Abstract
Understanding the factors that regulate hematopoiesis opens up the possibility of modifying these factors and their actions for clinical benefit. DEK, a non-histone nuclear phosphoprotein initially identified as a putative proto-oncogene, has recently been linked to regulation of hematopoiesis. DEK has myelosuppressive activity in vitro on proliferation of human and mouse hematopoietic progenitor cells and enhancing activity on engraftment of long term marrow repopulating mouse stem cells, has been linked in coordinate regulation with the transcription factor C/EBPα, for differentiation of myeloid cells, and apparently targets a long term repopulating hematopoietic stem cell for leukemic transformation. This review covers the uniqueness of DEK, what is known about how it now functions as a nuclear protein and also as a secreted molecule that can act in paracrine fashion, and how it may be regulated in part by Dipeptidylpeptidase 4, an enzyme known to truncate and modify a number of proteins involved in activities on hematopoietic cells. Examples are provided of possible future areas of investigation needed to better understand how DEK may be regulated and function as a regulator of hematopoiesis, information possibly translatable to other normal and diseased immature cell systems.
Keywords: DEK, Hematopoietic Stem Cells, Hematopoietic Progenitor Cells, Receptors, Chromatin, Dipeptidylpeptidase IV
Introduction
Hematopoietic stem (HSC) and progenitor (HPC) cells give rise to all blood forming elements and have been used to successfully treat non-malignant and malignant disorders [1,2]. However, much remains to be deciphered regarding regulation of HSC and HPC function and fate. In efforts to uncover key factors involved in HSC and HPC production and fate decisions, we identified DEK, a biochemically distinct mammalian nuclear phosphoprotein initially classified as a putative proto-oncoprotein [3], as a candidate for regulating hematopoiesis [4]. We noted that DEK had negative regulatory effects on proliferation of HPCs: granulocyte-macrophage (CFU-GM), erythroid (BFU-E), and multipotential (CFU-GEMM), but positive effects on engrafting HSC [4]. Others have linked DEK in coordinate regulation with the transcription factor C/EBPα on the differentiation of myeloid cells [5]. This complex of C/EBPα and DEK, whose assembly and disassembly is regulated by serine 21 phosphorylation of C/EBPα, enhanced the activation of the granulocyte-colony stimulating factor receptor 3 promotor. Knocking down expression of the DEK gene reduced the capacity of C/EBPα to drive granulocyte target gene expression. We recently reported that the cell surface enzyme CD26, a Dipeptidylpeptidase 4 (DPP4), truncates and changes the functional activities of cytokines such as the colony stimulating factors (CSF: granulocyte macrophage (GM)-CSF, granulocyte (G)-CSF, interleukin-3 (IL-3), and erythropoietin (EPO)) and of the chemokine, stromal derived factor-1 (SDF-1/CXCL12) [6, 7]. We now know that other proteins with cell regulatory activity have putative truncation sites for DPP4 [8,9]. As discussed below, DEK has a putative specific truncation site for DPP4. Based on this information, we hypothesize that DEK is a key and perhaps crucial regulatory determinant of HSC and HPC function and fate decisions in both steady-state and stressed hematopoiesis, effects that may be mediated or regulated by DPP4 truncation of DEK. There is still a paucity of information on DEK and its actions. This review covers current knowledge of DEK and its role in cell regulation and fate decisions, with a particular emphasis on HSC, HPC, and hematopoiesis. Examples are suggested for future studies in this area.
DEK Regulation and Activities
DEK bears little resemblance to other known proteins (Fig. 1A), and is the only representative of its own protein class. While DEK, an abundant non-histone chromosomal factor, is vital to global heterochromatin integrity [10] (Fig. 1B and 1C), it can be secreted by certain cells, sometimes in exosomes, or in its free form and subsequently be taken up as the same molecule in bioactive form in a heparin sulfate-dependent process by other cells where it, in turn, modulates global chromatin structure [11], a process similar to what is seen in a paracrine loop. Whether uptake of DEK can also take place through G-Protein coupled receptors, or whether DEK can act through stimulation of such receptors, is not clear, and this may vary between cell types and their maturational status.
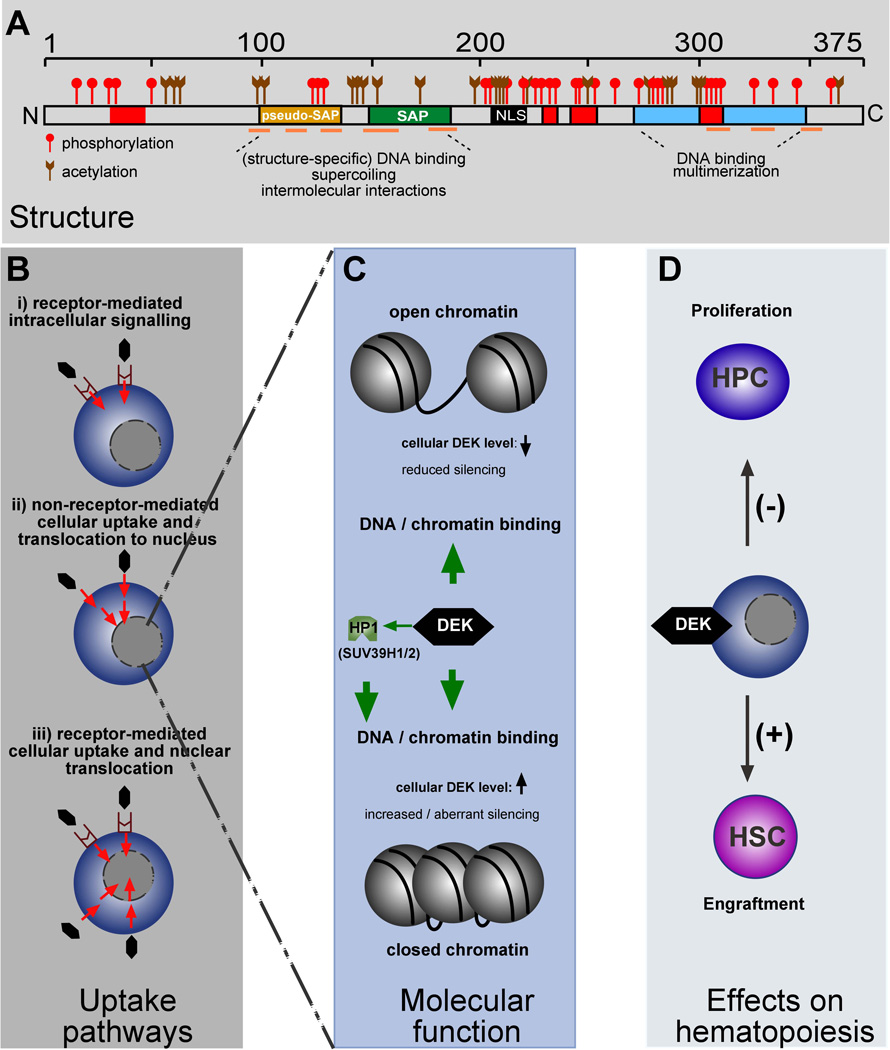
Figure 1.
