Abstract
Rationale
Fibroblast growth factor (FGF) homologous factors (FHFs, FGF11-14) are intracellular modulators of voltage-gated Na+ channels, but their cellular distribution in cardiomyocytes indicated that they performed other functions.
Objective
We aimed to uncover novel roles for FHFs in cardiomyocytes starting with a proteomic approach to identify novel interacting proteins.
Methods and Results
Affinity purification of FGF13 from rodent ventricular lysates followed by mass spectroscopy revealed an interaction with Junctophilin-2, a protein that organizes the close apposition of the L-type Ca2+ channel, CaV1.2, and the ryanodine receptor, RyR2, in the dyad. Immunocytochemical analysis revealed overall T-tubule structure and localization RyR2 were unaffected by FGF13 knockdown in adult ventricular cardiomyocytes, but localization of CaV1.2 was affected. FGF13 knockdown decreased CaV1.2 current density, and reduced the amount of CaV1.2 at the surface due to aberrant localization of the channels. CaV1.2 current density and channel localization were rescued by expression of an shRNA-insensitive FGF13, indicating a specific role for FGF13. Consistent with these newly discovered effects on CaV1.2, we demonstrated that FGF13 also regulated Ca2+-induced Ca2+ release, indicated by a smaller Ca2+ transient after FGF13 knockdown. Further, FGF13 knockdown caused a profound decrease in the cardiac action potential half width.
Conclusions
This study demonstrates that FHFs are not only potent modulators voltage-gated Na+ channels, but also affect Ca2+ channels and their function. We predict that FHF loss-of-function mutations would adversely affect currents through both Na+ and Ca2+ channels, suggesting that FHFs may be arrhythmogenic loci, leading to arrhythmias through a novel, dual-ion channel mechanism.
Keywords: Fibroblast growth factor; homologous factor; CaV1.2, Ca2+ channels; Junctophilin-2; arrhythmia; cardiac electrophysiology; heart
INTRODUCTION
Despite an ever-growing understanding of ion channel structure, function, and regulation, many components of the macromolecular complexes anchored by ion channels are not yet known or well characterized. Identification of these channel interacting proteins and discovery of their functions within the channelsome provides important insight into physiologic and pathologic function. Mutations in newly defined channel interacting proteins often explain genetic causes of arrhythmias in cases where mutations in known arrhythmia loci are not found. Fibroblast growth factor (FGF) homologous factors (FHFs), a subfamily of FGF proteins (FGF11-FGF14) expressed predominantly in excitable cells,1 are prime examples of channel interacting proteins for which cardiac functions are not well understood. Although part of the FGF superfamily, FHFs do not function as growth factors and are incapable of activating FGF receptors.2 Rather, FHFs remain intracellular and have been shown to bind and modulate voltage-gated Na+ channels.3, 4 Their roles as Na+ channel regulators have been studied most extensively in the brain, driven in large part by the identification of FGF14 as the locus for spinocerebellar ataxia and by observations that Fgf14−/− mice display an ataxia phenotype that correlates with decreased Na+ channel function and diminished neuronal excitability.5, 6
FHFs are also expressed in cardiomyocytes, but their roles in regulating cardiac function have heretofore received less attention. We showed that FGF13, the most highly expressed FHF in murine heart, directly binds NaV1.5, the predominant cardiac Na+ channel, and participates in trafficking NaV1.5 to the sarcolemmal membrane and modulating Na+ channel kinetics.7–9 Consistent with these regulatory roles for Na+ channels, knockdown of FGF13 led to a reduction in conduction velocity and maximum capture rate (the ability of the cells to recover from a stimulus of a specific speed) in a neonatal rat ventricular cardiomyocyte monolayer.7
Several lines of evidence suggest that the effects of FHFs in excitable cells extend beyond Na+ channel modulation. For example, the complex changes in synaptic physiology in Fgf14−/− mice are not consistent with a defect limited to Na+ channel dysfunction.10 Indeed, we recently found that FGF14 knockdown in cerebellar granule cells reduced presynaptic Ca2+ currents and synaptic transmission at the granule cell to Purkinje cell synapse.11 Moreover, as we reported,7 the cellular distribution of FHFs in ventricular cardiomyocytes extended beyond the distribution of NaV1.5 channels. We therefore aimed to determine novel roles for FHFs in cardiomyocytes by looking for new FHF interactors. Here we report the discovery that that junctophilin-2 (JPH2) interacts with FGF13 in rodent ventricular myocytes. JPH2 is a protein responsible for coordinating the interaction of the sarcolemma and sarcoplasmic reticulum in the dyad. We therefore investigated the role for FGF13 in regulating ionic currents in the dyad and showed that FGF13 has essential roles in regulating the L-type, voltage-gated Ca2+ channel (CaV1.2) currents in adult ventricular myocytes. These results provide to the best of our knowledge the first evidence that FHFs modulate ion channels other than voltage-gated Na+ channels in cardiomyocytes and highlight previously unknown modulatory roles for FHFs in cardiac physiology, such as regulation of Ca2+-induced Ca2+ release (CICR) and the integrated electrical activity of the ventricular action potential. Our results lead to the hypothesis that loss-of-function mutations in FHFs could underlie inherited cardiac arrhythmias.
METHODS
See Supplemental Material for complete details.
Adenovirus
The adenoviruses expressing FGF13 shRNA or scrambled shRNA with GFP have been previously described.7 FGF13 rescue viruses and the shRNA virus with the GFP removed were generated similarly using the AdEasy System (Agilent).
Cardiomyocyte isolation
Animals were handled according to National Institutes of Health Guide for the Care and Use of Laboratory Animals. The study was approved by Duke University Animal Care and Welfare Committee. Cardiomyocytes were isolated from 6–8 week old C57/Bl6 mice or Sprague Dawley rats and cultured as previously described.7
Electrophysiology
Ca2+ currents (ICa) were recorded using the whole-cell voltage-clamp technique as previously described.12 Cardiac action potentials were recorded in current clamp as previously described.13
Immunocytochemistry and T-tubule staining
Immunocytochemistry methods have been previously described.7 Imager/analyzer was blinded to the manipulation and all cells imaged were used for analysis. For T-tubule staining, cells were plated onto glass bottom plates (MatTek Corp.) and cultured as above. Cells were then incubated in 0.5 µM di-8-ANEPPS (Life Technologies) and were imaged live.
Image processing and Fast Fourier Transform
The experimenter analyzing images was blinded to treatment. Stacks were deconvolved using Hyugens software (Scientific Volume Imaging) and exported as Tiff files. Voxel colocalization was performed on deconvolved images using Pearson correlation coefficient. Channel localization analysis was performed in ImageJ (NIH) and fast Fourier transformed using OriginLab software.
Simultaneous patch clamping and Ca2+ transient recording
Cardiomyocytes were plated onto glass bottom plates (MatTek Corp.) and cultured as above. Viruses expressing GFP were not used as GFP interferes with the Fura-2 emission. Whole cell patch clamping was performed as above with with the addition of 150 µM Fura-2 pentapotassium salt (Life Technologies) and the removal of EGTA in the pipette solution. For Ca2+ transient peak measurement, autofluorescence and background emissions were first subtracted and the peak was measured as the difference from the baseline using IonWizard software (IonOptix). EC-coupling gain was defined as the Ca2+ transient peak divided by the Ca2+ current peak.
