1. Introduction
1.1. Summary
The amphibian embryo has classically been one of the best systems for elucidating the molecular mechanisms of early development, in particular for studies of mesodermal and neural induction. Amphibian embryos develop externally and are large and robust. Therefore tissues can be dissected, isolated or transplanted with high precision and ease in these embryos. In addition, it is relatively easy to manipulate the expression of gene products by injecting in vitro transcribed RNAs into developing embryos. However, since RNAs are translated soon after injection, this method has been used mainly for studying early stages of development. Manipulating genes specifically during later stages of development requires fine control over the time and place of expression, which can only be achieved through transgenic technology. In this chapter we describe a very efficient method of transgenesis developed for Xenopus laevis and Xenopus tropicalis.
1.2. Background
Understanding the molecular basis of pattern formation and differentiation in frog embryos was previously hindered by the lack of a system for temporal and tissue-specific expression of wild type and mutant forms of developmentally important genes. RNA injection, the most common transient expression method in Xenopus, has been effectively used to study maternally expressed genes. However, since RNAs are translated immediately after injection, this method is unfavorable for the study of zygotic gene products that are expressed only after the midblastula transition. Direct injection of DNA can be used to express genes behind temporal and tissue specific promoters after the mid-blastula transition. However, in frog embryos this approach has only been marginally successful for two reasons: (1) injected DNA does not integrate into the frog chromosomes during early cell cycles, and therefore, the embryo expresses the genes in a highly mosaic pattern, and (2) many promoters lack adequate temporal fidelity and tissue specificity of expression when the DNA is not integrated into the genome of the frog. To overcome these technical problems, we have developed a nuclear transplantation-based approach to transgenesis [1]. The approach enables stable expression of cloned gene products in Xenopus embryos, allowing a broader range of feasible experimentation than that previously possible by transient expression methods.
This technique has been used for many applications. For example, transgenesis has been used to express wild type and mutant forms of genes in specific regions of the embryo or at distinct times of development (for examples, see: [1–7]). In addition, transgenic lines that express fluorescent proteins ubiquitously have been established, and these have proved useful for lineage studies [8–12]. Transgenic lines that express fluorescent proteins in a tissue-specific manner have also been used to visualize an inductive response [13–15]. Transgenesis can also be used to study the regulation of genes in vivo (for examples, see: [16–27]). Regulatory elements from various species can drive appropriate expression in transgenic Xenopus embryos [28–34]. By comparison with mouse transgenesis, transgenic Xenopus embryos can be obtained rapidly, at low cost, and in large numbers. Thus transgenesis is useful for large-scale functional analysis of cis-regulatory regions of genes [33].
The transgenic technique has enormous potential for addressing the function of genes in late development and organogenesis. However, in cases where the effect of misexpression is subtle, it may be difficult to rely solely on F0 transgenic embryos for the analysis. The reason for this is that each F0 animal is unique; i.e. each carries a different copy number of transgenes and distinct sites of integration. The net result is that F0 embryos have an inherent variability that complicates the analysis of misexpression experiments. In addition, integration of plasmids into the embryonic genome occurs through chromosomal damage with this transgenic technique; therefore, a subset of the embryos develop with abnormalities. For these reasons, strategies for doing misexpression studies using established transgenic lines have been developed. One approach is to use the GAL4-UAS system, which has been very useful for misexpression studies in the fruitfly, Drosophila melanogaster [35, 36, 6]. Another approach is the use of the site specific recombination system, FLP/FRT (and CRE/LOX), in order to “FLP ON” genes of interest in restricted temporal and spatial patterns in established transgenic lines [37]. This system also offers the potential for cell-lineage studies in living embryos [38].
1.3. Overview of Transgenesis Procedure
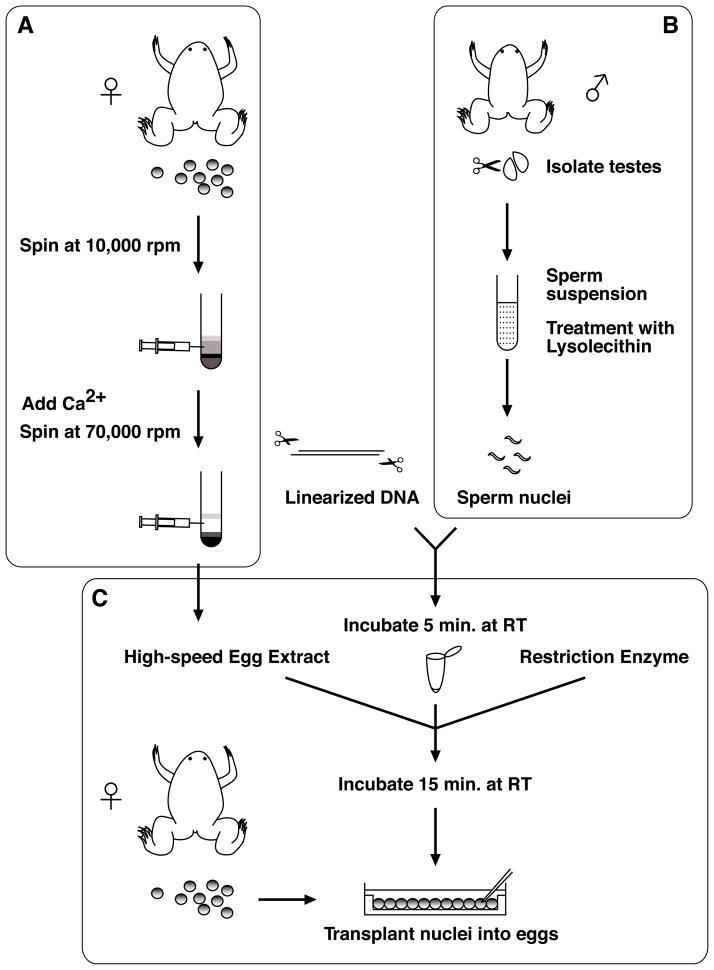
The transgenesis protocol described here can be divided into three parts, (A) preparation of egg extracts, (B) sperm nuclei preparation and (C) nuclear transplantation (Fig. 1). A crude egg extract is prepared using a low speed centrifugation step. These extracts are driven into the interphase stage of the cell cycle by addition of calcium. A high speed centrifugation is then performed to generate an interphase cytosolic fraction containing proteins required for the efficient decondensation of the sperm nuclei (Fig. 1A). In addition, sperm nuclei are prepared from isolated sperm by treatment with lysolecithin, which causes a gentle permeabilization of the sperm plasma membrane (Fig. 1B). The nuclear transplantation procedure involves (1) incubation of linearized plasmid DNA with sperm nuclei, (2) decondensation of sperm nuclei by addition of a high-speed egg extract containing a small amount of the restriction enzyme, and (3) the reaction mix is diluted and a dilute suspension of treated nuclei is used for transplantation into unfertilized eggs (Fig. 1C). The egg extract partially decondenses sperm chromatin and the restriction enzyme stimulates recombination by creating double-strand breaks, facilitating integration of DNA into the genome [1, 39–41]. Since the transgene integrates into the genome prior to fertilization, the resulting transgenic embryos are not chimeric and there is no need to breed to the next generation in order to obtain non-mosaic transgenic animals, as is the case with transgenesis procedures in mice and zebrafish.