Schematic depiction of the structure, functional domains and post-translational modifications of DEK, potential DEK uptake pathways, and effects of DEK on hematopoiesis. A) Linear sequence of DEK with DNA binding domains (yellow: pseudo-SAP-box; green: SAP-box; blue: C-terminal DNA-binding domain; orange lines: position of α-helices, as revealed by NMR), and other functional features (red: acidic regions; black: putative NLS) indicated [See references 25,41,42,44,45]. The positions of previously mapped phosphorylation and acetylation sites are marked [See references 12, 14 and 15]. B) Potential DEK receptor mediated and/or uptake pathways. C) Simplified depiction of DEK functions in the setting of chromatin. DEK interacts with, and augments binding of HP1 to H3K9Me3, thereby recruiting SUV39H1/2, thus further enhancing the deposition of H3K9Me3. In addition, DEK binds to DNA/chromatin via its DNA binding domains (see A). However, the precise consequences for open or closed chromatin currently remains elusive. D) Functional consequences of DEK in the setting of hematopoietic stem (HSC) and progenitor (HPC) cells. Not shown is the potential influence of Dipeptidylpeptidase 4 truncation of DEK on DEK functional activity.
DEK is heavily post-translationally modified. Regulation of the phosphorylation status of DEK by CK2 and protein phosphatase 2A [12,13], acetylation [14,15], and poly (ADP-ribosyl)ation [16,17] can regulate the function of DEK. Transcription of the DEK gene is controlled by YY1 and NF-Y [18], E2F [19], and the estrogen receptor α [20]. The DEK protein can be degraded by the F-box/WD repeat containing protein 7 (Fbxwt) [21], and microRNA-489, which is involved in the maintenance of muscle stem-cell quiescence, does so by targeting DEK [22]. DEK interacts with RelA/p65 [23] and represses gene expression in conjunction with SET and PARP1 [24]. Phosphorylation by protein kinase CK2 [12] in one way affects the function of the DNA-binding domains (SAP-box and C-terminal DNA binding domain) of DEK [25], and in another way allows for the histone chaperone functions of DEK that co-activate transcription of a nuclear receptor [26]. However, CK2 does not seem to play a role in regulating DEK’s function in chromatin integrity [12].
Blood cell production is regulated by cytokine and microenvironmental influences that direct HSC and HPC functions and fate decisions [2,27]. Knowledge of these influences and how they are mechanistically mediated is crucial to understanding hematopoiesis under steady-state and stress conditions, and eventually for correction of the abnormalities in hematopoiesis associated with disease, and for optimal efficacy in the use of HSC and HPC for hematopoietic cell transplantation. Our knowledge of the extracellular and intracellular factors influencing the proliferation, self-renewal, survival, differentiation and movement of HSC and HPC is increasing [2,27], but there is still much to be learned if we are to best use this information for improved health care. In continuing efforts to define new factors in the regulation of hematopoiesis, we focused on DEK, a non-histone phosphoprotein which was initially identified as a fusion protein resulting from a t(6;9) translocation in a rare subtype of acute myelogenous leukemia [3]. Furthermore, DEK is overexpressed and implicated in many malignancies [3,28–38], and exhibits critical functions in several central tumor-promoting pathways, e.g inhibition of apoptosis and senescence, among others [3,28,30,33,37,39,40]. DEK bears little resemblance to other known proteins, but is well-conserved among higher eukaryotes, as all DEK proteins share a unique conserved region, the “SAP-box” (SAP = Saf/Actinus/PARP) [41]. This motif is found in proteins such as DEK that are typically involved in DNA binding, chromatin remodeling, and/or RNA processing [41–43]. DEK is capable of binding to the TG-rich pets site in the human immunodeficiency virus type 2 (HIV-2) promoter, where it acts as a transcriptional repressor [13,44], although it appears that DEK primarily recognizes DNA on the basis of structure rather than sequence and thus might play an active role in maintaining higher-order chromatin architecture [42,43,45,46]. In addition to its DNA binding properties, DEK is found in association with mRNA splicing and export factors, as well as with spliced transcripts, where it influences 3’ splice fidelity [46–50]. Intense post-translational modification of DEK by phosphorylation [12], acetylation [14–16], and poly(ADP-ribosyl)ation [17] point to the importance of these post-translational modifications in regulating DEK’s multiple functions and sub-or extracellular localization. DEK antibodies are found in patients with juvenile idiopathic arthritis (JIA) and other auto-immune diseases [15,51,52], raising the question of why this nuclear protein is an autoantigen. Although DEK is primarily associated with chromatin throughout the cell cycle [53], two independent pathways, both involving post-translational modifications, were recently identified that result in DEK’s presence in the extracellular space. The first pathway implicated non-classical secretion of DEK by activated human monocyte-derived macrophages in both a free form and in exosomes [54]. In the second, passive release of poly(ADP-ribosyl)ated, hyperphosphorylated DEK by apoptotic T-lymphocytes was observed possibly occurring as a result of Fas-ligand-or stress-mediated apoptosis [16]. IL-8 induces the secretion of DEK, and DEK acts as a chemoattractant for peripheral blood leukocytes [54]. Of particular note in this context, secreted DEK can be taken up by other cells, move to the nucleus, and effectively carry out the intranuclear functions of DEK, including control of global heterochromatin integrity and DNA repair [11].
