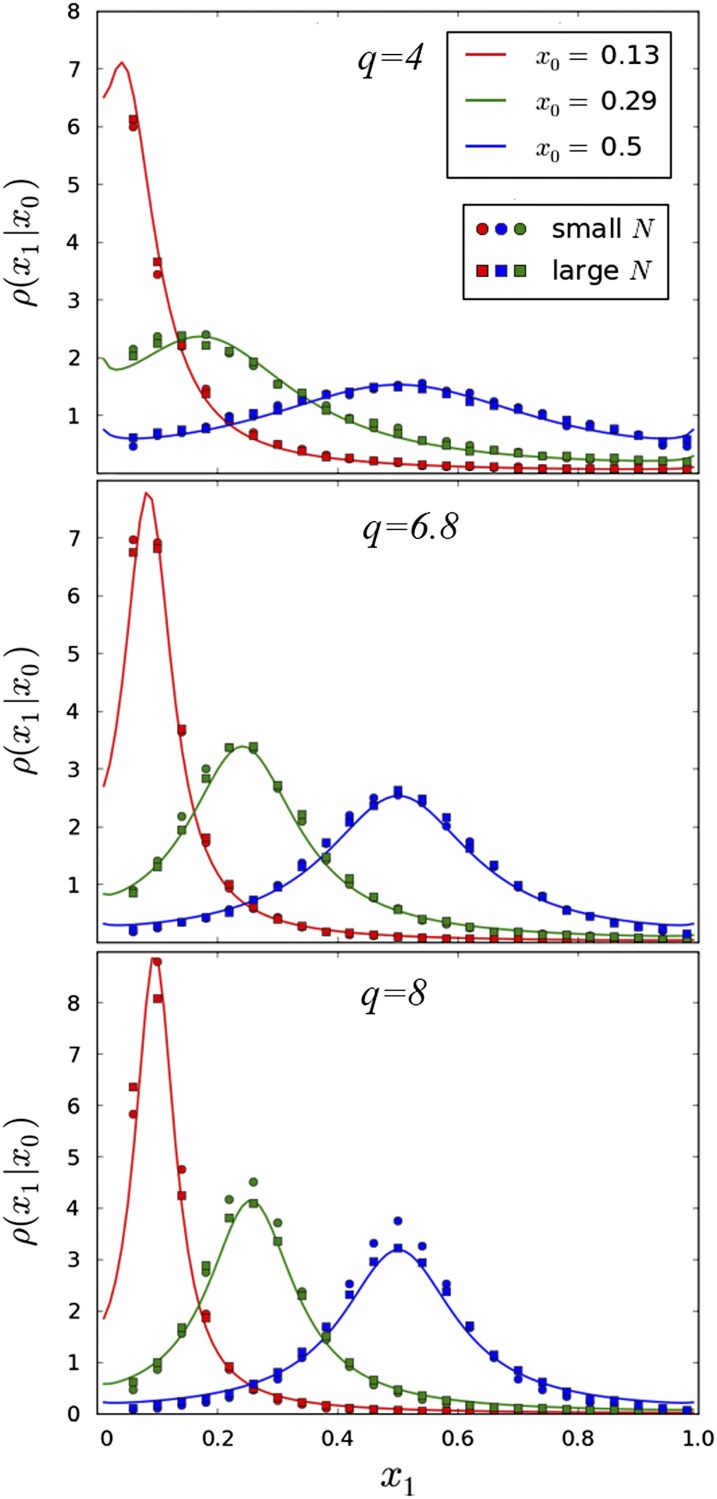
Figure 3.
The transition probability ρ(x1|x0) plotted for different values of x0, q, and other population parameters Ub, s, and N. Curves represent the theoretical prediction for the transition probabilities given in Equation 15. Circles and squares represent the results of forward-time simulations. The circular and square points have identical values of q, but differ in population size by a factor of 10. Circles (small N) represent simulations run with parameters s = 0.1, Ub = 0.001, and squares (large N) are run with parameters s = 0.01, Ub = 0.0001. The three plots represent different values of q, with q = 4 calculated from the approximate value in the text, and q = 6.8, 8 calculated with higher-order corrections given in Desai and Fisher (2007). These corrections become more important for higher q and are the reason for the slight discrepancy between theorized and predicted values for small N at q = 8. More information about the simulations may be found in File S1.
Note that while the result for one time step is exact, the generalization to several steps forward assumes that for 0 < k′ < k, or that the rate of establishment of each new class does not deviate too much from typical values. If this is the case, then , and each lineage may be assumed to grow independently of its competition in each new establishment. Errors introduced using this method start to become significant for k ≈ q, which is the timescale at which fluctuations in the establishment time of new lead classes begin to affect the rate of adaptation and mean fitness. We later demonstrate that calculations using ρ(xk|x0) yield robust estimates for a variety of diversity statistics and other population-wide properties, supporting our claim that the above assumption does not significantly change most quantitative predictions.
Although the analytic form of this jump probability is somewhat cryptic, some illuminating properties can be gleaned by considering the form of the distribution in the limit of infinite population size, q → ∞:
| (16) |
Because this distribution has a fat tail, large jumps may occasionally occur between adjacent fitness classes even in the limit of a massively large population, indicating that the dynamics described by this stochastic process are super-diffusive. This nondiffusive property of genetic draft has been observed many times for a number of different models of adaptation (Gillespie 2001; Neher and Shraiman 2011; Desai et al. 2013; Neher and Hallatschek 2013). One important, commonly stressed consequence of this nondiffusivity is that the stochastic dynamics of the population cannot be fully encapsulated by a single rescaled variance effective population size Ne.
Implications for Genetic Diversity
Whereas ρ is not directly measurable by itself—describing transition probabilities between fitness classes and not the population as a whole—genetic draft leaves distinct signatures on the diversity of rapidly adapting populations, which are both readily measurable and readily derived from ρ. In what follows we derive some implications of the stochastic jump process on the site frequency spectra of both beneficial and neutral mutations.
The site frequency spectrum of beneficial mutations
One statistic that is strongly affected by the stochastic jumps described above is the site frequency spectrum (SFS) of beneficial mutations f(x), the expected density of mutations between frequencies x and x + dx. In rapidly adapting populations, the SFS is partitioned into two regimes: on one hand, common mutations first arise at the distribution’s high fitness nose and are strongly affected by the process of stochastic jumps. On the other hand, nearly private variants, which constitute the majority of beneficial mutations, are overwhelmingly founded near the distribution’s bulk and are largely unaffected by this process. Thus, we expect the high- and low-frequency spectra to be qualitatively different. As a result, the derivation that follows is split into two segments: first, we derive the SFS of common alleles, which are founded at the exponentially expanding wavefront. Since our previous analysis describes only the dynamics of these exponentially expanding nose classes, it does not describe the frequency spectrum of extremely rare, nearly private alleles. Thus, in the second part of this section we make use of a different branching process method to derive the distribution of these plentiful but extremely rare variants.
We begin by deriving the site frequency spectrum of common alleles. For the moment, we may further simplify the problem by considering the site frequency spectrum of mutants in only one fitness class. Since the frequencies of common mutations freeze shortly after a given class establishes, we need only to calculate the distribution of frequencies in a class that is near the nose of the wave. Once the class leaves the nose, the frequencies of these common mutants will be frozen and the class will have the same SFS regardless of its position relative to other classes.
Now, as we have previously argued, almost every common polymorphism was once founded in a class that was at the population’s high-fitness nose. Thus, given a fitness class k that is near the distribution’s nose, we might decompose the frequency spectrum of mutants in class k according to which class they were founded in. Specifically, we consider mutations founded in class k from class k − 1 when class k was at the nose; these mutations originate in class k. Next, we may consider mutations that originated in class k − 1 from class k − 2 when class k − 1 was near the nose, whose lineages subsequently jumped into class k after acquiring more beneficial mutations; these mutations originate in class k − 1. Analogously, we can consider the distribution of mutants in class k that originated in classes k − 2, when these classes were at the distribution’s nose. The SFS f(x) in class k is then the sum over all of these distributions.
