Abstract
The formation of a single G-quadruplex structure adopted by a promising 25 nt G-rich vascular endothelial growth factor aptamer in a K+ rich environment was facilitated by locked nucleic acid modifications. An unprecedented all parallel-stranded monomeric G-quadruplex with three G-quartet planes exhibits several unique structural features. Five consecutive guanine residues are all involved in G-quartet formation and occupy positions in adjacent DNA strands, which are bridged with a no-residue propeller-type loop. A two-residue D-shaped loop facilitates inclusion of an isolated guanine residue into the vacant spot within the G-quartet. The remaining two G-rich tracts of three residues each adopt parallel orientation and are linked with edgewise and propeller loops. Both 5′ with 3 nt and 3′ with 4 nt overhangs display well-defined conformations, with latter adopting a basket handle topology. Locked residues contribute to thermal stabilization of the adopted structure and formation of structurally pre-organized intermediates that facilitate folding into a single G-quadruplex. Understanding the impact of chemical modifications on folding, thermal stability and structural polymorphism of G-quadruplexes provides means for the improvement of vascular endothelial growth factor aptamers and advances our insights into driving nucleic acid structure by locking or unlocking the conformation of sugar moieties of nucleotides in general.
INTRODUCTION
Vascular endothelial growth factor (VEGF) is a homodimeric protein considered to be the main regulator of angiogenesis, a process of development of new blood vessels. Its function is exhibited in vascular permeability and regulation of survival, proliferation and migration of endothelial cells. Apart from being a normal process in growth and wound healing, angiogenesis, together with its main protagonist VEGF, is involved in the development and progression of several diseases, such as age-related macular degeneration (1,2), diabetes (3,4), rheumatoid arthritis (5–7) and cancer (8–10). Owing to its numerous biological roles, VEGF can be exploited in diagnostics and as a drug target. Several different strategies have been used to target the VEGF signaling system, ranging from the development of VEGF-neutralizing antibodies, soluble VEGF receptors and inhibitors of VEGF receptors to the VEGF neutralizing aptamers (11–13). The latter approach resulted in the development of the first Food and Drug Administration (FDA)-approved aptamer drug, Macugen (Pegaptanib sodium), against age-related macular degeneration (13).
Aptamers are short nucleic acids with high affinities toward target molecules (14). Their generation involves several cycles of selection and enrichment using the so-called systematic evolution of ligands by exponential enrichment (SELEX) procedures starting from a random oligonucleotide sequence library (14,15). Aptamers display several advantages in comparison with antibodies, such as low toxicity, low immunogenicity, small size, ease of production and amenability to structural modification. Modifications introduced during or after the selection process are generally needed to optimize synthesis, stability, resistance toward nucleases and blood stream circulation time. Amongst numerous existing aptamer modifications, locked nucleic acids (LNA) (Figure 1a) contribute to increased binding affinity and higher nuclease resistance (16,17). LNA are widely exploited as a building block in therapeutic oligonucleotides (18,19). Sugar moieties of LNA nucleotides with their O2′-C4′-methylene bridge are locked in an N-type sugar conformation, mimicking that of RNA nucleotides. In Watson–Crick base-paired helices, LNA residues were shown to induce local reorganization of the sugar-phosphate backbone and to drive N↔S pseudorotational equilibria in neighboring residues toward N-type conformations. As a consequence, inclusion of LNA residues drives nucleic acid double helices toward an A form (20,21). On the other hand, a search for correlations between target binding and the propensity of aptamers to adopt secondary and tertiary structures has stimulated the introduction of other unnatural modified residues. Unlocked nucleic acid (UNA) in many ways represents an antipode to LNA (Figure 1a). The missing bond between the C2′ and C3′ atoms in the sugar moiety induces a high flexibility of UNA residues, which is reflected in destabilization of helices on UNA modifications (22). UNA modification was shown to improve nuclease resistance and to be valuable toward increasing target specificity (23–25). The applicability of UNA nucleotides in aptamer development was recently studied by systematically incorporating them in a known DNA-based thrombin-binding aptamer, whereby it was demonstrated that introduction of UNA residues at specific positions significantly increased the thrombin-binding affinity (26,27).
Figure 1.
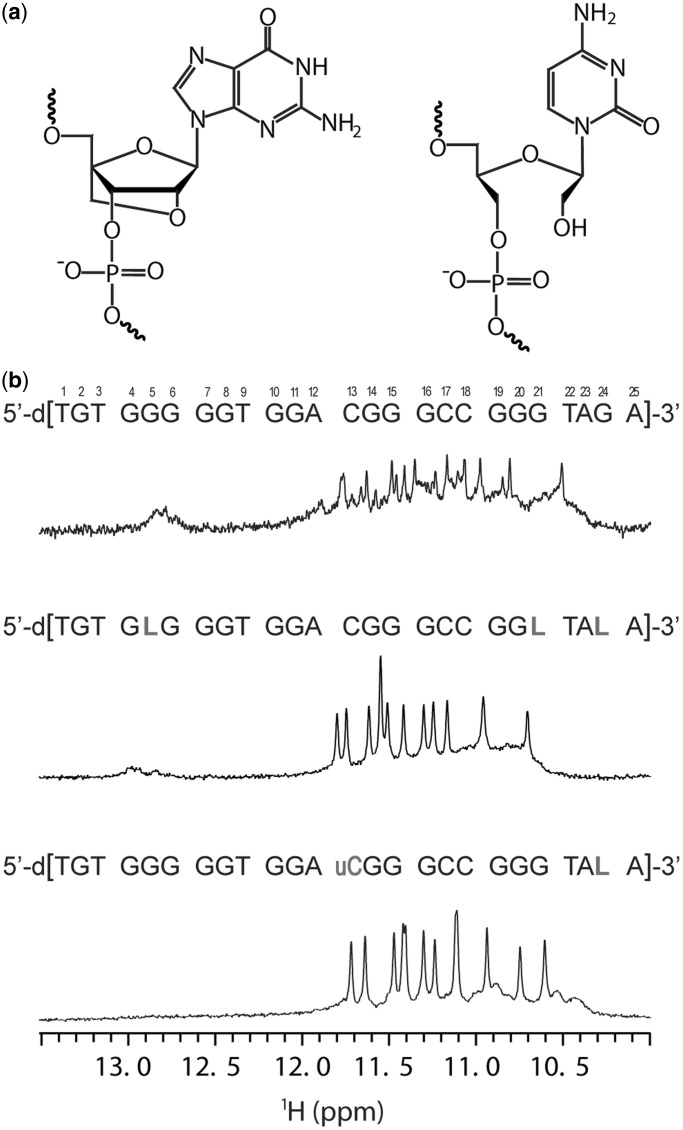
(a) Chemical structures of the LNA modified guanine residue (left) and UNA modified cytosine residue (right). (b) Sequences and the corresponding 1H NMR spectra of G-rich oligonucleotides V7t1 (top), RNV66 (middle) and RNV70 (bottom) in the presence of 100 mM concentration of K+ ions. 600 MHz 1H NMR spectra of ca. 1 mM oligonucleotide concentration per strand were acquired at 25°C in 10% 2H2O, 10 mM KPi buffer with pH 7. LNA and UNA nucleotides are designated with letter L and letter uC, respectively. Numbering of the residues is indicated.
One of the secondary structures often adopted by aptamers is a G-quadruplex, formed by four G-rich tracts within DNA and RNA sequences. The basic building blocks of G-quadruplex structures are four guanine residues arranged in coplanar G-quartets, which are stabilized with eight Hoogsteen hydrogen bonds. Stacked G-quartets form a G-quadruplex core, whereas extra-quartet residues connect nucleic acid strands in a form of different loops and represent one of the main contributions to the structural diversity and polymorphism of G-quadruplex structures (28,29).
