SUMMARY
We report the solution NMR structure of a designed dimetal-binding protein, di-Zn(II) DFsc, along with a secondary refinement step employing molecular dynamics techniques. Calculation of the initial NMR structural ensemble by standard methods led to distortions in the metal-ligand geometries at the active site. Unrestrained molecular dynamics using a non-bonded force field for the metal shell, followed by quantum mechanical/molecular mechanical dynamics of DFsc, were used to relax local frustrations at the dimetal site that were apparent in the initial NMR structure and provide a more realistic description of the structure. The MD model is consistent with NMR restraints, and in good agreement with the structural and functional properties expected for DF proteins. This work demonstrates that NMR structures of metalloproteins can be further refined using classical and first-principles molecular dynamics methods in the presence of explicit solvent to provide otherwise unavailable insight into the geometry of the metal center.
INTRODUCTION
It is estimated that approximately half of all known proteins bind metal ions or metal-containing cofactors (Thomson and Gray, 1998). Metalloproteins are functionally diverse and support numerous biological activities, including electron transfer, catalysis, and signal transduction processes. NMR is an excellent tool for probing the three-dimensional structure and dynamics of many proteins. However, NMR structure determination of metalloproteins typically requires a prior knowledge of the geometry of the metal coordination site owing to a lack of direct experimental metal ion-protein distance restraints. Typically, one imposes restraints on the metal center during initial structure calculation using a bonded approach to metal-ligand refinement. The restraints can be based on external information, such as a crystal structure of the protein or crystal structures of homologous proteins, or based on reasonable estimates of the bond lengths. These coordination-bonded restraints can often lead to poor description of the metal site, with inaccurate coordination geometry and orientation. Here we used a general method in which unrestrained molecular dynamics (MD) and quantum mechanical/molecular mechanical (QM/MM) dynamics are used in a secondary refinement step to help relax local frustrations apparent within the NMR structure of the metal-binding site of the de novo designed metalloprotein, due ferri single chain (DFsc).
The water-soluble diiron/dimanganese class of proteins represents a diverse group of structures that catalyze a wide range of oxygen-dependent processes including H2O2 dismutation, epoxidation, hydroxylation, and radical-forming reactions (Solomon et al., 2000; Wallar and Lipscomb, 1996). To help decipher how the protein matrix tunes the reactivity of the cofactor in these proteins, we have designed and structurally characterized a series of minimal models for this class of structures (Calhoun et al., 2005). The initial due ferri proteins (DF1, DF2) were antiparallel homodimers of helix-loop-helix motifs that bind the cofactor near the center of the structure (Lahr et al., 2005; Lombardi et al., 2000). More recently, we have designed a single-chain version of the protein, DFsc, which provides enhanced stability and allows one to make asymmetric changes to the binding site (Calhoun et al., 2003). Herein we report the di-Zn(II) DFsc structure, which is derived from a secondary refinement step employing classical MD and QM/MM MD calculations to optimize the metal-binding geometry of the calculated NMR structure.
RESULTS AND DISCUSSION
Structure Determination
The NMR solution structure of DFsc was solved using the di-Zn(II) derivative as a diamagnetic analog of the di-Fe(III) and di-Mn(II) protein. Previous studies of the dimeric DF2 proteins indicate that this substitution has minimal effect on structure (Maglio et al., 2005). The di-Zn(II) DFsc structure was calculated using a total of 2367 experimental restraints including 1731 nuclear Overhauser effect (NOE) -based distance restraints (488 intraresidue, 509 sequential, 489 medium-range, and 176 long-range), 205 dihedral angle restraints, 69 hydrogen bond restraints, 190 proton and 161 CαCβ chemical shift restraints, and 80 amide N-H residual dipolar coupling restraints. Stereospecific assignments of the methyl groups of leucine and valine were made using trace glucose labeling (Neri et al., 1989). A family of structures was calculated using the program CNS (Brunger et al., 1998), with the di-Zn(II) site treated essentially the same as in the DF2 structure (Lahr et al., 2005; Maglio et al., 2005).
