Atherosclerosis arises from an excessive fibroproliferative-inflammatory response to injury of the artery wall and typically evolves over decades (1). Smooth muscle cells play a central role in the development of atherosclerotic lesions. Their ability to migrate from the medial compartment of the artery wall to the intima, where they proliferate, accumulate lipid, and generate extracellular matrix, is key to the development of occlusive vascular lesions.
A common intervention for treating arterial blockage is percutaneous transluminal coronary angioplasty, in which an inflatable balloon is positioned at the site of arterial narrowing and used to physically disrupt the local atherosclerotic plaque. While angioplasty usually provides immediate symptomatic relief, its longer-term benefits are limited in a large proportion of cases (30–50% within 6 months) because the treated artery once again narrows at the site of inflation (2). This process, termed restenosis, involves elastic recoil, in the first instance, followed by extensive replication and remodeling — mainly by smooth muscle cells, which lay down matrix and eventually decrease luminal diameter at the site of ballooning. Most angioplasties are now performed in conjunction with the local deployment of metallic stents (3), which provide structural support in the reopened vessel by helping to prevent vascular recoil (4, 5).
Pharmacologic agents able to decrease the incidence of restenosis in human subjects are extremely limited, with antiplatelet glycoprotein IIb/IIIa receptor antagonists (inhibitors of platelet aggregation and thrombus formation) providing most utility. Intracoronary radiation, antioxidants, and growth factor antagonists have also shown promise as inhibitors of restenosis (6). The hunt for efficient inhibitors of restenosis remains an ongoing challenge. Here, I review the transcriptional control of specific genes that regulate smooth muscle cell biology, and I discuss the use of novel nucleic acid–based therapeutics that might be employed at the time of treatment to block restenosis over the long term.
Activation of Egr-1 in response to injury
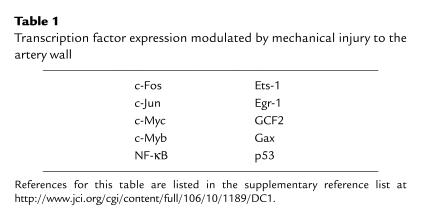
Cell movement, proliferation, and extracellular matrix deposition in the injured artery wall are mediated by the local activity of growth factors, cytokines, and other regulatory molecules. Mechanical injury activates signaling and transcriptional pathways that converge at the promoters of many genes whose products then influence the development of lesions. Insights into the molecular mechanisms associated with the transcriptional response to injury derive from a wealth of rat and other animal studies involving balloon catheter injury. As seen in these models, injury to the artery wall results in the rapid activation of mitogen-activated protein kinase subclasses, such as extracellular signal–regulated kinase (ERK) and stress-activated protein kinase/c-Jun NH2-terminal protein kinase (JNK), as well as a multitude of transcription factors (Table 1), including early growth response factor-1 (Egr-1) (7, 8).
Table 1.
Transcription factor expression modulated by mechanical injury to the artery wall
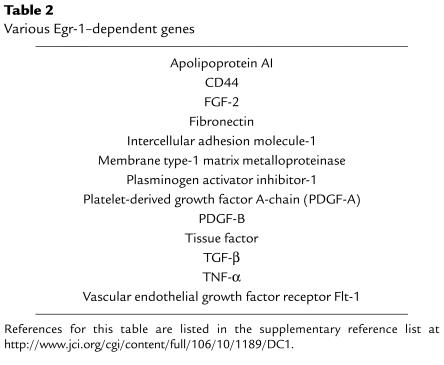
Egr-1 (also known as NGFI-A, zif268, Krox24, and TIS8) is an immediate-early gene product and transcription factor (9) expressed at low or undetectable levels in the normal artery wall (8, 10, 11). Within minutes of mechanical injury, Egr-1 is induced in vascular smooth muscle cells and endothelial cells (8, 10, 11), where it binds to specific G/C-rich nucleotide recognition elements in the promoter regions of certain genes, facilitating RNA polymerase II–dependent transcription. A growing list of pathophysiologically relevant genes are known to be targets of Egr-1 transactivation (see Table 2). The interaction of Egr-1 with several of these promoters involves the displacement of prebound Sp1 from common binding sites (8) (Figure 1). Interplay between Egr-1 and this “housekeeping” transcription factor may be a regulatory circuit common to the inducible expression of many genes. Activation of the Egr-1 promoter, like that of c-fos, depends on cooperative interactions between serum response and ternary complex factors at serum response elements (SREs), which are, in turn, governed by the activity of mitogen-activated protein kinases (12). The Egr-1 promoter bears six SREs, compared with one in the c-fos promoter (12).