Fig. 1.
Transgenesis procedure includes: (A) Preparation of egg extracts; (B) Sperm nuclei preparation; and (C) Nuclear transplantation. The egg extracts and sperm nuclei can be stored at −80°C. (A) Calcium is added to allow the crude egg extract (which are held in meiotic arrest) to progress to interphase, and a high-speed centrifugation is performed to obtain the cytosolic fraction. (B) Testes are macerated and filtered, and then the sperm suspension is treated with lysolecithin to disrupt the plasma membrane of the cells. (C) Sperm nuclei are incubated with linearized DNA for a brief period of time. High-speed egg extracts and a restriction enzyme are added. The egg extracts partially decondenses chromosomes and the restriction enzyme stimulates recombination by creating double-strand breaks, facilitating integration of DNA into the genome. Diluted nuclei are transplanted into unfertilized egg.
1.4. Efficiency of Transgenesis Procedure
One person can transplant sperm nuclei into several hundred to thousands of eggs in a typical experiment. About 30–40% of these transplanted eggs develop into normally cleaving 4-cell stage embryos. About 60–80% of these embryos proceed through gastrulation normally, while the other 20–40% exhibit gastrulation abnormalities resulting from chromosomal damage to the sperm nuclei or physical damage to the egg occurring during transplantation. Thus approximately 20–30% of the eggs initially injected with nuclei often proceed to post-gastrula stages in an experiment. Approximately 10–50% of these normally developing embryos show stable expression of transgenes.
One of the main factors affecting the efficiency of transgenesis is the amount of chromosomal damage sustained by sperm nuclei prior to transplantation. Chromosomal damage can occur during sperm nuclear preparation and is also deliberately induced by addition of restriction enzyme to the sperm, extract, and plasmid reaction mixture used for transplantations. Nuclei with very little chromosomal damage will promote a much higher frequency of normal embryonic development to late stages. However, nuclei that have sustained more chromosomal damage give rise to transgenic embryos at higher frequencies. Thus a balance must be achieved between attaining normal development of embryos versus promoting high frequency transgenesis. We generally use a procedure that results in a lower (10–30%) efficiency of transgenesis, but with improved overall survival of the embryos. Another variable that can affect efficiency is egg quality. Needles for nuclear transplantation are much larger than those used for RNA injections, in order to not damage swollen nuclei. Thus eggs need to be of high quality to recover after injection. Eggs used for nuclear transplantation should, in particular, have a firm cortex and maintain a regular round shape.
In this transgenesis method, the transgene integrates into the genome as a concatemer (see section 1.5). Therefore two different constructs mixed in the same reaction co-integrate into the same site of the genome at high frequency (i.e. 80–90%) [4]. To distinguish transgenic embryos, a marker gene, for example γcrystallin promoter driving GFP, can be co-integrated with a desired transgene. The γ-crystallin promoter drives expression specifically in the lens, making it very easy to detect, even in adult animals [42, 13, 9].
Generally the same restriction enzyme used to linearize the transgene is used in the transgenic reaction before the nuclear transplantation. However, a different enzyme, which was not used to linearize the transgene, can still be used for the reaction, as long as it does not cut within the transgene. To co-integrate two different constructs into the genome, both DNAs can be digested with either the same enzyme or with different enzymes, without loss of co-integration efficiency. Although it is possible to omit restriction enzyme from the transgenic reaction, this usually results in a lower efficiency of transgenesis [1]. This is desirable for some applications such as establishment of transgenic lines, as embryos generated from reactions that do not include restriction enzyme tend to develop to post-metamorphic and adult stages with fewer anomalies.
1.5. Analysis of DNA Integration in Transgenic Embryos
We have analyzed genomic DNA from transplantation-derived tadpoles to determine whether early integration of introduced plasmids into sperm or egg chromosomes is responsible for the nonmosaic expression observed [1]. The pCARGFP plasmid was introduced into embryos by transplantation of sperm nuclei, and tadpoles 2.5 wk to 1 mo old were scored for nonmosaic expression of GFP in the somites; the presence and arrangement of pCARGFP were then determined by probing Southern blots of genomic DNA from these tadpoles with a 1-kb probe consisting of GFP sequences from one end of the linearized pCARGFP plasmid. We found that transplantation-derived tadpoles that did not express pCARGFP did not contain the plasmid, whereas each tadpole that expressed GFP contained between 5 and 35 copies of the plasmid/cell. The probe recognized four to eight bands in each GFP-expressing tadpole which were of unique sizes and were not found in other GFP-expressing tadpoles. These fragments represent putative junction points at which pCARGFP was integrated into the genome of each tadpole. Additionally, the probe recognized two common bands in all of the GFP-expressing tadpoles, corresponding to products formed by tandem and back-to-back concatemerization of the pCARGFP plasmid. By comparing the intensity of these bands relative to the putative junction fragments, we estimate that pCARGFP was integrated into the genome as single copies in some instances and as short (two to six copies) concatemers in other instances. Since the plasmid is expressed in all expected cells in the embryo, it is likely that most of these integrations occurred prior to the first cleavage division, assuring that all cells of the embryo would inherit several copies of the plasmid.
2. Materials
2.1. High-Speed Egg Extract Preparation
1X Marc’s Modified Ringer (MMR): 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 5 mM HEPES, pH 7.5. Prepare a 10X stock, and adjust pH with NaOH to 7.5. Sterilize 10X solutions by autoclaving.
20X Extract buffer (XB) salt stock: 2 M KCl, 20 mM MgCl2, 2 mM CaCl2, filter-sterilize and store at 4°C.
Extract buffer (XB): 1X XB salts (100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2; from 20X XB salts stock solution), 50 mM sucrose (1.5 M stock; filter-sterilize and store in aliquots at −20°C), 10mM HEPES (1 M stock, titrated with KOH so that pH is 7.7 when diluted to 15 mM, should require about 5.5 ml of 10 N KOH for 100 ml, dilution drastically changes the pH of HEPES, so pH must be monitored after dilution; filter-sterilize, and store in aliquots at −20°C). Prepare about 100 ml.
2% (w/v) L-Cysteine hydrochloride 1-hydrate (BDH, 370555X): Made up in 1X XB salts before use and titrated to pH 7.8 with NaOH. Prepare about 300 ml.
CSF-XB: 1X XB salts (100 mM KCl, 0.1 mM CaCl2, 1 mM MgCl2), 1 mM MgCl2 (in addition to MgCl2 present in XB salts; final concentration 2 mM), 10 mM HEPES, pH 7.7, 50 mM sucrose, 5 mM EGTA, pH 7.7. Prepare 50 ml.
Protease inhibitors: Mixture of leupeptin (Roche, 1 017 101), chymostatin (Roche, 1004 638), and pepstatin (Roche 253 286), each dissolved to a final concentration of 10 mg/ml in dimethyl sulfoxide (DMSO). Store in small aliquots at −20°C.
1 M CaCl2 (filter-sterilize and store at 4°C).
Energy mix (store in aliquots at −20°C): 150 mM creatine phosphate (Roche, 621 714), 20 mM ATP (Roche, 519 979), 20 mM MgCl2, Store in 0.1 ml aliquots at −20°C.