Overall NMR Structure
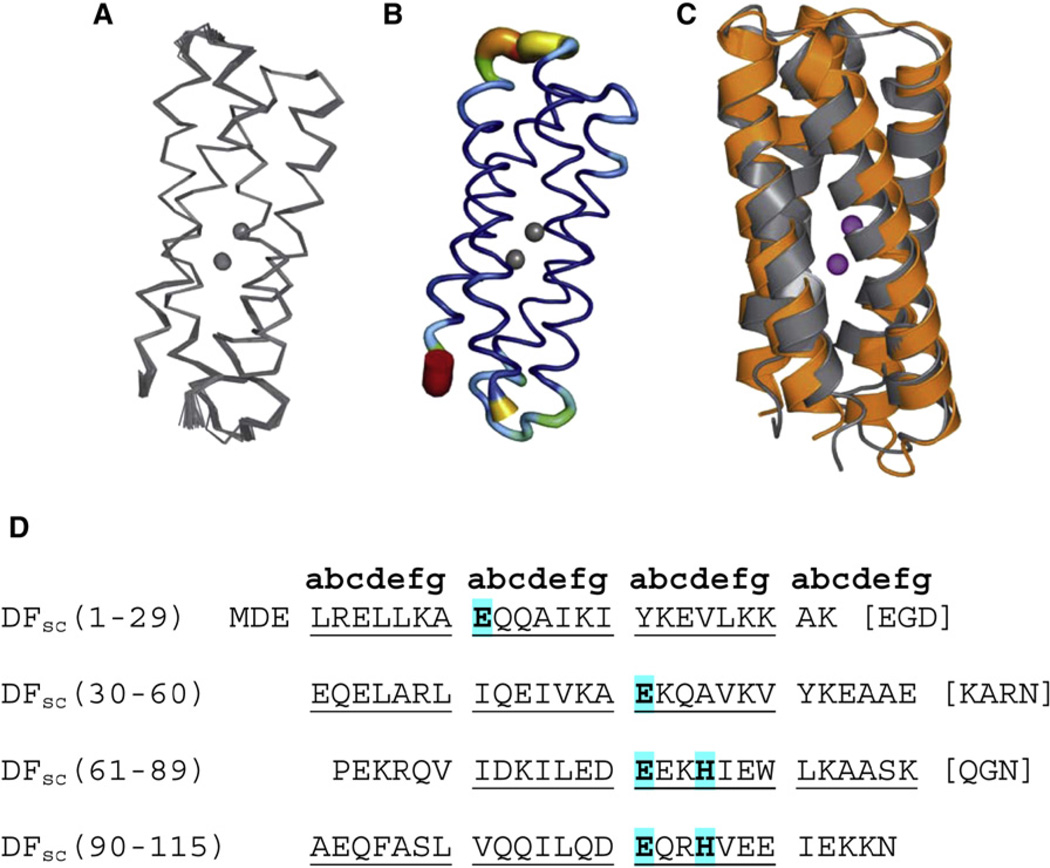
The structure was initially calculated by introducing metal-ligand bonding restraints intended to recapitulate the geometry in the structure of di-Zn(II) DF1. The dimetal site is derived from two copies of an EXXH motif (located on helices 3 and 4), each consisting of one Glu µ-1,3 bridging ligand and one terminally δN-ligated His ligand and two chelating Glu residues from helices 1 and 2. The resulting ensemble of 36 structures is tightly clustered, with a root-mean-square deviation (rmsd) of 0.33 ± 0.10 Å for the backbone atoms (residues 2–115) and 1.17 ± 0.13 Å for all atoms (residues 2–115) (Figure 1). The family of structures contained no violation of the distance constraints greater than 0.5 Å or of the dihedral angle constraints greater than 5°.
Figure 1. Di-Zn(II) DFsc NMR Structure.
(A) α carbon trace of the ensemble of di-Zn(II) DFsc NMR structures.
(B) Sausage representation of the 36 NMR structures of di-Zn(II) DFsc. The color and tube size are based on the B factor, where the color scale is from dark blue to red, with red corresponding to the highest B factor and dark blue to the lowest B factor.
(C) Target designed model (orange) of DFsc overlaid with the lowest-energy NMR structure (gray).
(D) Amino acid sequence of DFsc; blue-highlighted residues consist of the primary coordination sphere.
All figures were generated with PyMOL (DeLano, 2002) unless otherwise noted.