In 2010, Nonaka et al. (30) described a VEGF aptamer V7t1 with KD values in a low nanomolar range for the most abundant VEGF isoforms VEGF165 and VEGF121 (31,32). V7t1 comprises several G-rich tracts (Figure 1b) and was predicted to fold into a G-quadruplex structure that was shown to be essential for VEGF binding. However, V7t1 contains 15 guanine residues arranged in a manner that does not allow straightforward predictions of their inclusion into G-quartets and, furthermore, G-quadruplex topology. Four G-rich tracts consisting of five, two and two times three residues together with two additional isolated guanine residues offer several possibilities for G-quadruplex core formation, and one would expect to observe structural polymorphism. The most straightforward prediction of G-quadruplex core assembly would include G10-G11, G14-G16 and G19-G21 tracts with any combination of guanine residues from the first G-tract. Potential two-residue loops (A12-C13 and C17-C18) would in this case favor propeller-type orientations and lead to the formation of a presumably most stable parallel G-quadruplex fold. Although knowledge of existing topologies has been growing constantly (33–36), it is hard to predict a G-quadruplex fold or to presume which guanine residues will form a G-quadruplex core even in the case of less diverse sequences (36,37). Our preliminary nuclear magnetic resonance (NMR) measurements indicated that V7t1 folds into several G-quadruplex structures. Our quest to generate a single stable G-quadruplex structure and to significantly improve the aptamer’s nuclease resistance, required for therapeutic applications, led to systematic modifications of V7t1 by inserting LNA and UNA nucleotides at various positions. The binding affinity analysis for all these constructs was then performed by surface plasmon resonance using Biacore-3000 instrument. After screening the affinity, it was found that oligonucleotide RNV66 with LNA guanine nucleotides at positions 5, 21 and 24 and oligonucleotide RNV70 with LNA guanine and UNA cytosine residue at positions 24 and 13, respectively (Figure 1b), were the best in binding VEGF (data not shown, a separate manuscript is currently under preparation including the biological studies). From a structural point of view, because of their C3′-endo furanose ring conformation, steric interactions of the nucleobase prevent LNA residues to adopt a syn conformation. As a consequence, incorporation of the LNA residues, especially in the G-quadruplex core, may completely abolish G-quadruplex formation (38). At the outset of the study on RNV66, we could expect to observe everything from formation of a parallel structure to complete prevention of G-quadruplex formation. In addition, NMR study of oligonucleotide RNV70 was initiated to obtain better understanding of the effect of constrained and flexible nucleotide modifications on folding of the G-rich aptamer sequence.
MATERIALS AND METHODS
Sample preparation
The LNA and UNA oligonucleotides were synthesized on a DNA synthesizer via standard phosphoramidite chemistry in 10 µmol scale. The synthesized oligonucleotides were deprotected and cleaved from the solid support by treatment with NH4OH at 55°C overnight. The crude oligonucleotides were then purified by RP-HPLC and desalted, before use in NMR experiments. UNA phosphoramidites are commercially available from RiboTask ApS.
Samples were dissolved in H2O containing 10% of 2H2O and titrated with aliquots of 1 M KCl or NaCl solutions to the final concentration of 50 or 100 mM potassium or sodium ions. pH of the samples was adjusted to 7 with 10 mM TE or K+ phosphate buffer. The sample in 2H2O was prepared by lyophilization of the titrated sample and dissolution in 100% 2H2O. Strand concentration in the samples was 0.5 or 1.0 mM and was determined by UV absorption at 260 nm using UV/VIS Spectrophotometer Varian CARY-100 BIO UV-VIS. Titrated samples were annealed with fast and slow cooling procedures. After heating the sample up to the 95°C for 4 min, the sample was cooled at a rate of 0.1°C/s to 24°C and put on ice for 10 min (fast cooling) or left to cool to room temperature overnight (slow cooling). Ultrafiltration was used to remove cations from the titrated sample: sample was heated to 95°C, diluted to 10 ml with 0.5 M LiCl, cooled to room temperature and subjected to several cycles of ultrafiltration.
UV spectroscopy
Thermal stability measurements were performed on a Varian CARY-100 BIO UV-VIS spectrophotometer with the Cary Win UV Thermal program using a 1-cm path-length cell. Samples were heated at 0.1°C min−1 from 10 to 90°C. Mineral oil and fixed cuvette cap was used to prevent sample loss at high temperatures. Condensation at lower temperatures was prevented with a stream of nitrogen applied throughout the measurements. Samples were prepared at 5 μM oligonucleotide concentration in 10 mM potassium phosphate buffer containing 50 mM K+ ions. Tm was determined from the A295 versus temperature plot.
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were recorded on an Applied Photophysics Chirascan CD spectrometer at 25°C using a 0.1 cm path length quartz cell. The wavelength was varied from 200 to 320 nm. Five scans were averaged for each CD spectrum. Samples for CD measurements were prepared at 5 μM oligonucleotide concentration in 10 mM potassium phosphate buffer containing 50 mM K+ ions. A blank containing only buffer was used for baseline correction.
NMR spectroscopy
NMR spectra were recorded on Agilent VNMRS 600 and 800 MHz NMR spectrometers at 25 and 0°C. Double-pulsed field gradient spin echo (DPFGSE) pulse sequence was used to suppress the water signal. The translational diffusion coefficient was determined using pulse field gradient stimulated echo (PFG-STE) pulse sequence. The 2D nuclear overhauser effect spectroscopy (NOESY) spectra acquired at τm of 40, 80, 250 ms (samples in 10% 2H2O/90% H2O) and 150 ms (samples in 10% 2H2O/90% H2O and 100% 2H2O) were used to determine glycosidic torsion angle conformation, to establish oligonucleotide topology and consequently to assign exchangeable and non-exchangeable proton resonances. The 2D double quantum filtered correlation spectroscopy (DQF-COSY) and total correlation spectroscopy (TOCSY) (τm of 20, 60 and 80 ms) spectra were used to cross-check assignment of 2D NOESY spectra and to estimate sugar conformations.
Spectra were processed with programs VNMRJ (Agilent Technologies) and NMRPipe (39). Cross-peak assignment and integration with a Gaussian fit procedure was achieved using software SPARKY (UCSF). NOE distance restraints for non-exchangeable protons were obtained from 2D NOESY spectra (τm 150 ms) recorded at 25°C in 100% 2H2O and 10% 2H2O/90% H2O. Non-overlapping peaks only were used for the distance restraints calculations. H5-H6 cross-peaks could not be used as the distance reference owing to the spectral overlap and average volume of H7-H6 cross-peaks of T1, T3, T9 and T22 was used instead (reference distance of 3.0 Å). Cross-peaks were classified as strong (1.8–3.6 Å), medium (2.5–5.0 Å) and weak (3.6–6.0 Å). Certain cross-peaks in the aromatic–aromatic region of 2D NOESY spectra were only observed in 2D NOESY spectra with a mixing time of 250 ms. These cross-peaks were also used in structure calculations and were classified as weak. NOE distance restraints for exchangeable protons were obtained from 2D NOESY spectra recorded at 25 and 0°C in 10% 2H2O/90% H2O with mixing times of 150 and 250 ms. Cross-peaks that could be observed in 2D NOESY spectrum with a mixing time of 150 ms were classified as medium (2.5–5.0 Å). Cross-peaks that appeared in 2D NOESY spectrum with a mixing time of 250 ms were classified as weak (3.6–6.0 Å).
Structure calculations
Structure calculations were performed with AMBER 11 software (40) and parmbsc0 force field (41). Force field parameters for the LNA guanine residues were used from previous studies (42). The initial extended single-stranded DNA structure was obtained with tleap. The solution-state structure was calculated in three cycles of 100 ps NMR restrained simulated annealing (SA) simulations with the generalized Born implicit model to account for solvent effects. Topology was constructed in the first two SA cycles with help of hydrogen-bond and torsion angle restraints. NOE restraints were used in the third step of SA calculations after the first 7 ps. The cutoff for non-bonded interactions was 20 Å, and the SHAKE algorithm for hydrogen atoms was used with the tolerance of 0.0005 Å. After 5 ps at 300 K, the temperature was raised to 1000 K in the next 10 ps and held constant at 1000 K for 30 ps. Temperature was scaled down to 100 K in the next 45 ps and reduced to 0 K in the last 10 ps. Restraints used in the calculation were hydrogen bond (force constant 50 kcal mol−1 Å−2) and NOE-derived distance restraints (force constant 40 kcal mol−1 Å−2), torsion angle χ (anti region), ν2 and ε (region 150°–300°) restraints (force constant 250 and 200 kcal mol−1 rad−2, respectively) and planarity restraints for G-quartets (force constant 30 kcal mol−1 rad−2). Van der Waals, electrostatic components and force constants for restraints were linearly increased to their final value in the first 10 ps of SA. Structures were minimized with a maximum of 20 000 steps of energy minimization. Of 100 calculated structures, a family of 10 was selected based on the smallest number of restraint violations. The overlap area between nucleobases of successive residues was calculated with program 3DNA (43,44).
