Background: Drs2p was known to be stimulated by phosphatidylinositol 4-phosphate (PI(4)P), but the mechanism was unclear.
Results: Loss of the C-terminal tail, or presence of PI(4)P, stimulates the activity of Drs2p.
Conclusion: Inhibition of Drs2p activity by its C-terminal tail is relieved by PI(4)P.
Significance: This is the first evidence for an auto-inhibitory domain in a phospholipid flippase.
Keywords: ATPases, Membrane Lipids, Membrane Reconstitution, Membrane Trafficking, Phosphatidylserine, Protein Purification, Drs2, Membrane Asymmetry, P4-ATPase, Phosphatidylinositol 4-Phosphate
Abstract
Drs2p, a yeast type IV P-type ATPase (P4-ATPase), or flippase, couples ATP hydrolysis to phosphatidylserine translocation and the establishment of membrane asymmetry. A previous study has shown that affinity-purified Drs2p, possessing an N-terminal tandem affinity purification tag (TAPN-Drs2), retains ATPase and translocase activity, but Drs2p purified using a C-terminal tag (Drs2-TAPC) was inactive. In this study, we show that the ATPase activity of N-terminally purified Drs2p associates primarily with a proteolyzed form of Drs2p lacking the C-terminal cytosolic tail. Truncation of most of the Drs2p C-terminal tail sequence activates its ATPase activity by ∼4-fold. These observations are consistent with the hypothesis that the C-terminal tail of Drs2p is auto-inhibitory to Drs2p activity. Phosphatidylinositol 4-phosphate (PI(4)P) has been shown to positively regulate Drs2p activity in isolated Golgi membranes through interaction with the C-terminal tail. In proteoliposomes reconstituted with purified, N-terminally TAP-tagged Drs2p, both ATPase and flippase activity were significantly higher in the presence of PI(4)P. In contrast, PI(4)P had no significant effect on the activity of a truncated form of Drs2p, which lacked the C-terminal tail. This work provides the first direct evidence, in a purified system, that a phospholipid flippase is subject to auto-inhibition by its C-terminal tail, which can be relieved by a phosphoinositide to stimulate flippase activity.
Introduction
P-type ATPases are a large family of membrane pumps that transport various substrates, such as ions, against their chemical gradients across membranes. They are divided into five subfamilies (P1- to P5-ATPases) and further into smaller subgroups based on sequence homology and substrate specificity (3, 4). The P4-ATPases4 translocate specific phospholipid molecules from the extracellular leaflet of the plasma membrane, or luminal leaflet of internal organelles, to the cytosolic leaflet. This activity is crucial for both the establishment of plasma membrane asymmetry and vesicle-mediated protein transport in the secretory and endocytic pathways (5). The overall architecture of P-type ATPases is exemplified by the crystal structures of several P-type ATPases. They possess three cytosolic domains (A: actuator; N: nucleotide binding; P: phosphorylation) and a multispan transmembrane domain (6, 7). In addition to these common structural features, some P-type ATPases have an additional regulatory cytosolic domain, called an R domain, within the N- or C-terminal cytosolic tail of the protein (8–16). Some members of the P-type ATPase family also have additional β-subunits associated with them. There is evidence that these subunits, in varying degrees, aid in regulation, folding, and/or proper localization of the P-type ATPase (4, 5, 17).
The R domain serves as an auto-inhibitory element that limits the activity of the P-type ATPase. Removal of the R domain, by limited proteolysis or genetic engineering, activates the protein. Various regulatory activities can relieve the auto-inhibition caused by this R domain. For example, the P2B-ATPase subgroup, comprising the plasma membrane Ca2+ pumps, contains an R domain in the C-terminal tail (in animals) (8, 9, 18) or in the N-terminal tail (in plants) (13–16, 19). When calmodulin interacts with this R domain, it displaces the auto-inhibitory tail and activates the protein for Ca2+ transport. Phosphorylation of serine or threonine residues within the R domain of P2B-ATPases has also been found to regulate ATPase activity by disrupting calmodulin binding and resulting in protein activation (in animals) or inactivation (in plants) (18–21). The P3A-ATPase family, which comprises plasma membrane proton pumps from plants and fungi, also possesses a C-terminal R domain and utilizes a similar activation mechanism, which includes phosphorylation and binding of regulatory proteins to the R domain (10–12, 22–27). Evidence for the role of the N terminus in regulating P-type ATPase function has also been shown (28, 29).
Drs2p is a P4-ATPase from the budding yeast Saccharomyces cerevisiae. At steady state, Drs2p is localized primarily to the trans-Golgi network (TGN) and is involved in multiple protein trafficking pathways between the TGN, plasma membrane and endosomes (5). Drs2p is structurally similar to the other P-type ATPases with A, N, and P cytosolic domains and 10 transmembrane segments (5). Drs2p associates with a β-subunit, Cdc50p, which is its co-chaperone and a member of the Cdc50p family of proteins. Cdc50p is essential for proper localization of Drs2p within the cell (30–32). Drs2p catalyzes a phosphatidylserine (PS) flippase activity detected in isolated TGN membranes (33) and post-Golgi secretory vesicles (34), as well as in proteoliposomes reconstituted with purified Drs2p (1). Interestingly, like P2B- and P3A-ATPases, Drs2p appears also to have an R domain within its C-terminal tail. Features identified in the Drs2p R domain include a motif that binds Gea2p (an Arf guanine nucleotide exchange factor) (35) and a region that binds phosphatidylinositol 4-phosphate (PI(4)P) (2). Drs2p flippase activity was virtually abolished in TGN membranes isolated from yeast cells deficient in Gea2p and PI(4)P production, and addition of Gea2p and PI(4)P to these TGN membranes restored flippase activity (2). Additional evidence from the Lenoir laboratory sheds light on the role that PI(4)P plays in Drs2p regulation. It was found that, in crude membranes, PS inhibited the dephosphorylation of the Drs2p-Cdc50p complex. It was only in the presence of PI(4)P that PS was able to accelerate the dephosphorylation step associated with substrate transport (36). These results suggested that Drs2p is auto-inhibited by its C-terminal tail in the absence of Gea2p and PI(4)P, both of which are positive regulators that stimulate Drs2p activity by binding to its C-terminal R domain. In this study, we present evidence that directly supports this hypothesis.
EXPERIMENTAL PROCEDURES
Reagents
IgG-Sepharose 6 Fast Flow, calmodulin-Sepharose 4B, and ATP (>99% purity) were from GE Healthcare. ATPγS was from Sigma, and nickel-nitrilotriacetic acid-agarose was from Qiagen. AcTEV protease and SimplyBlue SafeStain (Coomassie G-250) were from Invitrogen. SM-2 Bio-Beads were from Bio-Rad, and Triton X-100, polyoxyethylene (9) dodecyl ether (C12E9), and polyoxyethylene (8) dodecyl ether (C12E8) were from Anatrace. Phospholipids and fluorescent derivatives were from Avanti Polar Lipids and were: DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), DOPS (1,2-dioleoyl-sn-glycero-3-phosphoserine), PI(4)P (l-α-phosphatidylinositol-4-phosphate from porcine brain), NBD-PC (1-palmitoyl-2-[6-(NBD-amino)hexanoyl]-sn-glycero-3-phosphocholine), C6 NBD-PS (1-palmitoyl-2-[6-(NBD-amino)hexanoyl]-sn-glycero-3-phosphoserine), and C12 NBD-PS (1-palmitoyl-2-[12-(NBD-amino)dodecanoyl]-sn-glycero-3-phosphoserine). Lipids were dissolved in chloroform except PI(4)P, which was dissolved in chloroform/methanol/water (20:9:1, v/v/v), and stored at −20 °C. Antibodies used in this study were rabbit primary antibodies against Drs2p, and an Alexa Fluor 680-labeled goat secondary antibody against rabbit IgG (Invitrogen).