Sarcoplasmic reticulum load measurements
SR load was measured using 0.25 µM Fura-2 AM loaded cardiomyocytes as previously described.14
Immunoprecipitation
Fresh adult mouse ventricular heart lysate was prepared by homogenizing tissue and immunoprecipitating FGF13 as previously described.7 After immunoprecipitation, samples were subjected to SDS-PAGE and co-immunoprecipitation was verified by western blot.
Proteomics
Antibody (20 µg of anti-FGF13 or control IgG) was irreversibly crosslinked to protein A/G agarose beads (Santa Cruz Biotechnology) and ventricular tissue lysate (~23 mg total protein) was added to cross-linked beads. The bound proteins were eluted in 400 µl of 0.2% Rapigest SF Surfactant (Waters) in 50 mM ammonium bicarbonate and subjected to an in-solution tryptic digestion at the Duke Proteomics Core Peptide. Identifications were determined using liquid chromatography/tandem mass spectrometry; following data acquisition, all spectra were searched against the SwissProt database with the mouse taxonomy selected.
Biotinylation and western blotting
Surface biotinylation and western blotting were performed as previously described.7
Statistical analyses
Results are presented as means ± standard error of the mean; statistical significance of differences between groups was assessed using one-way analysis of variance (ANOVA) and was set at P < 0.05.
RESULTS
Junctophilin-2 is in complex with FGF13
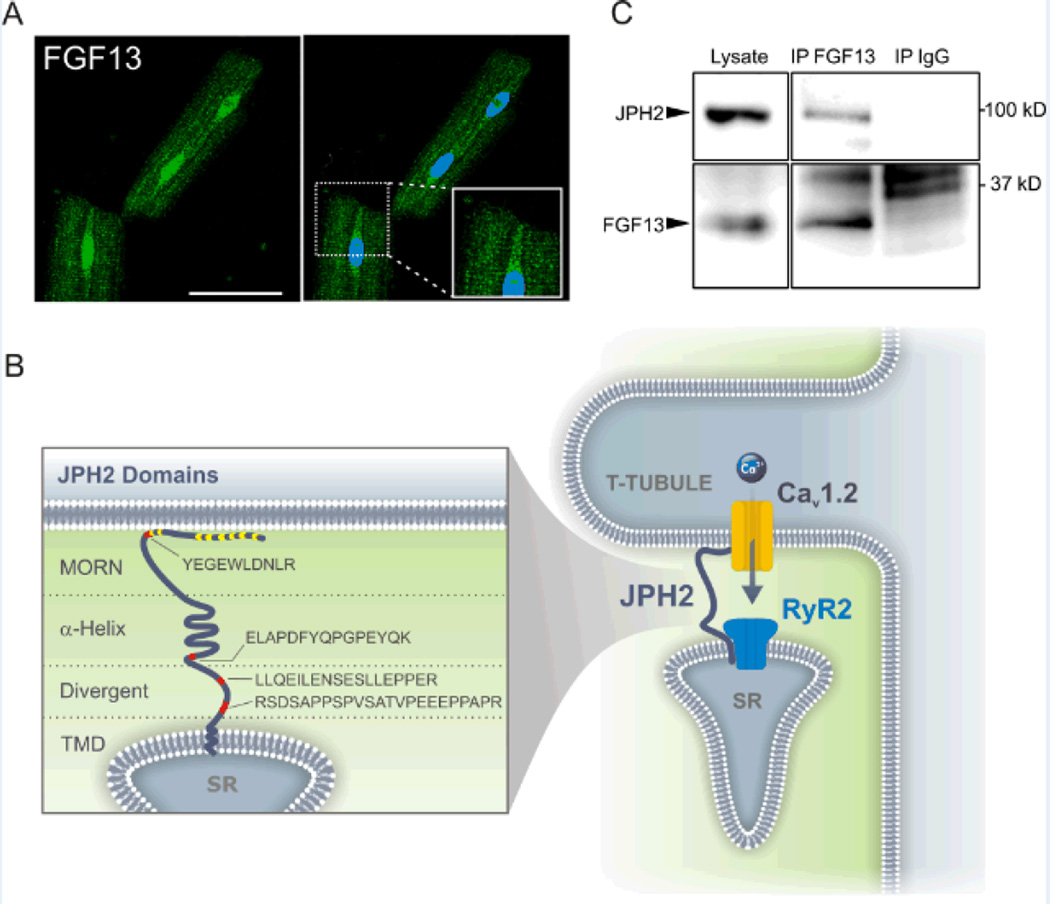
Although we previously showed that FGF13 is an intracellular modulator of voltage-gated Na+ channels,7 close analysis of FGF13 immunocytochemistry in adult rat cardiomyocytes revealed an overall distribution that extended beyond what has been reported for NaV1.5 in heart.15 Specifically, we observed a striated pattern similar to a T-tubule distribution (see Figure 1A, inset), in addition to the sarcolemmal and nuclear distribution we previously reported.7 This led us to hypothesize that FGF13 may have roles in cardiomyocyte physiology beyond NaV1.5 regulation. To identify other potential FGF13 interactors, we performed immunoprecipitation using a previously validated FGF13 antibody or an IgG (as a control) from adult ventricular tissue and liquid chromatography / mass spectrometry analysis of the immunoprecipitated FGF13 protein complex. One interesting candidate we identified was Junctophilin-2 (JPH2). As shown in Figure 1B, we identified 4 unique peptides spanning all soluble domains of JPH2. We found this candidate to be of interest because it is a protein responsible for properly juxtaposing CaV1.2 and the Ryanodine Receptor 2 (RyR2) in the T-tubule (Figure 1B), consistent with the T-tubular distribution of FGF13. The specificity of the mass spectrometry approach was confirmed through the identification of three FGF13 peptides after FGF13 immunoprecipitation, but none with the control IgG; and eight peptides (but none in the control IgG sample) from the known FGF13 binding partner NaV1.5 (see Supplemental Table I for all peptide data). We further validated that FGF13 was in complex with JPH2 through co-immunoprecipitation, detecting JPH2 in FGF13 immunoprecipitates but not in the IgG control (Figure 1C).
Figure 1. FGF13 associates with JPH2 as part of the dyad macromolecular complex.
A, immunocytochemical analysis shows FGF13 (green) in the nucleus, the sarcolemma and T-tubules, enlarged in the inset. DAPI is in blue. Scale bar 50 µm. B, schematic of JPH2 indicating the putative protein motifs and the location of the unique peptides identified by mass spectroscopy in red. MORN motifs are indicated in yellow. JPH2 is found in the dyad in which CaV1.2 is juxtaposed to RyR2. MORN, membrane occupation and recognition nexus; α-helix, alpha helical domain; divergent, divergent region; TMD, transmembrane domain; SR, sarcoplasmic reticulum. C, representative co-immunoprecipation and western blot to validate the interaction of JPH2 and FGF13, repeated three times.