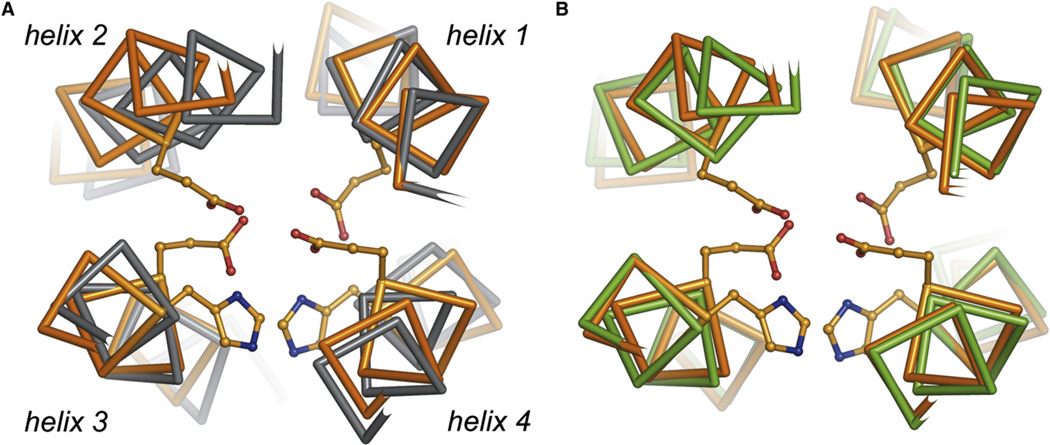
Although the calculated NMR structure of di-Zn(II) DFsc was of high precision, it displayed several questionable features. For example, helix 2 is displaced by 1–2 Å relative to the position expected from structures of earlier members of the DF family (Figure 2A). This relative movement of helix 2 results in compression of the helix 1, 2 interface and causes almost complete occlusion of the anticipated solvent/substrate access channel leading to the dimetal site (Figure 3). This occlusion of the active site appeared at odds with the reactivity of the di-Fe(II) form of the protein (unpublished results). The displacement of helix 2 also results in geometric parameters of the di-Zn(II) site that are highly distorted with respect to typical Zn(II) ligand environments and also inconsistent with known spectroscopic properties of di-Fe(II) derivatives (Wei et al., 2005), suggesting that further refinement of the metal center would be necessary. Indeed, the dimetal site exhibits a number of clashes between the chelating and bridging carboxylate groups of the Glu ligands, resulting in close contacts between the oxygen atoms (see Table S1 in the Supplemental Data available with this article online). These steric clashes can be rationalized through the inclusion of restraints on all metal ligand distances and the absence of explicit solvent, which leads to crowding at the dimetal site and an unphysical approach between helices 1 and 2 (Figure 2A). Also, in the absence of explicit restraints and nonbonded potentials, NMR-based structures tend to be distorted (Kuszewski et al., 1999). This tendency could easily explain the absence of the anticipated small active site solvent channel in the computed NMR structural model (Figure 3). These factors promoted a second phase of refinement employing explicit solvent molecular dynamics techniques.
Figure 2. Superposition of di-Zn(II) DFsc Models and Structure.
The di-Zn(II) DFsc target design model (orange) was superimposed versus (A) di-Zn(II) DFsc calculated NMR structure (gray), rmsd 1.2 Å and (B) average MD model of di-Zn(II) DFsc (green), rmsd 0.8 Å. The superpositions demonstrate the large movement of helix 2 in the di-Zn(II) DFsc calculated NMR structure relative to the targeted design and MD model. Helices 1, 3, and 4 are included in the superpositions: residues 6–16, 69–82, and 99–112; the loops are not included.
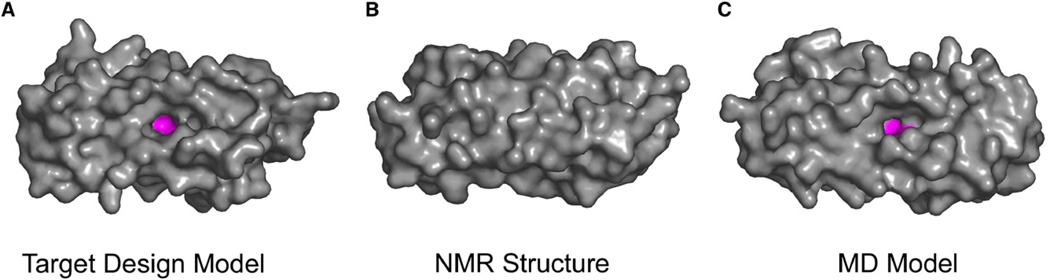
Figure 3. Surface Representation of di-Zn(II) DFsc Displaying the Accessibility to the Active Site.
(A) Target design model of di-Zn(II) DFsc.
(B) Calculated NMR structure of di-Zn(II) DFsc, where the metal site is inaccessible owing to the shift in helix 2.
(C) MD model of di-Zn(II) DFsc. All the atoms of the protein are colored gray and the metal ions and primary ligands are shown in magenta. Hydrogen atoms are not depicted in this figure.
Molecular Dynamics-Based Refinement
The di-Zn(II) DFsc protein was solvated in a water box and equilibrated by performing 10 ns classical MD (Cornell et al., 1995), followed by 5 ps of Car-Parrinello hybrid QM/MM dynamics (Car and Parrinello, 1985; Laio et al., 2002) to relax frustrations at the metal center from first principles. During initial force field-based MD, the di-Zn(II) cluster is treated without any additional bond and angle restraints on the metal-ligand interactions using a quantum-based atomic charge distribution (Dal Peraro et al., 2007), whereas in the QM/MM approach, the metal site is described at the DFT-BLYP (Becke, 1988; Lee et al., 1988) level of theory. The final MD computational model is represented by the QM/MM MD ensemble on the equilibrated trajectory, and a representative configuration is deposited in the Protein Data Bank (PDB ID code: 2HZ8).