Media and Strains
Yeast cells were grown in standard rich medium (1% yeast extract/2% peptone/2% dextrose) or synthetic minimal media containing required supplements (37), and nutrients were doubled in cell cultures for protein purification purposes.
The yeast strains used were XZY63b (MATα his3 leu2 ura3 lys2 atp2Δ::URA3 PGPD::CDC50 pRS425-PGPD::TAPN::DRS2), XZY51 (MATα his3 leu2 ura3 lys2 atp2Δ::URA3 PGPD::CDC50 pRS425-PGPD::DRS2:TAPC), XZY85 (MATα his3 leu2 ura3 lys2 atp2Δ::URA3 PGPD::CDC50 pRS425-PGPD::N::DRS2::TAPC), XZY94 (MATα his3 leu2 ura3 lys2 PGPD::CDC50 pRS425-PGPD::DRS2::(TEV)::TAPC2), XZY95 (MATα his3 leu2 ura3 lys2 PGPD::CDC50 pRS425-PGPD::DRS2(TEV)CT121:: (TEV)::TAPC2), XZY96 (MATα his3 leu2 ura3 lys2 PGPD::CDC50 pRS425-PGPD::DRS2(TEV)CT121::TAPC2), and XZY60m (MATα his3 leu2 ura3 lys2 atp2Δ::URA3 PGPD::TAPN::DRS2 PGPD::CDC50), where TAPN encodes the Protein A-TEV protease cleavage site-His10 tag, TAPC encodes the calmodulin-binding peptide (CBP)-TEV protease cleavage site-Protein A tag, (TEV) encodes the TEV protease cleavage site, TAPC2 encodes the Protein A-His10 tag, and DRS2(TEV) CT121 encodes a modified Drs2 protein that has the TEV protease cleavage site inserted in its C-terminal tail at the position that is 121-residue away from the C-terminal end of wild-type Drs2p.
Protein Purification
TAP-tagged Drs2p was affinity-purified using a TAP procedure described previously with some modifications (1). Briefly, yeast cells were cultured to 3–6 A600/ml in minimal medium or 5–10 A600/ml in rich medium at 30 °C, harvested, and lysed using an EmulsiFlex-C3 High Pressure Homogenizer (Avestin). The cell lysate was centrifuged at 15,000 × g for 12 min, and 20% Triton X-100, C12E9, or C12E8 was added to the supernatant to a final concentration of 1% to solubilize Drs2p. TAP-tagged Drs2p was then purified using an IgG column plus either a Ni2+ column (for TAPN and TAPC2) or a calmodulin column (for TAPC). In experiments where Drs2p was first purified using the TAPN tag and then different Drs2p populations were separated using the TAPC tag, the procedure was a combination of the TAPN purification plus the second affinity step (the calmodulin column) of the TAPC purification. Experiments where proteins were purified for the purposes of performing flippase assays were performed using C12E8 exclusively.
Purified Drs2p samples (except samples to be used in flippase assays) were centrifuged in a Microcon YM-100 filter (Millipore) at 13,000 × g for 15 min at 4 °C to near dryness and resuspended in the desired amount of storage buffer (40 mm Tris-HCl, pH 7.5, 150 mm NaCl, 40% glycerol, 0.1% Triton X-100) to store at −20 °C. Recovery of Drs2p was determined using Odyssey Infrared Imaging System (LI-COR) to quantify SimplyBlue-stained bands relative to a BSA standard curve. The recombinant Sec7 domain of Gea2p was purified as described previously (2).
Proteoliposome Formation
Purified Drs2p was reconstituted into proteoliposomes as described previously with some modifications (1). Briefly, 0.5 ml of single-step affinity-purified Drs2p using a nickel-nitrilotriacetic acid column was mixed with 2 mg of lipid mixture solubilized in 0.5 ml of 1% C12E9 for 30 min at 4 °C. After addition of 200 mg of extensively washed SM-2 Bio-Beads, and 12–15 h of incubation on an end-over-end rotator at 4 °C, the supernatant containing proteoliposomes was carefully removed and stored at 4 °C.
We found that proteoliposomes formed with Drs2p purified in C12E9 were often too “leaky” to use in flippase assays. The membrane seal was significantly improved when reconstitutions were performed using C12E8 solubilized protein. Therefore, the following changes were made during proteoliposome formation. 2 mg of lipids was solubilized in a 0.5-ml volume that contains 1.5% C12E8 and was incubated for 30 min at 4 °C. 150 mg of extensively washed SM-2 Bio-Beads was then added to this solution and incubated for 6 h on an end-over-end rotator at 4 °C. After this, another 300 mg of SM-2 beads was added, and the sample was incubated for a further 12–15 h with end-over-end rotation. The supernatant containing proteoliposomes was carefully removed and stored at 4 °C.
Flotation of Proteoliposomes
200 μl of the proteoliposome sample was first incubated with 10 mm dithionite, for 5 min at 4 °C, to pre-quench the fluorescence in the outer leaflet of the proteoliposomes. The sample was then mixed with 200 μl of 80% glycerol and placed at the bottom of a bipartite glycerol step gradient. 600 μl of 10% glycerol was laid on the top. The samples were centrifuged in a TLS-55 rotor (Beckman Coulter) at 50,000 rpm for 6 h at 4 °C, and 200 μl each, of the top two fractions, was collected by pipetting. These fractions contained pure Drs2p proteoliposomes.
ATPase Assay
Purified Drs2p was assayed for ATPase activity in ATPase buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 50 mm KCl, 1 mm NaN3, 0.1% Triton X-100, 4 mm Na+-ATP, pH 7.5, 10 mm MgCl2) at 37 °C for 1 h. For ATPase assays using proteoliposomes, detergent was omitted from ATPase buffer. Released phosphate was measured colorimetrically using modified protocols described previously (38–40). Briefly, the volume of the sample was brought to 275 μl with deionized water, and the ATPase reaction was stopped by addition of 150 μl of molybdate solution (2 m HCl/50 mm Na2MoO4) and 75 μl of malachite green solution (0.042% malachite green in 1% polyvinyl alcohol solution). The sample was mixed for 2 min before addition of 500 μl of citric acid (7%), and the optical density at 660 nm was read at 30 min after addition of the citric acid solution. The amount of released phosphate was determined by a phosphate standard curve constructed in the same ATPase buffer.
