Abstract
Despite substantial efforts to understand the interactions between nanoparticles and cells, the cellular processes that determine the efficiency of intracellular drug delivery remain largely unclear. Here we examined cellular uptake of siRNA delivered in lipid nanoparticles (LNPs) using cellular trafficking probes in combination with automated high-throughput confocal microscopy as well as defined perturbations of cellular pathways paired with systems biology approaches to uncover protein-protein and protein-small molecule interactions. We show that multiple cell signaling effectors are required for initial cellular entry of LNPs through macropinocytosis, including proton pumps, mTOR, and cathepsins. SiRNA delivery is substantially reduced as ≅70% of the internalized siRNA undergoes exocytosis through egress of LNPs from late endosomes/lysosomes. Niemann Pick type C1 (NPC1) is shown to be an important regulator of the major recycling pathways of LNP-delivered siRNAs. NPC1-deficient cells show enhanced cellular retention of LNPs inside late endosomes/lysosomes and increased gene silencing of the target gene. Our data suggests that siRNA delivery efficiency might be improved by designing delivery vehicles that can escape the recycling pathways.
Small interfering RNAs (siRNA) are a potential new class of medicines that can selectively silence disease-causing genes1–3. LNPs are the most advanced siRNA delivery system to date, having recently shown therapeutic efficacy in clinical trials4. Although efforts to define the physicochemical properties of nanoparticles that contribute to efficient cellular delivery are ongoing, the cellular factors that regulate nanoparticle-mediated siRNA delivery remain largely unclear5–7.
Nanoparticles utilize different endocytosis pathways for cellular entry5. Intracellular trafficking of nanoparticles is a dynamic process, which results in transport of macromolecules to different subcellular destinations, with the majority of material targeted to the lysosomes for degradation5. It has been hypothesized that the buffering capacity of nanoparticles can activate the proton pump that accentuates osmotic pressure inside an endosome, resulting in its swelling and subsequent endosomal escape of siRNA7,8. Alternatively, it has been suggested that cationic lipids in LNPs can interact with anionic lipids of the endosome, causing destabilization of the endosomal membrane9. Regardless of the release mechanism, imaging studies have shown that nucleic acids are largely trapped inside the endo- and lysosomes with only a small fraction being released to the cytoplasm10–12. Endosomal transport requires multiple heterotypic or homotypic endocytic fusion events with constantly changing vesicular milieu regulated by a diverse set of endosomal proteins and different cell signaling cascades13,14. The effect of this interplay between cell signaling and endocytosis on LNP trafficking needs to be further defined.
In this study, we utilized an automated, high-resolution microscopy-based screening approach with a library of small molecules that have a range of effects on cell signaling, endocytosis and autophagy to identify key regulators that are important for cellular uptake of cationic LNPs that can encapsulate siRNA (Supplemental Information-Small molecule library). These LNPs were formed by the complexation of siRNA with the cationic lipid, C12-200 and were further formulated with different excipients using a previously reported microfluidic process that generates slightly positively charged nanoparticles (ca. 70-80 nm in size)15; they have demonstrated unprecedented efficacy in the delivery of siRNA in vivo16,17. The potency of this delivery system tested in rodents and non-human primates is at a range similar to LNPs at the forefront of clinical trials4. In rodents no liver toxicity was observed at 1 mg/kg, a dose hundred fold above the efficacious range of silencing.16
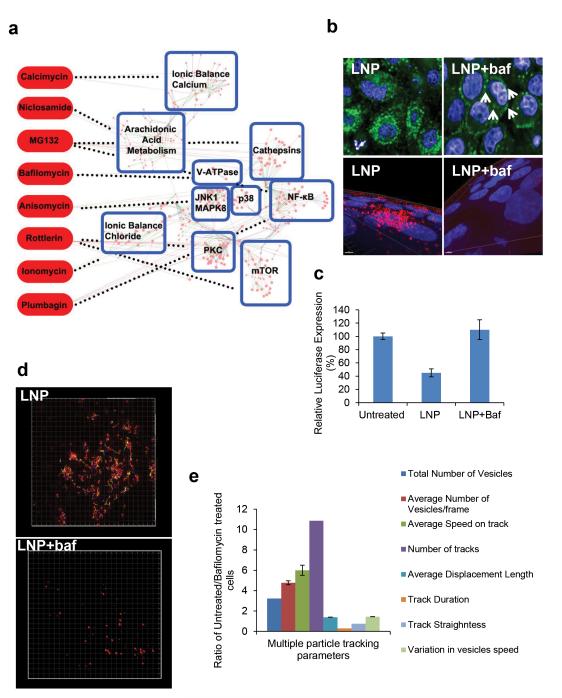