Consider first the SFS of sites in class k that originate in k, f1(x). We derive the SFS of these “new” mutations from the transition probability ρ as follows: first, we observe that class k − 1 first begins supplying mutants to class k that are destined to establish at a time of since its own establishment. Shortly before this time, we can decompose the growth of class k − 1 into 1/β independently growing blocks of frequency β such that β ≪ 1/q. If β is sufficiently large, so that , it is valid to model the growth of each block deterministically. In this case, the frequency x in class k of descendants of individuals in block β is distributed as ρ(x|β). Since , we require that for the jumps from each β-sized block into class k to be well described by ρ(x|β). Now, since β ≪ 1/q, it is unlikely that more than two founding mutants originate from the same block. In other words, the contribution of each block to the next class is dominated by the contribution of one most successful mutation, and the probability distribution of the frequency x of this most successful mutation is well approximated by ρ(x|β). The expected density of mutations between frequencies x and x + dx that are introduced from class k − 1 is then
| (17) |
Analogously, the distribution of mutations in class k originally arising from the (k − i)th class is obtained by the replacement α → αi, giving for the total SFS
| (18) |
Although straightforward to evaluate numerically, this integral has no simple closed-form expression. However, a first-order Taylor expansion in the sine is a reasonable approximation for 1 − x ≪ 1. This gives
| (19) |
For x → 1,
| (20) |
Note that this predicts that very-high-frequency mutations are actually more common than mutations at slightly lower frequencies. This upswing at very-high-frequency mutations is a widely recognized marker of selection for a number of different models, with and without linkage between sites (Wright 1938; Fay and Wu 2000; McVean and Charlesworth 2000; Neher and Hallatschek 2013). Importantly, it cannot be explained by many commonly studied forms of demographic history, such as population expansions. Recently, Neher and Hallatschek (2013) derived the upswing of the SFS arising from genealogies obeying the Bolthausen–Sznitman coalescent. However, to our knowledge, the excess of extremely common variants arising from linked beneficial mutations has not previously been derived directly from the forward-time dynamics of any model.
Another point worth mentioning is the invariance of the functional form of this distribution for different parameters N, s, and Ub. Given that our assumptions about the population hold, the spectra of different populations are identical up to a scaling factor that represents the different absolute numbers of common mutations in these populations, which itself is somewhat insensitive to the specific choice of parameters. This observation highlights an inherent limitation in inferring properties of adaptation through the functional form of the site frequency spectrum.
We now argue that the above distribution is a good approximation to the population-wide site frequency spectrum, instead of simply the frequency spectrum of mutations in a single class. First, we observe that we can arbitrarily set the distribution f(x) to describe the SFS of the mean class. The above approximation, then, is equivalent to the statement that the SFS of the mean class is a good approximation to the SFS of the entire population. If q is not too large (the relevant case for many biological populations), then the approximation holds because the vast majority of individuals reside in the mean class at any given time. The contribution of sites from other classes will then be a small perturbation on the SFS of the mean class, particularly relevant at low frequencies (where our approximation breaks down regardless, due to the contribution of mutants not founded at the nose). On the other hand, as q increases, the mean class constitutes a smaller and smaller fraction of the total population. However, the variance of the jumps in the frequencies of mutant sites also decreases in proportion to 1/q. Thus, while the mean class constitutes a smaller fraction of the total population, sites in adjacent classes tend to shift more slowly than for the case of smaller q (despite the fact that, as we have previously noted, large jumps may still occur occasionally). Thus the approximation should still be valid even in the limit that q is large. The strength of these arguments is corroborated by the close correlation of Equation 18 with site frequency spectra derived from our forward-time simulations (Figure 4).
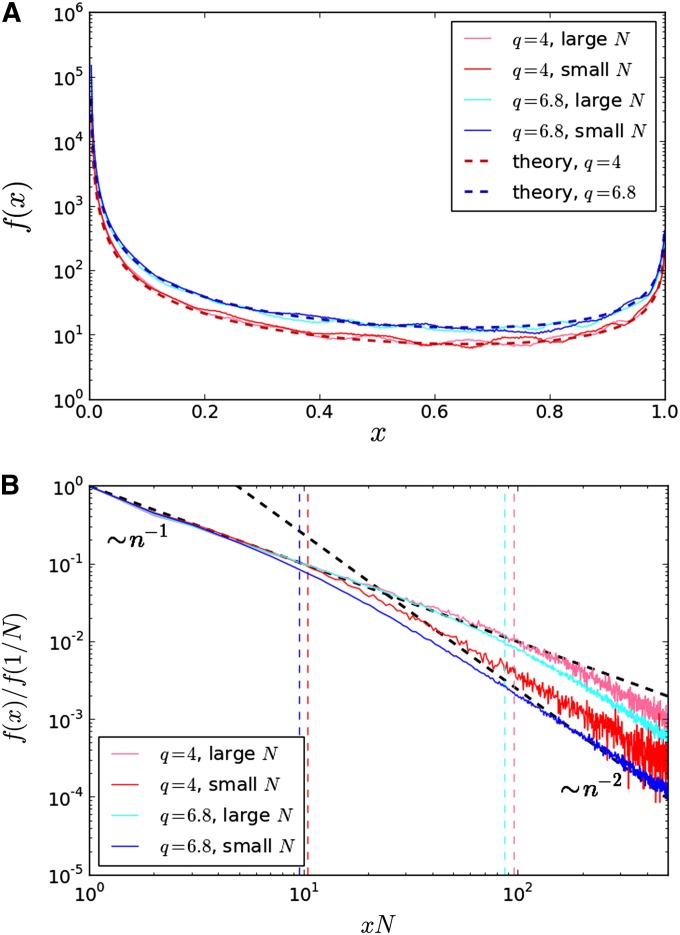
Figure 4.
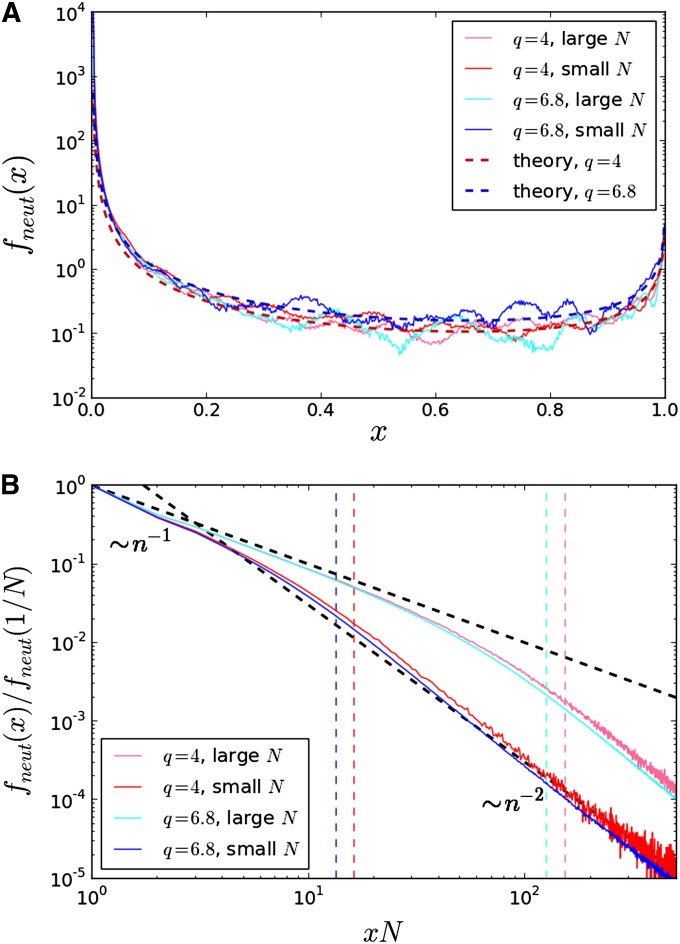
(A) The site frequency spectrum of common mutations. The dashed lines are the theoretical predictions from Equation 18 for the two tested values of q. Colored lines are frequency spectra derived from the results of 1000 forward-time simulations for each parameter set. For each evolved population, 1000 individuals were sampled from the final timepoint of an initially clonal population evolved for 15000 generations. s and Ub for small and large population sizes are the same as those quoted in Figure 3. (B) The site frequency spectrum of beneficial semiprivate variants (a magnification of A in the regime x ≪ 1). The site frequency spectrum for each parameter set is normalized by the number of purely private mutations (i.e., sites with lineage sizes n = 1). The 1/n dashed black line is the decay predicted from Equation 21 for small n, and the 1/n2 dashed black line indicates the crossover to the (approximately) 1/n2 decay predicted for more common alleles. Dashed vertical lines represent the predicted realm of validity of the 1/n decay for each parameter set, derived in File S1. Colored lines are frequency spectra derived from the results of 1000 forward-time simulations for each parameter set. For each evolved population, the total number of mutations carried by n individuals were explicitly tabulated from the final timepoint of an initially clonal population evolved for 15000 generations. Parameter sets are the same as those quoted in Figure 3.
