Abstract
Membrane proteins destined for the vacuolar or lysosomal lumen are typically ubiquitinated, the ubiquitin serving as a targeting signal for the multivesicular body pathway. The RING-domain ubiquitin ligase Tul1 is an integral membrane protein that modifies the yeast vacuolar enzyme carboxypeptidase S (Cps1), the polyphosphatase Ppn1/Phm5 and other proteins containing exposed hydrophilic residues within their transmembrane domains (TMDs). Here we show that Bsd2 provides an alternative ubiquitination mechanism for Cps1, Phm5 and other proteins. Bsd2 is a three-TMD protein with a PPXY motif that binds the HECT domain ubiquitin ligase Rsp5. It can thus act as a specific adaptor linking Rsp5 to its substrates. Like Tul1, the Bsd2 system recognises polar TMDs. Bsd2 also controls the vacuolar targeting of a manganese transporter and a mutant plasma membrane ATPase, and together with the ER retrieval receptor Rer1, it protects cells from stress. We suggest that Bsd2 has a wide role in the quality control of membrane proteins. Bsd2 is the yeast homologue of human NEDD4 binding protein N4WBP5, which may therefore have similar functions.
Keywords: Bsd2, multivesicular bodies, Rsp5, transmembrane domain, ubiquitin ligase
Introduction
Eukaryotic cells have several ways of destroying membrane proteins that are defective or no longer needed. Within the endoplasmic reticulum (ER), misfolded or unassembled proteins can be recognised by specialised machinery that ubiquitinates them, extracts them from the membrane and delivers them to the proteasome for degradation (for recent reviews see Ellgaard and Helenius, 2003; Jarosch et al, 2003; Trombetta and Parodi, 2003). Proteins that exit the ER can in some cases be retrieved by receptors, but the major destruction pathway involves their delivery to endosomes. Such proteins enter the internal membranes of multivesicular bodies (MVBs), which are formed by invagination of the endosomal membrane. After retrieval of component proteins from their surface membranes, MVBs fuse with lysosomes or, in yeast, the vacuole. The internal vesicles are thus delivered to the lumen of these organelles where they are digested by hydrolytic enzymes (Futter et al, 1996; Odorizzi et al, 1998; Katzmann et al, 2002).
Recent work has shown that many membrane proteins are marked for delivery into MVBs by ubiquitination of their cytoplasmic domains (Katzmann et al, 2001; Reggiori and Pelham, 2001; Urbanowski and Piper, 2001; reviewed by Hicke and Dunn, 2003). Modification by a single ubiquitin is sufficient, at least with some proteins, for diversion from the Golgi to endosomes (Reggiori and Pelham, 2002), a step mediated by the GGA clathrin-binding proteins (Black and Pelham, 2000), which can directly bind ubiquitin (Shiba et al, 2004). Monoubiquitination is also sufficient for internalisation from the plasma membrane (Shih et al, 2000), as well as for sorting by the MVB machinery (Katzmann et al, 2001; Reggiori and Pelham, 2001). MVB targeting is used not only for the removal of abnormal proteins but also for the selective downregulation of pheromone receptors and of small molecule transporters from the plasma membrane or directly from the biosynthetic pathway (Kolling and Hollenberg, 1994; Hein et al, 1995; Hicke and Riezman, 1996; Beck et al, 1999; Gitan and Eide, 2000). In addition, certain vacuolar enzymes, notably carboxypeptidase S (Cps1) and the polyphosphatase Ppn1/Phm5, are synthesised as precursors containing a transmembrane domain (TMD) and a short N-terminal cytoplasmic extension that becomes ubiquitinated, directing the proteins to the MVB pathway (Katzmann et al, 2001; Reggiori and Pelham, 2001). The ubiquitin tag is in turn recognised on endosomes by proteins that mediate MVB formation (Katzmann et al, 2001, 2003; Bilodeau et al, 2002).
Clearly, the selective targeting of proteins to MVBs requires their precise recognition by the ubiquitination machinery. Ubiquitination involves the ATP-dependent activation of a family of ubiquitin conjugating enzymes (E2) by a ubiquitin activating enzyme (E1), which transfers ubiquitin to a cysteine residue on the E2 protein. Substrate recognition is achieved by one of a series of ubiquitin protein ligases (E3), of which two main families have been studied. One family is characterised by the presence of a RING domain, which binds an E2 protein and is thought simply to bring it into proximity with the substrate, allowing direct transfer of ubiquitin to available lysines. Substrate recognition is achieved by other domains of the E3 proteins (for a review see Hershko and Ciechanover, 1998). The second family comprises the HECT domain proteins, which transiently accept ubiquitin from an E2 and transfer it to substrate. The HECT ligase Rsp5, and its mammalian relative Nedd4, has been implicated in several steps in endocytosis (Hicke and Dunn, 2003). In particular, yeast Rsp5 has been shown to be responsible for the modification and hence sorting to MVBs of several cell surface transporters (Hein et al, 1995; Galan et al, 1996; Hicke, 1999; Gitan and Eide, 2000). Rsp5 has other functions as well, and is essential for growth. It contains three WW domains, which recognise short amino-acid motifs typically containing the sequence PPXY (Sudol, 1996; Harty et al, 2000). Interactions between these domains and either substrates or other molecules such as Bul1 and Bul2 are thought to be important in controlling Rsp5 function (Yashiroda et al, 1996; Helliwell et al, 2001; Soetens et al, 2001).
Previously we have shown that the introduction of polar residues into otherwise hydrophobic TMDs results in ubiquitin-mediated sorting of the corresponding membrane proteins to MVBs (Reggiori et al, 2000; Reggiori and Pelham, 2002). We suggested that this represents a quality control mechanism to ensure the destruction of proteins that are misfolded within the lipid bilayer, since properly folded proteins do not usually expose polar residues to lipid. Using a derivative of the endosomal SNARE protein Pep12 with a glutamic acid residue inserted into its TMD, we identified a RING domain ubiquitin ligase, Tul1, which is required for sorting of this substrate into MVBs (Reggiori and Pelham, 2002). Tul1 is itself a membrane protein with seven TMDs, and thus has the potential to recognise the aberrant TMD directly and mediate ubiquitination of lysines on the unaltered cytoplasmic domain of Pep12. Surprisingly, we also found a requirement for Tul1 for the efficient sorting of Cps1 and Phm5 to MVBs, and suggested that these proteins, which have relatively polar TMDs, follow the MVB pathway because they mimic malfolded proteins. However, although removal of Tul1 reduced ubiquitination of Cps1 it did not abolish it, raising the possibility that other ubiquitin ligases also contribute to the recognition of this protein (Reggiori and Pelham, 2002).
