Abstract
A set of 37 doxycycline neoglycosides mediated via a C9 alkoxyamino-glycyl-based spacer reminiscent to that of tigecycline were prepared. The in vitro antibacterial assays against representative drug resistant Gram negative and Gram positive strains revealed a sugar-dependent activity profile and one doxycycline neoglycoside, the 2′-amino-α-D-glucoside conjugate, to rival that of the parent pharmacophore. In contrast, the representative tetracycline-susceptible strains E. coli 25922 was found to be relatively responsive to a range of doxycycline neoglycosides. This study also extends the use of aminosugars in the context of neoglycosylation via a simple two step strategy anticipated to be broadly applicable for neoglycorandomization.
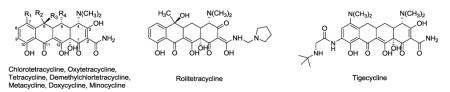
Tetracyclines are broad-spectrum antibiotics that have been in clinical use for over six decades and inhibit bacterial protein synthesis by binding bacterial 30S rRNA.1,2 Bacterial resistance to tetracyclines has spurred continual clinical development of analogues to circumvent primary resistance mechanisms,3 with tigecycline as the latest member approved for clinical use (Table 1). A semi-synthetic derivative of 9-amino minocycline, tigecycline is considered a new antibiotic class (the glycylcyclines) by virtue of its novel mode of 30S rRNA binding and extended spectrum of antibacterial activity.4-6 This unique activity derives from a key tert-butylation of the 9-glycylamino minocycline core and other short chain alkyl substitutions of the core architecture also provide antibacterial advantages.7-9 The success of glycylcyclines exemplifies the potential for antibiotic development through very subtle structural modifications of the privileged tetracycline pharmacophore.
Table 1.
Representative structures of known tetracycline analogues.
| Generic name | Chemical name | R4 | R3 | R2 | R1 | Trade Name | Yr of discovery |
Source |
|---|---|---|---|---|---|---|---|---|
| Chlorotetracycline | 7-Chlortetracycline | H | OH | CH3 | Cl | Aureomycin | 1948 | Natural occuring |
| Oxytetracycline | 5-Hydroxytetracycline | OH | OH | CH3 | H | Terramycin | 1948 | Natural occuring |
| Tetracycline | Tetracycline | H | OH | CH3 | H | Sumycin | 1953 | Natural occuring |
| Demethylchlortetracycline | 6-Demethyl-7-chlortetracycline | H | OH | H | Cl | Declomycin | 1957 | Natural occuring |
| Rolitetracycline | 2-N-Pyrrolidinomethyltetracycline | H | OH | CH3 | H | Colbiocin | 1958 | Semi-synthetic |
| Metacycline | 6-Methylene-7-chlortetracycline | OH | CH2 | H | Rondomycin | 1965 | Semi-synthetic | |
| Doxycycline | 6-Deoxy-5-hydroxytetracycline | OH | H | CH3 | H | Vibramycin | 1967 | Semi-synthetic |
| Minocycline | 7-Dimethylamino-6-demethyl-6- deoxytetracycline |
H | H | H | N(CH3)2 | Minocin | 1972 | Semi-synthetic |
| Tigecycline | 9-(t-butylglycylamido)-minocycline | H | H | H | N(CH3)2 | Tygacil | 1993 | Semi-synthetic |
In nature, such subtle modifications occur via a range of simple tailoring reactions including acylation,10 alkylation,11 and glycosylation.12 With respect to the latter, while naturally-occurring tetracycline glycosides have been reported including dactylocycline,13 TAN-1518,14 and SF2575,15 the systematic differential glycosylation of the tetracycline scaffold has not been pursued. Among emerging strategies to enable the rapid systematic differential glycosylation of complex natural products,12,16 neoglycosylation employs a mild chemo-selective reaction between free reducing sugars and alkoxyamine-bearing neoaglycons.17-20 As a result, neoglycosylation avoids the many protection/deprotections or anomeric activation manipulations typically required for glycoside synthesis by conventional methods.21 Inspired by both the impact of 9-glycylamino-tetracycline modification upon activity and the striking structural similarity between the tigecycline 9-glycylamino spacer and previously reported glycyl-based linkers for neoglycosylation,18,20b herein we report the synthesis of a set of differentially glycosylated 9-(methoxyglycyl)amino-doxycyclines as a simple tetracycline model. This study also highlights the first general application of aminosugars in the neoglycosylation reaction. Activity assessment of the doxycycline neoglycosides revealed the antibacterial potency of one specific aminosugar derived doxycycline neoglycoside to rival that of the parent pharmacophore. In addition, tetracycline sensitive E. coli was found to be relatively responsive to a range of doxycycline neoglycosides with a bias toward C-2′-substituted glucosides as most advantageous in this regard. Given the range of divergent activities reported for doxycycline analogues (including anticancer,22 anti-inflammatory,23 antiprotozoal,24 antihelminthic,25 multiple sclerosis,26 or neurodegenerative disease27), the diversification strategies and analogues highlighted herein may extend to other applications.
RESULTS AND DISCUSSION
The targeted doxycycline neoglycosides were specifically designed to incorporate a glycyl spacer reminiscent of tigecycline. The synthesis of the doxycycline neoaglycon 4 (Scheme 1, Figure 1) was initiated by regio-selective nitration of doxycycline 1.9 Subsequent reduction of 9-nitro-doxycycline afforded 9-amino-doxycycline (2, 99% in 2 steps from 1) and acylation of 2 with succinimidyl ester 318 followed by borane-trimethylamine-mediated reduction gave neoaglycon 4 (84% in 2 steps from 2) to set the stage for neoglycosylation. Cumulatively, 0.56 gm of neoaglycon 4 (4 steps, 84% overall yield) was produced to enable the synthesis of the diverse set of doxycycline neoglycosides as described below.
Scheme 1.
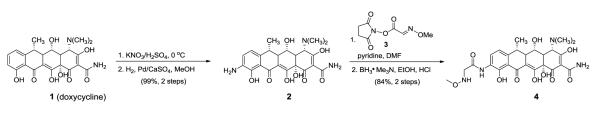
Synthesis of the 9-amino doxycycline-based neoaglycon
Figure 1.
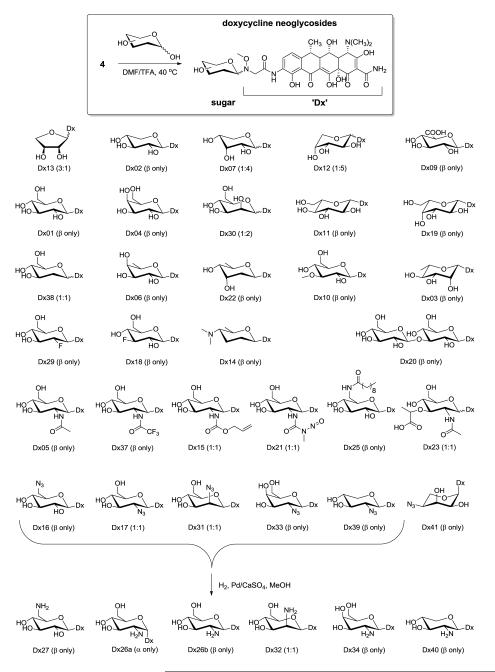
Synthesis of doxycycline neoglycosides with product anomeric ratios highlighted within parentheses (α:β). Neoglycosides are roughly organized by size (beginning with tetroses in upper left) and increasing alteration of monosaccharide (vertical). The syntheses of target aminosugar-bearing neoglycosides are also highlighted (lower). Doxycycline neoglycoside is designated as ‘Dx’.
Small scale optimization of 4 neoglycosylation with D-glucose revealed DMF/2.5% TFA, 40 °C, 12 h as the best conditions to afford desired neoglucoside while minimizing degradative side reactions (10 mg scale, 46%). Using these optimized conditions, the neoglycosylation of 4 using a diverse range of monosaccharides (5-40) was subsequently pursued (Figure 1 and Table 2). Saccharides employed in this endeavor included a representative D-tetrose (D-erythrose for Dx13), D-pentoses (D-xylose for Dx02, D-ribose for Dx07, D-arabinose for Dx12, 2-azido-D-xylose for Dx39, 4-azido-L-ribose for Dx41), D- and L-hexoses (L-rhamnose for Dx03, D-fucose for Dx06, forosamine for Dx14, digitoxose for Dx22, 2-deoxy-D-glucose for Dx38, D/L-glucose for Dx01/Dx11, D/L-galactose for Dx04/Dx19, D-mannose for Dx30, deoxy-fluoro-D-glucoses for Dx18/Dx29, azido-deoxy-D-glucoses for Dx16/Dx17, 2-azido-D-mannose for Dx31, 2-azido-D-galactose for Dx33, 3-O-methy-D-glucose for Dx10, N-acetyl-D-glucosamine for Dx05, N-trifluoroacetyl-D-glucosamine for Dx37, N-allyloxylcarbonyl-D-glucosamine for Dx15, streptozocin for Dx21, 6-N-decanoyl-D-glucosamine for Dx25, N-acetylmuramic acid for Dx23), acid-bearing sugars (D-glucuronic acid for Dx09) and a disaccharide (cellobiose for Dx20). The yield of neoglycosylation varied by sugar with an average isolated yield of 50% for most hexoses under standard conditions (DMF/2.5% TFA, 40 °C, 12 h). Using the same conditions, pentoses led to lower isolated yield (ranges from 11% to 41%) while decomposition was predominate in the tetrose-based reaction at longer reactions times. Milder conditions (3:1 DMF/AcOH, 40 °C, 6 h) enabled the desired tetrose neoglycoside in 44% isolated yield. Consistent with previous studies,17-20 the β-anomer was the predominate product with notable exceptions including D-erythroside, D-arabinoside, D-riboside, 2′-deoxy-D-glucoside, and D-mannoside (Table 2, Figure 1).
Table 2.