Flippase Assay
Floated proteoliposomes were assayed for flippase activity using previously published methods (1, 41). Briefly, 40 μl of floated, Drs2p-containing proteoliposomes were incubated, at 37 °C, with 5 mm MgCl2 and either 5 mm ATP or 5 mm ATPγS in a flippase buffer (40 mm Tris-HCl, pH 7.5, 200 mm NaCl). At time 0 min and 40 min, 20-μl samples were removed and mixed with 1 ml of flippase buffer in a cuvette. Base-line fluorescence readings were taken for 30 s on an AB2 fluorometer (SLM Instruments, Inc.) at λex = 460 nm, and λem = 534 nm. 10 μl of 1 m dithionite (dissolved in 1 m Tris, pH 9.4) was then added to the sample, and fluorescence was recorded for 120 s. Finally, the fluorescence was completely quenched by the addition of 50 μl of 10% Triton X-100, and background fluorescence was recorded for 30 s. The percentage of NBD-phospholipid in the outer bilayer of each of the proteoliposome samples (both ATP and the ATPγS control, at time t = 0 min and 40 min) was calculated using previously described formulas (1). To calculate the percentage of NBD-phospholipid flipped, the values for an ATPγS control were subtracted from the respective ATP-activated sample.
Western Blotting
Western blotting was performed as described previously (42). The primary antibody used was rabbit anti-Drs2p (1:2000). The secondary, Alexa Fluor 680-labeled goat anti-rabbit IgG antibody (Invitrogen), was used at 1:2000. Western blots were imaged with an Odyssey Infrared Imaging System.
RESULTS
Drs2p Retains ATPase Activity after Purification by the N-terminal TAPN Tag, but Not the C-terminal TAPC Tag
In a previous study, both TAPN-Drs2p and Drs2p-TAPC were expressed from chromosome-integrated expression cassettes and affinity-purified to comparable yields and purities (1). Although both TAP-tagged Drs2p forms appeared to be functional in vivo, only TAPN-Drs2p retained ATPase activity after purification, whereas purified Drs2p-TAPC was inactive. Interestingly, purified Drs2p-TAPC did not seem to be aggregated or denatured, suggesting that the differing activities between the two Drs2p proteins may have a biologically relevant cause (1).
To further address this issue and to more easily manipulate Drs2p, the expression cassettes for Drs2p were transferred to a multicopy plasmid (pRS425) to produce constructs (I) and (II), whereas the β-subunit Cdc50p was overexpressed using the strong GPD1 promoter integrated into the endogenous chromosomal locus (Fig. 1, A and B). Both TAPN-Drs2p and Drs2p-TAPC were successfully expressed and purified using this plasmid system (Fig. 1, B and C), and they still showed differential levels of ATPase activity, as observed previously (Fig. 1D). Cdc50p was recovered at low, substoichiometric levels in these preparations and was not visible in the Coomassie-stained gel. The band migrating just above the 28-kDa marker is the TEV protease used to elute the tagged proteins from an IgG column.
FIGURE 1.
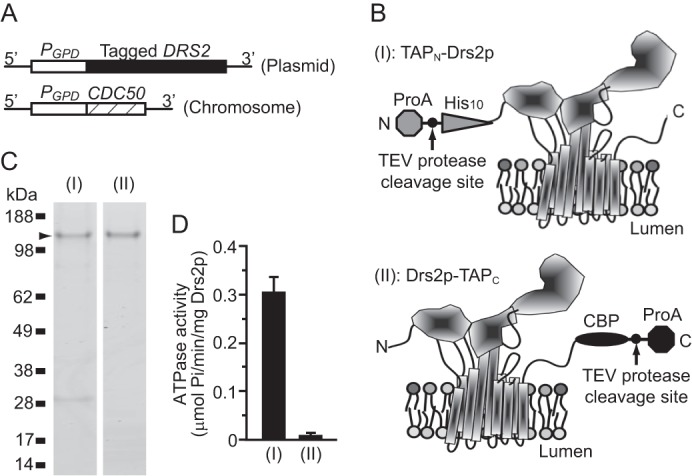
TAPN-Drs2p has ATPase activity. A, tagged DRS2 was expressed from a 2μ plasmid, and CDC50 was from the chromosome. B, schematic of TAPN-Drs2p (I) and Drs2p-TAPC (II) was modeled on the crystal structure of SERCA1 in the nucleotide-free E1·2Ca2+ conformation (7). The cytosolic domains are facing upward. C, purified TAPN-Drs2p (I) and Drs2p-TAPC (II) were subjected to SDS-PAGE, and the gel was stained with SimplyBlue. D, ATPase activity of purified TAPN-Drs2p and Drs2p-TAPC preparations. Samples were assayed with or without orthovanadate, and the orthovanadate controls were subtracted from the experimental value to obtain the nmol of orthovanadate released. ProA, protein A.
ATPase Activity Is Associated with a Faster-mobility Form of Drs2p in Purified Samples
The major molecular difference between purified TAPN-Drs2p and Drs2p-TAPC is different tag appendages (His10 versus CBP) to different positions of Drs2p (N versus C terminus) (Fig. 1B).
To test whether different tags at the termini caused varying levels of ATPase activity of purified Drs2p, a dual TAP-tagged construct of Drs2p (TAPN-Drs2p-TAPC (III)) was expressed (Fig. 2A). Cdc50p was, again, overexpressed using a strong GPD1 promoter. Purification of TAPN-Drs2p-TAPC, using either the TAPN tag or the TAPC tag, yielded purified protein levels that were comparable with the previous experiments (Fig. 2B). Theoretically, both preparations should contain Drs2p of the same molecular composition (His10-Drs2p-CBP), which indeed appeared as a single major band on Coomassie-stained gels for both preparations (Fig. 2B). Surprisingly, however, ATPase activity was only detected in samples of TAPN-Drs2p-TAPC purified using the TAPN tag, but not the TAPC tag (Fig. 2C), indicating some difference between these two preparations. Western blots revealed a minor Drs2p band with faster mobility in TAPN-Drs2p-TAPC samples purified with TAPN tag, but not with TAPC (Fig. 2D), suggesting a possible link between this faster-mobility band and ATPase activity.
FIGURE 2.
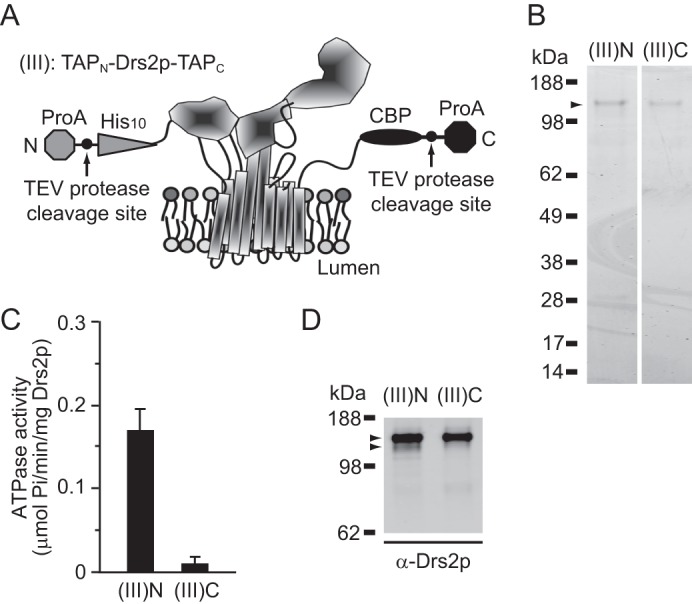
Differential activity of TAPN-Drs2-TAPC purified with either the N-terminal or C-terminal tag. A, schematic of TAPN-Drs2p-TAPC was modeled on the SERCA1 crystal structure. B, purified TAPN-Drs2p-TAPC samples from strain XZY85 (atp2Δ) using the TAPN tag ((III)N) or TAPC tag ((III)C) were subjected to SDS-PAGE and the gel was stained with SimplyBlue. C, ATPase activity of purified TAPN-Drs2p-TAPC preparations is shown. D, samples from B were electrotransferred to polyvinylidene difluoride (PVDF) membrane for Western blotting using Drs2 antibodies. The upper arrowhead indicates the intact TAPN-Drs2p-TAPC form, and the lower arrowhead indicates a minor band with faster mobility. Another faint band migrated below the two arrowheads, but was unlikely to contribute significantly to the ATPase activity. ProA, protein A.