FGF13 knockdown perturbs dyad organization by affecting CaV1.2 localization
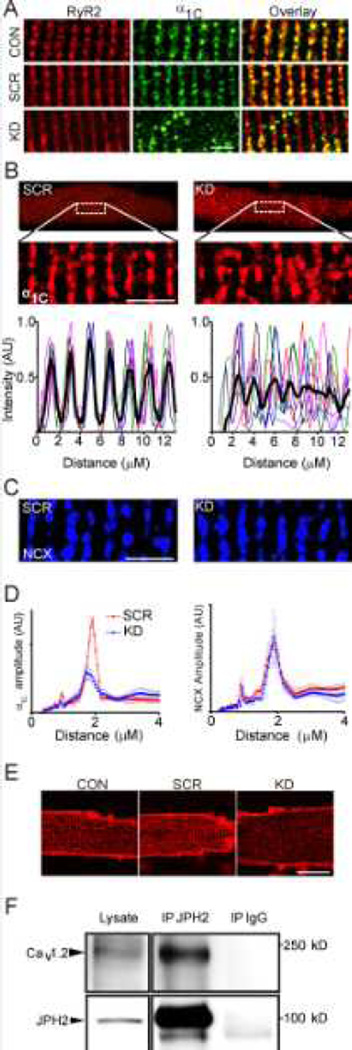
JPH2 is a protein responsible for coordinating the interactions between the sarcolemma and the sarcoplasmic reticulum (SR). JPH2 has a C-terminal transmembrane domain in the SR and its N-terminus interacts with the inner leaflet of the sarcolemma, providing a means to juxtapose the sarcolemma and SR so as to promote efficient CICR by apposing CaV1.2 and RyR2.16 Based on our discovery that FGF13 was a component of the JPH2 macromolecular complex, we tested whether FGF13 participated in dyad ion channel targeting. We performed immunocytochemistry on mouse ventricular myocytes that were uninfected (CON) or that had been infected with a FGF13 shRNA adenovirus targeting all FGF13 splice variants (FGF13 KD) or a scrambled shRNA adenovirus (SCR, Figure 2). As we previously showed (see also Figure 4), the KD adenovirus reduced FGF13 protein by ~ 90% and the SCR virus had no effect.7 In the CON and SCR, we observed α1C, the CaV1.2 pore-forming subunit, colocalized with RyR2 in a striated pattern. Little α1C or RyR2 was observed between the striations. After FGF13 KD, however, we saw a loss of colocalization and a large portion of α1C was now present between the RyR2 striations (Figure 2A, Pearson’s correlation co-efficient of 0.52 ± 0.03 versus 0.65 ± 0.03 and 0.62 ± 0.03 in CON and SCR cardiomyocytes, respectively; P < 0.05 for FGF13 KD versus CON or SCR cardiomyocytes, N=8 cells per group).
Figure 2. FGF13 perturbs dyad organization and CaV1.2 localization.
A, RyR2 is unaffected by FGF13 knockdown and CaV1.2 no longer colocalizes with RyR2 (p < 0.05, see text for numbers). B, intensity plots for α1C were drawn of a 15 µm × 5 µm area of the cell. Similar studies were done for sodium-calcium exchange (NCX), C. D, the interval distance was calculated using a FFT for α1C (left) and NCX (right). Amplitudes for α1C were 0.28 ± 0.02, 0.23 ± 0.01 and 0.12 ± 0.02 for control, scrambled and shRNA respectively. P < 0.001 for FGF13 KD vs. CON (not shown) or SCR. C (right), NCX is not affected by FGF13 knockdown, (p=0.40 for n=10 per group). Scale bar 5 µm for A, B, and C. E, Di-8- ANEPPS staining of T-tubules showed no change in morphology. Scale bar 20 µm, n=20, 10 and 14 for CON, SCR, and KD respectively. F, representative co-immunoprecipitation and western blot of the poreforming α1C subunit of CaV1.2 with JPH2 from mouse ventricular lysate from three independent experiments showing an interaction with an antibody specific to JPH2 but not the IgG control.
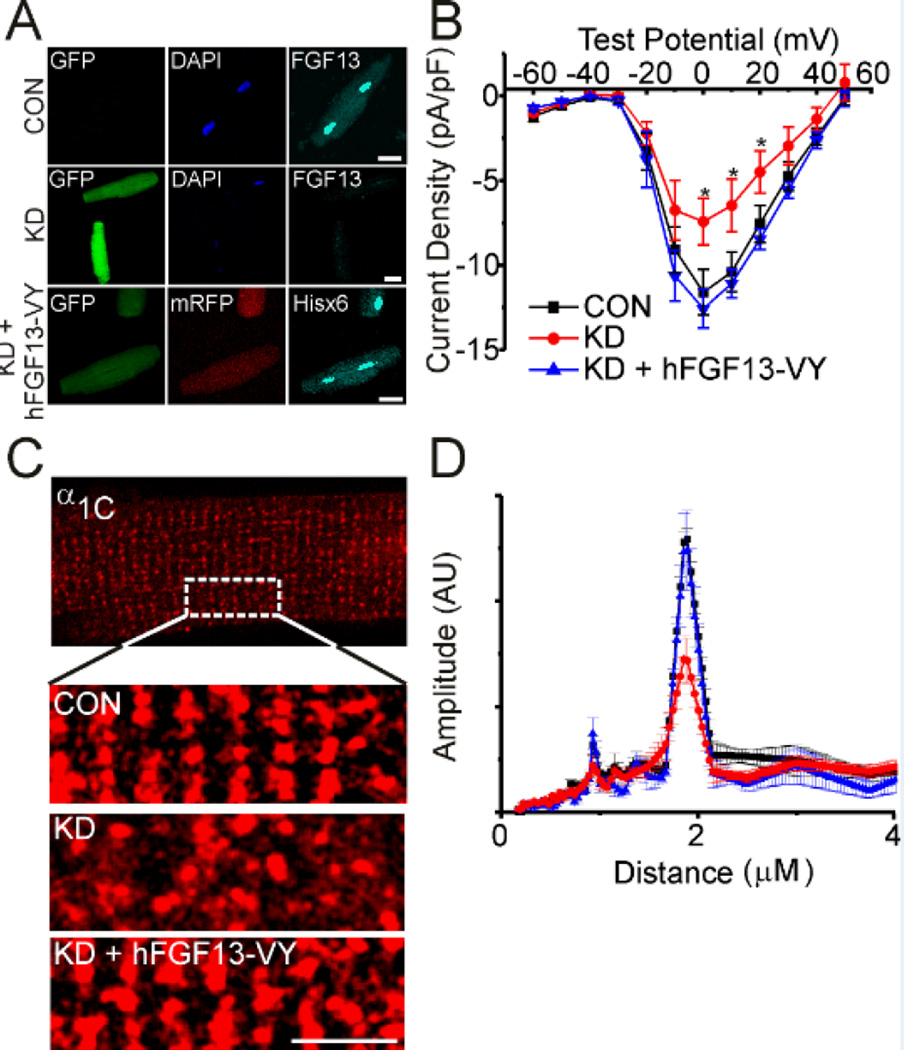
Figure 4.