As we have previously mentioned, this distribution describes only frequencies of mutations that are founded in the exponentially expanding, high-fitness front of the wave. As such, it fails for rare mutations, which are overwhelmingly dominated by mutations that are introduced when the class is near the mean of the distribution. A different approach is then necessary for understanding the spectrum of these extremely low-frequency mutations.
Fortunately, all the difficulties in accounting for effects of genetic draft and the stochasticity of the wavefront are no longer a factor when dealing with these rare variants. By definition, the lineage of a rare variant never comprises a substantial fraction of the population and only rarely acquires further (establishing) beneficial mutations. Thus, the site frequency spectrum of these individuals can be studied using standard branching process methods given some fixed death rate d = 1 and a diminishing birth rate b(t) = 1 + y0 − vt, where t is the time in generations, y0 is the initial (relative) fitness of the mutant, and v is the mean rate of adaptation of the population. Furthermore, because these mutants occur in the bulk of the fitness distribution, where the number of individuals is very large, we can assume that they occur deterministically at some rate Ubnk−1(t).
In supporting information, File S1, we show that as a consequence of these assumptions, the expected number of mutations with lineage sizes n in a class at fitness y = 0, Frare(n/N, 0), is given by
| (21) |
under the constraints that and . Significantly, the density of sites at the rare end of the frequency spectrum, frare(n/N) ∝ Frare(n/N, 0) ∝ 1/n. Note that in comparing to the Wright–Fisher results we must multiply Equation 21 by a factor of ∼2, which reflects the different stochastic dynamics of the branching process and Wright–Fisher model.
It is important to observe that these frequencies are at the extremely low end of what is colloquially considered to be a “rare” variant, and hence we dub these mutations more precisely as “nearly private” or “semiprivate.” For the frequencies commonly measured in a reasonably sized population sample, rare but non-singleton variants will still decay as 1/x2, and the effect of this skew for nearly private mutations will manifest itself as a smaller number of singletons than that predicted simply by the 1/x2 extrapolation.
The frequencies of mutant sites thus fall into two regimes. Common alleles founded at the wavefront have distributions similar to those of exponentially expanding populations. Conversely, semiprivate variants are largely founded recently in the past and in the bulk of the distribution and as a result exhibit a neutral SFS. This latter property is only to be expected, since a mutant landing in the mean fitness class has neutral relative fitness by definition, regardless of the specific fitness effects of the mutations it carries. These findings are supported by our forward-time simulations, demonstrated in Figure 4.
There is necessarily some crossover region between the regime of more common alleles and semiprivate variants, occurring in the region of landing fitness y0 ∼ (q − 2)s. Near these fitnesses, newly founded sites are no longer well described as originating in the expanding, high-fitness front of the wave; however, over the course of their existence a sufficiently significant number of them may reach large enough lineage sizes to be affected by draft. In this case, the distribution of site frequencies in fitness classes near the population’s bulk is skewed by mutations that were not founded near the nose, but still jumped into fitter classes. Such lineages will certainly contribute a potentially nonnegligible number of sites at the rare end of the spectrum (at lineage sizes larger than the semiprivate ones we have studied). However, because they do not change the results qualitatively, they are neglected in this work. This choice is supported by the rapid crossover between the 1/x rare variant decay and the (approximately) 1/x2 decay predicted for more common alleles, as exemplified by Figure 4.
The site frequency spectrum of neutral mutations
Our method is similarly well suited for calculating the SFS of neutral mutations. Once a mutation is present at some frequency in a given class, its frequency in subsequent classes is purely determined by draft. Thus, the only difference between the beneficial and neutral cases is in the distribution of mutations first introduced in a given class when that class was at the distribution’s nose, f1(x). The effect of draft on these mutations i classes later is then obtained by convolution with the jump probability,
| (22) |
where ρi−1(x|y) denotes the probability density of a mutation at frequency x in a class i − 1, given that it was at frequency y in class 0. The total site frequency spectrum is then obtained by summing over all timepoints, corresponding to all possible originating classes:
| (23) |
In File S1 we show that fneut,1(x) ≈ Un/((q − 1)sx2) to leading order, where Un is the per-genome neutral mutation rate. Similarly, fben,1(x), the distribution of newly introduced beneficial mutations in a given class, was derived in Equation 17 to be
| (24) |
for q → ∞. In this limit, it is true that
| (25) |
As a result,
| (26) |
and by extension (since this holds for each i),
| (27) |
in the limit q → ∞. Thus, in the limit of rapid adaptation, the neutral and beneficial site frequency spectra differ only by a scaling factor set by the rate of neutral mutation relative to the strength of selection. This relationship demonstrates the fact that, as beneficial mutations become more common, the relative importance of a mutation’s intrinsic fitness effect diminishes relative to the quality of its genetic background. Thus, the relative site frequency spectra are roughly set by the rate at which neutral mutations accrue in the nose classes relative to beneficial mutations. Although only strictly true in the infinite adaptation limit, our simulations demonstrate that this approximation is already accurate for q as small as 4 (Figure 5).
Figure 5.
(A) The site frequency spectrum of common neutral mutations. The dashed lines are the theoretical predictions using the exact scaling in Equation 27 for the two tested values of q. Colored lines are frequency spectra derived from the results of forward-time simulations for each parameter set. For each evolved population, 1000 individuals were sampled from the final timepoint of an initially clonal population evolved for 15000 generations. Parameter sets are the same as those quoted in Figure 3, and additionally Un = Ub for each parameter set. (B) The site frequency spectrum of neutral semiprivate variants (a magnification of A in the regime x ≪ 1). The site frequency spectrum for each parameter set is normalized by the number of purely private neutral mutations (i.e., neutral sites with lineage sizes n = 1). The 1/n dashed black line is the decay predicted from Equation 28 for small n, and the 1/n2 dashed black line indicates the crossover to the (approximately) 1/n2 decay predicted for more common neutral alleles. Dashed vertical lines represent the predicted realm of validity of the 1/n decay for each parameter set, derived in File S1. Colored lines are frequency spectra derived from the results of forward-time simulations for each parameter set. For each evolved population, the total number of mutations carried by n individuals were explicitly tabulated from the final timepoint of an initially clonal population evolved for 15000 generations. Parameter sets are the same as those quoted in Figure 3, and additionally Un = Ub for each parameter set.
To compute the site frequency spectrum of neutral semiprivate variants, the derivation follows identically as for beneficial mutations, with s → 0, Ub → Un. The result may then immediately be written down to be
| (28) |
which holds so long as . This 1/n dependence is demonstrated in Figure 5.
