Abstract
RNA interference (RNAi) is a highly specific gene-silencing mechanism triggered by small interfering RNA (siRNA). Effective intracellular delivery requires the development of potent siRNA carriers. Here, we describe the synthesis and screening of a series of siRNA delivery materials. Short polyethyleneimine (PEI, Mw 600) was selected as a cationic backbone to which lipid tails were conjugated at various levels of saturation. In solution these polymer–lipid hybrids self-assemble to form nanoparticles capable of complexing siRNA. The complexes silence genes specifically and with low cytotoxicity. The efficiency of gene knockdown increased as the number of lipid tails conjugated to the PEI backbone increased. This is explained by reducing the binding affinity between the siRNA strands to the complex, thereby enabling siRNA release after cellular internalization. These results highlight the importance of complexation strength when designing siRNA delivery materials.
Keywords: Lipid, Conjugation, Complexation, Affinity, Polyethyleneimine (PEI), Cationic
1. Introduction
Since its discovery by Fire and Mello [1], RNA interference (RNAi) has been studied pre-clinically for its potential to silence genes that are associated with cancer [2,3], infection and inflammation [4], respiratory and neurological disorders [5,6], and autoimmune diseases [7]. Clinical investigations of the therapeutic potential of RNAi are currently under evaluation [8,9]. However, additional advances are needed to make broad application of siRNA. This is explained by a series of hurdles that make delivery difficult. Carriers must bind sufficient amounts of siRNA, protect it from degradative enzymes, cross the anionic cell membrane and release the payload into the cytoplasm [3]. Moreover, the carrier must perform these tasks without damaging the cell. To overcome these hurdles, diverse delivery systems have been developed. Some of the most effective have been synthesized from cationic polymers, lipids, and lipid-like materials [10-13].
Amine-based cationic materials have been effective in binding and delivering nucleic acids. Of these, one of the most rigorously studied is polyethyleneimine (PEI). PEI was first used as a delivery vehicle for long nucleic strands due to its capacity to form condensed and stable structures with DNA and RNA [14]. These polyplexes are taken up by cells via endocytotic pathways [15,16] before being released to the cytosol [16,17]. However, toxicity and efficacy have both been reported to increase as the molecular weight of PEI increases [18-20]. More specifically, 25 kDa molecular weight PEI, which has been used in many studies, is effective, but is toxic to multiple cell lines [19]. Several approaches have been employed to reduce the toxicity associated with high molecular weight PEI [21,22]. For example, biodegradable bonds were inserted into the polymer backbone [23-25]. It was proposed that the long polymer would deliver nucleic payloads effectively and then degrade into well-tolerated short PEI fragments [26]. Another approach is to conjugate chemical groups, such as lipids, to high Mw PEI, thereby reducing toxicity [27-30].
Here, we sought to study the ability of low molecular-weight PEI (0.6 kDa) modified with lipid tails to act as siRNA delivery materials. Our hypothesis was that these short lipid-polymer hybrids would be well tolerated. Furthermore, we tested the effect of the carriers’ molecular structure on their capacity to induce RNAi. For this, lipid tails were conjugated to the amine backbone at increasing levels of saturation, and the expression of a targeted gene was measured. We found that increasing the degree of saturation, up to a point at which the complex was not stable in solution, was most effective for siRNA delivery.
2. Materials and methods
2.1. Lipid-modified PEI
Polyethyleneimine, ethylendiamine end capped, Mw 600 (Aldrich, Natick, MA) was conjugated to 1,2-Epoxyoctadecane, MW 268.49 (Alfa Aesar, Ward Hill, MA) via an epoxide ring-opening reaction as shown schematically in Fig. 1A and described previously in detail in [11]. In brief, epoxide-terminated C18 alkanes were dissolved in 200-proof ethanol and reacted with PEI at 90 °C for 48 h. The epoxide: PEI molar ratios were varied from 0:1 up to 3:1; above this ratio the product formed unstable complexes with siRNA in water. The product was dissolved in deuterated DMSO and characterized by 1H-NMR (400 MHZ, Bruker, Billerica, MA) (Fig. S2), structures coincide with previous publications on similar compounds [31-33]. In addition, the product was characterized using Ab42 matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry using a Microflex (Bruker Daltonics, Billerica, MA, USA) equipped with a pulsed nitrogen laser operating at 337 nm (Fig. S3). Small oligomer positive ion spectra were acquired in reflector mode over an m/z range from 500 to 6000 using a 20-kV accelerating voltage and a 150-ns delay extraction time. The spectrum for each spot was obtained by averaging the results of 200 laser shots. In brief, the compounds were dissolved in pure ethanol at 5 mg/mL, 1 μl was mixed with 100 μl matrix (25 mg dihydroxybenzoic acid (Sigma-Aldrich), 500 μl methanol, 500 μl H2O); 1 μl was spotted on the grid and let to dry.