Pregnant Mare Serum Gonadotropin (PMSG): 100 U/ml PMSG (P.G.600®, Intervet, Inc., 021825). Dissolve in water and stored at −20°C.
Human Chorionic Gonadotropin (HCG): 1000 U/ml HCG (CHORULON®, Intervet, Inc., 057176). Dissolve in water and stored at 4°C.
2.2. Sperm Nuclei Preparation
1X MMR. Prepare as described in Subheading 2.1., item 1.
0.1% Tricaine Methanesulfonate (MS222, aminobenzoic acid ethyl ester, Sigma A-5040), 0.1% sodium bicarbonate. Dissolve in water.
2X Nuclear Preparation Butter (NPB): 500 mM sucrose (1.5 M stock; filter-sterilize and store aliquots at −20°C), 30 mM HEPES (1 M stock; titrate with KOH so that pH 7.7 is at 15 mM, filter-sterilize, and store aliquots at −20°C), 1 mM spermidine trihydrochloride (Sigma S-2501; 10 mM stock; filter-sterilize and store aliquots at −20°C), 0.4 mM spermine tetrahydrochloride (Sigma S-1141; 10 mM stock; filter-sterilize and store aliquots at −20°C), 2 mM dithiothreitol (Sigma D-0632; 100 mM stock; filter-sterilize and store aliquots at −20°C), 2 mM EDTA (500 mM EDTA, pH 8.0 stock; autoclave and store at room temperature). On the day of the sperm nuclei preparation, make up 25 ml of 2X NPB for 1–2 males from the stock solutions.
1X NPB: Make up 30 ml of 1X NPB by mixing 15 ml of 2X NPB with 15 ml of water and store on ice.
Lysolecithin: 100 μl of 10 mg/ml L-α-lyso-Lecithin, Egg Yolk (Calbiochem, 440154); dissolve at room temperature just before use. Store solid stock at −20°C. Discard the stock powder if it becomes sticky. Digitonin can be used instead of Lysolecitin. Digitonin is more specific for the plasma membrane leaving the nuclear membranes intact.
Bovine serum albumin (BSA): 10% (w/v) BSA (fraction V, Sigma A-7906) Make up 5 ml in water on the day of the sperm nuclei preparation.
1X NPB, 3% BSA: Mix 5 ml 2X NPB, 3 ml 10% BSA and 2 ml water and store on ice.
1X NPB, 0.3% BSA: Mix 2.5 ml 2X NPB, 0.15 ml 10% BSA and 2.35 ml water and store on ice.
100% glycerol.
Sperm storage buffer: 1X NPB, 30% glycerol, 0.3% BSA. Make up by mixing the following solutions, 250 μl of 2X NPB, 15 μl of 10 % BSA, 150 μl of 100% glycerol and 85 μl water.
Sperm dilution buffer: 250 mM sucrose, 75 mM KCl, 0.5 mM spermidine trihydrochloride, 0.2 mM spermine tetrahydrochloride. Add about 80 μl of 0.1N NaOH per 20 ml solution to titrate to pH 7.3–7.5 and store 0.5–1 ml aliquots at −20°C.
Hoechst No. 33342 (Sigma B-2261): 10 mg/ml stock in dH2O, store in a light-tight vessel at −20°C.
2.3. Nuclear Transplantation Reagents and Equipment
2.5% Cysteine hydrochloride 1-hydrate in 1X MMR (titrate to pH 8.0 with NaOH). Make up freshly.
100 mM MgCl2.
Sperm dilution buffer. Prepare as described in Subheading 2.1., item 11.
0.4X MMR, 6% (w/v) Ficoll (Sigma, F-4375), 10 μg/ml gentamycin (a 10 mg/ml stock solution is purchased from Gibco-BRL 15710-015). Sterilize by filtration.
0.1X MMR, 6% (w/v) Ficoll, 10 μg/ml gentamycin. Sterilize by filtration.
0.1X MMR, 10 μg/ml gentamycin.
Linearized plasmid (100 ng/μl in water): Any enzyme can be used for linearization of plasmid. Digest DNA using standard conditions, and purify by phenol/chloroform extraction and ethanol precipitation. There is no need to gel purify the plasmid.
Restriction enzyme: Dilute in water before adding to the transplantation reaction. Although any enzyme can be used, do not use an enzyme that digests within regulatory or protein coding regions of the construct. We usually use enzymes, NotI, SalI or SfiI purchased from Roche. Some calibration may be required to determine the optimal amount of enzyme to add to each reaction, as too much enzyme may adversely affect the development of embryos derived from nuclear transplantations.
Agarose-coated injection dishes: 1.0% agarose in 0.1X MMR is poured into 60-mm Petri dishes. Before the agarose solidifies, a template is laid onto it. After the agarose has solidified and the templates are removed and the dishes are wrapped in parafilm and stored at 4°C until use. As a template we routinely use a 35-mm × 35-mm square, small weighing boat which holds about 400 X. laevis eggs.
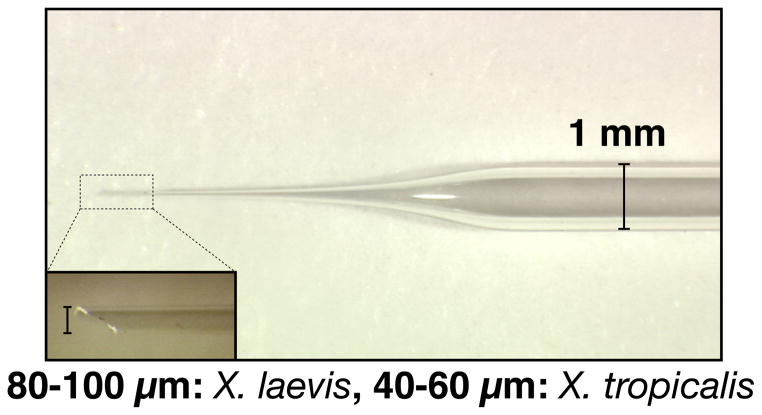
Transplantation needles: 30-μl Drummond MICROCAPS® (Cat. No. 1-000-0300) are pulled to produce large needles with long, gently sloping tips (Fig. 2). We use a Flaming/Brown Micropipet Puller Model P-87 (Sutter Instruments Co.) for pulling needles using a condition, p=50, v=100 and t=5. Needles are clipped with a forceps to produce a beveled tip of 80–100 μm diameter (40–60 μm for X. tropicalis), using the ocular micrometer of a dissecting microscope for measurement (Fig. 2).