Sojourn Times
So far, we have described the effect of genetic draft on the frequencies of lineages as they jump through fitter and fitter fitness classes and characterized the effects of these jumps in skewing the resulting beneficial and neutral SFS. Now, we are ready to make predictions regarding fates and trajectories of observed polymorphic sites in these populations. One of the most important predictions to be made is the time to fixation of a beneficial allele.
In contrast to the strong selection, weak mutation regime, in the strong selection, strong mutation regime, the lineage carrying a particular mutation usually jumps through many fitness classes before fixing in any one. Furthermore, it takes time for the class in which the mutation first fixed to traverse the length of the wave, adding ∼2q〈τk〉 generations before the mutation is fixed in the population. Thus, in studying the fates of mutations, there are two pertinent questions. First, given a mutation at some measured frequency, how long does it take before the mutation sweeps or goes extinct? Second, given a newly established fitness class, how long does it take any mutation introduced in this class to sweep (equivalently, what is the expected time to fixation of a new mutation that is destined to fix)?
Because our method assumes that lineages in the feeding class are frozen once they begin to feed establishing mutants into the next class, our method is poorly equipped to deal with lineages at very high or low frequencies that are strongly affected by drift. Furthermore, the pathologies of our distribution (which, treating nk as a continuous variable, allows for fractional numbers of individuals) introduces errors in the regime of nk ∼ 1. Nevertheless, we can calculate the sojourn time of these mutants by predicting when the frequency of a given mutation is expected to fall above or below a small threshold frequency ε. All the above problems may be circumvented if ε is taken to be small, but large enough for the lineage to be established in the feeding class when it begins supplying establishing mutants (roughly, this is fulfilled when ). When a lineage falls below frequency ε or rises above frequency 1 − ε in a given class, its probability of extinction or fixation is then 1 − ε, which is nearly certain if ε is sufficiently small. We note that this scenario more accurately imitates what one could measure in an experimental setting, with a sample that is much smaller than the total population size or (if performing whole population sequencing) some finite-sequencing read depth. In these experimental scenarios, the absence of a particular polymorphism in such a measurement may not mean that the polymorphism is extinct entirely, but rather that it is unlikely to be present in the population above a certain frequency.
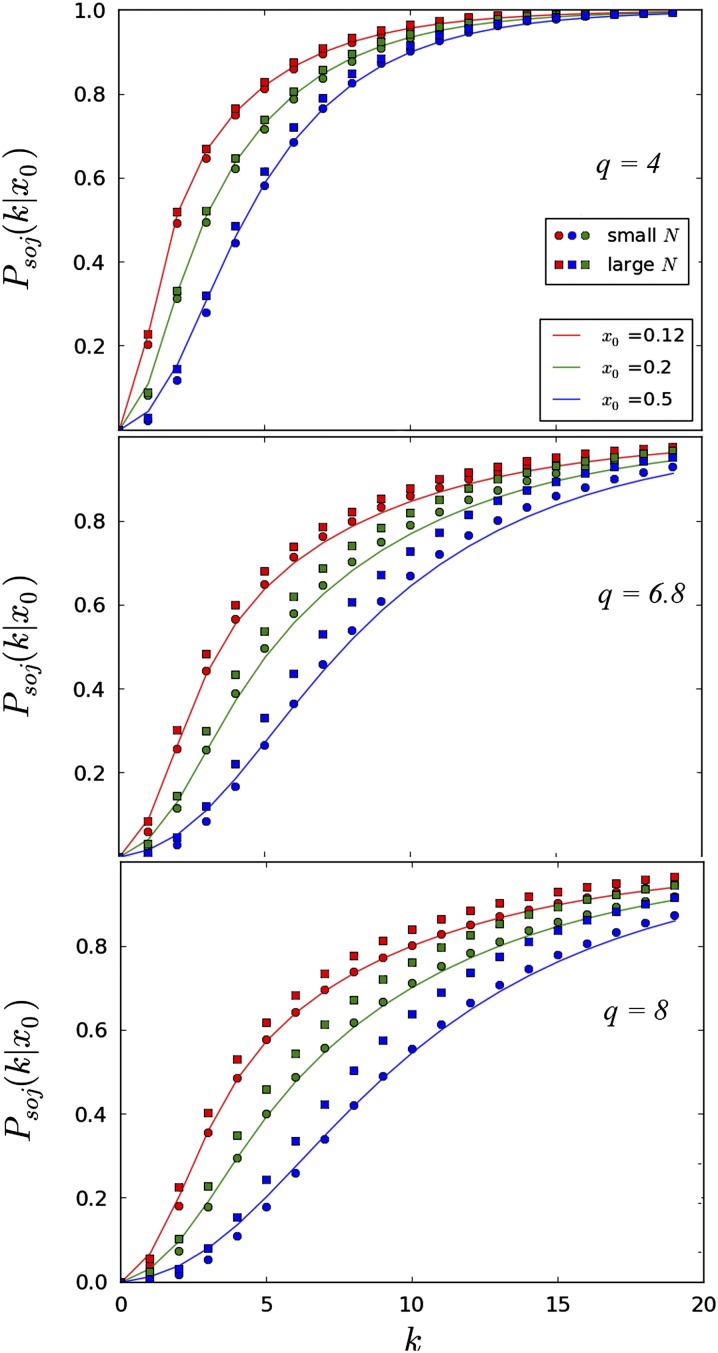
The distribution governing the sojourn time Psoj(k|x0)—the probability that a site at a frequency x0 in fitness class 0 has a frequency xk in fitness class k that freezes below the threshold ε (or above 1 − ε)—is calculated as
| (29) |
where ρ and Π are given in Equation 15 and Equation A7, respectively, and ζ = ε/(1 − ε). If the initial reference class is assumed to be the mean class, this is the probability that the mutation will be fixed or extinct in the mean class k〈τk〉 generations later. It will then require q〈τk〉 more generations before the mutation is fixed or extinct population-wide, corresponding to the time for the mean fitness class to go extinct. Given a mutation at a measured frequency x0 at t = 0, the true, population-wide sojourn time in generations is then soj((k + q)〈τk〉|x0) = Psoj(k|x0). Both simulated and predicted sojourn times for a number of different parameters are shown in Figure 6.
Figure 6.
The sojourn probability Psoj(k|x0) plotted for different values of x0, q, and other population parameters Ub, s, and N. Curves represent the theoretical prediction for the sojourn times given in Equation 29. Circles and squares represent the results of forward-time simulations. The circular and square points have identical values of q, but differ in population size by a factor of 10. Circles represent simulations run with parameters s = 0.1, Ub = 0.001, and squares are run with parameters s = 0.01, Ub = 0.0001. The three plots represent different values of q, with q = 4 calculated from the approximate value in the text, and q = 6.8, 8 calculated with higher-order corrections given in Desai and Fisher (2007). For both simulated and theoretical curves, mutations were considered extinct or fixed in a given class when they fell below or rose above a cutoff of ε = 0.03 or (1 − ε) = 0.97, respectively. More information about the simulations may be found in File S1.
A similar method is used to calculate the expected time for fixation of any lineage in a given reference fitness class, which we dub Pfix(k). Specifically, Pfix(k) is the probability that one of the mutations founded in class 0 is past the threshold 1 − ε, k fitness classes forward. Using the distribution of new mutations given in Equation 17, the probability that one is at frequency greater than (1 − ε) in class k is
| (30) |
for ε ≪ 1 and k large. In this case, the expected fixation time is
| (31) |
for q large, where the last relation holds for ε ∼ 10−2. Thus, it takes a time of about 2q〈τk〉 for a mutation destined to fix to sweep in any class and an additional time of about 2q〈τk〉 for that class to sweep through the population. The expected sweep time for successful mutations is then (4q〈τk〉) ∼ (4 log(s/Ub)/s) for q large.