When HeLa-GFP cells were incubated with LNPs containing Alexa-647-labelled siRNA against GFP for 3 hrs, the particles appear to localize at the perinuclear region within 1 hour post exposure (Fig S1 a-b). Subcellular localization of LNP-siRNA in the presence of small molecule inhibitors under similar conditions was visualized and the amount of internalized siRNA was quantified. The small molecule library consisted of molecules that affect cell signaling, intracellular trafficking as well as had the ability to either induce or inhibit autophagy (for a list of the inhibitors used and the quantification of their effect on LNP uptake see Supplemental Information Small Molecule Library). We considered the subset of small molecules that had substantial inhibition on LNP uptake and using databases (See Supplementary Information Methods) identified the proteins with which the small molecules interact with and created protein-protein and protein-small molecule interaction networks. This systems analysis identified several endocytic regulators required for LNP entry which includes VoATPase, mTOR (mammalian target of rapamycin), cathepsins, Protein Kinase C, NFκB, calcium channels, chloride channels and arachidonic acid metabolic pathways (Fig 1a). Previous work has identified, among other processes, signaling cascades such as mTOR and cathepsins, as regulators of the cellular uptake of glucose and viruses by macropinocytosis18,19. We have summarized known effectors of cellular trafficking in Table S1. Because previous studies show that interference with acidification impacts release of nucleic acids from endosomal compartments20, our results further corroborated the effects of proton pump on endocytosis of LNPs. We found that the VoATPase inhibitor, bafilomycin inhibits LNP entry inside cells (Fig 1b) whereas having no effect on the LNPs stability (FigS2a). Multiple LNPs with different cationic lipid compositions containing siRNA or Hydrogenated soybean phosphatidylcholine (HSPC)-based liposomes containing small molecules showed marked decrease in uptake in the presence of two different proton pump inhibitors, concanamycin and bafilomycin (FigS2b-c)21 . Furthermore, treatment with bafilomycin led to a decrease in siRNA-induced target gene silencing (Fig 1c).
Figure 1. Systems survey for endocytosis of lipid nanoparticles (LNP).
LNPs containing siAF647 were incubated with HeLa-GFP cells (50nM) for 3 hrs in the presence or absence of small molecules and imaged using an automated high throughput confocal microscope a. Small molecules that inhibit over 80% of internalization (with no more than 10%-15% loss of cell viability) and their network interactions are presented in the form of a systems diagram b. Localization of siAF647LNP in presence or absence of bafilomycin is represented as an individual image (arrows indicate peripheral localization) and a 3D z-stack. c. Dual HeLa cells were exposed to LNP (siLuc, 10nM) in the presence or absence of bafilomycin (1μM). Luciferase to Renilla levels was measured for silencing activity. Experiments were performed in triplicate; errors are plotted as standard error means (S.E.M.). (d-e). Time lapse Total internal reflection (TIRF) microscopy of cells exposed to siAF647-LNPs (50nM, 3hrs) in the presence or absence of bafilomycin was subjected to multiple particle tracking (MPT). A snapshot of vesicular tracks indicating the movement of LNPs in presence or absence of bafilomycin presented in (d). The ratio between the MPT parameters for untreated/bafilomycin treated cells was calculated to provide a quantitative measure of differences in LNP trafficking with or without bafilomycin (e). Errors of ratios are plotted through propagation of errors from division.
We further investigated the interactions of the LNPs with the cell membrane in the presence or absence of bafilomycin using time lapse total internal reflection fluorescence microscopy (TIRFM) that allows for high resolution imaging at the cell surface. Multiple particle tracking revealed that LNP-positive vesicles moved on average six times faster in untreated cells than in bafilomycin-treated cells. After bafilomycin treatment endosomes also appear to form in substantially lower numbers (3-fold less) and had a slower rate of disappearance (increased track duration) from the focal plane (Fig 1d-e and Movies S1-S2). Two-fold fewer LNP positive vesicles were observed after bafilomycin treatment and those that form get enlarged due to a block in transport of early endosomes to late endosome/lysosomes (as has been previously reported22).