It was possible that the faster-mobility band of Drs2p was a C-terminally cleaved form of Drs2p, which had lost some C-terminal sequences along with the TAPC tag during purification. This form would not be recovered during purifications using the TAPC tag because the tag was lost. To explore this idea and better determine the relationship between ATPase activity and the faster migrating (or cleaved) Drs2p, TAPN-Drs2p-TAPC purified by TAPN ((III)N) was further applied to a calmodulin column. Consistently, the unbound fraction was enriched with the faster-mobility form of Drs2p (Fig. 3A, column U), whereas the bound fraction was almost devoid of this minor band (Fig. 3A, column B). Furthermore, ATPase activity was significantly higher in the unbound fraction (U) and much lower in the bound fraction (B) (Fig. 3B). The simplest explanation for these data is that a C-terminally cleaved, faster-migrating Drs2p is primarily responsible for the ATPase activity observed in N-terminally purified Drs2p samples.
FIGURE 3.
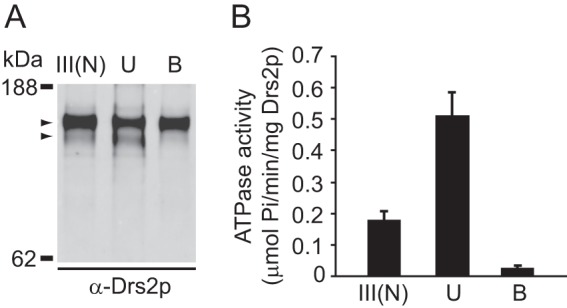
Drs2p ATPase activity associates with the faster mobility form. A, purified TAPN-Drs2p-TAPC before separation (III(N)), the unbound fraction from the calmodulin column (lane U), and the bound and eluted fraction (lane B) were subjected to SDS-PAGE and Western blotting with anti-Drs2p. The upper arrowhead indicates the intact TAPN-Drs2p-TAPC form, and the lower arrowhead indicates the faster mobility form. B, ATPase activity was assayed with purified TAPN-Drs2p-TAPC before separation and the two fractions after separation.
Removal of Drs2p C-terminal Tail Stimulates Drs2p ATPase Activity
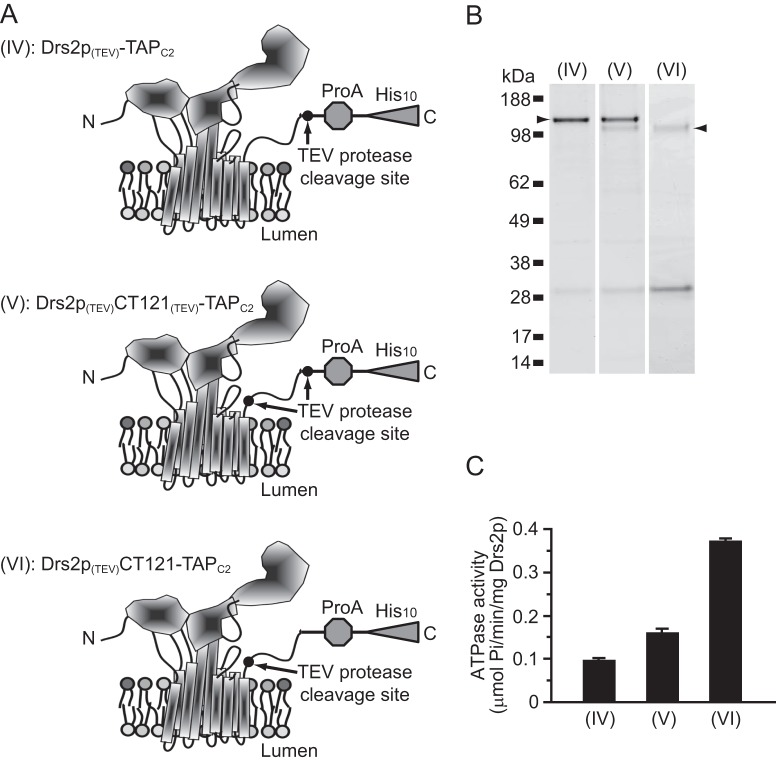
To test directly whether proteolytic cleavage within the C-terminal tail could stimulate Drs2p activity, the expression cassette was modified so that the C-terminal tail of Drs2p was either retained or removed during purification (Fig. 4, A and B). To do so, Drs2p was first tagged at the C terminus by the TAPC2 tag, which lacks the TEV protease cleavage site. Then, the TEV site was inserted immediately after the C terminus of Drs2p (Drs2p(TEV)-TAPC2 (IV)), or within the C terminus of Drs2p (Drs2p(TEV)CT121-TAPC2 (VI)), close to the end of transmembrane segment 10. Cleavage, at the latter TEV site, removes 121 residues from the predicted 137-amino acid C-terminal tail of Drs2p. An intermediate construct that contains both TEV sites was also constructed for analysis (Drs2p(TEV)CT121(TEV)-TAPC2 (V)). After purification using TAPC2 and final release by TEV cleavage, Drs2p was found to migrate at expected sizes with either full-length or almost no C-terminal tail (Fig. 4B). TEV cleavage within the tail was inefficient, as indicated by the doublet in the samples for (V) and the low recovery of the faster migrating form of Drs2p in both the (V) and (VI) preparations.
FIGURE 4.
Proteolytic removal of the C-terminal tail stimulates Drs2p activity. A, schematic shows three forms of Drs2p with TEV cleavage sites at different locations in the C-terminal tail. (IV) has a TEV cleavage site between the tail and the TAPC tag. (V) has two TEV cleavage sites: the first is between transmembrane segment 10 and the C-terminal tail, and the second is between the C-terminal tail and the TAPC tag. (VI) has a TEV cleavage site between transmembrane segment 10 and the C-terminal tail. B, these three variants were purified, and the samples were applied to an SDS-polyacrylamide gel and stained with SimplyBlue. Because of the poor recovery of (VI), the TEV protease near the 28-kDa marker is overrepresented in this sample. C, ATPase activity of (IV), (V), and (VI).
Unlike Drs2p-TAPC (Fig. 1D (II)), purified Drs2p(TEV)-TAPC2 (IV) displayed low, basal ATPase activity (Fig. 4C (IV)), indicating that the CBP moiety of TAPC inhibited Drs2p. More importantly, Drs2p(TEV)CT121-TAPC2 (VI), which has lost the majority of its C-terminal tail, exhibited an ∼3.5-fold higher specific activity than that of Drs2p with an intact C-terminal tail (Fig. 4C (IV) versus (VI)). Consistently, the Drs2p(TEV)CT121(TEV)-TAPC2 (V) preparation, containing a mixture of intact and CT121-cleaved forms, showed an intermediate ATPase-specific activity (Fig. 4C (V)). We conclude that the C-terminal tail of Drs2p contains an auto-inhibitory domain, and removal of the C-terminal tail activates Drs2p.