Table 2.
Various Egr-1–dependent genes
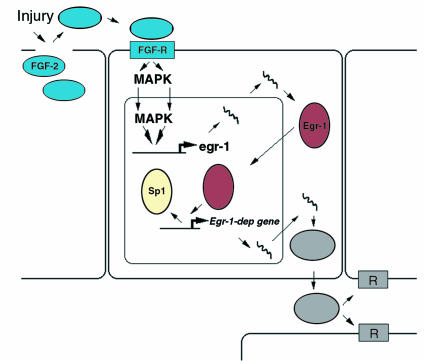
Figure 1.
Model of Egr-1–dependent response to injury. Injury to vascular cells releases preformed endogenous growth factors (such as FGF-2), which activate Egr-1 gene expression in a paracrine manner via mitogen-activated protein kinases (MAPKs). Egr-1 protein can, in certain promoters, displace Sp1 from overlapping binding sites and increase the expression of pathophysiologically relevant genes whose products contribute to neointima formation. Rec, receptor.
The activation of Egr-1 by injury may be the consequence of the local activity of factors resident in the artery wall (Figure 1). FGF-2, a mitogen and chemoattractant, is only inefficiently exocytosed and thus resides preformed in arterial cells. Levels of immunoreactive FGF-2, but not those of TGF-β or P-selectin, increase transiently in close proximity to atherosclerotic lesions in humans undergoing coronary stenting (13). Elevated FGF-2 levels have also been detected in the coronary sinus after angioplasty (14). Antibody blockade experiments indicate that the induction of Egr-1 after injury may be due to the release and local paracrine activity of endogenous FGF-2 (15).
Transcriptional regulators like Egr-1 are, in general, transiently expressed in the vasculature following injury. How these transcription factors and the genes they control are switched off after induction is poorly understood. The NGFI-A–binding proteins NAB1 (16) and NAB2 (17) repress Egr-1 activity via direct protein-protein interactions. NAB1 is constitutively expressed in most tissues, whereas NAB2 is weakly expressed and is induced by conditions that transiently upregulate Egr-1, including growth factor exposure (17) and vascular injury (18). In transient transfection experiments, overexpression of NAB2 inhibits Egr-1–dependent gene expression in vascular smooth muscle cells (18). The effects of Egr-1 can be offset by other transcriptional regulators that interact specifically with promoter elements in various Egr-1 target genes. For example, the transcription of PDGF-A, an Egr-1–dependent gene, is repressed by the inhibitory activity of GC factor 2 (GCF2), which is itself induced in the rat carotid artery wall following balloon injury (19). GCF2 competes with Egr-1, and other positive regulatory factors such as Sp1 and Sp3, for overlapping promoter binding sites in the PDGF-A promoter. Thus, GCF2 and the NABs, among other transcriptional repressors, may influence the course of gene expression in the artery wall after injury by antagonizing the activity of Egr-1.
Egr-1 as a therapeutic target in restenosis
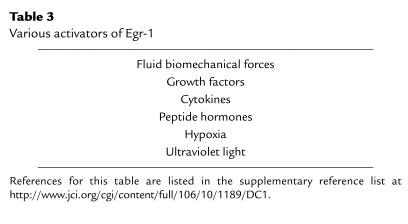
For a number of reasons, Egr-1 seems to offer an ideal target for therapeutic intervention to block restenosis. First, Egr-1 is poorly, if at all, expressed in the uninjured artery wall but is rapidly upregulated by mechanical injury (7, 8, 10). Second, Egr-1 controls the expression of a large number of genes whose products are implicated in the development of vascular lesions and associated complications — factors that modulate migration, proliferation, extracellular matrix production, intercellular adhesion, and thrombosis (Table 2). Third, besides injury, Egr-1 is activated by multiple external stimuli that may contribute to the pathogenesis of atherosclerotic and postangioplasty restenotic lesions (Table 3). These diverse influences may affect vascular remodeling and hyperplasia in atherogenesis, restenosis, hypertension, and ischemia reperfusion. Finally, Egr-1 and a large number of Egr-1–dependent genes have recently been detected in human atherosclerotic lesions. Immunohistochemical analysis revealed that Egr-1 is expressed primarily in smooth muscle cells in the fibrous cap mostly in areas of macrophage infiltration, as well as in endothelial cells (20).
Table 3.