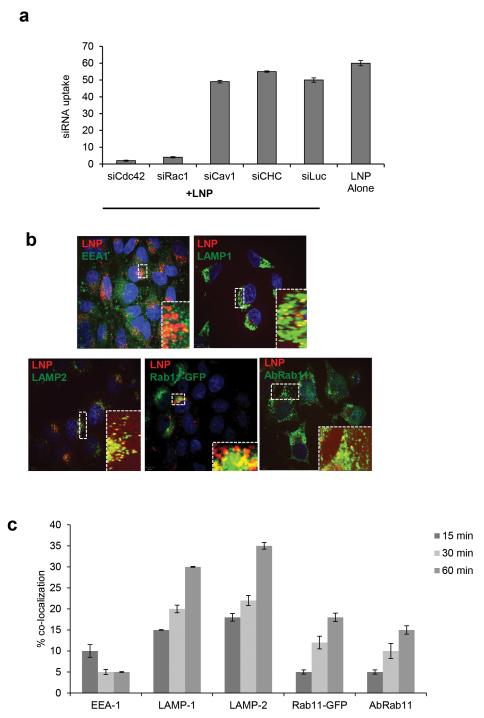
To identify internalization pathways that are required for cationic LNP entry into cells we depleted key endocytic regulators using siRNA in HeLa cells. Down-regulation of Cdc42 and Rac1 (regulators of macropinocytosis) led to ca. 80% decrease in LNP uptake whereas inhibition of clathrin heavy chain-1 and caveolin-1 (regulators of clathrin and caveolae mediate endocytosis, respectively) had little impact on LNP entry (Fig 2a). Further transport of LNPs to select endocytic compartments in these cells was analyzed through co-localization studies based on endocytic markers. First, LNP initial entry through macropinocytosis was confirmed through strong localization with Cdc42-GFP and ovalbumin positive vesicles (both markers for macropinocytosis) as compared to that with Arf6-GFP positive vesicles (clathrin-and dynamin-independent pathways) (Fig S3a). Second, image based kinetics of LNP delivery to the general endo/lysosomal system revealed little co-localization of LNPs with EEA-1 or Rab5-RFP (early endosome markers) (post 3 hr incubation, at multiple time chase points, ca. 5-10% co-localization) but a steady increase in co-localization with LAMP-1, LAMP-2, Rab7-GFP and Lysotracker positive vesicles (late endosome/lysosome marker) starting as early as after 15 min chase and showing a steady increase to around 50% localization with late endosomes/lysosomes after 60 min (Fig 2b-c, Fig S3b-c). Notably, after 60 min a fraction of the labeled siRNA starts to show co-localization with markers of the tubulovesicular endocytic recycling compartment (ERC) (Rab11-GFP, antibody against Rab-11, transferrin) whereas a decrease in localization with lysosomal positive vesicles was observed (Fig 2b-c, Fig S3c). Inhibition of Rab11 (Rab11 siRNA and dominant negative (DN)-Rab11) to interfere with early endocytic recycling causes a 1.5 fold reduction in LNP internalization (Fig S3d-e). This decreased internalization may be a result of reduced endocytic recycling of trafficking regulators required for LNP entry.
Figure 2. Cellular trafficking of LNPs.
a. Quantitative image analysis of siAF647-LNP cellular uptake (3 hrs) in HeLa cells silenced with siRNA against key endocytic regulators (Cdc42, Rac-1, Clathrin heavy chain (CHC), caveolin-1 (Cav-1). siRNA against luciferase serve as a negative control. (b-c) Image based quantitative analysis of siAF647-LNPs co-localization (3 hrs pulse, 15, 30, 60 min. chase) with markers of endocytosis, anti-EEA-1 (early endosomes), anti-LAMP-1 (late endosome/lysosomes), anti-LAMP-2 (late endosomes/lysosomes), Rab11-GFP and anti-Rab11 (both mark endocytic recycling compartment). High-resolution z-stack confocal images representative of 60 min chase are presented in different panels (b). Quantification of LNP-siRNA cellular uptake after indicated times (a, c).
Autophagy, another catabolic process which routes cytosolic proteins to the lysosomes23,24, was apparently not involved in LNP-mediated gene silencing as LNPs showed little co-localization with markers of autophagy (Fig S4a-b). Autophagy deficient cells (Atg5−/−)24 showed little if any differences in LNP mediated gene silencing when compared to autophagy competent (Atg5+/+) cells (Fig S4c).
Overall, C12-200-based cationic LNPs utilize macropinocytosis to enter cells and bypass the early endosomes; a small fraction is directed to the ERC while most LNPs traffic towards the late endosome from where they are routed to the lysosomes. Our results are consistent with previous studies that cationic LNPs utilize distinct mechanisms of entry when compared to ionizable LNPs, which internalize through ApoE dependent entry mechanisms16,25.