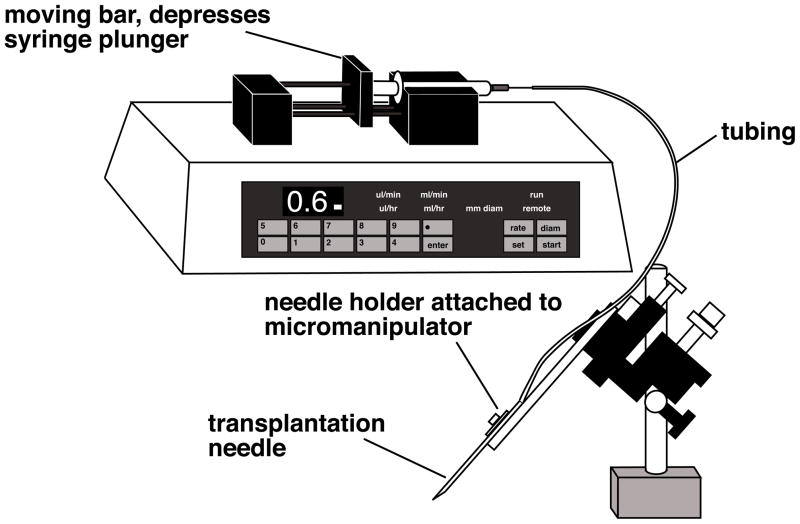
Transplantation apparatus: Most commercial injection apparatus used for RNA and DNA injections which are based on air pressure are unsuitable for nuclear transplantation, due to the difference in needle tip size. Therefore we use an oil-filled injection system, Harvard apparatus 22 syringe pump (NP 55-2222) with two 2.5 ml Hamilton Gas Tight Syringes (two 0.1ml Hamilton Gas Tight Syringes for X. tropicalis) and plastic tubing (ID=0.7 mm, OD=2.4 mm) (Fig. 3). Two people can transplant nuclei at the same time. We use Mineral Oil (Sigma M-8410) in the system. The infusion pump allows us to control flow rate easily. We set the speed of flow at 0.6 μl/min (0.2 μl/min for X. tropicalis).
Fig. 2.
Transplantation needle has a gently sloping tip and is clipped with forceps to produce a beveled, 80–100 μm wide tip for X. laevis and a 40–60 μm wide tip for X. tropicalis.
Fig. 3.
We use an oil-filled injection system for the nuclear transplantations. The syringe and tubing are filled with mineral oil and the infusion pump depresses the syringe plunger, resulting in a constant, desirable flowrate.
3. Methods
3.1. Transgenesis Method
3.1.1. High-Speed Extract Preparation
This protocol is an adaptation of Murray [39]. Briefly, a crude cytostatic factor (CSF) arrested egg extract (cytoplasm arrested in meiotic metaphase) is prepared. Calcium is then added to allow the extract to progress into interphase, and a high-speed spin is performed to obtain a purer cytoplasmic fraction. Cytochalasin is omitted from the protocol, since carryover of cytochalasin into the final extract used for sperm incubations interferes with normal development of transplant embryos. Use of high-speed rather than crude cytoplasmic extracts is advantageous, because high-speed extracts promote swelling of added sperm nuclei (and some chromatin decondensation), but do not promote DNA replication. Replication of sperm DNA incubated in these extracts occurs after transplantation of the nucleus into the egg rather than in the extract. High-speed extract can be stored frozen in small aliquots (at −80°C) and thawed before use.
Prime 8–12 female adult X. laevis about 3–5 days prior to HCG injection by injecting 50 units of PMSG into the dorsal lymph sac. The evening before the extract preparation begins, inject each frog with 500 units HCG and place 2 frogs/container into 2 L 1X MMR. Since one frog with lysing or activating eggs can compromise the whole extract preparation, we prefer to separate the frogs into pairs for the ovulation. The frogs are then placed at 15–18°C overnight (12–14 h). On the next morning, the egg quality from each container is screened before mixing all the eggs and starting the extract preparation. All the eggs released in a container with mottled, lysing, or dying eggs are left out of the extract preparation.
-
All solutions should be prepared before beginning the extract preparation, since the procedure should be carried through all steps promptly once it is initiated; optimally, the high-speed spin should begin within 45–60 min of dejellying the eggs.
Gently, manually expel eggs from each frog into large beakers containing 1X MMR, and collect unbroken eggs with even pigmentation. Good eggs can also be collected from the 1X MMR in the frog buckets. Total volume of eggs should be 100 ml or greater before dejellying.
Remove as much MMR as possible from the eggs. Dejelly eggs in 2% cysteine in XB salts (no HEPES/sucrose). Add a small amount at a time, swirl eggs, and partially replace with fresh cysteine several times during dejellying. Remove broken eggs with a pipet during dejellying. Dejellying should be initiated separately for different batches of eggs, and batches that show breakage or egg activation are discarded. The rest of the eggs can then be combined.
Wash eggs in XB (with HEPES/sucrose). We use about 35 ml for each wash, and do four washes.
Wash eggs in CSF-XB with protease inhibitors. We do two 25 ml washes.
Using a wide-bore Pasteur pipet, transfer eggs into Beckman ultraclear tubes. For these volumes, we typically use 14 × 95 mm tubes (Beckman, 344060). If multiple tubes will be used, try to transfer an equal volume of eggs per tube. Allow the eggs to settle and remove as much CSF-XB as possible.
Spin for about 60 seconds at 1000 rpm (150g) in a Beckman SW 40 Ti Rotor (or similar rotor) in an ultracentrifuge. Remove the excess CSF-XB and then balance the tubes.
Spin the tubes for 10 min at 16,000g (10,000 rpm) at 2°C in a Beckman SW 40 Ti Rotor (or similar rotor) in an ultracentrifuge to crush the eggs. The eggs should be separated into three layers: lipid (top), cytoplasm (center), and yolk (bottom). Collect the cytoplasmic layer from each tube with an 18-gage needle by inserting the needle at the base of the cytoplasmic layer and withdrawing slowly. Transfer cytoplasm to a fresh Beckman tube on ice. If large volumes of darkly pigmented eggs are used, the cytoplasmic layer may be grayish rather than golden at this step. After a second spin to clarify this extract, it should be golden.
Add protease inhibitors to the isolated cytoplasm (do not add cytochalasin); re-centrifuge the cytoplasm in Beckman tubes for an additional 10 min at 16,000g to clarify, again using a swinging bucket rotor. Collect the clarified cytoplasm as before. Expect to get obtain 0.75–1 ml cytoplasm/batch of eggs collected from one frog.
Add 1/20 vol of the ATP-regenerating system (energy mix). Transfer the clarified cytoplasm into TL100.3 thick-wall polycarbonate tubes (Beckman, 349622). Tubes hold about 3 ml each and should be at least half full.
Add CaCl2 to each tube to a final concentration of 0.4 mM; this inactivates CSF and pushes the extract into interphase. Incubate at room temperature for 15 min and then balance for the high-speed spin.
Spin tubes in a Beckman tabletop TL-100 ultracentrifuge in a TL100.3 rotor (gold top; fixed angle) at 70,000 rpm for 1.5 h at 4°C.
The cytoplasm will fractionate into four layers, top to bottom: lipid, cytosol, membranes/mitochondria, and glycogen/ribosomes. Remove the cytosolic layer from each tube (about 1 ml if 2–3 ml were loaded into the tube) by inserting a syringe into the top of the tube through the lipid layer. Transfer this fraction to fresh TL-100 tubes, and spin again at 70,000 rpm for 20 min at 4°C.
Aliquot the high-speed cytosol supernatant into 25 μl aliquots in 0.5-ml Eppendorf tubes. Quick-freeze aliquots in liquid nitrogen, and store at −80°C until use. We typically obtain 1–2 ml of high-speed cytosol from preparations of this scale. Sperm nuclei should be incubated in an aliquot of extract and stained with Hoechst as previously described to determine whether extract is effective. If active, interphase extract should cause nuclei to swell visibly (thicken and lengthen) within 10 min of addition at room temperature.