Human FGF13-VY rescues CaV1.2 current density and localization. A, immunocytochemistry showing endogenous FGF13 and the level of knockdown with adenovirus expressing GFP and shRNA to all FGF13 splice variants (top two panels). Overexpression of shRNA-resistant, hFGF13-VY (also expressing mRFP) in the bottom panel. Scale bar 40 µm. hFGF13-VY rescued decreased current density (B) and the change in α1C localization (C, D) seen with FGF13 knockdown (KD). Scale bar 5 µm. Fast Fourier transform amplitudes were CON, 0.27 ± 0.01 (n=5), FGF13 KD, 0.15 ± 0.24 (n=6) and FGF13 KD + hFGF13-VY, 0.27 ± 0.04 (n=5), P < 0.001 for FGF13 KD compared to CON and FGF13 KD + hFGF13-VY. *P < 0.05 for FGF13 KD vs. control and FGF13 KD + hFGF13-VY.
We then performed quantitative analysis on the pattern of CaV1.2. In CON adult mouse cardiomyocytes (Figure 2A) or after infection with the SCR virus (Figure 2B), α1C displayed the expected striated pattern with a periodicity of ~ 2 µm, consistent with a T-tubular distribution.17 In contrast, the pattern in FGF13 KD cells was discontinuous with multiple punctae found between the residual striations (Figure 2B). With the analyzer blinded to treatment status, we quantified the change in distribution with intensity profiles of α1C in confocal z-stacks of non-nuclear areas, as shown in Figure 2B. In SCR cardiomyocytes, we detected a regular pattern of intense α1C staining at ~ 2 µm intervals (analyzed by fast Fourier transform, Figure 2D), with almost no signal in the intervening intervals. In contrast, after FGF13 KD, the peak amplitude of α1C staining at the ~ 2 µm interval was reduced by more than 50% (FFT amplitudes (in arbitrary units): CON 0.284 ± 0.017 (n=8), SCR 0.234 ± 0.009 (n=8), FGF13 KD 0.116 ± 0.020 (n=12); P < 0.0001 for FGF13 KD vs. SCR and CON), and significant α1C signal was observed in between the peaks (Figure 2B, D).
We tested whether this altered α1C distribution was secondary to a general defect in T-tubule architecture or was more specific to α1C using two separate analyses. First, we performed immunocytochemistry for the sodium-calcium exchanger (NCX), another T-tubule-localized protein, and observed that FGF13 KD did not affect NCX distribution (Figure 2C, D). Second, the membrane-binding dye, di-8-ANEPPS, revealed that T-tubular distribution was grossly unaffected by FGF13 KD (Figure 2E).
The specific change in CaV1.2 localization after FGF13 KD led us to hypothesize that FGF13, as a member of the JPH2 macromolecular complex anchored by JPH2, affected CaV1.2 targeting to Ttubules. We therefore tested whether CaV1.2 was a component of the JPH2 complex. Indeed, by coimmunoprecipitation, we were able to pull down the α1C pore-forming subunit of CaV1.2 with an antibody to JPH2 but not a control IgG (Figure 2F). These data are consistent with a previous observation that JPH2 interacts with Cav1.1 in skeletal muscle,18 but are, to the best of knowledge, the first demonstration of an interaction between JPH2 and an L-type Ca2+ channel in in cardiac muscle. Together, these data indicated that FGF13 KD did not affect T-tubule or SR architecture, but FGF13 has a specific role in targeting CaV1.2 to its proper T-tubular location.
FGF13 modulates CaV1.2 trafficking to the surface and current density
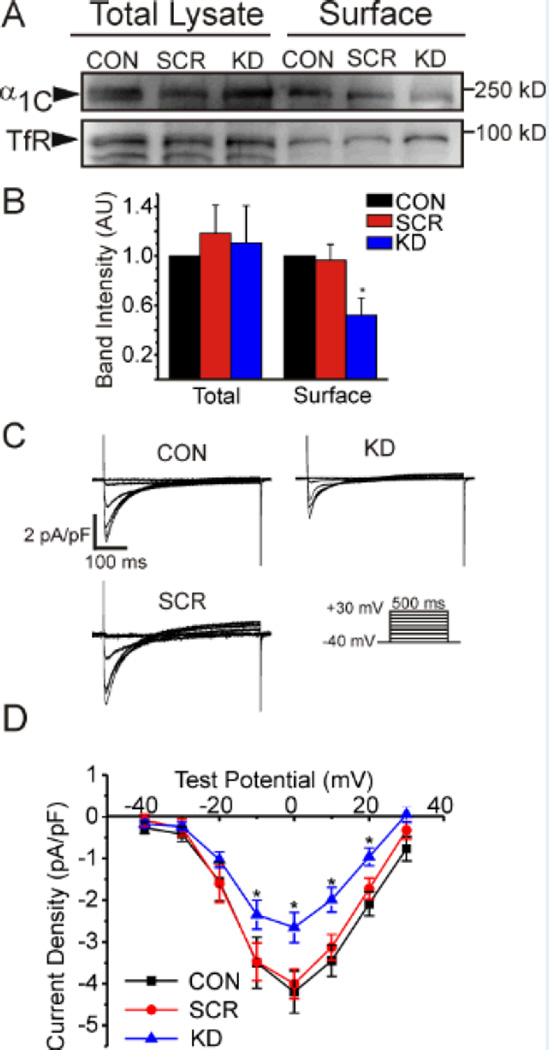
We hypothesized that this aberrant CaV1.2 localization would lead to a decrease in CaV1.2 at the surface. We therefore quantified the relative amount of CaV1.2 at the sarcolemma after FGF13 KD by labeling surface proteins with biotin and capturing them with avidin beads after cell lysis. Both the surface fraction and the whole cell lysate of adult mouse ventricular myocytes were then probed with an antibody against the pore-forming α1C subunit of CaV1.2. These biotinylation experiments revealed a decrease in amount of α1C at the surface after FGF13 KD (Figure 3A–B). The total amount of α1C (in the cellular lysate) was unchanged. To test whether this decreased CaV1.2 at the surface, led to physiologic changes, we measured Ca2+ channel current density using whole-cell patch clamp. In FGF13 KD cardiomyocytes, but not the SCR cardiomyocytes, Ca2+ channel current density was reduced by 35% compared to CON cardiomyocytes (Figure 3C, D). Channel availability, voltage-dependence of activation and steady-state inactivation, were unaffected (Table I). Together, these results suggest that FGF13 knockdown reduced Ca2+ current density by affecting the number of channels at the sarcolemma.
Figure 3.
FGF13 affects CaV1.2 surface expression and current density. Surface expression of CaV1.2 is reduced with FGF13 KD while total protein remains unchanged in surface biotinylation and western blotting, A. Summarized data of four independent biotinylation experiments in B. C, representative ICa traces from voltage-clamp of mouse ventricular cardiomyocytes (voltage protocol shown below). D, summarized IV curve. * P < 0.05 for n=11, 6 and 14 for CON, SCR, and KD respectively.
Table 1.