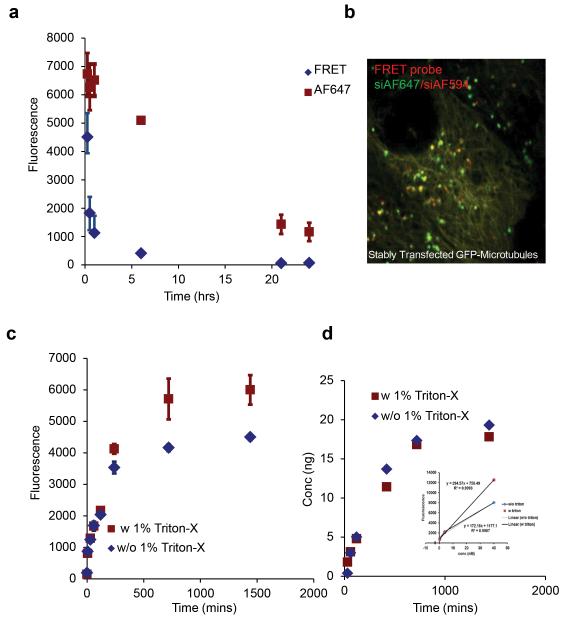
In addition to the moderate co-localization of LNPs with ERC, our imaging studies also indicated a considerable redistribution of LNPs in the presence of wide variety of compounds from the screen that cause stress on the endoplasmic reticulum. (Supplemental Information Small Molecule Library). Therefore, we wanted to quantify the kinetics of LNP disassembly and the relative contributions of internalization and recycling on siRNA delivery using a FRET probe26. The probe contains two identical siRNA’s, each labelled with a different fluorophore that together form a FRET pair. The siRNA-bound fluorophores aggregate with the cationic lipids in the nanoparticle mixture and undergo FRET, which is indicative of intact LNPs. Flow cytometery data shows a time dependent decrease in FRET based fluorescence signal within 1 hour of internalization, which was indicative of LNP disassembly inside cells (Fig 3a). However, the fluorescence emanating from a single fluorophore (AF647) remained constant during this time. A decline in AF647 signal was observed only after 1-4 hrs which substantially diminished after 24 hrs. Therefore, LNPs disassemble rapidly within the first hour of entry followed by degradation or recycling of siRNA within 24 hrs (Fig 3a). Labelled siRNA was located in vesicular compartments alongside microtubules 1 hour post incubation; indicating that, most siRNA’s remain trapped inside endosomes after nanoparticle disassembly (Fig 3b).
Figure 3. Quantitative analysis for disassembly and recycling of LNPs.
a. HeLa cells were exposed to LNP with the FRET pair (AF647/AF594 siRNA), washed and replenished with media. Changes in FRET (excitation-561 nm, emission-641 nm) intensity were monitored at different time points using flow cytometery to quantify LNP disassembly. The emission from a single fluorophore (excitation-633 nm, emission-641 nm) was used to measure intracellular siRNA at these time points. b. Cellular uptake of FRET probe-LNP (siAF647/siAF594, 3hrs) was imaged in stably transfected GFP-tubulin cells 1 hour post incubation c. siAF647-LNP was pulsed for 3 hrs, washed and incubated with fresh media to remove non internalized particles. Media was removed at multiple time points and analyzed using a fluorescent reader to determine amount recycled. 1% Triton-X was later added (red curve) to the media and fluorescence was re-measured. d. Amount of siRNA was obtained from fluorescence values from (c) that were extrapolated using the standard curve (inset).
To show that the reduction of intracellular siRNA-linked fluorescence was due to recycling of the siRNA to the extracellular media, we measured the fluorescence in the supernatant at multiple time points post transfection. We found an increase in the fluorescence intensity that appeared within 6 hrs post incubation and reached saturation at 24 hrs in the extracellular milieu (Fig 3c). Because intact nanoparticles partially quench fluorescence from the siRNA encapsulated inside them, we exposed the supernatant to Triton-X to dissociate any intact LNPs. Exposure to Triton-X resulted in little changes in fluorescence. This suggested that most LNPs had, in fact, disassembled inside cells, confirming our FRET based data. Approximately 36% of the initial amount of siRNA incubated with the cells recycled into the supernatant and 2.5 times (70%) the amount of the stable siRNA was present in the supernatant relative to inside cells after 24 hrs (Fig 3d, Fig S5).