Fig. 1.
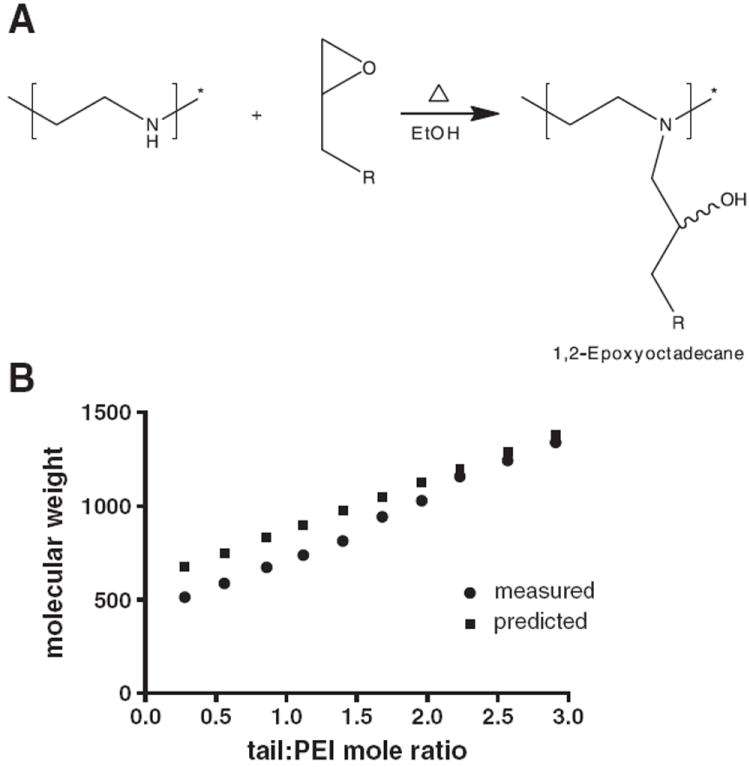
Conjugating lipid tails to PEI via an epoxide ring opening reaction. (A) A schematic representation of the chemical reaction. Epoxide terminated alkane was dissolved in ethanol and reacted with PEI at 90 °C for 48 h to form lipid–PEI hybrids. (B) The molecular weight of alkane-modified polyethyleneimine as a function of lipid:PEI mole ratio was determined using matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-ToF MS).
Upon hydration, these compounds self-assembled to form nanoparticles in solution. Particles were sized using dynamic light scattering with a ZetaPALS Zetasizer (Brookhaven Instruments, Vienna, Austria). Zeta potential was measured in either 25 mM sodium acetate, pH 5.3, or 0.1X PBS, pH 7.4, using a 90Plus particle size analyzer (Brookhaven Instruments).
TEM images were obtained with a JEOL 200CX electron microscope operated at 150 kV. Samples were prepared on carbon film coated TEM grid and subsequently stained with a 1% sodium phosphotungstate (PTA) aqueous solution.
2.2. RNAi screening
RNAi was tested using HeLa cells transfected to express both Renilla and firefly luciferase (a gift from Alnylam Pharmaceuticals, Cambridge, MA) [12].
The compounds were complexed with anti-firefly luciferase siRNA (siLuc) (Dharmacon, Boulder, Colorado) at an N/P ratio of 14 by incubating with siRNA in 25 mM Na-acetate (pH 5.2).
Knockdown was measured using the Dual Glo Assay (Promega, Madison, WI), which quantifies the expression of firefly (target gene) and Renilla (control gene) separately, thereby calibrating for off-target effects and toxicity [12]. In brief, 15,000 HeLa cells were plated in a 96-well plate and cultured in 90% DMEM (high glucose, w/ l-glutamine, 25 mM HEPES, w/o sodium pyruvate; Invitrogen, Carlsbad, CA) 10% FBS (Invitrogen) at 37 °C under 5% CO2. Twentyfour hours later 20 nM siRNA complexes was added to each well. The system was incubated overnight and firefly and Renilla gene expression were quantified.