Summary table of neoglycosides synthesized and tested.
| Neoglycoside (see Figure 1) | Sugars utilized | Yield | α/β ratio |
|---|---|---|---|
| Dx01 | D-glucose (5)b | 46% | β only |
| Dx02 | D-xylose (6)b | 69% | β only |
| Dx03 | L-rhamnose (7)b | 66% | β only |
| Dx04 | D-galactose (8)b | 38% | β only |
| Dx05 | N-acetyl-D-glucosamine (9)b | 30% | β only |
| Dx06 | D-fucose (10)b | 25% | β only |
| Dx07 | D-ribose (11)b | 14% | α/β=1/4 |
| Dx08 | D-glucosamine hydrochloride (12)b | -a | -a |
| Dx09 | D-glucuronic acid (13)b | 45% | β only |
| Dx10 | 3-O-methyl-D-glucose (14)b | 51% | β only |
| Dx11 | L-glucose (15)b | 34% | β only |
| Dx12 | D-arabinose (16)b | 32% | α/β=1/5 |
| Dx13 | D-erythrose (17)b | 44% | α/β=3/1 |
| Dx14 | forosamine (18)c | 41% | β only |
| Dx15 | 2-deoxy-2-N-alloc-D-glucosamine (19)28 | 47% | α/β=1/1 |
| Dx16 | 6-deoxy-6-azido-D-glucose (20)b | 38% | β only |
| Dx17 | 2-deoxy-2-azido-D-glucose (21)b | 41% | α/β=1/1 |
| Dx18 | 3-deoxy-3-fluoro-D-glucose (22)b | 48% | β only |
| Dx19 | L-galactose (23)b | 48% | β only |
| Dx20 | D-cellobiose (24)b | 23% | β only |
| Dx21 | streptozocin (25)b | 11% | α/β=1/1 |
| Dx22 | digitoxose (26)b | 28% | β only |
| Dx23 | N-acetylmuramic acid (27)b | 32% | α/β=1/1 |
| Dx24 | 3-deoxy-3-N-decanoyl-D-glucosamine (28)20a | -a | -a |
| Dx25 | 6-deoxy-6-N-decanoyl-D-glucosamine (29)20a | 18% | β only |
| Dx26a | 2-deoxy-2-azido-D-glucose (21)b | 20% 2 steps | α only |
| Dx26b | 2-deoxy-2-azido-D-glucose (21)b | 22% 2 steps | β only |
| Dx27 | 6-deoxy-6-azido-D-glucose (20)b | 25% 2 steps | β only |
| Dx28 | 2,3,4,6-tetraacetyl-D-glucose (30)c | -a | -a |
| Dx29 | 2-deoxy-2-fluoro-D-glucose (31)b | 21% | β only |
| Dx30 | D-mannose (32)b | 27% | α/β=1/2 |
| Dx31 | 2-deoxy-2-azido-D-mannose (33) 29 | 32% | α/β=1/1 |
| Dx32 | 2-deoxy-2-azido-D-mannose (33) 29 | 35% 2 steps | α/β=1/1 |
| Dx33 | 2-deoxy-2-azido-D-galactose (34) 29 | 31% | β only |
| Dx34 | 2-deoxy-2-azido-D-galactose (34) 29 | 31% 2 steps | β only |
| Dx35 | 2-deoxy-2-N-methyl-D-glucosamine (35)30 | -a | -a |
| Dx36 | 2-deoxy-2-N-dimethyl-D-glucosamine (36)30 | -a | -a |
| Dx37 | 2-deoxy-2-N-trifluoroacetyl-D-glucosamine (37)30 | 22% | β only |
| Dx38 | 2-deoxy-D-glucose (38)b | 37% | α/β=1/1 |
| Dx39 | 2-deoxy-2-azido-D-xylose (39)c | 37% | β only |
| Dx40 | 2-deoxy-2-azido-D-xylose (39)c | 30% 2 steps | β only |
| Dx41 | 4-deoxy-4-azido-L-ribose (40)c | 33% | β only |
no reaction
commercially available
synthesized in this work
The incompatibility of unprotected aminosugars with neoglycosylation has dramatically restricted their prior use in neoglycorandomization.18-20 Specifically, while amine-bearing neoaglycons can be compensated for via additional monosaccharide, an excess of aminosugar is believed to compete for the initial oxime-forming stage of the chemoselective neoglycosylation reaction. To address this limitation, the current study employed a small set of free azidosugars described in the previous paragraph (Figure 1, Dx16, Dx17, Dx31, Dx33, Dx39, Dx41) to enable the synthesis of the corresponding neoglycosides as requisite precursors of aminosugar-bearing analogues. For this work, 2-azido-D-glucose (21), 2-azido-D-mannose (33), and 2-azido-D-galactose (34) were synthesized from the corresponding D-glucosamine hydrochloride, D-mannosamine hydrochloride, and D-galactosamine hydrochloride, respectively via amino-azide interconversion using perfluorobutylsulfonyl azide as the diazo transfer agent.29 In addition, two azido pentoses (39 and 40) were synthesized via selective protection of D-lyxose (See Figure S3, supporting information) and subsequent azide installation as an alternative to previously reported strategies.31 Neoglycosylation using this azidosugar set was accomplished with an overall average isolated yield of 36% under standard conditions. In all cases, post-neoglycosylation reduction (Pd/CaSO4, H2) furnished the desired aminosugar products (Figure 1, Dx26, Dx27, Dx32, Dx34, Dx40) with an overall average yield of 26% (two steps from neoaglycon 4). Intriguingly, while the doxycycline 2′-azido-D-neoglucoside anomers (Dx17, 1:1 α/β) could not be resolved chromatographically, the corresponding α (Dx26a) and β (Dx26b) anomers of the 2′-amino-D-neoglucoside product were obtained with an isolated yield of 20% and 22%, respectively, after preparative HPLC.
The set of 37 doxycycline neoglycosides were tested for antibacterial activity using a panel of four bacterial strains comprised of a tetracycline susceptible Gram negative strain (E. coli 25922) and two drug-resistant clinical isolates (the Gram positive S. aureus R2507 and Gram negative E. coli 1-849) (Table 3 and Table S2, supporting information). While the mechanisms of drug resistance in these latter two strains have not been determined, both are known to display ~10-fold tetracycline/doxycycline resistance compared to their wild-type counterparts. This cumulative assessment enabled the following observations. First, consistent with previous studies,9 9-amino-doxycycline (2) and doxycycline (1) were nearly equipotent with 2 MIC values of 1 μg/ml, 2 μg/ml, 4 μg/ml versus 1 values of 1 μg/ml, 8 μg/ml, 2 μg/ml against E. coli 25922, E. coli 1-849, and S. aureus R2507, respectively (Table S2, supporting information). In comparison, neoaglycon 4 displayed a slight reduction in potency depending upon strain tested (ranging from 2-4 fold). While a general trend of further reduced potency upon neoglycosylation of 4 was observed, the 2′-amino-α-D-neoglucoside (Dx26a, MIC 4 μg/ml) afforded a slight improvement over 1 or neoaglycon 4 against the tetracycline resistant strain E. coli. 1-849 (Table S2, supporting information). In addition, the response of the tetracycline sensitive E. coli 25922 was more tolerant to sugar conjugate variation, with four additional neoglycosides (Dx13, Dx14, Dx32, Dx37) displaying similar activities to the neoaglycon 4 (MIC 4 μg/ml). Finally, while neoglycosylation generally reduced potency in the context of the tetracycline resistant strain S. aureus R2507, the conjugation to 2′-deoxy-2′-substituted glucoside analogues were found to be the most active conjugates. The overall trend of reduced activity observed upon neoglycosylation may derive from disfavored interactions with key contributors to binding short chain alkylated glycylcylines – specifically, helices H34/H18 and/or C1054 of 30S rRNA5,6 – where contacts afforded by the aminosugar substitution in Dx26a may partially compensate for unfavored interactions.
Table 3.
Antibacterial activity and cytotoxicity of selected doxycycline neoglycosides
| Neoglycoside (see Figure 1) |
Sugar utilized |
E. coli 25922 (μg/mL) |
E. coli 1-849 (μg/mL) |
S. aureus R2507 (μg/mL) |
A549 viability (%) |
IMR 90 viability (%) |
|---|---|---|---|---|---|---|
| 4 | none | 4 | 8 | 4 | 95.9 | 99.2 |
| Dx26a | 2-deoxy-2-azido-D-glucose (21) | 4 | 4 | 4 | 99.1 | 94.6 |
| Dx15 | 2-deoxy-2-N-alloc-D-glucosamine (19) | 8 | 16 | 8 | 98.8 | 97.6 |
| Dx14 | forosamine (18) | 4 | 32 | 16 | 96.9 | 100.7 |
| Dx37 | 2-deoxy-2-N-trifluoroacetyl-D-glucosamine (37) | 4 | 32 | 16 | 98.7 | 109.1 |
| Dx23 | N-acetylmuramic acid (27) | 8 | 32 | 16 | 97.7 | 104.4 |
| Dx32 | 2-deoxy-2-azido-D-mannose (33) | 4 | 32 | 32 | 97.1 | 98.9 |
| Dx26b | 2-deoxy-2-azido-D-glucose (21) | 8 | 32 | 32 | 95.6 | 101.7 |
| Dx12 | D-arabinose (16) | 8 | 64 | 32 | 97.2 | 101.7 |
| Dx13 | D-erythrose (17) | 4 | 64 | 32 | 98.2 | 99.3 |
The cytotoxicity of doxycycline neoaglycon 4 and the nine most potent antibacterial neoglycosides was subsequently assessed using both the non-small cell cancer cell line A549 and a comparator normal lung fibroblast cell line IMR 90 (Table 3). This analysis revealed no statistically significant cytotoxicity at 10 μM - a dose well above the typical serum concentration used to treat bacterial infections with existing clinical tetracyclines (0.2 to 5 μg/ml).32 This preliminary analysis suggests the general toxicity of the parent 1 and corresponding neoglycosides as potentially similar.
In summary, this study highlights the first systematic differential glycosylation of the privileged tetracycline scaffold. While neoglycosylation at C9 was predominately detrimental, one analogue, the 2′-amino-α-D-glucoside conjugate Dx26a, afforded a slight antibacterial benefit over the parental doxycycline against the tetracycline resistant strain E. coli 1-849 and corresponding low general cytotoxicity. While the influence of sugar conjugation upon in vivo drug properties (ADMET) remains to be determined, this study highlights the amenability of this complex scaffold to neoglycosylation and opens the door to similar modifications at other key positions of the tetracycline architecture, particularly those known to be influenced via glycosylation. In addition, this study extends the use of aminosugars in the context of neoglycosylation via a simple two step strategy anticipated to be broadly applicable for neoglycorandomization.
EXPERIMENTAL SECTION
General Experimental Procedures
Reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and were used without purification unless otherwise noted. Dichloromethane was freshly distilled from calcium hydride under nitrogen atmosphere. Pyridine and triethylamine were distilled and stored over 4 Å molecular sieves. Analytical TLC was performed using Sorbent Technologies silica gel glass TLC plates (EMD chemical Inc, PA, USA). Visualization was accomplished with UV light (254 nm) followed by staining with diluted sulfuric acid (5% in ethanol) solution and heating. Mass spectrometric data were obtained on a Waters (Milford, MA) LCT time-of-flight spectrometer for electrospray ionization (ESI). NMR spectra were obtained on either a Varian Unity Inova 400 or 500 MHz instrument (Palo Alto, CA) using 99.8% CDCl3 with 0.05% v/v TMS or 99.8% CD3OD from Cambridge Isotopes (Cambridge Isotope Laboratories , MA, USA). 1H and 13C chemical shifts were referenced to TMS for both CDCl3 and CD3OD. Multiplicities are indicated by s (singlet), d (doublet), t (triplet), q (quartet), and m (multiplet). Chemical shifts are reported in parts per million (ppm) and coupling constants J are given in Hz.