Various activators of Egr-1
Antisense oligonucleotides
Strategies that selectively target and inhibit regulatory transcription factors associated with the increased expression of pathophysiologic genes may be therapeutically useful in vascular disease. Antisense oligonucleotides represent useful molecular tools to investigate the roles played by specific gene products in certain disease settings. These molecules hybridize with their complementary target site in mRNA, blocking translation to protein either by sterically inhibiting ribosome movement along the mRNA or by triggering cleavage by endogenous RNase H (21). Adsorptive endocytosis and pinocytosis likely account for oligodeoxynucleotide internalization in most cell types (22).
Antisense oligonucleotides, especially those that are stabilized by the use of phosphorothioate, rather than phosphodiester linkages, have been used to block neointima formation in various vascular models. Intimal growth following injury is suppressed by oligonucleotides targeting c-Myb, c-Myc, NF-κB p65, cdk2, and cdc2 kinase/proliferating-cell nuclear antigen (23). A major concern in the use of phosphorothioate-linked antisense oligonucleotides is nonspecific biological effects. Oligomers carrying stretches of G nucleotides form an unusual structure called a G-quartet, which can interact in a sequence-independent manner with diverse proteins, including growth factors, signaling molecules, enzymes, and components of the extracellular matrix (22). Clearly, however, such interactions do not account for all of the biological effects of phosphorothioate oligonucleotides, since oligonucleotides that do not form G-quartets can inhibit intimal thickening following balloon injury in animal models. Another concern with phosphorothioate-containing oligonucleotides is that, although they increase resistance to nucleases, they decrease the thermodynamic stability of RNA-DNA hybrids and may compromise biological efficacy. For example, phosphodiester antisense c-myb oligonucleotides inhibit smooth muscle cell proliferation in vitro 30-fold more potently than their phosphorothioate counterparts.
A single endoluminal delivery of the native oligonucleotide inhibits neointimal hyperplasia after angioplasty to the pig coronary artery (24), but efforts to reduce the extent of intimal hyperplasia using the antisense oligonucleotide approach in humans have so far been disappointing. For instance, in the randomized ITALICS trial (25), antisense c-myc oligonucleotides delivered endoluminally had no significant effect on the incidence of in-stent restenosis.
Catalytic RNAs
The capacity of RNAs to catalyze chemical reactions in conformation-dependent manner has been known for almost two decades (ref. 26; and see Usman and Blatt, this Perspective series, ref. 27). Currently there are five major RNA catalytic motifs derived from naturally occurring ribozymes: the hammerhead, hairpin, group I intron, ribonuclease P, and hepatitis delta viral ribozymes. Ribozymes can hydrolyse, transfer, or form RNA phosphodiester linkages (28). Ribozymes may be introduced exogenously or produced by target cells after gene transfer.
Ribozymes have been assessed for their ability to block intimal thickening. For example, c-myb ribozymes delivered to the rat carotid artery via a replication-deficient adenovirus inhibit neointima formation after balloon angioplasty (29). The wider use of ribozymes as therapeutic agents in the vessel wall has been limited by their susceptibility to enzymatic hydrolysis in biological media, although the capacity to introduce site-specific modifications into chemically synthesized ribozymes improves stability. For example, Frimmerman et al. (30) found that the serum half-life of a chimeric DNA-RNA hammerhead ribozyme, designed to inhibit neointimal thickening upon coronary stenting by targeting proliferating cell nuclear antigen, was 4 minutes, compared with one composed entirely of RNA, which had a half-life of only a few seconds. Other modifications that enhance the nuclease resistance of synthetic ribozymes include substitution of the 2′-hydroxyl groups with 2′-amino, 2′-fluoro, or 2′-O-methyl moieties (see Usman and Blatt, this Perspective series, ref. 27), but these may compromise catalytic activity.
