Abstract
We adapted the CRISPR–Cas9 system for template-mediated repair of targeted double-strand breaks via homologous recombination in Caenorhabditis elegans, enabling customized and efficient genome editing. This system can be used to create specific insertions, deletions, and base pair changes in the germline of C. elegans.
Keywords: CRISPR–Cas9, germline, Caenorhabditis elegans, homologous recombination, genome editing
GENOME engineering has proven to be a useful tool in biological research. Specific and accurate insertions, deletions, and replacements allow for numerous invaluable ways to examine gene function. Yet, the accurate and efficient insertion or deletion of large, specific sequences has been challenging in most metazoans (Gaj et al. 2013). In the nematode Caenorhabditis elegans, creating heritable mutations had until recently involved random mutagenesis, and the insertion of genes into the genome had been limited to random loci, often resulting in the silencing of the inserted transgene in the germline due to repetitive insertions (Kelly et al. 1997; Jackstadt et al. 1999; Wilm et al. 1999). In the past few years new methods for genome editing have become available for this model system, all of which involve the creation of a specific double-strand break in the genome. The most widely applied method uses the Mos1 transposon and transposase (Robert and Bessereau 2007; Frokjaer-Jensen et al. 2008, 2010, 2012). Although this method enables precise genome editing, it is limited by the availability of a transposon at the desired locus.
In bacteria and archaea, CRISPRs (clustered regularly interspaced short palindromic repeats) and Cas (CRISPR-associated) proteins are used to protect the cell against invading viruses and exogenous DNA (Terns and Terns 2011; Sorek et al. 2013; Wiedenheft et al. 2012). In the type II CRISPR systems, the Cas9 endonuclease specifically cleaves the exogenous DNA by interacting with two types of RNAs, tracrRNA (trans-activating CRISPR RNA), and crRNA (CRISPR RNA), which contain sequences complementary to the invading element and are generated from the CRISPR loci. Given the potential for RNA-mediated programmable DNA cleavage, this system was pared down to two components by using Cas9 from Streptococcus pyogenes and an engineered single-guide RNA (sgRNA) (Jinek et al. 2012). Recently, this simplified system was transferred to several eukaryotic cell cultures and was shown to actively create specific double-strand breaks, enabling changes in the genomes of those cells (Cho et al. 2013a; Cong et al. 2013; Dicarlo et al. 2013; Jinek et al. 2013; Mali et al. 2013). Moreover, this system can now be used for genome editing in metazoan organisms such as mice, flies, and fish (Gratz et al. 2013; Hwang et al. 2013; Shen et al. 2013; Yu et al. 2013).
We have recently reported the successful use of the CRISPR–Cas9 system in the nematode C. elegans, establishing a robust strategy for creating random insertion and deletion (indel) mutations in the germline of the worm by expressing Cas9 and sgRNA targeted to the desired site (Friedland et al. 2013). Although this method provides an efficient and facile strategy to create loss-of-function gene mutations, it involves nonhomologous end joining and therefore does not allow for user-specified changes, the complete removal of genes, or the introduction of tags. Here we report the use of this method for template-mediated repair of targeted double-strand breaks via homologous recombination, enabling customized and efficient genome editing. This article is one of six companion articles (Chiu et al. 2013; Cho et al. 2013b; Katic and Grosshans 2013; Lo et al. 2013; Waaijers et al. 2013) that present different approaches to and features of CRISPR–Cas9 genome editing in C. elegans.
Results
Site-directed gene insertion using the CRISPR–Cas9 system
We hypothesized that targeted DNA double-strand breaks created by the CRISPR–Cas9 system can also be repaired by the homologous recombination pathway and that specific insertions and/or deletions can be engineered into the germline using a donor vector (Figure 1A). To test this hypothesis, we injected young adult worms with a mixture of four plasmids (see Supporting Information, File S1) containing our codon-optimized Cas9 driven by the eft-3 promoter, a klp-12 targeting sgRNA driven by a U6 snRNA promoter (Friedland et al. 2013), an mCherry reporter driven by a body wall muscle promoter to label F1 progeny and serve as a marker for extrachromosomal array formation, and a donor vector containing an eft-3 promoter-driven GFP transgene flanked on either side by 1.5 kb of sequence homologous to regions upstream and downstream of the Cas9-induced cleavage site at the klp-12 locus (Figure 1B and Figure 2A). We isolated F1 progeny expressing GFP and mCherry and then screened for F2 animals maintaining broad GFP but not mCherry expression, because these animals may have lost the extrachromosomal array and integrated the GFP transgene into the klp-12 locus. We additionally surveyed these GFP-expressing F2 animals by amplifying regions of genomic DNA specific for the recombinant product (see Supporting Information Table S1, Figure 2B). Using this combined screening approach, 1/72 (1.3%) isolated F1 animals generated progeny producing the expected PCR amplicon, indicating that homologous recombination occurred (Table 1). Consistent with the heritable nature of this recombination event, we obtained homozygous lines by isolating F2 worms producing 100% GFP-expressing F3 and subsequent progeny (Figure 2C). We further verified that the transgene was inserted seamlessly by sequencing our recombinant-specific PCR amplicon (Figure S1).