RESULTS
Pre-formed intermediates are crucial for a single parallel G-quadruplex structure formation
The lyophilized sample of RNV66 was dissolved in water and titrated with NaCl and KCl solutions while its folding was followed by NMR spectroscopy. Several broad signals of low intensity were observed in the imino region of the 1D proton spectrum already before the addition of any cations (Supplementary Figure S1), indicating the presence of pre-formed secondary structure(s). Our further experiments have shown that structures resulting from the self-organization of RNV66 before addition of cations appear to be essential for folding into predominantly one G-quadruplex structure. Three of the imino signals at δ ∼13 ppm designated presence of Watson–Crick hydrogen-bonded base pairs. CD spectra suggested nucleobase stacking interactions in the pre-formed state(s) at low cation concentrations. Although it is clear that RNV66 is not unfolded completely before addition of potassium ions, but rather adopts some sort of a globular structure with Watson–Crick base pairs, prediction of a structure(s), which is highly dynamic based on the fingerprint imino region in 1D 1H spectrum and CD data, would be highly speculative. On addition of potassium ions, the intensity of these three signals decreased, and 11 reasonably well-resolved signals started to appear in the region between δ 10.5 and 11.8 ppm (Supplementary Figure S1a). The signal at δ 11.5 ppm was of double intensity compared with the other signals in the imino region and corresponded to signals of two Hoogsteen hydrogen-bonded imino protons. The presence of altogether 12 signals in the imino region of the 1D proton spectrum were in agreement with the formation of a G-quadruplex structure with three G-quartets.
Titration experiments showed folding to be complete at 100 mM K+ concentration when three imino signals at δ 13 ppm disappeared. The translation diffusion coefficient of 1.48·10−6 ± 0.05 cm2 s−1 measured with PFG-STE pulse sequence was in agreement with a monomeric structure (36,37). Both slow and fast annealing resulted in the unfolding of the G-quadruplex structure and formation of what appeared to be intermediates with different partially refolded topologies, as indicated by a broad signal at δ 10.5–11.0 ppm and confirmed by CD experiments (vide infra). The slow annealing procedure was, when combined with dilution, more efficient in refolding RNV66 into a single G-quadruplex structure in the presence of K+ ions (Supplementary Figures S2a and b). Interestingly, titration with sodium ions failed to induce folding of RNV66 into a G-quadruplex (Supplementary Figure S1b).
We wanted to identify intermediates with different partially refolded topologies that were presumably formed after the annealing procedure and rule out the possibility of formation of aggregates. Therefore, cations were removed from the sample by ultrafiltration and, a 1D 1H NMR spectrum was recorded under low salt conditions (Supplementary Figure S2c). The resulting spectrum was similar to the 1D 1H NMR spectrum of RNV66 before addition of cations, and folding into a single G-quadruplex structure in the presence of K+ ions with a well-resolved set of 1H NMR signals could be reproduced. This procedure showed that RNV66 fails to refold into a single G-quadruplex structure after annealing but can be refolded when folding starts from the initial pre-formed structure in the absence of cations.
We noticed that dynamics of the folding is rather slow; when an aliquot of 100 mM K+ ion concentration was added at once, formation of several intermediates was observed. It appears that after addition of potassium ions, the apparently slow formation of the G-quadruplex is overtaken by generation of faster forming structures (Supplementary Figure S2c). These fast forming species do not (re)fold into a single G-quadruplex structure even after a prolonged time (up to several months). When potassium ions were added gradually, a single G-quadruplex structure was preferentially formed.
The CD spectrum of RNV66 G-quadruplex in the presence of K+ ions with a minimum at ∼240 nm and a maximum at ∼260 nm suggested its topology to be parallel. Interestingly, there seems to be no difference in the spectral characteristics before and after annealing (Supplementary Figure S3a), which is in concordance with our NMR data: structures after annealing retain the major characteristics of a G-quadruplex present before annealing and can be interpreted as partially refolded intermediates. The CD spectrum of RNV66 before addition of cations displays characteristics of a parallel topology, confirming that pre-formed structure(s), which seem crucial for folding, are intermediates with features of a parallel G-quadruplex.
Novel topology of RNV66 G-quadruplex
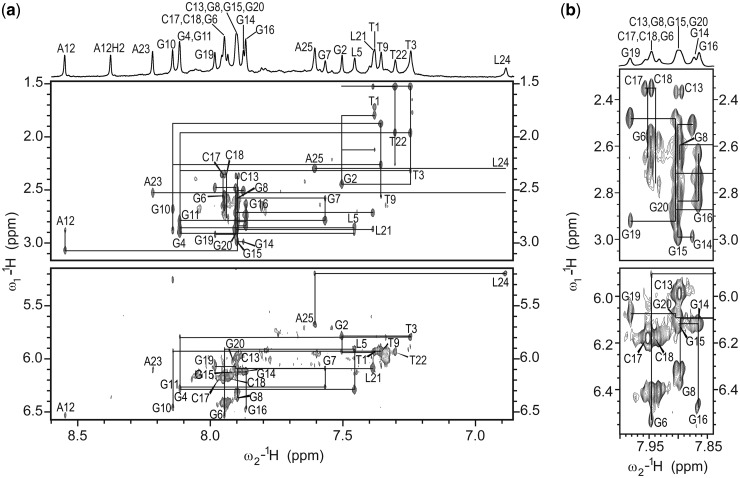
A 2D NOESY spectrum with a mixing time of 40 ms showed three strong cross-peaks in the aromatic–anomeric region that were assigned to H5–H6 correlations of three cytosine residues. No signals that could correspond to guanine residues in syn conformation were observed, which was in agreement with the CD spectrum, suggesting a parallel topology with all residues in anti conformation. Interpretation of NMR data relied on spectral resolution of 1H resonances alone as 15N/13C isotopically labeled oligonucleotides with LNA residues that would assist our assignment process were not available. Topology of a G-quadruplex was therefore determined as follows: (i) assignment of non-exchangeable aromatic and sugar proton resonances through a sequential walk; (ii) construction of a G-quadruplex core model(s) through NOE correlations between pairs of imino protons; and (iii) combination of sequential segments and model(s) with the help of inter-residual imino–aromatic NOE connectivities within and between neighboring G-quartets.
Inter-nucleotide connectivities were simultaneously followed through searching for sequential walks both in H8-H1′ and H8-H2′/H2′′ regions of 2D NOESY spectra (Figure 2) to prevent erroneous interpretation of the cross-peaks. Comparison of 2D NOESY spectra recorded in 2H2O and 2H2O/H2O helped to assign cross-peaks in a severely overlapped region from δ 7.86 to 7.98 ppm. G11-G7-G8 and G14-G15-G16 segments exhibited no NOE contacts with loop-forming residues and were identified after establishing their positions in the core of a G-quadruplex. On the other hand, cytosine, thymine and LNA residues with their specific spectral features enabled uninterrupted and unambiguous sequential walks in the T1-G6, T9-G10, A12-C13, C17-C18 and G19-A25 segments. Cytosine residues were identified through strong H5-H6 cross-peaks in the aromatic–anomeric region of the 2D NOESY spectrum. Thymine residues could be distinguished from the other residues, as they gave rise to rather strong H6-H7 cross-peaks in 2D NOESY spectrum. In addition, unless thymine residues were involved in sharp turns of the DNA strand, it was possible to observe sequential H7n-H6/8n-1 and H7n-H1′n-1 cross-peaks that complemented classical sequential connectivities. The presence of LNA residues, on the other hand, resulted in the interruption of a sequential walk in the H8-H2′/H2′′, but not in the H6/8-H1′ region, as H2′ protons of the LNA residues overlap with H3′/H4′ protons. It should be possible to distinguish LNA residues through strong H3′-H8 cross-peaks in a 2D NOESY spectrum. In the case of RNV66, however, H3′ protons resonated at δ ∼4.7 ppm and were therefore distorted by the water signal. In addition, several cross-peaks of the non-LNA (i.e. standard) residues in H6/8-H3′/H4′ region were overlapped, which further impeded distinction of H3′-H8 cross-peaks of LNA residues from the other cross-peaks in the H6/8-H3′/H4′ region of 2D NOESY spectra.