DEK and Hematopoiesis
Extracellular and intracellular DEK expression modulates hematopoiesis [4]. It remains to be determined exactly how DEK is mediating its different effects on hematopoiesis. This may be through receptor mediated cytokine-like activities in which a sequence of intracellular signals are induced, or perhaps by cellular uptake through a specific receptor, and/or non-receptor mediated DEK uptake. Uptake of DEK through receptor-mediated or non-receptor-mediated events may involve chromatin regulation (Fig. 1B and C). These different possibilities require additional investigation. Exploring these possibilities are important for understanding the effects of DEK and perhaps for casting new light on regulation of hematopoiesis during health and disease.
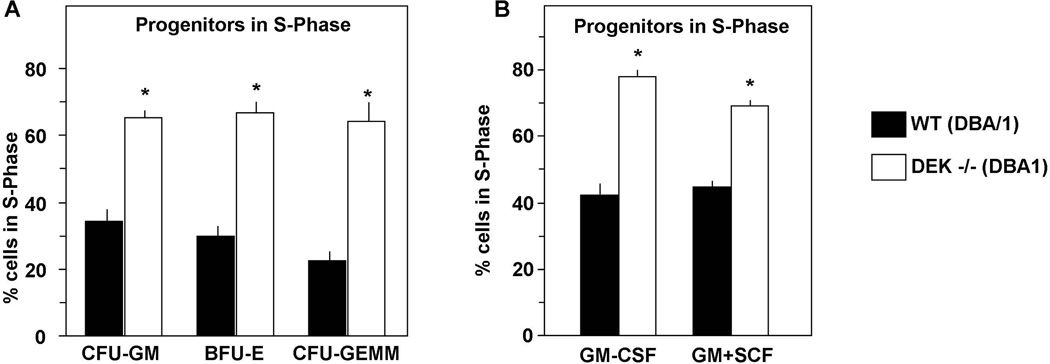
We found that DEK, in purified recombinant human form was myelosuppressive for colony formation by CFU-GM, BFU-E, and CFU-GEMM from C57Bl/6 mouse BM and human CB cells, effects that were dose-dependent [4]. The suppressive effects of DEK on colony formation were apparent when assayed on single isolated CD34+ CB cells, suggesting direct acting effects of DEK on HPC although how these effects are mediated is not known. This negative regulatory role of DEK was consistent with the enhanced numbers and cycling status of CFU-GM, BFU-E, and CFU-GEMM in the BM and spleen of C57Bl/6 DEK−/−, compared to C57Bl/6+/+, mice [4]. Some of this effect may reflect the reports of others showing that DEK works in concert with C/EBPα to regulate differentiation of myeloid cells [5]. Most recently, we found similar effects using cells from DEK−/− mice on another mouse strain background. DBA/1 DEK−/− HPC were at a higher cell cycling rate than DBA/1 DEK+/+ HPC (Fig. 2). Moreover, purified recombinant human DEK inhibited colony formation of DBA/1+/+ CFU-GM, BFU-E, and CFU-GEMM in dose-response fashion (Broxmeyer unpublished observations). These results demonstrate that the findings are not confined to effects on only one mouse strain. In contrast to the inhibition of CFU proliferation by DEK, BM cells from C57Bl/6 DEK−/− mice manifested decreased longer-, but not short-, term competitive repopulation capability in lethally-irradiated congenic mice, in addition to even greater decreases in repopulation of lethally-irradiated secondary mouse recipients in a non-competitive assay [4]. This suggested that DEK was important for the positive engrafting capability of a long-term, but not short-term, repopulating and self-renewing HSC (schematically shown in Fig. 1D). The results suggest that DEK may be necessary for maintenance of HSC, consistent with reports that DEK is expressed in immature cell populations and that this expression decreases with the differentiation and maturing of the cells [55]. Interestingly, the DEK/CAN fusion protein resulting from the t(6;9) chromosomal translocation [3] targets a long-term repopulating HSC for leukemic transformation [56].
Figure 2.
Effects of DEK knockout (−/−) on hematopoietic progenitor cells. Results are shown for these cells in S-phase of the cell cycle, as determined by high specific activity tritiated thymidine kill technique, when stimulated with 5% pokeweed mitogen spleen cell conditioned medium (PWMSCM), SCF (50ng/ml), and EPO (1U/ml) in methylcellulose culture (A) and when stimulated with GM-CSF (10ng/ml) alone and in combination with SCF in agar culture (B) [6]. Results are given as mean±1SEM of 3 mice per group. *, significantly different from control medium (p<0.05), as assessed by 2 tailed t test.
Still to be determined are effects of exogenously added DEK on HSC and HPC numbers and functional activity, and hematopoiesis in vivo, and possible effects of DEK on ex-vivo expansion of HSC and HPC, information that could possibly be of therapeutic use. The functional DEK domains that may be involved in DEK receptor-binding and/or non-receptor mediated uptake and translocation to the nucleus remain to be determined, although DEK does have a putative nuclear localization domain (Figure 1).
DEK and DPP4
We recently found that in addition to the homing and chemotactic protein SDF-1/CXCL12 [7, 57], GM-CSF, G-CSF, IL-3, and EPO have truncation sites for the enzyme DPP4 [6]. DPP4 is found on the surface of many cells as CD26 and is present within cells expressing CD26. It is also found as a soluble enzyme in serum and plasma. DPP4 treatment of SDF-1/CXCL12 produces a molecule in which the two N-terminal amino acids have been removed. Truncated SDF-1/CXCL12 is inactive as an HPC chemotactic molecule and as an HPC survival enhancing factor, and this truncated SDF-1/CXCL12 can block the chemotactic and survival enhancing effect of the full length SDF-1/CXCL12 molecule [6, 7]. DPP4 treatment of GM-CSF, G-CSF, IL-3, and EPO produced truncated forms of these CSFs that had greatly decreased CSF activity, but blocked the activity of the full length forms of their respective CSF, for colony formation in vitro, and with human GM-CSF for intracellular signaling (phosphorylation) of JAK2 and STAT5 using TF1, a human factor dependent cell line, and primary CD34+ CB cells [6]. These in vitro effects on HPC proliferation were duplicated in vivo in mice given exogenously added full length, truncated, or the combination of full-length and truncated GM-CSF, and also for full-length and truncated EPO [6]. The effects were most apparent on target cells pretreated with an inhibitor of DPP4 (e.g. Diprotin A; a tripeptide: ILE-PRO-ILE) or in CD26−/− mice. Both radiolabeled full length- and truncated-human GM-CSF demonstrated receptor binding to the GM-CSF receptor of TF1 cells and primary CD34+ CB cells, but the truncated GM-CSF bound to the GM-CSF receptor with higher affinity (had a lower dissociation constant), and truncated GM-CSF blocked receptor binding of full-length GM-CSF at concentrations of truncated GM-CSF one-eighth that of full length GM-CSF [6]. Thus, DPP4-treated and truncated SDF-1/CXCL12, GM-CSF, G-CSF, IL-3 and EPO act as dominant negative or competitive molecules for the actions of their respective full-length proteins, and offer a potential means for regulating specific protein actions, and it may be that DEK works in a similar fashion.