Summary of Electrophysiological Data
| ICa peak at 0 mV (pA/pF) |
V1/2 of activation (mV) |
k of activation (pA/mV) |
V1/2 of inactivation (mV) |
k of inactivation (pA/mV) |
|
|---|---|---|---|---|---|
| Mouse Cardiomyocytes | |||||
| Control | −4.20 ± 0.51 (11) | −10.27 ± 1.51 (11) | 5.43 ± 0.21 (11) | −29.12 ± 1.21 (6) | 5.59 ± 0.20 (6) |
| Scrambled shRNA | −4.01 ± 0.36 (6) | −12.16 ± 1.84 (6) | 5.02 ± 0.37 (6) | −30.80 ± 1.06 (11) | 5.44 ± 0.14 (11) |
| FGF13 shRNA | −2.66 ± 0.36 (14) * | −11.50 ± 1.14 (14) | 5.46 ± 0.15 (14) | −28.73 ± 1.74 (13) | 5.39 ± 0.25 (13) |
| Rat Cardiomyocytes | |||||
| Control | −11.61 ± 1.34 (14) | −12.01 ± 1.56 (12) | 4.48 ± 0.36 (12) | −27.24 ± 1.23 (9) | 4.09 ± 0.16 (9) |
| FGF13 shRNA | −7.44 ± 1.38 (11) * | −12.43 ± 1.24 (9) | 4.71 ± 0.29 (9) | −29.83 ± 1.85 (9) | 4.42 ± 0.16 (9) |
| FGF13 shRNA + hFGF13-VY | −12.57 ± 1.15 (10) | −11.65 ± 1.81 (9) | 4.42 ± 0.41 (9) | −35.98 ± 1.91 (7) * | 4.88 ± 0.21 (7) * |
ICa indicates calcium current
The number of cells analyzed for each parameter is in parentheses
P < 0.05 by ANOVA
Human FGF13VY can rescue CaV1.2 current density and localization
To confirm a role for FGF13 in targeting CaV1.2 to the T-tubules and regulating L-type Ca2+ channel current density, we performed “rescue” experiments on adult rat ventricular cardiomyocytes treated with FGF13 shRNA by co-expressing human, shRNA-insensitive and Hisx6-tagged, FGF13-VY (hFGF13-VY). FGF13 is differentially spliced at its N-terminus1 and we chose this FGF13 splice variant for rescue because it is the most highly expressed isoform in mouse and rat cardiomyocytes.7 The adenovirus expressing FGF13 shRNA also expressed GFP (via a CMV promoter) and the adenovirus expressing hFGF13-VY also expressed mRFP, allowing us to identify cells in which hFGF13-VY was expressed in the context of endogenous FGF13 knockdown (Figure 4A). Immunostaining for endogenous FGF13 (with an anti-FGF13 antibody) and hFGF13-VY (with an anti-Hisx6 antibody) demonstrated not only effective knockdown of endogenous FGF13 but also that the expressed hFGF13-VY recapitulated the overall cellular distribution of endogenous FGF13 (Figure 4A), although the striated pattern was even more obvious when compared to endogenous FGF13. We suspected that the subtle differences in pattern reflected the specific distribution of hFGF13-VY compared to the distribution of all endogenous splice variants recognized by the antibody in CON cells.
Having established the efficacy of the hFGF13-VY rescue, we examined CaV1.2 Ca2+ current density in adult rat ventricular myocytes using whole-cell patch clamp. As in mouse cardiomyocytes (Figure 3C), FGF13 knockdown reduced CaV1.2 Ca2+ channel current density (Figure 4B, Table I). Expression of hFGF13-VY restored Ca2+ current density to wild-type levels (Figure 4B). We also noted a hyperpolarizing shift in steady-state inactivation compared to CON cells (Table I). The reasons for this are unclear, but may result from the overexpression of the specific FGF13-VY splice variant in the context of the knockdown of other FGF13 splice variants, thus altering any counterbalancing effects imparted by these absent variants. Additionally, the cellular distribution of α1C was restored with FGF13-VY overexpression, as indicated by the overall pattern and by Fourier transform (Figure 4C, D). This rescue strategy therefore firmly established a role for FGF13 in targeting CaV1.2 to T-tubules and provided a potent confirmation of the specificity of the FGF13 KD virus.
FGF13 affects Ca2+ transients but preserves excitation-contraction coupling gain
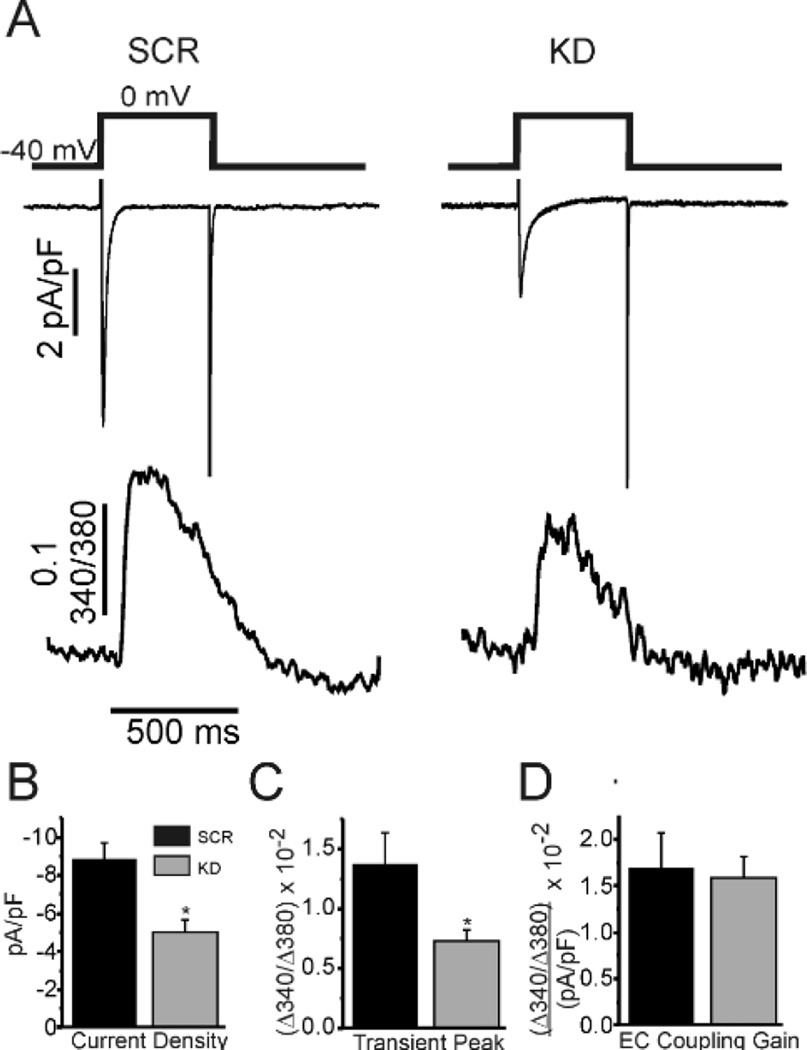
The mislocalization of CaV1.2 after FGF13 knockdown, the newly described interaction between FGF13 and JPH2, and previous studies that defined clear roles for JPH2 in Ca2+ induced Ca2+ release (CICR)14 prompted us to query whether FGF13 influenced excitation-contraction (EC) coupling gain. We therefore simultaneously recorded Ca2+ currents and Ca2+ transients in CON, SCR, and FGF13 KD rat ventricular myocytes (Figure 5). Following a previously described protocol to ensure steady-state CaV1.2 function,14 we held the cells at −40 mV to inactivate Na+ channels and gave a 500 ms pulse to 0 mV to induce influx of Ca2+ through CaV1.2 and recorded it via whole-cell patch clamp. We simultaneously recorded Ca2+ transients using Fura-2 in the pipette internal solution. Representative traces are shown in Figure 5A. Knockdown of FGF13 not only reduced Ca2+ current through CaV1.2 (Figure 5B), as previously shown, but also led to decreased Ca2+ release from the sarcoplasmic reticulum (SR, Figure 5C). Interestingly, it appeared that those channels that were at the surface were coupling with RyR2 appropriately as EC-coupling gain was not different between the groups. These changes were not due to decreased SR load as measured by rapid application of caffeine in Fura-2 loaded cells (peak height 0.28 ± 0.05 Δ340/380 (n=5), 0.32 ± 0.03 Δ340/380 (n=12), and 0.29 ± 0.03 Δ340/380 (n=14) for CON, SCR, and KD respectively, p=0.66). These data indicated that decreased Ca2+ transients after FGF13 KD were at least partially due to decrease current through CaV1.2 and are consistent with its discovered interaction with JPH2, a known regulator of this function.