Figure 2.
(a) The aromatic-H2′/H2′′ and aromatic-anomeric regions of 2D NOESY spectrum (τm = 150 ms) of oligonucleotide RNV66. The straight lines denote aromatic H6/H8-sugar H1′ and aromatic H6/H8-sugar H2′/H2′′ cross-peaks in a sequential walk. For T3 and T22 connections between H7n-H8n-1 are also shown. For clarity, the central part of the spectrum with severe cross-peak overlap is shown separately in panel (b). A 1D 1H NMR spectrum of aromatic region with resonance assignment is shown above the 2D NOESY spectrum. NOESY spectrum was recorded at 800 MHz, 25°C in 10% 2H2O, 100 mM K+, 10 mM KPi buffer with pH 7 and oligonucleotide concentration of 0.5 mM.
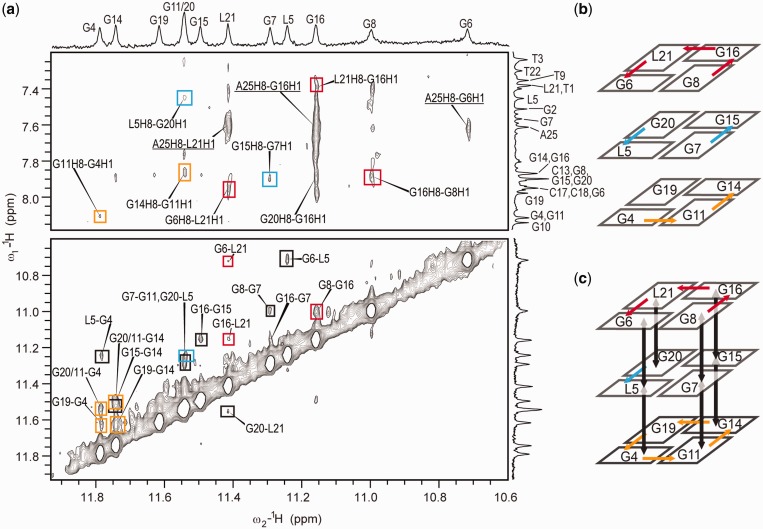
A model of a G-quadruplex core was built through careful analysis of the NOE correlations between imino protons. Our analysis of several structures deposited in PDB has shown that the expected distance between imino protons of sequential residues in ‘anti-anti’ steps is below 4.0 Å, the distance between neighboring imino protons within a G-quartet is between 4.0 and 4.5 Å, and the distance between non-sequential imino protons of neighboring G-quartets is also between 4.0 and 4.5 Å. Cross-peaks of medium intensity in the imino–imino region of 2D NOESY spectra were therefore assigned to correlations between sequential G-quartets forming residues. Distinction between weak and medium cross-peaks was facilitated by a 2D NOESY spectrum (τm of 250 ms) recorded at 0°C, which enabled observation of medium intensity cross-peaks (Supplementary Figure S4). Residues involved in a G-quadruplex core were identified through NOE correlations of imino and aromatic protons (Figure 3a) that connected cross-peaks in the imino region with assignment of residues in the aromatic region (Figure 3b and c). Identification of G4-G6 and G19-L21 tracts, which were unambiguously identified through a sequential walk, formed basis to build a topology model. NOE correlations in the imino–aromatic region have proved unequivocally that G4-G6 and G19-L21 tracts are neighboring strands in a G-quadruplex structure. Subsequently, G14-G16 was recognized as the neighboring strand to G19-L21. As G2, G10 and L24 exhibited no NOE correlations in the imino–aromatic region, and as their imino protons were involved in fast exchange, it was concluded that these residues do not participate in the formation of G-quartets. Consequently, one of the DNA strands in the G-quadruplex core had to comprise residues G7, G8 and G11. Their relative positions were established with help of a crucial albeit very weak G8 H1′-T9 H7 cross-peak, which identified G8 as member of the G6-G8-G16-L21 quartet. Furthermore, G8 H1-G16 H8 and G8 H1-G16 H1 cross-peaks affirmed hydrogen-bond directionalities within the G6-G8-G16-L21 quartet. Even though G11–G7 is not a sequential step, classical H1′-H8 and H2′/H2′′-H8 NOE connectivities could be observed for these two residues (Figure 2). G7 and G11 were determined to be positioned in the L5-G7-G15-G20 and G4-G11-G14-G19 quartets, respectively, on the basis of the intra-quartet imino–imino and inter-quartet imino–imino and imino–aromatic contacts (Figure 3).
Figure 3.
(a) Imino–aromatic and imino–imino regions of 2D NOESY spectrum (τm = 250 ms) of RNV66 with resonance assignment indicated along 1D traces. Cross-peaks labeled with red, blue and orange rectangles correspond to the G6-G8-G16-L21, L5-G7-G15-G20 and G4-G11-G14-G19 quartets, respectively. Cross-peaks labeled with black rectangles correspond to correlations between sequential residues within a G-quadruplex core. Underlined assignments correspond to the correlations of residues involved in loop and flanking regions. Spectrum was recorded at 800 MHz, 25°C in 10% 2H2O, 50 mM K+, 10 mM KPi buffer with pH 7 and oligonucleotide concentration of 0.5 mM. Schematic presentations of observed NOE connections for imino–aromatic and imino–imino regions are shown in (b) and (c), respectively. Colors of the arrows correspond to the color of the NOE cross-peaks in panel (a).
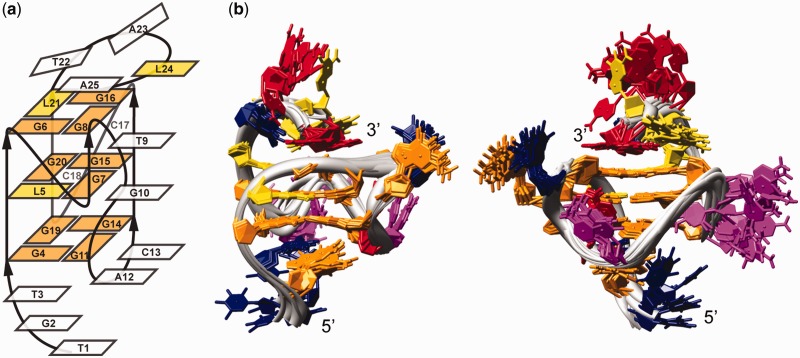
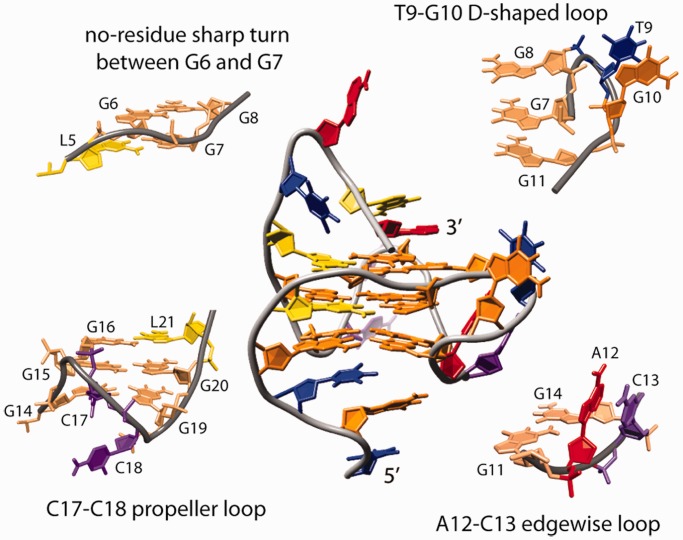
The proposed topology of a G-quadruplex that satisfies all NMR and other spectroscopic data is parallel with four loops, and three and four residue overhangs at the 5′ and 3′ ends, respectively (Figure 4a). One of the loops, which exhibits a propeller-type topology, but with no residues involved, spans the two G-quartets and connects G6 with G7. The most unique feature of RNV66 G-quadruplex is involvement of G11 in the G-quartet formation. G7–G8 and G11 can be considered parts of two DNA strands that form the same edge of G-quadruplex core. As G8 and G11 are on the opposite ends of a three G-quartet core they are connected with a novel type of D-shaped loop consisting of two-residues T9-G10 (Figure 4a). G11 and G14 belonging to the same G-quartet are linked by a two-residue (A12-C13) edgewise loop (Figure 4a). The last propeller loop consists of two residues (C17-C18). Hydrogen-bond directions in the G-quartets are as follows: G4→G11→G14→G19, L5→G7→G15→G20 and G6→G8→G16→L21. Several NOE contacts involving residues in T1-T3 and T22-A25 segments indicate that flanking regions are reasonably well ordered. A high number of NOE correlations of residues T3 and A25 (Supplementary Figures S4 and S5) demonstrates that their position is well defined by stacking on the G4-G11-G14-G19 and G6-G8-G16-L21 quartets, respectively. NOE contacts of protons H1′, H6 and H7 of T3 with the imino protons of all four residues of the G4-G11-G14-G19 quartet (Supplementary Figure S4) affirm that these residues compose one of the outer G-quartets. It is particularly interesting for A25 as the residue at the 3′ end of the molecule to exhibit high number of NOE contacts and consequently to adopt well-defined conformation. Position of A25 leads to the formation of the T22-A25 capping structure on the G6-G8-G16-L21 quartet, resembling a basket handle.