Chromatography Methods
Method A: Normal phase flash chromatography was performed using 40-63 m particle-sized silica gel using ethyl acetate and hexane or methanol and methylene chloride as the mobile phase. Method B: Pilot purification was conducted by flash column chromatography using an Alltech C18 Extrac-Clean column (10000 mg, 75 mL, Alltech Associates, Deerfield, IL, USA) with gradient elution water/acetonitrile 100/0 to 50/50 containing 0.1% TFA. Method C: Semi-preparative reverse-phase HPLC was conducted using a Gemini C18 (5 μm, 250 × 10 mm, Phenomenex, Torrance, California, USA) using a gradient of 5% B to 55% B over 27 min, 55% B to 100% B over 1 min, 100% B for 5 min, 100% B to 5% B over 1 min, 5% B for 4 min (A = dH2O with 0.1% TFA; B = acetonitrile; flow rate = 5 mL min−1; A254 nm). The desired fractions were collected and concentrated under reduced pressure, frozen at −80 °C, and lyophilized. Method D: Analytical reverse-phase HPLC was conducted with a Luna C-18 (4.6 mm × 250 mm, Phenomenex, Torrance, California, USA) with a gradient of 5% B to 55% B over 20 min, 55% B to 100% B over 1 min, 100% B for 5 min, 100% B to 5% B over 1 min, 5% B for 3 min (A = dH2O with 0.1% TFA; B = acetonitrile; flow rate = 1 mL min−1; A254 nm). HPLC peak areas were integrated with Star Chromatography Workstation software (from Varian, Palo Alto, CA, USA) and the percent conversion calculated as a percent of the total peak area.
Cytotoxicity assays
A resazurin-based cytotoxicity assay, also known as AlamarBlue assay, was used to assess the cytotoxicity of agents against the human lung non-small cell carcinoma cell line A549 cell line and normal fibroblast IMR 90 cell lines where degree of cytotoxicity was based upon residual metabolic activity as assessed via reduction of resazurin (7-hydroxy-10-oxido-phenoxazin-10-ium-3-one) to its fluorescent product resorufin. A549 and IMR 90 cells, purchased from ATCC (Manassas, VA, USA), were grown in DMEM/F-12 Kaighn’s modification and MEM/EBSS media, respectively (Thermo scientific HyClone, Logan, UT, USA), with 10% heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine. Cells were seeded at density 2 × 103 cells per well onto 96-well culture plates with a clear bottom (Corning, NY, USA), incubated 24 hours at 37 °C in a humidified atmosphere containing 5% CO2 and were exposed to standard toxin (positive controls - 1.5 mM hydrogen peroxide, 10 μg/ml actinomycine D) and test compounds (selected doxycycline neoglycosides at final concentration of 10 μM) for 2 days. Resazurin (150 μM final concentration) was subsequently added to each well, plates were shaken briefly for 10 seconds and were incubated for another 3 h (A549 cells) and 5 h (IMR 90 cells) at 37 °C to allow viable cells to convert resazurin into resorufin. The fluorescence intensity for resorufin was detected on a scanning microplate spectrofluorometer FLUOstar Omega (BMG LABTECH GmbH, Ortenberg, Germany) using an excitation wavelength of 560 nm and an emission wavelength of 590 nm. The assay was repeated in 3 independent experimental replications. In each replication, the resorufin values of treated cells were normalized to, and expressed as percent of, the mean resorufin values of untreated, metabolically active cells (100%, all cells are viable).
Antibacterial assays
Antibacterial assays using the two community acquired clinically-resistant isolates S. aureus R2507 and E. coli 1-849 (JMI Laboratories, North Liberty Iowa), and the standard E. coli 25922 model (ATCC, Manassas, VA), were conducted as previously described.20g
Stability of Doxycycline Neoglycoside Stock Solutions
A 3 mM DMSO solution of Dx26a was stored at −20 °C for 12 months. Subsequent purity assessment by HPLC revealed no change under these storage conditions over time.
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (2)
To a solution of doxycyline hyclate (2.00 g, 3.9 mmol) in concentrated H2SO4 solution was added KNO3 (0.46 g, 4.6 mmol) and the reaction was stirred under argon and monitered by HPLC (method D) for consumption of starting material. After 3 h, the reaction solution was added to ether at 0 °C in a dropwise fashion to afford a heavy yellow precipitate that was subseqeuntly collected by vacumm filtration and washed with cold ether (20 mL, x3). The yellow precipitate was dissolved in 40 mL degassed MeOH and to this 20 mg Pd/CaSO4 was added. The flask was sealed, degassed and exposed to a H2 balloon with stirring for 12 hours. The reaction was subseqeuntly filtered through celite, the collected residue washed with MeOH (10 mL, x3) and the combined filtrate concentrated to 10 mL. The concentrated MeOH solution was subsequently added to the solution of cold EtOAc/hexane (1/1) in a dropwise fashion to afford a heavy precipitate that was subsequently collected via filtration, washed with cold hexane and dried under vacuum. Compound 2 (2.16 g, 3.9 mmol) was obtained in a yield of 99% after two steps as a yellow powder: 1H NMR (CD3OD, 500 MHz) δ 8.50 (s, 1 H), 4.15 (s, 1 H), 3.3 (m, 1 H), 3.23 (m, 6 H), 3.0 - 3.1 (m, 3 H), 3.0 (m, 3H), 2.59 (dd, J = 15.7, 13.5 Hz, 2 H), 2.4 (m, 2 H), 1.7 (m, 1 H); 13C NMR (CD3OD, 100 MHz) δ 192.8, 172.4, 159.7, 154.6, 151.3, 142.7, 137.5, 130.6, 124.8, 116.6, 107.9, 95.8, 94.6, 73.8, 53.6 (2 carbons), 45.9, 41.6, 38.9, 31.2, 16.4, 15.8; HRESIMS m/z 460.17299 [M + H]+ (Calcd for C22H26N3O8 460.1714).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxyglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (4)
To a solution of 2 (0.56 g, 1.2 mmol) in 4 mL DMF was added 0.5 mL pyridine and succinimidyl ester 3 (0.48 g, 2.4 mmol)18 and the reaction was stirred at room temperature for 24 h. The reaction was quenched by removing the solvent in vacuo and then the residue was dissolved with 3 mL MeOH to which was added 40 mL EtOAc followed by 20 mL hexane to afford a heavy yellow precipitate. The precipitate was filtered, washed with cold hexane (10 mL, x3) and dried under vacuum. The yellow solid was dissolved with water and the pH of the mixture was adjusted to 7 by adding 5% NH4OH in a dropwise fashion. The solvent was removed under vacuum and the residue lyophilized overnight to give a yellow solid. The corresponding solid was dissolved in 20 mL EtOH to which was added BH3•Me3N (0.88 g, 12 mmol) and 2 mL 50% HCl in ethanol. The reaction was stirred under room temperature for 6 h (until the reaction was complete based upon HPLC - method D), solvent removed under vacuum, and the residue dissolved in 3 mL MeOH. Addition of 40 mL EtOAc followed by 10 mL hexane afforded a heavy precipitate which was collected via filtration and purified by flash reverse-phase (method B). Purified doxycycline neoaglycon 4 (0.56 g, 1.0 mmol) was obtained with a yield of 84% after two steps from 2 as a yellow powder: 1H NMR (CD3OD, 500 MHz) δ 8.49 (d, J = 8.5 Hz, 1 H), 7.01 (d, J = 8.8 Hz, 1 H), 4.43 (s, 1 H), 3.67 (s, 5 H), 3.6 (m, 1 H), 2.98 (s, 6 H), 2.9 (m, 1 H), 2.8 (m, 1 H), 2.6 (m, 1 H), 1.59 (d, J = 8.1 Hz, 3 H); 13C NMR (CD3OD, 125 MHz) δ 172.9, 168.3, 160.4, 160.1, 151.6, 142.8, 126.7, 125.2, 117.4, 115.5, 115.2, 107.4, 95.0, 73.5, 68.8, 65.9, 63.9, 61.2 (2 carbons), 54.1, 46.7, 44.2, 41.8, 38.6, 15.0; HRESIMS m/z 547.20165 [M + H]+ (Calcd for C25H31N4O10 547.2035).
General procedure of neoglycosylation
Neoaglycon 4 (10–40 mg) and reducing sugar (1.2 - 2.0 eq) were dissolved in 1 mL anhydrous DMF and to this was added 25 μL TFA. The reaction was stirred at 40 °C for 24 h and monitored by HPLC (method D). Upon completion, solvent was removed under vacuum and the residue was dissolved in 300 μL MeOH containing 0.01% NH4OH. The solution was centrifuged and the collected supernatant was purified via semi-preparative HPLC (method C). The final purified doxycycline neoglycosides were obtained in a yield ranging from 20%-69% from doxycycline neoaglycon 4.
General hydrogenation procedure for azidosugar appended doxycycline neoglycosides
Doxycycline neoglycoside (Dx16, 17, 31, 33, 39) was dissolved into 10 mL degassed MeOH to which was added 1 mg Pd/CaSO4. The vial was sealed, degassed and exposed to a H2 balloon for 12 h with stirring. The reaction was subsequently filtered over celite, washed with 10 mL MeOH and the filtrate dried under vacuum. The residue was dissolved in 300 μL MeOH containing 0.01% NH4OH, centrifuged and the collected supernatant was purified via semi-preparative HPLC (method C). The final purified aminosugar-appended doxycycline neoglycosides were obtained with an isolated yield ranging from 20%-31% after two steps from the doxycycline neoaglycon 4.
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx01)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.31 (d, J = 8.3 Hz, 1 H), 6.99 (d, J = 8.1 Hz, 1 H), 4.26 (d, J = 8.7 Hz, 1 H), 3.6 – 3.9 (m, 10 H), 3.74 (s, 3 H), 2.89 (s, 6 H), 2.80 (dd, J = 12.1, 7.4 Hz, 1 H), 2.7 (m, 1H), 2.58 (dd, J = 12.4, 8.3 Hz, 1 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 709.25484 [M + H]+ (Calcd for C31H41N4O15 709.2563).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-D-xylosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx02)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.31 (d, J = 8.5 Hz, 1 H), 6.98 (d, J = 8.3 Hz, 1 H), 4.41 (s, 2 H), 4.19 (d, J = 8.8 Hz, 1 H), 3.92 (dd, J = 11.2, 5.4 Hz, 1 H), 3.3 – 3.8 (m, 3 H), 3.73 (s, 3 H), 3.1 - 3.2 (m, 3 H), 3.01 (s, 6 H), 2.94 (dd, J = 12.0, 9.8 Hz, 1 H), 2.8 (m, 1 H), 2.59 (dd, J = 12.2, 8.3 Hz, 1 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 679.24703 [M + H]+ (Calcd for C30H39N4O14 679.2457) .