We note that the time for any mutation to cross some threshold frequency 1 − ε is closely related to the time it takes a sample of n ≫ 1 individuals to coalesce within some fitness class, with ε ∝ 1/n. In this case, for the coalescence time we recover the formula q(log(log(n)) + (1)) derived by both Desai et al. (2013) and Neher and Hallatschek (2013) for the corresponding backward process (for the latter work, the factor of q must be replaced by the model independent Tc, the coalescence time of two individuals in the high-fitness nose).
Discussion
We have used a simple, infinite-sites model of adaptation featuring a single beneficial selection coefficient to carefully account for the effects of genetic drift, mutation, selection, and by extension, genetic draft in determining the evolutionary dynamics of polymorphic sites. In what follows we discuss the connection between the dynamics of rapid adaptation and the Bolthausen–Sznitman coalescent, and comment on applications of our results for interpreting frequency trajectories and inferring the action of positive selection on sequence data. We also discuss pertinent similarities between our model and other models of genetic draft. Finally, we note that our method might be applied toward understanding evolution on populations with strong epistasis between evolutionary trajectories.
Rapid adaptation and the Bolthausen–Sznitman coalescent
Having established the effect of genetic draft on the frequencies of polymorphic sites, we then explored the effect that the stochastic jumps characterizing the process have on genetic diversity. In particular, we derived the site frequency spectrum of both beneficial and neutral mutations. As expected, we found that nearly private variants—which overwhelmingly occur and drift near the mean class—decay according to the well-known 1/x behavior predicted for unlinked neutral sites, whereas common mutations exhibit site frequency spectra more closely related to those of exponentially expanding clones, with the additional feature of an upswing at high frequency that is characteristic of adaptation for many models (Messer and Petrov 2013; Neher and Hallatschek 2013).
Of particular interest is the diversity of populations in the limiting case of q → ∞, whose genealogies obey the Bolthausen–Sznitman coalescent. Note that in this case, the Bolthausen–Sznitman coalescence rates apply for individuals in the lead, with each “generation” taken over the time of a new fitness class establishment (elaboration on this setup can be found in Desai et al. 2013). Although this genealogical structure was observed previously in Desai et al. (2013), in the context of these stochastic jumps its origin is intuitive: in the high-q limit, the contribution of one individual at the wavefront to the next class falls off as 1/x2, which can be seen by taking xk−1 → 0 in Equation 16. As proven by Schweinsberg (2003), the genealogies of populations with offspring distributions that decay as x−2 converge to the Bolthausen–Sznitman coalescent in the limit of N → ∞. The 1/x2 decay is also well known to describe the frequency spectrum of exponentially expanding populations and in the case of adapting populations results from the exponential expansion of new mutants at the front of the wave. The Bolthausen–Sznitman coalescent has been associated with a growing number of different adaptive models (Brunet and Derrida 2012, 2013; Desai et al. 2013; Neher and Hallatschek 2013), suggesting that such genealogies may be a universal limiting feature of rapid adaptation.
Quasi-neutrality over long timescales
When many beneficial mutations segregate simultaneously, frequency dynamics of mutations begin to exhibit a qualitatively different behavior than those in the strong selection, weak mutation regime. After the introduction and establishment of a mutation in the fittest class, a mutation’s fixation probability is equal to its frequency in that class, regardless of its intrinsic fitness effect. To see this, assume a mutation first freezes to some frequency x0 in its founding class. Then, since xk→∞ ∈ {0, 1} (i.e., at long times the allele is either fixed or extinct), this implies that the fixation probability
| (32) |
Essentially, this means that once a mutation founded at the high-fitness wavefront freezes to a particular frequency in a class, its likelihood of success depends only on its frequency in that class (at least, without more information about its frequency in fitter classes). Note, however, that the above formula technically holds only for mutations at high enough frequencies to have necessarily been founded near the distribution’s nose class, since mutations founded away from the wavefront have fixation probabilities that are virtually zero. However, since these latecomers never reach more than a negligible frequency in their class, the above formula still describes the fixation probability of a randomly selected mutant reasonably well.
Now, once a mutation freezes in the fittest class, it will typically be present in the mean fitness class a time q〈τk〉 later. This corresponds to the timescale for the fittest class to become the mean class. Since the mean class consists of the majority of individuals in the population, the measured frequency of the mutant at this time is a good approximation to its frequency in the mean class. Barring any information about the frequency of the mutation in fitter classes, its measured frequency at this time is then roughly its fixation probability. (A better approximation is straightforwardly obtained by taking into account the fact that the mutation’s frequency in classes below the mean is identically 0 at this time.) This equality becomes (nearly) exact at a time of (2q〈τk〉), when the founding fitness class becomes the least fit in the population, and the mutation’s frequency in all classes is determined solely by draft.
These considerations have important consequences for predicting the fate of polymorphic sites in experimentally evolving populations. In a population that is adapting rapidly enough for a stratification of fitnesses to always be present, sites that are polymorphic for sufficiently long have dynamics that are indistinguishable from neutral mutations at comparable frequencies in these populations, because they are determined only by genetic draft. Once the frequency of a mutation is frozen in a particular fitness class, its fitness effect alone is no longer important in determining its future success. This is because it is only the net fitness of the mutant and its genetic background that is important in determining its dynamics, and not the fitness effect of the mutation itself. Because it takes a time of (2q〈τk〉) for a mutation founded at the nose to occupy all strata of fitnesses in the population, common mutations in these rapidly adapting populations can be thought to have a relaxation time of 2q〈τk〉, beyond which their population-wide frequency approaches their frequency in the mean class, and the fitness of their founding haplotype decouples from their population-wide dynamics.
In short, the significance of this finding is simple: if a mutation—regardless of its fitness effect—has been measurable in the population for longer than 2q〈τk〉 generations, its fixation probability is equal to its frequency in the population. This approximation should already be quite good (albeit consistently too low due to the inclusion of less-fit classes where the mutation is absent) at a time q〈τk〉.
These considerations provide a parallel to Haldane’s formula for the fixation probability of a beneficial mutation in the successive sweep regime, Pfix = s. Whereas this formula gives the likelihood of a mutation to fix despite the stochastic effects of genetic drift, the probability of a long-standing polymorphism to fix despite the randomizing effect of draft is simply Pfix = x, where x is the frequency of the mutation in the population. The two forces are similar in that they stochastically amplify the fluctuations in the trajectories of polymorphic sites, but the length- and timescale of the fluctuations caused by draft are much greater.
Implications for the McDonald–Kreitman test
Another useful application of our findings is the ability to analytically correct for the effect of genetic draft on the results of tests for signals of adaptation, such as the McDonald–Kreitman test (McDonald and Kreitman 1991). This test, along with many other widely used tests for selection, assumes that beneficial mutations are rare and segregate independently. However, both assumptions are invalid for rapidly adapting populations, and new analytical predictions are needed. Fortunately, we demonstrate that our method provides a simple way to correct for the effect of linkage between beneficial mutations. For the case of the McDonald–Kreitman test, this correction is straightforwardly obtained by accounting for the extra heterozygosity contributed by many simultaneously segregating beneficial mutations.
The McDonald–Kreitman test approximates the fraction αMK of nucleotide substitutions that are adaptive by considering relative quantities of fixed and polymorphic, synonymous and nonsynonymous sites between two diverged populations. The fraction of adaptive substitutions is simply
| (33) |
where d+, ds, and dn are the adaptive, synonymous, and nonsynonymous substitution rates, and pn, ps are the numbers of nonsynonymous and synonymous polymorphisms in one of the sampled populations, respectively. The last approximation arises from the assumption that
| (34) |
where is the substitution rate of nonadaptive, nonsynonymous mutations and is the number of nonadaptive, nonsynonymous polymorphic sites in the sample. Implicit in Equation 34 are several assumptions: first, the rate of nonadaptive nonsynonymous substitutions in the sample is equal to the rate of synonymous substitutions, scaled by the relative frequencies of nonadaptive nonsynonymous to synonymous polymorphisms. This implicitly assumes that deleterious mutations do not fix, that deleterious mutations do not significantly contribute to the measured numbers of nonsynonymous polymorphisms in the sample, and that the population has not undergone any demographic change to skew the distributions of polymorphic sites. Second, it is assumed that the number of nonadaptive, nonsynonymous polymorphisms is precisely equal to the measured numbers of nonsynonymous polymorphisms. In other words, beneficial mutations are rare and fix quickly upon arising; thus, they are rarely present in the population as polymorphisms.