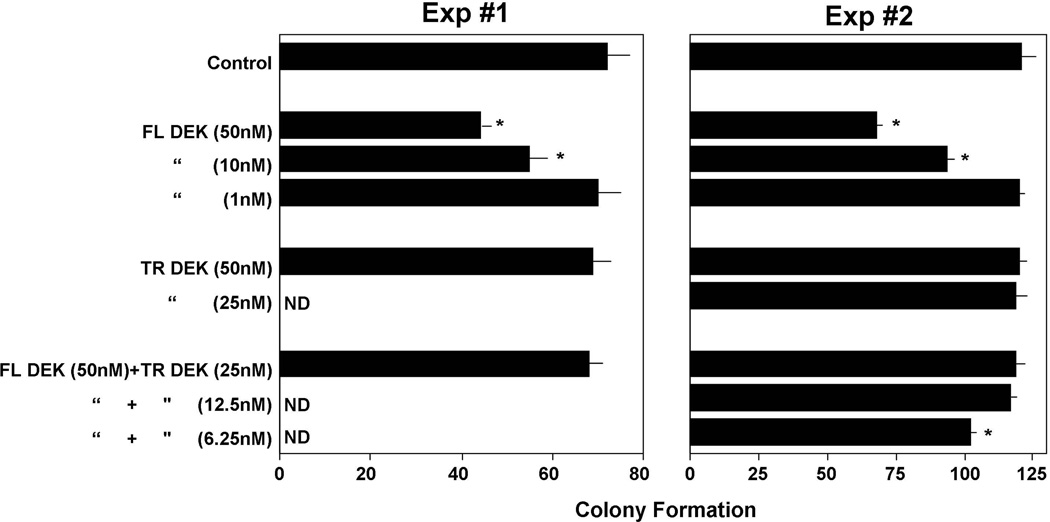
DPP4 can truncate proteins at a penultimate alanine or proline at the N-terminus and also when serine or other amino acids are at this penultimate site [8,9]. Most recently, we found that the N-terminus start site of the DEK protein contained a putative DPP4 truncation site (MSASAPAAEGEGTPTQP…) in which serine (the second amino acid in this sequence), rather than alanine or proline, served as a potential site for DPP4 activity, unless in this case methionine is pre-clipped off and it is the alanine (the second amino acid in the following N-terminus sequence) that serves as a DPP4 truncation site (SASAPAAEGEGTPTQP…). As shown in Fig. 3, DPP4-treated DEK did not manifest inhibitory activity against GM-CSF- or GM-CSF plus SCF stimulated colony formation of mouse BM cells pretreated with Diprotin A, an inhibitor of DPP4, and at one-fourth to one-eighth the concentration of untreated DEK, the DPP4-treated DEK blocked the inhibitory activity of full-length DEK, suggesting that DPP4-treated DEK can act as a dominant negative effector molecule for full-length DEK, perhaps at the level of specific DEK receptor binding or non-receptor mediated uptake. Whether DPP4 truncates at the second amino acid (serine) or if the methionine is pre-clipped and DPP4 truncates at the alanine that follows the serine, this would leave the next sets of amino acids open, with potential additional truncation sites (serine, alanine, or proline) available, so that DPP4 may be able to continue to truncate DEK. It remains to be determined by mass spectrometry or other analysis what the effects of DPP4 are on the DEK protein itself and where exactly the DPP4 may be acting. Such analysis needs to be linked to functional effects of DEK. These results, which need to be pursued in greater depth both in vitro and in vivo, suggest a potentially strong and modifying influence of DPP4 on DEK activity, which may be of physiological relevance, and of potential clinical interest.
Figure 3.
Influence of DPP4 on DEK activity. Shown are effects of full length (FL) – and DPP4-treated (=likely truncated, TR) DEK, alone and in combination, on 5×104 C57Bl/6 mouse BM cells/ml pretreated with Diprotin A, an inhibitor of DPP4, and stimulated with GM-CSF (Exp#1) or GM-CSF+SCF (Exp#2) (See reference 6 for details of such studies done with other growth modulating proteins). Results are given as mean±1SEM. *, significantly different from control medium (p<0.05); ND= not done
Future efforts to more precisely define hematopoiesis in DEK−/− mice seem reasonable in the context of steady-state hematopoiesis, and especially for hematopoiesis in mice subjected to different stresses. We recently evaluated the effects of radiation and drugs in CD26−/− mice [6]. Stress situations can help define the relevance of DEK in a way that may not be picked up using untreated mice. The stresses to be evaluated for hematological recovery can be low and higher non-lethal γ-irradiation, and non-lethal doses of drugs such as 5 Fluorouracil, and cyclophosphamide (Cytoxan®).