3.1.2. Sperm Nuclei Preparation
We generally follow the standard protocol of Murray [39], but omit the protease inhibitors leupeptin and phenylmethylsulfonyl fluoride from all steps to avoid transfer into the final mixture, which is diluted for egg injections. We always obtain better sperm nuclei when males are injected with hormones.
Prime one or two males about 3–5 days prior to HCG injection with 50 units of PMSG.
Inject the males 12–15 hours before the preparation with 500 units of HCG.
-
Dissect and isolate the testes from the male:
Anesthetize a male by immersion in 0.1% Tricaine Methanesulfonate/0.1% sodium bicarbonate for at least 20 min (immersion of the animal in ice water for 20 min may also be used), and pith it.
Cut through the ventral body wall and musculature, and lift the yellow fat bodies to isolate the two testes, which are attached to the base of the fat bodies, one on each side of the midline.
Remove the testes with dissecting scissors, and roll them on a paper towel to remove the blood, blood vessels and fat body.
Wash the testes briefly in a 60-mm Petri dish containing cold 1X MMR, removing any attached pieces of fat body or debris with a forceps. Take care not to puncture the testes, as this releases the sperm. This is particularly true with testes from X. tropicalis.
Rinse the testes in cold 1X NPB.
Move the cleaned testes to a dry 60-mm Petri dish, and macerate the testes well (until clumps are no longer visible to the naked eye) with a pair of clean forceps.
Add 2 ml cold 1X NPB and mix well by pipetting the solution up and down with a 10 ml plastic pipet.
Squirt the sperm suspension through four thicknesses of cheesecloth placed into a funnel, and collect the solution in a 14-ml tube (we use Falcon 2059).
Rinse the dish with an additional 3 ml of cold 1X NPB, and force this through the cheesecloth into the 14-ml tube.
Add 5 ml of cold 1X NPB and squeeze the cheesecloth by hand, wearing gloves, to get any remaining liquid through the funnel into the 14-ml tube.
Pellet the sperm by centrifugation at 3000 rpm for 10 min at 4°C (we use a Sorvall HB-4 or similar swinging bucket rotor with the appropriate adapters). During spin, allow 1 ml of 1X NPB to equilibrate to room temperature.
Decant the supernatant and resuspend the sperm in 9 ml 1X NPB using a 10-ml plastic pipet and repellet by centrifugation at 3000 rpm for 10 min at 4°C.
Decant the supernatant and resuspend the pellet with a 1 ml blue tip in the 1 ml 1X NPB that has equilibrated at room temperature.
Add 50 μl of 10 mg/ml lysolecithin. Mix gently and incubate for 5 min at room temperature.
Add 10 ml of cold 3% BSA/1X NPB to the suspension to stop the reaction, and centrifuge at 3000 rpm for 10 min at 4°C.
Decant the supernatant and resuspend the pellet in 5 ml cold 0.3% BSA/1X NPB. Mix well by pipetting with 5-ml plastic pipet and centrifuge at 3000 rpm for 10 min at 4°C.
Take supernatant carefully and resuspend the pellet in 500 μl of sperm storage buffer, and transfer suspension into a 1.5-ml Eppendorf tube.
Count the number of sperm nuclei using a hemacytometer: Cut off the end of a yellow tip with a razor blade and mix the sperm nuclei well by pipetting. Dilute 1 μl of the sperm nuclei with 100 μl of sperm dilution buffer, and add 1μl of 1:100 diluted Hoechst stock to visualize the sperm nuclei under a fluorescence microscope. For a 1:100 dilution of our sperm stock, we typically obtain counts of 100–200 (×104 nuclei/ml) in 1-mm × 1-mm × 0.1-mm square of an improved Neubauer hemacytometer. At this concentration, the undiluted stock contains 1–2×105 nuclei/μl. If your sperm stock is substantially less concentrated (i.e., a count of <50 for a 1:100 dilution), let the sperm settle for a few hours or overnight, and remove some of supernatant. We leave fresh nuclei overnight at 4°C to allow the penetration of glycerol for best cryopreservation of the sperm. Then next day the sperm is aliquoted (20μl/aliquot) and fast frozen in liquid nitrogen. The frozen aliquots are then stored at −80°C.
3.1.3. Transgenesis by Sperm Nuclear Transplantation into Unfertilized Eggs
Prime two females 3–5 days before HCG injection with 50 units of PMSG.
Inject the females with 500 units of HCG, 12–15 hours before they are needed.
Allow an aliquot of Sperm Dilution buffer (SDB) to equilibrate to room temperature, and make up 2.5% Cysteine in 1X MMR, pH 8.0.
Turn on the switch of the Harvard Apparatus and start the infusion pump to get the flow to stabilize before injection.
Set up a reaction using a clipped yellow tip: mix 4 μl sperm stock (~4–8 × 105 nuclei) and 1–2 μl linearized plasmid (100 ng/μl), and incubate for 5 min.
Dilute 0.5 μl of a restriction enzyme in 4.5 μl of water, and mix 1 μl of the diluted enzyme with 18 μl of SDB, 2 μl of 100 mM MgCl2 and 2 μl of high-speed egg extract.
Add the mixture to the sperm/DNA and mix well by gentle pipetting (using a clipped yellow tip). Incubate for 15 min at room temperature; sperm nuclei should visibly swell if diluted into Hoechst as before and observed with a 10X–20X objective.
During the reaction, collect eggs in a beaker by squeezing frogs and dejelly them in 2.5% cysteine/1X MMR, pH 8.0. This usually takes about 10 minutes, so by the time the eggs are ready, the reaction is nearly complete. Squeeze eggs directly into a dry beaker and add cysteine immediately to keep egg quality. You may need to dejelly eggs from different females separately, as dejellying times vary among females or poor quality eggs make the solution dirty.
Wash the dejellied eggs with 1X MMR at least three times, and transfer the eggs to injection dishes containing 0.4% MMR/6% Ficoll using a wide-bore Pasteur pipet. We generally fill the square space with eggs so that no gap is left between the eggs. After about 5 min in 0.4X MMR, 6% Ficoll, the eggs will pierce easily. Transplantation should be performed at around 16°C. To achieve this we place the injection dish on the plastic box half-filled with ice and place the box and eggs under the injection microscope.
After the incubation with extracts, mix the sperm nuclei gently by pipetting with a clipped yellow tip. Then transfer 5 μl of the reaction into 150 μl of SDB that has equilibrated at room temperature.
