Abstract
Deacetylases of the Sir2 or sirtuin family are thought to regulate life cycle progression and life span in response to nutrient availability. This family has undergone successive rounds of duplication and diversification, enabling the enzymes to perform a wide variety of biological functions. Two evolutionarily conserved functions of yeast Sir2 proteins are the generation of repressive chromatin in subtelomeric domains and the suppression of unbalanced recombination within the tandem rDNA array. Here, we describe the function of the Sir2 ortholog ClHst1 in the yeast Clavispora lusitaniae, an occasional opportunistic pathogen. ClHst1 was localized to the non-transcribed spacer regions of the rDNA repeats and deacetylated histones at these loci, indicating that, like other Sir2 proteins, ClHst1 modulates chromatin structure at the rDNA repeats. However, we found no evidence that ClHst1 associates with subtelomeric regions or impacts gene expression directly. This surprising observation highlights the plasticity of sirtuin function. Related yeast species, including Candida albicans, possess an additional Sir2 family member. Thus, it is likely that the ancestral Candida SIR2/HST1 gene was duplicated and subfunctionalized, such that HST1 retained the capacity to regulate rDNA whereas SIR2 had other functions, perhaps including the generation of subtelomeric chromatin. After subsequent species diversification, the SIR2 paralog was apparently lost in the C. lusitaniae lineage. Thus, C. lusitaniae presents an opportunity to discover how subtelomeric chromatin can be reconfigured.
Author Summary
The Sir2 deacetylases or sirtuins are an important family of proteins thought to regulate life cycle progression and life span according to nutrient availability. These proteins have diversified over the course of evolution such that humans possess seven family members and the laboratory yeast S. cerevisiae possesses five. By comparing the functions of sirtuins in various yeast species, we are gaining insight into how this family has diversified. Here, we studied the sirtuin called ClHst1 from C. lusitaniae, a yeast species known to cause infections in immunocompromised people. We demonstrate that although ClHst1 shares one function with its relatives in other yeast species, it has lost the capacity to act at telomeres, which are the ends of chromosomes. Thus, we have identified an opportunity to discover how organisms adapt to the loss of an evolutionarily conserved function.
Introduction
The Sir2, or sirtuin, family of proteins is ubiquitous throughout all kingdoms of life [1], [2]. Members of this family catalyze the deacetylation of lysines, coupled to the consumption of the cofactor NAD+ to release nicotinamide, 2′-O-acetyl-ADP-ribose and the unacetylated lysine residue [3], [4]. The dependence of sirtuin activity on NAD+ has suggested a link between deacetylation and the metabolic state of a cell [5], [6]. Moreover, the biological consequences of sirtuin-mediated deacetylation imply a role for sirtuins in connecting life cycle progression and life span with nutrient availability [7], [8].
The genomes of most prokaryotic species encode a single member of the sirtuin family, whereas eukaryotic genomes encode multiple paralogs. For example, humans have seven sirtuins (SirT1-SirT7), and the yeast Saccharomyces cerevisiae encodes five (Sir2, Hst1-Hst4). This expansion can reasonably be explained by gene duplication followed by functional divergence of the duplicated genes. Based on phylogenetic analysis, sirtuins can be grouped into several subfamilies [1], [2]. The most-studied subfamily contains the founding member, yeast Sir2, as well as its ortholog, the mammalian SirT1. These proteins are nuclear and regulate chromatin structure and gene expression through the deacetylation of histones and transcription factors. A different subfamily contains SirT6 and SirT7, which are also nuclear and have chromatin-associated functions that overlap with those of the Sir2/SirT1 subfamily [9], [10], [11], [12]. However, S. cerevisiae lacks a member of the SirT6/7 subfamily. Other sirtuins are localized in the cytoplasm or mitochondria, where they deacetylate metabolic enzymes and other proteins.
The varied biological functions of sirtuins illustrate the diversity that can be achieved through gene duplication. One interesting observation is that the biological functions of sirtuins have shifted over evolutionary time. For example, the budding yeast ScSir2 and the fission yeast SpSir2 are associated with subtelomeric heterochromatin [13], [14], but their mammalian ortholog SirT1 is not a major driver of subtelomeric chromatin structure. Instead, a different mammalian sirtuin, SirT6, is associated with subtelomeric heterochromatin [12]. Similarly, the roles of sirtuins in the nucleolus have shifted. Budding yeast ScSir2 localizes to the nucleolus [15], where it suppresses unequal recombination of the tandemly repeated rRNA genes [16], [17]. In contrast, the mammalian SirT1 displays little nucleolar localization [18] although it does repress rRNA transcription through several mechanisms [19], [20], [21]. Another sirtuin, SirT7, is predominantly nucleolar in localization [18] and acts in opposition to SirT1 by promoting rRNA transcription [9], [10]. These distinct nucleolar roles of ScSir2, SirT1 and SirT7 indicate that functions have shifted during the course of evolution. Thus, sirtuins have likely adapted and redistributed functions as genes have been duplicated and lost. These features make the Sir2 family interesting and useful for studying the expansion of a gene family.
To understand how sirtuin functions evolve, we have undertaken a comparative study of Sir2/SirT1 subfamily members in various yeast species [22], [23], [24]. As the founding member of the family, ScSir2 has been extensively studied for its roles in heterochromatin formation and suppression of recombination. ScSir2 acts with other Sir, or silent information regulator, proteins to generate a special chromatin structure that is restrictive to transcription [25]. This chromatin represses the cryptic mating-type cassettes to maintain haploid cell identity. It also occurs in subtelomeric regions of the genome where it serves a structural role and represses subtelomeric genes. For the formation of this repressive chromatin, histone H4-K16 is a particularly important target of the ScSir2 deacetylase [3], [26], and deacetylation of this lysine promotes binding of Sir3 to nucleosomes. A second role of ScSir2 is to suppress unbalanced recombination of the tandemly repeated rRNA genes, minimizing the expansion or contraction of the array [16], [27]. ScSir2 is recruited to the non-transcribed spacer regions of the rDNA repeats through its interaction with ScNet1 in the RENT complex [28], [29]. At these locations, ScSir2 promotes the association of cohesin by repressing transcription from a bidirectional RNA polymerase II promoter whose activity would otherwise displace cohesin. The presence of cohesin keeps sister chromatids aligned, minimizing unequal sister chromatid exchange and intrachromosomal recombination among repeats [27], [30], [31].
ScSIR2 has a paralog, ScHST1 (homolog of Sir two), which arose through gene duplication approximately 100 million years ago [32]. ScHst1 also represses transcription. However, instead of silencing genes through a long-range spreading mechanism, ScHst1 represses single genes in a promoter-specific manner [33]. It is recruited to target genes, including middle sporulation and NAD+ biosynthetic genes [33], [34], through the DNA-binding protein ScSum1 [35], [36].
The budding yeast Kluyveromyces lactis diverged from the S. cerevisiae lineage before the duplication of SIR2/HST1 occurred. Consequently, K. lactis has a single ortholog of ScSir2 and ScHst1. KlSir2 shares functions with both ScSir2 and ScHst1 [24], [37], [38], indicating that the ancestral gene that became duplicated likely had multiple functions and that subfunctionalization occurred after duplication. Characterization of the interaction domains for the proteins that recruit Sir2 and Hst1 to chromatin revealed that subfunctionalization occurred through the acquisition of complementary inactivating mutations in these domains [23]. It is less well understood how the ancestral Sir2 acquired its multiple functions.
Sir2 has also been studied in the fission yeast S. pombe, which diverged from the Saccharomycetaceae budding yeast lineage about one billion years ago. Like ScSir2, SpSir2 is involved in the formation of heterochromatin at the subtelomeric regions and cryptic mating-type cassettes, as well as peri-centromeric domains [13], [39], [40]. However, the consequences of histone deacetylation by Sir2 are different in these two species due to the distinct molecular composition of heterochromatin. In particular, SpSir2 targets H3-K9 for deacetylation, enabling the subsequent methylation of this lysine residue, which is then recognized by the heterochromatin proteins Swi6 and Chp2 [39], [40], [41]. SpSir2 is also associated with the rDNA repeats [41], but promoter-specific repression by SpSir2 has not been documented. Thus, given that Sir2 acts in heterochromatin formation and rDNA maintenance in both S. pombe and S. cerevisiae, these are likely to be ancient functions of Sir2.
An important question is when Sir2 acquired a role in promoter-specific repression mediated by a Sum1-like complex, as occurs for ScHst1 and KlSir2. Therefore, we sought to characterize the functions of Sir2 homologs in species that diverged from S. cerevisiae after S. pombe but before K. lactis. The “CTG” or Candida clade of yeast falls in this position, and the genomes for several species in the clade are sequenced and annotated [42], [43]. Although most experimental work has focused on the opportunistic pathogen C. albicans, other species in the clade have garnered attention due to their status as emerging pathogens [42], [44], [45]. Clavispora lusitaniae, aka Candida lusitaniae, was selected for this study because of its phylogenetic position and experimental tractability. C. lusitaniae propagates in the haploid state, has a defined sexual cycle including meiosis, and is amenable to genetic manipulation [46], [47], [48]. We used a combination of genetic, biochemical, and high-throughput sequencing approaches to study the function of the C. lusitaniae Sir2 ortholog, called ClHst1. ClHst1 is localized to the non-transcribed spacer regions of the rDNA repeats and deacetylates histones at these loci, indicating that ClHst1 acts at the rDNA repeats. However, we found no evidence that ClHst1 associates with subtelomeric regions or impacts gene expression directly. This surprising observation highlights the plasticity of sirtuin function.
Results
Identification of Sir2 family members in C. lusitaniae
We detected four genes encoding sirtuins in the sequenced genome of C. lusitaniae. BLAST analysis and tree construction revealed that the product of gene CLUG_01277 is most similar in sequence to ScSir2, ScHst1 and KlSir2 and falls into the Sir2/SirT1 subfamily (Figure S1). None of the other sirtuins from C. lusitaniae was associated with this subfamily. CLUG_04582 clustered with HST2/SirT2, CLUG_04799 fell in the HST3/4 subfamily found only in fungi, and CLUG_02628 is related to SirT5. An examination of gene synteny using tools on the CoGe Comparative Genomics website [49] was also consistent with CLUG_01277 being the ortholog of ScSIR2 and KlSIR2 (data not shown). Therefore we focused our studies on CLUG_01277.
For reference, we also identified C. albicans genes encoding sirtuins. Interestingly, this species possesses two proteins that fall into the Sir2/SirT1 subfamily (Figure S1). Of the two, the product of orf 19.4761, which is annotated as HST1 in the Candida genome database [50], is more similar in sequence to CLUG_01277. Indeed, orf 19.4761 and CLUG_01277 are identified as orthologs by the Candida gene order browser [51]. The other C. albicans protein in the Sir2/SirT1 subfamily is encoded by orf 19.1992, annotated as SIR2. This protein is less similar to CLUG_01277 and has no identified ortholog in C. lusitaniae according to the gene order browser. Therefore, in keeping with the C. albicans nomenclature, we refer to CLUG_01277 as ClHST1. However, it is critical to note that the SIR2 and HST1 genes from C. albicans and S. cerevisiae arose through distinct gene duplications, and consequently there is no reason to expect CaHST1 to be more similar in function to ScHST1 than to ScSIR2.