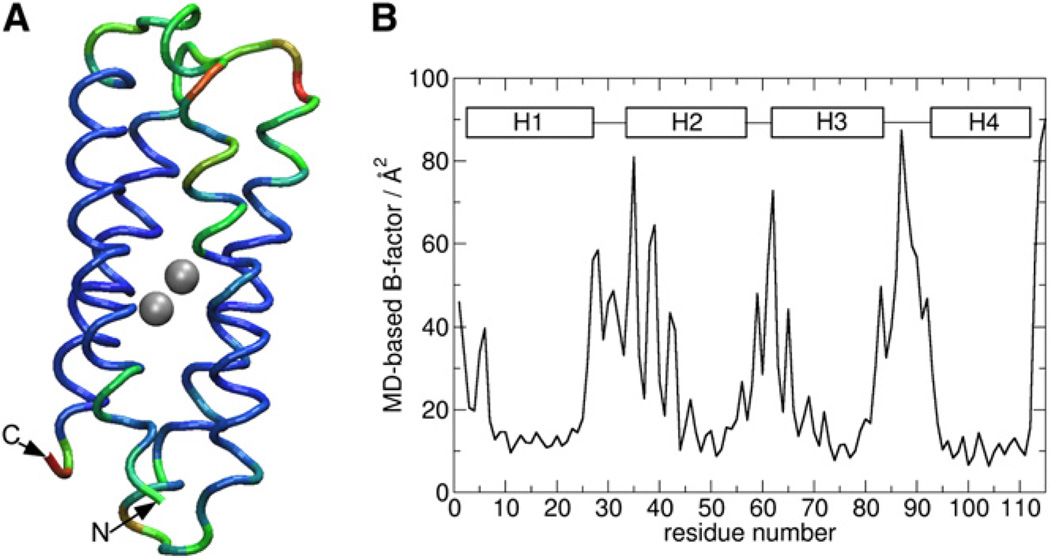
The MD model appears to be consistent with NMR restraints as demonstrated by the low number of NOE violations, which occurred only in surface-exposed or highly dynamic regions of the protein (Table S6). The MD model is also dynamically consistent with the NMR ensemble as seen from the comparison of experimental and calculated B factors (Figures 1B and 4). Most importantly, the MD model resolves all major discrepancies between the target designed model and experimental structures. The backbone rmsd for the target designed model and the initially calculated NMR structure is 1.2 Å. The backbone rmsd for the target designed model and the final unrestrained MD model is 0.8 Å. The backbone rmsd of the MD model versus the target designed model agrees well with typical values of less than 1 Å that are calculated from designed proteins in our laboratory versus experimental structures for this class of proteins (Calhoun et al., 2005; Lombardi et al., 2000). Much of the improvement within the MD model versus calculated NMR structure is localized to helix 2, which moved to the expected orientation during MD simulation (Figure 2B). Helices 1 and 2 are thus farther apart, presumably driven by relief of steric clashes in the dimetal site and the infiltration of water into the access channel.
Figure 4. B Factors of the MD Model of di-Zn(II).
(A) MD model of di-Zn(II) DFsc. The color scale is from blue to red, where red corresponds to the highest B factor and blue to the lowest B factor.
(B) Plot of MD-based B factor/Å2 versus residue number.
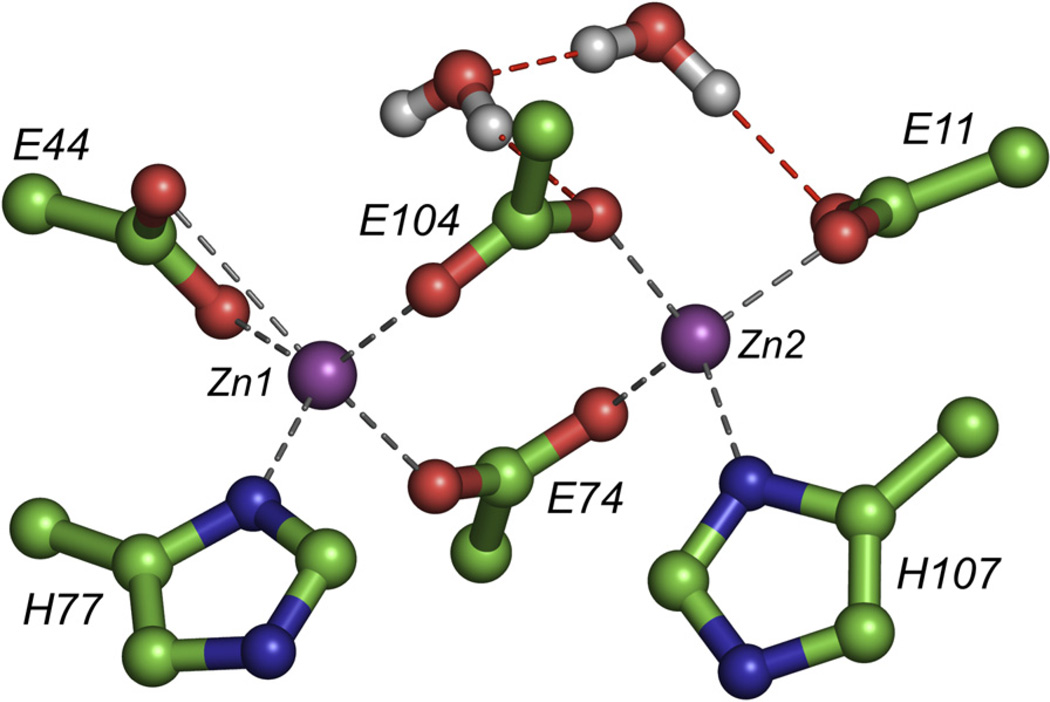
Accompanying the observed shift of helix 2 in the MD model are changes in ligand geometry to a 4-coordinate/5-coordinate geometry (the initial ensemble was calculated using the bis-5-coordinated geometry observed in previous crystal structures of DF family members) (Calhoun et al., 2005; Lombardi et al., 2000). In the final MD model, Zn1 adopts an approximately square pyramidal coordination geometry, whereas Zn2 is approximately tetrahedral (Table 1; Figure 5). The terminally bound E11 ligand undergoes a carboxylate shift from chelating to monodentate, forming a tight OE2-Zn2 bond, whereas OE1 is no longer in bonding distance of the metal ion but instead forms hydrogen bonds to Tyr51 and one water molecule (Figure 5). It is also noteworthy that the bridging ligands E74 and E104 are less distorted owing to the additional space created between helices 1 and 2 which results from the previously mentioned shift of helix 2. The bridging carboxylates as well as the two His ligands show negligible variation in metal-ligand distances in the MD model.