Here we show that an evolutionarily conserved three-TMD protein, Bsd2, is also implicated in the sorting of Cps1 and Phm5. Bsd2 was previously found to control the vacuolar targeting of a metal ion transporter (Liu et al, 1997; Liu and Culotta, 1999b). We show that it binds the WW domains in Rsp5 via a PPXY motif, and that this motif is required for the efficient ubiquitination and sorting of Cps1 and Phm5. Like Tul1, Bsd2 modifies proteins with polar TMDs, although with different specificity. Our data confirm that the TMDs of Cps1 and Phm5 can direct them into the MVB pathway, and show that recognition of these can be achieved by at least two distinct mechanisms. Furthermore, Bsd2, together with the ER retrieval receptor Rer1, helps to protect cells against the toxic effects of the arginine analogue canavanine, or high temperature. We suggest that Bsd2 contributes generally to the recognition and removal of misfolded membrane proteins, as well as sorting specific substrates to the MVB pathway.
Results
Bsd2 is required for the efficient sorting of Cps1 and Phm5
To investigate the possible contribution of Bsd2 to MVB sorting, we compared bsd2, tul1, bsd2 tul1 double mutants and wild-type cells for their ability to sort proteins to the vacuolar lumen. The markers were green fluorescent protein (GFP)-tagged versions of Cps1, Phm5 and Pep3D, a derivative of Pep12 with a glutamate residue at position 3 of the TMD, which we have previously characterised as a Tul1 substrate. If not ubiquitinated, all three of these proteins reach the vacuole but are found on the outer vacuolar membrane (Reggiori and Pelham, 2002). Thus, the extent of their modification can be deduced by the relative amounts of GFP visualised within and around the vacuole.
As shown in Figure 1, MVB sorting of Pep3D was greatly reduced in tul1 mutant cells, but largely unaffected in the bsd2 mutant. In contrast, Phm5 and Cps1 were strongly affected in bsd2 cells, and showed a more variable dependence on tul1, depending on growth conditions (see Methods). The bsd2 tul1 double mutant showed the strongest effect with these proteins, very little GFP fluorescence being visible in the vacuole lumen. Nevertheless, a ubiquitin-Phm5 conjugate (Ub-Phm5) was still sorted correctly in bsd2 and bsd2 tul1 cells (Figure 1), indicating that Bsd2 is not required for the MVB sorting machinery to function, but rather inhibits the efficient ubiquitination of substrates. This suggests that Bsd2 and Tul1 both contribute to the modification of Phm5 and Cps1.
Figure 1.
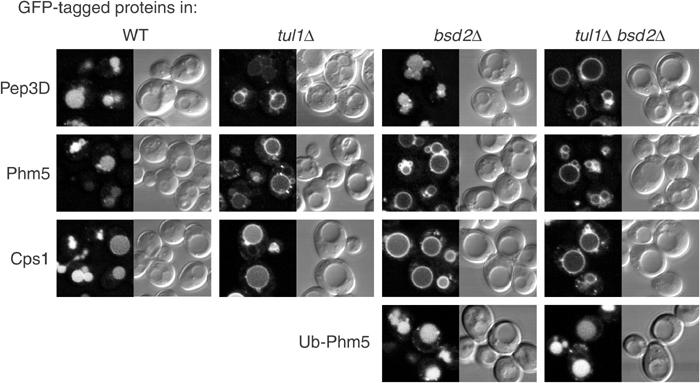
Bsd2 contributes to the sorting of proteins into MVBs. GFP-tagged proteins were expressed in the indicated strains and examined by confocal microscopy. Representative examples of the GFP fluorescence and the corresponding interference contrast images are shown.
Bsd2 contains a cytoplasmic PPTY motif
Bsd2 is a three-TMD protein with a relatively long N-terminal domain containing the sequence PPTY (Figure 2A). An N-terminal GFP fusion, expressed from the BSD2 promoter, showed very weak fluorescence within the vacuole (data not shown; see also below). To confirm this localisation, we compared the fate of the protein in a wild-type strain and in one deficient in vacuolar proteases. Immunoblotting showed that most of the protein was cleaved in the wild-type strain to yield free GFP, which is relatively protease-resistant, and that this cleavage was dependent on vacuolar proteases (Figure 2B). Thus, Bsd2 evidently reaches the vacuole.
Figure 2.
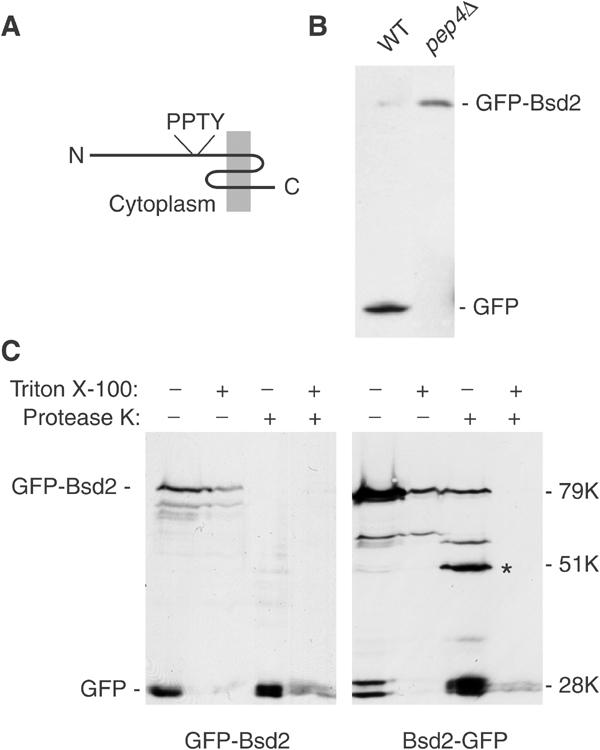
Orientation and fate of Bsd2. (A) Predicted structure of Bsd2, with the position of the PPTY motif indicated. The protein is 321 residues long. (B) Immunoblotting with anti-GFP of an N-terminal GFP fusion protein expressed from its own promoter in bsd2Δ cells that were wild type for protease activity (WT), and in a strain deficient in vacuolar proteases (pep4Δ). (C) Membranes from cells expressing Bsd2 tagged with GFP at the N terminus (GFP-Bsd2) or the C terminus (Bsd2-GFP) were digested with protease K with or without 1% Triton X-100 as indicated, and the fragments were detected by immunoblotting. Numbers indicate the apparent sizes of the proteins relative to globular protein markers. Note that Bsd2 and its fragments appear larger than they really are. The asterisk indicates a prominent proteolytic fragment with an apparent size of 51 kDa.
To determine the orientation of Bsd2 within membranes, we performed protease protection assays on membranes from cells expressing the protein from the relatively strong TPI promoter. This allowed us to focus on the full-length molecules that have not yet reached the vacuole. When the GFP tag was on the N terminus, it was readily removed by proteinase K even in the absence of detergent, indicating a cytoplasmic location (Figure 2C). In contrast, a C-terminal GFP tag was much more resistant to protease, and yielded a fragment intermediate in size between free GFP and the intact fusion protein. We observed that the fusion protein migrated with an apparent molecular weight larger than expected (approximately 79 kDa instead of 63 kDa), a property that was also apparent when the N-terminal domain was expressed as a fusion protein in Escherichia coli (data not shown). Taking this into account, the mobility of the major protected fragment was consistent with loss of most of the N-terminal domain. Detergent treatment resulted in complete protease sensitivity. These data thus indicate that the N terminus of Bsd2, containing the PPTY motif, is cytoplasmic and the C terminus luminal, as depicted in Figure 2A.