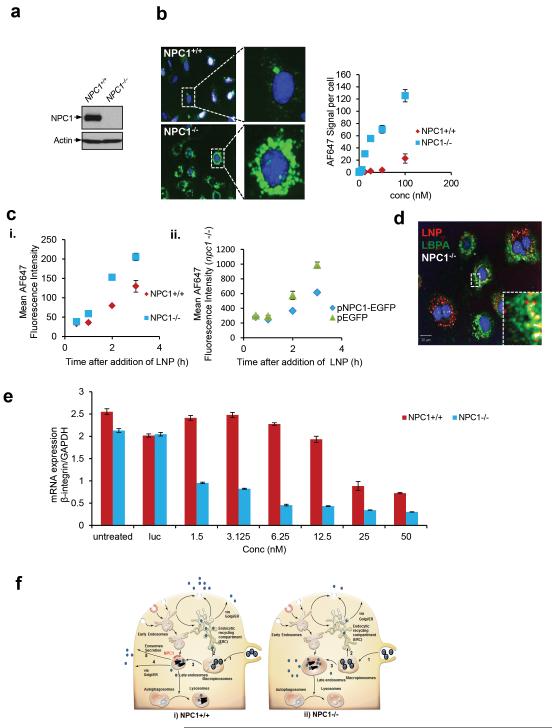
Subcellular trafficking of lipids (e.g. cholesterol) from late endosomes/lysosomes towards the extracellular milieu has been reported to utilize the thirteen transmembrane glycoprotein NPC1 which is located on the surface of multivesicular late endosomes. Absence of NPC1 causes late endo/lysosomal dysfunction, cholesterol accumulation and is implicated in a lysosomal storage disease that causes liver and neural degeneration in human patients and animal models27–29. As the components of an LNP show some similarities to the endogenous lipids that use NPC1 as a receptor for recycling, we compared LNP retention in mouse embryonic fibroblasts (MEFs) devoid of NPC1 (NPC1−/−) with their wild type counterparts (NPC1+/+ MEFs) (Fig 4a). NPC1 deficient cells accumulate cholesterol due to defects in recycling (Fig S6). A markedly increased level (approximately 15 fold) of AF647-siRNA was observed in perinuclear enlarged endosomes in NPC1−/− compared to NPC1+/+ MEFs at 24 hrs post incubation, presumably due to enhanced cellular retention at a wide range of LNP concentrations (Fig 4a-b). Furthermore, enhanced cellular retention was observed in multiple cell types deficient in NPC1 including those isolated from human patients (FigS7a-b). We further tested for differences in the kinetics of LNP uptake in these cells. NPC1−/− cells show increased accumulation of siRNA after 2 hour of incubation as compared to NPC1+/+ cells where at early time points the amount of internalization is not effected. Moreover, a rescue experiment where we transfected an NPC1-GFP plasmid into NPC1−/− cells reduced the amount of intracellular LNP-siRNA to that of wild type cells (4c (i-ii)). We conclude that the increased level of siRNA in late endosomes is based on accumulation of LNPs due to defects in constitutively active NPC1 mediated recycling. Small molecules that impair cholesterol metabolism28 and increase cholesterol accumulation in cells failed to yield a significant increase in LNP accumulation (Fig S7c). Thus, the increased accumulation of LNPs is not due to just increased endosomal cholesterol but rather due to lack of direct interaction with putative intraluminal domains of NPC1 required for egress of LNPs. NPC1−/− cells have been reported to have high numbers of enlarged late endosomes, containing intraluminal vesicles enriched in lysobisphosphatidic acid (LBPA)29. LNPs accumulate in these structures and showed co-localization with antibody against LBPA in these cells (Fig 4d).
Figure 4. Enhanced cellular retention and efficacy of siAF647-LNPs in NPC1 deficient MEFs.
a. Immunoblot analysis with anti-NPC1 antibody was used to validate wild type and NPC1 deficient MEFs. b. Automated confocal microscopy on NPC1+/+ or NPC1−/− MEFs exposed to different concentrations of labeled LNPs and imaged 24 hrs post incubation. Inset from a representative image (100 nM) shows siRNA accumulation in individual cells. c. Flow cytometery analysis on LNP-siRNA uptake in (i) NPC1+/+ or NPC1−/− cells or in (ii) NPC1−/− cells transfected with pEGFP or pNPC1-GFP. The mean fluorescent intensity represents LNP uptake. d. NPC1−/− cells treated with LNP-AF647-siRNA (red) (3 hrs pulse, 30 min chase) and immuno-stained with anti-LBPA antibody (green) e. LNPs containing siRNA against β integrin were added to wild type or NPC1 deficient MEFs as in (a), mRNA levels were quantitated at 24 hrs post incubation using branched DNA analysis. The experiment was done in triplicate and the errors are reported as S.E.M. f. A schematic representation of LNP trafficking (i) in NPC1+/+ and (ii) NPC1−/− cells. Intact cationic LNPs enter through macropinocytosis (1); a small fraction of LNPs transport from macropinosomes to the endocytic recycling compartment (ERC) (2) while the majority is directed to late endosomes (3). Late endosome sort LNPs to lysosomes for degradation or utilize multiple recycling pathways to traffic them to the extracellular milieu. These mechanisms include recycling through transport to the ER-Golgi route (4) or direct fusion of late endosomes containing multivesicular bodies, with the plasma membrane (Exosomes secretion) (5). In NPC1 deficient cells the late endosome recycling mechanisms are impaired causing LNP-siRNA to accumulate in enlarged late endosomes leading to persistent escape of siRNA that improves gene silencing. (Intact nanoparticle- , siRNA complex-
, siRNA complex- )
)
To study the broader potential relationship between lipid structure and NPC1-dependent LNP retention, we tested cationic siAF647-LNPs formulated with a small set of lipidoids consisting of two different amine cores, each with lipid tails of increasing chain lengths (Fig S-Reaction Scheme). Different LNPs containing related cationic lipids also showed increased retention in NPC1-deficient cells. We also found that increased carbon chain lengths of these cationic lipids led to better uptake and retention in both NPC1+/+ and NPC1 −/− MEFs (Fig S8a-b). This marked retention is consistent with studies that show that lipids with higher chain length can insert inside NPC1 carrying late endosome membranes30. Notably, liposomes containing small molecules showed similar increases in cellular retention in knock out cells (Fig S8c).