Figure 4.
(a) RNV66 G-quadruplex topology. Residues forming the G-quadruplex core and LNA residues are shown in orange and yellow rectangles, respectively. (b) Superimposition of the family of the 10 best structures. The two views are rotated with respect to one another to offer better overview of the structures. Guanine, LNA, adenine, thymine and cytosine residues are colored orange, yellow, red, blue and pink, respectively.
It was previously observed that LNA residues in G-quadruplexes promote ‘C3′-endo’ conformation in neighboring nucleotides, especially residues in the 3′ direction (33,45). However, this could not be unequivocally verified for residues of the LNA-modified RNV66 G-quadruplex. In general, an estimate of JH1’-H2’ coupling constant values and thus a predominant sugar pucker could be established on the basis of the sums of JH1’-H2’ and JH1’-H2” coupling constants determined from 2D DQF-COSY spectrum (Supplementary Figure S7). The values of JH1′-H2′ coupling constants were in the range from 7 to 10 Hz for residues T1, T9, G10, G11, C13, C17, C18, T22 and A25, which is consistent with their high population of a S-type sugar conformers. In comparison, JH1’-H2’ coupling constants from 1 to 3 Hz were determined for residues G4 and G19, suggesting predominance of N-type sugar conformation. Intermediate JH1′-H2′ coupling constants ranging from 4 to 6 Hz were observed for residues G2, G7, G8, A12, G15, G16, G20 and A23. Accordingly, these residues were considered to be involved in an unbiased N↔S pseudorotational equilibrium. The values of JH1′-H2 coupling constants could not be assessed for T3, G6 and G14 residues due to spectral overlap.
LNA and UNA modifications promote formation of a single G-quadruplex
Characterization of oligonucleotides V7t1 and RNV70 was performed to evaluate the effect of substitutions of a few residues with locked or unlocked sugar conformation on the formation and structure of a G-quadruplex. Numerous signals in the imino region of the 1D 1H NMR spectra of oligonucleotide V7t1 recorded at increasing K+ ion concentration indicated formation of several structures; some of them stabilized by Watson–Crick hydrogen-bonded GC base pairs (Supplementary Figure S6a). Twelve signals with the highest intensity in the region between δ 10.5 and 11.8 ppm indicate that the predominant structure adopted by V7t1 is a G-quadruplex with three G-quartets. Incorporation of LNA residues in the same oligonucleotide sequence has evidently led to the stabilization of a single G-quadruplex with parallel topology described in the current study. We could therefore presume that other G-quadruplex structures of V7t1 were of antiparallel or mixed-type topologies, and that the modification with LNA residues was responsible for folding into a single G-quadruplex. Additionally, LNA modifications were expected to have greater impact on the stabilization of a parallel structure if placed in the G-quadruplex core. For RNV70, LNA modification at residue G24 was expected to restrict flexibility of the 3′ flanking region and destabilize the G-quadruplex structure. Moreover, UNA modification at residue uC13 of RNV70 could contribute to additional flexibility of the loop and lead to the formation of structurally less constrained and consequently thermally destabilized G-quadruplexes. As uC13 was expected to have a stabilizing effect not in only one, but in a set of different structures, structural polymorphism observed for V7t1 should be preserved. However, titration of RNV70 with potassium ions resulted in the formation of a single G-quadruplex structure (Supplementary Figure S6b) with the same parallel topology adopted by RNV66, which highlighted the importance of L24 modification for G-quadruplex stabilization. Comparison of melting temperatures determined with UV spectroscopy (Supplementary Figure S8) has shown higher thermal stability of modified oligonucleotides RNV66 (Tm ∼52°C) and RNV70 (Tm ∼46°C) compared with unmodified V7t1 (Tm ∼40°C).
The CD spectra of RNV66 before and after addition of cations are similar (Supplementary Figure 3a), indicating that modification of G5 and G21 with LNA residues locks the oligonucleotide in the conformation that will most easily fold into a parallel G-quadruplex. The suggested pre-organized structure, however, is absent in the case of RNV70 and V7t1 (Supplementary Figures S3b and c). Addition of potassium ions induces formation of a predominant parallel G-quadruplex for all three oligonucleotides. In the case of V7t1 in the presence of K+ ions, a broad signal of low intensity at ∼290 nm in the CD spectra before and after annealing indicates formation of G-quadruplex topologies of antiparallel nature, in agreement with both numerous signals in the imino region of 1D 1H NMR spectrum and predicted effect of the LNA modifications. It is clear that inclusion of a single LNA residue (L24) in RNV70 drives equilibrium toward a parallel topology, which shows that the T22-A25 capping structure is important for formation and stabilization of the peculiar topology adopted by RNV66 and RNV70. Further LNA modifications of residues in the G-quadruplex core (L5 and L21) of RNV66 have two effects: first, they stabilize pre-formed structures that more efficiently lead to the formation of a single G-quadruplex structure, and second, they stabilize intermediates that are formed after annealing and retain certain characteristics of G-quadruplex topology. In concordance, signals in the CD spectra after annealing exhibit reduced intensity in comparison with the signals before annealing for RNV70. The reduction is even more pronounced for V7t1, but there is hardly any decrease for RNV66 (Supplementary Figure S3), showing that the LNA-modified G-quadruplex core forms more efficiently in the case of RNV66.
RNV66 adopts a parallel G-quadruplex with well-defined loops and flanking regions
Structure calculation was based on a large number of NOE distance and other types of restraints (Table 1). We have refrained from using ambiguous NOE restraints in structural calculations. As a consequence, only 248 NOE distance restraints were used of ∼360 assigned NOE cross-peaks in 2D NOESY spectra. Ambiguous NOE restraints were, however, used in the evaluation process, and structures not in agreement with the observed NOEs were discarded. Superimposition of the 10 lowest energy structures with the lowest number of restraint violations is presented in Figure 4b, and representative structure in Figure 5.
Table 1.
Structural statistics of the family of 10 structures
| NMR distance and torsion angle restraints | |
|---|---|
| NOE-derived distance restraints | |
| Total | 248 |
| Intra-residue | 143 |
| Inter-residue | 105 |
| Sequential | 54 |
| Long-range | 51 |
| NOEs not observed | 2 |
| Hydrogen bond restraints | 24 |
| Torsion angle restraints | 59 |
| G-quartet planarity restraints | 36 |
| Structure statistics | |
| Violations | |
| Mean NOE restraint violation (Å) | 0.078 ± 0.0005 |
| Max. NOE restraint violation (Å) | 0.176 |
| Max torsion angle restraint violation (°) | 4.270 |
| Deviation from idealized geometry | |
| Bonds (Å) | 0.012 ± 0.000 |
| Angles (°) | 2.731 ± 0.001 |
| Pairwise heavy atom RMSD (Å) | |
| Overall | 2.310 |
| G-quartets | 0.622 |
| G-quartets and G2, T3, T9, G10, A12, C13, T22 | 0.894 |
Figure 5.