Of course, these assumptions break down when deleterious or beneficial mutations significantly contribute to the number of polymorphic sites and when linkage between sites skews the relative frequencies of mutations. These issues usually result in a measured αMK that severely underestimates the true fraction of adaptive substitutions. A large body of work has been put forward in an effort to correct for this skew (Andolfatto 2008; Charlesworth and Eyre-Walker 2008; Eyre-Walker and Keightley 2009; Messer and Petrov 2013). For example, introducing a low-frequency cutoff for measured polymorphisms significantly improves estimates of αMK for the case of many weakly deleterious mutations (Fay et al. 2001; Charlesworth and Eyre-Walker 2008), since in the absence of genetic linkage few deleterious mutations will ever reach high frequencies. However, few studies have carefully analyzed the effect of linkage on αMK, particularly the effect of linked beneficial mutations. One notable exception is the work of Messer and Petrov (2013), who used a sophisticated extension of the McDonald–Kreitman test accounting for demographic history and distributions of fitness effects to infer αMK from simulated rapidly adapting populations. The authors found that the inference of a massive population expansion (derived from the site frequency spectrum of the sample) resulted in superior estimates of αMK, although no such expansion ever occurred in the simulation. The intuition guiding this finding is that the site frequency spectrum of a population undergoing rapid adaptation resembles that of a population undergoing an exponential expansion. Regardless, analytic corrections for the effect of genetic draft have yet to be derived and would provide for a more straightforward way of accounting for this confounding factor.
Our results provide a simple analytical correction to αMK for the case of tightly linked sections of the genome that accounts for genetic draft. First, we note that the number of polymorphic sites is closely related to heterozygosity π, the average number of nucleotide differences between two randomly drawn individuals, through
| (35) |
Thus, the ratio pn/ps is simply
| (36) |
In general, the moments of the frequencies of synonymous and nonsynonymous sites may be measured straightforwardly from the site frequency spectrum of the sample. However, as we have already demonstrated, in the rapid adaptation limit the SFS of neutral and beneficial mutations is similar in form. Thus, the heterozygosities of beneficial and neutral mutations are largely determined by different numbers of neutral and selected sites—i.e., by pn and ps—rather than significantly different frequency distributions.
The heterozygosity for beneficial mutations is calculated from the moments of ρ(xk|x0) (using the same method used in the calculation of the beneficial SFS) to be π ≈ 2(q − 1), with higher-order corrections given in Desai et al. (2013). The neutral heterozygosity is simply 2UnT2, where T2 is the expected coalescence time for two randomly chosen individuals. The rate of beneficial substitutions is 1/〈τk〉, and the rate of neutral substitutions is Un. Thus, given the rate of synonymous mutations, Un,s, and the rate of neutral nonsynonymous mutations, Un,n, the expected, measured value of αMK (by naively plugging in each measured value in Equation 33) will be
| (37) |
Clearly, only the first term of πn corresponds to nonadaptive sites. Thus, we have
| (38) |
This gives for the fraction of adaptive substitutions
| (39) |
In practice, the parameter q is measurable from estimates of N, s, and Ub or the distribution of fitnesses within the population.
The interpretation of this correction is intuitively simple. In populations where adaptation is rapid, the assumption that beneficial mutations do not contribute significantly to measured polymorphism breaks down. As a result, ascribing all measured nonsynonymous polymorphism pn to neutral (or deleterious) mutations results in an underestimate of d+/dn. In fact, in the limit of infinitely rapid adaptation, q → ∞, αMK,meas → 1/2, in contrast to the true fraction of adaptive substitutions, d+/dn → 1. Our model, then, provides a simple correction for this underestimate by predicting the expected fraction of observed nonsynonymous polymorphism (relative to synonymous polymorphism) that arises from beneficial mutations.
Relation to other models of genetic draft
There is a natural connection between our model and the classic model of genetic draft between a neutral locus and a strongly selected locus, first studied by Gillespie (2000, 2001). In these seminal works, the diffusive random walk of a neutral allele is coupled with the stochastic process of a hitchhiking event occurring at rate R, which drives the neutral allele to either fixation or extinction. In Gillespie’s model, the time required to fix the strongly selected allele relative to the time between substitutions is small enough that fixation/extinction is assumed to occur instantaneously. One basic result derived under the assumptions of such a model are the first two moments of the stochastic jump process: 〈Δx〉 = 0, 〈Δx2〉 = Rx0(1 − x0), for Δx = x1 − x0 [here, x1 denotes the frequency of the neutral allele after some time of (〈τfix〉) for the strongly selected locus]. It turns out that the stochastic process described by Gillespie emerges naturally as the weak mutation limit of the model studied in this work.
To demonstrate this, we observe that the first two moments of our jump distribution ρ(x1|x0) are 〈Δx〉 = 0 and 〈Δx2〉 = x0(1 − x0)/q (the derivation, along with a general formula for higher order moments, is given in File S1). The similar functional dependence on x0 between the two models is not particularly surprising, since it arises from a general property of any exchangeable coalescent (because the jump process describes the changing composition of each new lead class, and because all individuals within the lead have the same fitness, our model is exchangeable). However, the prefactor for each moment depends strongly on the stochastic dynamics predicted by different models. To compare the stochastic dynamics described by the different works, we first note that all of our results take time in units of fitness-class establishments, which is equal to the substitution time of beneficial mutations. In the weak mutation limit, we have q → 1, so that 〈Δx2〉 → x0(1 − x0) = Rx0(1 − x0), where R is the substitution rate of beneficial mutations. Higher-order moments of the two distributions are also similarly related, so that Gillespie’s results emerge from our model in the limit that q → 1. Thus, our results generalize the stochastic process describing genetic draft to the regime where the time between beneficial mutations is no longer large.
Next, we might ask how generalizable our results are when compared with genetic draft arising from different sources. For example, Neher and Shraiman (2011) recently analyzed the effect of draft arising from infrequent recombination events instead of de novo mutations. They introduced an approach similar to ours to study how the frequency of an allele changes over the course of a “meta-generation” set by a typical turnover timescale. The resulting dynamics are qualitatively similar to many features of our model. Specifically, Neher and Shraiman (2011) found that the origination of high-fitness clonal bubbles determines the long-term frequency dynamics of an allele and showed that these dynamics can be encapsulated in an effective offspring distribution for recombinant genotypes, which decays asymptotically as x−2. They then linked this effective offspring distribution to genealogies described by multiple merger coalescents. As a consequence, they found that the asymptotic form of the site frequency spectrum also decays asymptotically as x−2 for both neutral and beneficial alleles, analogous to our results here. The parallels between these two models reflect some general effects of draft on the dynamics of linked sites, which are robust to the specific source of the randomizing effect.
Applications to forks and more complicated fitness landscapes
Finally, because of the flexibility of our model with respect to the underlying fitness landscape, it is straightforward to extend our results to a small class of more general models involving complex interactions between sites. Specifically, we might consider the case in which two or more disjoint and incompatible evolutionary pathways are possible. That is, each individual can acquire one of two or more distinct sets of beneficial mutations, but mutations from different sets are incompatible. Note here that different evolutionary pathways are effectively incompatible if there is sign epistasis between them that is at least as strong as ∼s, so that any individual that acquires mutations from two or more pathways must leave the lead of the wave and is unlikely to fix as a result. In this scenario, we can apply our approach to analyzing the jumps from class to class for each different pathway separately. Different relative mutation rates for each pathway simply contribute a different prefactor in the exponent of the generating function for each contributing lineage (i.e., replacing the xℓ,k−1 with some relative rate μℓ in Equation 12). This allows us to apply our method to answer questions about the probability that the population will follow any given path, its time to fixation, and the degree to which the population will simultaneously explore the other pathways.