The Bsd2 PPTY motif binds to the WW domains of Rsp5
A number of PPXY motifs have been identified as ligands for one or more of the WW domains of Rsp5 or its mammalian homologues. An obvious possibility was therefore that Bsd2 recruits the ubiquitin ligase Rsp5 to Cps1 and Phm5, using the PPTY sequence. As a direct test of this possible interaction between Bsd2 and Rsp5, we expressed in bacteria each of the WW domains of Rsp5 fused to glutathione-S-transferase (GST), and the N-terminal cytoplasmic domain of Bsd2 fused to protein A. The GST-WW chimeras were bound to beads and incubated with bacterial extracts containing the Bsd2 fusion protein. As shown in Figure 3, the Bsd2 fragment bound specifically to each of the WW domains, the highest yield being obtained with WW2. This binding largely resisted washing with 1.5 M KCl. Mutation of the Bsd2 PPTY sequence to PPTA abolished binding to WW1 and WW3, and greatly reduced binding to WW2. Thus, Bsd2 does indeed have the ability to recruit Rsp5, binding in particular to its WW2 domain.
Figure 3.
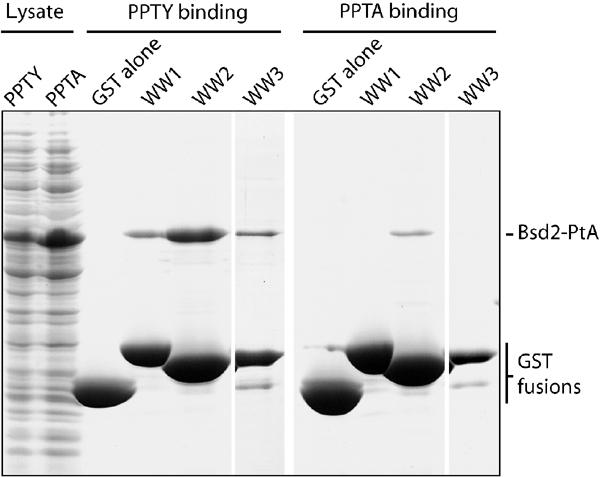
Binding of the Bsd2 N terminus to the WW domains of Rsp5. A Coomassie-stained gel is shown. E. coli lysates (left lanes) containing either the PPTY or PPTA versions of the Bsd2 N terminus fused to protein A (Bsd2-PtA) were incubated with beads containing GST alone or fused to one of the three WW domains of Rsp5. After extensive washing, the GST fusions and bound proteins were eluted with SDS sample buffer and subjected to gel electrophoresis.
Bsd2 is required for efficient ubiquitination of Phm5
Our data suggest strongly that Bsd2 mediates ubiquitination of Phm5. To facilitate detection of this, we first reduced the size of GFP-Phm5 by deleting the luminal domain. As discussed below, this did not affect MVB sorting of the construct. Immunoisolation of the truncated GFP chimera and blotting with antiubiquitin antibodies showed that modified forms could readily be detected in wild-type cells (Figure 4A). As ubiquitin is absent from cargo molecules inside MVBs (Dupre and Haguenauer-Tsapis, 2001; Katzmann et al, 2001), this assay detects ubiquitinated molecules in transit to MVBs. In agreement with this, more ubiquitinated protein could be detected in pep12 cells, in which delivery to endosomes is blocked, and in vps27 cells, in which MVB formation cannot occur (Figure 4A).
Figure 4.
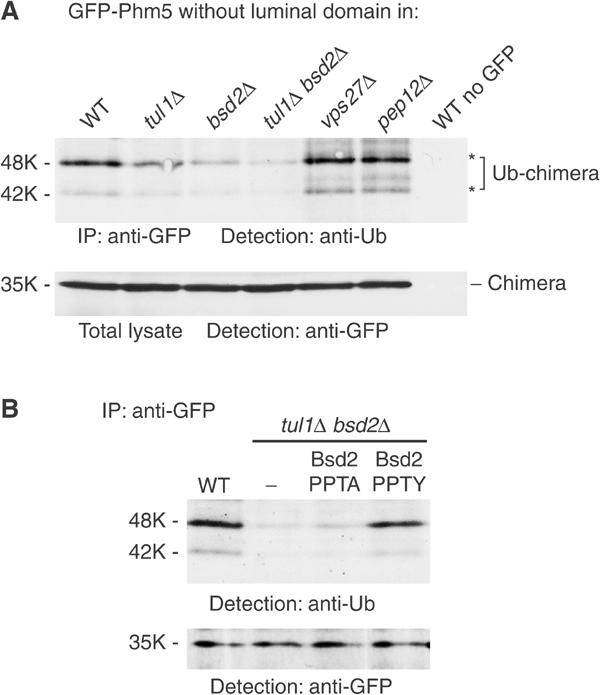
Bsd2 affects ubiquitination of Phm5. (A) A GFP-tagged variant of Phm5 lacking the luminal domain was immunoprecipitated from the indicated strains, a strain not expressing the GFP-tagged protein serving as a control. Ubiquitinated species were then identified by immunoblotting with anti-ubiquitin. Asterisks indicate the two major ubiquitinated species. These species could also be detected, although inefficiently, by blotting with anti-GFP (not shown). Samples of the total cell lysates were run in parallel and immunoblotted to show the equivalent amounts of intact GFP-Phm5 chimera in each sample. (B) As (A) except that tul1 bsd2 double mutant cells contained either an empty vector plasmid (−), or plasmids expressing untagged wild-type (PPTY) or point mutant (PPTA) Bsd2 from its own promoter. Samples of the immunoprecipitates were also blotted with anti-GFP to show that they contained equivalent amounts of GFP-Phm5.
Using this assay, we found that ubiquitination of the GFP chimera was reduced by about 40–50% in tul1 cells, and by 60–80% in bsd2 cells (Figure 4A). It was reduced still further in the bsd2 tul1 double mutant, in agreement with the fluorescence localisation data. However, even in the double mutant, 10–20% of the wild-type ubiquitination level could still be observed, suggesting the existence of additional ubiquitination mechanisms, independent of both Tul1 and Bsd2.
Ubiquitination of the Phm5 construct by Bsd2 depended on the PPTY motif: in a bsd2 tul1 double mutant, ubiquitination could be restored by expression of wild-type levels of untagged Bsd2, but not by comparable expression of the PPTA mutant (Figure 4B). Furthermore, the PPTA mutant was unable to mediate the entry of GFP-Cps1 into vacuoles by the MVB pathway (Figure 5A). These results, together with parallel studies showing that WW domains 2 and 3 of Rsp5 are required for ubiquitination of Cps1 (Katzmann et al, 2004), provide strong evidence that Bsd2 mediates the ubiquitination of Cps1 and Phm5 by recruiting Rsp5.
Figure 5.