Catalytic DNAs
In recent years, catalytic nucleic acids composed entirely of DNA have been generated by in vitro selection strategies (see Hicke and Stephens, this Perspective series, ref. 31; and White et al., this series, ref. 32). These molecules ideally combine the catalytic activity of ribozymes with the stability of oligodeoxynucleotides. Among the first deoxyribozymes, or “DNAzymes,” were a Pb2+-dependent ribonuclease, a DNA ligase, Mn2+-, Mg2+-, and Zn2+-dependent ribonucleases, a Ca2+-dependent ribonuclease, and a porphyrin metallation catalyst (33). Most of these DNAzyme subtypes proved inefficient at cleaving target sequences consisting of a more biologically relevant all-RNA substrate, rather than a chimeric DNA-RNA substrate. Against this background, a highly efficient RNA-cleaving phosphodiester-linked DNAzyme was identified from a population of 1014 different DNA molecules after multiple successive rounds of selective amplification in vitro (34). The “10-23” DNAzyme (Figure 2), so called because it was the 23rd clone of the 10th cycle of in vitro selection, comprises a Mg2+-dependent catalytic domain composed of 15 deoxynucleotides, flanked by two substrate recognition arms each composed of 7–8 bases that bind to target RNA through Watson-Crick base-pairing. The DNAzyme cleaves a specific phosphodiester linkage between an unpaired purine (A, G) and a paired pyrimidine (C, U), producing two products, one containing a 2′,3′-cyclic phosphate terminus and the other a 5′-hydroxyl terminus. DNAzymes based on the 10-23 design may be “custom-made” to target specific sites in virtually any messenger RNA, with sequence specificity conferred by the choice of bases in its arms. Such DNAzymes can discriminate between even subtle differences in RNA sequence (35). Kuwabara et al. compared the catalytic activity of DNAzymes and hammerhead ribozymes targeting bcr-abl RNA and found greater target flexibility and sequence specificity of DNAzymes in this system (36).
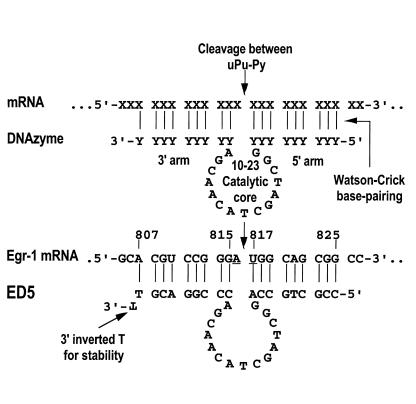
Figure 2.
Schematic representation of a 10-23 DNAzyme with 9×9 arms and modified 3′ terminus. The top panel demonstrates how a DNAzyme can be targeted to cleave the phosphodiester linkage between a selected unpaired purine (uPu) and a paired pyrimidine (Py) by virtue of the nucleotide sequence in its 5′ and 3′ arms, which bind the mRNA through Watson-Crick pairing. The 3′,3′-linked inverted T significantly improves resistance to degradation in biological media. The bottom panel shows the DNAzyme targeting the translational start site of rat Egr-1 mRNA. DNAzyme-dependent cleavage of this sequence can inhibit vascular smooth muscle cell growth in vitro and in vivo (11) and may be of clinical use in blocking neointimal formation in injured arteries, such as occurs in restenosis following angioplasty.
DNAzymes are extremely versatile and efficient catalytic molecules. For example, DNAzymes with the same catalytic domain, but with different, 7-nucleotide-long arms of equal length (i.e., 7×7 DNAzymes), cleave synthetic RNA bearing the translational initiation sites of HIV-1 gag/pol, env, nef, tat, and vpr (34). Under optimized reaction conditions, the gag/pol DNAzyme exhibited a catalytic rate constant (kcat) of approximately 0.1 min–1 with Michaelis-Menten constant (Km) values in the subnanomolar range and catalytic efficiencies (kcat/Km) of about 109 M–1min–1 (34). These values compare favorably with those of ribonuclease A (as measured using UpA as a substrate) and various hairpin and hammerhead ribozymes, whose catalytic efficiencies fall in the range of 107–108 M–1min–1 (34). The catalytic rate of 10-23 improved with pH and increasing concentrations of divalent metal cations (34, 37–39). These ions may serve to hold the DNAzyme in its active conformation, stabilize the transition state of the reactants, and/or shield the negative charges on the oligonucleotide, allowing more efficient duplex formation. The rate-limiting step of DNAzyme catalysis, determined by turnover experiments, is the rate of RNA-DNA hybridization, rather than the release of the cleavage products (34).
DNAzyme catalytic efficiency and target specificity depend upon the length and nature of the substrate-recognition arms (34, 37). For example, the catalytic efficiencies of 10-23 DNAzymes targeting the AUG codon of c-myc mRNA can be improved by employing an 8×8 or 9×9 structure in place of a 7×7 type (40). A 3′,3′-linked base inversion, which has previously been used to increase the half-life of hammerhead ribozymes in serum (41), reduces cleavage efficiency of the 7×7 c-Myc subtype (11, 40) but increases efficiency in the 9×9 subtype (40). Sioud and Leirdal (42) report that the stability in serum of a 10-23 DNAzyme (8×8) containing a 3′ inverted T targeting protein kinase C-α is six times greater than that of its unmodified counterpart, despite similar kcat values. A complete phosphorothioate version of this DNAzyme enjoys superior stability but shows >100-fold lower cleavage activity than its counterpart carrying phosphodiester linkages.