Representative structure and detailed views of four different fragments connecting G-rich strands in the G-quadruplex core. Guanine, LNA, adenine, thymine and cytosine residues are colored orange, yellow, red, blue and purple, respectively. Guanine residues of the G-quadruplex core are shown in light orange in the views of loop fragments.
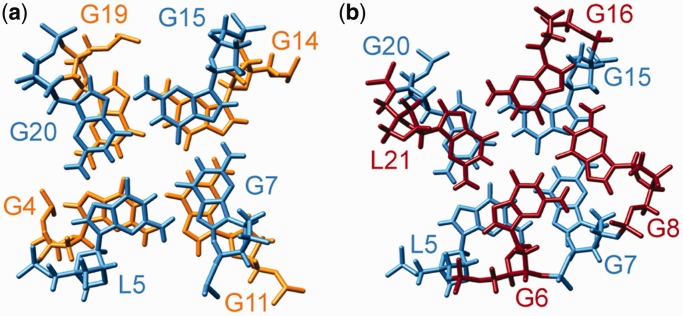
The RNV66 G-quadruplex structure is parallel with three stacked G-quartets. Loop regions comprises a no-residue propeller-type loop between G6 and G7, a T9-G10 D-shaped loop connecting outermost residues G8 and G11 within the same strand, an A12-C13 edgewise loop and a most flexible C17-C18 propeller loop (Figure 5). Long flanking regions are well-ordered; we can observe stacking in the T1-T3 segment and in the capping structure formed by T22-A25 (Figure 4b). The DNA backbone is in an extended conformation in the G6-G8 tract, which causes displacement of G6 and G8 from what is considered their ideal stacking position (Figure 6), as reflected in the small overlap of the aromatic rings of the residues L5-G6 (∼1.0 A2) and G7-G8 (∼0 A2). In comparison, the average overlap area between polygons defined by nucleobase ring atoms of the successive residues of G4-G11-G14-G19 and L5-G7-G15-G20 quartets is 3.7 A2. Helical twist of 18.7° and 26.8° for the two quartet–quartet steps denote underwinding of the helical architecture. All four grooves are of medium width (14–16 Å) and are extensively protected from solvent by loop segments, except for the groove defined by G4-G6 and G19-L21 strands, which is available for potential interactions. Substantial solvent protection of the core of RNV66 G-quadruplex is reflected in the slow disappearance of imino protons in 1D 1H NMR spectrum after solvent exchange to 2H2O. Hydrogen–deuterium exchange rates are uniform for all three G-quartets, although the central G-quartets are usually better protected from solvent (Supplementary Figure S9).
Figure 6.
Stacking interactions between (a) G4-G11-G14-G19 (orange) and L5-G7-G15-G20 (light blue) quartets and between (b) L5-G7-G15-G20 (light blue) and G6-G8-G16-L21 (red) quartets.
The DNA strand must adopt a sharp turn to connect residues G8 and G11 through the T9-G10 D-shaped loop (Figure 5) and enable G11 to adopt anti-conformation along the glycosidic torsion angle. Anti-conformation of G11 is in concordance with the observed medium intensity of a G11 H1′-H8 cross-peak in the 2D NOESY spectrum. Syn conformation along the glycosidic bond angle, however, would be expected for G11 considering solely the direction of the DNA strand, as the relative orientations of the strands are related to the glycosidic conformation of the guanine residues (46). Owing to the abrupt change in DNA strand directionality followed by the T9-G10 loop, no sequential NOE contacts between G8 and T9 were observed. On the other hand, stacking interactions between T9 and G10 could be observed in practically all calculated structures (Figures 4b and 5), in complete agreement with T9 H1′-G10 H8 and T9 H6-G10 H8 NOE correlations. A12 and C13 form a peculiar type of edgewise loop that connects residues in anti-conformation in the neighboring DNA strands (Figures 4b and 5). A12 is held in the center of the groove between G11-G7-G8 and G14-G15-G16 DNA strands, whereas C13 stacks with A12 on the outer side of the G-quadruplex (Figures 4b and 5).
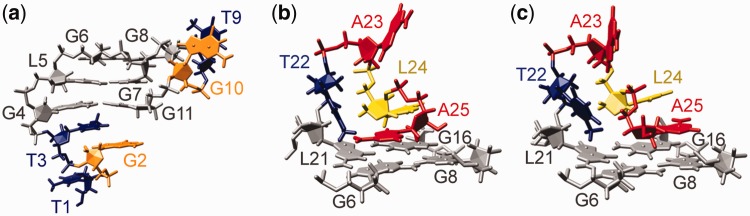
Residues of the T1-T3 and T22-A25 regions exhibit several inter-residual NOE contacts and are well defined in the calculated structures. The T1-T3 flanking region participates in continuous stacking of a G4-G6 strand of the G-quadruplex core (Figures 4b and 7a). T3 displays one of the highest numbers of NOE restraints per residue (Supplementary Figure S5), and its stacked position on G4-G11-G14-G19 quartet is very well defined. Although stacking interactions with T3 keep G2 in relatively restrained conformational space, T1 exhibits the greatest level of flexibility in the T1-T3 segment (Figure 4b).
Figure 7.
Close-up views of (a) stacking of T1-T3 on G4-G11-G14-G19 quartet and (b-c) two distinct conformations of A25 in the T22-A25 capping structure stacked over the G6-G8-G16-L21 quartet. Guanine, LNA, adenine, thymine and cytosine residues of loops are depicted in orange, yellow, red, blue and pink, respectively; G-quartet forming residues are shown in gray.
Position of T22 is well defined, with a methyl group protruding over residues L21 and G6. A hydrogen bond could be formed between residues T22 and A25. However, there is no affirmative experimental data supporting T22-A25 base pair formation. A25 exhibits several NOEs with guanine residues in the G6-G8-G16-L21 quartet. During SA simulations, two sets of A25 conformations were found that agree with experimental data; in the first one, A25 is positioned over the center of the G6-G8-G16-L21 quartet pointing toward G6 (Figure 7b), whereas in the second, A25 is shifted above residues G16 and G8 (Figure 7c).
We have observed that all resonances of residue L24 exhibit a substantial upfield shift (45) (Figure 2). Such chemical shift changes are usually correlated with residues well protected from solvent. Few and weak NOE contacts of L24 with other residues offer insufficient structural restraints for L24 adopting a well-defined solvent-protected position within the T22-A25 flanking region. However, structural calculations have shown that L24 adopts a relatively fixed position in most of the structures (Figure 4b). A23 is thus the only residue of the T22-A25 capping region that displays a higher level of flexibility, in agreement with its scarce NOE contacts.
DISCUSSION
A VEGF aptamer V7t1 with unmodified residues was proposed to fold into a G-quadruplex with a topology similar to a thrombin binding aptamer (30). In such a hypothetical structure with two G-quartets, seven guanine residues were not involved in hydrogen bond formation and belonged to the loop regions. It seems highly unlikely, though, that an oligonucleotide with 15 guanine residues will form a structure with only two G-quartets, especially as the last two G-rich tracts in the sequence comprise three guanine residues. Accordingly, formation of a G-quadruplex with three G-quartets was anticipated and subsequently observed as described herein for V7t1 and its modified analogues RNV66 and RNV70 in a K+ rich environment. As both CD and NMR data show that the predominant G-quadruplex structure adopted by V7t1, RNV66 and RNV70 exhibits parallel strand orientation, it could be envisaged that LNA modifications selectively stabilize a parallel topology described in the current study and do not lead to the re-organization into a completely different topology. Introduction of LNA residues in RNV66 and RNV70 therefore destabilizes structures with Watson–Crick hydrogen-bonded GC base pairs formed by V7t1 and results in formation of a single G-quadruplex, allowing characterization of a predominant G-quadruplex formed by the parent oligonucleotide V7t1.