Knockdown was compared to the levels of luciferase expression in untreated cells. Lipofectamine-RNAiMax (Invitrogen) at was used as a positive control.
Cellular uptake was examined using Cy5 labeled siRNA (AllStars-Negative Control, Qiagen, Valencia, CA) with a Perkin Elmer Spinning Disk Confocal Microscope. For this, 50,000 HeLa cells per well were plated in chambered glass coverslips overnight. Cells were then exposed to labeled nanoparticle/siRNA complexes for 1 h, washed with PBS, and incubated with Hoescht (2 μg/ml) for nuclear stain.
2.3. siRNA binding affinity
Anti-luciferase siRNA was complexed with the compounds, as described above. Then, Na-heparin (Sigma) was added at various concentrations, from 0.0002 to 0.075 mg/mL, and incubated with the complexes for 10 min. The solutions were developed using a 4% agarose EGel system (Invitrogen) to detect complexation affinity.
2.4. DNA transfection
After aspirating conditioned medium, cells were washed with PBS and detached using 25 μL 0.25% trypsin-EDTA (Invitrogen) per well. 50 μL of FACS running buffer, consisting of 98% PBS, 2% FBS, and 1:200 propidium iodide solution (Invitrogen), was added to each well. Cells were mixed thoroughly and then transferred to a 96-well round-bottom plate. GFP expression was measured using FACS on a BD LSR II (Becton Dickinson, San Jose, CA, USA). Propidium iodide (PI) staining was used to exclude dead cells from the analysis. 2D gating was used to separate increased auto-fluorescence signals from increased GFP signals to more accurately count positively expressing cells. Gating and analysis were performed using FlowJo v8.8 software (TreeStar, Ashland, OR, USA).
3. Results
This study examines the potential of alkane-modified low molecular weight PEI as siRNA delivery materials.
Sixteen-carbon-long lipid tails were conjugated to PEI at increasing levels of saturation. When immersed in aqueous solutions these compounds self-assembled to form nanoparticles. As the number of tails conjugated to the PEI backbone increased, the size of the self-assembled particles increased too (Fig. 2A). When having, on average, between one and two conjugated lipid tails per PEI molecule the compounds self-assembled into ~10-nm particles in solution. These particles were capable of encapsulating the water-insoluble hydrophobic drug sorafenib at a 1:5 wt ratio (sor:PEI), thereby suggesting that the particles have a lipid core [34]. Above two lipid tails conjugated per PEI backbone, the particles assume a larger size in solution. Conjugating 3 or more lipid tails per PEI molecule resulted in compounds that aggregated and precipitated in solution. At pH 5.3, from a lipid:PEI mole ratio of 0.56 the particles had a similar positive zeta potential (~52 mV), enabling efficient siRNA complexation, while at physiological pH, the zeta increased slightly around neutrality as the number of lipid tails increased (Fig. 2B).
Fig. 2.
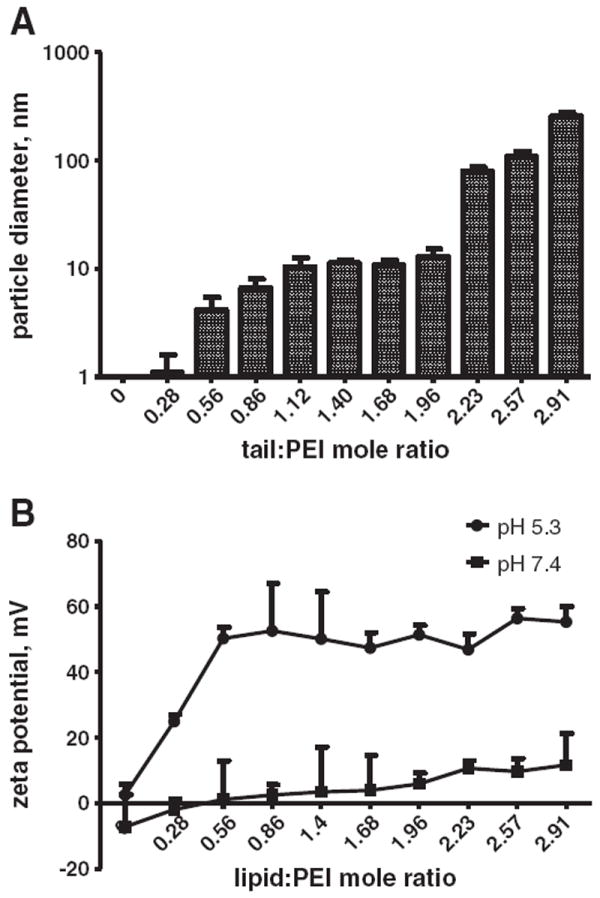
The spontaneous self-assembled mean particle size and zeta potential of lipid–PEI compounds, as a function of the lipid-tail:PEI mole ratio. (A) As the number of conjugated lipid tails increases — the particle size increases. The polydispersity of the particles was <0.1. (B) The Zeta potential of the compounds was measured at pH 7.4 and 5.3.