To investigate whether the faster migrating (clipped) form of Drs2p was detectable in vivo, whole cell extracts were prepared, and Western blotting was performed (Fig. 5). A wild-type (WT) strain was grown under a variety of conditions (low or high temperature, rich or minimal media) to determine whether cleavage events would occur physiologically. Drs2p mobility was also examined in a strain lacking three other P4-ATPases (dnf1,2,3Δ), which, along with DRS2, constitutes an essential group of genes with overlapping function. We thought that deleting three of the four members of this group might cause the cells to express a more active form of Drs2p (possibly a cleaved form). However, in all conditions tested, Drs2p migrated slightly slower than the 160-kDa marker. We could not detect a form that migrated slightly faster than the 160-kDa marker at the same position of purified tail-less Drs2p, even after overexposing the blot (Fig. 5). A minor band was detected that migrated at ∼130 kDa (asterisk in Fig. 5), which could represent a proteolyzed form, although the functional significance of this form is uncertain. Although we cannot rule out the possibility that Drs2p activity can be activated physiologically by proteolytic cleavage, we suspect that this cleavage event only occurs after the cells are lysed.
FIGURE 5.
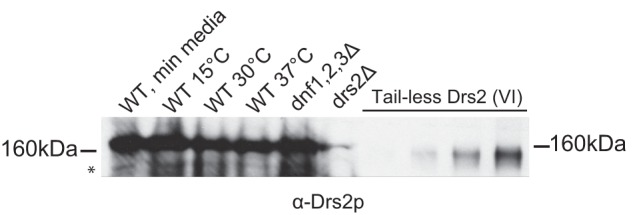
The proteolyzed form of Drs2p is not readily detected in cells. Western blotting was performed with whole cell extracts (1 A600 of cells) from wild-type strain (BY4742) grown in minimal media at 30 °C, rich media at 15 °C, rich media at 30 °C, and rich media at 37 °C. dnf1,2,3Δ and drs2Δ whole cell extracts were also blotted, as well as aliquots (in increasing concentrations) of purified tail-less Drs2p (Fig. 4 (VI)). A band that migrates faster than the tail-less form of Drs2p is marked with an asterisk. The membrane was blotted with α-Drs2p.
PS Stimulates and PI(4)P Activates Drs2p Activity
Following observations made previously, we decided to determine the effect that increasing levels of PS had on the ATPase activity of Drs2p. Phosphatidylserine is the preferred substrate of ATPase II/ATP8a1, which has 67% amino acid sequence similarity to Drs2p (43). PS strongly stimulates the ATPase activity of Atp8a1 and Atp8a2 in detergent micelles or when reconstituted into phospholipid bilayers (39, 44, 45). In contrast, when assayed in detergent micelles, the ATPase activity of Drs2p is weakly stimulated by PS (by ∼20%) (1). However, TAPN-Drs2 displayed ∼3-fold higher specific activity when reconstituted into PC liposomes containing 20% PS compared with PC liposomes (Fig. 6A). Both basal and substrate-stimulated activities were inhibited by vanadate as expected. Thus, the substrate seems to stimulate Drs2p ATPase activity more effectively when presented in a phospholipid bilayer.
FIGURE 6.
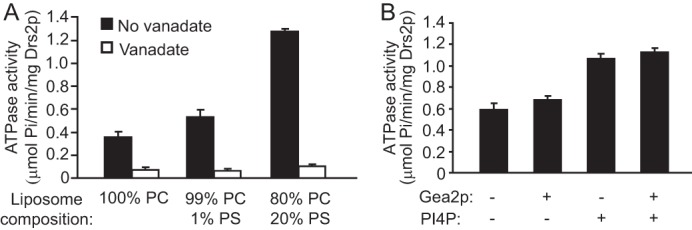
PS and PI(4)P stimulate ATPase activity of Drs2p. A, purified TAPN-Drs2p was reconstituted into proteoliposomes containing either 100 mol % DOPC, 99 mol % DOPC with 1 mol % DOPS, or 80 mol % DOPC with 20 mol % DOPS. These proteoliposomes were assayed for ATPase activity either in the presence or absence of 100 μm orthovanadate. B, TAPN-Drs2p was reconstituted into proteoliposomes comprising a DOPC backbone with 1 mol % DOPS in the presence or absence of 1 mol % PI(4)P. For each of these proteoliposome samples, ATPase activity was measured in the presence or absence of Gea2p (Sec7 domain) at a ratio of ∼1:20 mol:mol Drs2p:Gea2p. The proteoliposomes were incubated with Gea2p for 15 min on ice before assaying for activity.
Natarajan and colleagues have demonstrated that, in isolated Golgi membranes, Drs2p flippase activity is positively regulated by PI(4)P and Gea2p (Sec7 domain), both of which were shown to bind the C-terminal tail of Drs2p (2). These observations suggest that PI(4)P and Gea2p regulate Drs2p activity by directly binding its C-terminal tail. Alternatively, they may act indirectly through other factors that were present in the membrane samples. To distinguish these two possibilities, we tested whether PI(4)P and/or Gea2p could stimulate the ATPase activity of purified Drs2p reconstituted in proteoliposomes (also containing 2.5% C6 NBD-PS). PI(4)P was added at 1 mol % during proteoliposome formation, and Gea2p (recombinant Sec7 domain at a Drs2p:Gea2p ratio of ∼1:20, mol:mol) was added directly to proteoliposomes containing TAPN-Drs2p. However, whereas addition of PI(4)P enhanced the ATPase activity of Drs2p, Gea2p had no effect on overall ATPase activity (Fig. 6B). Furthermore, the presence of both PI(4)P and Gea2p did not improve the ATPase activity over the levels observed by PI(4)P alone. These data suggest that PI(4)P is directly influencing Drs2p activity, whereas the Gea2p Sec7 domain alone is insufficient for Drs2p activation (see “Discussion”).
To further examine the influence of PI(4)P on Drs2p activity, TAPN-Drs2p was reconstituted into POPC proteoliposomes containing NBD-PS and 0, 1, 2.5, or 5% PI(4)P. Stimulation of ATPase activity was observed at all concentrations of PI(4)P with a peak at 2.5% (Fig. 7A). Phospholipid translocase activity was also measured in proteoliposomes containing TAPN-Drs2p using a dithionite quenching assay to monitor ATP-dependent translocation of NBD-PS from the inner leaflet to the outer leaflet. In this case, the amount of NBD-PS flipped by TAPN-Drs2p also increased with increasing concentrations of PI(4)P (Fig. 7B). Proteoliposomes containing 5% PI(4)P did not form a tight seal to dithionite and were, therefore, not assayed. Whereas PI(4)P appeared to stimulate TAPN-Drs2p flippase activity, there was substantial variability between replicates leading to a large standard deviation for each data set.
FIGURE 7.