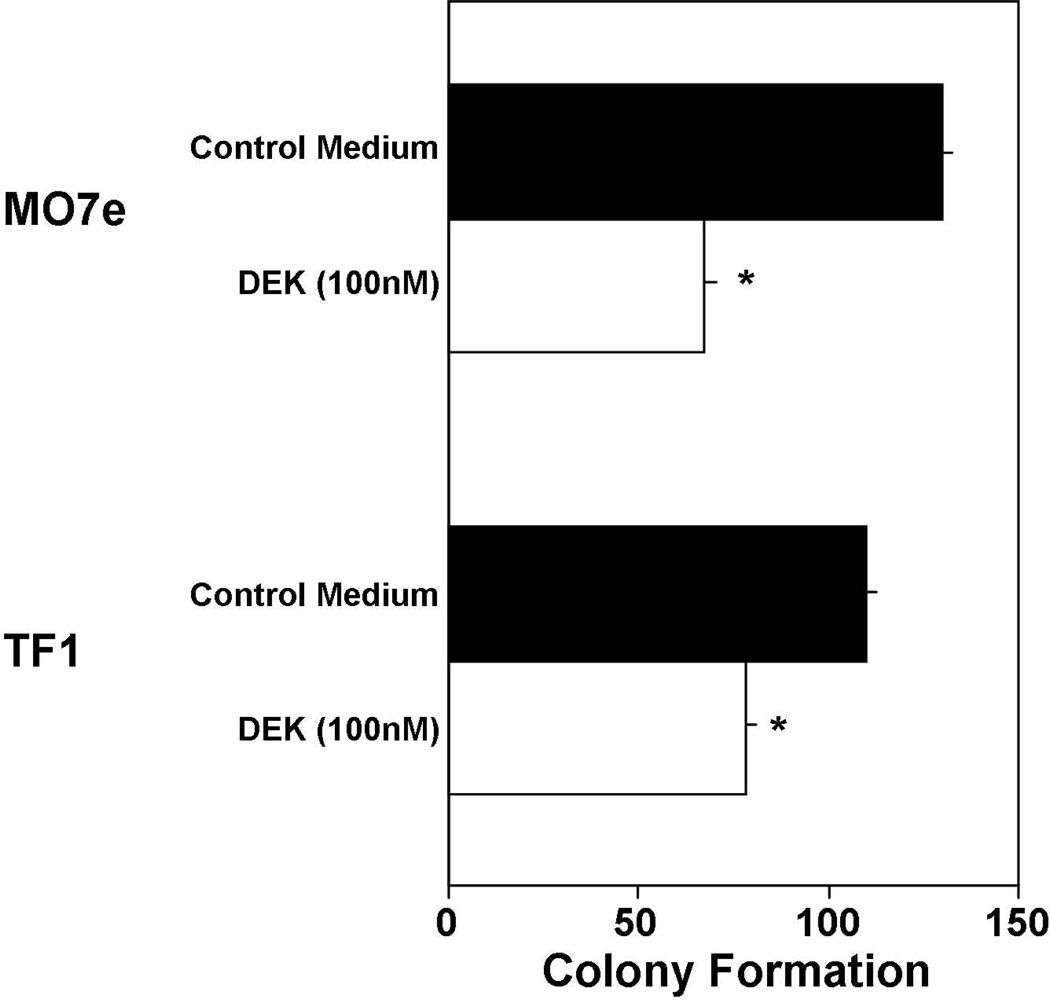
In depth studies on mechanisms of DEK activities might best be done on relevant established cell lines with confirming studies on primary target cells, such as we recently published for GM-CSF [6]. We have identified the human factor dependent cell lines, TF1 (responsive to stimulation of proliferation by either GM-CSF, IL-3 or EPO, and when used in combination with SCF eliciting a synergistic stimulating effect) and M07e (responsive to GM-CSF or IL-3, and synergistically to either GM-CSF or IL-3 with SCF) as responding to the inhibitory effects of DEK with a one hour pulse exposure of these cell lines to DEK (Fig. 4). TF1 [6] and M07e cells express active DPP4 on their cell surface as CD26, (O’Leary and Broxmeyer, unpublished studies), as do HPC and HSC in primary mouse and human BM and human CB [6,7]. Although the effects of cytokines with DPP4 truncation sites are more potent on CD26-expressing cells that have been pretreated to inhibit DPP4 [6 and Broxmeyer unpublished observations], the activities of these cytokines can still be detected although at a lesser effect if the CD26-expressing cells are not pretreated to inhibit DPP4. Much work remains to elucidate a role for DPP4 on protein activity [8,9]. Hence, even though the TF1 and M07e cells were not first pretreated to inhibit DPP4, we did detect DEK inhibition of colony formation by these cell lines. Since both cell lines have been used by us as models for stimulating and negatively acting cytokines at a cell and intracellular level [58–68], these growth factor dependent human cell lines can be used as models to initiate intracellular studies on DEK signal transduction, effects which can then be verified in less extensive studies using, for example, primary CD34+ CB cells, as we previously reported for GM-CSF [6].
Figure 4.
Influence of DEK on colony formation by human factor dependent cell lines MO7e and TF1. These cells were not pretreated with a DPP4 inhibitor. Results are shown as mean±1SEM for 1000 MO7e and 500 TF1 cells/ml −/+ 1 hour pulse treatment of these cell lines with control medium or 50nM DEK prior to their stimulation with GM-CSF plus SCF (See reference 6 for details of such studies done with the TF1 human growth factor dependent cell line assessing the effects of other proteins on colony formation). *, significantly different from control medium (p<0.05)
Since communication between BM microenvironmental cells and the HSC and HPC that reside in the BM are involved in HSC and HPC function and fate decisions, and DEK is expressed in osteoblasts, endothelial cells and BM stromal cells [69–71; these authors deposited their gene expression data sets into the GEO profiles], DEK may be involved in microenvironmental cell-HSC/HPC interactions, perhaps with regards to positive effects we have noted with DEK on engrafting capability of HSC and DEK negative regulation of HPC. Such possible influences can be investigated in a stem cell transplant model that can distinguish donor (e.g. CD45.2+) from recipient (e.g. CD45.1+/CD45.2+ F1) from competitor (e.g. CD45.1+) cells in a competitive HSC setting, and also donor from recipient cells in a non-competitive HSC transplant assay. DEK−/− mice (on a C57Bl/6 mouse strain background) can be used alternatively as recipients and/or as a source of donor cells.