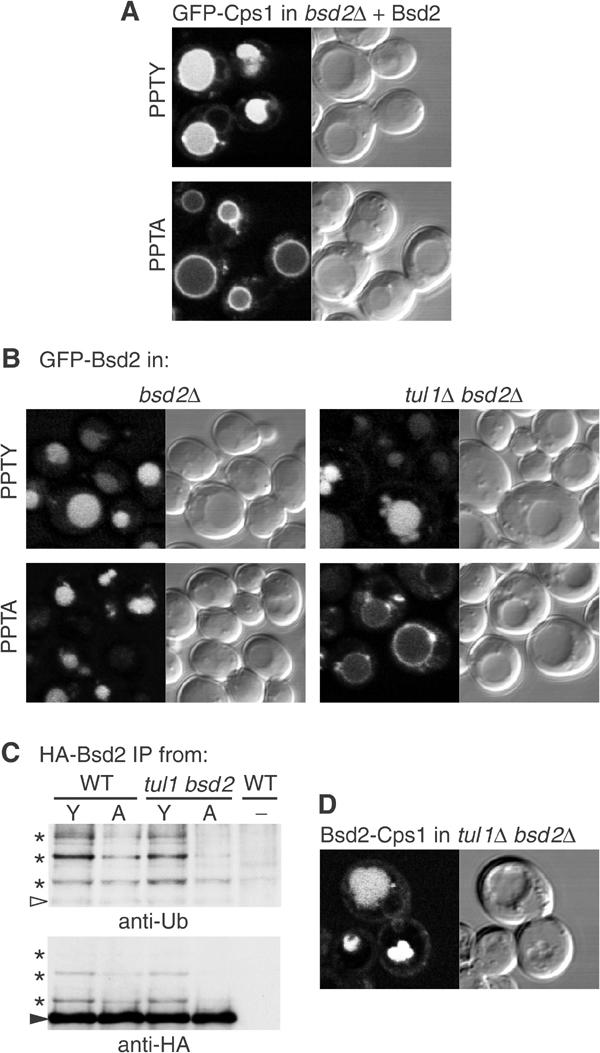
Effects of the PPTY motif on sorting of Cps1 and Bsd2. (A) Complementation of a bsd2 null mutant by expression of untagged wild-type (PPTY) Bsd2 restores sorting of Cps1 into vacuoles, whereas the point mutant (PPTA) does not. (B) Expression of GFP-Bsd2 with wild-type (PPTY) or mutant (PPTA) sequences in the indicated strains. Note that entry into vacuoles requires either the PPTY sequence or Tul1. (C) HA-tagged Bsd2 containing the normal (Y) or mutated (A) PPTY motif was immunoprecipitated from wild-type or tul1 bsd2 cells and probed with antiubiquitin; the blot was then re-probed with anti-HA. The arrowhead indicates the position of full-length HA-Bsd2 (apparent size 54 kDa), and the asterisks indicate ubiquitinated forms. Control cells (−) expressed no HA-tagged protein. (D) A chimera consisting of the N-terminal cytoplasmic domain of Bsd2 fused to GFP-Cps1 is sorted into vacuoles in a tul1 bsd2 double mutant.
Bsd2 is ubiquitinated and enters MVBs
The fact that GFP on the N terminus of Bsd2, initially cytoplasmic, is subsequently removed by vacuolar proteases (Figure 2B) indicates that GFP-Bsd2 must itself be a substrate for the MVB pathway. As Bsd2 can mediate ubiquitination of other substrates, an obvious possibility is that it also facilitates its own modification by Rsp5. When GFP-Bsd2 was expressed from the strong TPI promoter in bsd2 cells, internal vacuolar fluorescence was readily visible (Figure 5B). Surprisingly, the same was true for the PPTA mutant, even though it should not be capable of recruiting Rsp5. However, in a tul1 bsd2 double mutant a significant amount of the PPTA form of Bsd2 was on the vacuolar membrane (Figure 5B), implying that it is a substrate for Tul1. Tul1 is evidently not needed for sorting of the active PPTY form of Bsd2, since this entered the vacuole efficiently in tul1 cells.
To confirm ubiquitination of Bsd2, HA epitope-tagged versions of the wild-type and PPTA mutant were immunoprecipitated and probed with antiubiquitin antibodies (Figure 5C). Forms of Bsd2 with sizes corresponding to addition of one, two or possibly three ubiquitins could be detected. They were less prominent with the PPTA mutant and, as predicted from the GFP data were further reduced when the tul1 gene was mutated. Thus Bsd2 not only mediates its own ubiquitination and MVB sorting via the PPTY motif but also has features that are recognised by Tul1.
That the N-terminal cytoplasmic domain of Bsd2 is indeed sufficient for MVB targeting was shown by fusing it to GFP-Cps1. The resultant chimera entered vacuoles efficiently in the bsd2 tul1 double mutant (Figure 5D). Indeed, its sorting was more efficient than that of Cps1 in wild-type cells, suggesting that cis-ubiquitination is more reliable than trans-ubiquitination.
Bsd2-dependent ubiquitination depends on TMD sequences
We have previously demonstrated that Tul1 can modify Pep12 derivatives whose only deviation from wild-type sequence is within the TMD (Reggiori and Pelham, 2002). However, Bsd2 was not required for sorting of Pep3D, and it has been reported that ubiquitination is specified by a short sequence in the cytoplasmic domain of Cps1 (Katzmann et al, 2001). We therefore investigated further the features required for Bsd2-dependent ubiquitination and sorting. As mentioned above, removal of the entire luminal domain of Phm5 did not affect its ubiquitination (Figure 4), nor its Bsd2- and Tul1-dependent entry into vacuoles, as assayed by fluorescence (Figure 6A). Hence, features recognised by the Bsd2 system must lie in the cytoplasmic and/or TMDs of Phm5.
Figure 6.
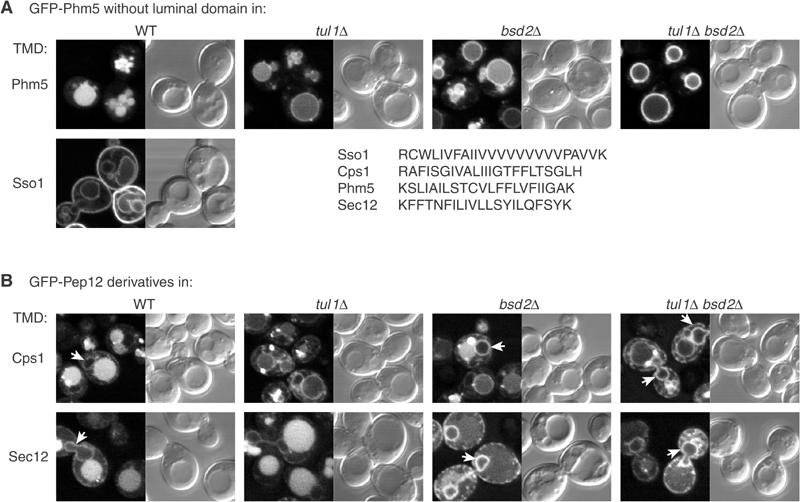
Sorting depends on TMDs. (A) GFP-Phm5 constructs in which the entire luminal domain was deleted were expressed in the indicated strains. The constructs had either the normal Phm5 TMD or the Sso1 TMD as indicated. The sequences of these, and the Cps1 and Sec12 TMDs for comparison, are shown. (B) GFP-Pep12 constructs in which the Pep12 TMD had been replaced with that from Cps1 or Sec12 were expressed in the indicated strains. Note that Pep12 itself is never found inside vacuoles, and thus entry of these constructs is dictated entirely by their TMDs. Arrows indicate nuclear envelope and peripheral ER.