Figure 5.
FGF13 knockdown reduces Ca2+ transients but preserves EC-coupling gain. A, representative traces of simultaneous whole-cell voltage clamp of Ca2+ current and Ca2+ transient recordings. B-D, summarized data for the labeled measurements. * P < 0.05 for KD vs SCR (n=10 per group).
FGF13 affects multiple phases of the cardiac action potential
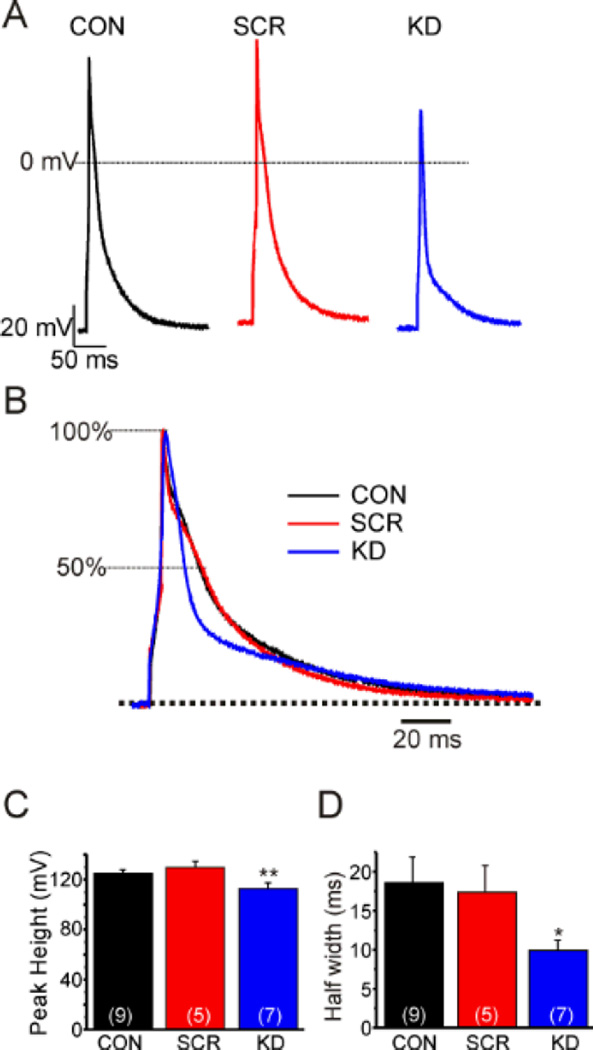
Having established that FGF13 modulates not only voltage-gated Na+ channels,7 but also CaV1.2 Ca2+ channels and Ca2+ transients, we hypothesized that loss of FGF13 would have measurable affects on the cardiac action potential. Therefore, using current clamp, we recorded evoked action potentials in CON, SCR, or KD adult rat ventricular cardiomyocytes (Figure 6A–D). In FGF13 KD cells, the action potential peak amplitude decreased by about 20% (Figure 6A, B), consistent with the previously defined effects of FGF13 on the cardiac Na+ channel current.7 Additionally, , we also observed a shortening of the action potential half-width (Figure 6A, C) (control 18.60 ± 3.34 ms (n=7), scrambled shRNA 17.41 ± 3.40 ms (n=5), FGF13 shRNA 9.85 ± 1.34 ms (n=7), p < 0.05 for FGF13 KD vs. SCR and CON). These data are consistent with changes in phase 2 of the cardiac action potential that is predominantly mediated by CaV1.2 and implicate FGF13 as a potent regulator of Na+ and Ca2+ channels in cardiac myocytes, leading to changes in the cardiac action potential.
Figure 6.
FGF13 affects the cardiac action potential. A, induced action potentials in rat ventricular cardiomyocytes in current clamp mode at 1 Hz. B, normalized induced action potentials to emphasize the change in half width with FGF13 shRNA. C, D, Summarized data of action potential amplitude and half width. The number of cells analyzed is indicated in parentheses. * P < 0.05, ** P < 0.01.
DISCUSSION
Since the initial identification of FHFs1 their complete physiological roles have remained shrouded. Originally hypothesized to act as extracellular growth factors similar to canonical FGFs, these non-secreted FHF proteins do not appear capable of activating FGF receptors.2 The subsequent discovery that FHFs are binding partners for the intracellular C-terminus of voltage-gated Na+ channels3 provided context to appreciate how knockout of FGF14 might reduce neuronal excitability and cause ataxia in mice19 and to hypothesize mechanisms for spinocerebellar ataxia 27, for which FGF14 was identified as the genetic locus.5 Our recent identification of a role for FHFs in the regulation of cardiac Na+ channel function and conduction7 demonstrated cardiac specific roles. Still, the cellular distribution of FGF13 in cardiomyocytes hinted at additional functions distinct from Na+ channel regulation.
Here, we show for the first time that FHFs regulate cardiac ion channels other than voltage-gated Na+ channels. As such, these results fit well with our recent demonstration that FGF14 regulates presynaptic Ca2+ channels in cerebellar neurons.11 Specifically, we demonstrated here that FHFs are part of the dyad macromolecular complex with JPH2 and CaV1.2. Through electrophysiologic and molecular biological methods we demonstrated that FGF13 affects Ca2+ current density and targeting of CaV1.2 to T-tubules. This effect on targeting is specific to CaV1.2, and not a general effect on T-tubule structure, as NCX and RyR localization were not altered, nor was overall T-tubule structure perturbed by FGF13 knockdown. We hypothesize that the abnormal localization of CaV1.2 after FGF13 knockdown represents a defect in intracellular sorting of these proteins in cardiomyocytes. Missorting of CaV1.2 was rescued by overexpression of a human FGF13 splice variant, demonstrating the specificity of the FGF13 knockdown shRNA. Not only did these data provide the first evidence that FGF13 is required for proper targeting of CaV1.2 to the T-tubule in cardiac muscle, but they also show that FGF13 knockdown had profound physiologic effects on Ca2+ current density and Ca2+ cycling, culminating in a decrease in cardiac action potential amplitude and duration and a reduced Ca2+ transient amplitude in parallel with its reduction in CaV1.2 Ca2+ channel current.