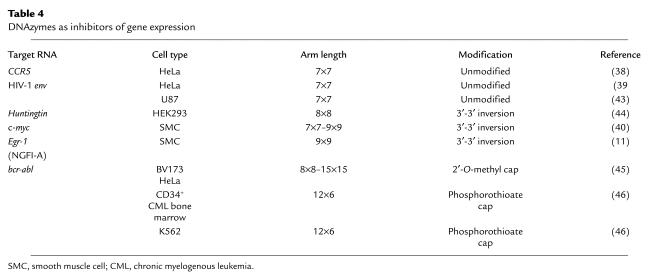
The performance of 10-23 DNAzymes has been demonstrated in a number of in vitro biological systems (Table 4). First, a 7×7 DNAzyme targeting the A916UG in the mRNA for the HIV-1 chemokine coreceptor CCR5 cleaved its target in vitro and inhibited cell membrane fusion activity (38). Second, 7×7 DNAzymes directed at A6339UG (39) and A7212UU (43) in HIV-1 env cleaved synthetic mRNA and inhibited cell fusion activity (39) as well as HIV-1 virus infection and replication (43). Third, 8×8 DNAzymes targeting G9CC and A421UG in the Huntingtin mRNA cleaved in vitro transcribed mRNA and decreased cytomegalovirus-driven Huntingtin protein expression in a cotransfection system (44). Fourth, a 9×9 DNAzyme directed to the AUG start codon of human c-Myc cleaved its target mRNA, blocked c-Myc expression, and inhibited proliferation of an arterial smooth muscle cell line (40). Finally, several DNAzymes targeting variants of bcr-abl mRNA inhibited reporter expression in HeLa cells (45), and suppressed protein expression and growth of CD34+ bone marrow cells from patients with chronic myeloid leukemia (46). 10-23 has recently been exploited in PCR-based strategies for the homogeneous amplification and quantification of nucleic acids in clinical specimens. Todd et al. (47) have developed a method to assay levels of serum K-ras DNA in patients with gastrointestinal malignancies using a real-time approach in which reporter substrates are cleaved by amplicons bearing active DNAzymes (47). These studies demonstrate the therapeutic and diagnostic potential of DNAzymes in diverse pathologic settings.
Table 4.
DNAzymes as inhibitors of gene expression
Egr-1 DNAzymes
Suppression of gene expression via antisense (catalytic or noncatalytic) means would be predicted to be more efficient if the target mRNA and protein are short-lived. Egr-1 mRNA and protein have relatively short half-lives of less than 1 and 2 hours, respectively (48). Insights into the functional roles of Egr-1 in arterial smooth muscle cells came initially from experiments using conventional antisense oligonucleotides directed to the Egr-1 start codon or to a sequence encoding one of the crucial zinc fingers that are required for Egr-1 DNA-binding (49). These oligomers, which do not form G-quartets, block the inducible synthesis of Egr-1 protein in a sequence-specific manner (49), and they inhibit DNA synthesis and cell replication, apparently without cytotoxicity (49). We used these oligonucleotides to show that Egr-1 is required for smooth muscle cell and endothelial cell recovery after mechanical injury to these cells in culture (15, 49).
We recently generated a DNAzyme targeting the A816U817 junction at the start codon of rat Egr-1. This DNAzyme was of the 9×9 subtype and carried a 3′,3′-linked inverted thymidine for improved stability (Figure 2). The Egr-1 DNAzyme, as expected, cleaved a short synthetic Egr-1 RNA substrate and in vitro transcribed mRNA of various length in a sequence-specific manner (11). Confocal microscopy using a fluorescently tagged form of this DNAzyme revealed that it localized within smooth muscle cell nuclei in a sequence-independent fashion. The DNAzyme inhibited endogenous Egr-1 mRNA and protein synthesis and blocked smooth muscle cell proliferation in a reversible manner (11). Substitution of the G at position 6 with a C in the catalytic core, a mutation that abolishes its enzymatic activity (37), reduced its ability to inhibit proliferation. The residual activity was most likely due to noncatalytic antisense mechanisms.