Concluding Comments
In summary, DEK is an apparently unique molecule whose functional activity in the normal regulation of hematopoietic stem and progenitor cells [4, 5], and in leukemogenesis [56], is only just beginning to be elucidated. It is likely that DEK will be found to have controlling roles in other stem and progenitor cell types in addition to those in muscle [22], including embryonic stem cells, induced pluripotent stem cells, and mesenchymal stem/stromal cells. Abnormalities in DEK and its actions may be associated with cancer and cancer stem/progenitor cells [28,72]. How DEK fits in with other chromatin and intracellular molecules involved in regulation of HSC and HPC [2,27] is of great interest, and remains to be determined. DEK modulates global heterochromatin integrity in human cells and in a Drosophila model [10]. Interference with DEK expression in human cells induces a phenotype indicative of more accessible chromatin organization. DEK is a Su(var), meaning that it is a positive modulator of heterochromatin as shown using the model of white-mottled variegation in the eyes of Drosophila [10] (Fig. 1C). Loss of DEK is accompanied by loss of the key heterochromatic marker H3K9Me3, and both phosphorylated and unphosphorylated DEK interact with the vital heterochromatin factor HP1α, thus bringing HP1α to the H3K9 mark. DEK also plays a key role in epigenetic silencing by bringing the KMT1 A/B methylase (which adds the third methyl group to H3K9), along with HP1α, to what becomes the H3K9Me3 mark, a hallmark of heterochromatin [10]. This was shown by CHIP analysis, co-immunoprecipitation, and biochemical interactions. Thus, it is of interest to determine if uptake of DEK into HSCs and HPCs leads to changes in chromatin architecture with the subsequent effects on gene expression leading to effects on hematopoiesis. To further understand the mechanism of action of DEK in hematopoiesis, it will be essential to determine if DEK effects are mediated via specific receptors with subsequent induction of intracellular signaling and/or by uptake and translocation of DEK into the cytoplasm and subsequently into the nucleus (Fig. 1B and 1C). It will be important to identify the domains of DEK that are responsible for the effects on HSC and HPC, and if they are the same domains that are involved in DNA binding and heterochromatin integrity [23]. A search for a specific receptor or receptors for DEK is currently ongoing. In the context of tumor cell sensitivity to chemotherapy, up-regulation of DEK expression may enhance cell survival and chemoresistance, while decreasing DEK expression in such cells may enhance the sensitivity of the tumor cells to kill by specific chemotherapeutic agents [33,39,73,74]. Since DEK expression is higher in immature compared to more mature blood cells such as those present in the CD34+ cell population [55], and leukemia (LIC) or tumor (TIC) initiating cells are present in the CD34+ cell population [2,27], DEK could be one of the reasons that LIC or TIC populations survive chemotherapy. Also of interest is the effect of DPP4 truncation of DEK on these functional activities of DEK. Therefore, further understanding the function and molecular mechanisms of action of this biochemically distinct protein, including assessment of protein partners, DNA and RNA targets and the secretion and uptake by neighboring cells, is likely to yield information relevant to clinically important questions.
Acknowledgements
Studies referenced from the Broxmeyer laboratory, and original data in Figures 2–4 were generated with support from Public Health Service Grants from the NIH to HEB: R01 HL056416, R01 HL67384, R01 HL112669, and P01 DK090948. H.A.O. is supported by a post-doctoral stipend from NIH T32 DK07519 to H.E.B. F.K. is supported by the START program of the Faculty of Medicine, RWTH Aachen. N.M.V. is supported by NIH grant K01-AR-055620. Work on DEK in D.M.M.’s laboratory is supported in part by NIH grant R01-AI087128.
Footnotes
Author Contributions
Hal E. Broxmeyer: Conception and design, Financial Support, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Nirit Mor-Vaknin: Conception and design, Manuscript writing
Ferdinand Kappes: Conception and design, Manuscript writing
Maureen Legendre: Conception and design.
Anjan K. Saha: Conception and design, Manuscript writing
Xuan Ou: Conception and design, Collection and/or assembly of data, Data analysis and interpretation
Heather O’Leary: Conception and design, Collection and/or assembly of data, Data analysis and interpretation
Maegan Capitano: Conception and design, manuscript writing
Scott Cooper: Conception and design, Collection and/or assembly of data, Data analysis and interpretation
David M. Markovitz: Conception and design, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Disclosure of potential conflicts of interest
Hal E. Broxmeyer is a member of the Medical Scientific Advisory Board of Corduse, a cord blood banking company based in Orlando, Florida.
References
- 1.Broxmeyer HE, Smith FO. Cord Blood Hematopoietic Cell Transplantation. In: Applelbaum FR, Forman SJ, Negrin RS, Blume KG, editors. Thomas' Hematopoietic Cell Transplantation. 4th ed. West Sussex, United Kingdom: Wiley-Blackwell; 2009. pp. 559–576. [Google Scholar]
- 2.Shaheen M, Broxmeyer HE. The humoral regulation of hematopoiesis. In: Hoffman R, Benz EJ Jr, Shattil SJ, Furie B, Silberstein LE, McGlave P, Heslop H, Anastasi J, editors. Hematology: Basic Principles and Practice. Philadelphia: Elsevier Churchill Livingston; 2009. pp. 253–275. [Google Scholar]
- 3.von Lindern M, Fornerod M, van Baal S, et al. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broxmeyer HE, Kappes F, Mor-Vaknin N, et al. DEK regulates hematopoietic stem engraftment and progenitor cell proliferation. Stem Cells Dev. 2012;21:1449–1454. doi: 10.1089/scd.2011.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koleva RI, Ficarro SB, Radomska HS, et al. C/EBPα and DEK coordinately regulate myeloid differentiation. Blood. 2012;119:4878–4888. doi: 10.1182/blood-2011-10-383083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broxmeyer HE, Hoggatt J, O'Leary HA, et al. Dipeptidylpeptidase 4 negatively regulates colony-stimulating factor activity and stress hematopoiesis. Nat Med. 2012;18:1786–1796. doi: 10.1038/nm.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christopherson KW, 2nd, Hangoc G, Broxmeyer HE. Cell surface peptidase CD26/dipeptidylpeptidase IV regulates CXCL12/stromal cell-derived factor-1 alpha-mediated chemotaxis of human cord blood CD34+ progenitor cells. J Immunol. 2002;169:7000–7008. doi: 10.4049/jimmunol.169.12.7000. [DOI] [PubMed] [Google Scholar]