Table 1.
MD Average Structure Distances for Zn(II)-Ligand
| Zn 1 | Distance (Å) | Zn 2 | Distance (Å) |
|---|---|---|---|
| E44-OE1 | 2.22 ± 0.26 | E11-OE1 | 3.12 ± 0.27 |
| E44-OE2 | 2.36 ± 0.27 | E11-OE2 | 2.03 ± 0.05 |
| E74-OE1 | 2.13 ± 0.07 | E74-OE2 | 2.01 ± 0.04 |
| H77-ND1 | 2.09 ± 0.07 | H107-ND1 | 2.10 ± 0.07 |
| E104-OE2 | 2.07 ± 0.07 | E104-OE1 | 2.03 ± 0.05 |
Zn1-Zn2 bond length is 4.16 Å.
Figure 5. Di-Zn(II) Cluster in the Average MD Model.
Dashed lines indicate hydrogen bonding. The Zn(II)-Zn(II) distance is 4.2 Å. Compared to the calculated NMR structure, E11 undergoes a carboxylate shift to monodentate and two water molecules solvate the dimetal site. Hydrogen bonds between the water molecules and ligands are shown as red dashed lines.
The 4- and 5-coordinate geometry of the metal center are in complete agreement with known solution properties of diferrous DF derivatives. Previous investigations with the homodimeric protein DF2t (Wei et al., 2005) show 4- and 5-coordinate geometry in solution. Striking 4- and 5-coordinate geometry also exists between di-Fe(II) DFsc as determined by circular dichroism/magnetic circular dichroism (C. Bell and E.I. Solomon, personal communication). Also of note, the rearrangement to 4-coordinate and 5-coordinate geometry was previously observed from Car-Parrinello MD and DFT studies of minimal models for the di-Zn(II) site in DF1 (Magistrato et al., 2003; Papoian et al., 2003).
The presence of a solvent channel in our DF family of proteins is extremely important for their metal-binding and catalytic properties. As previously seen in the homodimeric protein DF1 (Di Costanzo et al., 2001), the lack of a solvent channel rendered the metal-binding site inaccessible, and hence the protein had to be denatured to bind metal ions. The solvent channel also needs to be large enough to accommodate substrates for catalytic activity (Kaplan and DeGrado, 2004). Classical MD calculations allow two ordered nonligand water molecules to solvate the solvent/substrate access channel lying above the dimetal site. One of these water molecules hydrogen bonds to OE1 of E11, possibly stabilizing the monodentate binding mode of this residue while also hydrogen bonding to the second water molecule. The second water molecule also interacts with the bridging E104 ligand (Figure 5).