As with Egr-1 antisense oligonucleotides, this DNAzyme suppressed the reparative response to injury in vitro. Fluorescently tagged DNAzymes localized within the rat carotid artery wall after adventitial delivery and inhibited the induction of Egr-1 in the vessel wall shortly after injury without affecting levels of Sp1 (11). Moreover, the DNAzyme blocked neointimal thickening 14 days after balloon injury and adventitial delivery in pluronic gel (11). This study was the first to demonstrate DNAzyme efficacy in vivo, and it confirmed the significance of Egr-1 as a target in vascular disease, as well as the promise of catalytic DNAs for probing the functions of genes in vivo. Since smooth muscle cell hyperplasia underlies in-stent restenosis in humans (50), Egr-1 DNAzymes, re-engineered to cleave and block human EGR-1 expression, may be useful as inhibitors of smooth muscle cell proliferation in endovascular strategies adjunct to stents. Similar strategies should allow catalytic DNA to be applied in other pathologic settings.
Concluding remarks
Catalytic molecules composed entirely of DNA represent a new class of sequence-specific molecular tools with gene therapeutic potential based on Watson-Crick base-pairing. DNAzymes have considerable advantages over ribozymes in that they are easier to synthesize and less sensitive to chemical and enzymatic degradation than RNA-based reagents. They also exhibit greater catalytic efficiency than conventional hairpin and hammerhead ribozymes. Their size and capacity to form stable DNA-RNA heteroduplexes facilitate efficient target accessibility and efficacy, given the complex secondary structure of RNA. Factors likely to influence the eventual therapeutic use of DNAzymes include its efficient cellular uptake, subcellular localization, stability, as well as local divalent cation concentration. An important challenge in antirestenotic strategies using DNAzymes is the efficient delivery of the molecule to its intended site of action. The focal nature of restenosis and the capacity to deliver agents endoluminally via catheters and nucleic acid–coated or –impregnated stents provide a useful starting point for the clinical use of DNAzymes.
Supplementary Material
Acknowledgments
I thank past and present colleagues for their contribution to the work described in this review and regret that, due to strict space constraints, only a limited number of references could be cited. Readers are urged to see the supplementary reference list accompanying this article, at http://www.jci.org/cgi/content/full/106/10/1189/DC1. These studies were supported by grants from Johnson & Johnson Research Pty. (Sydney, Australia), the Australian Research Council, and the National Health and Medical Research Council of Australia.
References
- 1.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 2.Nikod P, Scherrer U. Explosive growth of coronary angioplasty: success story of a less than perfect procedure. Circulation. 1993;87:1749–1751. doi: 10.1161/01.cir.87.5.1749. [DOI] [PubMed] [Google Scholar]
- 3.Brophy JM. Improved coronary outcome after widespread use of coronary-artery stenting in Canada. N Engl J Med. 2000;342:1448–1449. doi: 10.1056/NEJM200005113421913. [DOI] [PubMed] [Google Scholar]
- 4.Serruys PW, et al. A comparison of balloon-expandable-stent implantation with balloon angioplasty in patients with coronary artery disease. N Engl J Med. 1994;331:489–495. doi: 10.1056/NEJM199408253310801. [DOI] [PubMed] [Google Scholar]
- 5.Fischman DL, et al. A randomized comparison of coronary-stent placement and balloon angioplasty in the treatment of coronary artery disease. N Engl J Med. 1994;331:496–501. doi: 10.1056/NEJM199408253310802. [DOI] [PubMed] [Google Scholar]
- 6.Casterella PJ, Teirstein PS. Prevention of coronary restenosis. Cardiol Rev. 1999;7:219–231. doi: 10.1097/00045415-199907000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, et al. Angiotensin II type 1 receptor blockade inhibits the expression of immediate-early genes and fibronectin in rat injured artery. Circulation. 1995;92:88–95. doi: 10.1161/01.cir.92.1.88. [DOI] [PubMed] [Google Scholar]