ClHst1 associates with the rDNA repeats and causes deacetylation
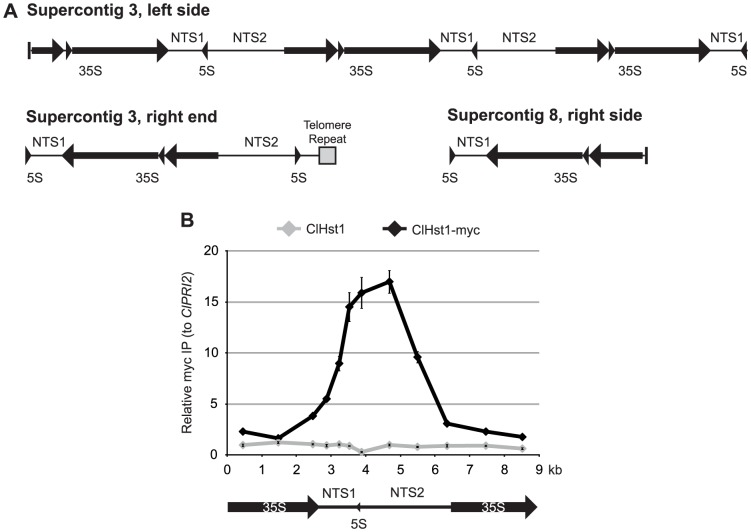
Based on the properties of Sir2/Hst1 in S. cerevisiae, K. lactis, and S. pombe, it is expected that ClHst1 associates with the genomic loci at which it acts. Candidate loci include the rDNA repeats, subtelomeric regions, and promoters of repressed genes. C. lusitaniae lacks cryptic mating-type loci, which are sites of Sir2-mediated silencing in other species. To examine the candidate loci, the association of myc-tagged ClHst1 was examined by chromatin immunoprecipitation (IP). We first examined the repeated rRNA genes, which are annotated in three locations in the sequenced genome of C. lusitaniae. Three successive repeats are found on the left end of supercontig 3, one repeat with an additional 5S gene occurs on the right end of supercontig 3, and one repeat lacking a non-transcribed spacer is on the right end of supercontig 8 (Figure 1A). It is expected that the actual number of repeats at these loci is much greater, as S. cerevisiae encodes 100–200 tandem rRNA genes. Alignment of the three loci revealed significant variation in the non-transcribed spacers, NTS1 and NTS2. Because the rRNA genes at supercontig 3L are repeated in the assembly and display similarity among the non-transcribed spacers, this locus was selected as the template for primer design.
Figure 1. ClHst1 associates with the rDNA repeats.
(A) Features of the three rDNA loci in C. lusitaniae are illustrated. The Pol I-transcribed 35S gene is processed to generate the 18S, 5.8S, and 25S rRNAs (arrows). The Pol III-transcribed 5S gene divides the non-transcribed spacer. All three loci occur near the ends of supercontigs, but only the right end of supercontig 3 has a telomere repeat (gray box), indicating the end of the chromosome. Vertical bars indicate the beginning (3L) or end (8R) of the supercontig sequence. (B) The association of ClHst1 across an rDNA repeat was examined by chromatin IP. Anti-myc antibody was used to immunoprecipitate proteins from ClHST1 (LRY2826) or ClHST1-MYC (LRY2858) yeast. The relative enrichment of each probe compared to a control locus, ClPRI2, is indicated.
In S. cerevisiae, ScSir2 displays peaks of enrichment over the replication fork barrier in NTS1 and the polymerase I promoter in NTS2 [28]. Similarly, in C. lusitaniae we observed ClHst1-myc enriched across NTS1 and NTS2 and less abundant over the transcribed region (Figure 1B). The similar distributions of ClHst1 and ScSir2 across an rDNA repeat are consistent with a conserved role for these proteins in shaping chromatin structure at this locus.
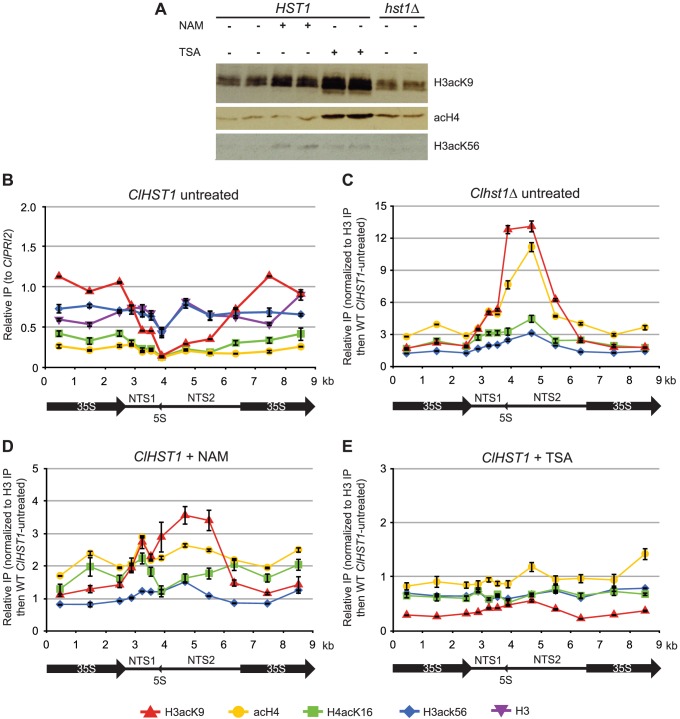
The presence of ClHst1 at the rDNA locus is expected to result in deacetylation of histones. Therefore, we compared the levels of histone acetylation in wild-type and hst1Δ cells, as well as in cells treated with deacetylase inhibitors. Nicotinamide inhibits sirtuin deacetylases such as ClHst1, and trichostatin A inhibits non-sirtuin deacetylases. If ClHst1 causes histone deacetylation, the level of acetylation within the rDNA array should be greater in hst1Δ and nicotinamide-treated cells compared to untreated wild-type cells. The efficacy of the inhibitors at their experimental concentrations was assessed by immunoblotting whole-cell lysates with antibodies against acetylated histones. An increase in acetylation of H3K56 was observed upon treatment with nicotinamide (Figure 2A), presumably due to inactivation of Hst3/4, the orthologs of which target histone H3-K56 globally in S. cerevisiae [52], [53]. An increase in H3-K9 and H4 acetylation was observed upon treatment with trichostatin A (Figure 2A). Therefore, the inhibitors did enter the cells and act upon deacetylases.
Figure 2. ClHst1 causes deacetylation at the rDNA repeats.
(A) The relative abundance of acetylated histones was assessed by immunoblotting. Whole-cell lysates were prepared from replicate samples of ClHST1 yeast (LRY2544) untreated or treated with 25 mM nicotinamide (NAM) or 10 µM trichostatin A (TSA) and from Clhst1Δ yeast (LRY2671). Proteins were electrophoretically separated, transferred to membranes, and probed with antibodies against tetra-acetylated H4, H3K9Ac, or H3K56Ac. (B) The acetylation of histones H3 and H4 across the rDNA repeat was examined by chromatin IP. Total H3, tetra-acetylated H4, H4-K16Ac, H3-K9Ac, or H3-K56Ac was immunoprecipitated from ClHST1 (LRY2544) yeast. For each immunoprecipitation, the recovery of probes was normalized to the control locus, ClPRI2. (C–E) The change in acetylation in deacetylase-deficient yeast relative to wild-type untreated yeast was determined. H3-K9Ac, tetra-acetylated H4, H4-K16Ac, or H3-K56Ac was immunoprecipitated from yeast. For each probe, the enrichment was determined relative to that of total H3. This value was then compared to wild-type untreated cells. Yeast strains used were (C) Clhst1Δ (LRY2671), (D) ClHST1 (LRY2544) treated with 25 mM nicotinamide (NAM), or (E) ClHST1 (LRY2544) treated with 10 µM trichostatin A (TSA).
The distribution of acetylation marks on histones was examined across the rDNA repeat by chromatin IP using antibodies specific for acetylation of histone H3-K9 or K56, histone H4-K16, or tetra acetylated histone H4 (K5,8,12,16). In wild-type cells, most acetylation marks were fairly constant across the repeat (Figure 2B). However, H3K9 acetylation was noticeably lower over NTS1 and NTS2, the same region in which ClHst1 is enriched. We then compared the levels of acetylation across the rDNA locus in wild-type and hst1Δ cells and observed increases in H3-K9 acetylation and H4-tetraacetylation across NTS1 and NTS2 in hst1Δ cells (Figure 2C). Treatment with nicotinamide had a modest effect upon H3-K9 acetylation, but little impact on other lysines (Figure 2D). In contrast, treatment with TSA had little effect on acetylation (Figure 2E). These results are consistent with ClHst1 deacetylating histones in the non-transcribed spacers and being particularly active towards H3K9 acetylation.
ClHst1 acts with ClNet1 at the rDNA repeats
Previously studied deacetylases from the Sir2/SirT1 subfamily do not bind directly to DNA. Rather, they are targeted to particular genomic loci through other proteins. For example, ScSir2 is recruited to the rDNA repeats as part of the RENT complex containing the Sir2-interacting protein ScNet1 [29]. However, BLAST analysis of the C. lusitaniae genome failed to identify orthologs of known Sir2 interacting proteins from other yeast species. Therefore, we employed mass spectrometry to identify the interacting partners of ClHst1. Myc-tagged ClHst1 was partially purified from a clarified lysate using anti-c-myc agarose resin under conditions that should permit retention of associated proteins. A parallel purification from an untagged strain served as a negative control. Protein samples were separated on an SDS-PAGE gel, and successive gel slices were analyzed by LC-MS/MS. The resulting spectra were searched against the NCBI database to identify proteins. From this analysis, 18 proteins from C. lusitaniae were identified as potential interactors with ClHst1 based on their increased enrichment in the myc-tagged ClHst1 sample compared to the untagged ClHst1 strain (Froyd and Rusche, unpublished data). Importantly, one of the identified proteins, the product of CLUG_02406, has similarity to ScNet1. We had not initially identified CLUG_02406 as the ortholog of ScNET1 because the BLAST score is low and only a small segment of the proteins align. However, both proteins return one another as top BLAST hits.
If CLUG_02406 is indeed the ortholog of ScNET1, the protein should associate with the rDNA repeats. We therefore generated an HA-tagged allele of this protein and assessed by chromatin IP whether it associates with the non-transcribed spacer. ClNet1-HA and ClHst1-myc were separately precipitated from a doubly tagged strain and displayed similar distributions across the rDNA repeat (Figure 3). The enrichment of ClNet1 was likely higher than that of ClHst1 because it is closer to the DNA and more efficiently crosslinked. The similar distributions of the two proteins are consistent with both being recruited to the repeat as part of the same complex, as described for the RENT complex in S. cerevisiae. These results imply that ClHst1 and ScSir2 act by similar mechanisms to shape chromatin structure at the rDNA locus.