In summary, refinement of an initial NMR structural model for di-Zn(II) DFsc with classical and QM/MM MD simulations results in a structure that is globally consistent with (1) the experimental restraints available for the protein from NMR, (2) experimental data on DFsc (unpublished data), and (3) the target designed model, providing otherwise unavailable insight into the geometry of the dimetal center. Clearly, NMR-based structure calculations of metalloproteins can be further refined using MD-based techniques (Hsiao et al., 2005; Ryde, 2007); our approach is based on a nonbonded force-field description of the metal-ligand interactions followed by quantum MD in the presence of explicit water. This is particularly suited for the determination of designed metalloproteins whose structure determinants are not available to aid in NMR structure determination. Improved results apparently arise from the flexibility of the metal site and inclusion of water molecules. Having obtained structural information for the designed DFsc protein provides us with an excellent opportunity to study the reaction mechanism with ferrous ions and oxygen to explore the species that form in this system. DFsc along with the other subset of DF proteins can further be used to provide models that help elucidate the assembly and catalytic pathway of natural systems.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Protein was overexpressed in Escherichia coli BL21 (DE3) from a pet-28a vector containing the gene encoding DFsc. Uniformly 15N- and 15N/13C-labeled samples were prepared by growing bacteria in M9 minimal media supplemented with 1 g/l 15NH4Cl, 2 g/L [U-13C]glucose (Cambridge Isotope Laboratories, Andover, MA, USA). Fractional 13C labeling was achieved with 10% [U-13C]glucose (420 mg/l) for stereospecific assignment of methylene protons. The protein was purified as described in Calhoun et al. (2003). Sample purity and molecular weight were confirmed by SDS-PAGE and MALDI-TOF mass spectroscopy.
NMR Data Collection and Analysis
All samples were prepared in Shigemi NMR tubes, with a metal ion:protein ratio of 2.5 Zn(II):1 DFsc in H2O solution containing 50 mM deuterated sodium acetate, 10 mM NaCl, and 5% D2O (pH 6.0). NMR experiments were recorded at 308K on a Varian INOVA 750 MHz or Varian INOVA 500 MHz NMR spectrometer equipped with four radio frequency channels, a pulsed-field gradient accessory, and a Nalorac HCN triple-resonance 750 MHz, 5 mm inverse probe with a z gradient coil. Backbone amide 1H and 15N, Cα, C = O, and side-chain Cβ resonances were assigned by using CT-HNCO, HNCACB, CBCA(CO)NH, and H(CCO)NH. Side-chain assignments were obtained by C(CCO)NH, HCCH-TOCSY, and HCCH3-TOCSY. Aromatic side-chain assignments were made using 2D and 3D aromatic experiments: hbcbcgcdhd_aro, hbcbcgcdehe_aro, and HCCH-TOCSY. Data were processed using FELIX (Molecular Simulations, San Diego, CA, USA).
Structure Calculation
Distance constraints were derived from 3D 15N-, 13C-separated NOE, and 4D 15N/13C/13C-separated NOE experiments with mixing times of 100 ms. NOE restraints were classified into four distance ranges: strong, 1.8–2.9 Å; medium, 1.8–3.5 Å; weak, 1.8–5.0 Å; and very weak, 1.8–6.0 Å. Backbone φ torsion angle restraints were derived from 3JHNHα coupling values obtained from a 3D HNHA experiment. Talos was used to generate ψ torsion angle restraints (Cornilescu et al., 1999). H-bond restraints were derived from hydrogen exchange data. Proton and CαCβ chemical shift restraints were derived from backbone assignments. Residual dipolar coupling restraints were derived from an IPAP-HSQC experiment. During structure calculation, the metal site was treated essentially the same as previous dimeric DF structures: the bonds and angles of the ligands were restrained by upper and lower limits (Table S2). The structures were calculated from 2367 experimental restraints by simulated annealing using CNS (Brunger et al., 1998). Out of 300 calculated structures, the 36 with the lowest target functions were used for structural analyses. The quality of the structures was evaluated with PROCHECK (Laskowski et al., 1996). A list of experiments and references is available under accession number 7247 in the BioMagResBank.
Classical Molecular Dynamics Simulation
To allow for maximal flexibility in the bimetal cluster, within the AMBER force field (Cornell et al., 1995) we adopt a nonbonded model (Dal Peraro et al., 2007) in which no explicit Zn(II)-ligand bonded interactions are present but the metal-ligand geometry depends only on Lennard-Jones and electrostatic interactions. Lennard-Jones parameters are adopted from Stote and Karplus (1995). The charges of the metal shell have been obtained from a Bader analysis (Bader, 1990) of the electronic density within the active site calculated at the DFT/BLYP level (Becke, 1988; Lee et al., 1988). Integration of the charge density over each atomic basin (Henkelman et al., 2006), as defined by zero-flux hypersurfaces of the electron density with respect to the coordinates, gives the effective point charge belonging to each atom (Dal Peraro et al., 2007).