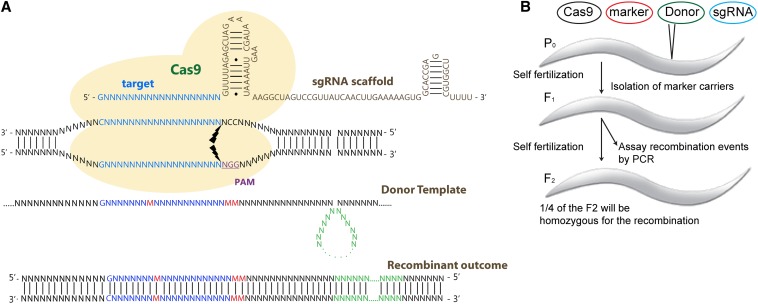
Figure 1.
Rationale and design applied in adapting the CRISPR–Cas9 system for homologous recombination-mediated genome engineering in C. elegans. (A) Schematic representation of the elements generating the engineered product. Cas9 (yellow) interacts with the sgRNA carrying a G/A(N)19NGG sequence, where G/A(N)19 corresponds to 20 nucleotides complementary to the homologous genomic target, cleaving the target double-stranded DNA (bolts) at the 3′ NGG sequence that corresponds to the essential protospacer-associated motif (PAM). A donor vector that shares homology with the genomic locus provides a template for repairing the break, thus inserting custom mutations (red), deletions, and insertions (green) to the recombinant outcome. (B) Experimental design to generate and screen for worms carrying engineered genomes. Adult worms were injected with CRISPR–Cas9 expression vectors containing Cas9, a mCherry marker, donor template sequence, and the targeting sgRNA.
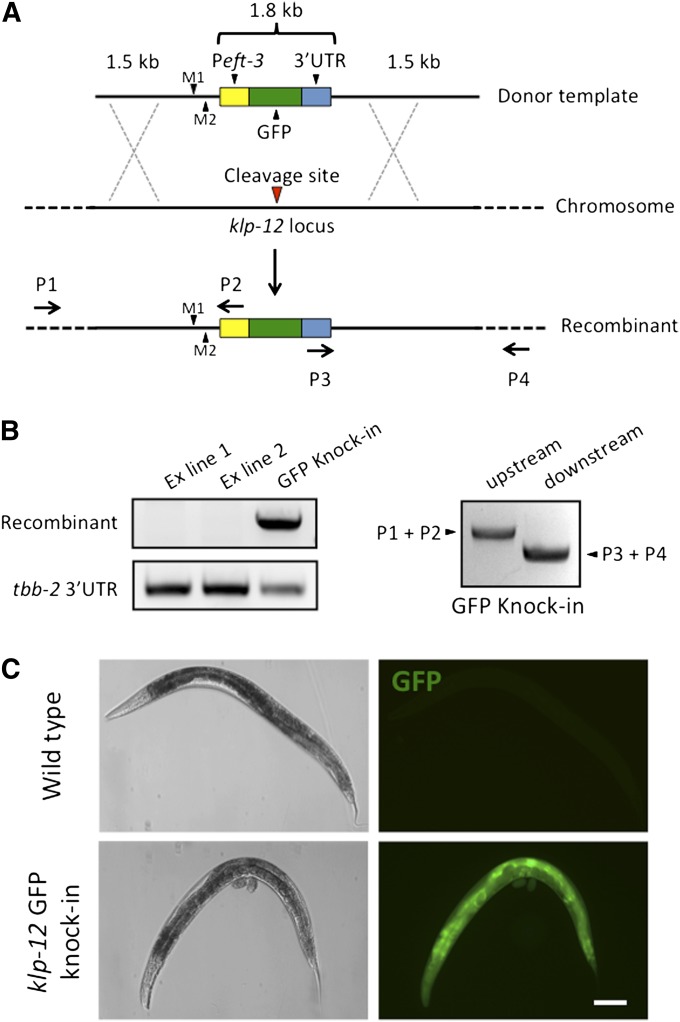
Figure 2.