Figure 3. ClHst1 co-localizes with ClNet1 at the rDNA repeats.
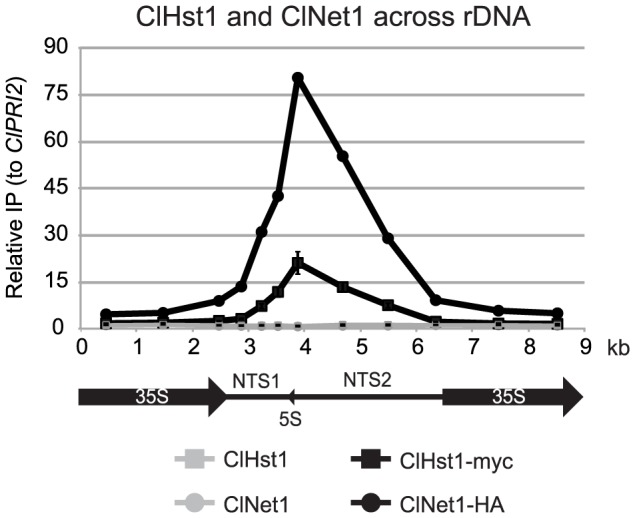
The association of ClNet1 and ClHst1 across an rDNA repeat was examined by chromatin IP. Anti-HA and anti-myc antibodies were used to immunoprecipitate proteins from ClHST1 ClNET1 (LRY2826) or ClHST1-MYC ClNET1-HA (LRY2917) yeast. The relative enrichment of each probe compared to the control locus, ClPRI2, is indicated.
ClHst1 was not associated with telomeres
An evolutionarily conserved role for ClHst1 in regulating the chromatin structure of the tandem rDNA array is indicated by the co-localization of ClHst1 and ClNet1 at the rDNA locus and by the ClHst1-dependent deacetylation of histones in this region. To determine whether ClHst1 also has a conserved function at subtelomeric regions, we investigated the association of ClHst1 with these loci. Telomeres were identified by BLAST using the previously identified C. lusitaniae telomere repeat sequence AAAGAACATCAGTACCTCCCT [42]. This sequence returned hits at six different supercontig ends: 2L, 3R, 4R, 5L, 7L, and 7R. In addition, one hit occurred at an internal location, which is likely the telomerase RNA gene.
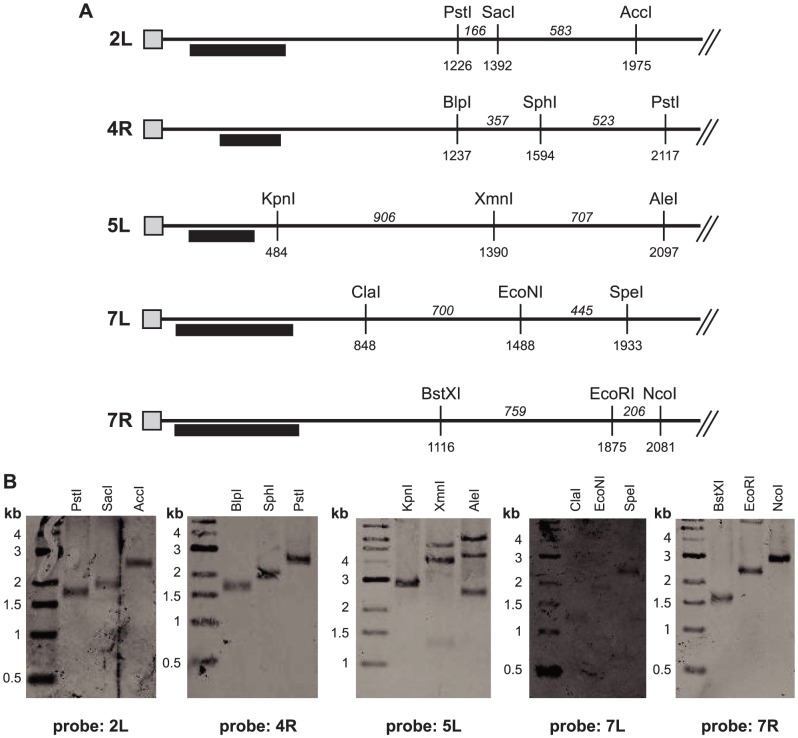
To confirm that the identified telomere repeat sequences are indeed located at the ends of chromosomes, Southern blot analysis was performed. Genomic DNA was digested separately with three restriction enzymes that cut 0.5–2.5 kb from each telomere repeat sequence (Figure 4A). The cut DNA was separated electrophoretically, transferred to membranes, and analyzed using probes located just inside the telomere repeat. If the identified sequences are indeed at the ends of the chromosomes, the three digestions should yield bands whose sizes differ by the distances between the restriction sites. In contrast, if the repeats are actually internal sequences and additional unknown sequence occurs beyond the repeats, the sizes of the restriction products will be inconsistent with the prediction. For telomere repeats 2L, 4R, 7L, and 7R, the observed restriction fragments were consistent with the repeat sequence being at the end of the chromosome (Figure 4B). In all four cases, the telomere repeat length can be deduced to be 500–600 bp. Putative telomere 3R was not examined because it is associated with an rDNA array (Figure 1A). For contig 5L, multiple bands were observed, some of which differed in length by the expected amounts. However, these restriction fragments were longer than expected, suggesting that there is approximately 500 bp of additional unknown sequence between the restriction sites and the chromosome end. Nevertheless, four bona fide telomeres were identified.
Figure 4. Telomere repeats are located at ends of chromosomes.
(A) Features at the ends of five supercontigs are illustrated. Telomere repeats are represented by gray boxes. Restriction sites are marked with their distance from the telomere repeat, and the distance between sites is indicated in italics. Probes used for Southern analysis are shown as black rectangles. (B) Digested genomic DNA (LRY2826) was separated electrophoretically, transferred to membranes, and hybridized with the indicated probes located near telomere repeat sequences.
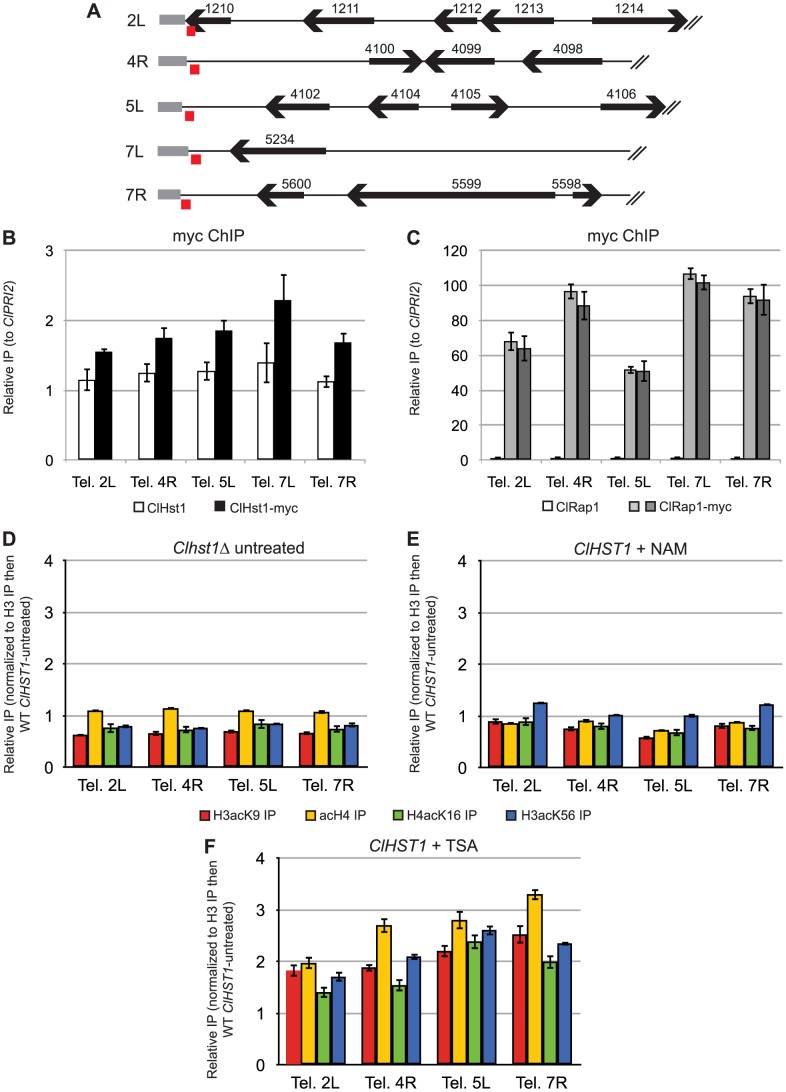
The same chromatin IP samples that were analyzed for association of ClHst1 with the rDNA repeat (Figure 1B) were examined to determine whether ClHst1 also associates with telomeres. For this analysis, primers were designed to amplify the unique sequences immediately adjacent to the telomere repeats (Figure 5A). Telomere 3R was excluded because it contains an rDNA repeat, which is expected to associate with ClHst1. Surprisingly, ClHst1-myc showed no enrichment at any of the examined telomeres (Figure 5B), implying that ClHst1 is not associated with subtelomeric chromatin. To determine whether the loci being examined had other telomere-like properties, we determined whether they associated with the telomere-binding protein Rap1. Rap1 has multiple functions in S. cerevisiae, including recruiting the SIR complex and other proteins to the telomere to regulate chromatin structure and telomere length [54], [55], [56], [57]. Rap1 also associates with telomeres in other species, including K. lactis, C. albicans, S. pombe, and humans [58], [59], [60], [61]. Indeed, we found that ClRap1-myc was enriched 50 to 100-fold at all five queried telomeres (Figure 5C), supporting the conclusion that the telomeres have been properly identified. Moreover, the presence of ClRap1, whose S. cerevisiae homolog recruits the SIR complex to telomeres, indicates that these are loci that would be associated with ClHst1 if its mode of recruitment resembled that of ScSir2.
Figure 5. ClHst1 is not associated with telomeres.