These considerations provide a different perspective on the fates of populations evolving on rugged fitness landscapes, and particularly on the effect of a larger population size in avoiding local fitness peaks. Previous works have suggested that over certain timescales, smaller populations may have an advantage in adapting on these rugged landscapes, because their trajectories are more heterogeneous, whereas larger populations have an increased tendency to get stuck on local fitness peaks (Rozen et al. 2008; Handel and Rozen 2009; Jain et al. 2010; Szendro et al. 2013). However, our analysis suggests that if the landscape is dominated by several distinct uphill trajectories featuring mutational steps of similar size, large populations may be capable of traveling for many steps down multiple paths, effectively exploring the surrounding landscape before settling upon one particular uphill trajectory. For example, in the case of a simple fork with equal mutation rates down each pathway, a large, rapidly adapting population will typically explore about 2q mutational steps forward before a particular pathway is closed off. The transition between this behavior and that considered in the work cited above evidently occurs between classical clonal interference, in which adaptation is dominated by the rare emergence of extremely fit mutants, and the multiple mutations regime, in which most fixed mutations are of roughly the same size. This, in turn, strongly depends on the distribution of fitness effects of mutations, with long-tailed or short-tailed distributions giving rise to dynamics dominated by clonal interference or multiple mutations, respectively (Desai and Fisher 2007; Fogle et al. 2008). The different outcomes predicted by these two regimes could explain the lack of experimental consensus on the effect of population size on outcomes of adaptation (Rozen et al. 2008; Schoustra et al. 2009; Miller et al. 2011).
Conclusions and Future Work
By using a simple model, we have made considerable headway in understanding how genetic draft affects the frequencies of mutations through a series of stochastic jumps, how these jumps affect genetic diversity, sojourn times, and fixation times of mutations, and why these statistics resemble those derived from the Bolthausen–Sznitman coalescent. We then showed how our method leads to a simple correction to the McDonald–Kreitman test that accounts for linkage between beneficial mutations. Finally, we discussed how our analysis might be extended to describe evolution on certain classes of rugged fitness landscapes, which—although admittedly very simple—nonetheless describe limiting behavior for sign epistasis between multiple evolutionary pathways.
Still, our model has some shortcomings. First, we neglect recombination, making our results applicable only to the evolution of microbial populations and tightly linked regions of the genomes of sexually reproducing organisms. Naturally, in cases where recombination is no longer rare, the effects of genetic draft are tempered as competing beneficial mutations recombine onto a single genetic background. Fitness classes that evolve disjointly in the asexual model are then allowed to mingle at each reproductive step, meaning that a series of stochastic jumps between classes no longer correctly describe the dynamics. Fortunately, the work of Neher and Shraiman (2011), which accounts for the effects of occasional (facultative) outcrossing of clones provides a framework for combining these two sources of genetic draft. In particular, since common mutations must at one point propagate near the most-fit class, evolutionary dynamics in these populations are still largely informed by the distribution of haplotypes in the nose. This distribution would then obtain contributions from both mutations from the adjacent class and recombined haplotypes obtained from mating between less-fit clones.
We also make use of the assumption of a single selection coefficient. Indeed, two facets of our model that are key in deriving analytical results—the organization of clones according to fitness classes and the asymptotic freezing of frequencies in each class—both break down when a single selection coefficient is replaced with some distribution of fitness effects. However, several works (Desai and Fisher 2007; Good et al. 2012) have shown that even in populations with a distribution of fitness effects, evolutionary dynamics are well described by the use of an effective, or predominant selection coefficient, which coincides exactly with the most common fixed mutational effect. Still, the inclusion of a distribution of fitness effects, and the resulting unified understanding of the effects of multiple mutations and mutations of varying effect sizes in driving evolutionary dynamics, remains a promising subject of future work.
Supplementary Material
Acknowledgments
We thank Benjamin Good, Sergey Kryazhimskiy, Richard Neher, and an anonymous reviewer for their helpful comments and suggestions. This work was supported by a National Science Foundation Graduate Research Fellowship (K.K.) and by the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, and grant GM104239 from the National Institutes of Health (M.M.D.).
Appendix: Dynamics of the Transition Process
In this appendix we show that frequencies of mutant lineages are frozen when a class begins feeding mutants to the lead that are destined to establish. We then explicitly derive the probability distribution ρk of a transition in a mutation’s frequency from a starting class 0 to some final fitness class k.
To prove that frequencies of mutant lineages are frozen when a class begins supplying establishing mutants to the next class, we first note that the Lth establishing mutant in a given fitness class typically occurs at time tL such that
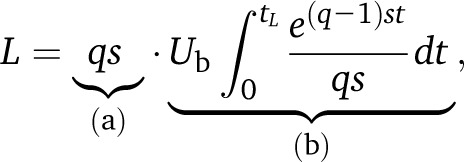 |
(A1) |
where (a) is the establishment probability of one mutant and (b) is the total number of mutants introduced into the lead class by time tL. Thus, using the same argument as in Desai et al. (2013), the amount that the Lth establishing lineage contributes to a fitness class as a fraction of the first lineage is
| (A2) |
Note that this is an upper limit on the contribution of ηL,k, since the growth of each subsequent lineage actually decreases according to the rate of adaptation v. A fitness class typically establishes in a time 〈τk〉 and generates its first establishing mutant a time t1 after that. If we neglect the decreasing growth rate due to adaptation of the population (valid for large q), then at this point, the class below it has supplied
| (A3) |
establishing mutants.
In practice, however, few biological populations evolve with q > 4, in which case the diminishing growth rate becomes important in the above calculation. A simple modification of the analysis gives the number of establishing lineages to be roughly
for q > 3, which, although considerably smaller than the asymptotic value of s/Ub is offset by the more rapid decay of ηL,k/η1,k for smaller q. Extensions for q = 2, 3 are likewise straightforward.
Thus, we have demonstrated that the contribution of subsequent mutations diminishes rapidly, and by the time a fitness class begins feeding establishing mutants to the next class, it has (s/Ub) ≫ 1 mutations that are destined to establish in it. As a result, the contribution of subsequent mutations after this time is already very small, meaning that the frequencies of common lineages in the fitness class at this time may safely be treated as frozen.
The astute reader might also note that mutant lineages that are destined to go extinct may also contribute to shifting the frequencies, which is certainly a contributing factor close to the time that the class establishes. Although not as straightforward a calculation, if s ≫ Ub then it is still true that by the time a class begins feeding establishing mutations, the contribution of these “doomed lineages” is also small (File S1). Thus we are justified in treating the process as a Markov chain.
Now we are ready to derive the transition probability of a mutation at frequency x0 in class 0 to some frequency xk in class k. Desai and Fisher (2007, Equation 29) showed that the contribution to class k, νk, from a lineage at frequency xk−1 in class k − 1 is described by the generating function
| (A4) |
Thus, after class k establishes, the lineage grows as
| (A5) |
The generating function for the contribution νk+1 of the mutation 2 fitness steps forward is then obtained by averaging over the intermediate value νk,
| (A6) |
where the step in line 2 makes use of the standard formula for inverting the Laplace transform. Although technically the average should be taken over the frequency xk (which is analytically intractable), an average over νk is a reasonable approximation if σk, the growth of the entire class, does not deviate too much from its typical value, and if the growth of the lineage νk in future classes may be taken independently of the growth of other lineages. Because the effect of this approximation is compounded at each step forward, it introduces significant deviations at timescales of roughly q fitness steps forward, which is the timescale over which fluctuations in the advance of the fitness wave become significant. Accepting this approximation, it is then straightforward to show that
| (A7) |
Without loss of generality, we can index the class in which the mutant begins to be tracked to 0. The fitness class at k = 1 can then be divided into those individuals descended from a particular lineage in class 0, at frequency x0, and those not descended from that lineage, at frequency (1 − x0). Since, for each subsequent fitness class, new frequencies are frozen near the nose, where the two sets of individuals proliferate independently, there are two independent variables encoding the fate of the lineage: νk, denoting the contribution of the lineage to a class with k more beneficial mutations, and , denoting the contribution of individuals not derived from that lineage, so that .