When complexed with anti-luciferase siRNA the compounds induced RNAi in HeLa cells expressing both Renilla and firefly luciferase (Fig. 3A). Compounds having less than two conjugated lipid tails per PEI molecule were significantly less effective in inducing RNAi (<30% knockdown) in comparison to those having more conjugated lipid tails (~80% knockdown). Using an unpaired t-test to compare each of the lipids that have more than two tails to those that have less than two tails, as well as using one-way ANOVA analysis of that variance, a statistically significant difference of p<0.001 was found. All the compounds were well tolerated by cells (<20% cell death or off-target silencing). The compounds were well tolerated by the cells (Fig. 3B).
Fig. 3.
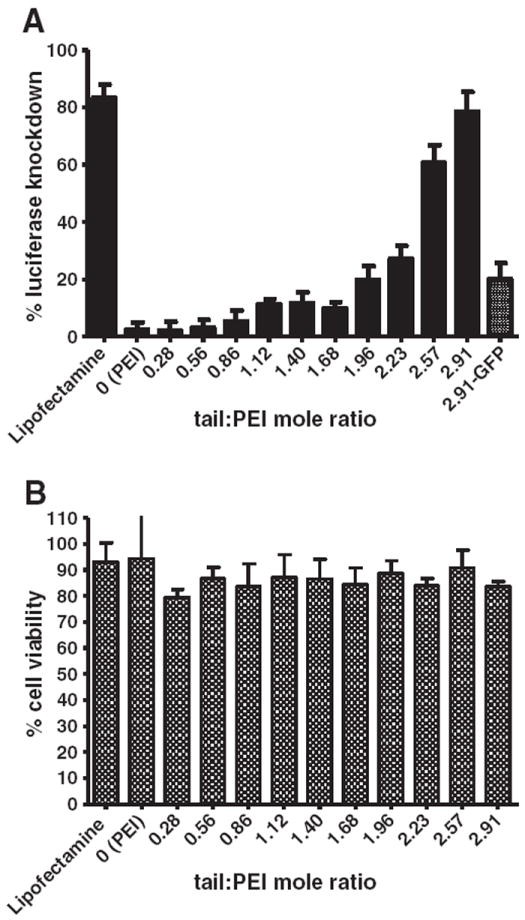
Gene knockdown capacity of alkylated PEI compounds. RNAi and cell viability were tested in HeLa cells that express both Renilla and firefly luciferase. (A) Lipid–PEI compounds were complexed with anti-firefly siRNA and then incubated with the cells. Mean knockdown levels increased as the number of lipid tails conjugated to the PEI increased. (B) Cell viability was measured by recording the expression of an off-target gene — Renilla luciferase and is relative to non-treated cells. In both graphs error bars represent the standard deviation (N=8).
Our initial hypothesis was that compounds that did not induce significant knockdown were not entering the cells effectively; however, confocal microscopy conducted using fluorescently labeled siRNA indicated that all the compounds were capable of delivering siRNA intracellularly (Fig. 4).
Fig. 4.