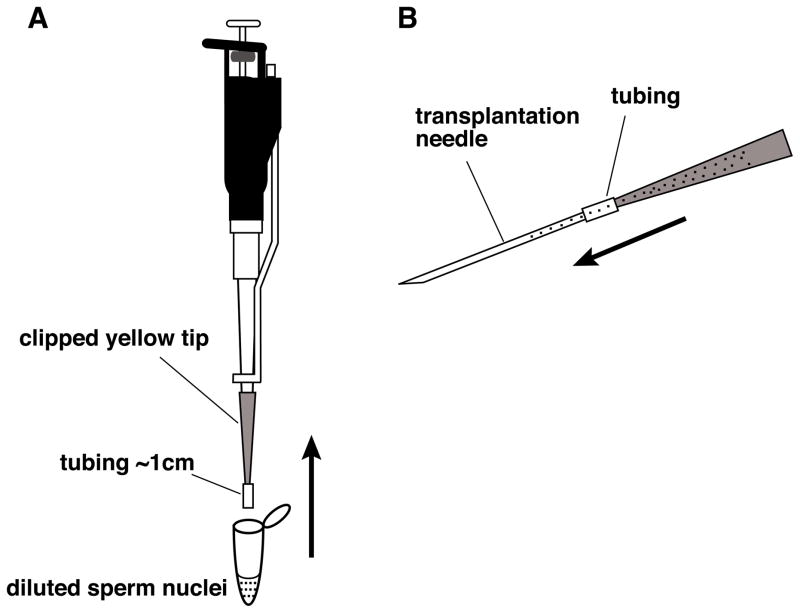
Mix well but avoid making bubbles, using a clipped yellow tip with a piece of plastic tube attached (Fig. 4A). Fill the clipped yellow tip with the diluted sperm suspension, carefully detach the clipped yellow tip, keeping the tip horizontal and backfill a transplantation needle by attaching it to the tube (Fig. 4B). You can keep the yellow tip with the remaining nuclei, by placing it horizontally, in case you need to load another needle. Keep decondensed sperm nuclei at room temperature and transplant them within an hour, but preferably within 30 minutes.
Attach the needle to the tube filled with mineral oil that is connected to the syringe on the Harvard Apparatus.
-
Check the flow and start injecting. Keep the needle inside each egg for approximately 0.5 second, and move the needle fairy rapidly from egg to egg piercing the plasma membrane of each egg with single, sharp motion. We usually transplant for about 15–20 min. If needle is blocked by debris during transplantation, change needle or try to fix by pinching the tube or cutting the tip of needle using forceps.
You can determine whether the sperm dilution and the flow rate used for injections were appropriate by watching the first cleavage of the transplanted eggs. If few of the eggs received a nucleus, the frequency of cleavage will be low; one-third of our transplantations typically result in normally cleaving embryos. Eggs that were injected with more than one nucleus will divide at the time of first cleavage abnormally into three or four (or more) cells. Many of these embryos will develop to blastula stages, but most fail during gastrulation; in some, a region of the embryo will fail to cellularize and die. Eggs injected with multiple nuclei that do gastrulate usually do so abnormally; typically, blastopore closure is incomplete, resulting in embryos that form two wings of somites and neural tissue on each side of the exposed yolky tissue lying in the center of the trunk. This type of gastrulation failure is common to stressed or unhealthy embryos (particularly embryos derived from “soft” eggs).
After injection, incubate embryos at 16°C.
When the embryos reach the 4-cell stage (about 3–4 hours after injection at 16°C), gently transfer normally dividing embryos to 10-cm Petri dish containing 0.1X MMR, 6%Ficoll, 10 μg/ml gentamycin using a wide-bore Pasteur pipet.
The next day, when embryos are around stage 12, transfer healthy embryos to a new 10-cm Petri dish containing 0.1X MMR, 10 μg/ml gentamycin without Ficoll. Because of the large needle tip used for transplantations, embryos often develop large blebs at the site of injection. These blebs occur when cells are forced out of the hole left in the vitelline membrane at the injection site, but they generally do not affect development. The blebs usually fall off at the neurula or tailbud stages.
Incubate embryos until they reach the stage that you need to analyze at 14-22°C.
Fig. 4.
Method for backfilling nuclei into a transplantation needle. (A) The diluted reaction mix is drawn into a clipped yellow tip containing about 1cm of Tygon tubing. (B) The yellow tip containing the Tygon tubing and dilute sperm nuclei is carefully detached from the pipetman and connected to the back of needle using the tubing. The needle is gently loaded with the dilute sperm nuclear reaction by gravity. This is done by slowly increasing the angle of the yellow tip/tubing/needle so that the mixture flows gently into the needle. Once the needle is complete filled, the needle is detached from the yellow tip and the needle is ready to connect to the infusion pump. The remaining sperm mixture can be set aside horizontally and used to reload another needle, if two people are injecting simultaneously or if a needle is accidentally damaged or blocked.
3.2. Transgenesis Method for Xenopus tropicalis
Xenopus tropicalis has a diploid genome and shorter generation time than X. laevis, but retains many of the advantages of X. laevis [43] [44]. In X. tropicalis transgenesis can be combined with genetic experiments. The basic protocol for transgenesis in X. tropicalis is very similar to the procedure used for X. laevis. In this section we point out the differences in X. tropicalis procedure relative to the procedure already describe for X. laevis. We use a high-speed egg extract from X. laevis in the protocol for X. tropicalis.
3.2.1. Reagents and Equipment
X. tropicalis eggs/embryos appear to be more sensitive to salt concentration than X. laevis. Therefore we use less concentrated MMR solutions for dejellying and culture, and MOH buffer instead of SDB for the nuclear transplantation [13]. We also use L-Cysteine (free base) for dejellying X. tropicalis eggs instead of L-Cysteine hydrochloride 1-hydrate used to dejelly X. laevis eggs. The reason for this is the higher sensitivity of X. tropicalis eggs/embryos to high salt concentrations. The acidic version of cysteine requires considerable amounts of NaOH to bring the pH up to 8.0, thus bringing the NaCl concentration to a high level. The free base version of cysteine requires much less NaOH to bring the pH up to 8.0, and thus results in lower final concentration of NaCl.
-
a
2% Cysteine 0.1X MMR (titrate to pH 8.0 with NaOH). Make up freshly.
-
b
0.1X MMR, 6% (w/v) Ficoll, 10 μg/ml gentamycin. Sterilize by filtration.
-
c
0.01X MMR, 6% (w/v) Ficoll, 10 μg/ml gentamycin. Sterilize by filtration.
-
d
0.01X MMR without gentamycin. Since the development of X. tropicalis embryos in later stages seems to be affected by gentamycin, we do not use gentamycin for long-term culture.
-
e
Sperm Injection Buffer modified for Xenopus tropicalis (MOH): 10 mM KPO4, pH 7.2, 125 mM potassium D-gluconate (Sigma, G-4500), 5 mM NaCl, 0.5 mM MgCl2, 250 mM Sucrose, 0.25 mM spermidine trihydrochloride, 0.125 mM spermine tetrahydrochloride.
-
f
Agarose-coated 24 well dishes: 1.0% agarose in 0.1X MMR is poured into 24 well dishes. We use agarose-coated dishes to culture X. tropicalis embryos until they hatch at around stage 28, because their vitellin membrane is quite sticky. We also use 24 well dishes in which 1–2 embryos are cultured per one well until the neurula stage.
-
g
Transplantation needles: Needles are clipped with a forceps to produce a beveled tip of 40–60 μm diameter.
-
h
Transplantation apparatus: 0.1 ml Hamilton Gas Tight Syringes on the system. Set the flow rate at 0.2 μl/min.
3.2.2. Sperm Nuclei Preparation
-
i
Amount of hormone: Prime two males the day before the sperm nuclei preparation with 15 units of PMSG. Inject the males 3 hours before the nuclei preparation with 150 units of HCG.