Perusal of the oligonucleotide sequence suggested that RNV66 adopts unusual topology; five consecutive guanine residues (G4-G8) could not form two complete columns of a G-quadruplex core consisting of three G-quartets established by the presence of 12 imino protons and would require insertion of residues G2, G10, G11 or L24 into a G-quartet. Interruption of the G-rich tract with T3 or T9 could be accommodated by a bulge as seen in the dimeric G-quadruplex formed by HIV-1 integrase inhibitor (47). Long G-tracts were also observed to form no-residue propeller-type loops that can span three G-quartets (33,36,48). In the RNV66 G-quadruplex, a no-residue propeller-type loop facilitates involvement of five sequential (G4-G8) guanine residues in a G-quartet formation. However, no bulge is formed in the RNV66 G-quadruplex structure, and a new element is observed instead: a two-residue propeller-type loop within the DNA strand connecting residues G8 and G11 of the outer G-quartets within the same column of a G-quadruplex core. G11 adopts anti-conformation along its glycosidic bond, and G7-G8 and G11 segments can be considered parts of two DNA strands oriented in a parallel fashion. Propeller loops generally connect adjacent parallel DNA strands. In this respect, the T9-G10 loop in the RNV66 G-quadruplex represents a new kind of D-shaped intra-strand propeller-type loop orientation within a single-column of a G-quadruplex core. The subsequent edgewise (A12-C13) and propeller (C17-C18) loops in the RNV66 structure are more commonly observed in G-quadruplexes. Flanking regions were reported to importantly contribute to the stabilization of G-quadruplex folds (49–53). Likewise, stabilization of the capping structure formed by the T22-A25 overhang containing a LNA residue contributes to the formation of a single structure for RNV70 and RNV66 G-quadruplexes.
Analysis of NMR data clearly shows that G11 adopts an anti-conformation in the RNV66 G-quadruplex. Nevertheless, during structure calculations, it was observed that it could easily switch to a syn conformation, even though a force constant of 250 kcal mol−1 rad−2 was used to restrain it to the anti-region. In most of the structures that were calculated during the first two steps of SA, the G11 glycosidic bond angle was in a ‘high-anti’ or classical syn (∼60°) region. Anti-conformation of G11 could only be achieved if the sugar-phosphate backbone of the T9-G10 segment adopted extended conformation. This conformation, in turn, leads to the displacement of G8 and to a smaller extent G6 from the position considered to be optimal for the stacking of the guanine bases, which is reflected in reduction of the overlap area between residues L5-G6 and G7-G8 according to the assessment by 3DNA program (43,44). As stacking of the nucleobases is recognized as one of the most important factors contributing to the stability of G-quadruplex structures, this displacement either causes destabilization of the structure or must be compensated for by other stabilizing interactions. Relatively low melting temperatures established for all three oligonucleotides—considering parallel organization of the strands—might consequently reflect less efficient stabilization of the parallel topology due to the reduced stacking interactions.
Differences in the melting temperatures of RNV66 and RNV70 with respect to V7t1 can be attributed to the thermal stabilization of their structures by LNA and UNA modifications. RNV66 with three LNA residues is more stable than RNV70 with one LNA and one UNA residue, whereas unmodified V7t1 displays the lowest thermal stability. In general, it was reported that LNA modifications contribute to the thermal stabilization of parallel G-quadruplexes (38,45,54), whereas impact on the stability of non-parallel G-quadruplexes depends on the position of the modification in the sequence. It is clear that LNA modification of a guanine residue adopting syn conformation in a certain G-quadruplex structure will prevent that structure to be formed (33,55,56), whereas LNA modifications of the residues adopting anti-conformation may lead to thermal stabilization of the overall fold. In concordance, LNA modifications of RNV66 singled out and thermally stabilized parallel G-quadruplex structure. As RNV66 adopts a parallel topology and LNA inclusion leads to the stabilization of the single structure, we expect to detect improved binding affinity of RNV66 in comparison with the V7t1.
For thrombin-binding aptamer, it was shown that crucial interactions of loop regions with thrombin contribute to the biological activity, and therefore preserving the flexibility of the loops is of great importance to achieve binding (57). Stabilization of the loops and flanking regions by introduction of LNA residue(s) into an oligonucleotide sequence can accordingly compromise flexibility of the structure and decrease binding affinity. As the 3′ flanking region of RNV66 is not flexible, it can be expected that structural changes in this region on binding to VEGF will not be substantial. Moreover, LNA modification in this part might stabilize conformation of the T22-A25 capping structure that resembles a bound state. In such a case, stabilization could lead to increased biological activity as seen, for example, in the transition state analogues that bind enzymes. However, the proposed structural implications for binding of RNV66 to VEGF clearly need experimental validation, which is currently on-going in our laboratories.
CONCLUSIONS
We have determined the solution state structure of a G-quadruplex adopted by LNA-modified VEGF aptamer RNV66 in the presence of K+ ions using NMR spectroscopy. RNV66 folds into a parallel G-quadruplex with three G-quartet planes and unique loop orientations. Inclusion of five consecutive guanine residues in the G-quadruplex core is accommodated by a no-residue propeller-type loop, whereas a two-residue D-shaped intra-strand propeller-type loop connects residues of the outer G-quartets within the same G-rich strand and is followed by edgewise and propeller loops. The 5′ and 3′ flanking regions assume well-defined conformations and contribute to G-quadruplex stability. Although RNV66 comprised LNA residues as well as RNV70 with LNA and UNA modifications adopt a single G-quadruplex structure, their counterpart V7t1 with natural nucleotides forms several structures, some of them stabilized with Watson–Crick hydrogen bonds. LNA modifications were shown to stabilize a single G-quadruplex structure and importantly affect its thermal stability. Moreover, inclusion of LNA residues contributes to the structural pre-organization of the oligonucleotide before addition of cations that are essential for G-quadruplex folding.
ACCESSION NUMBERS
pdb 2M53, BMRB 19035.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Slovenian Research Agency [ARRS, P1-0242 and J1-4020] and EU FP7 project Bio-NMR [261863]; the UQ Fellowship Scheme (to R.N.V.); acknowledges support from the Danish National Research Foundation and EU FP7 [ERC grant no. 268776 to J.W.]. Funding for open access charge: Slovenian Research Agency (ARRS).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.D’Amore PA, Glaser BM, Brunson SK, Fenselau AH. Angiogenic activity from bovine retina: partial purification and characterization. Proc. Natl Acad. Sci. USA. 1981;78:3068–3072. doi: 10.1073/pnas.78.5.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller JW, Adamis AP, Shima DT, D’Amore PA, Moulton RS, O’Reilly MS, Folkman J, Dvorak HF, Brown LF, Berse B, et al. Vascular endothelial growth factor/vascular permeability factor is temporally and spatially correlated with ocular angiogenesis in a primate model. Am. J. Pathol. 1994;145:574–584. [PMC free article] [PubMed] [Google Scholar]
- 3.Sivalingam A, Kenney J, Brown GC, Benson WE, Donoso L. Basic fibroblast growth factor levels in the vitreous of patients with proliferative diabetic retinopathy. Arch. Ophthalmol. 1990;108:869–872. doi: 10.1001/archopht.1990.01070080113046. [DOI] [PubMed] [Google Scholar]
- 4.Adamis AP, Miller JW, Bernal MT, D’Amico DJ, Folkman J, Yeo TK, Yeo KT. Increased vascular endothelial growth factor levels in the vitreous of eyes with proliferative diabetic retinopathy. Am. J. Ophthalmol. 1994;118:445–450. doi: 10.1016/s0002-9394(14)75794-0. [DOI] [PubMed] [Google Scholar]
- 5.Fassbender HG, Simmling-Annefeld M. The potential aggressiveness of synovial tissue in rheumatoid arthritis. J. Pathol. 1983;139:399–406. doi: 10.1002/path.1711390314. [DOI] [PubMed] [Google Scholar]