To determine whether the absence of NPC1 also leads to increased retention with polymeric nanoparticles, we tested polystyrene nanoparticles of different sizes and charges. We found a moderate increase in retention and perinuclear distribution with positively and negatively charged particles at sizes of 100 or 200 nm in NPC1 −/− cells whereas negatively charged particles 20 nm in size (known to use caveolae mediated pathway31) showed similar retention when compared to wild type cells (Fig S8d). We found that dextran, a marker for macropinocytosis, shows increased retention in KO cells as compared to markers of caveolae or clathrin-dependent endocytosis (data not shown). Thus, 20 nm particles that can access 80 nm caveolae5,31 due their smaller size are able to be retained inside both cell types (NPC1 −/− and NPC1 +/+) when compared to larger particles that may only utilize macropinocytosis to gain entry. It is possible that macropinosomes directly fuse with NPC1 positive late endosomes and are recycled to the extracellular milieu, in the absence of which the cargo remains diverted to the endolysosomal system. It is believed that in the case of certain types of cargo, NPC1 independent recycling allows for partial escape from the endo-lysosomal system30. This may not be the case with longer chain lipids that require interaction with NPC1 for cellular exit30. This may be the reason for the major retention of LNPs as compared to polystyrene nanoparticles.
We hypothesized that enhanced cellular retention could translate to improved gene silencing. LNPs containing β-integrin siRNA were added to either NPC1+/+ or NPC1−/− MEFs at different concentrations. A substantial improvement in LNP potency was observed in NPC1−/− cells in comparison to wild type cells (Fig 4e). The IC50 of 1.5 nM in NPC1 deficient cells was roughly 10 times lower than in wild type MEFs, and the improved potency was seen across all LNP doses tested (Fig 4e). To further measure cellular retention and siRNA efficacy, NPC1+/+ and NPC1−/− MEFs (stably transfected with GFP) were treated with LNPs containing GFP siRNA (siAF647-GFP); high-throughput microscopy was used to simultaneously measure cellular retention and GFP fluorescence 72 hrs post incubation in both cell types. Enhanced cellular retention of siAF647-GFP and a decrease in normalized GFP fluorescence in NPC1 −/− compared to that in NPC1+/+ cells were observed confirming our previous results (Fig S9a-c). Next, we utilized siRNA against NPC1 to experimentally lower NPC1 protein levels in wild type cells and a 60% reduction in protein level was achieved (Fig S9d-e). A 4-fold increase in LNP efficacy was observed in wild type cells treated with NPC1 siRNA when compared to the luciferase control (Fig S9f). Although this LNP efficacy is lower than what is achieved in NPC1−/− cells, 40% NPC1 protein still remains in NPC1+/+ cells treated with siNPC1 and this amount may still sufficient to recycle some LNPs, albeit at a less efficient rate. The use of lipids for transfection of NPC1 siRNA may alter intracellular trafficking or recycling and further explain why these effects were partial with siRNA knockdown versus the effect seen in complete NPC1−/− MEFs.