-
j
Concentration of lysolecithin: Dilute 20 μl of 10 mg/ml lysolecithin with 980 μl of water (0.2 mg/ml) and add 50 μl of that diluted lysolecithin to the sperm suspension (thus 50 times less lysolecithin than what is used for X. laevis). Mix gently and incubate for 5 min at room temperature.
-
k
Count of the sperm nuclei: After final spin, resuspend the pellet in 100 μl of sperm storage buffer. For a 1:100 dilution of our sperm stock, we typically obtain counts of ~80 (×104 nuclei/ml) in 1-mm × 1-mm × 0.1-mm square of an improved Neubauer hemacytometer. At this concentration, the undiluted stock contains 8 ×104 nuclei/μl, and we use 10 μl of that for a reaction. If the sperm stock is substantially less concentrated, let the sperm settle for a few hours or overnight, and remove some of supernatant. If the count is still low, you can use up to 30 μl of the sperm nuclei per reaction. Sperm nuclei can be frozen in aliquots at −80°C.
3.2.3. Nuclear transplantations
-
l
Amount of hormone: Prime two females the day before the nuclear transplanations with 15 units of PMSG. Inject the females 3–4 hours before the nuclear transplantation with 150 units of HCG and leave frogs at 22–24°C. We use two females for 2 rounds of the nuclear transplantation a day. If more rounds of transgenesis are needed in the same day, inject another two females 1 or 2 hours after the first pair. This will allow for four rounds of transgenics, each around 1 hour apart.
-
m
Reaction of the sperm nuclei: Use 10 μl of sperm nuclei (~8 × 105 nuclei) per reaction. After the incubation with egg extract, dilute all the reaction mix with 130 μl of MOH that has equilibrated at room temperature. Mix well and backfill a needle using a clipped yellow tip with a piece of tube. If you use more than 10 μl of the sperm (up to 30 μl), amount of MOH buffer should be reduced. The concentration of nuclei must be considerably higher with X. tropicalis, since the volume injected into each egg is about 1/5 the amount used for X. laevis.
-
n
Injection: After dejellying eggs in 2% Cysteine/0.1X MMR, pH 8.0, transfer the eggs to injection dishes containing 0.1% MMR/6% Ficoll. We generally fill the half of square space with eggs, because we need to inject eggs within 30 min. Transplantation is performed at room temperature (22–24°C).
-
o
Screening: Embryos reach the 4-cell stage approximately 1.5 hr after injection at 22–24°C, therefore it may be necessary for one person to sort the embryos while others continue later rounds of injections (i.e. 3rd and 4rth rounds of injections). Collect normally dividing embryos and transfer them to agarose-coated 24 well dishes (1–2 embryo/well) containing 0.01X MMR/6% Ficoll/10 μg/ml gentamycin. Culture embryos overnight at 22°C.
-
p
The next day transfer healthy embryos to an agarose-coated 10-cm Petri dish containing 0.01X MMR without Ficoll or gentamycin, and culture embryos at 22°C.
Acknowledgments
We would like to thank Jennifer Taylor and Roz Friday for comments on the manuscript. We would also like to thank Odile Bronchain, who helped modify the protocol for Xenopus tropicalis. EA is a Welcome Trust Senior Research Fellow. This work was funded by The Wellcome Trust.
References
- 1.Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- 2.Huang H, Marsh-Armstrong N, Brown DD. Metamorphosis is inhibited in transgenic Xenopus laevis tadpoles that overexpress type III deiodinase. Proc Natl Acad Sci U S A. 1999;96:962–967. doi: 10.1073/pnas.96.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breckenridge RA, Mohun TJ, Amaya E. A role for BMP signalling in heart looping morphogenesis in Xenopus. Dev Biol. 2001;232:191–203. doi: 10.1006/dbio.2001.0164. [DOI] [PubMed] [Google Scholar]
- 4.Hartley KO, Hardcastle Z, Friday RV, Amaya E, Papalopulu N. Transgenic Xenopus embryos reveal that anterior neural development requires continued suppression of BMP signaling after gastrulation. Dev Biol. 2001;238:168–184. doi: 10.1006/dbio.2001.0398. [DOI] [PubMed] [Google Scholar]
- 5.Schreiber AM, Das B, Huang H, Marsh-Armstrong N, Brown DD. Diverse developmental programs of Xenopus laevis metamorphosis are inhibited by a dominant negative thyroid hormone receptor. Proc Natl Acad Sci U S A. 2001;98:10739–10744. doi: 10.1073/pnas.191361698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartley KO, Nutt SL, Amaya E. Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc Natl Acad Sci U S A. 2002;99:1377–1382. doi: 10.1073/pnas.022646899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh-Armstrong N, Cai L, Brown DD. Thyroid hormone controls the development of connections between the spinal cord and limbs during Xenopus laevis metamorphosis. Proc Natl Acad Sci U S A. 2004;101:165–170. doi: 10.1073/pnas.2136755100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh-Armstrong N, Huang H, Berry DL, Brown DD. Germ-line transmission of transgenes in Xenopus laevis. Proc Natl Acad Sci U S A. 1999;96:14389–14393. doi: 10.1073/pnas.96.25.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nutt SL, Bronchain OJ, Hartley KO, Amaya E. Comparison of morpholino based translational inhibition during the development of Xenopus laevis and Xenopus tropicalis. Genesis. 2001;30:110–113. doi: 10.1002/gene.1042. [DOI] [PubMed] [Google Scholar]
- 10.Byrne JA, Simonsson S, Gurdon JB. From intestine to muscle: nuclear reprogramming through defective cloned embryos. Proc Natl Acad Sci U S A. 2002;99:6059–6063. doi: 10.1073/pnas.082112099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gargioli C, Slack JM. Cell lineage tracing during Xenopus tail regeneration. Development. 2004;131:2669–2679. doi: 10.1242/dev.01155. [DOI] [PubMed] [Google Scholar]
- 12.Kuroda H, Wessely O, Robertis EM. Neural Induction in Xenopus: Requirement for Ectodermal and Endomesodermal Signals via Chordin, Noggin, beta-Catenin, and Cerberus. PLoS Biol. 2004;2:E92. doi: 10.1371/journal.pbio.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Offield MF, Hirsch N, Grainger RM. The development of Xenopus tropicalis transgenic lines and their use in studying lens developmental timing in living embryos. Development. 2000;127:1789–1797. doi: 10.1242/dev.127.9.1789. [DOI] [PubMed] [Google Scholar]
- 14.Chalmers AD, Welchman D, Papalopulu N. Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev Cell. 2002;2:171–182. doi: 10.1016/s1534-5807(02)00113-2. [DOI] [PubMed] [Google Scholar]