Insertion of the Peft-3::GFP::tbb-2 3′-UTR transgene into the klp-12 locus by CRISPR–Cas9 mediated homologous recombination. (A) Design of donor repair template and diagram of expected recombinant product. The donor template (top image) contains 1.5 kb of homology (solid black lines) flanking the Cas9 cleavage site at the klp-12 locus (middle image, red triangle). The donor also carries a GFP transgene (top and bottom images, green box) flanked by the eft-3 promoter (yellow box), and the tbb-2 3′-UTR (blue box), and mutations to introduce a HindIII recognition sequence (M1) and to destroy the protospacer-associated motif (PAM) of the sgRNA target sequence (M2). Any recombination events (bottom image) can be screened using PCR primers complementary to the transgene (arrows P2 or P3) and complementary to genomic regions outside the homology arms of the donor (arrows P1 or P4; dashed lines mark outer genomic regions). (B) PCR confirmation of recombinant animals. Left top: PCR assay using primers specific for a recombination event (P3 and P4 from A) sampling lysates of pooled F2 animals from two different lines carrying the transgene in an extrachromosomal array along with our body wall muscle mCherry marker (lanes 1 and 2) and a candidate recombinant line (lane 3) identified by fluorescence microscopy. Left bottom: PCR assay using primers complementary to the tbb-2 3′-UTR to serve as a loading control. Right: PCR assays testing the recombinant line from the left, amplifying recombinant-specific PCR products spanning upstream (lane 1, primers P1 and P2) and downstream (lane 2, primers P3 and P4) of the cleavage site. (C) GFP expression in a strain in which a GFP transgene was inserted into the klp-12 locus. Bar, 100 μm.
Table 1. Summary of experiments creating homologous recombination-mediated mutations.
| Experiment | Gene | Injected worms | F1 worms | Recombinant worms | Frequency (%) |
|---|---|---|---|---|---|
| A | klp-12 | 13 | 72 | 1 | 1/72 (1.3) |
| B | lab-1 | 9 | 40 | 1 | 1/40 (2.5) |
| C | lab-1 | 7 | 24 | 4 | 4/24 (16.7) |
A one-step CRISPR–Cas9-mediated gene replacement strategy
Next, we tested whether we could use our expression vector system to knock out a complete gene and replace it with a GFP transgene. We generated an sgRNA targeting the lab-1 gene and verified its effectiveness in generating indel mutations at the appropriate locus by direct sequencing when injected with Cas9 (data not shown; Friedland et al. 2013). We then injected this sgRNA expression vector, our Cas9 expression vector, the mCherry reporter vector, and a donor vector containing a GFP transgene driven by the baf-1 promoter, flanked by 1020 and 1029 bp upstream and downstream of the lab-1 coding region, respectively (File S1 and Figure 3A). This donor vector was designed to completely remove the 652 bp of lab-1 and replace it with Pbaf-1::GFP. We screened for successfully engineered genomes in two independent experiments by using a similar PCR assay as described above, and we isolated F1 worms that had recombinant progeny in 1/40 (2.5%) and 4/24 (16.7%) of the worms (Table 1 and Figure 3B). Again, isolated from the original insertion strain were homozygous recombinant progeny that express GFP through multiple generations, both in somatic and germline tissues (Figure 3C and Figure S2). Immunostaining using LAB-1-specific antibodies confirmed that the protein was absent in our recombinant line (Figure 3D). Sequencing of a PCR product spanning the recombinant locus confirmed the excision of the lab-1 gene and its replacement with the GFP transgene (Figure S3). Consistent with the proposed functions for lab-1 in promoting sister chromatid cohesion and accurate meiotic chromosome segregation (de Carvalho et al. 2008), we observed increased embryonic lethality (45%, n = 651; Emb) and a high incidence of males (11%, n = 361; Him) in our lab-1 knock-out GFP knock-in line. These levels are similar to those reported for the lab-1 RNAi-depleted worms (57% Emb and 6% Him), and higher than those observed for the lab-1(tm1791) hypomorph mutant (22% Emb and 4% Him) (de Carvalho et al. 2008), underscoring the importance of generating full knock-outs for assaying the null phenotypes of genes. We did not observe any new or unexpected phenotypes for either of the strains reported here.