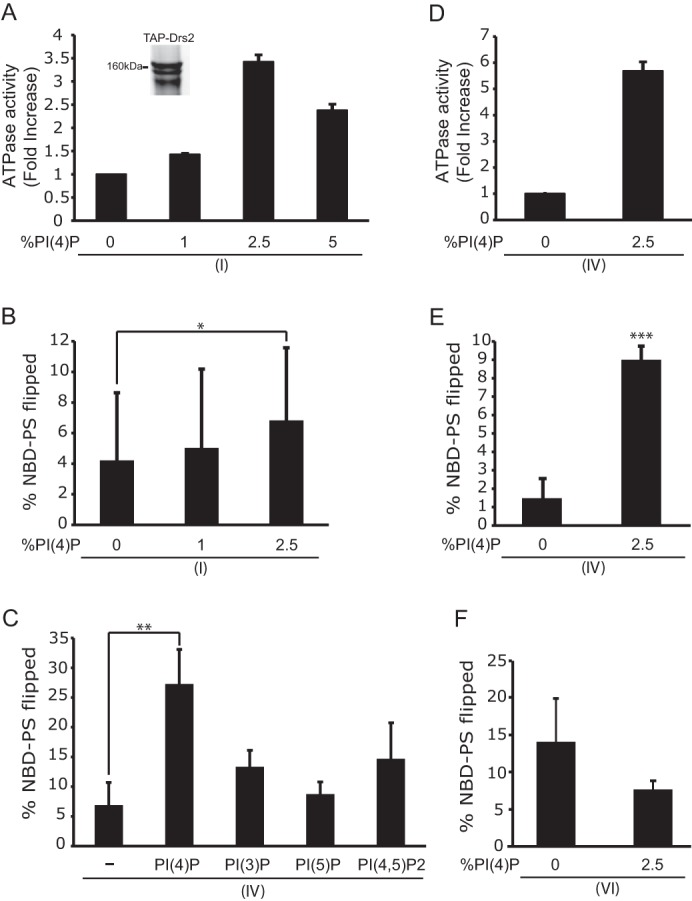
PI(4)P directly stimulates Drs2p ATPase and NBD-PS flippase activity. TAPN-Drs2p (I) was reconstituted into proteoliposomes comprising a DOPC backbone with 2.5 mol % C12 NBD-PS and the indicated amount of PI(4)P. A, ATPase activity of TAPN-Drs2p (I) after subtracting the orthovanadate controls. A Coomassie-stained sample of purified TAPN-Drs2p shows a faster migrating band, which is possibly the proteolyzed form of Drs2p (inset). B, Drs2p proteoliposomes assayed for NBD-PS flippase activity. The data are averages from 14 experiments ± S.D. (error bars). C, Drs2p(TEV)-TAPC2 (Fig. 4 (IV)) purified and reconstituted into proteoliposomes containing DOPC, 2.5 mol % C12 NBD-PS, and 2.5 mol % PI(4)P, PI(3)P, PI(5)P, or PI(4,5)P2. The sample marked with a minus sign contains no phosphoinositide. D and E, Drs2p(TEV)-TAPC2 (IV) purified, reconstituted, and assayed for ATPase activity (D) and NBD-PS flippase activity (E) (n = 3 independent experiments). F, Drs2p(TEV)CT121-TAPC2 (Fig. 4 (VI)) purified, reconstituted, and assayed for NBD-PS flippase activity (n = 3 independent experiments). *, p < 0.05; **, p < 0.01; ***, p < 0.0001.
We hypothesized that the variability in translocase activity with TAPN-Drs2p likely arose from differing extents of C-terminal tail proteolysis in each preparation of enzyme. For instance, the proteoliposomes may have either the full-length form of TAPN-Drs2p, the clipped form, or a mixture of the two forms, which could lead to variability in the activity observed. One of the Drs2p preparations used in the assays shown in Fig. 7, A and B, clearly displayed the full-length and clipped forms (Fig. 7A, inset) To overcome this limitation, Drs2p(TEV)-TAPC2 (Fig. 4 (IV)), which yields a full-length form of Drs2p with an intact C-terminal tail, was purified and reconstituted into proteoliposomes in the presence or absence of 2.5 mol % PI(4)P. The ATPase activity of the full-length Drs2p was stimulated almost 6-fold in the presence of PI(4)P (Fig. 7D). The NBD-PS flippase activity was similarly enhanced by PI(4)P (Fig. 7E). Accordingly, the variation in flippase activity that was observed with TAPN-Drs2p was no longer observed with the full-length form of Drs2p. Furthermore, Drs2p(TEV)CT121-TAPC2 (Fig. 4 (VI)) was purified and reconstituted into proteoliposomes. These samples, when assayed for flippase activity, were found to be slightly more active in the absence of PI(4)P, as they were in proteoliposomes that contained 2.5% PI(4)P (Fig. 7F). Thus, tail-less Drs2 is no longer responsive to PI(4)P, and these data are consistent with the proposed model that PI(4)P activates Drs2p through direct interaction with the Drs2p C-terminal tail.
To determine whether the C-terminal tail of Drs2p has a preference for a specific phosphoinositide, we purified the full-length Drs2p (Fig. 4 (IV)) and reconstituted it into PC/PS proteoliposomes containing different phosphoinositides. The proteoliposomes contained either no phosphoinositides or 2.5 mol % of PI(4)P, PI(3)P, PI(5)P, or PI(4,5)P2. PI(4)P significantly stimulated the flippase activity of Drs2p compared with the other phosphoinositides (Fig. 7C). Thus, PI(4)P preferentially interacts with the C-terminal tail of Drs2p to stimulate Drs2p activity.
DISCUSSION
A Model for Drs2p C-terminal Tail Auto-inhibition and Regulation
When this study and published data are taken together, a model for the regulation of Drs2p can be proposed. Drs2p has a C-terminal cytosolic tail predicted to be 137 amino acids long. The tail likely contains an auto-inhibitory domain (an R domain) that keeps Drs2p activity at a low, basal level within membranes that lack PI(4)P (e.g. the ER and early Golgi). Upon arrival in the TGN, Drs2p would be activated by the high levels of PI(4)P in this compartment. We show that the TAPN-Drs2p samples, purified with the N-terminal tag, and which contain a faster-migrating, clipped form of Drs2p, have a higher level of ATPase activity compared with preparations of Drs2p-TAPC, which lack the cleaved form of Drs2p. Thus, it appears that a cleavage event within the C-terminal tail of Drs2p enhances activity of TAPN-Drs2p. A Western blot of whole cell extracts of wild-type strains (grown under various conditions) and mutant strains did not show substantial cleavage of Drs2p. The cleavage event observed in the purified protein samples most likely occurs after cell lysis, and there is no indication that proteolysis is a physiologically relevant mechanism for activating Drs2p.
Furthermore, when Drs2p is purified with the full-length tail intact, the ATPase activity levels of Drs2p are quite low but still detectable (Fig. 4C (IV)). Reconstituted full-length Drs2p also has low levels of flippase activity (Fig. 7D, 0% PI(4)P). However, once the tail is cleaved off during purification using an engineered TEV site, we observe a significant increase in Drs2p specific activity (Fig. 4C (VI)). Therefore, removal of the C-terminal tail stimulates activity. As in the case of the other P-type ATPases, the R domain (as described in the Introduction) had the ability to self-regulate the activity of the P-type ATPase. Relieving the interaction between the R domain and the P-type ATPase stimulated the activity of the protein (18, 19, 22, 23). This study, similarly, provides direct evidence for a role of the C-terminal tail (and an R domain) in regulating the activity of Drs2p.