- 15.Geng X, Xiao L, Lin GF, Hu R, Wang JH, Rupp RA, Ding X. Lef/Tcf-dependent Wnt/beta-catenin signaling during Xenopus axis specification. FEBS Lett. 2003;547:1–6. doi: 10.1016/s0014-5793(03)00639-2. [DOI] [PubMed] [Google Scholar]
- 16.Casey ES, Tada M, Fairclough L, Wylie CC, Heasman J, Smith JC. Bix4 is activated directly by VegT and mediates endoderm formation in Xenopus development. Development. 1999;126:4193–4200. doi: 10.1242/dev.126.19.4193. [DOI] [PubMed] [Google Scholar]
- 17.Mani SS, Besharse JC, Knox BE. Immediate upstream sequence of arrestin directs rod-specific expression in Xenopus. J Biol Chem. 1999;274:15590–15597. doi: 10.1074/jbc.274.22.15590. [DOI] [PubMed] [Google Scholar]
- 18.Hyde CE, Old RW. Regulation of the early expression of the Xenopus nodal-related 1 gene, Xnr1. Development. 2000;127:1221–1229. doi: 10.1242/dev.127.6.1221. [DOI] [PubMed] [Google Scholar]
- 19.Lerchner W, Latinkic BV, Remacle JE, Huylebroeck D, Smith JC. Region-specific activation of the Xenopus brachyury promoter involves active repression in ectoderm and endoderm: a study using transgenic frog embryos. Development. 2000;127:2729–2739. doi: 10.1242/dev.127.12.2729. [DOI] [PubMed] [Google Scholar]
- 20.Ryffel GU, Lingott A. Distinct promoter elements mediate endodermal and mesodermal expression of the HNF1alpha promoter in transgenic Xenopus. Mech Dev. 2000;90:65–75. doi: 10.1016/s0925-4773(99)00230-0. [DOI] [PubMed] [Google Scholar]
- 21.Sparrow DB, Cai C, Kotecha S, Latinkic B, Cooper B, Towers N, Evans SM, Mohun TJ. Regulation of the tinman homologues in Xenopus embryos. Dev Biol. 2000;227:65–79. doi: 10.1006/dbio.2000.9891. [DOI] [PubMed] [Google Scholar]
- 22.Polli M, Amaya E. A study of mesoderm patterning through the analysis of the regulation of Xmyf-5 expression. Development. 2002;129:2917–2927. doi: 10.1242/dev.129.12.2917. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Mei W, Otto A, Xiao L, Tao Q, Geng X, Rupp RA, Ding X. Repression through a distal TCF-3 binding site restricts Xenopus myf-5 expression in gastrula mesoderm. Mech Dev. 2002;115:79–89. doi: 10.1016/s0925-4773(02)00121-1. [DOI] [PubMed] [Google Scholar]
- 24.Small EM, Krieg PA. Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev Biol. 2003;261:116–131. doi: 10.1016/s0012-1606(03)00306-3. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka T, Kubota M, Shinohara K, Yasuda K, Kato JY. In vivo analysis of the cyclin D1 promoter during early embryogenesis in Xenopus. Cell Struct Funct. 2003;28:165–177. doi: 10.1247/csf.28.165. [DOI] [PubMed] [Google Scholar]
- 26.Karaulanov E, Knochel W, Niehrs C. Transcriptional regulation of BMP4 synexpression in transgenic Xenopus. Embo J. 2004;23:844–856. doi: 10.1038/sj.emboj.7600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sipe CW, Gruber EJ, Saha MS. Short upstream region drives dynamic expression of hypoxia-inducible factor 1alpha during Xenopus development. Dev Dyn. 2004:229–238. doi: 10.1002/dvdy.20049. [DOI] [PubMed] [Google Scholar]
- 28.Beck CW, Slack JM. Gut specific expression using mammalian promoters in transgenic Xenopus laevis. Mech Dev. 1999;88:221–227. doi: 10.1016/s0925-4773(99)00217-8. [DOI] [PubMed] [Google Scholar]
- 29.Gottgens B, Barton LM, Gilbert JG, Bench AJ, Sanchez MJ, Bahn S, Mistry S, Grafham D, McMurray A, Vaudin M, Amaya E, Bentley DR, Green AR, Sinclair AM. Analysis of vertebrate SCL loci identifies conserved enhancers. Nat Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- 30.Rodriguez TA, Casey ES, Harland RM, Smith JC, Beddington RS. Distinct enhancer elements control Hex expression during gastrulation and early organogenesis. Dev Biol. 2001;234:304–316. doi: 10.1006/dbio.2001.0265. [DOI] [PubMed] [Google Scholar]
- 31.Zhang T, Tan YH, Fu J, Lui D, Ning Y, Jirik FR, Brenner S, Venkatesh B. The regulation of retina specific expression of rhodopsin gene in vertebrates. Gene. 2003;313:189–200. doi: 10.1016/s0378-1119(03)00680-2. [DOI] [PubMed] [Google Scholar]
- 32.Lim W, Neff ES, Furlow JD. The mouse muscle creatine kinase promoter faithfully drives reporter gene expression in transgenic Xenopus laevis. Physiol Genomics. 2004;18:79–86. doi: 10.1152/physiolgenomics.00148.2003. [DOI] [PubMed] [Google Scholar]
- 33.Stapleton T, Luchman A, Johnston J, Browder L, Brenner S, Venkatesh B, Jirik FR. Compact intergenic regions of the pufferfish genome facilitate isolation of gene promoters: characterization of Fugu 3′-phosphoadenosine 5′-phosphosulfate synthase 2 (fPapss2) gene promoter function in transgenic Xenopus. FEBS Lett. 2004;556:59–63. doi: 10.1016/s0014-5793(03)01353-x. [DOI] [PubMed] [Google Scholar]
- 34.Warkman AS, Atkinson BG. Amphibian cardiac troponin I gene’s organization, developmental expression, and regulatory properties are different from its mammalian homologue. Dev Dyn. 2004;229:275–288. doi: 10.1002/dvdy.10434. [DOI] [PubMed] [Google Scholar]
- 35.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 36.Chae J, Zimmerman LB, Grainger RM. Inducible control of tissue-specific transgene expression in Xenopus tropicalis transgenic lines. Mech Dev. 2002;117:235–241. doi: 10.1016/s0925-4773(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 37.Werdien D, Peiler G, Ryffel GU. FLP and Cre recombinase function in Xenopus embryos. Nucleic Acids Res. 2001;29:E53–53. doi: 10.1093/nar/29.11.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryffel GU, Werdien D, Turan G, Gerhards A, Goosses S, Senkel S. Tagging muscle cell lineages in development and tail regeneration using Cre recombinase in transgenic Xenopus. Nucleic Acids Res. 2003;31:e44. doi: 10.1093/nar/gng044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray AW. Cell cycle extracts. Meth in Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 40.Schiestl RH, Petes TD. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci U S A. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bronchain OJ, Hartley KO, Amaya E. A gene trap approach in Xenopus. Curr Biol. 1999;9:1195–1198. doi: 10.1016/S0960-9822(00)80025-1. [DOI] [PubMed] [Google Scholar]
- 43.Amaya E, Offield MF, Grainger RM. Frog genetics: Xenopus tropicalis jumps into the future. Trends Genet. 1998;14:253–255. doi: 10.1016/s0168-9525(98)01506-6. [DOI] [PubMed] [Google Scholar]
- 44.Hirsch N, Zimmerman LB, Grainger RM. Xenopus, the next generation: X. tropicalis genetics and genomics. Dev Dyn. 2002;225:422–433. doi: 10.1002/dvdy.10178. [DOI] [PubMed] [Google Scholar]