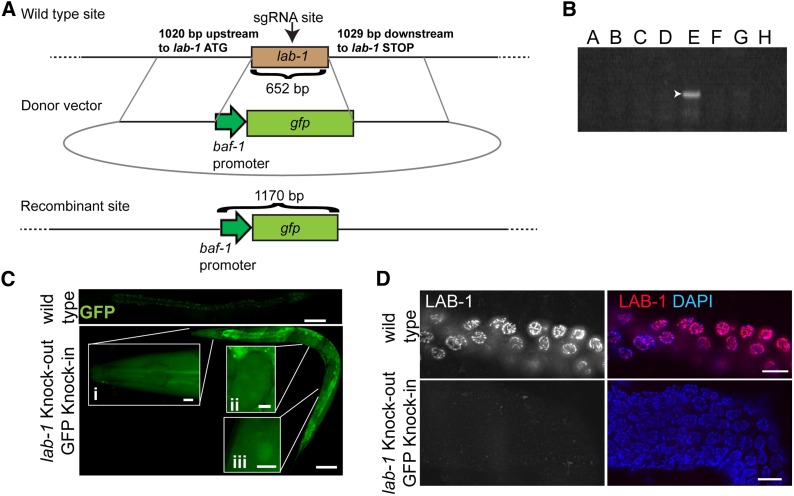
Figure 3.
Insertion of the Pbaf-1::GFP transgene into the lab-1 locus and excision of the lab-1 gene by CRISPR–Cas9 mediated homologous recombination. (A) Design of donor repair template and diagram of expected recombinant product. The donor template (middle image) contains 1020 bases upstream to the lab-1 ATG and 1029 downstream of the lab-1 STOP codon regions of homology (solid black lines). The Cas9 cleavage site at the lab-1 locus (652 bp) is located 310 base downstream of the ATG (arrow, top image). The donor also possesses a GFP transgene (884 bp, green box) flanked by the baf-1 promoter (286 bp, green arrow). (B) PCR detection of recombinant specific species. PCR amplicon generated by using primers annealing 1167 bp upstream of the lab-1 ATG and to the baf-1 promoter. A–D and F–H, negative clones. E, a positive clone. Arrowhead, ∼1200-bp fragment. (C) GFP expression in a strain in which a GFP transgene was inserted and lab-1 was deleted. Bars, 100 μm. Insets depict GFP expression in the head (i), embryo (ii) and oocyte (iii). Bars, 10 μm. (D) Anti-LAB-1 (red) and DAPI (blue) costaining of late pachytene nuclei in the germlines of (top)wild-type, (bottom) lab-1-deleted, and gfp-inserted worms. Bars, 10 μm.
Finally, to test whether the inserted promoter might regionally perturb gene expression, we assessed by RT–qPCR the expression of asfl-1 and T05F1.11, which flank lab-1 (Figure S4). We found no significant change in the expression of T05F1.11 (P = 0.153 by the two-tailed Mann–Whitney test, 95% C.I.), but the expression of asfl-1, which lies upstream of lab-1, exhibited a small yet significant change (P = 0.046 by the two-tailed Mann-Whitney test, 95% C.I.). Further experiments will be required to determine whether this increase is due to a polar position effect exerted by the inserted promoter-gfp fusion or a secondary effect of the lab-1 knock-out. Altogether, these results demonstrate that CRISPR–Cas9 is a useful system for the seamless replacement of genes in the C. elegans genome.
Discussion
The results presented here show the powerful use of the CRISPR–Cas9 system to accurately engineer the C. elegans genome, thereby enabling the creation of almost any custom mutation desired. We were able to insert and delete genes, as well as do both in one step. The transgenes were seamlessly inserted and were expressed both in the soma and the germline, indicating the functionality and specificity of this method. Although we did not observe any off-target effects, it would be a good practice to outcross any mutation created using the CRISPR–Cas9 system. This method (also see accompanying C. elegans studies by Chiu et al. 2013; Cho et al. 2013b; Katic et al. 2013; Lo et al. 2013; Waaijers et al. 2013, in this issue of GENETICS) now paves the way to various in vivo applications, including the replication of mutations identified in other organisms and the assessment of protein–protein binding sites as well as the amino acids predicted to undergo post-translational modifications. Tagging of genes in their own natural sites will become easier, thus enabling scientists to monitor their expression more accurately and facilitating immunoprecipitation experiments. Different gene mutation projects have provided the worm community with a growing collection of invaluable base change and deletion mutants. The CRISPR–Cas9 system now provides the community the ability to expand this collection and obtain precise gene deletions or targeted changes in a simple and rapid manner.