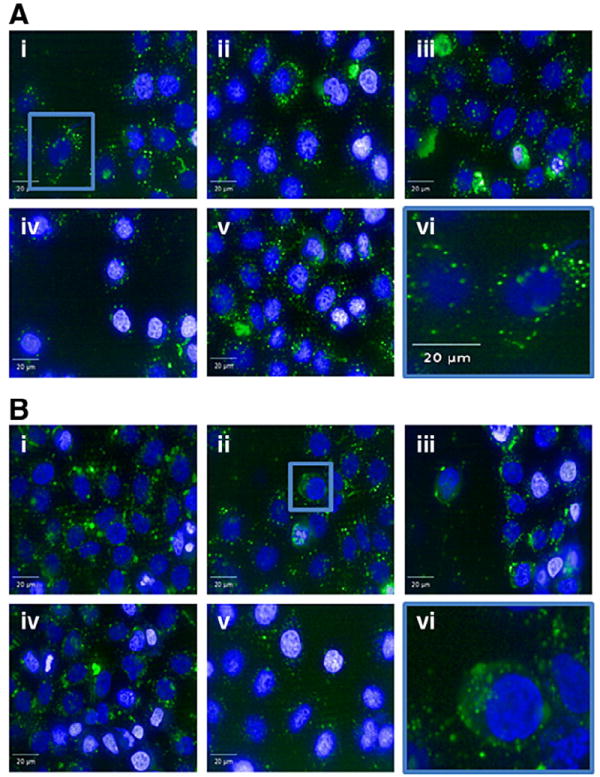
Cellular uptake of lipid–PEI–siRNA complexes. The uptake of lipid–PEI compounds complexed with Cy5-labled siRNA by HeLa cells was imaged using an Opera spinning disk confocal microscope. The uptake of the lipid:PEI mole ratio of 0.56 (A) is compared to that of lipid:PEI mole ratio of 2.91 (B). Five different representative images are presented for each compound (i–v) and image (vi) represents an enlarged image. Punctate structures can be noticed in panel A (vi), while cytoplasmic as well as punctuate structures can be seen in panel B (vi). Blue is Hoechst dye.
A different mechanism seemed to govern the superiority of some of the complexes in their ability to induce RNAi. We screened the different compounds for their binding affinity to siRNA. For this, each of the siRNA–PEI–lipid complexes was treated with Na-heparin (Fig. 5). Heparin encompasses the highest negative charge density of any known natural compound. When added to dispersions with nucleic polyplexes heparin can replace the nucleic acid, and thereby be used to titer the affinity between the gene and the complex [35]. Interestingly, as the degree of lipid-tails per PEI increased the affinity of siRNA to the complex decreased, suggesting that high affinity may inhibit siRNA release from the complex intracellularly, subsequently inhibiting RNAi (Fig. 6).
Fig. 5.
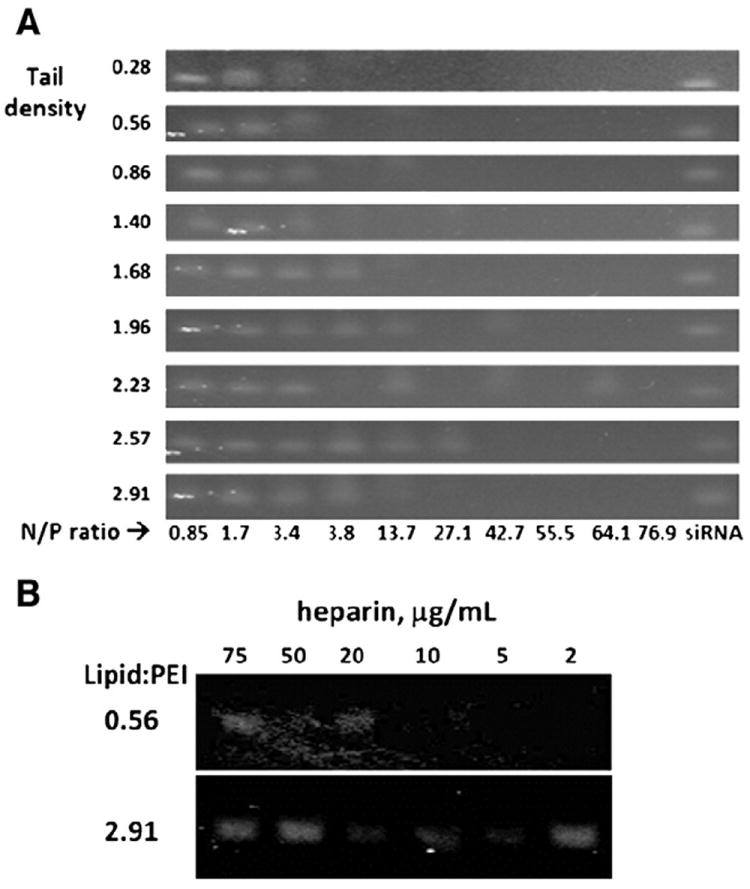
Binding affinity between alkane-modified PEI to siRNA. PEI with higher conjugation levels had weaker binding affinity to siRNA in comparison to lower conjugation levels. This was noticed when testing the affinity between modified PEI to siRNA at various N/P ratios (A), and after incubating siRNA-complexed particles with heparin sodium salt (Sigma) at various concentrations (B). Bands represent free (non-bound) siRNA as detected on an agarose gel.