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-L-rhamnosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx03)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.41 (d, J = 8.3 Hz, 1 H), 6.97 (d, J = 9.0 Hz, 1 H), 4.44 (d, J = 1.0 Hz, 1 H), 4.3 (m, 1H), 4.20 (t, J = 3.1 Hz, 1 H), 4.06 (d, J = 2.7 Hz, 1 H), 3.9 – 4.0 (m, 1 H), 3.73 (s, 3 H), 3.72 (s, 2 H), 3.5 (m, 1 H), 3.5 (m, 1 H), 2.8 (m, 7 H), 2.5 (m, 2 H), 1.55 (d, J = 6.8 Hz, 3 H), 1.32 (d, J = 5.9 Hz, 3 H); HRESI m/z 693.26213 [M + H]+ (Calcd for C31H41N4O14 693.2614).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-D-galactosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx04)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.32 (d, J = 8.4 Hz, 1 H), 6.99 (d, J = 8.5 Hz, 1 H), 4.36 (s, 1 H), 4.26 (d, J = 9.0 Hz, 1 H), 3.8 - 4.0 (m, 2 H), 3.6 (s, 3 H), 3.4 - 3.8 (m, 7 H), 3.93 (s, 6 H), 2.8 (m, 2 H), 2.58 (dd, J = 12.0, 8.2 Hz, 1 H), 1.56 (d, J = 6.7 Hz, 3 H); HRESI m/z 709.25597 [M + H]+ (Calcd for C31H41N4O15 709.2563).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-N’-acetylamino-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx05)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.49 (d, J = 8.3 Hz, 1 H), 6.98 (d, J = 8.3 Hz, 1 H), 4.42 (d, J = 10.0 Hz, 1 H), 4.32 (s, 1 H), 3.9 (m, 2 H), 3.7 (m, 3 H), 3.63 (s, 3 H), 3.6 (m, 6 H), 3.50 (t, J = 8.8 Hz, 1 H), 2.92 (s, 6 H), 2.7 - 2.8 (m, 2 H), 2.58 (dd, J = 12.1, 8.2 Hz, 1 H), 2.08 (s, 3 H), 1.55 (d, J = 6.8 Hz, 3 H). HRESI m/z 750.28313 [M + H]+ (Calcd for C33H44N5O15 750.2828).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-D-fucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx06)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.30 (d, J = 8.4 Hz, 1 H), 6.96 (d, J = 8.4 Hz, 1 H), 4.22 (d, J = 8.6 Hz, 1 H), 3.8 - 3.9 (m, 3 H), 3.72 (s, 3 H), 3.5 - 3.7 (m, 5 H), 2.80 (br. s, 6 H), 2.6 (m, 3 H), 1.55 (d, J = 6.6 Hz, 3 H), 1.28 (d, J = 6.4 Hz, 3 H); HRESI m/z 693.26212 [M + H]+ (Calcd for C31H41N4O14 693.2641).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-D-ribosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx07)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.35 (d, J = 8.3 Hz, 1 H), 6.99 (d, J = 8.1 Hz, 1 H), 4.59 (m, 1 H), 4.54 (d, J = 8.8 Hz, 1 H), 4.19 (m, 1 H), 4.15 (m, 2 H), 4.08 (t, J = 5.5 Hz, 1 H), 3.8 - 3.9 (m, 1 H), 3.75 (s, 3 H), 3.6 - 3.7 (m, 2 H), 3.59 (dd, J = 9.2, 2.8 Hz, 1 H), 3.47 (s, 1 H), 2.8 (s, 6 H), 2.6 (m, 3 H), 1.58 (d, J = 7.1 Hz, 3 H); HRESI m/z 679.24632 [M + H]+ (Calcd for C30H39N4O14 679.2457).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-D-glucuronosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx09)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.24 (d, J = 8.4 Hz, 1 H), 6.97 (d, J = 8.3 Hz, 1 H), 4.7 (m, 1 H), 4.56 (d, J = 6.1 Hz, 1 H), 4.51 (m, 1 H), 4.1 (m, 1 H), 4.0 (m, 1 H), 3.68 (s, 3 H), 3.6 – 3.7 (m, 3 H), 3.5 (m, 1 H), 3.4 (m, 1 H), 3.2 (m, 1 H), 2.7 (s, 6 H), 2.6 (m, 2 H), 2.58 (dd, J = 11.6, 7.4 Hz, 1 H), 1.56 (d, J = 7.3 Hz, 3 H). HRESI m/z 737.24934 [M + H]+ (Calcd for C32H41N4O16 737.2512).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-3′-O-methyl-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx10)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.31 (d, J = 8.5 Hz, 1 H), 6.98 (d, J = 8.3 Hz, 1 H), 4.27 (d, J = 9.3 Hz, 1 H), 3.8 (m, 2 H), 3.78 (s, 2 H), 3.73 (s, 3 H), 3.67 (s, 3 H), 3.6 - 3.7 (m, 2 H), 3.5 (m, 1 H), 3.4 (m, 1 H), 3.3 (m, 1 H), 3.1 (m, 1 H), 2.84 (s, 6 H), 2.8 (m, 1 H), 2.7 (m, 1 H), 2.58 (dd, J = 12.1, 7.9 Hz, 1 H), 1.56 (d, J = 7.1 Hz, 3 H); HRESI m/z 723.27208 [M + H]+ (Calcd for C32H43N4O15 723.2719).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-L-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx11)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.31 (d, J = 8.6 Hz, 1 H), 6.98 (d, J = 8.6 Hz, 1 H), 4.26 (d, J = 8.2 Hz, 1 H), 4.2 (m, 1H), 3.8 (m, 1 H), 3.86 (s, 2 H), 3.8 (m, 1 H), 3.75 (s, 3 H), 3.72 (m, 1 H), 3.67 (m, 1 H), 3.60 (t, J = 8.2 Hz, 1 H), 3.3 - 3.5 (m, 2 H), 2.87 (s, 6 H), 2.79 (dd, J = 12.8, 6.7 Hz, 1 H), 2.6 (m, 1 H), 2.57 (dd, J = 12.4, 8.1 Hz, 1 H), 1.56 (d, J = 6.6 Hz, 3 H); HRESI m/z 709.25706 [M + H]+ (Calcd for C31H41N4O15 709.2563).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-D-arabinosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx12)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.32 (d, J = 8.3 Hz, 1 H), 6.99 (d, J = 8.1 Hz, 1 H), 4.3 (m, 1 H), 4.18 (d, J = 9.0 Hz, 1 H), 3.7 - 4.0 (m, 3 H), 3.72 (s, 6 H), 3.5 – 3.6 (m, 2 H), 2.93 (s, 6 H), 2.8 (m, 2 H), 2.58 (dd, J = 12.2, 8.1 Hz, 1 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 679.24591 [M + H]+ (Calcd for C30H39N4O14 679.2457).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-D-erythrosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx13)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.38 (d, J = 8.5 Hz, 1 H), 6.98 (d, J = 8.1 Hz, 1 H), 4.67 (m, 1 H), 4.57 (s, 1 H), 4.22 (d, J = 4.5 Hz, 1 H), 4.0 (m, 1 H), 3.73 (s, 3 H), 3.6 - 3.7 (m, 2 H), 3.44 (m, 2 H), 3.2 (m, 1 H), 2.77 (s, 6 H), 2.6 (m, 1 H), 2.5 (m, 2 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 649.23770 [M + H]+ (Calcd for C29H37N4O13 649.2352).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′,3′,4′,6′-tetradeoxy-4-N’-dimethylamino-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx14)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.41 (d, J = 8.5 Hz, 1 H), 6.99 (d, J = 8.3 Hz, 1 H), 4.50 (d, J = 9.9 Hz, 1 H), 4.40 (s, 2 H), 3.8 - 3.9 (m, 2 H), 3.70 (s, 3 H), 3.6 - 3.7 (m, 1 H), 3.63 (s, 1 H), 3.57 (dd, J = 11.4, 8.7 Hz, 1 H), 2.94 (s, 6 H), 2.89 (s, 6 H), 2.7 – 2.8 (m, 2 H), 2.59 (dd, J = 12.3, 8.4 Hz, 1 H), 2.3 (m, 1 H), 2.1 (m, 1 H), 1.9 (m, 2 H), 1.56 (d, J = 6.8 Hz, 3 H), 1.38 (d, J = 5.9 Hz, 3 H); HRESI m/z 688.31860 [M + H]+ (Calcd for C33H46N5O11 688.3188).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-N’-allyloxycarbonylamino-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx15)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.48 (d, J = 8.3 Hz, 0.5 H), 8.47 (d, J = 8.3, 0.5 H), 7.02 (d, J = 7.8 Hz, 0.5 H), 7.00 (d, J = 6.4 Hz, 0.5H), 6.93 (m, 1 H), 5.33 (dd, J = 11.0, 0.9 Hz, 1 H), 5.14 (d, J = 10.6 Hz, 1 H), 4.7 (m, 1 H), 4.69 (d, J = 4.5 Hz, 0.5 H), 5.0 (m, 1 H), 4.46 (d, J = 9.4 Hz, 0.5 H), 4.2 (m, 1 H), 3.9 (m, 1 H), 3.7 – 3.8 (m, 2 H), 3.70 (s, 2 H), 3.67 (s, 3 H), 3.5 (m, 2 H), 2.89 (s, 6 H), 2.7 (m, 2 H), 2.60 (d, J = 12.1, 7.9 Hz, 1 H), 1.59 (d, J = 6.6 Hz, 3 H). HRESI m/z 792.29846 [M + H]+ (Calcd for C35H46N5O16 792.2934).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-6′-deoxy-6′-azido-β-D glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx16)