The resulting charges on Zn(II) are typically lower than the formal 2+ charge, and the charges on the ligands needed only small modifications (Dal Peraro et al., 2007) (Table S4). The system is solvated in a box of TIP3P water (11 Å between solute and box edge), and 11 Na+ and 10 Cl− ions are added, leading to roughly the same ionic strength as in the experiment. Because of the high concentration of charged side chains, the presence of counterions is especially important to prevent side chains from distorting in an unphysical manner. The system is first minimized in 40,000 steps of steepest descent using the AMBER MD engine. Next, the system is equilibrated stepwise by applying a constraint on the solute of 5 kcal/mol·Å2, which is slowly switched off during the initial first ns of MD. Ten nanoseconds of constant-pressure (1 atm), constant-temperature (298K) simulations are then performed coupling the system with a Nosé-Hoover Langevin barostat and thermostat. A conservative time step of 1 fs is used to solve Newton’s equation, and hydrogen-involving bonds of water molecules are constrained by the LINKS algorithm.
First-Principles Molecular Dynamics Simulations
After 10 ns of classical MD, the system is simulated for 5 ps by Car-Parrinello MD (Car and Parrinello 1985; Laio et al., 2002), using the QM/MM interface developed by Laio et al. (2002) implemented in the CPMD code (http://www.cpmd.org/). In this approach, the metal center of the complex, namely the two Zn(II) ions and their ligands—four glutamates (cut at Cβ atom), two his-tidines (cut at Cα), and two water molecules solvating the metal center after MD calculations—is treated at the quantum level (DFT-BLYP; Becke, 1988; Lee et al., 1988). The remaining part of the protein and the solvent is treated at the classical level using the AMBER force field (Cornell et al., 1995). The boundary QM atoms are saturated by hydrogen capping (Laio et al., 2002), and the nonbonded interactions between the MM and QM regions are treated in a fully Hamiltonian coupling scheme (Laio et al., 2002). The valence electrons are described by a plane wave basis set to a cutoff of 70 Ry. The interactions between valence electrons and ionic cores are described with norm-conserving Martins-Troullier pseudopotentials. Car-Parrinello molecular dynamics simulations are carried out with a time step of 0.12 fs (totally for 5 ps) and a fictitious electron mass of 500 au; constant-temperature (298K) simulations are achieved by coupling the system with a Nosé-Hoover thermostat at 500 cm−1 frequency. This protocol has shown to be successful for a variety of metalloproteins, and specifically for the DF family (Magistrato et al., 2003).
Measurement of Distances between Helices
Local helix axes, of each helix, in the four-helix bundles were calculated with HELANAL (Bansal et al., 2000), which calculates the local helix axis based on the Cα atoms of four consecutive residues. The end and loop regions from this analysis were excluded. Analysis for DFsc includes only residues 4–21 (helix 1), 37–54 (helix 2), 67–84 (helix 3), and 95–112 (helix 4) (Figure S2). The distances between these local helix-axis points were measured and the pairs of axis points closest in the NMR model were identified. Distances in the experimental structure to the corresponding distances from the simulation (average over selected snapshots) were compared (Table S5).
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by NIH grants GM 35940 and GM 54616, and NSF MRSEC award DMR05-200020. None of the authors have financial interest related to this work.
Footnotes
Supplemental Data
Supplemental Data include two figures and six tables and can be found with this article online at http://www.structure.org/cgi/content/full/16/2/210/DC1/.
Accession Numbers
The NMR QM/MM MD refined structure (MD model) has been deposited in the RCSB Protein Data Bank under ID code 2HZ8. Chemical shifts and NMR constraints have been deposited in the BioMagResBank under accession number 7247.