- 8.O’Leary H, Ou X, Broxmeyer HE. The role of dipeptidylpeptidase 4 in hematopoiesis and transplantation. Current Opinions in Hematopoiesis. 2013 doi: 10.1097/MOH.0b013e32836125ac. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou X, O’Leary H, Broxmeyer HE. Implications of DPP4 modifications of proteins that regulate stem/progenitors and more mature cell types. Blood. 2013 doi: 10.1182/blood-2013-02-487470. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kappes F, Waldmann T, Mathew V, et al. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 2011;25:673–678. doi: 10.1101/gad.2036411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha AK, Kappes F, Mundade A. Intercellular trafficking of the nuclear oncoprotein DEK. Proc. Natl. Acad. Sci. USA. 2013 doi: 10.1073/pnas.1220751110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kappes F, Damoc C, Knippers R, et al. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faulkner NE, Hilfinger JM, Markovitz DM. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J Biol Chem. 2001;276:25804–25812. doi: 10.1074/jbc.M006454200. [DOI] [PubMed] [Google Scholar]
- 14.Cleary J, Sitwala KV, Khodadoust MS, et al. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280:31760–31767. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 15.Mor-Vaknin N, Kappes F, Dick AE, et al. DEK in the synovium of patients with juvenile idiopathic arthritis. Arthritis & Rheumatism. 2011;63:556–567. doi: 10.1002/art.30138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kappes F, Fahrer J, Khodadoust MS, et al. DEK is a poly(ADP-Ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28:3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahrer J, Popp O, Malanga M, et al. High affinity interaction of poly (ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010;49:7119–7130. doi: 10.1021/bi1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sitwala KV, Adams K, Markovitz DM. YY1 and NF-Y binding sites regulate the transcriptional activity of the dek and dek-can promoter. Oncogene. 2002;21:8862–8870. doi: 10.1038/sj.onc.1206041. [DOI] [PubMed] [Google Scholar]
- 19.Carro MS, Spiga FM, Quarto M, et al. DEK expression is controlled by E2F and degraded in diverse tumor types. Cell Cycle. 2006;5:1202–1207. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 20.Privette Vinnedge LM, Ho S-K. The DEK oncogene is a target of steroid hormone receptor signaling in breast cancer. Plos One. 2012;7:e46985. doi: 10.1371/journal.pone.0046985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babaei-Jadidi R, Li N, Saadeddin A. FBXW7 influences murine intestinal homeostasis and cancer, targeting, Notch, Jun, DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung TH, Quach NL, Charville GW, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–530. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sammons M, Wan SS, Vogel NL, et al. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J Biol Chem. 2006;281:26802–26812. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- 24.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nature Struct & Mol Biol. 2007;14:548–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 25.Kappes F, Scholten I, Richter N, et al. Functional domains of the ubiuitous chromatin protein DEK. Mol Cell Biol. 2004;24:6000–6010. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawatsubashi S, Murata T, Lim J, et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes & Dev. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaheen M, Broxmeyer HE. Hoffman R, Benz EJ, Jr, Silberstein LE, Heslop H, Weitz J, Anastasi J. Hematology: Basic Principles and Practice. 6th Edition. Chapter 14. Philadelphia: Elsevier Churchill Livingston; 2012. Principles of Cytokine Signaling; pp. 136–146. [Google Scholar]
- 28.Privette Vinnedge LM, McClaine R, Wagh PK, et al. The human DEK oncogene stimulates β-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 2011;30:2741–2752. doi: 10.1038/onc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise-Draper TM, Allen HV, Jones EE, et al. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26:7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wise-Draper TM, Allen HV, Thobe MN, et al. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol. 2005;79:14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise-Draper TM, Mintz-Cole RA, Morris TA, et al. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69:1792–1799. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wise-Draper TM, Morreale RJ, Morris TA, et al. DEK proto-oncogene expression interferes with the normal epithelial differentiation program. Amer J Pathol. 2009;174:71–81. doi: 10.2353/ajpath.2009.080330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khodadoust MS, Verhaegen M, Kappes F, et al. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69:6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riveiro-Falkenbach E, Soengas MS. Control of tumorigenesis and chemoresistance by the DEK oncogene. Clin Cancer Res. 2010;16:2932–2938. doi: 10.1158/1078-0432.CCR-09-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kappes F, Khodadoust MS, Yu L, et al. DEK expression in melanocytic lesions. Hum Pathol. 2011;42:932–938. doi: 10.1016/j.humpath.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel RM, Walters LL, Kappes F, et al. DEK expression in merkel cell carcinoma and small cell carcinoma. J Cutan Pathol. 2012;39:753–757. doi: 10.1111/j.1600-0560.2012.01941.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu S, Wang X, Sun F, et al. DEK overexpression is correlated with the clinical features of breast cancer. Pathol Int. 2012;62:176–181. doi: 10.1111/j.1440-1827.2011.02775.x. [DOI] [PubMed] [Google Scholar]
- 38.Liu K, Feng T, Liu J, et al. Silencing of the DEK gene induces apoptosis and senescence in CaSki cervical carcinoma cells via the up-regultaion of NF-κB p65. Biosci Res. 2012;32:323–332. doi: 10.1042/BSR20100141. [DOI] [PubMed] [Google Scholar]
- 39.Kavanaugh GM, Wise-Draper TM, Morreale RJ, et al. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Research. 2011;39:7465–7476. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim DW, Chae JI, Kim JY, et al. Proteomic analysis of apoptosis related proteins regulated by proto-oncogene protein DEK. J Cell Biochem. 2009;106:1048–1059. doi: 10.1002/jcb.22083. [DOI] [PubMed] [Google Scholar]