yellow powder; 1H NMR (CD3OD, 400 MHz) δ 8.36 (d, J = 8.5 Hz, 1 H), 7.01 (d, J = 8.3 Hz, 1 H), 4.45 (s, 2 H), 4.36 (d, J = 8.5 Hz, 1 H), 3.90 (s, 1 H), 3.8 (m, 1 H), 3.77 (s, 3 H), 3.7 - 3.8 (m, 2 H), 3.7 (m, 1 H), 3.6 (m, 2 H), 3.4 - 3.5 (m, 2 H), 2.98 (s, 6 H), 2.87 (t, J = 9.3 Hz, 1 H), 2.8 (m, 1 H), 2.61 (dd, J = 12.2, 8.3 Hz, 1 H), 1.59 (d, J = 6.8 Hz, 3 H); HRESI m/z 734.26611 [M + H]+ (Calcd for C31H40N7O14 734.2628).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-azido-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx17)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.46 (d, J = 8.5 Hz, 0.5 H), 8.45 (d, J = 8.5 Hz, 0.5 H), 6.98 (d, J = 8.2 Hz, 0.5 H), 6.97 (d, J = 8.0 Hz, 0.5 H), 4.80 (s, 0.5 H), 4.38 (s, 1 H), 4.27 (d, J = 9.5 Hz, 0.5 H), 3.8 - 4.0 (m, 2 H), 3.76 (s, 2 H), 3.7 (m, 1H), 3.64 (s, 3 H), 3.57 (dd, J = 11.5, 8.3 Hz, 1 H), 3.49 (t, J = 9.4 Hz, 1 H), 3.4 (m, 1 H), 3.2 (m, 1 H), 2.94 (s, 6 H), 2.8 (m, 2 H), 2.58 (dd, J = 12.2, 8.3 Hz, 1 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 734.26129 [M + H]+ (Calcd for C31H40N7O14 734.2628).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-3′-deoxy-3′-fluoro-β-D glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx18)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ ppm 8.32 (d, J = 8.3 Hz, 1 H), 6.99 (d, J = 7.8 Hz, 1 H), 4.37 (t, J = 8.7 Hz, 1 H), 4.29 (d, J = 9.3 Hz, 1 H), 3.9 (m, 2 H), 3.61 (s, 2 H), 3.77 (s, 1 H), 3.74 (s, 3 H), 3.5 - 3.7 (m, 3 H), 3.5 (m, 1 H), 3.44 (dt, J = 3.4, 1.6 Hz, 1 H), 3.2 (m, 1 H), 2.90 (s, 6 H), 2.8 (m, 2 H), 2.59 (dd, J = 12.5, 8.3 Hz, 1 H), 1.56 (d, J = 6.9 Hz, 3 H); HRESI m/z 711.25146 [M + H]+ (Calcd for C31H40FN4O14 711.2520).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-L-galactosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx19)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.33 (d, J = 8.3 Hz, 1 H), 6.98 (d, J = 8.3 Hz, 1 H), 4.39 (s, 1 H), 4.26 (d, J = 9.3 Hz, 1 H), 3.9 (m, 1 H), 3.87 (d, J = 3.2 Hz, 1 H), 3.8 (m, 1 H), 3.76 (s, 2 H), 3.73 (s, 3 H), 3.7 (m, 1 H), 3.6 (m, 1 H), 3.60 (dd, J = 9.4, 4.5 Hz, 1 H), 3.6 (m, 2 H), 2.95 (s, 6 H), 2. 8 (m, 2 H), 2.58 (dd, J = 12.0, 8.3 Hz, 1 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 709.25671 [M + H]+ (Calcd for C31H41N4O15 709.2563).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-D-cellobiosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx20)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.32 (d, J = 8.3 Hz, 1 H), 6.99 (d, J = 8.3 Hz, 1 H),4.42 (d, J = 8.0 Hz, 1 H), 4.41 (s, 1 H), 4.30 (d, J = 9.2 Hz, 1 H), 3.9 (m, 2 H), 3.89 (s, 2 H), 3.8 (m, 1 H), 3.75 (s, 3 H), 3.66 (dd, J = 12.0, 5.9 Hz, 1 H), 3.6 (m, 3 H), 3.4 (m, 3 H), 3.3 (m, 1 H), 3.22 (t, J = 8.6 Hz, 1 H), 3.2 (m, 1 H), 2.95 (s, 6 H), 2.8 (m, 1 H), 2.7 (m, 1H), 2.59 (dd, J = 12.2, 8.3 Hz, 1 H), 1.55 (d, J = 6.8 Hz, 3 H); HRESI m/z 871.31004 [M + H]+ (Calcd for C37H51N4O20 871.3091).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-({[methyl(nitroso)amino]carbonyl}amino-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx21)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.51 (d, J = 8.4 Hz, 0.5 H), 8.50 (d, J = 8.5 Hz, 0.5 H), 6.98 (d, J = 8.8 Hz, 1 H), 4.79 (s, 0.5 H), 4.65 (d, J = 10.0 Hz, 0.5 H), 4.4 (s, 2 H), 4.07 (dd, J = 11.3, 9.1 Hz, 0.5 H), 3.9 – 4.0 (m, 2 H), 3.75 (t, J = 5.7 Hz, 0.5 H), 3.70 (s, 1 H), 3.7 (m, 2 H), 3.63 (s, 3 H), 3.5 – 3.6 (m, 2 H), 3.4 (m, 1 H), 3.39 (d, J = 8.5 Hz, 1 H), 3.17 (s, 3 H), 3.06 (s, 3 H), 3.03 (s, 3 H), 2.8 (m, 2 H), 2.6 (m, 1 H), 1.57 (d, J = 6.8 Hz, 1.5 H), 1.54 (d, J = 6.6, 1.5 H); HRESI m/z 794.28488 [M + H]+ (Calcd for C33H44N7O16 794.2839).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-β-D-digitoxosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx22)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.42 (d, J = 8.3 Hz, 1 H), 6.98 (d, J = 8.5 Hz, 1 H), 4.37 (s, 2 H), 4.10 (dd, J = 2.7 Hz, 2.2 Hz, 1 H), 4.00 (dd, J = 6.1, 2.2 Hz, 1 H), 3.7 - 3.8 (m, 4 H), 3.69 (s, 3 H), 3.5 - 3.7 (m, 3 H), 3.4 (m, 1 H), 3.2 (m, 1 H), 2.94 (s, 6 H), 2.7 – 2.8 (m, 2 H), 2.6 (m, 1 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 677.26693 [M + H]+ (Calcd for C31H41N4O13 m/z 677.2665).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-acetylamino-3′-O-(1″-carboxyethyl)-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx23)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.50 (d, J = 8.4 Hz, 0.5 H), 8.49 (d, J = 8.5 Hz, 0.5 H), 6.98 (d, J = 7.6 Hz, 0.5 H), 6.97 (d, J = 7.5 Hz, 0.5 H), 4.74 (d, J = 3.2 Hz, 0.5 H), 4.7 (m, 1 H), 4.64 (dd, J = 13.1, 6.3 Hz, 1 H), 4.59 (d, J = 6.9 Hz, 1 H), 4.56 (d, J = 9.7 Hz, 0.5 H), 4.43 (d, J = 9.7 Hz, 1 H), 4.41 (s, 2 H), 3.6 – 4.0 (m, 3 H), 3.74 (s, 1 H), 3.63 (s, 3 H), 3.5 – 3.6 (m, 2 H), 2.95 (s, 6 H), 2.83(t, J = 11.1 Hz, 1 H), 2.8 (m, 1 H), 2.59 (dd, J = 11.9, 8.3 Hz, 1 H), 2.08 (s, 1.5 H), 2.07 (s, 1.5 H), 1.55 (d, J = 6.9 Hz, 1.5 H), 1.49 (d, J = 7.0 Hz, 1.5 H), 1.41 (d, J = 6.9 Hz, 1.5 H), 1.38 (d, J = 6.9 Hz, 1.5 H); HRESI m/z 822.30525 [M + H]+ (Calcd for C36H48N5O17 822.3040).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-6′-deoxy-6′-N’-decanoylamino-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx25)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.32 (d, J = 8.3 Hz, 1 H), 6.98 (d, J = 8.3 Hz, 1 H), 4.45 (d, J = 7.8 Hz, 1 H), 4.26 (d, J = 8.8 Hz, 1 H), 3.8 (m, 2 H), 3.74 (s, 3 H), 3.6 (m, 3 H), 3.3 - 3.5 (m, 2 H), 3.1 - 3.2 (m, 2 H), 2.90 (s, 6 H), 2.8 (m, 1 H), 2.7 (m, 1 H), 2.59 (dd, J = 11.7, 8.3 Hz, 1 H), 2.21 (q, J = 7.1 Hz, 2 H), 1.6 (m, 2 H), 1.56 (d, J = 6.8 Hz, 3 H), 1.3 (m, 12 H), 0.85 (t, J = 6.8 Hz, 3 H); HRESI m/z 862.41238 [M + H]+ (Calcd for C41H60N5O15 862.4080).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-amino-α-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx26a)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.28 (d, J = 8.4 Hz, 1 H), 6.96 (d, J = 8.6 Hz, 1 H), 5.30 (d, J = 3.5 Hz, 1 H), 4.1 (m, 1 H), 3.95 (s, 3 H), 3.7 – 3.8 (m, 3 H), 3.74 (dd, J = 11.7, 5.4 Hz, 1 H), 3.6 – 3.7 (m, 3 H), 3.4 (m, 1 H), 3.2 (m, 1 H), 3.04 (dd, J = 10.6, 4.0 Hz, 1 H), 2.86 (s, 3 H), 2.85 (s, 3 H), 2.7 (m, 1 H), 2.6 (m, 1 H), 2.56 (dd, J = 12.0, 7.7 Hz, 1 H), 1.55 (d, J = 6.8 Hz, 3 H); HRESI m/z 708.27445 [M + H]+ (Calcd for C31H42N5O14 708.2723).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-amino-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx26b)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.21 (d, J = 8.3 Hz, 1 H), 7.00 (d, J = 8.3 Hz, 1 H), 4.55 (d, J = 10.0 Hz, 1 H), 4.3 (s, 1 H), 3.94 (s, 2 H), 3.91 (dd J = 11.0, 1.5 Hz, 1 H), 3.8 (m, 1 H), 3.7 (m, 2 H), 3.64 (s, 3 H), 3.56 (dd, J = 9.8, 8.4 Hz, 1 H), 3.44 (dt, J = 3.5, 1.6 Hz, 1 H), 3.2 (m, 1H), 3.07 (t, J = 10.0 Hz, 1 H), 2.90 (s, 6 H), 2.8 (m, 2 H), 2.59 (dd, J = 12.1, 8.2 Hz, 1 H), 1.57 (d, J = 6.8 Hz, 3 H); HRESI m/z 708.27352 [M + H]+ (Calcd for C31H42N5O14 708.2723).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-6′-deoxy-6′-amino-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx27)