The identification of JPH2 as a FGF13 interacting protein provides mechanistic insight to this effect. JPH2 is a structural protein found in the cardiac dyad, where the T-tubule is juxtaposed to one terminal cisterna of the sarcoplasmic reticulum. The structure of JPH2 fixes the distance between the plasma membrane and SR for efficient CICR (Figure 1C). JPH2 contains a cytosolic alpha helical domain that is capped on either side by membrane interaction motifs. On the N-terminus are multiple ‘membrane occupation and recognition nexus’ (MORN) motifs that interact with the inner leaflet of the sarcolemma. At the C-terminus is a transmembrane domain that anchors JPH2 to the sarcoplasmic reticulum.16 Reminiscent of our results after FGF13 knockdown, JPH2 knockdown in cardiomyocytes affects CICR. This provides further support that the interaction between JPH2 and FGF13 that we observed is functionally relevant. Moreover, analogous to loss-of-function mutations in JPH2, we hypothesize that FGF13 mutations may lead to “orphaned ryanodine receptors” that are no longer apposed to CaV1.2, and thus may be associated with heart failure.20 Further studies in the appropriate model are necessary to test that hypothesis.
Nevertheless, the phenotypes after FGF13 and JPH2 knockdown are not identical. JPH2 knockdown did not decrease CaV1.2 Ca2+ current and has no reported effect on voltage-gated Na+ currents. Moreover, JPH2 is observed only in a striated pattern in cardiomyocytes, while we observed additional FGF13 throughout the cytoplasm and in the nucleus. Thus, the fraction of FGF13 that interacts with JPH2 likely represents only one component of the overall FGF13 pool, and is likely also distinct from the fraction interacting with NaV1.5. Other potential FGF13 interactors have previously been reported, such as microtubules in neurons,21 likely indicating additional FGF13 pools. The tubulin interaction site on FGF13 maps to a region in close proximity to where NaV1.5 interacts in our recent crystal structure.9 Thus, we predict that any FGF13 interacting with microtubules would be unable to bind NaV1.5 simultaneously, further underlining the concept of distinct FGF13 pools. While we have so far been unable to observe high affinity binding between tubulin and FGF13, the possibility of such an interaction in cardiomyocytes is attractive, particularly in light of the demonstration that CaV1.2 is trafficked along microtubules via BIN1 in ventricular myocytes.22
In summary, this study identified FHFs as novel modulators of the cardiac L-type CaV1.2 Ca2+ channel and thereby significantly expands our understanding of the roles of these proteins. Our data have important clinical implications. FGF12, the dominant FHF in human heart (data not shown), is greater than 60% homologous with mouse FGF13. Our data suggest that FGF12 loss-of-function mutations would decrease both Ca2+ and Na+ channel currents. Since loss-of-function mutations in NaV1.5 or CaV1.2 have been reported in Brugada syndrome (BrS),23, 24 we specifically hypothesize that FGF12 should be explored as a candidate locus for BrS. Only ~ 30% of BrS patients have an identified mutation,25 so loss-of-function mutations in FGF12 may underlie the mechanism in at least some of the remaining cases by nature of its ability to affect both Na+ and Ca2+ currents.
Supplementary Material
Novelty and Significance.
What Is Known?
Fibroblast growth factor (FGF) homologous factors (FHFs) bind to and modulate the voltagegated Na+ channels.
FGF13 is the predominant adult murine cardiac FHF that modulates NaV1.5 channel current density and kinetics.
FGF13 has widespread subcellular distribution within ventricular cardiac myocytes.
What New Information Does This Article Contribute?
FGF13 is in complex with Junctophilin-2 (JPH2) and a pool of FGF13 localizes to the t-tubules in adult ventricular cardiac myocytes.
Knockdown of FGF13 in mouse and rat ventricular cardiac myocytes leads to mis-localization of the CaV1.2 L-type Ca2+ channel, but it does not affect the localization of the sodium-calcium exchanger or ryanodine receptor or the t-tubule structure.
FGF13 knockdown decreases CaV1.2 current density (ICa) in adult mouse and rat ventricular cardiac myocytes due to a decrease in the number of channels at the surface, leading to reduced Ca2+-induced Ca2+ release (CICR).
FGF13 knockdown reduces peak amplitude and half-width of the action potential in adult rat ventricular cardiac myocytes.
FGF13 modulates voltage-gated Na+ channels in cardiac myocytes, but its widespread distribution, specifically in a striated pattern, indicates that it might regulate other ion channels as well. We discovered that FGF13 resides in the cardiac dyad in complex with the structural protein - JPH2. Without disturbing the t-tubule structure or other ion channels in the dyad, FGF13 knockdown specifically reduces CaV1.2 channels density at the sarcolemma due to mis-localization, leading to reduced ICa and decreased CICR. Cardiac myocytes from FGF13 knockdown show a decrease in the peak amplitude and the half width of the action potential. These findings reveal a novel modulatory pathway through which FGF13 regulates Ca2+ channel function and suggests that mutations in FHFs could underlie cardiac arrhythmias characterized by the loss-of-function of Na+ or Ca2+ channels.
ACKNOWLEDGMENTS
We would like to thank the Duke Proteomics Core Facility for their help in the affinity purification / mass spectroscopy and G. Vann Bennett (Duke University) for the antibody to sodium-calcium exchanger.
SOURCES OF FUNDING
This work was supported by NHLBI R01 HL71165 and R01 HL112918 (G.S.P.), and the Gertrude Elion Mentored Medical Student Award and NHLBI F30 HL112540-01 (J.A.H.).
Nonstandard Abbreviations and Acronyms
- FGF
fibroblast growth factor
- FHF
fibroblast growth factor homologous factor
- CaV1.2 L-type
voltage-gated Ca2+ channel
- α1C
pore-forming subunit of CaV1.2
- FFT
fast Fourier transform
- CICR
Ca2+-induced Ca2+ release
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
None.