(A) Diagrams represent the genetic features of 10 kbp at ends of contigs bearing telomere repeat sequences. The positions of genes (black arrows), telomere repeat sequences (gray boxes), and PCR products used for chromatin IP analysis (red boxes) are shown. Two dubious ORFs have been omitted. CLUG_04101 overlaps the telomere repeat sequence of contig 5L, and CLUG_04103 is antisense to CLUG_4102. (B) The association of ClHst1 with telomeres was examined by chromatin IP. Anti-myc antibody was used to immunoprecipitate proteins from ClHST1 (LRY2826) or ClHST1-MYC (LRY2858) yeast. The relative enrichment of each probe compared to the control locus, ClPRI2, is indicated. (C) The association of ClRap1 with telomeres was examined by chromatin IP. Anti-myc antibody was used to immunoprecipitate proteins from ClRAP1 (LRY2826) or ClRAP1-MYC (LRY2859, LRY2860) yeast. The relative enrichment of each probe compared to the control locus, ClPRI2, is indicated. (D–F) The change in acetylation of histones H3 and H4 at the telomeres was examined in deacetylase-deficient yeast relative to wild-type. Tetra-acetylated H4, H4-K16Ac, H3-K9Ac, or H3-K56Ac was immunoprecipitated from yeast. For each immunoprecipitation, the enrichment was determined relative to that of total H3, and this value was compared to the value for wild-type untreated cells. Yeast strains used were (D) Clhst1Δ (LRY2671), (E) ClHST1 (LRY2544) treated with 25 mM nicotinamide (NAM), or (F) ClHST1 (LRY2544) treated with 10 µM trichostatin A (TSA).
As an alternate approach to detect ClHst1 at telomeres, the level of histone acetylation at these loci was compared in wild-type and Clhst1Δ cells. If ClHst1 is present even transiently at telomeres and deacetylates histones, acetylation should increase in Clhst1Δ cells. However, no change in acetylation was observed upon deletion of ClHST1 (Figure 5D). In contrast, when these same samples were analyzed with rDNA-specific primers, histone acetylation levels increased significantly across the non-transcribed spacer in Clhst1Δ cells (Figure 2C). These results suggest that ClHst1 is not a major deacetylase at the C. lusitaniae telomeres.
The lack of ClHst1 at the telomeres raised the question of whether another deacetylase acts in the subtelomeric regions. Therefore, the level of histone acetylation at these loci was compared in wild-type and deacetylase-deficient cells, as described above for the rDNA locus (Figure 2D, E). If an inhibitable deacetylase is present at the telomeres, acetylation will increase upon treatment with the appropriate inhibitor. Although no change in acetylation was observed upon nicotinamide treatment (Figure 5E), acetylation did increase slightly upon TSA treatment (Figure 5F). Thus, non-sirtuin deacetylases may play a role in telomere structure in C. lusitaniae.
The loss of ClHst1 did not alter expression of most subtelomeric genes
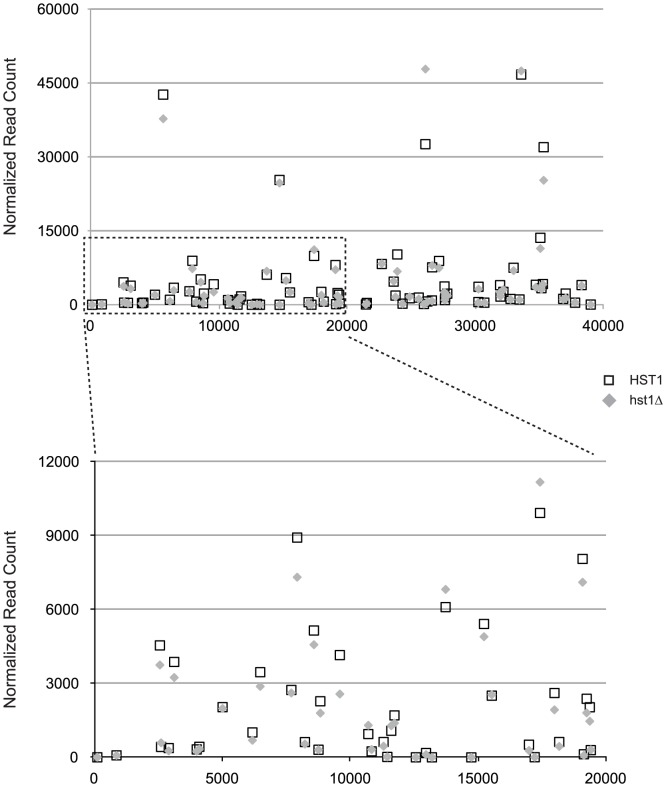
Unlike ClHst1, Sir2 orthologs in S. cerevisiae and K. lactis are associated with telomeres, and consequently subtelomeric genes are induced in the absence of these proteins. To confirm the absence of ClHst1 at telomeres, we compared the expression of subtelomeric genes in wild-type and hst1Δ C. lusitaniae cells. If ClHst1 is indeed absent from the telomeres, no change in expression of subtelomeric genes is expected. RNA was isolated from two independently constructed Clhst1Δ strains as well as the parental wild-type ClHST1 strain, and cDNA was prepared for Illumina sequencing. The normalized number of reads derived from each gene was compared in Clhst1Δ and wild-type strains. All genes within 40 kbp of a telomere repeat sequence were examined, and none were found to differ by more than two-fold (Figure 6). Of the seven genes located elsewhere that were induced, CLUG_02300 is 2 kbp from the end of a supercontig lacking telomere repeats, and CLUG_01197 is 24 kbp from an end. However, neither gene displayed properties consistent with being silenced by block of Hst1-mediated heterochromatin. CLUG_01197 was well-expressed even in the presence of ClHst1, whereas CLUG_02300 remained poorly expressed in the absence of ClHst1 (Figure S2). Moreover, neighboring genes did not share the expression patterns of these two genes. Therefore, the gene expression data are consistent with the absence of ClHst1 at telomeres.
Figure 6. Expression of subtelomeric genes did not change in the absence of ClHst1.
Average normalized read counts are plotted for all genes within 40,000 base pairs of a telomere repeat sequence. These genes are located on contigs 2L, 4R, 5L, 7L, and 7R. For each gene, the distance of the start codon from the telomere repeat sequence is plotted on the x-axis. The dashed rectangle in the upper panel is expanded in the lower panel. Strains examined were wild-type (LRY2544; open boxes) and hst1Δ (LRY2623 and LRY2671; shaded diamonds).
ClHst1 has a limited and likely indirect impact on gene expression
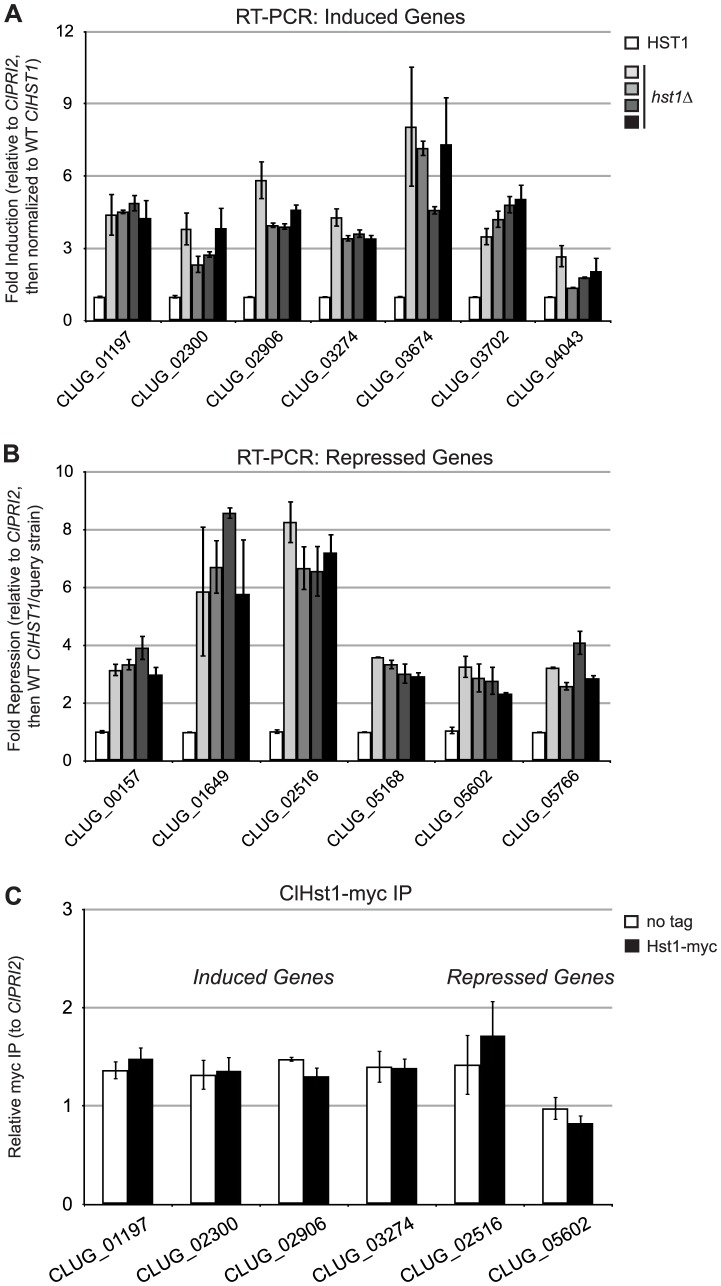
Outside of roles at the rDNA array and subtelomeric loci, ScHst1 and KlSir2 repress transcription of particular genes through a promoter-specific mechanism. Consequently, deletion of ScHST1 or KlSIR2 results in the induction of scores of genes [[24], [35], [62]; Hasan and Rusche, unpublished]. To investigate the possibility that ClHst1 has a similar function and to determine in an unbiased way which genes are targets of ClHst1, the RNA-seq data described above were used to identify genes whose expression changed 3-fold or more when ClHST1 was deleted. These genes were re-analyzed by quantitative reverse transcriptase PCR (Figures 7A and B). A total of 13 genes changed upon the deletion of ClHST1 and are summarized in Table 1. This number was unexpectedly low, given the number of genes regulated by ScSir2, ScHst1, and KlSir2. In addition, the expression of some of these genes was inconsistent in subsequent RT-PCR analyses (Figure S3), suggesting they are not direct targets of ClHst1. Moreover, although seven of the genes were induced in the absence of ClHst1, as expected for targets of a transcriptional repressor, the remaining six genes were repressed. The functions of the genes were also surprising. ScHst1 and KlSir2 repress genes such as the middle sporulation genes and NAD+ biosynthetic genes [24], [33], [34]. In contrast, genes affected by ClHst1 are related to flocculation or adhesion, cell wall biogenesis, and membrane transport. Some genes are also involved in stress response.
Figure 7. ClHst1 has a limited and indirect impact on gene expression.
(A) Expression levels were assessed for genes identified by RNA-seq as induced in Clhst1Δ strains. Expression was assessed by quantitative RT-PCR in ClHST1 (LRY2544, white) and four independently constructed Clhst1Δ strains (LRY2623, light gray; LRY2671; medium gray; LRY2672, dark gray; LRY2673, black). Levels of mRNA for each gene were first normalized to ClPRI2 (CLUG_00368) and then expressed relative to the WT ClHST1 strain. (B) Expression of genes repressed in hst1Δ strains was measured as in part A. For fold repression, the inverse of the fold expression was calculated. (C) The association of ClHst1 with promoters of candidate genes was examined by chromatin IP. Anti-myc antibody was used to immunoprecipitate proteins from ClHST1 (LRY2826) or ClHST1-MYC (LRY2858) yeast.