The Role of Effectors in Regulating the Activity of Drs2p
In purified TGN membranes, it has been shown that the addition of Gea2p (Sec7 domain) stimulates the phospholipid translocase activity of Drs2p (2). Furthermore, the addition of both PI(4)P and Gea2p to the samples stimulated the flippase activity of Drs2p to greater levels than either of them did individually. In this study, we attempted to recapitulate these observations in the reconstituted system; and, whereas addition of PI(4)P to Drs2p-proteoliposomes activated the P4-ATPase (Figs. 6B and 7), the Gea2p Sec7 domain had little effect on the specific activity of Drs2p, even when added in concert with PI(4)P (Fig. 6B).
During the course of our studies, work published recently from the laboratory of Fang-Jen Lee provides a likely explanation for these observations (46). Arl1p (Arf-like protein 1), is a member of the small GTP-binding protein family that cycles, like Arf, between active GTP-bound and inactive GDP-bound forms. Like Drs2p, Arl1p also localizes to the TGN in yeast (47, 48). The Lee group was able to show that all three proteins, Drs2p, Gea2p, and Arl1p, form a stable ternary complex through direct interactions with each other. This complex is important for stimulating the flippase activity of Drs2p at the Golgi. Furthermore, Arl1p function at the Golgi also requires this complex. To our reconstituted system, we added the recombinantly purified Sec7 domain of Gea2p. However, it is the N-terminal portion of Gea2p (upstream of the Sec7 domain) that is responsible for interacting with Arl1p (46). Additionally, Arl1p was absent from our reconstituted reactions. However, in purified Golgi membranes, it is likely that Arl1p-GTP co-purifies with the membrane (2). As a result, in the presence of Gea2p, the activity of Drs2p was stimulated. Our data imply that the interaction of Gea2p Sec7 domain is insufficient to activate Drs2p when Arl1p-GTP is absent. More work will be needed to determine the temporal and spatial organization of these various effectors and regulators during the formation of transport vesicles at the Golgi.
Although a previous study was able to show that PI(4)P stimulates the flippase activity of Drs2p in purified Golgi membranes (2), this study provides the first evidence, in a reconstituted system, that PI(4)P directly regulates the activity of Drs2p. When the full-length form of Drs2p (Fig. 4 (IV)) is reconstituted, PI(4)P is able to stimulate a 6-fold increase in the ATPase and flippase activities (Fig. 7, D and E). In contrast, tail-less Drs2p (Fig. 4 (VI)) does not display a difference in lipid translocase activity when reconstituted in the presence or absence of PI(4)P (Fig. 7F).
The step at which PI(4)P exerts its influence seems to be the dephosphorylation step of the Drs2p catalytic cycle (36). P4-ATPases do not require the presence of substrate for phosphorylation (E1 to E2-P), although for the mammalian orthologs of Drs2p, ATP8A1, and ATP8A2, the PS substrate potently stimulates dephosphorylation (E2-P to E1) (44, 49). In contrast, PS slows down the ability of Drs2p to undergo dephosphorylation, whereas addition of PI(4)P to the reaction strongly accelerates this step. Addition of PI(4)P in the absence of PS, meanwhile, only had a small effect on dephosphorylation (36). So, the major role that PI(4)P plays is at the dephosphorylation step, the point at which the phospholipid substrate has been loaded into the flippase, presumably at a site we described as the exit gate formed from residues in the first four transmembrane segments (50–52). Displacement of the C-terminal R domain by interaction with PI(4)P would then allow dephosphorylation and ejection of substrate into the cytosolic leaflet.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM062367 (to T. R. G.).
- P4-ATPases
- P-type ATPase of subfamily 4
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- C12E8
- polyoxyethylene (8) dodecyl ether
- C12E9
- polyoxyethylene (9) dodecyl ether
- CBP
- calmodulin-binding peptide
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phosphoserine
- NBD-PC
- 1-palmitoyl-2-[6-(NBD-amino)hexanoyl]-sn-glycero-3-phosphocholine
- C6 NBD-PS
- 1-palmitoyl-2-[6-(NBD-amino)hexanoyl]-sn-glycero-3-phosphoserine
- C12 NBD-PS
- 1-palmitoyl-2-[12-(NBD-amino)dodecanoyl]-sn-glycero-3-phosphoserine
- PI(4)P
- phosphatidylinositol 4-phosphate
- PS
- phosphatidylserine
- TAPC
- C-terminal tandem affinity purification tag
- TAPN
- N-terminal tandem affinity purification tag
- TEV protease
- tobacco etch virus protease
- TGN
- trans-Golgi network.
REFERENCES
- 1. Zhou X., Graham T. R. (2009) Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl. Acad. Sci. U.S.A. 106, 16586–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Natarajan P., Liu K., Patil D. V., Sciorra V. A., Jackson C. L., Graham T. R. (2009) Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat. Cell Biol. 11, 1421–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Axelsen K. B., Palmgren M. G. (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101 [DOI] [PubMed] [Google Scholar]
- 4. Kühlbrandt W. (2004) Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295 [DOI] [PubMed] [Google Scholar]
- 5. Sebastian T. T., Baldridge R. D., Xu P., Graham T. R. (2012) Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 1821, 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Toyoshima C. (2009) How Ca2+-ATPase pumps ions across the sarcoplasmic reticulum membrane. Biochim. Biophys. Acta 1793, 941–946 [DOI] [PubMed] [Google Scholar]
- 7. Toyoshima C., Nakasako M., Nomura H., Ogawa H. (2000) Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 [DOI] [PubMed] [Google Scholar]
- 8. Au K. S. (1987) Activation of erythrocyte membrane Ca2+-ATPase by calpain. Biochim. Biophys. Acta 905, 273–278 [DOI] [PubMed] [Google Scholar]
- 9. James P., Maeda M., Fischer R., Verma A. K., Krebs J., Penniston J. T., Carafoli E. (1988) Identification and primary structure of a calmodulin binding domain of the Ca2+ pump of human erythrocytes. J. Biol. Chem. 263, 2905–2910 [PubMed] [Google Scholar]
- 10. Palmgren M. G., Larsson C., Sommarin M. (1990) Proteolytic activation of the plant plasma membrane H+-ATPase by removal of a terminal segment. J. Biol. Chem. 265, 13423–13426 [PubMed] [Google Scholar]
- 11. Palmgren M. G., Sommarin M., Serrano R., Larsson C. (1991) Identification of an auto-inhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J. Biol. Chem. 266, 20470–20475 [PubMed] [Google Scholar]
- 12. Portillo F., de Larrinoa I. F., Serrano R. (1989) Deletion analysis of yeast plasma membrane H+-ATPase and identification of a regulatory domain at the carboxyl terminus. FEBS Lett. 247, 381–385 [DOI] [PubMed] [Google Scholar]
- 13. Rasi-Caldogno F., Carnelli A., De Michelis M. I. (1992) Plasma membrane Ca-ATPase of radish seedlings. II. Regulation by calmodulin. Plant Physiol. 98, 1202–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rasi-Caldogno F., Carnelli A., De Michelis M. I. (1993) Controlled proteolysis activates the plasma membrane Ca2+ pump of higher plants: a comparison with the effect of calmodulin in plasma membrane from radish seedlings. Plant Physiol. 103, 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rasi-Caldogno F., Carnelli A., De Michelis M. I. (1995) Identification of the plasma membrane Ca2+-ATPase and of its auto-inhibitory domain. Plant Physiol. 108, 105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hwang I., Harper J. F., Liang F., Sze H. (2000) Calmodulin activation of an endoplasmic reticulum-located calcium pump involves an interaction with the N-terminal auto-inhibitory domain. Plant Physiol. 122, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geering K. (2001) The functional role of β-subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 33, 425–438 [DOI] [PubMed] [Google Scholar]