If we denote the probability densities of νk and by Pk(νk|x0) and , respectively, then
| (A8) |
where ε ∈ ℝ, ε → 0−, and step 4 to 5 makes use of a transformation z = −iz1 + iε. Simplifying gives
| (A9) |
Finally, we perform a change of variables to , giving the jump distribution,
| (A10) |
This function denotes the probability density of observing a mutation at frequency xk in class k, given that it was at frequency x0 in class 0. The function cited in the text is ρ(x1|x0) = ρ1(x1|x0).
Footnotes
Communicating editor: L. M Wahl
Literature Cited
- Andolfatto P., 2008. Controlling type-i error of the McDonald–Kreitman test in genomewide scans for selection on noncoding DNA. Genetics 180: 1767–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet E., Derrida B., 2012. How genealogies are affected by the speed of evolution. Philos. Mag. 92: 255–271 [Google Scholar]
- Brunet E., Derrida B., 2013. Genealogies in simple models of evolution. J. Stat. Phys. P01006 [Google Scholar]
- Brunet E., Rouzine I. M., Wilke C. O., 2008. The stochastic edge in adaptive evolution. Genetics 179: 603–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth J., Eyre-Walker A., 2008. The McDonald–Kreitman test and slightly deleterious mutations. Mol. Biol. Evol. 25: 1007–1015 [DOI] [PubMed] [Google Scholar]
- de Visser J. A., Rozen D. E., 2006. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics 172: 2093–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A., Zeyl C. W., Gerrish P. J., Blanchard J. L., Lenski R. E., 1999. Diminishing returns from mutation supply rate in asexual populations. Science 283: 404–406 [DOI] [PubMed] [Google Scholar]
- Der R., Epstein C., Plotkin J. B., 2012. Dynamics of neutral and selected alleles when the offspring distribution is skewed. Genetics 191: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Fisher D., 2007. Beneficial mutation–selection balance and the effect of linkage on positive selection. Genetics 176: 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Walczak A., Fisher D., 2013. Genetic diversity and the structure of genealogies in rapidly adapting populations. Genetics 193: 565–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldon B., Wakeley J., 2006. Coalescent processes when the distribution of offspring number among individuals is highly skewed. Genetics 172: 2621–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2009. Estimating the rate of adaptive molecular evolution in the presence of slightly deleterious mutations and population size change. Mol. Biol. Evol. 26: 2097–2108 [DOI] [PubMed] [Google Scholar]
- Fay J. C., Wu C.-I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wyckoff G. J., Wu C.-I., 2001. Positive and negative selection on the human genome. Genetics 158: 1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D. S., 2013. Asexual evolution waves: fluctuations and universality. J. Stat. Mech. P01011 [Google Scholar]
- Fogle C. A., Nagle J. L., Desai M. M., 2008. Clonal interference, multiple mutations and adaptation in large asexual populations. Genetics 180: 2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish P. J., Lenski R. E., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102–103: 127–144 [PubMed] [Google Scholar]
- Gillespie J. H., 2000. Genetic drift in an infinite population: the pseudohitchhiking model. Genetics 155: 909–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 2001. Is the population size of a species relevant to its evolution? Evolution 55: 2161–2169 [DOI] [PubMed] [Google Scholar]
- Good B. H., Rouzine I. M., Balick D. J., Hallatschek O., Desai M. M., 2012. Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proc. Natl. Acad. Sci. USA 109: 4950–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallatschek O., 2011. The noisy edge of traveling waves. Proc. Natl. Acad. Sci. USA 108: 1783–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel A., Rozen D. E., 2009. The impact of population size on the evolution of asexual microbes on smooth vs. rugged fitness landscapes. BMC Evol. Biol. 9: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R. D., Kelley J. L., Elyashiv E., Melton S. C., Auton A., et al. , 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain K., Krug J., Park S.-C., 2010. Evolutionary advantage of small populations on complex fitness landscapes. Evolution 65: 1945–1955 [DOI] [PubMed] [Google Scholar]
- Kao K. C., Sherlock G., 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40: 1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T., Messer P. W., Petrov D. A., 2010. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 6: e1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Orr H. A., 2005. Adaptation in sexuals vs. asexuals: clonal interference and the Fisher–Muller model. Genetics 171: 1377–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2003. Selective sweeps in the presence of interference among partially linked loci. Genetics 164: 389-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Botstein D., Desai M. M., 2011. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics 188: 647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Rice D. P., Hickman M. J., Sodergren E., Weinstock G. M., et al. , 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500: 571–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. H., Kreitman M., 1991. Adaptive protein evolution at the ADH locus in Drosophila. Nature 351: 652–654 [DOI] [PubMed] [Google Scholar]
- McVean G. A. T., Charlesworth B., 2000. The effects of Hill–Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155: 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer P. W., Petrov D. A., 2013. Frequent adaptation and the McDonald–Kreitman test. Proc. Natl. Acad. Sci. USA 110: 8615–8620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. R., Joyce P., Wichman H. A., 2011. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187: 185–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles R., Gerrish P. J., Moya A., Elena S. F., 1999. Clonal interference and the evolution of RNA viruses. Science 285: 1745–1747 [DOI] [PubMed] [Google Scholar]
- Muller H., 1932. Some genetic aspects of sex. Am. Nat. 66: 118–138 [Google Scholar]
- Neher R. A., Hallatschek O., 2013. Genealogies of rapidly adapting populations. Proc. Natl. Acad. Sci. USA 110: 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R., Shraiman B., 2011. Genetic draft and quasi-neutrality in large facultatively sexual populations. Genetics 188: 975–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R. A., Shraiman B. I., Fisher D. S., 2010. Rate of adaptation in large sexual populations. Genetics 184: 467–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Krug J., 2007. Clonal interference in large populations. Proc. Natl. Acad. Sci. USA 104: 18135–18140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-C., Simon D., Krug J., 2010. The speed of evolution in large asexual populations. J. Stat. Phys. 138: 381 [Google Scholar]
- Pritchard J. K., Pickrell J. K., Coop G., 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I., Wakeley J., Coffin J. M., 2003. The solitary wave of asexual evolution. Proc. Natl. Acad. Sci. USA 100: 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine I. M., Brunet E., Wilke C. O., 2008. The traveling wave approach to asexual evolution: Muller’s ratchet and speed of adaptation. Theor. Popul. Biol. 73: 24–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., Habets M. G., Handel A., de Visser J. A. G. M.2008. Heterogeneous adaptive trajectories of small populations on complex fitness landscapes. PLOS ONE 3:e1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoustra S. E., Bataillon T., Gifford D. R., Kassen R., 2009. The properties of adaptive walks in evolving populations of fungus. PLoS Biol 7: e1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg J., 2003. Coalescent processes obtained from supercritical Galton-Watson processes. Stochast. Process. Appl 160: 107–139 [Google Scholar]
- Sella G., Petrov D. A., Przeworski M., Andolfatto P., 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet 5: e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. M., Haigh J., 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35 [PubMed] [Google Scholar]
- Szendro I. G., Franke J., de Visser J. A. G. M., Krug J., 2013. Predictability of evolution depends nonmonotonically on population size. Proc. Natl. Acad. Sci. USA 110: 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsimring L., Levine H., Kessler D., 1996. RNA virus evolution via a fitness-space model. Phys. Rev. Lett. 76: 4440–4443 [DOI] [PubMed] [Google Scholar]
- Wright S., 1938. The distribution of gene frequencies under irreversible mutation. Proc. Natl. Acad. Sci. USA 24: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.