Table 1. Genes whose expression changed in hst1Δ compared to wild-type strains.
| C. lusitaniae Gene | RT-PCR | normalized read count | kbp from contig end | S. cerevisiae Homolog | C. albicans Homolog | Function in S. cerevisiae or C. albicans | ||
| fold change | wt | hst1 | ||||||
| INDUCED | CLUG_01197 | 4.5 | 6403 | 17414 | 23.8 | MUC1/CCW12 | PGA6/YWP1 | GPI-anchored cell surface glycoprotein involved in adhesion |
| CLUG_02300 | 3.2 | 46 | 124 | 2.1 | FLO9/5/1 | Lectin-like cell wall protein that binds mannose, involved in flocculation | ||
| CLUG_02906 | 4.6 | 626 | 2379 | 646.5 | FLO5/9/1 | Lectin-like cell wall protein that binds mannose, involved in flocculation | ||
| CLUG_03274 | 3.7 | 4213 | 11457 | 166.2 | SAG1/MUC1 | ALS4/ALS2 | GPI-anchored cell surface glycoprotein involved in adhesion | |
| CLUG_03674 | 6.8 | 1049 | 3554 | 848.4 | BIO2 | BIO2 | Biotin synthase, catalyzes the conversion of dethiobiotin to biotin | |
| CLUG_03702 | 4.4 | 13 | 39 | 798.1 | STP1/STP2 | orf19.3643 | Transcription factor activated by proteolysis, activates amino acid permease genes | |
| CLUG_04043 | 2 | 28 | 108 | 120.0 | YMR244W | orf19.5896 | SUN family protein of unknown function | |
| REPRESSED | CLUG_00157 | 3.4 | 1065 | 202 | 300.5 | HSP21 | Small heat shock protein (sHSP) with chaperone activity | |
| CLUG_01649 | 6.7 | 1871 | 596 | 865.3 | PUT1 | PUT1 | Proline oxidase involved in utilization of proline as sole nitrogen source | |
| CLUG_02516 | 7.2 | 336 | 90 | 409.8 | VBA5/AZR1/VBA2 | orf19.7554 | Transporter of the major facilitator superfamily | |
| CLUG_05168 | 3.2 | 489 | 73 | 127.6 | ESBP6 | orf19.4337 | Transporter of the major facilitator superfamily | |
| CLUG_05602 | 2.8 | 22 | 4 | 13.2 | unknown | |||
| CLUG_05766 | 3.2 | 9645 | 2323 | 362.2 | CTA1 | CAT1 | Catalase, breaks down hydrogen peroxide | |
For each gene, the difference in expression between wild-type and hst1 cells is provided, based on both RT-PCR and RNA-seq experiments. In addition, the table includes the distance of the start codon from the end of the supercontig and a description of homologous genes in S. cerevisiae and C. albicans.
To determine whether ClHst1 directly influences the transcription of these genes, chromatin IP was used to assess whether ClHst1 was enriched at their promoters (Figure 7C). However, none of the six examined promoters was enriched for ClHst1. This analysis included the three genes closest to chromosome ends, and the lack of ClHst1 association with these genes is consistent with the absence of ClHst1 near telomere repeats. It was theoretically possible that the myc tag disrupts the interaction of ClHst1 with a protein that targets Hst1 to the gene promoters. If this technical artifact were responsible for the lack of ClHst1-myc association with induced promoters, the expression of these genes would be altered in an HST1-myc strain as in hst1Δ strains. However, RT-PCR analysis revealed that the pattern of gene expression in the HST1-myc cells matched that of wild-type and not hst1Δ cells (Figure S3). Therefore, Hst1-myc must be functional. Thus, ClHst1 does not directly regulate the transcription of the genes whose expression changed in its absence, and ClHst1 is unlikely to repress transcription through a promoter-specific mechanism.
Discussion
Functions of ClHst1
This study demonstrates that the single member of the Sir2/SirT1 subfamily in C. lusitaniae, ClHst1, possesses only a subset of the activities observed for its orthologs in other yeast species. ClHst1 has retained the ability to associate with the non-transcribed portions of rDNA repeats, where it causes deacetylation. It is likely that ClHst1 acts through a similar mechanism as ScSir2 to regulate chromatin structure and hence repress unbalanced recombination within the tandem rDNA array. These findings support the widely held notion that homologous proteins have similar functions from species to species.
In contrast, no evidence was obtained supporting a role for ClHst1 in the subtelomeric regions of C. lusitaniae. Although the genome of this species has not been as extensively annotated as the genomes of other yeast, we believe we have identified bona fide telomeres. First, the telomere repeat sequence is found primarily at the ends of supercontigs, where it is reiterated 16–24 times, as would be expected for a telomere. The only exception is the likely gene encoding telomerase RNA, for which 1.3 units of the repeat occur at an internal location. Thus, there is no evidence that internal tracts of telomere repeat sequence are found in C. lusitaniae. Second, Southern blot analysis is consistent with the identified tracts of telomere repeats being at the ends of chromosomes. Finally, the telomere binding protein Rap1 is associated with the presumed telomeres, as expected. Nevertheless, our observations are inconsistent with ClHst1 being associated with these telomeres. Subtelomeric DNA was not enriched in ClHst1-myc chromatin IP samples that did show enrichment of rDNA. In addition, no change in acetylation of subtelomeric histones was observed upon deletion of ClHST1 or treatment with the deacetylase inhibitor nicotinamide, although both conditions resulted in changes in histone acetylation at the rDNA repeats. Moreover, the expression of subtelomeric genes did not change upon deletion of ClHST1. Thus, ClHst1 appears to lack the ability to promote heterochromatin formation and silencing of subtelomeric genes. This finding is surprising given the presumed importance of maintaining proper telomere structure. Moreover, these results highlight the limitations of extrapolating the function of a protein from orthologous proteins in other species.
ClHst1 was also found not to have an impact on gene expression through a promoter-specific mechanism, as has been observed for KlSir2 and ScHst1. Deletion of ClHST1 caused few changes in gene expression, and ClHst1 was not present at the promoters of those genes whose expression did change. Therefore, ClHst1 does not appear to be directly responsible for their regulation. Instead, deleting ClHST1 may cause mild stress, which in turn alters gene expression. Indeed many of the genes whose expression does change respond to stress in C. albicans. Alternatively, ClHst1 may deacetylate other proteins that regulate these genes.
Our data also provide insight into the in vivo substrates of the ClHst1 deacetylase. Of the histone modifications examined, acetylation of H3-K9 was most affected by the absence of ClHst1. H3-K9 is also a documented substrate of Sir2 in S. pombe. A significant increase in H4 acetylation at the rDNA non-transcribed spacer was also detected using an antibody against tetra-acetylated H4. In addition, acetylation of total cellular H4 increased in hst1Δ compared to wild-type cells (Figure 2A). Thus, ClHst1 must deacetylate at least one of the lysines in the N-terminus of H4. H4-K16 is a key target of Sir2 in S. cerevisiae, but only modest changes in acetylation of this lysine were detected using a specific antibody.
Duplication and loss of SIR2 genes in yeast species
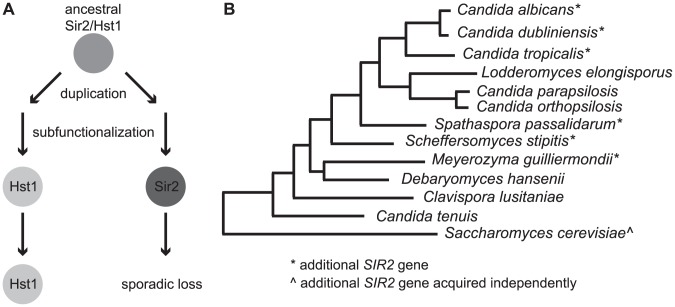
The observation that ClHst1 performs only a subset of the activities possessed by its Saccharomyces orthologs raises the question of how this loss occurred. One possibility (Figure 8A) is a multi-step process, in which duplication and subfunctionalization led to the partitioning of functions between two Sir2 paralogs, one of which was subsequently lost. This model is consistent with the observation that other species of the CTG (Candida) clade, including C. albicans, have a second gene from the Sir2/SirT1 subfamily (Figures 8B, S4). This gene is annotated as SIR2 in C. albicans. Phylogenetic analysis of Sir2 and Hst1 proteins from CTG clade species reveals that these two paralogs cluster separately (Figure S4), consistent with a duplication having occurred in a common ancestor of these species. The sporadic presence of SIR2 throughout the clade (Figure 8B), suggests that SIR2 was lost independently in several lineages. Prior to these losses, the duplicated genes would have subfunctionalized, such that HST1 retained the capacity to regulate rDNA whereas SIR2 had other functions, perhaps including the generation of subtelomeric chromatin. It is noteworthy that SIR2 rather than HST1 has been lost in all Candida lineages bearing a single SIR2/HST1 gene, implying that HST1 is more critical for survival.
Figure 8. The ancestral SIR2 gene may have duplicated and subfunctionalized prior to loss of one paralog.
(A) A model is shown depicting the hypothesized evolutionary history of the SIR2 gene within the CTG clade. An early duplication followed by subfunctionalization partitioned the ancestral functions. The product of one gene (HST1) acted at the rDNA locus, whereas the other (SIR2) possessed different functions, perhaps including a role at telomeres. In some lineages, including C. lusitaniae, only the HST1 gene was retained. (B) A species tree indicates with an asterisk (*) organisms whose genomes encode the additional SIR2 gene. The maximum likelihood tree is based on [43].
Although phylogenetic analysis suggests that the duplication of the Candida SIR2 and HST1 genes occurred in a common ancestor of the CTG clade species, it is unclear exactly when this event happened. Duplication at the base of the CTG clade is consistent with the absence of genes similar to CaSIR2 in non-CTG clade species. However, an earlier timing for the duplication is suggested by the clustering of CaHst1 with Saccharomyces Sir2 and Hst1 and not CaSir2 (Figure S1).
The loss of sirtuin genes has occurred repeatedly during the course of evolution, as particular lineages often lack one or another subfamily. It is also likely that subsequent to these losses, other sirtuins have acquired compensatory functions. Consequently, the biological functions of sirtuins shift from species to species. In humans, SIRT6 and SIRT7 have functions related to those of yeast Sir2, although they are not orthologous. In particular, SIRT6 is associated with subtelomeric heterochromatin and influences transcription at other loci as well [11], [12]. SIRT7 is associated with the rDNA, although its ability to repress recombination between the rDNA repeats has not been examined [9], [10]. In contrast, SirT1, the ortholog of Sir2, has its major functions away from the telomeres and nucleolus. Given that S. cerevisiae and related yeast species do not encode orthologs of either SIRT6 or SIRT7, it is possible that yeast Sir2 acquired functions previously associated with these sirtuins.
This work demonstrates that, although homologous proteins in two species can have similar functions, this is not always the case. It is therefore prudent to acquire experimental support rather than extrapolating function from one species to another.