yellow powder; 1H NMR (CD3OD, 400 MHz) δ 8.30 (d, J = 9.0 Hz, 1 H), 7.00 (d, J = 8.4 Hz, 1 H), 4.37 (d, J = 8.8 Hz, 1 H), 4.21 (dd, J = 5.8, 3.3 Hz, 1 H), 3.7 -3.8 (m, 2 H), 3.77 (s, 2 H), 3.7 (m, 1 H), 3.68 (s, 1 H), 3.64 (s, 3 H), 3.3 – 3.4 (m, 3 H), 3.2 (m, 1 H), 2.8 (s, 6 H), 2.6 (m, 3 H), 1.41 (d, J = 7.5 Hz, 3 H); HRESI m/z 708.27284 [M + H]+ (Calcd for C31H42N5O14 708.2723).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-fluoro-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx29)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.43 (d, J = 8.3 Hz, 1 H), 6.98 (d, J = 8.5 Hz, 1 H), 4.48 (dd, J = 9.0, 2.0 Hz, 1 H), 4.40 (d, J = 9.0 Hz, 1 H), 4.28 (t, J = 8.9 Hz, 1 H), 4.2 (m, 1 H), 3.8 - 3.9 (m, 3 H), 3.75 (s, 3 H), 3.7 (m, 1 H), 3.6 (m, 1 H), 3.3 (m, 1 H), 3.2 (m, 1 H), 2.87 (s, 6 H), 2.78 (dd, J = 12.8, 6.7 Hz, 1 H), 2.7 (m, 1 H), 2.57 (dd, J =12.5, 8.1 Hz, 1 H), 1.55 (d, J = 7.1 Hz, 3 H); HRESI m/z 711.25609 [M + H]+ (Calcd for C31H40FN4O14 711.2520).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-D-mannosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx30)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ ppm 8.39 (d, J = 8.3 Hz, 1 H), 6.99 (d, J = 7.1 Hz, 1 H), 4.47 (d, J = 1.0 Hz, 1 H), 4.34 (d, J = 2.3 Hz, 1 H), 4.2 (m, 2 H), 4.21 (t, J = 3.4 Hz, 1 H), 4.1 (m, 1 H), 4.0 (m, 1 H), 3.90 (dd, J = 9.2, 2.4 Hz, 1 H), 3.75 (s, 2 H), 3.74 (s, 1 H), 3.73 (s, 1 H), 3.5 – 3.7 (m, 2 H), 2.89 (s, 3 H), 2.88 (s, 3 H), 2.7 (m, 2 H), 2.6 (m, 1 H), 1.55 (d, J = 6.9 Hz, 3 H); HRESI m/z 709.25859 [M + H]+ (Calcd for C31H41N4O15 709.2563).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-azido-D-mannosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx31)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ ppm 8.40 (d, J = 8.4 Hz, 0.5 H), 8.36 (d, J = 8.4 Hz, 0.5 H), 6.99 (d, J = 8.3 Hz, 1 H), 4.77 (d, J = 0.5 Hz, 0.5 H), 4.69 (s, 0.5 H), 4.54 (s, 0.5 H), 4.44 (d, J = 2.9 Hz, 0.5 H), 4.2 - 4.3 (m, 3 H), 4.16 (t, J = 3.2 Hz, 1 H), 4.0 - 4.1 (m, 1 H), 3.8 (m, 2 H), 3.75 (s, 3 H), 3.5 - 3.7 (m, 2 H), 2.91 (s, 6 H), 2.8 (m, 1 H), 2.6 (m, 2 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 734.26404 [M + H]+ (Calcd for C31H40N7O14 734.2628).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-amino-D-mannosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx32)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.28 (d, J = 8.8 Hz, 1 H), 6.99 (d, J = 7.6 Hz, 1 H), 4.96 (d, J = 0.5 Hz, 0.5 H), 4.76 (s, 1 H), 4.68 (s, 1 H), 4.65 (d, J = 3.5 Hz, 0.5 H), 4.5 (m, 1 H), 4.41 (s, 1 H), 4.18 (dd, J = 7.7, 4.5 Hz, 0.5 H), 3.8 – 4.1 (m, 3.5 H), 3.73 (s, 1.5 H), 3.72 (s, 1.5 H), 3.70 (s, 1 H), 3.6 (m, 1 H), 2.8 (m, 6 H), 2.7 (m, 2 H), 2.6 (m, 1 H), 1.56 (d, J = 6.8 Hz, 3 H); HRESI m/z 708.27411 [M + H]+ (Calcd for C31H42N5O14 708.2723).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-azido-β-D galactosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx33)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.47 (d, J = 8.5 Hz, 1 H), 6.99 (d, J = 8.5 Hz, 1 H), 4.39 (m, 1 H), 4.24 (d, J = 9.3 Hz, 1 H), 4.0 - 4.1 (m, 1 H), 3.94 (s, 1 H), 3.85 (d, J = 1.2 Hz, 1 H), 3.75 (s, 3 H), 3.7 (m, 3 H), 3.5- 3.6 (m, 4 H), 2.94 (s, 6 H), 2.8 (m, 2 H), 2.59 (dd, J = 12.2, 8.3 Hz, 1 H), 1.58 (d, J = 7.1 Hz, 3 H); HRESI m/z 734.26589 [M + H]+ (Calcd for C31H40N7O14 734.2628).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-amino-β-D-galactosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx34)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.20 (d, J = 8.3 Hz, 1 H), 6.97 (d, J = 8.3 Hz, 1 H), 4.57 (s, 1 H), 4.50 (d, J = 9.9 Hz, 1 H), 3.9 (m, 3 H), 3.7 - 3.8 (m, 4 H), 3.64 (s, 3 H), 3.4 (m, 1 H), 3.59 (t, J = 6.2 Hz, 1 H), 2.77 (s, 6 H), 2.5 (m, 3 H), 1.56 (d, J = 6.6 Hz, 3 H); HRESI m/z 708.27445 [M + H]+ (Calcd for C31H42N5O14 708.2723).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-N-trifluoroacetylamino-β-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx37)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.46 (d, J = 8.5 Hz, 1 H), 6.98 (d, J = 7.8 Hz, 1 H), 4.73 (d, J = 7.9 Hz, 1 H), 4.40 (d, J = 5.1 Hz, 1 H), 4.20 (dd, J = 12.0, 4.9 Hz, 1 H), 4.1 (m, 1 H), 4.0 (m, 3 H), 3.7 (m, 1 H), 3.64 (s, 3 H), 3.57 (m, 1 H), 2.95 (s, 6 H), 2.87 (m, 1 H), 2.78 (dd, J = 12.5, 6.8 Hz, 1 H), 2.59 (dd, J = 12.2, 8.3 Hz, 1 H), 1.55 (d, J = 6.5 Hz, 3 H); HRESI m/z 804.25463 [M + H]+ (Calcd for C33H41F3N5O15 804.2546).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-D-glucosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx38)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.43 (d, J = 8.5 Hz, 0.5 H), 8.42 (d, J = 8.6 Hz, 0.5 H), 7.02 (d, J = 8.3 Hz, 0.5 H), 7.00 (d, J = 9.2 Hz, 0.5 H), 5.20 (t, J = 6.3 Hz, 0.5 H), 4.71 (s, 1 H), 4.63 (t, J = 4.8 Hz, 0.5 H), 4.38 (s, 2 H), 4.39 (s, 1 H), 4.20 (m, 1 H), 3.74 (s, 1.5 H), 3.67 (s, 1.5 H), 3.6 (m, 3 H), 3.5 (m, 1 H), 2.96 (s, 6 H), 2.8 (m, 2 H), 2.61 (dd, J = 4.9, 1.1 Hz, 1 H), 2.4 (m, 1 H), 2.3 (m, 1 H), 1.59 (t, J = 6.7 Hz, 3 H); HRESI m/z 693.26248 [M + H]+ (Calcd for C31H41N4O14 693.2641).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-azido-β-D-xylosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx39)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ 8.47 (d, J = 8.5 Hz, 1 H), 6.98 (d, J = 8.3 Hz, 1 H), 4.38 (s, 1 H), 4.19 (d, J = 9.0 Hz, 1 H), 3.94 (t, J = 5.4 Hz, 1 H), 3.91 (s, 1 H), 3.88 (s, 1 H), 3.77 (s, 1 H), 3.74 (s, 3 H), 3.57 (dd, J = 11.4, 8.6 Hz, 1 H), 3.5 (m, 1 H), 3.4 (m, 1 H), 3.21 (d, J = 10.9 Hz, 1 H), 3.2 (m, 1 H), 2.94 (s, 6 H), 2.81 (d, J = 11.4 Hz, 1 H), 2.76 (t, J = 5.8 Hz, 1H), 2.58 (dd, J = 12.3, 8.3 Hz, 1 H), 1.55 (d, J = 6.9 Hz, 3 H); HRESI m/z 704.25084 [M + H]+ (Calcd for C30H38N7O13 704.2528).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-2′-deoxy-2′-amino-β-D-xylosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx40)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ ppm 8.16 (d, J = 8.1 Hz, 1 H), 6.96 (d, J = 8.1 Hz, 1 H), 4.57 (s, 1 H), 4.47 (d, J = 10.0 Hz, 1 H), 3.99 (dd, J = 11.6, 5.0 Hz, 1 H), 3.8 - 3.9 (m, 3 H), 3.7 (m, 1 H), 3.63 (s, 3 H), 3.4 - 3.6 (m, 2 H), 3.03 (t, J = 9.9 Hz, 1 H), 2.78 (s, 6 H), 2.6 (m, 3 H), 1.56 (d, J = 6.6 Hz, 3 H); HRESI m/z 678.25815 [M + H]+ (Calcd for C30H40N5O13 678.2617).
(4S,4aR,5S,5aR,6R,12aS)-4-(dimethylamino)-9-(N-methoxy-N-4′-deoxy-4′-azido-β-L-ribosylglycyl)amino-3,5,10,12,12a-pentahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide (Dx41)
yellow powder; 1H NMR (CD3OD, 500 MHz) δ ppm 8.49 (d, J = 8.3 Hz, 1 H), 6.99 (d, J = 8.5 Hz, 1 H), 4.2 (m, 1 H), 4.22 (d, J = 4.4 Hz, 1 H), 4.01 (t, J = 11.5 Hz, 1 H), 3.95 (dd, J = 10.6, 5.2 Hz, 2 H), 3.90 (s, 1 H), 3.8 (m, 1 H), 3.78 (s, 3 H), 3.6 (m, 2 H), 3.5 (m, 1 H), 3.45 (t, J = 4.2 Hz, 1 H), 2.92 (s, 6 H), 2.8 (m, 1 H), 2.7 (m, 1 H), 2.57 (dd, J = 12.2, 8.1 Hz, 1 H), 1.57 (d, J = 6.8 Hz, 3 H); HRESI m/z 704.25589 [M + H]+ (Calcd for C30H38N7O13 704.2522).
Supplementary Material
ACKNOWLEDGMENT
We thank the School of Pharmacy Analytical Instrumentation Center (University of Wisconsin-Madison) for analytical support, Dr. Pauline Peltier-Pain for her assistance in HPLC analysis and Prof. Shanteri Singh for her advice on this manuscript. This work was supported by NIH R37 AI52218 (JST) and the National Center for Advancing Translational Sciences (UL1TR000117).
Footnotes
Supporting Information Synthetic procedures of sugars, 1H, 13C, and related spectra of the synthesized compounds, activity assessment results. This material is available free of charge via the internet at http://pubs.acs.org.
Notes The authors declare the following competing financial interests: J.S.T. is a co-founder of Centrose (Madison, WI, USA).