Fusion of the Phm5 cytoplasmic domain to the TMD of the plasma membrane SNARE Sso1 resulted in transport of much of the chimera to the plasma membrane, but some reached the vacuolar membrane (Figure 6A). The essentially complete absence of GFP from the vacuole lumen indicates that this construct can pass through the Golgi and endosomes without being modified by either Tul1 or Bsd2, and hence that TMD sequences are important for recognition by either of these. It also shows that the cytoplasmic domain of Phm5 is not a sufficient signal for modification.
As an alternative approach, we examined derivatives of Pep12 in which only the TMD had been altered. Pep12 itself contains no signals for ubiquitination and does not enter MVBs, and thus provides a neutral test molecule (Reggiori et al, 2000; Reggiori and Pelham, 2002). Introduction of the Cps1 TMD induced MVB sorting, and this was reduced in both tul1 and bsd2 mutants (Figure 6B). Thus the Cps1 TMD is a sufficient signal not only for Tul1-mediated but also for Bsd2-mediated ubiquitination.
In addition to MVB targeting, the Cps1 TMD also caused a small amount of the Pep12 to be retained in the nuclear envelope and peripheral ER. When MVB sorting was reduced, especially in the tul1 bsd2 double mutant, there was a corresponding increase in the amount of ER fluorescence (arrows in Figure 6B). Thus ER retention and MVB targeting seem to be alternative and competing fates for polar TMDs.
A more extreme example of this was provided by the strongly polar Sec12 TMD. This is a well-characterised target for a receptor, Rer1, which mediates its retrieval from the Golgi to the ER (Sato et al, 1997; Sato et al, 2001). Despite this, Pep12 with the Sec12 TMD mostly escapes the ER and instead is targeted to the MVB pathway (Figure 6B; Reggiori et al, 2000). As shown in Figure 6B, mutation of bsd2 almost completely inhibited transport of the Pep12-Sec12 chimera to vacuoles. Instead, it accumulated in the ER, indicating that in wild-type cells Bsd2 effectively competes with Rer1 to force a different fate on the Sec12 TMD. Strikingly, with this construct MVB targeting was mediated entirely by Bsd2: a tul1 mutation had no discernable effect either alone or in combination with bsd2.
We conclude from these experiments that Tul1 and Bsd2 both respond to polar TMDs, and have distinct but overlapping specificities. In particular, while both influence the fate of Phm5 and Cps1, Tul1 can efficiently sort Pep3D in the absence of Bsd2 (Figure 1), and likewise Bsd2 can sort Pep12-Sec12 in the absence of Tul1 (Figure 6B). These distinct specificities show that Tul1 and Bsd2 act independently, rather than in concert or sequentially. The simplest interpretation is that they each directly recognise the TMDs of their substrates. Tul1 binds the ubiquitin-conjugating enzyme Ubc4 (Reggiori et al, 2000; Reggiori and Pelham, 2002), whereas Bsd2 binds Rsp5, which is also thought to receive ubiquitin from Ubc4 (Gitan and Eide, 2000). In each case, the net result is the transfer of ubiquitin from Ubc4 to cytoplasmic lysines on the substrate. Thus marked, the substrate then enters the MVB pathway.
The PPTY motif is required for Bsd2 to regulate a metal ion transporter
Bsd2 has previously been shown to be required for downregulation of the metal ion transporter Smf1. In the presence of heavy metals, Smf1 is transported to the vacuole and degraded. The consequent loss of activity protects cells from heavy-metal poisoning. This downregulation depends on Bsd2, and as a result bsd2 mutants grow very poorly on plates containing cadmium (Figure 7A; Liu et al, 1997). The mechanism of Smf1 sorting has not previously been described, but it is thought to be triggered by a metal-induced conformational change in the transmembrane portion of the molecule (Liu and Culotta, 1999a). We found that the growth of bsd2 cells on cadmium could be restored by expression of wild-type or GFP-tagged Bsd2, but not by the PPTA mutant (Figure 7A). This suggests that Bsd2 controls Smf1 by mediating Rsp5-dependent ubiquitination of the metal-bound state.
Figure 7.
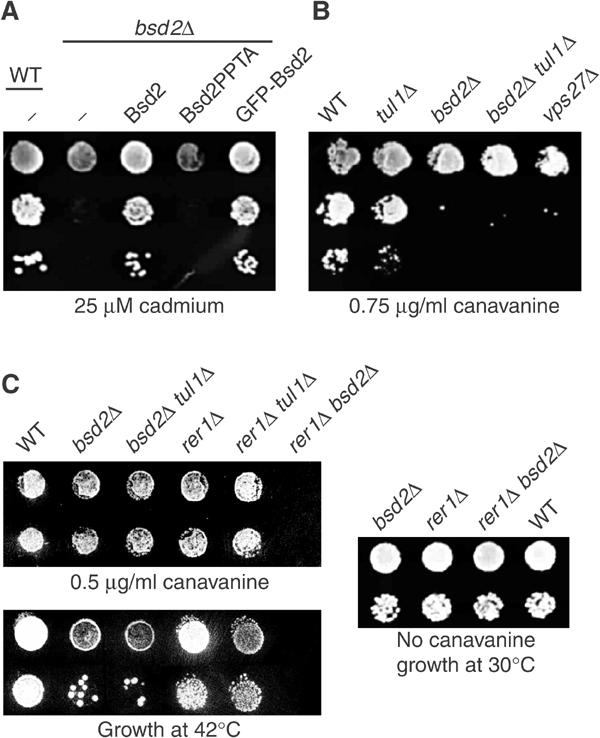
Stress sensitivity of bsd2 mutants. (A) Serial dilutions of cells were grown on plates containing 25 μM cadmium chloride. The bsd2Δ cells contained either an empty vector plasmid (−), or one expressing untagged wild-type Bsd2 or the PPTA mutant (Bsd2PPTA) from the BSD2 promoter, or a plasmid expressing GFP-Bsd2. (B) Serial dilutions of strains with the indicated genotypes were grown on plates lacking arginine and containing 0.75 μg/ml canavanine. Note that growth strongly depends on the number of cells plated, since uptake of canavanine by the cells depletes it from the plate. All strains were plated at the same densities. (C) Cells were grown on plates containing 0.5 μg/ml canavanine, or on normal plates that were incubated either at 30 or 42°C.