This led us to further investigate which endocytic recycling pathways participate in exocytosis of LNPs, and in particular whether inhibition of these pathways could enhance cellular retention and LNP mediated gene silencing. Late endosomes have been shown to exchange material with the Golgi/ER from where the cargo can be secreted to the extracellular milieu32,33 and/or multivesicular bodies that can fuse with the plasma membrane to release their intracellular content as exosomes34. Therefore, we examined the effects of siRNA that blocks secretion from the Golgi/ER (Rab8a)32 or exosomes secretion and/or fusion with the plasma membrane (Rab27a and Rab27b)34. Rab8a and Rab27b siRNA treatment led to significant increase in retention of LNP in cells. A moderate increase in LNP retention was observed after reduction of Rab27a expression (Fig S10 a-f). Notably, only in the case of Rab27b depletion did we detect a substantial improvement in LNP mediated gene silencing (Fig S10 g). Recent reports have shown that Rab27b inhibition leads to redistribution late endosomes in the perinuclear region34. Concurrently, in our experiments both Rab27b-depleted and NPC1-deficient cells show similarly increased perinuclear retention of LNPs which contains functional siRNA that contributes to enhanced gene silencing. On the other hand siRNA against Rab11 did not improve retention and gene silencing (Fig S10 h-i), these results are consistent with previous reports that NPC1-mediated recycling of endogenous lipids is independent of Rab11 function even though they show some co-localization with Rab11 positive ERC35.
In summary, to enter cells, cationic LNPs require Cdc42 dependent macropinocytosis which is shown to be regulated through VoATPase, once internalized remain largely trapped within endosomes with little escape to the cytosol. LNP-mediated siRNA delivery can be impaired by either lack of LNP disassembly inside endosomes or their subsequent degradation in the lysosomes as well as recycling from the cell. In the absence of NPC1, due to decrease in motility of late endosomes29, LNPs remain in enlarged late endosomes/lysosomes and are not recycled back out of the cell. One hypothesis to explain how this leads to a greater efficiency in siRNA-mediated gene silencing is that the increased residence time of more siRNA inside cells results in a slow controlled diffusion of the siRNA into the cytoplasm (Fig 4f (i-ii)). It appears that late endosome/multivesicular late endosomes may serve as a transient reservoir where LNP recycle through multiple pathways. Prevention of LNPs direct egress from the late endosomes leads to enhanced retention and perinuclear positioning of these vesicular structures may serve as a persistent site for endosomal escape. Future studies that target proteins and lipids involved in biogenesis, trafficking and recycling of the late endosomes may provide further clues into the cellular factors that may improve drug delivery. Finally, the RNA-induced silencing complex (RISC) has been shown to be associated with membranes of late endosomes, and certain RISC complex components can be seen inside intraluminal vesicles of late endosomes/exosomes as well as the cytosolic side of the perinuclear rough endoplasmic reticulum 36–39. It is possible that, in NPC1 deficient as well as Rab27b depleted cells, LNPs remain stalled in a manner that affects siRNA delivery to RISC. From a therapeutics perspective, it is tempting to speculate that LNPs may demonstrate enhanced siRNA/drug delivery in lysosomal storage diseases where NPC1 function is impaired.
Supplementary Material
Acknowledgments
The authors wish to dedicate this work in the memory of MIT police Officer Sean Collier who valiantly gave his life for the protection of MIT community. The authors would like to thank Dr. Peter Lobel and Liang Huang from Rutgers University for NPC1 primary and immortalized cell lines. We would also like to thank Dr. Dennis Brown (Harvard MGH), Dr. Kathryn Whitehead, Dr. Roman Bogorad (MIT) and Dr. Hao Yin (MIT) for healthy discussion. We would also like to thank Dr. John Maraganore (Alnylam) and Dr. Michael Invernale (MIT) for critical reading of the manuscript. Special thanks to Daria Alakhova at University of Nebraska Medical Center for her help with graphic design. We would also like to thank Alnylam Pharmaceuticals, Control release grant EB000244 for funding. This work was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health, as a Program of Excellence in Nanotechnology (PEN) Award, Contract #HHSN268201000045C.
Footnotes
Authors Contribution: G.S, R.L and D.G.A conceived the idea, G.S, W.Q, R.L. and D.G.A designed research, G.S, W.Q and C.Z performed high throughput microscopy. G.S, C.A, A.E, K.L., D.C and A.S designed LNPs and their probes, G.S, C.A, A.E, S.S. and Y.B performed studies with NPC1 deficient and competent cells, G.S and S.S performed studies with autophagy based studies, G.S and R.Z performed TIRF microscopy, G.S. and E.K performed systems biology, G.S, W.Q, R.L. and D.G.A wrote the manuscript.
Conflict of interest statement: R.L. is a shareholder and member of the Scientific Advisory Board of Alnylam. D.G.A. is a consultant with Alnylam. R.L and D.G.A have sponsored research grants from Alnylam. W.Q. and C.Z. are employed by Alnylam.
Material and Methods: Included in the supplemental section of the paper.