Potential absence of heterochromatin in C. lusitaniae
The absence of ClHst1 in the subtelomeric regions of C. lusitaniae leaves unresolved the nature of chromatin at these loci. In most eukaryotic species, subtelomeric domains are associated with heterochromatin, generated in part by Sir2 family deacetylases. Heterochromatin formation can also be initiated by siRNA-containing RISC-like complexes or by methylation of histone H3-K9. However, neither mechanism is likely in C. lusitaniae, given that the genome does not encode either the siRNA-presenting protein Argonaute or the methyltransferase specific for H3-K9. It does remain possible that a deacetylase(s) other than ClHst1 acts at telomeres. Thus, C. lusitaniae presents an opportunity to discover how subtelomeric chromatin can be reconfigured.
Materials and Methods
Yeast growth and transformation
Clavispora lusitaniae strains were grown at 30°C in YPD (1% yeast extract, 2% peptone, 2% glucose). Electroporation conditions were derived from previous protocols [46], [63], [64]. Briefly, 75 OD of cells were harvested at an optical density between 1.5 and 1.8 and resuspended in 10 mL 0.1 M lithium acetate, 1× TE, and 10 mM DTT. Cells were shaken one hour at room temperature, collected, washed in 25 mL cold sterile water, and suspended in 0.5 mL cold 1 M sorbitol. Electroporation reactions were set up in 0.2 mm cuvettes using 40 µL cells, 2 µL 10 mg/mL salmon sperm DNA, and 1–5 µg DNA in a volume no more than 5 µL. Electroporation conditions were 1800 V, 200 Ω, and 25 µF. After electroporation, cells were incubated in YPD four hours at 30°C and then spread on selective media (YPD + 200 µg/ml CloNat or supplemented yeast minimal medium lacking uracil or histidine). Cells to be plated on 5-FOA medium were first incubated overnight at 30°C.
Strain construction
Strains used in this study were derived from CL143 (Table 2), which is congenic to the sequenced strain ATCC 42720. Genomic sequence was obtained from the Candida database hosted by the Broad Institute (http://www.broadinstitute.org/annotation/genome/candida_albicans/MultiHome.html), and orthologs of S. cerevisiae and C. albicans genes were identified by BLAST analysis (Table 3). To aid in the genetic manipulation of C. lusitaniae, genes were first subcloned into S. cerevisiae shuttle vectors. Each gene, including 800–1000 bp upstream and downstream of the open reading frame, was amplified by PCR from genomic DNA. The PCR products were then ligated into the shuttle vector pRS415 [65]. These plasmids were subsequently modified to generate deletion constructs, in which the open reading frame was replaced by a selectable marker, or tagging constructs, in which an epitope tag and selectable marker were inserted just after the last non-stop codon of the open reading frame. Finally, the deletion or tagging cassettes were integrated into the C. lusitaniae genome through homologous recombination.
Table 2. Yeast strains used in this study.
| Strain | Genotype | Source |
| CL143 | MAT a chxr (congenic to ATCC 42720) | Reedy and Heitman |
| LRY2544 | CL143 ura3Δ | |
| LRY2623 | CL143 ura3Δ hst1Δ::ClURA3 | |
| LRY2671 | CL143 ura3Δ hst1Δ::ClURA3 | |
| LRY2672 | CL143 ura3Δ hst1Δ::ClURA3 | |
| LRY2673 | CL143 ura3Δ hst1Δ::ClURA3 | |
| LRY2826 | CL143 ura3Δ his1Δ::FRT | |
| LRY2858 | CL143 ura3Δ his1Δ::FRT HST1::MYC-ClHIS1 | |
| LRY2859 | CL143 ura3Δ his1Δ::FRT RAP1:: MYC -ClHIS1 | |
| LRY2860 | CL143 ura3Δ his1Δ::FRT RAP1:: MYC -ClHIS1 | |
| LRY2917 | CL143 ura3Δ his1Δ::FRT HST1:: MYC -ClHIS1 NET1::HA-ClURA3 |
The genotypes of yeast strains used in this study are given.
Table 3. Genes modified in this study.
| Common Name | C. lusitaniae Systematic Name | S. cerevisiae Homolog | Biological Function in S. cerevisiae |
| ClHST1 | CLUG_01277 | ScSIR2 | Silences HML, HMR, and telomeres; represses unequal recombination of rDNA repeats |
| ScHST1 | Represses gene expression in promoter-specific manner | ||
| ClNET1 | CLUG_02406 | ScNET1 | subunit of the RENT complex; involved in nucleolar silencing and telophase exit |
| ClRAP1 | CLUG_02967 | ScRAP1 | Telomere maintenance; recruits Sir complex to telomeres; transcriptional regulator |
| ClURA3 | CLUG_04406 | ScURA3 | Biosynthesis of uracil |
| ClHIS1 | CLUG_02145 | ScHIS1 | Biosynthesis of histidine |
Genes modified in this study are listed with their common and systematic gene names.
Strains auxotrophic for uracil or histidine were generated by deleting the complete open reading frames of URA3 or HIS1. To create the Clura3Δ deletion cassette, plasmid pLR787 containing ClURA3 was amplified by PCR using oligos that bridge and excise the open reading frame to create pLR810. Because loss of URA3 confers resistance to 5-FOA, no selectable marker replaced URA3. To generate the Clhis1Δ deletion cassette, the caSAT1 flipper cassette from pSFS2 [64] was used. This cassette confers resistance to ClonNat and is flanked by FRT sites that allow it to be excised from the genome upon induction of a FLP recombinase gene located in the cassette. The caSAT1 flipper cassette was amplified by PCR using primers whose 5′ ends matched sequence immediately upstream and downstream of the ClHIS1 open reading frame. The amplified cassette was then incorporated into pLR891 in place of the HIS1 open reading frame by homologous recombination in S. cerevisiae to generate pLR909. The resulting Clhis1Δ::FRT-caFLP-caSAT1-FRT cassette was then integrated into the genome of C. lusitaniae. Before a stable integrant was isolated, the caSAT1 flipper excised itself, leaving a single FRT site in place of the ClHIS1 gene. We surmise that the maltose-inducible FLP gene was expressed at a low level in the absence of maltose.
As another tool, we generated a plasmid containing a myc tag linked to the ClHIS1 gene because we suspected that the C. albicans HIS1 gene in the tagging plasmid pFA-MYC-HIS1 [66] was not expressed in C. lusitaniae. The ClHIS1 gene, including its promoter and downstream sequence, was amplified from pLR891 using primers whose 5′ ends matched sequence flanking CaHIS1 in pFA-MYC-HIS1. The ClHIS1 PCR product was then incorporated into pFA-MYC-HIS1 in place of the CaHIS1 gene by a PCR stitching strategy to create pLR950.
To create the Clhst1Δ deletion cassette, the ClURA3 gene was amplified using primers whose 5′ ends matched sequence immediately upstream and downstream of the ClHST1 open reading frame. This PCR product was incorporated into pLR786 in place of ClHST1 by homologous recombination to generate pLR811. To add a myc tag to the C-terminus of ClHST1, the myc tag and ClHIS1 marker were amplified from pLR950. The resulting PCR product was inserted at the 3′ end of the ClHST1 gene by PCR stitching to generate pLR956. Using a similar strategy, a myc tag was added to the C-terminus of ClRap1, to generate pLR958. To add the HA tag to the C-terminus of ClNET1, the HA tag and CaURA3 marker were amplified from pFA-HA-URA3 [66], and this PCR product was incorporated at the 3′ end of the ClNET1 gene by PCR stitching to generate pLR963. The CaURA3 gene and promoter were then replaced with ClURA3 to generate pLR973. Correct integration of all tags was confirmed by sequencing.
Phylogenetic analysis of sirtuin genes
Sequences of sirtuin proteins were obtained from Genbank, the yeast gene order browser [32], and the Candida gene order browser [43], [51]. Phylogenetic trees of the core catalytic domains were constructed using the CLUSTAL_W algorithm of MEGA version 4.0 [67], and distances were calculated with default parameters of the neighbor-joining algorithm applying amino: poisson correction [68] in a pair-wise deletion procedure. The robustness of tree topologies was evaluated by 500 bootstrap replications.
We considered the possibility that a second SIR2 gene does exist in C. lusitaniae but is not represented in the sequenced genome. We first searched for SIR2 at its syntenic position. In the closest species that contains SIR2, M. guillermondii, SIR2 is in the center of a block of seven genes, the other six of which are arranged in the same order in C. lusitaniae. The position where the absent SIR2 gene should be is 1.1 kb in length and contains no missing sequence. Thus, if SIR2 is present in C. lusitaniae, it must have relocated. To search for a partial, unannotated reading frame for SIR2, we performed TBLASTN analysis comparing the M. guillermondii Sir2 protein to the C. lusitaniae genomic sequence. Four hits were returned, corresponding to the four known sirtuin genes. Thus, the available genomic sequence for C. lusitaniae lacks the SIR2 gene.
DNA blotting and analysis of telomere repeat locations
To verify the accuracy of the publically available genomic sequence adjacent to C. lusitaniae telomere repeats, PCR was performed using sets of overlapping primers spanning 3 kb from the telomere repeats. The size of each PCR product was examined, and, with the exception of telomere 4R, all were consistent with the published sequence. Some PCR products were sequenced to obtain approximately one kb of additional sequence at 4R and small annotated gaps in the sequence for telomeres 2L and 5L. These sequences were deposited in GenBank (telomere 2L: KF015764; telomere 4R: KF015766; telomere 5L: KF015765).
For Southern blotting, 150 µg genomic DNA (LRY2826) was treated with restriction enzymes, electrophoretically separated on 1% agarose gels, and transferred to Biodyne B Nylon membrane in 10× SSC buffer (3 M NaCl, 0.3 M trisodium citrate) by capillary transfer. The membrane was rinsed in 2× SSC, air dried, and crosslinked by exposure to uv light. Probes were generated by PCR using oligonucleotides listed in Table S1. PCR products were purified using a QIAquick PCR purification kit and labeled with Biotin-High Prime (Roche Diagnostics, Cat # 11585649910) according to manufacturer's instructions. Blots were hybridized overnight in 5× SSPE , 10% dextran sulfate, 2% (w/v) SDS, 1× Denhardt's solution, 10 µg/ml denatured salmon sperm DNA at 65°C. Blots were washed twice for five minutes with 2× SSPE at 50°C, twice in 2× SSPE, 1% (w/v) SDS, and twice in 0.1× SSPE for 15 minutes at 60°C. The membrane was blocked in Odyssey Blocking Buffer containing 1% SDS for 30 minutes at room temperature and developed in streptavidin IRDye 800CW solution (1∶10,000 in Blocking Buffer plus 1% SDS) in the dark at room temperature for 30 minutes. The blot was washed 3 times in 1× PBST, 0.1% Tween-20, once in 1× PBS, and scanned using the Odyssey Infrared Imaging System, Licor Biosciences.