A general role for Bsd2
Although Bsd2 has specific roles in the sorting of Smf1 and of vacuolar enzymes, its ability to recognise relatively polar TMDs suggests that it may contribute more generally to the recognition of misfolded membrane proteins. To explore this possibility, we exposed bsd2 cells to the arginine analogue canavanine. Figure 7B shows that at a critical concentration of canavanine, bsd2 cells grew poorly. In contrast, tul1 cells were relatively unaffected by these treatments. Interestingly, a vps27 mutant, in which the MVB pathway cannot operate efficiently, showed the same sensitivity to canavanine as the bsd2 mutant (Figure 7B). This suggests that the MVB pathway plays a significant role in the disposal of misfolded proteins, and that Bsd2 is responsible for targeting at least some of them to this pathway.
There are two principal means for a cell to dispose of aberrant membrane proteins. They can be retained in and degraded from the ER, or they can be delivered to the vacuole. Rer1 and Bsd2 appear to recognise similar substrates, and contribute to ER and vacuolar sorting, respectively. Thus, removal of both might be expected to have a particularly strong additive effect. In agreement with this, we found that bsd2 rer1 cells were completely unable to grow on levels of canavanine that the bsd2 single mutant could easily tolerate (Figure 7C). In contrast, there was no interaction between rer1 and tul1. This is consistent with previous findings that Tul1 substrates tend to be retained in the ER in an Rer1-independent manner (Reggiori et al, 2000; Reggiori and Pelham, 2002).
Although the transporter responsible for canavanine uptake, Can1, is expressed constitutively in growing cells, it is difficult to exclude completely the possibility that changes to its trafficking contribute to canavanine sensitivity. To avoid this, we instead stressed cells by growth at high temperature. As shown in Figure 7C, the results were very similar to those obtained with canavanine. The bsd2 single mutant was slightly temperature-sensitive, and the bsd2 rer1 double mutant could not grow at all at 42°C, although it was perfectly viable under normal conditions. We conclude that Bsd2 is involved not only in specific functions but also more generally in the quality control of membrane proteins. The results also demonstrate that Rer1 can contribute to the protection of cells from damaged proteins.
Discussion
In this paper, we have established that Bsd2 contributes to the ubiquitination and sorting of membrane proteins into the MVB pathway. It does so in a manner that depends on features of the substrate within the membrane bilayer, and the data are most consistent with direct interaction of Bsd2 with substrate TMDs. Bsd2 possesses a PPTY motif that binds to the WW domains of the ubiquitin ligase Rsp5, and formation of a substrate–Bsd2–Rsp5 complex can readily explain how the substrate becomes ubiquitinated on cytoplasmic lysines.
We have previously reported that a distinct transmembrane ubiquitin ligase, Tul1, is required for the MVB sorting of Cps1 and Phm5. Under many growth conditions, it appears that both Tul1 and Bsd2 contribute to the effective sorting of these substrates. However, they appear to act independently and although both recognise polar TMDs they show different specificities, with Tul1 preferentially recognising the Pep3D construct and Bsd2 the Sec12 TMD. With Cps1 in particular, Bsd2 seemed to make more of a contribution than Tul1. However, the strongest and most reproducible effects were always seen with the bsd2 tul1 double mutant.
In parallel with this study, others have found that certain rsp5 mutants are defective in Cps1 sorting (Katzmann et al, 2004). In particular, the second and third WW domains of Rsp5 are required for ubiquitination of Cps1, in excellent agreement with the predictions of our work. Interestingly, removal of the putative lipid-binding C2 domain of Rsp5 also affects Cps1 modification. It is possible that recruitment of Rsp5 by Bsd2 involves not only the WW domain/PPTY interaction but also C2 domain/lipid interactions, which could help to restrict recruitment to specific membranes. Some residual ubiquitination of a Phm5 construct could be observed even in the bsd2 tul1 double mutant, suggesting that there may be additional mechanisms for recruiting Rsp5, or other ubiquitin ligases that can substitute. It may be that proteins such as Phm5 and Cps1 have evolved ubiquitination sites that are particularly good substrates, thus aiding their modification and sorting.
Our findings suggest that Bsd2 is one of the several receptors that are able to detect misfolded membrane proteins because they expose to the lipid bilayer polar residues that are normally buried. Such proteins may be ubiquitinated and transported to vacuoles (Reggiori et al, 2000; Reggiori and Pelham, 2002), or returned to the ER for refolding or degradation (Sato et al, 2003), and we have shown that with some substrates there is competition between these fates. As with MVB sorting, TMD-dependent ER retention appears to be mediated by multiple receptors, Rer1 being responsible for some but not all substrates (Rayner and Pelham, 1997; Sato et al, 1997). Bsd2 and Rer1 appear to have similar specificities and, when both are absent cells become sensitive to protein-damaging stresses, such as incorporation of the amino-acid analogue canavanine or growth at high temperature. This suggests that they represent two protective systems: misfolded proteins are retrieved to the ER, but if this fails a second defence is for them to be tagged with ubiquitin for destruction in the vacuole. Vacuolar enzymes such as Cps1 and Phm5 have evidently evolved to escape the first of these quality controls, but use the second to reach their destination.
A role for Bsd2, and by inference Rsp5, in the disposal of damaged proteins in the vacuole is consistent with other observations, including the stress sensitivities of an rsp5 allele (Hoshikawa et al, 2003) and of many vps mutants that lack a functional MVB pathway. Most strikingly, mutants lacking Bsd2 were isolated multiple times in a screen for suppression of a temperature-sensitive allele of the plasma membrane ATPase Pma1 (Luo and Chang, 1997). The mutant protein is transported to the vacuole, but in bsd2 cells it is stabilised, suggesting that Bsd2 is required to recognise the aberrant ATPase and target it for destruction.
Bsd2 also has a more subtle sorting function. It is required for movement of the manganese transporter Smf1 from a punctate distribution, probably corresponding to Golgi and early endosomes, into vacuoles (Liu and Culotta, 1999b). This protects cells against heavy-metal poisoning, and such protection requires the PPXY motif of Bsd2. Thus it is very likely that, as with other transporters, vacuolar targeting of Smf1 involves Rsp5-mediated ubiquitination. Interestingly, mutational studies indicate that degradation of Smf1 is abolished when its transport activity is blocked by a mutation within one of the TMDs, and it has been suggested that Bsd2 recognises a conformation of Smf1 induced by binding of the transported ions (Liu and Culotta, 1999a). As we have shown, Bsd2 has exactly the properties needed to recognise a conformational change within or close to the membrane, ubiquitinate Smf1, and thus target it to the vacuole. Moreover, transporters and channels are quite often regulated by their substrates and it is possible that this mechanism, which exploits the conformational changes inherent in the transport process, is used more generally.
An additional property of Bsd2 is that it mediates its own ubiquitination and thus is efficiently targeted to the MVB pathway. It is also a substrate for Tul1, perhaps because its role in recognising polar TMDs requires it to have polar TMDs itself. The resultant rapid turnover may help to maintain Bsd2 protein at a low and constant concentration, as befits a reagent that is potentially able to mark even a transiently unfolded protein for destruction. An added advantage may be that even if a Bsd2 molecule were to become tightly entangled with an unfolded protein that lacks convenient lysine residues, ubiquitination of Bsd2 itself would ensure destruction of the complex. This does not, however, appear to be the major mechanism by which Bsd2 acts, at least with the naturally occurring single TMDs that we have used—we have not so far been able to detect stable complexes with substrates by crosslinking or coimmunoprecipitation.