Other methods have been previously implemented in C. elegans to create similar engineering capabilities like ZFNs (zinc-finger nucleases), TALENs (transcription activator-like effector nucleases), MosSCI (Mos1-mediated single copy insertion), and MosDel (Mos1-mediated deletion) (Frokjaer-Jensen et al. 2008, 2010, 2012; Wood et al. 2011). The CRISPR–Cas9 method offers comparable and sometimes superior efficiency, along with three additional advantages: (1) It does not require the complex engineering of a special nuclease for every project, (2) it does not require any positive genetic selection marker, and (3) it can be applied to practically any locus given that the only requirement for the targeting site is the presence of G/A(N)19NGG. These advantages, combined with the fact that this method calls for no specific genetic background, should now make it possible to sequentially create lines harboring multiple mutations in tightly linked genes that would otherwise require difficult genetic crosses. Finally, our results not only more generally demonstrate the versatility of the CRISPR–Cas9 system for customized engineering, now reported for several metazoan species, but also more specifically represent a significant step forward in facilitating a wide range of gene function and regulatory studies in C. elegans.
Supplementary Material
Acknowledgments
We thank Yoav Mayshar, John Aach and Elisabeth Altendorfer for valuable suggestions. pAD010 was a kind gift from Yosef Gruenbaum. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by U.S. National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was supported by NIH grant R01GM072551 to M.P.C., by an NIH Early Independence Award (1DP5OD009153) and additional support from Harvard University to J.A.C., and by an NIH National Human Genome Research Institute Center of Excellence in Genome Sciences grant (P50 HG005550) to G.M.C. A.E.F. is supported by a Ralph Ellison/American Federation for Aging Research postdoctoral fellowship.
Footnotes
Communicating editor: O. Hobert
Literature Cited
- Chiu H., Schwartz H. T., Antoshechkin I., Sternberg P. W., 2013. Transgene-free genome editing in Caenorhabditis elegans using CRISPR-Cas. Genetics 195: 1167–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M., Kim J. S., 2013a Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 230–232 [DOI] [PubMed] [Google Scholar]
- Cho S. W., Lee J., Carroll D., Kim J.-S., Lee J., 2013b Heritable gene knockout in C. elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics 195: 1177–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho C. E., Zaaijer S., Smolikov S., Gu Y., Schumacher J. M., et al. , 2008. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 22: 2869–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo J. E., Norville J. E., Mali P., Rios X., Aach J., et al. , 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41: 4336–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., et al. , 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Hollopeter G., Taylor J., Harris T. W., et al. , 2010. Targeted gene deletions in C. elegans using transposon excision. Nat. Methods 7: 451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C., Davis M. W., Ailion M., Jorgensen E. M., 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9: 117–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., 3rd, 2013. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz S. J., Cummings A. M., Nguyen J. N., Hamm D. C., Donohue L. K., et al. , 2013. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194: 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Kaini P., et al. , 2013. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS ONE 8: e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackstadt P., Wilm T. P., Zahner H., Hobom G., 1999. Transformation of nematodes via ballistic DNA transfer. Mol. Biochem. Parasitol. 103: 261–266 [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., et al. , 2013. RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Grosshans H., 2013. Targeted heritable mutation and gene conversion by Cas9-CRISPR in Caenorhabditis elegans. Genetics 195: 1173–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Xu S., Montgomery M. K., Fire A., 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo T., Pickle C. S., Lin S., Ralston E. J., Gurling M., et al. , 2013. Heritable genome editing using TALENs and CRISPR/Cas9 to engineer precise insertions and deletions in evolutionarily diverse nematode species. Genetics 195: 331–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V., Bessereau J. L., 2007. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 26: 170–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Zhang J., Wu H., Wang J., Ma K., et al. , 2013. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 23: 720–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Lawrence C. M., Wiedenheft B., 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82: 237–266 [DOI] [PubMed] [Google Scholar]
- Terns M. P., Terns R. M., 2011. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 14: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waaijers S., Portegijs V., Kerver J., Lemmens B.B.C.G., Tijsterman M., et al. , 2013. Crispr/Cas9-targeted mutagenesis in C. elegans. Genetics 195: 1187–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A., 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338 [DOI] [PubMed] [Google Scholar]
- Wilm T., Demel P., Koop H. U., Schnabel H., Schnabel R., 1999. Ballistic transformation of Caenorhabditis elegans. Gene 229: 31–35 [DOI] [PubMed] [Google Scholar]
- Wood A. J., Lo T. W., Zeitler B., Pickle C. S., Ralston E. J., et al. , 2011. Targeted genome editing across species using ZFNs and TALENs. Science 333: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.