REFERENCES
- Bader RFW. Atoms in Molecules—A Quantum Theory. Volume 22. Oxford: Oxford University Press; 1990. [Google Scholar]
- Bansal M, Kumar S, Velavan R. HELANAL: a program to characterize helix geometry in proteins. J. Biomol. Struct. Dyn. 2000;17:811–819. doi: 10.1080/07391102.2000.10506570. [DOI] [PubMed] [Google Scholar]
- Becke AD. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Calhoun JR, Kono H, Lahr S, Wang W, DeGrado WF, Saven JG. Computational design and characterization of a monomeric helical dinuclear metalloprotein. J. Mol. Biol. 2003;334:1101–1115. doi: 10.1016/j.jmb.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Calhoun JR, Nastri F, Maglio O, Pavone V, Lombardi A, DeGrado WF. Artificial diiron proteins: from structure to function. Biopolymers. 2005;80:264–278. doi: 10.1002/bip.20230. [DOI] [PubMed] [Google Scholar]
- Car R, Parrinello M. Unified approach for molecular dynamics and density functional theory. Phys. Rev. Lett. 1985;55:2471–2474. doi: 10.1103/PhysRevLett.55.2471. [DOI] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Dal Peraro M, Spiegel K, Lamourex G, De Vivo M, DeGrado WF, Klein ML. Modeling the charge distrubution at metal sites in proteins for molecular dynamics simulations. J. Struct. Biol. 2007;157:444–453. doi: 10.1016/j.jsb.2006.10.019. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Di Costanzo L, Wade H, Geremia S, Randaccio L, Pavone V, DeGrado WF, Lombardi A. Toward the de novo design of a catalytically active helix-bundle: a substrate accessible carboxylate-bridged dinuclear metal center. J. Am. Chem. Soc. 2001;123:12749–12757. doi: 10.1021/ja010506x. [DOI] [PubMed] [Google Scholar]
- Henkelman G, Arnaldsson A, Jonsson H. A fast and robust algorithm for Bader decomposition of charge density. Comput. Mater. Sci. 2006;36:354–360. [Google Scholar]
- Hsiao YW, Drakenberg T, Ryde U. NMR structure determination of proteins supplemented by quantum chemical calculations: detailed structure of the Ca2+ sites in the EGF34 fragment of protein S. J. Biomol. NMR. 2005;31:97–114. doi: 10.1007/s10858-004-6729-7. [DOI] [PubMed] [Google Scholar]
- Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc. Natl. Acad. Sci. USA. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuszewski J, Gronenborn AM, Clore GM. Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J. Am. Chem. Soc. 1999;121:2337–2338. [Google Scholar]
- Lahr SJ, Engel D, Stayrook SE, Maglio O, North B, Geremia S, Lombardi A, DeGrado WF. Analysis and design of turns in α-helical hairpins. J. Mol. Biol. 2005;346:1441–1454. doi: 10.1016/j.jmb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Laio A, VandeVondele J, Rothlisberger U. A Hamiltonian electrostatic coupling scheme for hybrid Car-Parrinello molecular dynamics simulations. J. Chem. Phys. 2002;116:6941–6947. [Google Scholar]
- Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- Lombardi A, Summa C, DeGrado WF. Retrostructural analysis of metalloproteins: de novo design of a model for diiron proteins. Proc. Natl. Acad. Sci. USA. 2000;97:6298–6305. doi: 10.1073/pnas.97.12.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistrato A, DeGrado WF, Laio A, Rothlisberger U, VandeVondele J, Klein ML. Characterization of the dizinc analogue of the synthetic diiron protein DF1 using ab initio and hybrid quantum/classical molecular dynamics simulations. J. Phys. Chem. B. 2003;107:4182–4188. [Google Scholar]
- Maglio O, Nastri F, Calhoun JR, Lahr S, Wade H, Pavone V, DeGrado WF, Lombardi A. Artificial di-iron proteins: solution characterization of four helix bundles containing two distinct types of inter-helical loops. J. Biol. Inorg. Chem. 2005;10:539–549. doi: 10.1007/s00775-005-0002-8. [DOI] [PubMed] [Google Scholar]
- Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- Papoian GA, DeGrado WF, Klein ML. Probing the configurational space of a metalloprotein core: an ab initio molecular dynamics study of Duo Ferro 1 binuclear Zn cofactor. J. Am. Chem. Soc. 2003;125:560–569. doi: 10.1021/ja028161l. [DOI] [PubMed] [Google Scholar]
- Ryde U. Accurate metal-site structures in proteins obtained by combining experimental data and quantum chemistry. Dalton Trans. 2007:607–625. doi: 10.1039/b614448a. [DOI] [PubMed] [Google Scholar]
- Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee SK, Lehnert N, Neese F, Skulan AJ, Yang YS, Zhou J. Geometric and electronic structure/function correlations in non-heme iron enzymes. Chem. Rev. 2000;100:235–350. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- Stote RH, Karplus M. Zinc binding in proteins and solution: a simple but accurate nonbonded representation. Proteins. 1995;23:12–31. doi: 10.1002/prot.340230104. [DOI] [PubMed] [Google Scholar]
- Thomson AJ, Gray HB. Bio-inorganic chemistry. Curr. Opin. Chem. Biol. 1998;2:155–158. doi: 10.1016/s1367-5931(98)80056-2. [DOI] [PubMed] [Google Scholar]
- Wallar BJ, Lipscomb JD. Dioxygen activation by enzymes containing binuclear non-heme iron clusters. Chem. Rev. 1996;96:2625–2657. doi: 10.1021/cr9500489. [DOI] [PubMed] [Google Scholar]
- Wei PP, Skulan AJ, Wade H, DeGrado WF, Solomon EI. Spectroscopic and computational studies of the de novo designed protein DF2t: correlation to the biferrous active site of ribonucleotide reductase and factors that affect O2 reactivity. J. Am. Chem. Soc. 2005;127:16098–16106. doi: 10.1021/ja053661a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.