If Bsd2, like its substrates, is synthesised in the ER and travels to the vacuole, where does it actually act? It has been shown that Cps1 does not become ubiquitinated in a sec7 mutant, which restricts both enzyme and substrate to the ER and possibly early Golgi membranes (Katzmann et al, 2001). On the other hand, the apparent competition between Rer1 and Bsd2 for some substrates suggests that Bsd2 normally acts at a point that is still accessible to Rer1, probably no later than an early Golgi compartment. It makes sense for Bsd2 to be inactive in the ER, since proteins are still folding in this environment and indiscriminate ubiquitination of folding intermediates would be counterproductive. We therefore favour the idea that Rsp5 does not associate with Bsd2 in the ER, either because binding requires additional membrane features that the ER lacks, or because of some more elaborate control mechanism. Taken together, the data suggests that Bsd2 exerts its quality control in the Golgi, and perhaps also in endosomes. We have found no evidence that it can reach the plasma membrane.
Quality control of TMDs is likely to be a universal requirement, although it may be performed by different proteins in different organisms. Tul1 homologues are restricted to fungi, plants and protozoa. In contrast, proteins related to Bsd2 are present in many higher eukaryotes, including at least two distinct variants in humans, named N4WBP5 and N4WBP5A. Both are found in the Golgi, and contain PPXY motifs that bind to the WW domains of the Rsp5 homologue, Nedd4 (Harvey et al, 2002; Cristillo et al, 2003). Furthermore, ubiquitination of N4WBP5 has been detected. Although as yet no substrates have been described, it seems plausible that these proteins perform functions in mammalian cells similar to those of Bsd2 in yeast. There is good evidence that in mammalian cells, as in yeast, some mutant or partially assembled membrane proteins escape from the ER but are detected as abnormal and are ultimately routed to lysosomes (Trombetta and Parodi, 2003). These include proteins bearing mutations within a TMD (Zaliauskiene et al, 2000; Wilson et al, 2001; Fayadat and Kopito, 2003). It will be interesting to see whether the Bsd2 homologues play a role in such quality control, or perhaps in more specific types of membrane protein downregulation.
Materials and methods
Yeast strains and plasmids
The strains used in this study were derivatives of B4742 from the EUROSCARF consortium, containing complete deletions of bsd2 or tul1. The bsd2 tul1 double mutant was made by replacement of the complete TUL1 reading frame with Schizosaccharomyces pombe HIS5 in the bsd2 mutant. The protease deficient strain used in Figure 2 was pep4 prb1 prc1 cps1 ura3 leu2 (Heinemeyer et al, 1991). URA3 centromere plasmids (derived from pRS416) expressing GFP-tagged derivatives of Pep12 with the 3D TMD mutation, or the TMD of Sso1, Cps1 or Sec12 have been described previously (Reggiori et al, 2000), as have GFP-tagged Cps1, Phm5 and the ubiquitin-Phm5 chimera (Reggiori and Pelham, 2001). Plasmids expressing BSD2 were based on Ycplac111 (Gietz and Sugino, 1988) containing the BSD2 reading frame under the control of its own promoter. The Y140A mutation in BSD2 was introduced by PCR. SacI and BamHI sites were introduced just after the ATG for insertion of N-terminal GFP, HA and protein A tags. In some constructs, the BSD2 promoter was changed for the TPI1 promoter. The Bsd2-GFP-Cps1 chimera was generated by fusion of the N-terminus (residue 1–175) of Bsd2 up to the first TMD to the N terminus of GFP-Cps1 in pRS416. Deletion of the luminal domain of GFP-Phm5 (Phm5Δlum) was achieved by PCR, leaving three residues after the TMD. The resultant construct was expressed from the TPI promoter in a pRS416 derivative. To express the Phm5 cytosolic domain fused to the SSO TMD, a HindIII site was introduced upstream of the Phm5 TMD by PCR, allowing fusion to the Sso1 TMD present as a HindIII–BamHI fragment in plasmid GSSSO (Lewis et al, 2000).
Binding of Bsd2 to WW domains of Rsp5
For expression in E. coli, the N-terminal domain of Bsd2 (residues 1–169, with or without the Y140A mutation) was cloned into pET30a containing a protein A tag at the C terminus. The WW domains of RSP5 were cloned into pGEX6 (WW1, residues 221–273; WW2, residues 324–372; WW3 residues 382–428). E. coli-produced GST fusion proteins were bound to glutathione-Sepharose beads, washed in binding buffer (20 mM Tris–HCl, pH 8.0, 150 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mM PMSF, 0.05% Triton X-100) and incubated for 2 h with E. coli cytosol prepared from cells expressing the N terminus of Bsd2, lysed in binding buffer. Beads were subsequently spun down and washed extensively with binding buffer and once with binding buffer containing 1.5 M KCl. Proteins were eluted from beads with sample buffer and analysed by SDS polyacrylamide gel electrophoresis.
Immunoprecipitation and detection of ubiquitinated proteins
Spheroplasts from 50 OD units of cells (grown to an OD600 of 0.5–1.0) were resuspended in 1.8 ml of lysis buffer: 20 mM Tris–HCl, pH 7.4, 100 mM KCl, 1% Triton X-100, 2 mM EDTA, 10 mM N-ethyl maleimide, 1 mM PMSF and protease inhibitor cocktail (Roche; one tablet per 25 ml of buffer). Lysis was aided by passing the lysate through a fine needle 3–4 times. The lysate was centrifuged for 10 min at 13 000 g. The supernatant (1.4 ml) was transferred to a 1.5 ml Eppendorf tube and 20 μl of anti-GFP (Santa Cruz Biotech.) or anti-HA (Sigma) monoclonal antibodies coupled to agarose beads were added. After incubation for 2 h at 4°C, the beads were washed twice with 1 ml of lysis buffer and eluted with 40 μl of sample buffer without mercaptoethanol for 10 min at 50°C. The eluate was analysed by SDS–PAGE and immunoblotting. After blocking with 0.05% Tween 20 and 5% dried skimmed milk in phosphate-buffered saline (PBS), the ubiquitinated proteins were detected with antiubiquitin antibody coupled to HRP (Santa Cruz Biotech), diluted 1:500. Antibody incubation was carried out in 0.05% Tween 20 in PBS with 1% bovine serum albumin instead of milk, which allowed for a slight increase in sensitivity. One-tenth of the eluate was run separately and detected using anti-GFP antibody 1/2000 (Roche) to determine the total Phm5Δlum input.