Fig. 6.
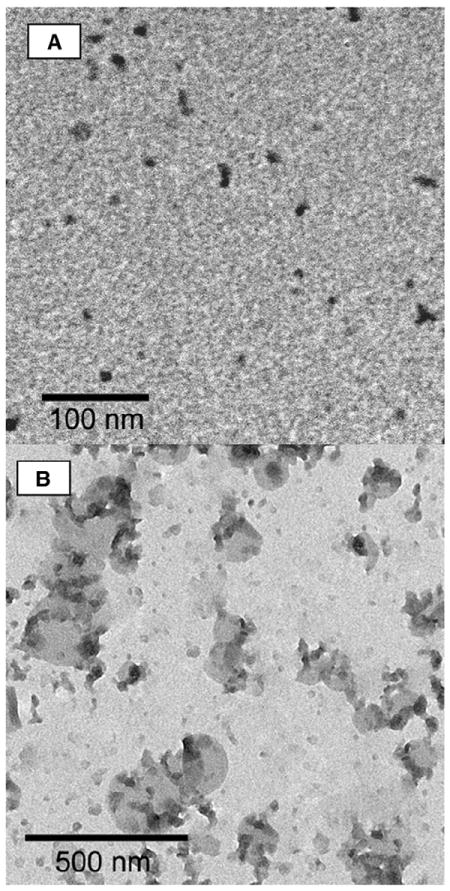
EM images of alkane-modified PEI particles. Alkane-modified PEI particles were stained with phosphotungstate and imaged by EM. (A) particles at a lipid:PEI ratio of 0.56 were clearly noticed on the grid; on the other hand, particles with a lipid:PEI ratio of 2.91 (B) were unstable during the imaging preparation procedure, although silhouettes of the particles could be noticed on the grid, and excess stained material was removed from the figure by false coloring.
Finally, we tested the ability of the most efficacious compounds to deliver green fluorescent protein (GFP)-encoding DNA plasmid. Transfection efficacy reached only 16.4±4.5%, suggesting that these compounds are more effective for siRNA delivery, rather than nuclear delivery of DNA. This was also suggested by the confocal microscopy studies that showed much of the carrier in cytoplasm (Fig. 4B).
4. Discussion
RNAi has broad clinical applications. Realizing the full potential of RNAi will require tailoring new delivery materials. Here, we tested the ability of short lipid-modified PEI to act as siRNA delivery materials.
Sixteen-carbon-long lipid tails were conjugated to a 0.6 kDa PEI backbone at various levels of saturation. While the compounds were well tolerated, capable of complexing siRNA and facilitated cellular uptake, we found a structure–function relationship to occur. As the number of lipid tails attached to the PEI backbone increased, RNAi increased. The increased number of lipid tails per PEI backbone also translated into lower binding affinity between siRNA and the complex. Contrarily, compounds that were non-efficacious in inducing RNAi had high binding affinity to the complex. These data suggest that binding affinity between siRNA to the delivery material plays a role in facilitating RNAi. High binding affinity may not enable release of the nucleic cargo after entering the cell, compromising its ability to take part in the RNAi pathway.
The dependence of the size of the particles on the number of lipid tails conjugated to the PEI backbone was previously explained by different packing configurations of such polymer–lipid compounds [36-38]. Having one lipid tail per PEI backbone, the compounds self-assemble into micelle like structures [36-39] (Fig. 6A). At higher saturation levels the structure may be liposomal or more complex [36-39] (Fig. 6B).
In conclusion, this study suggests that engineering short amine-based polymer chains modified with lipid-like materials may prove to be a promising siRNA delivery strategy.
Supplementary Material
Acknowledgments
AS was supported by the Misrock Foundation. This work was supported by MIT-Harvard Center for Cancer Nanotechnology Excellence Grant U54 CA151884 from the National Cancer Institute and by NIH grant # EB000244. JD thanks the NDSEG, NSF and MIT Presidential Fellowships. We thank Natalia Schiller from the Biopolymers and Proteomics Core Facility at the David H. Koch Institute for Integrative Cancer Research at MIT for helping with mass spectrometry. The authors thank Alnylam Pharmaceuticals for its kind support.