- 8.Khachigian LM, Lindner V, Williams AJ, Collins T. Egr-1-induced endothelial gene expression: a common theme in vascular injury. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 9.Sukhatme VP, et al. A zinc-finger encoding gene corregulated with c-Fos during growth and differentiation and after depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 10.Silverman ES, Khachigian LM, Lindner V, Williams AJ, Collins T. Inducible PDGF A-chain transcription in vascular smooth muscle cells is mediated by Egr-1 displacement of Sp1 and Sp3. Am J Physiol. 1997;42:H1415–H1426. doi: 10.1152/ajpheart.1997.273.3.H1415. [DOI] [PubMed] [Google Scholar]
- 11.Santiago FS, et al. New DNA enzyme targeting Egr-1 mRNA inhibits vascular smooth muscle proliferation and regrowth factor injury. Nat Med. 1999;11:1264–1269. doi: 10.1038/15215. [DOI] [PubMed] [Google Scholar]
- 12.Gashler A, Sukhatme V. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 13.Lowe, H.C., Chesterman, C.N., Hopkins, A., Juergens, C.P., and Khachigian, L.M. Acute local release of fibroblast growth factor-2 but not transforming growth factor-beta1 following coronary stenting. Thromb. Haemost. In press. [PubMed]
- 14.Caplice NM, et al. Growth factors released into the coronary circulation after vascular injury promote proliferation of human vascular smooth muscle cells in culture. J Am Coll Cardiol. 1997;29:1536–1541. doi: 10.1016/s0735-1097(97)00076-4. [DOI] [PubMed] [Google Scholar]
- 15.Santiago FS, Lowe HC, Day FL, Chesterman CN, Khachigian LM. Egr-1 induction by injury is triggered by release and paracrine activation by fibroblast growth factor-2. Am J Pathol. 1999;154:937–944. doi: 10.1016/S0002-9440(10)65341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russo MW, Sevetson BR, Milbrandt J. Identification of NAB-1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svaren J, et al. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol. 1996;16:3545–3553. doi: 10.1128/mcb.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silverman ES, et al. Vascular smooth muscle cells express the transcriptional corepressor NAB2 in response to injury. Am J Pathol. 1999;155:1311–1317. doi: 10.1016/S0002-9440(10)65233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khachigian LM, et al. GC factor 2 represses platelet-derived growth factor A-chain transcription and is itself induced by arterial injury. Circ Res. 1999;84:1258–1267. doi: 10.1161/01.res.84.11.1258. [DOI] [PubMed] [Google Scholar]
- 20.McCaffrey TA, et al. High-level expression of Egr-1 and Egr-1–inducible genes in mouse and human atherosclerosis. J Clin Invest. 2000;105:653–662. doi: 10.1172/JCI8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett MR, Schwartz SM. Antisense therapy for angioplasty restenosis: some critical considerations. Circulation. 1995;92:1981–1993. doi: 10.1161/01.cir.92.7.1981. [DOI] [PubMed] [Google Scholar]
- 22.Stein CA. Controversies in the cellular pharmacology of oligodeoxynucleotides. Ciba Found Symp. 1997;209:79–89. [PubMed] [Google Scholar]
- 23.Topol EJ, Serruys PW. Frontiers in interventional cardiology. Circulation. 1998;98:1802–1820. doi: 10.1161/01.cir.98.17.1802. [DOI] [PubMed] [Google Scholar]
- 24.Gunn J, et al. The effect of oligonucleotides to c-myb on vascular smooth muscle cell proliferation and neointima formation after porcine coronary angioplasty. Circ Res. 1997;80:520–531. doi: 10.1161/01.res.80.4.520. [DOI] [PubMed] [Google Scholar]
- 25.Serruys PW, et al. Antisense oligonucleotoide against c-myc administered with the Transport delivery catheter for the prevention of in-stent restenosis: results of the randomized ITALICS trial. Circulation. 1998;98:I363. (Abstr.) [Google Scholar]
- 26.Kruger K, Grabowski PJ, Zaug AJ, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequencing of tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 27.Usman N, Blatt LM. Nuclease-resistant synthetic ribozymes: developing a new class of therapeutics. J Clin Invest. 2000;106:1197–1202. doi: 10.1172/JCI11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Long DL, Uhlenbeck OC. Self-cleaving catalytic RNA. FASEB J. 1993;7:25–30. doi: 10.1096/fasebj.7.1.8422971. [DOI] [PubMed] [Google Scholar]
- 29.Macejak DG, et al. Adenovirus-mediated expression of a ribozyme to c-myb mRNA inhibits smooth muscle cell proliferation and neointima formation in vivo. J Virol. 1999;73:7745–7751. doi: 10.1128/jvi.73.9.7745-7751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frimmerman A, et al. Chimeric DNA-RNA hammerhead ribozyme to proliferating cell nuclear antigen reduces stent-induced stenosis in a porcine coronary model. Circulation. 1999;99:697–703. doi: 10.1161/01.cir.99.5.697. [DOI] [PubMed] [Google Scholar]