Chromatin immunoprecipitation
Chromatin IPs were performed by harvesting approximately 100 OD of logarithmically growing cells at an OD600 of 3.0–4.0. For deacetylase inhibition, overnight cultures were diluted and grown for 6 hours at 30°C with no inhibitor, 25 mM nicotinamide, or 10 µM trichostatin A. Cells were crosslinked in two ways. For histone immunoprecipitations, cells were rocked at room temperature in 1% formaldehyde for 30 minutes. For Hst1-myc, Rap1-myc, and Net1-HA immunoprecipitations, cells were washed twice with PBS, resuspended in DMA (10 mM dimethyl adipimidate, 0.1% DMSO, 1× PBS) and rocked for 45 minutes, washed again in PBS, resuspended in 1% formaldehyde in PBS and rocked for 45 minutes. The preparation of soluble chromatin and immunoprecipitation were performed as previously described [69], except that cells were lysed by vortexing for 45 minutes. Immunoprecipitations were performed using 5–10 µl of α-myc (Millipore 06-549), α-HA (Sigma H6908), α-histone H3 (Upstate 06-755), α-histone H3 acetyl-lysine 9 (Upstate 06-942), α-histone H3 acetyl-lysine 56 (Upstate 07-677), α-tetraacetyl histone H4 (Millipore 06-866), or α-histone H4 acetyl-lysine 16 (Active Motif 39167) antibody. Chromatin IP samples were analyzed by real-time PCR using a standard curve prepared from input DNA and oligonucleotides listed in Table S2. The amounts of the immunoprecipitated DNA at experimental loci and a control locus, ClPRI2, were determined relative to the standard curve, and then the relative enrichment of the experimental loci compared to the control locus was calculated. To assess the change in acetylation, the amount of immunoprecipitated DNA in the acetyl IP was normalized to total histone H3, and this value was compared to the value for wild-type untreated cells. Results represent IPs from two independent cultures of each transformed strain, and the SEM was calculated from the differences in the relative enrichment from the mean.
Analysis of bulk histone acetylation
To assess whether treatment with deacetylase inhibitors was effective, 25 OD equivalents of treated cells were resuspended in 400 µl lysis buffer (50 mM HEPES-KOH, pH 7.5, 0.5 M NaCl, 10% glycerol, 0.5% NP-40, 1 mM EDTA, 10 mM DTT, 1× Complete Protease Inhibitors (Roche), 1 mM PMSF, 1 µg/mL pepstatin A, 2 mM benzamidine) and approximately 0.5 mL 0.5 mm Zirconia/Silica beads (Biospec). Cells were lysed by vortexing 30 minutes at 4°C. Samples were spun 10 minutes at 13,200 rpm at 4°C. Supernatants were collected, combined with 3× protein sample buffer, and heated 5 minutes at 95°. Samples were fractionated on 12% polyacrylamide-SDS gels, transferred to nitrocellulose membranes, and probed with either α-histone H3 acetyl-lysine 9 (Upstate 06-942), α-histone H3 acetyl-lysine 56 (Upstate 07-677), or α-tetraacetyl histone H4 (Millipore 06-866) and detected by chemiluminescence (GE RPN2135).
Gene expression analysis
RNA was isolated from 20 OD logarithmically growing cells with optical densities between 3 and 4 [70]. DNA was removed from up to 100 µg RNA using the RNase-Free DNase kit (QIAGEN) and further purified using the RNeasy Mini Kit (QIAGEN). To confirm that the DNase treatment was complete, approximately 100 ng of DNase-treated RNA was used in a PCR reaction containing primers for the ClACT1 transcript.
For RNA-seq analysis, RNA samples were processed for Illumina sequencing by the Duke IGSP sequencing facility. Sequencing reads were mapped to known C. lusitaniae open reading frames using BWA [71] , and the normalized number of hits per gene was calculated. To identify genes whose expression changed in hst1Δ strains, the normalized numbers of hits were compared for wild-type and hst1Δ strains.
For RT-PCR analysis, 1 µg of DNA-free RNA was used for cDNA synthesis as previously described [38]. To quantify the relative amount of mRNA transcripts, cDNA was analyzed by real-time PCR in the presence of SYBR green using a Bio-Rad iCycler. Oligonucleotide sequences are provided in Table S3. A standard curve was generated with genomic DNA isolated from CL145. Transcript levels of queried genes were first normalized to ClPRI2 (CLUG_00368) for each sample. The fold-change was then calculated by normalizing to the strain expressing wild-type ClHST1. Results represent the average fold change of two independent cultures. The standard error measurement (SEM) was calculated from the differences in fold induction of two independent cultures from the mean.
Protein purification and mass spectrometry
To identify proteins associated with ClHst1, the protein was affinity purified from yeast, and co-purifying proteins were identified by mass spectrometry. Six liters of cells were harvested at an OD600 around 3. Cells were washed with ice cold water, then ice cold lysis buffer (50 mM HEPES-KOH, pH 7.5, 0.5 M NaCl, 10% glycerol, 0.5% NP-40, 1 mM EDTA, 1 mM PMSF, 1 µg/mL pepstatin A, 2 mM benzamidine, 2 µg/mL leupeptin). Cell pellets were stored at −80°C. For lysis, cell pellets from 3 liters were resuspended in ice cold lysis buffer containing 100 µl Yeast Protease Inhibitor (Sigma P8215) and placed in a 50 mL BeadBeater small chamber assembly (Biospec cat. no. 110803-50SS) containing 0.5 mm glass beads as indicated by the manufacturer. Additional lysis buffer was added to fill the chamber, and cells were homogenized by alternately beating 30 seconds then resting 90 seconds for 45 minutes at 4°C. The lysates from each assembly were split into 2 tubes and rocked with 50 µl 10 mg/mL heparin and 50 µl 2.5 U/mL benzonase per tube for 15 minutes at room temperature. Lysates were centrifuged for 20 minutes at 14,000×g at 4°C. Supernatants were then spun in an ultracentrifuge for 90 minutes at 45,000 rpm at 4°C. After ultracentrifugation, supernatants were combined (two into one 50 mL tube), and protein concentrations were determined using the BioRad assay (BioRad 500-0006).
Lysates containing 580–750 mg of total protein were incubated overnight at 4°C with 600 µl of a 50% slurry of pre-washed anti-c-myc agarose resin (Sigma A7470). The resin was collected and washed once with 10 mL lysis buffer, once with 10 mL PBS+0.5% NP-40 with protease inhibitor cocktail, then twice with 10 mL PBS with protease inhibitor cocktail. The resin was resuspended in 500 µl PBS with protease inhibitor cocktail and transferred to a microcentrifuge tube and collected. The resin was then submitted to the Duke Proteomics Core Facility, where the sample was eluted by Rapigest and run on an SDS-PAGE gel. Bands were selected for isolation by in-gel digestion and LC-MS/MS analysis by FastMS2.
Supporting Information
Phylogenetic tree of yeast and metazoan sirtuin proteins. A phylogenetic tree represents the relationships of the core catalytic domains of sirtuin proteins from yeast and metazoan species. The tree was constructed using the CLUSTAL_W algorithm of MEGA version 4.0 [67], and distances were calculated with default parameters of the neighbor-joining algorithm applying amino: poisson correction [68] in a pair-wise deletion procedure. The robustness of tree topologies was evaluated by 500 bootstrap replications. All known sirtuins from the following species were included: Scer (Saccharomyces cerevisiae), Zrou (Zygosaccharomyces rouxii), Klac (Kluyveromyces lactis), Calb (Candida albicans), Clus (Clavispora lusitaniae), Spom (Schizosaccharomyces pombe), Hsap (Homo sapiens), and Dmel (Drosophila melanogaster).
(PDF)
Induced genes near chromosome ends. (A) Diagrams display the genetic features surrounding two genes induced in hst1Δ cells compared to wild-type cells (indicated with stars). Genes marked with asterisks are dubious because they lack similarity to known proteins in Genbank and are antisense to bona fide genes. (B) Average normalized read counts derived from RNA-seq experiment are plotted for genes located near the right end of supercontig 2. (C) Normalized read counts are plotted for genes located near the right end of supercontig 1.
(PDF)
The expression of Hst1-influenced genes is the same in wild-type and HST1-myc strains. (A) Expression was assessed by quantitative RT-PCR in ClHST1 (LRY2826, white), ClHST1-myc (LRY2858, black) and two independently constructed Clhst1Δ strains (LRY2671; medium gray; LRY2672, dark gray). Levels of mRNA for each gene were first normalized to ClPRI2 (CLUG_00368) and then expressed relative to the WT ClHST1 strain. (B) Expression of genes repressed in hst1Δ strains was measured as in part A. For fold repression, the inverse of the fold expression was calculated.
(PDF)
Phylogenetic tree of Sir2 and Hst1 proteins from CTG clade species. A phylogenetic tree represents the relationships of the core catalytic domains of Sir2 and Hst1 proteins from CTG clade species. The tree was constructed using the CLUSTAL_W algorithm of MEGA version 4.0 [67], and distances were calculated with default parameters of the neighbor-joining algorithm applying amino: poisson correction [68] in a pair-wise deletion procedure. The robustness of tree topologies was evaluated by 500 bootstrap replications. Sir2 and Hst1 from the following species were included: Calb (Candida albicans), Cdub (Candida dubliniensis), Ctro (Candida tropicalis), Spas (Spathaspora passalidarum), Ssti (Schefferomyces stipitis), Cpar (Candida parapsilosis), Cort (Candida orthopsilosis), Lelo (Lodderomyces elongisporus), Cten (Candida tenuis), Dhan (Debaryomyces hansenii), Mgui (Meyerozyma guilliermondii), and Clus (Clavispora lusitaniae).
(PDF)
Oligonucleotides used for ChIP. The sequences of oligonucleotides used for chromatin IP experiments are provided.
(PDF)
Oligonucleotides used for RT-PCR. The sequences of oligonucleotides used for reverse transcriptase- qPCR analyses are provided.
(PDF)
Oligonucleotides used to generate probes. The sequences of oligonucleotides used to generate probes for Southern analysis are provided.