References
- 1.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheridan C. Proof of concept for next-generation nanoparticle drugs in humans. Nature Biotechnology. 2012;30:471–473. doi: 10.1038/nbt0612-471. [DOI] [PubMed] [Google Scholar]
- 5.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release. 2010;145:182–195. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Byrne JD, Napier ME, DeSimone JM. More effective nanomedicines through particle design. Small. 2011;7:1919–1931. doi: 10.1002/smll.201100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juliano RL, Ming X, Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug. Chem. 2012;23:147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonawane ND, Szoka FC, Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. J. Biol. Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 9.Basha G, et al. Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells. Mol Ther. 2011;19:2186–200. doi: 10.1038/mt.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suh J, et al. Real-time gene delivery vector tracking in the endo-lysosomal pathway of live cells. Microsc. Res. Tech. 2012;75:691–697. doi: 10.1002/jemt.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu JJ, Langer R, Chen J. A novel mechanism is involved in cationic lipid-mediated functional siRNA delivery. Mol. Pharm. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen J, Szoka FC. Nucleic Acid Delivery: The Missing Pieces of the Puzzle? Accounts of Chemical Research. 2012 doi: 10.1021/ar3000162. doi:10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collinet C, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- 14.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 15.Chen D, et al. Rapid Discovery of Potent siRNA-Containing Lipid Nanoparticles Enabled by Controlled Microfluidic Formulation. J. Am. Chem. Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 16.Love KT, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leuschner F, et al. Therapeutic siRNA silencing in inflammatory monocytes. Nat Biotechnol. 2011;29:1005–1010. doi: 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zoncu R, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011 doi: 10.1038/nature10348. doi:10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalil IA, Kogure K, Akita H, Harashima H. Uptake Pathways and Subsequent Intracellular Trafficking in Nonviral Gene Delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 21.Huss M, et al. Concanamycin A, the specific inhibitor of V-ATPases, binds to the V(o) subunit c. J. Biol. Chem. 2002;277:40544–40548. doi: 10.1074/jbc.M207345200. [DOI] [PubMed] [Google Scholar]
- 22.Bayer N, et al. Effect of bafilomycin A1 and nocodazole on endocytic transport in HeLa cells: implications for viral uncoating and infection. J. Virol. 1998;72:9645–9655. doi: 10.1128/jvi.72.12.9645-9655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravikumar B, et al. Regulation of Mammalian Autophagy in Physiology and Pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 24.Sarkar S, et al. Complex Inhibitory Effects of Nitric Oxide on Autophagy. Molecular Cell. 2011;43:19–32. doi: 10.1016/j.molcel.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akinc A, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alabi CA, et al. FRET-labeled siRNA probes for tracking assembly and disassembly of siRNA nanocomplexes. ACS Nano. 2012;6:6133–6141. doi: 10.1021/nn3013838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nature Reviews Molecular Cell Biology. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 28.Ko DC, Gordon MD, Jin JY, Scott MP. Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol. Biol. Cell. 2001;12:601–614. doi: 10.1091/mbc.12.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang M, et al. Cessation of Rapid Late Endosomal Tubulovesicular Trafficking in Niemann–Pick Type C1 Disease. PNAS. 2001;98:4466–4471. doi: 10.1073/pnas.081070898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koivusalo M, Jansen M, Somerharju P, Ikonen E. Endocytic Trafficking of Sphingomyelin Depends on Its Acyl Chain Length. Mol. Biol. Cell. 2007;18:5113–5123. doi: 10.1091/mbc.E07-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai SK, Hida K, Chen C, Hanes J. Characterization of the intracellular dynamics of a non-degradative pathway accessed by polymer nanoparticles. Journal of Controlled Release. 2008;125:107–111. doi: 10.1016/j.jconrel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linder MD, et al. Rab8-dependent Recycling Promotes Endosomal Cholesterol Removal in Normal and Sphingolipidosis Cells. Mol Biol Cell. 2007;18:47–56. doi: 10.1091/mbc.E06-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-5-reviews3007. REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostrowski M, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 35.Hölttä-Vuori M, Tanhuanpää K, Möbius W, Somerharju P, Ikonen E. Modulation of cellular cholesterol transport and homeostasis by Rab11. Mol. Biol. Cell. 2002;13:3107–3122. doi: 10.1091/mbc.E02-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 37.Lee YS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat. Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siomi H, Siomi MC. RISC hitches onto endosome trafficking. Nat. Cell Biol. 2009;11:1049–1051. doi: 10.1038/ncb0909-1049. [DOI] [PubMed] [Google Scholar]
- 39.Stalder L, et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013 doi: 10.1038/emboj.2013.52. doi:10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.