- 41.Aravind L, Koonin EV. SAP-a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 42.Böhm F, Kappes F, Scholten I, et al. The SAP-box domain of chromatin protein DEK. Nucleic Acids Res. 2005;33:1101–1110. doi: 10.1093/nar/gki258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devany M, Kappes F, Chen KM, et al. Solution NMR structure of the N-terminal domain of the human DEK protein. Protein Sci. 2008;17:205–215. doi: 10.1110/ps.073244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fu GK, Grosveld G, Markovitz DM. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc Natl Acad Sci U S A. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldmann T, Baack M, Richter N, et al. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–7010. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waldmann T, Eckerich C, Baack M, et al. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277:24988–24994. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- 47.Le Hir H, Gatfield D, Izaurralde E, et al. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Hir H, Izaurralde E, Maquat LE, et al. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGarvey T, Rosonina E, McCracken S, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150:309–320. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soares LM, Zanier K, Mackereth C, et al. Intron removal requires proofreading of U2AF/3' splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 51.Szer IS, Sierakowska H, Szer W. A novel autoantibody to the putative oncoprotein DEK in pauciarticular onset juvenile rheumatoid arthritis. J Rheumatol. 1994;21:2136–2142. [PubMed] [Google Scholar]
- 52.Murray KJ, Szer W, Grom AA, et al. Antibodies to the 45 kDa DEK nuclear antigen in pauciarticular onset juvenile rheumatoid arthritis and iridocyclitis: selective association with MHC gene. J Rheumatol. 1997;24:560–567. [PubMed] [Google Scholar]
- 53.Kappes F, Burger K, Baack M, et al. Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001;276:26317–26323. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- 54.Mor-Vaknin N, Punturieri A, Sitwala K, et al. The DEK nuclear autoantigen is a secreted chemotactic factor. Mol Cell Biol. 2006;26:9484–9496. doi: 10.1128/MCB.01030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ageberg M, Gullberg U, Lindmark A. The involvement of cellular proliferation status in the expression of the human proto-oncogene DEK. Haematologica. 2006;91:268–269. [PubMed] [Google Scholar]
- 56.Oancea C, Rüster B, Henschler R, et al. The t(6;9) associated DEK/CAN fusion protein targets a population of long-term repopulating hematopoietic stem cells for leukemogenic transformation. Leukemia. 2010;24:1910–1919. doi: 10.1038/leu.2010.180. [DOI] [PubMed] [Google Scholar]
- 57.Christopherson KW, 2nd, Hangoc G, Mantel CR, et al. Modulation of hematopoietic stem cell homing and engraftment by CD26. Science. 2004;305:1000–1003. doi: 10.1126/science.1097071. [DOI] [PubMed] [Google Scholar]
- 58.Gotoh A, Takahira H, Geahlen RL, et al. Cross-linking of integrins induces tyrosine phosphorylation of the proto-oncogene product Vav and the protein tyrosine kinase Syk in human factor-dependent myeloid cells. Cell Growth Differ. 1997;8:721–729. [PubMed] [Google Scholar]
- 59.Gotoh A, Takahira H, Mantel C, et al. Steel factor induces serine phosphorylation of Stat3 in human growth factor-dependent myeloid cell lines. Blood. 1996;88:138–145. [PubMed] [Google Scholar]
- 60.Hendrie PC, Broxmeyer HE. Myeloid cell proliferation stimulated by Steel factor is pertussis toxin sensitive and enhanced by cholera toxin. Int J Immunopharmacol. 1994;16:547–560. doi: 10.1016/0192-0561(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 61.Hendrie PC, Miyazawa K, Yang YC, et al. Mast cell growth factor (c-kit ligand) enhances cytokine stimulation of proliferation of the human factor-dependent cell line, M07e. Exp Hematol. 1991;19:1031–1037. [PubMed] [Google Scholar]
- 62.Horie M, Broxmeyer HE. Involvement of immediate-early gene expression in the synergistic effects of steel factor in combination with granulocyte-macrophage colony-stimulating factor or interleukin-3 on proliferation of a human factor-dependent cell line. J Biol Chem. 1993;268:968–973. [PubMed] [Google Scholar]
- 63.Lee Y, Gotoh A, Kwon HJ, et al. Enhancement of intracellular signaling associated with hematopoietic progenitor cell survival in response to SDF-1/CXCL12 in synergy with other cytokines. Blood. 2002;99:4307–4317. doi: 10.1182/blood.v99.12.4307. [DOI] [PubMed] [Google Scholar]
- 64.Mantel C, Luo Z, Hendrie P, et al. Steel factor and granulocyte-macrophage colony stimulating factor act together to enhance choline-lipid turnover during synergistically stimulated proliferation of the human factor dependent cell line, M07E. Biochem Biophys Res Commun. 1993;197:978–984. doi: 10.1006/bbrc.1993.2575. [DOI] [PubMed] [Google Scholar]
- 65.Miyazawa K, Hendrie PC, Kim YJ, et al. Recombinant human interleukin-9 induces protein tyrosine phosphorylation and synergizes with steel factor to stimulate proliferation of the human factor-dependent cell line, M07e. Blood. 1992;80:1685–1692. [PubMed] [Google Scholar]
- 66.Miyazawa K, Hendrie PC, Mantel C, et al. Comparative analysis of signaling pathways between mast cell growth factor (c-kit ligand) and granulocyte-macrophage colony-stimulating factor in a human factor-dependent myeloid cell line involves phosphorylation of Raf-1, GTPase-activating protein and mitogen-activated protein kinase. Exp Hematol. 1991;19:1110–1123. [PubMed] [Google Scholar]
- 67.Miyazawa K, Toyama K, Gotoh A, et al. Ligand-dependent polyubiquitination of c-kit gene product: a possible mechanism of receptor down modulation in M07e cells. Blood. 1994;83:137–145. [PubMed] [Google Scholar]
- 68.Ritchie A, Gotoh A, Gaddy J, et al. Thrombopoietin upregulates the promoter conformation of p53 in a proliferation-independent manner coincident with a decreased expression of Bax: potential mechanisms for survival enhancing effects. Blood. 1997;90:4394–4402. [PubMed] [Google Scholar]
- 69.Patel MJ, Liu W, Sykes MC, et al. Identification of mechanosensitive genes in osteoblasts by comparative microarray studies using the rotating wall vessel and the random positioning machine. J Cell Biochem. 2007;101:587–599. doi: 10.1002/jcb.21218. [DOI] [PubMed] [Google Scholar]
- 70.Johnson LA, Clasper S, Holt AP, et al. An inflammation-induced mechanism for leukocyte transmigration across lymphatic vessel endothelium. J Exp Med. 2006;203:2763–2777. doi: 10.1084/jem.20051759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larson BL, Ylöstalo J, Prockop DJ. Human multipotent stromal cells undergo sharp transition from division to development in culture. Stem Cells. 2008;26:193–201. doi: 10.1634/stemcells.2007-0524. [DOI] [PubMed] [Google Scholar]
- 72.Shibata T, Kokubu A, Miyamato M, et al. DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendicrine carcinoma of the lung. Oncogene. 2010;29:4671–4681. doi: 10.1038/onc.2010.217. [DOI] [PubMed] [Google Scholar]
- 73.Riveiro-Falkenbach E, Soengas MS. Control of tumorigenesis and chemoresistance by the DEK oncogene. Clin. Cancer Res. 2010;16:2932–2938. doi: 10.1158/1078-0432.CCR-09-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sansing HA, Sarkeshik A, Yates JR, et al. Integrin αβ1, αvβ, α6β effectors p130Cas, Src and talin regulate carcinoma invasion and chemoresistance. Biochem. Biophys. Res. Commun. 2011;406:171–176. doi: 10.1016/j.bbrc.2011.01.109. [DOI] [PMC free article] [PubMed] [Google Scholar]