- 18. Carafoli E. (1994) Biogenesis: plasma membrane calcium ATPase: 15 years of work on the purified enzyme. FASEB J. 8, 993–1002 [PubMed] [Google Scholar]
- 19. Sze H., Liang F., Hwang I., Curran A. C., Harper J. F. (2000) Diversity and regulation of plant Ca2+ pumps: insights from expression in yeast. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 433–462 [DOI] [PubMed] [Google Scholar]
- 20. Caroni P., Carafoli E. (1981) Regulation of Ca2+-pumping ATPase of heart sarcolemma by a phosphorylation-dephosphorylation Process. J. Biol. Chem. 256, 9371–9373 [PubMed] [Google Scholar]
- 21. James P. H., Pruschy M., Vorherr T. E., Penniston J. T., Carafoli E. (1989) Primary structure of the cAMP-dependent phosphorylation site of the plasma membrane calcium pump. Biochemistry 28, 4253–4258 [DOI] [PubMed] [Google Scholar]
- 22. Palmgren M. G. (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845 [DOI] [PubMed] [Google Scholar]
- 23. Portillo F. (2000) Regulation of plasma membrane H+-ATPase in fungi and plants. Biochim. Biophys. Acta 1469, 31–42 [DOI] [PubMed] [Google Scholar]
- 24. Fuglsang A. T., Visconti S., Drumm K., Jahn T., Stensballe A., Mattei B., Jensen O. N., Aducci P., Palmgren M. G. (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr946-Thr-Val and requires phosphorylation of Thr947. J. Biol. Chem. 274, 36774–36780 [DOI] [PubMed] [Google Scholar]
- 25. Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. (2000) A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275, 17762–17770 [DOI] [PubMed] [Google Scholar]
- 26. Olsson A., Svennelid F., Ek B., Sommarin M., Larsson C. (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 118, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piotrowski M., Morsomme P., Boutry M., Oecking C. (1998) Complementation of the Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase generates a highly abundant fusicoccin binding site. J. Biol. Chem. 273, 30018–30023 [DOI] [PubMed] [Google Scholar]
- 28. Cornelius F., Mahmmoud Y. A., Meischke L., Cramb G. (2005) Functional significance of the shark Na,K-ATPase N-terminal domain: is the structurally variable N terminus involved in tissue-specific regulation by FXYD proteins? Biochemistry 44, 13051–13062 [DOI] [PubMed] [Google Scholar]
- 29. Ekberg K., Palmgren M. G., Veierskov B., Buch-Pedersen M. J. (2010) A novel mechanism of P-type ATPase auto-inhibition involving both termini of the protein. J. Biol. Chem. 285, 7344–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen S., Wang J., Muthusamy B. P., Liu K., Zare S., Andersen R. J., Graham T. R. (2006) Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7, 1503–1517 [DOI] [PubMed] [Google Scholar]
- 31. Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., Tanaka K. (2007) Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell 18, 295–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. (2004) Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 3418–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Natarajan P., Wang J., Hua Z., Graham T. R. (2004) Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. U.S.A. 101, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alder-Baerens N., Lisman Q., Luong L., Pomorski T., Holthuis J. C. (2006) Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell 17, 1632–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chantalat S., Park S. K., Hua Z., Liu K., Gobin R., Peyroche A., Rambourg A., Graham T. R., Jackson C. L. (2004) The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J. Cell Sci. 117, 711–722 [DOI] [PubMed] [Google Scholar]
- 36. Jacquot A., Montigny C., Hennrich H., Barry R., le Maire M., Jaxel C., Holthuis J., Champeil P., Lenoir G. (2012) Phosphatidylserine stimulation of Drs2p: Cdc50p lipid translocase dephosphorylation is controlled by phosphatidylinositol 4-phosphate. J. Biol. Chem. 287, 13249–13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 38. Carter S. G., Karl D. W. (1982) Inorganic phosphate assay with malachite green: an improvement and evaluation. J. Biochem. Biophys. Methods 7, 7–13 [DOI] [PubMed] [Google Scholar]
- 39. Paterson J. K., Renkema K., Burden L., Halleck M. S., Schlegel R. A., Williamson P., Daleke D. L. (2006) Lipid specific activation of the murine P4-ATPase Atp8a1 (ATPase II). Biochemistry 45, 5367–5376 [DOI] [PubMed] [Google Scholar]
- 40. Zimmerman M. L., Daleke D. L. (1993) Regulation of a candidate aminophospholipid-transporting ATPase by lipid. Biochemistry 32, 12257–12263 [DOI] [PubMed] [Google Scholar]
- 41. McIntyre J. C., Sleight R. G. (1991) Fluorescence assay for phospholipid membrane asymmetry. Biochemistry 30, 11819–11827 [DOI] [PubMed] [Google Scholar]
- 42. Chen C. Y., Ingram M. F., Rosal P. H., Graham T. R. (1999) Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147, 1223–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang X., Halleck M. S., Schlegel R. A., Williamson P. (1996) A subfamily of P-type ATPases with aminophospholipid transporting activity. Science 272, 1495–1497 [DOI] [PubMed] [Google Scholar]
- 44. Ding J., Wu Z., Crider B. P., Ma Y., Li X., Slaughter C., Gong L., Xie X. S. (2000) Identification and functional expression of four isoforms of ATPase II, the putative aminophospholipid translocase: effect of isoform variation on the ATPase activity and phospholipid specificity. J. Biol. Chem. 275, 23378–23386 [DOI] [PubMed] [Google Scholar]
- 45. Coleman J. A., Kwok M. C., Molday R. S. (2009) Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 284, 32670–32679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsai P. C., Hsu J. W., Liu Y. W., Chen K. Y., Lee F. J. (2013) Arl1p regulates spatial membrane organization at the trans-Golgi network through interaction with Arf-GEF Gea2p and flippase Drs2p. Proc. Natl. Acad. Sci. U.S.A. 110, E668–E677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peyroche A., Paris S., Jackson C. L. (1996) Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384, 479–481 [DOI] [PubMed] [Google Scholar]
- 48. Peyroche A., Courbeyrette R., Rambourg A., Jackson C. L. (2001) The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J. Cell Sci. 114, 2241–2253 [DOI] [PubMed] [Google Scholar]
- 49. Coleman J. A., Vestergaard A. L., Molday R. S., Vilsen B., Andersen J. P. (2012) Critical role of a transmembrane lysine in aminophospholipid transport by mammalian photoreceptor P4-ATPase ATP8A2. Proc. Natl. Acad. Sci. U.S.A. 109, 1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baldridge R. D., Graham T. R. (2013) Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 110, E358–E367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baldridge R. D., Graham T. R. (2012) Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 109, E290–E298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baldridge R. D., Xu P., Graham T. R. (2013) Type IV P-type ATPases distinguish mono- versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J. Biol. Chem. 288, 19516–19527 [DOI] [PMC free article] [PubMed] [Google Scholar]