Footnotes
Contributions: A.S., J.D. and D.G.A conceived the idea and wrote the manuscript.; A.S., J.D., K.T.L., C.G.L. performed chemical synthesis and analysis; K.T.L, Y.W., A.S. performed siRNA transfection and data analysis; A.A.E. performed DNA transfection, FACS and data analysis; and G.S., S.J. EM, performed microscopy and data analysis.
Appendix A. Supplementary data
Supplementary data to this article can be found online at doi:10. 1016/j.jconrel.2011.11.030.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689–696. doi: 10.1111/j.1349-7006.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponnappa BC. siRNA for inflammatory diseases. Curr Opin Investig Drugs. 2009;10:418–424. [PubMed] [Google Scholar]
- 5.Garbuzenko OB, Saad M, Betigeri S, Zhang M, Vetcher AA, Soldatenkov VA, Reimer DC, Pozharov VP, Minko T. Intratracheal versus intravenous liposomal delivery of siRNA, antisense oligonucleotides and anticancer drug. Pharm Res. 2009;26:382–394. doi: 10.1007/s11095-008-9755-4. [DOI] [PubMed] [Google Scholar]
- 6.Davidson BL, Paulson HL. Molecular medicine for the brain: silencing of disease genes with RNA interference. Lancet Neurol. 2004;3:145–149. doi: 10.1016/S1474-4422(04)00678-7. [DOI] [PubMed] [Google Scholar]
- 7.Prud’homme GJ. Gene Therapy of Autoimmune Diseases. Kluwer Academic / Plenum Publishers; New Delhi: 2005. [Google Scholar]
- 8.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cervantes A, Alsina M, Tabernero J, Infante JR, LoRusso P, Shapiro G, Paz-Ares LG, Falzone R, Hill J, Cehelsky J, White A, Toudjarska I, Bumcrot D, Meyers R, Hinkle G, Svrzikapa N, Sah DW, Vaishnaw A, Gollob J, Burris HA. Phase I dose-escalation study of ALN-VSP02, a novel RNAi therapeutic for solid tumors with liver involvement. J Clin Oncol. 2011;29:3025. [Google Scholar]
- 10.Schroeder A, Levins CG, Cortez C, Langer R, Anderson DG. Lipid-based nanotherapeutics for siRNA delivery. J Intern Med. 2010;267:9–21. doi: 10.1111/j.1365-2796.2009.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, Cantley W, Qin LL, Racie T, Frank-Kamenetsky M, Yip KN, Alvarez R, Sah DW, de Fougerolles A, Fitzgerald K, Koteliansky V, Akinc A, Langer R, Anderson DG. Lipidlike materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, Bacallado SA, Nguyen DN, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin JR, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev KG, Sah DW, Soutschek J, Toudjarska I, Vornlocher HP, Zimmermann TS, Langer R, Anderson DG. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko YT, Falcao C, Torchilin VP. Cationic liposomes loaded with proapoptotic peptide D-(KLAKLAK)(2) and Bcl-2 antisense oligodeoxynucleotide G3139 for enhanced anticancer therapy. Mol Pharm. 2009;6:971–977. doi: 10.1021/mp900006h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunlap DD, Maggi A, Soria MR, Monaco L. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res. 1997;25:3095–3101. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu JJ, Langer R, Chen J. A novel mechanism is involved in cationic lipid-mediated functional siRNA delivery. Mol Pharm. 2009;6:763–771. doi: 10.1021/mp900023v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 17.Sonawane ND, Szoka FC, Jr, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine–DNA polyplexes. J Biol Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 18.Godbey WT, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 19.Grayson AC, Doody AM, Putnam D. Biophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitro. Pharm Res. 2006;23:1868–1876. doi: 10.1007/s11095-006-9009-2. [DOI] [PubMed] [Google Scholar]
- 20.Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;26:6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 21.Merkel OM, Beyerle A, Librizzi D, Pfestroff A, Behr TM, Sproat B, Barth PJ, Kissel T. Nonviral siRNA delivery to the lung: investigation of PEG–PEI polyplexes and their in vivo performance. Mol Pharm. 2009;6:1246–1260. doi: 10.1021/mp900107v. [DOI] [PubMed] [Google Scholar]