REFERENCES
- (1) (a).Chopra I, Roberts M. Microbiol. Mol. Biol. R. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhanel GG, Homenuik K, Noreddin A, Vercaigne L, Embil J, Gin A, Karlowsky JA, Hoban DJ. Drugs. 2004;64:63–68. doi: 10.2165/00003495-200464010-00005. [DOI] [PubMed] [Google Scholar]
- (2).Brodersen DE, Clemon WM, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- (3) (a).Koza DJ, Nsiah YA. Bioorg. Med. Chem. Lett. 2002;12:2163–2165. doi: 10.1016/s0960-894x(02)00369-4. [DOI] [PubMed] [Google Scholar]; (b) Chopra I. Drug Resistance Updates. 2002;5:119–125. doi: 10.1016/s1368-7646(02)00051-1. [DOI] [PubMed] [Google Scholar]; (c) Nelson ML, Ismail MY, McIntyre L, Bhatia B, Viski P, Hawkins P, Rennie G, Andorsky D, Messersmith D, Stapleton K, Dumornay J, Sheahan P, Verma AK, Warchol T, Levy SB. J. Org. Chem. 2003;68:5838–5851. doi: 10.1021/jo030047d. [DOI] [PubMed] [Google Scholar]; (d) Charest MG, Siegel DR, Myers AG. J. Am. Chem. Soc. 2005;127:8292–8293. doi: 10.1021/ja052151d. [DOI] [PubMed] [Google Scholar]; (e) Roberts MC. FEMS Microbiol. Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]; (f) Charest MG, Lerner CD, Brubaker JD, Siegel DR, Myers AG. Science. 2005;308:395–398. doi: 10.1126/science.1109755. [DOI] [PubMed] [Google Scholar]; (g) Sum PE, Ross AT, Petersen PJ, Testa RT. Bioorg. Med. Chem. Lett. 2006;16:400–403. doi: 10.1016/j.bmcl.2005.09.078. [DOI] [PubMed] [Google Scholar]; (h) Clark RB, He M, Fyfe C, Lofland D, O’Brien WJ, Plamondon L, Sutcliffe JA, Xiao X. J. Med. Chem. 2011;54:1511–1528. doi: 10.1021/jm1015389. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Sun C, Hunt DK, Clark RB, Lofland D, O’Brien WJ, Plamondon L, Xiao X. J. Med. Chem. 2011;54:3704–3731. doi: 10.1021/jm1015395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4) (a).Felmingham D. J. Chemother. 2005;17:5–11. doi: 10.1179/joc.2005.17.Supplement-1.5. [DOI] [PubMed] [Google Scholar]; (b) Nathwani D. Int. J. Antimicrob. Agents. 2005;25:185–192. doi: 10.1016/j.ijantimicag.2004.11.006. [DOI] [PubMed] [Google Scholar]; (c) Bradford PA, Petersen PJ, Young M, Jones CH, Tischler M, O’Connell J. Antimicrob. Agents Chemother. 2005;49:3903–3909. doi: 10.1128/AAC.49.9.3903-3909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Doan T, Fung HB, Mehta D, Riska PF. Clin. Ther. 2006;28:1079–1104. doi: 10.1016/j.clinthera.2006.08.011. [DOI] [PubMed] [Google Scholar]; (e) Hylands J. Intensive and Critical care Nursing. 2008;24:260–261. doi: 10.1016/j.iccn.2008.03.006. [DOI] [PubMed] [Google Scholar]; (f) Peterson LR. Int. J. Antimicrob. Agents. 2008;32:S215–S222. doi: 10.1016/S0924-8579(09)70005-6. [DOI] [PubMed] [Google Scholar]
- (5) (a).Petersen PJ, Jacobus NV, Weiss WJ, Sum PE, Testa RT. Antimicrob. Agents Chemother. 1999;43:738–744. doi: 10.1128/aac.43.4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rubinstein E, Vaughan D. Drugs. 2005;65:1317–1336. doi: 10.2165/00003495-200565100-00002. [DOI] [PubMed] [Google Scholar]; (c) Katsandri A, Avlamis A, Pantazatou A, Petrikkos GL, Legakis NJ, Papaparaskevas J. Diagn. Microbiol. Infect. Dis. 2006;55:231–236. doi: 10.1016/j.diagmicrobio.2006.01.022. [DOI] [PubMed] [Google Scholar]; (d) Fraise AP. J. Infect. 2006;53:293–300. doi: 10.1016/j.jinf.2006.05.014. [DOI] [PubMed] [Google Scholar]; (e) Garrison MW, Nuemiller JJ. Int. J. of Antimicrob. Agents. 2007;29:191–196. doi: 10.1016/j.ijantimicag.2006.08.048. [DOI] [PubMed] [Google Scholar]; (f) Moet GJ, Dowzicky MJ, Jones RN. Diagn. Microbiol. Infect. Dis. 2007;57:333–336. doi: 10.1016/j.diagmicrobio.2006.08.001. [DOI] [PubMed] [Google Scholar]; (g) Insa R, Cercenado E, Goyanes MJ, Morente A, Bouza E. J. Antimicrob. Chemother. 2007;9:583–585. doi: 10.1093/jac/dkl496. [DOI] [PubMed] [Google Scholar]; (h) Borbone S, Lupo A, Mezzatesta ML, Campanile F, Santagati M, Stefani S. Int. J. Antimicrob. Agents. 2008;31:209–215. doi: 10.1016/j.ijantimicag.2007.03.014. [DOI] [PubMed] [Google Scholar]; (i) McConeghy KW, LaPlante KL. Diagn. Microbiol. Infect. Dis. 2010;68:1–6. doi: 10.1016/j.diagmicrobio.2010.04.011. [DOI] [PubMed] [Google Scholar]
- (6) (a).Bergeron J, Ammirati M, Danley D, James L, Norcia M, Retsema J, Strick CA, Su W, Sutcliffe J, Wondrack L. Antimicrob. Agents and Chemother. 1996;40:2226–2228. doi: 10.1128/aac.40.9.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bauer GB, Berens C, Projan SJ, Hillen W. J. Antimicrob. Chemother. 2004;53:592–599. doi: 10.1093/jac/dkh125. [DOI] [PubMed] [Google Scholar]; (c) Olsen MW, Ruzin A, Feyfant E, Rush TS, O’Connel J, Bradford PA. Antimicrob. Agents. Chemother. 2006;50:2156–2166. doi: 10.1128/AAC.01499-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Sum PE, Lee VJ, Testa RT, Hlavka JJ, Ellestad GA, Bloom JD, Gluzman Y, Tally FP. J. Med. Chem. 1994;37:184–188. doi: 10.1021/jm00027a023. [DOI] [PubMed] [Google Scholar]
- (8) (a).Wenzel R, Bate G, Kirkpatrick P. Nat. Rev. Drug Discov. 2005;4:809–810. doi: 10.1038/nrd1857. [DOI] [PubMed] [Google Scholar]; (b) Livermore DM. J. Antimicrob. Chemother. 2005;56:611–614. doi: 10.1093/jac/dki291. [DOI] [PubMed] [Google Scholar]
- (9) (a).Barden TC, Buckwalter BL, Testa RT, Petersen PJ, Lee VJ. J. Med. Chem. 1994;37:3205–3211. doi: 10.1021/jm00046a003. [DOI] [PubMed] [Google Scholar]; (b) Sum PE, Petersen P. Bioorg. Med. Chem. Lett. 1999;9:1459–1462. doi: 10.1016/s0960-894x(99)00216-4. [DOI] [PubMed] [Google Scholar]
- (10).Gonzalez-Sabin J, Moran-ramallal R, Rebolledo F. Chem. Soc. Rev. 2011;40:5321–5335. doi: 10.1039/c1cs15081b. [DOI] [PubMed] [Google Scholar]
- (11) (a).Nandy JP, Prakesch M, Khadem S, Reddy T, Sharma U, Arya P. Chem. Rev. 2009;109:1999–2060. doi: 10.1021/cr800188v. [DOI] [PubMed] [Google Scholar]; (b) Blunt JW, Copp BR, Keyzers RA, Munro MHG, Prinsep MR. Nat. Prod. Rep. 2013;30:237–323. doi: 10.1039/c2np20112g. [DOI] [PubMed] [Google Scholar]
- (12) (a).Salas JA, Mendez C. Trends microbial. 2007;15:119–232. [Google Scholar]; (b) Zhu X, Schmidt RR. Angew. Chem. Inter. Ed. 2009;48:1900–1934. doi: 10.1002/anie.200802036. [DOI] [PubMed] [Google Scholar]; (c) Singh S, Phillips GN, Thorson JS. Nat. Prod. Rep. 2012;29:1201–1237. doi: 10.1039/c2np20039b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Hager D, Mayer P, Paulitz C, Tiebes J, Trauner D. Angew. Chem. Inter. Ed. 2012;51:6525–6528. doi: 10.1002/anie.201201826. [DOI] [PubMed] [Google Scholar]
- (13) (a).Wells JS, O’Sullivan J, Aklonis C, Ax HA, Tymiak AA, Kirsch DR, Trejo WH, Principe P. J. Antibiot. 1992;45:1892–1898. doi: 10.7164/antibiotics.45.1892. [DOI] [PubMed] [Google Scholar]; (b) Tymiak AA, Ax HA, Bolgar MS, Kahle AD, Porubcan MA, Andersen NH. J. Antibiot. 1992;45:1899–1906. doi: 10.7164/antibiotics.45.1899. [DOI] [PubMed] [Google Scholar]; (c) Tymiak AA, Akkibus C, Bolgar MS, Kahle AD, Kirsch DR, O’sullivan J, Porubcan MA, Principe P, Trejo WH, Ax HA, Wells JS, Andersen N,H, Devasthale PV, Telikepalli H, Velde DV, Zou J, Mitscher LA. J. Org. Chem. 1993;58:535–537. [Google Scholar]
- (14).Horiguchi T, Hayashi K, Tsubotani S, Iinuma S, Harada S, Tanida S. J. Antibiot. 1994;47:545–556. doi: 10.7164/antibiotics.47.545. [DOI] [PubMed] [Google Scholar]
- (15).Pickens LB, Kim W, Wang P, Zhou H, Watanabe K, Gomi S, Tang Y. J. Am. Chem. Soc. 2009;131:17677–17689. doi: 10.1021/ja907852c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16) (a).Blanchard S, Thorson JS. Curr. Opin. Chem. Biol. 2006;10:263–271. doi: 10.1016/j.cbpa.2006.04.001. [DOI] [PubMed] [Google Scholar]; (b) Thibodeaux CJ, Melancon CE, Liu H–W. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]; (c) Thibodeaux CJ, Melancon CE, Liu H. -w. Angew. Chem. Inter. Ed. 2008;41:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Schmaltz RM, Hanson SR, Wong C–H. Chem. Rev. 2011;111:4259–4307. doi: 10.1021/cr200113w. [DOI] [PubMed] [Google Scholar]; (e) Gantt RW, Peltier-Pain P, Thorson JS. Nat. Prod. Rep. 2012;28:1811–1853. doi: 10.1039/c1np00045d. [DOI] [PubMed] [Google Scholar]
- (17) (a).For recent examples see: Langenhan JM, Engle JM, Slevin LK, Fay LR, Lucker RW, Smith KR, Endo MM. Bioorg. Med. Chem. Lett. 2008;18:670–673. doi: 10.1016/j.bmcl.2007.11.058. Carrasco MR, Alvarado CI, Dashner ST, Wong AJ, Wong MA. J. Org. Chem. 2010;75:5757–5759. doi: 10.1021/jo101066c. Iyer AK, Zhou M, Azad N, Elbaz H, Wang L, Rogalsky DK, Rojanasakul Y, O’Doherty GA, Langenhan JM. ACS Med. Chem. Lett. 2010;12:326–330. doi: 10.1021/ml1000933. Cipolla L, Peri F. Mini-Rev. Med. Chem. 2011;11:39–54. doi: 10.2174/138955711793564060. Langenhan JM, Endo MM, Engle JM, Fukumoto LL, Rogalsky DR, Slevin LK, Fay LR, Lucker RW, Rohlfing JR, Smith KR, Tjaden AE, Werner HM. Carbohydr. Res. 2011;346:2663–2676. doi: 10.1016/j.carres.2011.09.019. Langenhan JM, Mullarky E, Rogalsky DK, Rohlfing JR, Tjaden AE, Werner HM, Rozal LM, Loskot SA. J. Org. Chem. 2013;78:1670–1676. doi: 10.1021/jo302640y.