Protease protection assay
Spheroplasted yeast cells were resuspended in lysis buffer (20 mM Tris, pH 7.4, 100 mM KCl, 2 mM EDTA, 1 mM DTT, 1 mM PMSF and protease inhibitor cocktail as for immunoprecipitation) and further lysed with a dounce homogeniser. The homogenate was centrifuged at 4°C for 5 min at 300 g. The supernatant was centrifuged at 13 500 g for 10 min. Pelleted membranes (approx. 50 μg of protein) were incubated with proteinase K (1 mg/ml final concentration), in the presence or absence of 1% Triton X-100, for 30 min at 37°C in a final volume of 50 μl. The reaction was stopped by addition of one volume of 30% (wt/vol) trichloroacetic acid and left for 10 min on ice. Precipitated proteins were collected by centrifugation and resuspended in sample buffer for SDS–PAGE.
Microscopy
Cells were grown to early log phase and imaged in water with a BioRad Radiance confocal microscope, using a single slow scan. We observed some variation between experiments in the efficiency with which some constructs entered vacuoles, as mentioned in the Results. In general, cultures that were denser or later in the logarithmic phase tended to show more of the marker proteins within vacuoles. As a result of this variation, all critical comparisons were made between cultures grown in parallel on a single day.
Acknowledgments
We are particularly grateful to Mike Lewis for data on the growth of the rer1 bsd2 double mutant, and for useful discussions. We also thank Sean Munro for originally pointing out Bsd2 to us. EHH was supported in part by a long-term fellowship from the Human Frontiers Science Program Organisation.
References
- Beck T, Schmidt A, Hall MN (1999) Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J Cell Biol 146: 1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC (2002) The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol 4: 534–539 [DOI] [PubMed] [Google Scholar]
- Black MW, Pelham HR (2000) A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J Cell Biol 151: 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristillo AD, Nie L, Macri MJ, Bierer BE (2003) Cloning and characterization of N4WBP5A, an inducible, cyclosporine-sensitive, Nedd4-binding protein in human T lymphocytes. J Biol Chem 278: 34587–34597 [DOI] [PubMed] [Google Scholar]
- Dupre S, Haguenauer-Tsapis R (2001) Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol Cell Biol 21: 4482–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Helenius A (2003) Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol 4: 181–191 [DOI] [PubMed] [Google Scholar]
- Fayadat L, Kopito RR (2003) Recognition of a single transmembrane degron by sequential quality control checkpoints. Mol Biol Cell 14: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futter CE, Pearse A, Hewlett LJ, Hopkins CR (1996) Multivesicular endosomes containing internalized EGF–EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol 132: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JM, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R (1996) Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem 271: 10946–10952 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Gitan RS, Eide DJ (2000) Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem J 346: 329–336 [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP (2000) A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci USA 97: 13871–13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Shearwin-Whyatt LM, Fotia A, Parton RG, Kumar S (2002) N4WBP5, a potential target for ubiquitination by the Nedd4 family of proteins, is a novel Golgi-associated protein. J Biol Chem 277: 9307–9317 [DOI] [PubMed] [Google Scholar]
- Hein C, Springael JY, Volland C, Haguenauer-Tsapis R, Andre B (1995) NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol 18: 77–87 [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH (1991) Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J 10: 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell SB, Losko S, Kaiser CA (2001) Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J Cell Biol 153: 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hicke L (1999) Gettin' down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol 9: 107–112 [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19: 141–172 [DOI] [PubMed] [Google Scholar]
- Hicke L, Riezman H (1996) Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84: 277–287 [DOI] [PubMed] [Google Scholar]
- Hoshikawa C, Shichiri M, Nakamori S, Takagi H (2003) A nonconserved Ala401 in the yeast Rsp5 ubiquitin ligase is involved in degradation of Gap1 permease and stress-induced abnormal proteins. Proc Natl Acad Sci USA 100: 11505–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch E, Lenk U, Sommer T (2003) Endoplasmic reticulum-associated protein degradation. Int Rev Cytol 223: 39–81 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD (2002) Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol 3: 893–905 [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD (2004) Multivesicular Body Sorting: Ubiquitin Ligase Rsp5 is Required for the Modification and Sorting of Carboxypeptidase S. Mol Biol Cell 15: 468–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Stefan CJ, Babst M, Emr SD (2003) Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol 162: 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolling R, Hollenberg CP (1994) The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J 13: 3261–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR (2000) Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell 11: 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XF, Culotta VC (1999a) Mutational analysis of Saccharomyces cerevisiae Smf1p, a member of the Nramp family of metal transporters. J Mol Biol 289: 885–891 [DOI] [PubMed] [Google Scholar]
- Liu XF, Culotta VC (1999b) Post-translation control of Nramp metal transport in yeast. Role of metal ions and the BSD2 gene. J Biol Chem 274: 4863–4868 [DOI] [PubMed] [Google Scholar]
- Liu XF, Supek F, Nelson N, Culotta VC (1997) Negative control of heavy metal uptake by the Saccharomyces cerevisiae BSD2 gene. J Biol Chem 272: 11763–11769 [DOI] [PubMed] [Google Scholar]
- Luo W, Chang A (1997) Novel genes involved in endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant. J Cell Biol 138: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD (1998) Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95: 847–858 [DOI] [PubMed] [Google Scholar]
- Rayner JC, Pelham HR (1997) Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J 16: 1832–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Black MW, Pelham HR (2000) Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol Biol Cell 11: 3737–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR (2001) Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J 20: 5176–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Pelham HR (2002) A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat Cell Biol 4: 117–123 [DOI] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A (1997) Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proc Natl Acad Sci USA 94: 9693–9698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A (2001) Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J Cell Biol 152: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A (2003) Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol Biol Cell 14: 3605–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y, Katoh Y, Shiba T, Yoshino K, Takatsu H, Kobayashi H, Shin HW, Wakatsuki S, Nakayama K (2004) GAT (GGA and Tom1) domain responsible for ubiquitin binding and ubiquitination. J Biol Chem, In press [DOI] [PubMed] [Google Scholar]
- Shih SC, Sloper-Mould KE, Hicke L (2000) Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J 19: 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soetens O, De Craene JO, Andre B (2001) Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J Biol Chem 276: 43949–43957 [DOI] [PubMed] [Google Scholar]
- Sudol M (1996) Structure and function of the WW domain. Prog Biophys Mol Biol 65: 113–132 [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Parodi AJ (2003) Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol 19: 649–676 [DOI] [PubMed] [Google Scholar]
- Urbanowski JL, Piper RC (2001) Ubiquitin sorts proteins into the intralumenal degradative compartment of the late-endosome/vacuole. Traffic 2: 622–630 [DOI] [PubMed] [Google Scholar]
- Wilson MH, Highfield HA, Limbird LE (2001) The role of a conserved inter-transmembrane domain interface in regulating alpha(2a)-adrenergic receptor conformational stability and cell-surface turnover. Mol Pharmacol 59: 929–938 [DOI] [PubMed] [Google Scholar]
- Yashiroda H, Oguchi T, Yasuda Y, Toh EA, Kikuchi Y (1996) Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae. Mol Cell Biol 16: 3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaliauskiene L, Kang S, Brouillette CG, Lebowitz J, Arani RB, Collawn JF (2000) Down-regulation of cell surface receptors is modulated by polar residues within the transmembrane domain. Mol Biol Cell 11: 2643–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]