REFERENCES
- 1.Smallwood PM, Munoz-Sanjuan I, Tong P, Macke JP, Hendry SH, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. Fibroblast growth factor (fgf) homologous factors: New members of the fgf family implicated in nervous system development. Proc Natl Acad Sci U S A. 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast growth factor (fgf) homologous factors share structural but not functional homology with fgfs. J Biol Chem. 2003;278:34226–34236. doi: 10.1074/jbc.M303183200. [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Dib-Hajj SD, Waxman SG. Fibroblast growth factor homologous factor 1b binds to the c terminus of the tetrodotoxin-resistant sodium channel rnav1.9a (nan) J Biol Chem. 2001;276:18925–18933. doi: 10.1074/jbc.M101606200. [DOI] [PubMed] [Google Scholar]
- 4.Wittmack EK, Rush AM, Craner MJ, Goldfarb M, Waxman SG, Dib-Hajj SD. Fibroblast growth factor homologous factor 2b: Association with nav1.6 and selective colocalization at nodes of ranvier of dorsal root axons. J Neurosci. 2004;24:6765–6775. doi: 10.1523/JNEUROSCI.1628-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Swieten JC, Brusse E, de Graaf BM, Krieger E, van de Graaf R, de Koning I, Maat-Kievit A, Leegwater P, Dooijes D, Oostra BA, Heutink P. A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebral ataxia. Am J Hum Genet. 2003;72:191–199. doi: 10.1086/345488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q, Bardgett M, Wozniak D, Lou J, McNeill B, Chen C, Nardi A, Reid D, Yamada K, Ornitz DM. Ataxia and paroxysmal dyskinesia in mice lacking axonally transported fgf14. Neuron. 2002;35:25–38. doi: 10.1016/s0896-6273(02)00744-4. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Hennessey JA, Kirkton RD, Wang C, Graham V, Puranam RS, Rosenberg PB, Bursac N, Pitt GS. Fibroblast growth factor homologous factor 13 regulates na+ channels and conduction velocity in murine hearts. Circ Res. 2011;109:775–782. doi: 10.1161/CIRCRESAHA.111.247957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Wang C, Hoch EG, Pitt GS. Identification of novel interaction sites that determine specificity between fibroblast growth factor homologous factors and voltage-gated sodium channels. J Biol Chem. 2011;286:24253–24263. doi: 10.1074/jbc.M111.245803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Chung BC, Yan H, Lee SY, Pitt GS. Crystal structure of the ternary complex of a nav c-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure. 2012;20:1167–1176. doi: 10.1016/j.str.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao M, Xu L, Laezza F, Yamada K, Feng S, Ornitz DM. Impaired hippocampal synaptic transmission and plasticity in mice lacking fibroblast growth factor 14. Mol Cell Neurosci. 2007;34:366–377. doi: 10.1016/j.mcn.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Yan H, Pablo JL, Pitt GS. Fgf14 regulates presynaptic ca2+ channels and synaptic transmission. Cell Reports. doi: 10.1016/j.celrep.2013.06.012. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomsen MB, Wang C, Ozgen N, Wang HG, Rosen MR, Pitt GS. Accessory subunit kchip2 modulates the cardiac l-type calcium current. Circ Res. 2009;104:1382–1389. doi: 10.1161/CIRCRESAHA.109.196972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren M, Spitzer KW, Steadman BW, Rees TD, Venable P, Taylor T, Shibayama J, Yan P, Wuskell JP, Loew LM, Zaitsev AV. High-precision recording of the action potential in isolated cardiomyocytes using the near-infrared fluorescent dye di-4-anbdqbs. Am J Physiol Heart Circ Physiol. 2010;299:H1271–H1281. doi: 10.1152/ajpheart.00248.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Oort RJ, Garbino A, Wang W, Dixit SS, Landstrom AP, Gaur N, De Almeida AC, Skapura DG, Rudy Y, Burns AR, Ackerman MJ, Wehrens XH. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation. 2011;123:979–988. doi: 10.1161/CIRCULATIONAHA.110.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier SK, Westenbroek RE, McCormick KA, Curtis R, Scheuer T, Catterall WA. Distinct subcellular localization of different sodium channel alpha and beta subunits in single ventricular myocytes from mouse heart. Circulation. 2004;109:1421–1427. doi: 10.1161/01.CIR.0000121421.61896.24. [DOI] [PubMed] [Google Scholar]
- 16.Garbino A, Wehrens XHT. Emerging role of junctophilin-2 as a regulator of calcium handling in the heart. Acta Pharmacologica Sinica. 2010;31:1019–1021. doi: 10.1038/aps.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia J, Tanabe T, Beam KG. Relationship of calcium transients to calcium currents and charge movements in myotubes expressing skeletal and cardiac dihydropyridine receptors. J Gen Physiol. 1994;103:125–147. doi: 10.1085/jgp.103.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golini L, Chouabe C, Berthier C, Cusimano V, Fornaro M, Bonvallet R, Formoso L, Giacomello E, Jacquemond V, Sorrentino V. Junctophilin 1 and 2 proteins interact with the l-type ca2+ channel dihydropyridine receptors (dhprs) in skeletal muscle. J Biol Chem. 2011;286:43717–43725. doi: 10.1074/jbc.M111.292755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Bardgett ME, Wong M, Wozniak DF, Lou J, McNeil BD, Chen C, Nardi A, Reid DC, Yamada K, Ornitz DM. Ataxia and paroxysmal dyskinesia in mice lacking axonally transported fgf14. Neuron. 2002;35:25–38. doi: 10.1016/s0896-6273(02)00744-4. [DOI] [PubMed] [Google Scholar]
- 20.Song LS, Sobie EA, McCulle S, Lederer WJ, Balke CW, Cheng H. Orphaned ryanodine receptors in the failing heart. Proc Natl Acad Sci U S A. 2006;103:4305–4310. doi: 10.1073/pnas.0509324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu QF, Yang L, Li S, Wang Q, Yuan XB, Gao X, Bao L, Zhang X. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell. 2012;149:1549–1564. doi: 10.1016/j.cell.2012.04.046. [DOI] [PubMed] [Google Scholar]
- 22.Hong TT, Smyth JW, Gao D, Chu KY, Vogan JM, Fong TS, Jensen BC, Colecraft HM, Shaw RM. Bin1 localizes the l-type calcium channel to cardiac t-tubules. PLoS Biol. 2010;8:e1000312. doi: 10.1371/journal.pbio.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burashnikov E, Pfeiffer R, Barajas-Martinez H, Delpon E, Hu D, Desai M, Borggrefe M, Haissaguerre M, Kanter R, Pollevick GD, Guerchicoff A, Laino R, Marieb M, Nademanee K, Nam GB, Robles R, Schimpf R, Stapleton DD, Viskin S, Winters S, Wolpert C, Zimmern S, Veltmann C, Antzelevitch C. Mutations in the cardiac l-type calcium channel associated with inherited j-wave syndromes and sudden cardiac death. Heart Rhythm. 2010;7:1872–1882. doi: 10.1016/j.hrthm.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paula LH, Poul J, Sarah S, Johanna M-S, Jørgen KK, Valerie AC, Michael C. The genetic basis of brugada syndrome: A mutation update. Human Mutation. 2009;30:1256–1266. doi: 10.1002/humu.21066. [DOI] [PubMed] [Google Scholar]
- 25.Antzelevitch C, Brugada P, Borggrefe M, Brugada J, Brugada R, Corrado D, Gussak I, LeMarec H, Nademanee K, Perez Riera AR, Shimizu W, Schulze-Bahr E, Tan H, Wilde A. Brugada syndrome: Report of the second consensus conference: Endorsed by the heart rhythm society and the european heart rhythm association. Circulation. 2005;111:659–670. doi: 10.1161/01.CIR.0000152479.54298.51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.