- (18).Goff RD, Thorson JS. Org. Lett. 2009;11:461–464. doi: 10.1021/ol8025704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19) (a).Peluso S, Imperiali B. Tetrahedron Lett. 2001;42:2085–2087. [Google Scholar]; (b) Carrasco MR, Nguyen MJ, Burnell DR, MacLaren MD, Hengel SM. Tetrahedron Lett. 2002;43:5727–5729. [Google Scholar]; (c) Carrasco MR, Brown RT, Serafimova IM, Silva O. J. Org. Chem. 2003;68:195–197. doi: 10.1021/jo026641p. [DOI] [PubMed] [Google Scholar]; (d) Carrasco MR, Brown RT. J. Org. Chem. 2003;68:8853–8858. doi: 10.1021/jo034984x. [DOI] [PubMed] [Google Scholar]; (e) Filira F, Biondi B, Biondi L, Giannini E, Gobbo M, Negri L, Rocchi R. Org. Biomol. Chem. 2003;1:3059–3063. doi: 10.1039/b306142f. [DOI] [PubMed] [Google Scholar]; (f) Matsubara N, Oiwa K, Hohsaka T, Sadamoto R, Niikura K, Fukuhara N, Takimoto A, Kondo H, Nishimura S-I. Chem. -Eur. J. 2005;11:6974–6981. doi: 10.1002/chem.200500531. [DOI] [PubMed] [Google Scholar]
- (20) (a).For recent examples see: Ahmed A, Peters NR, Fitzgerald MK, Watson JA, Hoffmann FM, Thorson JS. J. Am. Chem. Soc. 2006;128:14224–14225. doi: 10.1021/ja064686s. Griffith BR, Krepel C, Fu X, Blanchard S, Ahmed A, Edmiston CE, Thorson JS. J. Am. Chem. Soc. 2007;129:8150–8155. doi: 10.1021/ja068602r. Goff RD, Thorson JS. J. Med. Chem. 2010;53:8129–8139. doi: 10.1021/jm101024j. Goff RD, Singh S, Thorson JS. Chem. Med. Chem. 2011;6:774–776. doi: 10.1002/cmdc.201100028. Peltier-Pain P, Timmons SC, Grandemange A, Benoit E, Thorson JS. Chem. Med. Chem. 2011;6:1347–1350. doi: 10.1002/cmdc.201100178. Goff RD, Thorson JS. Org. Lett. 2012;14:2454–2457. doi: 10.1021/ol300703z. Peltier-Pain P, Marchillo K, Zhou M, Andes DR, Thorson JS. Org. Lett. 2012;14:5086–5089. doi: 10.1021/ol3023374.
- (21) (a).Galoni DP, Gin DY. Nature. 2007;446:1000–1007. doi: 10.1038/nature05813. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Crich D. Acc. Chem. Res. 2010;43:1144–1153. doi: 10.1021/ar100035r. [DOI] [PubMed] [Google Scholar]; (c) Hsu C–H, Hung S–C, Wu C–Y, Wong C–H. Angew. Chem. Inter. Ed. 2011;50:11872–11923. doi: 10.1002/anie.201100125. [DOI] [PubMed] [Google Scholar]; (d) McKay MJ, Nguyen HM. ACS Catal. 2012;2:1563–1595. doi: 10.1021/cs3002513. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Daude D, Remaud-Simeon M, Andre I. Nat. Prod. Rep. 2012;29:945–960. doi: 10.1039/c2np20054f. [DOI] [PubMed] [Google Scholar]; (e) Shen Z, Wang S–Y, Chok Y–K, Xu Y–H, Loh T–P. Chem. Rev. 2013;113:271–401. doi: 10.1021/cr300051y. [DOI] [PubMed] [Google Scholar]
- (22) (a).Fife RS, Sledge GW. Adv. Dent. Res. 1998;12:94–96. doi: 10.1177/08959374980120012801. [DOI] [PubMed] [Google Scholar]; (b) Cox CA, Amaral J, Salloum R, Guedez L, Reid TW, Jaworski C, John-Aryankalayil M, Freedman KA, Campos M, Martinez A, Becerra S, Carper DA. Ophthalmology. 2010;117:1782–1791. doi: 10.1016/j.ophtha.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Shieh J-M, Huang T, Hung C, Chou K, Tsai Y, Wu W. Br J. Pharmacol. 2010;160:1171–1184. doi: 10.1111/j.1476-5381.2010.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23) (a).Sweet RL, Schachter J, Landers DV, Ohm-Smith M, Robbie MO. Am. J. Obstet. Gynecol. 1988;158:736–741. doi: 10.1016/s0002-9378(16)44537-0. [DOI] [PubMed] [Google Scholar]; (b) Martens MG, Gordon S, Yarborough DR, Faro S, Binder D, Berkeley A. South. Med. J. 1993;86:604–610. doi: 10.1097/00007611-199306000-00002. [DOI] [PubMed] [Google Scholar]; (c) Lauhio A, Salo T, Ding Y, Konttinen YT, Nordstrom D, Twschesche H, Lahdeyirta J, Golub LM, Sorsa T. Clin. Exp. Immunol. 1994;98:21–28. doi: 10.1111/j.1365-2249.1994.tb06601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lee H–M, Ciancio SG, Tuter G, Ryan ME, Komaroff E, Golub LM. J. Periodontol. 2004;75:453–463. doi: 10.1902/jop.2004.75.3.453. [DOI] [PubMed] [Google Scholar]; (e) De Paiva CS, Corrales RM, Villarreal AL, Farley WJ, Li DQ, Stern ME, Pflugfelder SC. Exp. Eye Res. 2006;83:526–535. doi: 10.1016/j.exer.2006.02.004. [DOI] [PubMed] [Google Scholar]
- (24) (a).Dahl EL, Shock JL, Shenai BR, Gut J, DeRisi JL, Rosenthal PJ. Antimicrob. Agents Chemother. 2006;50:3124–3131. doi: 10.1128/AAC.00394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lalloo DG, Shingadia D, Pasvol G. J. Infect. 2007;54:111–121. doi: 10.1016/j.jinf.2006.12.003. [DOI] [PubMed] [Google Scholar]
- (25) (a).Hoerauf A, Mand S, Fischer K, Kruppa T, Marfo-Debrekyei Y, Yaw Debrah A, Pfarr KM, Adjei O, Buttner DW. Med. Microbiol. Immunol. 2003;192:211–216. doi: 10.1007/s00430-002-0174-6. [DOI] [PubMed] [Google Scholar]; (b) Taylor MJ, Makunde WH, McGarry HF, Turner JD, Mand S, Hoerauf A. Lancet. 2005;365:2116–2121. doi: 10.1016/S0140-6736(05)66591-9. [DOI] [PubMed] [Google Scholar]
- (26) (a).Gould DJ, Berenstein M, Dreja H, Ledda F, Podhajcer OL, Chernajovsky Y. Gene Therapy. 2000;7:2061–2070. doi: 10.1038/sj.gt.3301354. [DOI] [PubMed] [Google Scholar]; (b) Minagar A, Alexander JS, Schwendimann RN, Kelley RE, Gonzalez-Toledo E, Jimenez JJ, Mauro L, Jy W, Smith S. J. Arch Neurol. 2008;65:199–204. doi: 10.1001/archneurol.2007.41. [DOI] [PubMed] [Google Scholar]
- (27) (a).Tremblay P, Meiner Z, Galou M, Heinrich C, Petromilli C, Lisse T, Cayetano J, Torchia M, Mobley W, Buiard H, DeArmond SJ, Prusiner SB. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12580–12585. doi: 10.1073/pnas.95.21.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Corti O, Sabate O, Horellou P, Colin P, Dumas S, Buchet D, Buc-Caron M, Mallet J. Nat. Biotechnol. 1999;17:349–354. doi: 10.1038/7901. [DOI] [PubMed] [Google Scholar]; (c) Aebischer P, Ridet J–L. Trends Neurosci. 2001;24:533–540. doi: 10.1016/s0166-2236(00)01899-3. [DOI] [PubMed] [Google Scholar]; (d) Domercq M, Matute C. Trends Pharmaco. Sci. 2004;25:609–612. doi: 10.1016/j.tips.2004.10.001. [DOI] [PubMed] [Google Scholar]
- (28).Kamst E, Zegelaar -JK, van der Marel GA, van Boom JH, Lugtenberg BJ, Spaink HP. Carbohydr. Res. 1999;321:176–189. [Google Scholar]
- (29) (a).Zhu S. Tetrahedon Lett. 1992;33:6503–6504. [Google Scholar]; (b) Suarez JR, Trastoy B, Perrez-Ojeda ME, Marin-arrios R, Chiara JL. Adv. Synth. Catal. 2010;352:2515–2520. [Google Scholar]
- (30).Myszka H, Bednarezyk D, Najder M, Laca W. Carbohydr. Res. 2003;338:133–141. doi: 10.1016/s0008-6215(02)00407-x. [DOI] [PubMed] [Google Scholar]
- (31) (a).Wolfrom ML, Olin SM, Polglase WJ. J. Am. Chem. Soc. 1950;72:1724–1729. [Google Scholar]; (b) Wolfrom ML, Anno K. J. Am. Chem. Soc. 1953;75:1038–1039. [Google Scholar]; (c) Gigg R, Warren CD. J. Chem. Soc. 1965:1351–1355. doi: 10.1039/jr9650002975. [DOI] [PubMed] [Google Scholar]
- (32) (a).Saux MC, Mosser J, Pontagnier H, Leng B. Eur. J. Drug Metab. Pharmacokinet. 1982;7:123–130. doi: 10.1007/BF03188729. [DOI] [PubMed] [Google Scholar]; (b) Stroller NH, Johnson LR, Trapnell LR, Trapnell S, Harrold CQ, Garrett S. J. Perodontol. 1998;69:1085–1091. doi: 10.1902/jop.1998.69.10.1085. [DOI] [PubMed] [Google Scholar]; (c) Rolain J–M, Boulos A, Mallet M–N, Raoult D. Antimicrob. Agents Chemother. 2005;49:2673–2676. doi: 10.1128/AAC.49.7.2673-2676.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lecaillet A, Mallet M-N, Raoult D, Rolain L-M. J. Antimicrob. Chemother. 2009;63:771–774. doi: 10.1093/jac/dkp013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.