- 6.Rooney M, Condell D, Quinlan W, Daly L, Whelan A, Feighery C, Bresnihan B. Analysis of the histologic variation of synovitis in rheumatoid arthritis. Arthritis Rheum. 1988;31:956–963. doi: 10.1002/art.1780310803. [DOI] [PubMed] [Google Scholar]
- 7.Koch AE, Harlow LA, Haines GK, Amento EP, Unemori EN, Wong WL, Pope RM, Ferrara N. Vascular endothelial growth factor. A cytokine modulating endothelial function in rheumatoid arthritis. J. Immunol. 1994;152:4149–4156. [PubMed] [Google Scholar]
- 8.Folkman J, Cotran R. Relation of vascular proliferation to tumor growth. Int. Rev. Exp. Pathol. 1976;16:207–248. [PubMed] [Google Scholar]
- 9.Folkman J. Anti-angiogenesis: new concept for therapy of solid tumors. Ann. Surg. 1972;175:409–416. doi: 10.1097/00000658-197203000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo P, Fang Q, Tao HQ, Schafer CA, Fenton BM, Ding I, Hu B, Cheng SY. Overexpression of Vascular Endothelial Growth Factor by MCF-7 Breast Cancer Cells Promotes Estrogen-independent Tumor Growth in Vivo. Cancer Res. 2003;63:4684–4691. [PubMed] [Google Scholar]
- 11.Folkman J. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1995;1:27–31. doi: 10.1038/nm0195-27. [DOI] [PubMed] [Google Scholar]
- 12.Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat. Rev. Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 13.Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling: in control of vascular function. Nat. Rev. Mol. Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 14.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, Friebe M, Dinkelborg L, Erdmann VA. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lebars I, Richard T, Di Primo C, Toulmé JJ. LNA derivatives of a kissing aptamer targeted to the trans-activating responsive RNA element of HIV-1. Blood Cells. Mol. Dis. 2007;38:204–209. doi: 10.1016/j.bcmd.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Veedu RN, Wengel J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol. 2009;6:321–323. doi: 10.4161/rna.6.3.8807. [DOI] [PubMed] [Google Scholar]
- 19.Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem. Biodiv. 2010;7:536–542. doi: 10.1002/cbdv.200900343. [DOI] [PubMed] [Google Scholar]
- 20.Petersen M, Bondensgaard K, Wengel J, Jacobsen JP. Locked nucleic acid (LNA) recognition of RNA: NMR solution structures of LNA:RNA hybrids. J. Am. Chem. Soc. 2002;124:5974–5982. doi: 10.1021/ja012288d. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen KE, Rasmussen J, Kumar R, Wengel J, Jacobsen JP, Petersen M. NMR Studies of Fully Modified Locked Nucleic Acid (LNA) Hybrids: solution structure of an LNA:RNA hybrid and characterization of an LNA:DNA hybrid. Bioconjugate Chem. 2004;15:449–457. doi: 10.1021/bc034145h. [DOI] [PubMed] [Google Scholar]
- 22.Langkjær N, Pasternak A, Wengel J. UNA (unlocked nucleic acid): a flexible RNA mimic that allows engineering of nucleic acid duplex stability. Bioorg. Med. Chem. 2009;17:5420–5425. doi: 10.1016/j.bmc.2009.06.045. [DOI] [PubMed] [Google Scholar]
- 23.Itkes AV, Karpeisky MY, Kartasheva ON, Mikhailov SN, Moiseyev GP, Pfleiderer W, Charubala R, Yakovlev GI. A route to 2′,5′-oligoadenylates with increased stability towards phosphodiesterases. FEBS Lett. 1988;236:325–328. doi: 10.1016/0014-5793(88)80048-6. [DOI] [PubMed] [Google Scholar]
- 24.Bramsen JB, Laursen MB, Nielsen AF, Hansen TB, Bus C, Langkjær N, Babu BR, Højland T, Abramov M, Van Aerschot A, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bramsen JB, Pakula MM, Hansen TB, Bus C, Langkjær N, Odadzic D, Smicius R, Wengel SL, Chattopadhyaya J, Engels JW, et al. A screen of chemical modifications identifies position-specific modification by UNA to most potently reduce siRNA off-target effects. Nucleic Acids Res. 2010;38:5761–5773. doi: 10.1093/nar/gkq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasternak A, Hernandez FJ, Rasmussen LM, Vester B, Wengel J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011;39:1155–1164. doi: 10.1093/nar/gkq823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen TB, Henriksen JR, Rasmussen BE, Rasmussen LM, Andresen TL, Wengel J, Pasternak A. Thermodynamic and biological evaluation of a thrombin binding aptamer modified with several unlocked nucleic acid (UNA) monomers and a 2′-C-piperazino-UNA monomer. Bioorg. Med. Chem. 2011;19:4739–4745. doi: 10.1016/j.bmc.2011.06.087. [DOI] [PubMed] [Google Scholar]
- 28.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Nonaka Y, Sode K, Ikebukuro K. Screening and Improvement of an Anti-VEGF DNA Aptamer. Molecules. 2010;15:215–225. doi: 10.3390/molecules15010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- 32.Keck PJ, Hauser SD, Krivi G, Sanzo K, Warren T, Feder J, Connolly DT. Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science. 1989;246:1309–1312. doi: 10.1126/science.2479987. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen JT, Arar K, Petersen M. Solution structure of a locked nucleic acid modified quadruplex: introducing the V4 folding topology. Angew. Chem. Int. Ed. 2009;48:3099–3103. doi: 10.1002/anie.200806244. [DOI] [PubMed] [Google Scholar]
- 34.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crnugelj M, Sket P, Plavec J. Small change in a G-rich sequence, a dramatic change in topology: new dimeric G-quadruplex folding motif with unique loop orientations. J. Am. Chem. Soc. 2003;125:7866–7871. doi: 10.1021/ja0348694. [DOI] [PubMed] [Google Scholar]
- 37.Marušič M, Šket P, Bauer L, Viglasky V, Plavec J. Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic Acids Res. 2012;40:6946–6956. doi: 10.1093/nar/gks329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randazzo A, Esposito V, Ohlenschläger O, Ramachandran R, Mayol L. NMR solution structure of a parallel LNA quadruplex. Nucleic Acids Res. 2004;32:3083–3092. doi: 10.1093/nar/gkh629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, et al. 2010 AMBER 11, University of California, San Francisco. [Google Scholar]
- 41.Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, III, Laughton CA, Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aduri R, Psciuk BT, Saro P, Taniga H, Schlegel HB, SantaLucia J. AMBER force field parameters for the naturally occurring modified nucleosides in RNA. J Chem. Theory Comput. 2007;3:1464–1475. doi: 10.1021/ct600329w. [DOI] [PubMed] [Google Scholar]
- 43.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protocols. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen JT, Arar K, Petersen M. NMR solution structures of LNA (locked nucleic acid) modified quadruplexes. Nucleic Acids Res. 2006;34:2006–2014. doi: 10.1093/nar/gkl144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webba da Silva M. Geometric formalism for DNA quadruplex folding. Chem. Eur. J. 2007;13:9738–9745. doi: 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- 47.Mukundan VT, Do NQ, Phan AT. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011;39:8984–8991. doi: 10.1093/nar/gkr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuryavyi V, Patel DJ. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure. 2010;18:73–82. doi: 10.1016/j.str.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 51.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaynutdinov TI, Neumann RD, Panyutin IG. Structural polymorphism of intramolecular quadruplex of human telomeric DNA: effect of cations, quadruplex-binding drugs and flanking sequences. Nucleic Acids Res. 2008;36:4079–4087. doi: 10.1093/nar/gkn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedersen EB, Nielsen JT, Nielsen C, Filichev VV. Enhanced anti-HIV-1 activity of G-quadruplexes comprising locked nucleic acids and intercalating nucleic acids. Nucleic Acids Res. 2011;39:2470–2481. doi: 10.1093/nar/gkq1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pradhan D, Hansen LH, Vester B, Petersen M. Selection of G-quadruplex folding topology with LNA-modified human telomeric sequences in K+ solution. Chem. Eur. J. 2011;17:2405–2413. doi: 10.1002/chem.201001961. [DOI] [PubMed] [Google Scholar]
- 56.Randazzo A, Esposito V, Ohlenschläger O, Ramachandran R, Virgilio A, Mayol L. Structural studies on LNA quadruplexes. Nucleos. Nucleot. Nucl. 2005;24:795–800. doi: 10.1081/ncn-200060279. [DOI] [PubMed] [Google Scholar]
- 57.Bonifacio L, Church FC, Jarstfer MB. Effect of locked-nucleic acid on a biologically active G-quadruplex. A structure-activity relationship of the thrombin aptamer. Int. J. Mol. Sci. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.