(PDF)
Funding Statement
This work was supported by the National Science Foundation Grant MCB-1306862. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Frye RA (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273: 793–798. [DOI] [PubMed] [Google Scholar]
- 2. Greiss S, Gartner A (2009) Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol Cells 28: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800. [DOI] [PubMed] [Google Scholar]
- 4. Jackson MD, Denu JM (2002) Structural identification of 2′- and 3′-O-acetyl-ADP ribose as novel metabolites derived from the Sir2 family of beta-NAD+-dependant histone/protein deacetylases. J Biol Chem 277: 18535–18544. [DOI] [PubMed] [Google Scholar]
- 5. Guarente L (2000) Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 14: 1021–1026. [PubMed] [Google Scholar]
- 6. Lu SP, Lin SJ (2010) Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim Biophys Acta 1804: 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donmez G, Guarente L (2010) Aging and disease: connections to sirtuins. Aging Cell 9: 285–290. [DOI] [PubMed] [Google Scholar]
- 8. Yu J, Auwerx J (2009) The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci 1173 Suppl 1: E10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ford E, Voit R, Liszt G, Magin C, Grummt I, et al. (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 20: 1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, et al. (2009) Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci 122: 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, et al. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 136: 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, et al. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freeman-Cook LL, Gomez EB, Spedale EJ, Marlett J, Forsburg SL, et al. (2005) Conserved locus-specific silencing functions of Schizosaccharomyces pombe sir2+. Genetics 169: 1243–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hecht A, Strahl-Bolsinger S, Grunstein M (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96. [DOI] [PubMed] [Google Scholar]
- 15. Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, et al. (1997) Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J 16: 3243–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gottlieb S, Esposito RE (1989) A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56: 771–776. [DOI] [PubMed] [Google Scholar]
- 17. Smith JS, Boeke JD (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev 11: 241–254. [DOI] [PubMed] [Google Scholar]
- 18. Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16: 4623–4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, et al. (2008) Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133: 627–639. [DOI] [PubMed] [Google Scholar]
- 20. Muth V, Nadaud S, Grummt I, Voit R (2001) Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J 20: 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou Y, Schmitz KM, Mayer C, Yuan X, Akhtar A, et al. (2009) Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol 11: 1010–1016. [DOI] [PubMed] [Google Scholar]
- 22. Hickman MA, Froyd CA, Rusche LN (2011) Reinventing heterochromatin in budding yeasts: Sir2 and the origin recognition complex take center stage. Eukaryot Cell 10: 1183–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Froyd CA, Rusche LN (2011) The duplicated deacetylases Sir2 and Hst1 subfunctionalized by acquiring complementary inactivating mutations. Mol Cell Biol 31: 3351–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hickman MA, Rusche LN (2009) The Sir2-Sum1 complex represses transcription using both promoter-specific and long-range mechanisms to regulate cell identity and sexual cycle in the yeast Kluyveromyces lactis . PLoS Genet 5: e1000710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rusche LN, Kirchmaier AL, Rine J (2003) The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae . Annu Rev Biochem 72: 481–516. [DOI] [PubMed] [Google Scholar]
- 26. Johnson LM, Kayne PS, Kahn ES, Grunstein M (1990) Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae . Proc Natl Acad Sci USA 87: 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobayashi T, Ganley AR (2005) Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584. [DOI] [PubMed] [Google Scholar]
- 28. Huang J, Moazed D (2003) Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev 17: 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, et al. (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245–256. [DOI] [PubMed] [Google Scholar]
- 30. Huang J, Brito IL, Villen J, Gygi SP, Amon A, et al. (2006) Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev 20: 2887–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M (2004) SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453. [DOI] [PubMed] [Google Scholar]
- 32. Byrne KP, Wolfe KH (2005) The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15: 1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, et al. (1999) Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae . EMBO J 18: 6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bedalov A, Hirao M, Posakony J, Nelson M, Simon JA (2003) NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae . Mol Cell Biol 23: 7044–7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCord R, Pierce M, Xie J, Wonkatal S, Mickel C, et al. (2003) Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol Cell Biol 23: 2009–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pierce M, Benjamin KR, Montano SP, Georgiadis MM, Winter E, et al. (2003) Sum1 and Ndt80 proteins compete for binding to middle sporulation element sequences that control meiotic gene expression. Mol Cell Biol 23: 4814–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Astrom SU, Kegel A, Sjostrand JO, Rine J (2000) Kluyveromyces lactis Sir2p regulates cation sensitivity and maintains a specialized chromatin structure at the cryptic alpha-locus. Genetics 156: 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hickman MA, Rusche LN (2007) Substitution as a mechanism for genetic robustness: the duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae . PLoS Genetics 3: e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shankaranarayana GD, Motamedi MR, Moazed D, Grewal SI (2003) Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol 13: 1240–1246. [DOI] [PubMed] [Google Scholar]
- 40. Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, et al. (2013) Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J 32: 1250–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wiren M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, et al. (2005) Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J 24: 2906–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, et al. (2009) Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maguire SL, Oheigeartaigh SS, Byrne KP, Schroder MS, O'Gaora P, et al. (2013) Comparative genome analysis and gene finding in Candida species using CGOB. Mol Biol Evol 30: 1281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen SC, Marriott D, Playford EG, Nguyen Q, Ellis D, et al. (2009) Candidaemia with uncommon Candida species: predisposing factors, outcome, antifungal susceptibility, and implications for management. Clin Microbiol Infect 15: 662–669. [DOI] [PubMed] [Google Scholar]
- 45. Krcmery V, Barnes AJ (2002) Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J Hosp Infect 50: 243–260. [DOI] [PubMed] [Google Scholar]
- 46. Reedy JL, Floyd AM, Heitman J (2009) Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol 19: 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Young LY, Lorenz MC, Heitman J (2000) A STE12 homolog is required for mating but dispensable for filamentation in Candida lusitaniae . Genetics 155: 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Francois F, Noel T, Pepin R, Brulfert A, Chastin C, et al. (2001) Alternative identification test relying upon sexual reproductive abilities of Candida lusitaniae strains isolated from hospitalized patients. J Clin Microbiol 39: 3906–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lyons E, Freeling M (2008) How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J 53: 661–673. [DOI] [PubMed] [Google Scholar]
- 50. Arnaud MB, Costanzo MC, Skrzypek MS, Shah P, Binkley G, et al. (2007) Sequence resources at the Candida Genome Database. Nucleic Acids Res 35: D452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fitzpatrick DA, O'Gaora P, Byrne KP, Butler G (2010) Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics 11: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, et al. (2006) The sirtuins Hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr Biol 16: 1280–1289. [DOI] [PubMed] [Google Scholar]
- 53. Maas NL, Miller KM, DeFazio LG, Toczyski DP (2006) Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol Cell 23: 109–119. [DOI] [PubMed] [Google Scholar]
- 54. Conrad MN, Wright JH, Wolf AJ, Zakian VA (1990) RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63: 739–750. [DOI] [PubMed] [Google Scholar]
- 55. Longtine MS, Wilson NM, Petracek ME, Berman J (1989) A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet 16: 225–239. [DOI] [PubMed] [Google Scholar]
- 56. Luo K, Vega-Palas MA, Grunstein M (2002) Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev 16: 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lustig AJ, Kurtz S, Shore D (1990) Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science 250: 549–553. [DOI] [PubMed] [Google Scholar]
- 58. Krauskopf A, Blackburn EH (1996) Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383: 354–357. [DOI] [PubMed] [Google Scholar]
- 59. Li B, Oestreich S, Lange Td (2000) Identification of human Rap1: Implications for telomere evolution. Cell 101: 471–483. [DOI] [PubMed] [Google Scholar]
- 60. Longtine MS, Enomoto S, Finstad SL, Berman J (1992) Yeast telomere repeat sequence (TRS) improves circular plasmid segregation, and TRS plasmid segregation involves the RAP1 gene product. Mol Cell Biol 12: 1997–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu EY, Yen WF, Steinberg-Neifach O, Lue NF (2010) Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol Cell Biol 30: 1254–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, et al. (1999) Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402: 418–421. [DOI] [PubMed] [Google Scholar]
- 63. Kohler GA, White TC, Agabian N (1997) Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol 179: 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reuss O, Vik A, Kolter R, Morschhauser J (2004) The SAT1 flipper, an optimized tool for gene disruption in Candida albicans . Gene 341: 119–127. [DOI] [PubMed] [Google Scholar]
- 65. Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lavoie H, Sellam A, Askew C, Nantel A, Whiteway M (2008) A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans . BMC Genomics 9: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 68. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 69. Rusche LN, Rine J (2001) Conversion of a gene-specific repressor to a regional silencer. Genes Dev 15: 955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae . Nucleic Acids Res 18: 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of yeast and metazoan sirtuin proteins. A phylogenetic tree represents the relationships of the core catalytic domains of sirtuin proteins from yeast and metazoan species. The tree was constructed using the CLUSTAL_W algorithm of MEGA version 4.0 [67], and distances were calculated with default parameters of the neighbor-joining algorithm applying amino: poisson correction [68] in a pair-wise deletion procedure. The robustness of tree topologies was evaluated by 500 bootstrap replications. All known sirtuins from the following species were included: Scer (Saccharomyces cerevisiae), Zrou (Zygosaccharomyces rouxii), Klac (Kluyveromyces lactis), Calb (Candida albicans), Clus (Clavispora lusitaniae), Spom (Schizosaccharomyces pombe), Hsap (Homo sapiens), and Dmel (Drosophila melanogaster).
(PDF)
Induced genes near chromosome ends. (A) Diagrams display the genetic features surrounding two genes induced in hst1Δ cells compared to wild-type cells (indicated with stars). Genes marked with asterisks are dubious because they lack similarity to known proteins in Genbank and are antisense to bona fide genes. (B) Average normalized read counts derived from RNA-seq experiment are plotted for genes located near the right end of supercontig 2. (C) Normalized read counts are plotted for genes located near the right end of supercontig 1.
(PDF)
The expression of Hst1-influenced genes is the same in wild-type and HST1-myc strains. (A) Expression was assessed by quantitative RT-PCR in ClHST1 (LRY2826, white), ClHST1-myc (LRY2858, black) and two independently constructed Clhst1Δ strains (LRY2671; medium gray; LRY2672, dark gray). Levels of mRNA for each gene were first normalized to ClPRI2 (CLUG_00368) and then expressed relative to the WT ClHST1 strain. (B) Expression of genes repressed in hst1Δ strains was measured as in part A. For fold repression, the inverse of the fold expression was calculated.
(PDF)
Phylogenetic tree of Sir2 and Hst1 proteins from CTG clade species. A phylogenetic tree represents the relationships of the core catalytic domains of Sir2 and Hst1 proteins from CTG clade species. The tree was constructed using the CLUSTAL_W algorithm of MEGA version 4.0 [67], and distances were calculated with default parameters of the neighbor-joining algorithm applying amino: poisson correction [68] in a pair-wise deletion procedure. The robustness of tree topologies was evaluated by 500 bootstrap replications. Sir2 and Hst1 from the following species were included: Calb (Candida albicans), Cdub (Candida dubliniensis), Ctro (Candida tropicalis), Spas (Spathaspora passalidarum), Ssti (Schefferomyces stipitis), Cpar (Candida parapsilosis), Cort (Candida orthopsilosis), Lelo (Lodderomyces elongisporus), Cten (Candida tenuis), Dhan (Debaryomyces hansenii), Mgui (Meyerozyma guilliermondii), and Clus (Clavispora lusitaniae).
(PDF)
Oligonucleotides used for ChIP. The sequences of oligonucleotides used for chromatin IP experiments are provided.
(PDF)
Oligonucleotides used for RT-PCR. The sequences of oligonucleotides used for reverse transcriptase- qPCR analyses are provided.
(PDF)
Oligonucleotides used to generate probes. The sequences of oligonucleotides used to generate probes for Southern analysis are provided.
(PDF)