- 22.Beyerle A, Braun A, Merkel O, Koch F, Kissel T, Stoeger T. Comparative in vivo study of poly(ethylene imine)/siRNA complexes for pulmonary delivery in mice. J Control Release. 2011;151:51–56. doi: 10.1016/j.jconrel.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 23.Tarcha PJ, Pelisek J, Merdan T, Waters J, Cheung K, von Gersdorff K, Culmsee C, Wagner E. Synthesis and characterization of chemically condensed oligoethylenimine containing beta-aminopropionamide linkages for siRNA delivery. Biomaterials. 2007;28:3731–3740. doi: 10.1016/j.biomaterials.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 24.Breunig M, Hozsa C, Lungwitz U, Watanabe K, Umeda I, Kato H, Goepferich A. Mechanistic investigation of poly(ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. J Control Release. 2008;130:57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Breunig M, Lungwitz U, Liebl R, Goepferich A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc Natl Acad Sci U S A. 2007;104:14454–14459. doi: 10.1073/pnas.0703882104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjug Chem. 2008;19:1448–1455. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]
- 27.Furgeson DY, Chan WS, Yockman JW, Kim SW. Modified linear polyethylenimine–cholesterol conjugates for DNA complexation. Bioconjug Chem. 2003;14:840–847. doi: 10.1021/bc0340565. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Zheng M, Meng F, Zhang J, Peng R, Zhong Z. Branched polyethylenimine derivatives with reductively cleavable periphery for safe and efficient in vitro gene transfer. Biomacromolecules. 2011;12:1032–1040. doi: 10.1021/bm101364f. [DOI] [PubMed] [Google Scholar]
- 29.Neamnark A, Suwantong O, Bahadur RK, Hsu CY, Supaphol P, Uludag H. Aliphatic lipid substitution on 2 kDa polyethylenimine improves plasmid delivery and transgene expression. Mol Pharm. 2009;6:1798–1815. doi: 10.1021/mp900074d. [DOI] [PubMed] [Google Scholar]
- 30.Ko YT, Kale A, Hartner WC, Papahadjopoulos-Sternberg B, Torchilin VP. Self-assembling micelle-like nanoparticles based on phospholipid–polyethyleneimine conjugates for systemic gene delivery. J Control Release. 2009;133:132–138. doi: 10.1016/j.jconrel.2008.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vasylyev MV, Maayan G, Hovav Y, Haimov A, Neumann R. Palladium nanoparticles stabilized by alkylated polyethyleneimine as aqueous biphasic catalysts for the chemoselective stereocontrolled hydrogenation of alkenes. Org Lett. 2006;8:5445–5448. doi: 10.1021/ol062052k. [DOI] [PubMed] [Google Scholar]
- 32.Simons MJ. Hydrophobically Modified Polyethyleneimines and Ethoxylated Polyethyleneimines. Dept of Chemistry, Wright State University, USA. 2007 [Google Scholar]
- 33.Zorvaryan A, Inceoglu S, Acar MH. Alkylation of polyethyleneimine for homogeneous ligands in ATRP. Polymer. 2011;52:617–621. [Google Scholar]
- 34.Torchilin VP. Targeted polymeric micelles for delivery of poorly soluble drugs. Cell Mol Life Sci. 2004;61:2549–2559. doi: 10.1007/s00018-004-4153-5. [DOI] [PubMed] [Google Scholar]
- 35.Won YW, Kim HA, Lee M, Kim YH. Reducible poly(oligo-D-arginine) for enhanced gene expression in mouse lung by intratracheal injection. Mol Ther. 2010;18:734–742. doi: 10.1038/mt.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi H, Wang H, Xin JH, Zhang X, Wang D. Nanoconfinement crystallization of frustrated alkyl groups: crossover of mesophase to crystalline structure. Chem Commun (Camb) 2011;47:3825–3827. doi: 10.1039/c0cc05245k. [DOI] [PubMed] [Google Scholar]
- 37.Kuo PL, Chen CC, Jao MW. Effects of polymer micelles of alkylated polyethylenimines on generation of gold nanoparticles. J Phys Chem B. 2005;109:9445–9450. doi: 10.1021/jp050136p. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Qu XZ, Gray AI, Tetley L, Uchegbu IF. Self-assembly of cetyl linear polyethylenimine to give micelles, vesicles, and dense nanoparticles. Macromolecules. 2004;37:9114–9122. [Google Scholar]
- 39.Pshezhetskii VS, Nikolaev GM, Lukjanova AP. Structure, conformation and molecular mobility of modified polyethyleneimine — functional and structural analogs of alpha-chymotrypsin. Eur Polym J. 1977;13:423–429. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