- 31.Hicke BJ, Stephens AW. Escort aptamers: a delivery service for diagnosis and therapy. J Clin Invest. 2000;106:923–928. doi: 10.1172/JCI11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White RR, Sullenger BA, Rusconi CP. Developing aptamers into therapeutics. J Clin Invest. 2000;106:929–934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sen D, Geyer CR. DNA enzymes. Curr Opin Chem Biol. 1998;2:680–687. doi: 10.1016/s1367-5931(98)80103-8. [DOI] [PubMed] [Google Scholar]
- 34.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cairns MJ, King A, Sun L-Q. Nucleic acid mutation analysis using catalytic DNA. Nucleic Acids Res. 2000;28:E9. doi: 10.1093/nar/28.3.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuwabara T, et al. Comparison of the specificities and catalytic activities of hammerhead ribozymes and DNA enzymes with respect to the cleavage of BCR-ABL chimeric L6 (b2a2) mRNA. Nucleic Acids Res. 1997;25:3074–3091. doi: 10.1093/nar/25.15.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santoro SW, Joyce GF. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry. 1998;37:13330–13342. doi: 10.1021/bi9812221. [DOI] [PubMed] [Google Scholar]
- 38.Goila R, Banerjea AC. Sequence-specific cleavage of the HIV-1 coreceptor CCR5 gene by a hammer-head ribozyme and a DNA-enzyme: inhibition of the coreceptor function by DNA-enzyme. FEBS Lett. 1998;436:233–238. doi: 10.1016/s0014-5793(98)01137-5. [DOI] [PubMed] [Google Scholar]
- 39.Dash BC, et al. Targeted cleavage of HIV-1 envelope gene by a DNA enzyme and inhibition of HIV-1 envelope-CD4 mediated cell fusion. FEBS Lett. 1998;431:395–399. doi: 10.1016/s0014-5793(98)00799-6. [DOI] [PubMed] [Google Scholar]
- 40.Sun L-Q, et al. Suppression of smooth muscle cell proliferation by a c-myc RNA-cleaving deoxyribozyme. J Biol Chem. 1999;274:17236–17241. doi: 10.1074/jbc.274.24.17236. [DOI] [PubMed] [Google Scholar]
- 41.Beigelman L, et al. Chemical modification of hammerhead ribozymes. J Biol Chem. 1995;270:25702–25708. doi: 10.1074/jbc.270.43.25702. [DOI] [PubMed] [Google Scholar]
- 42.Sioud M, Leirdal M. Design of nuclease resistant protein kinase C-alpha DNA enzymes with potential therapeutic application. J Mol Biol. 2000;296:937–947. doi: 10.1006/jmbi.2000.3491. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Xu Y, Ling H, Hattori T. Inhibition of infection of incoming HIV-1 virus by RNA-cleaving DNA enzyme. FEBS Lett. 1999;458:151–156. doi: 10.1016/s0014-5793(99)01149-7. [DOI] [PubMed] [Google Scholar]
- 44.Yen L, Strittmatter SM, Kalb RG. Sequence-specific cleavage of Huntingtin mRNA by catalytic DNA. Ann Neurol. 1999;46:366–373. doi: 10.1002/1531-8249(199909)46:3<366::aid-ana12>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 45.Warashina M, Kuwabara T, Nakamatsu Y, Taira K. Extremely high and specific activity of DNA enzymes in cells with a Philadelphia chromosome. Chem Biol. 1999;6:237–250. doi: 10.1016/S1074-5521(99)80039-8. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, McMahon R, Rossi JJ, Forman SJ, Snyder DS. Inhibition of bcr-abl oncogene by novel deoxyribozymes (DNAzymes) Hum Gene Ther. 1999;10:2847–2857. doi: 10.1089/10430349950016573. [DOI] [PubMed] [Google Scholar]
- 47.Todd AV, Fuery C, Impey HL, Applegate TL, Haughton MA. DzyNA-PCR: use of DNAzymes to detect and quantify nucleic acid sequences in a real-time fluorescent format. Clin Chem. 2000;46:625–630. [PubMed] [Google Scholar]
- 48.Huang RP, Fan Y, Boynton AL. UV irradiation upregulates Egr-1 expression at transcription level. J Cell Biochem. 1999;73:227–236. doi: 10.1002/(sici)1097-4644(19990501)73:2<227::aid-jcb9>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 49.Santiago FS, Atkins DA, Khachigian LM. Vascular smooth muscle cell proliferation and regrowth after injury in vitro is dependent upon NGFI-A/Egr-1. Am J Pathol. 1999;155:897–905. doi: 10.1016/S0002-9440(10)65189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kearney M, et al. Histopathology of in-stent restenosis in patients with peripheral artery disease. Circulation. 1997;95:1998–2002. doi: 10.1161/01.cir.95.8.1998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.