Background: Abortive cycling is a key feature of RNA polymerases.
Results: Nicks and mismatches have little effect on abortive probabilities.
Conclusion: The energetics of the hybrid pushing on the protein, rather than that of bubble expansion, is the primary contributor to abortive cycling.
Significance: Abortive cycling arises from the need to couple RNA growth to promoter release in RNA polymerases.
Keywords: DNA-Protein Interaction, Enzyme Mechanisms, Nucleic Acid Enzymology, RNA, RNA Polymerase, Transcription, Enzymology
Abstract
It has long been known that during initial transcription of the first 8–10 bases of RNA, complexes are relatively unstable, leading to the release of short abortive RNA transcripts. An early “stressed intermediate” model led to a more specific mechanistic model proposing “scrunching” stress as the basis for the instability. Recent studies in the single subunit T7 RNA polymerase have argued against scrunching as the energetic driving force and instead argue for a model in which pushing of the RNA-DNA hybrid against a protein element associated with promoter binding, while likely driving promoter release, reciprocally leads to instability of the hybrid. In this study, we test these models in the structurally unrelated multisubunit bacterial RNA polymerase. Via the targeted introduction of mismatches and nicks in the DNA, we demonstrate that neither downstream bubble collapse nor compaction/scrunching of either the single-stranded template or nontemplate strands is a major force driving abortive instability (although collapse from the downstream end of the bubble does contribute significantly to the instability of artificially halted complexes). In contrast, pushing of the hybrid against a mobile protein element (σ3.2 in the bacterial enzyme) results in substantially increased abortive instability and is likely the primary energetic contributor to abortive cycling. The results suggest that abortive instability is a by-product of the mechanistic need to couple the energy of nucleotide addition (RNA chain growth) to driving the timed release of promoter contacts during initial transcription.
Introduction
Abortive cycling, marked by the premature release of short RNA products from initially transcribing complexes, is a common feature in all RNA polymerases (1–3) and is known to be a point of cellular regulation at some promoters (4). Over the last decade, abortive cycling has been extensively studied, and various models have been proposed to explain its occurrence (5–7).
Prior to the initiation of transcription, an RNA polymerase must melt open the promoter DNA to form an open complex. The initial bubble in the open complex is 13–14 base pairs (bp) long for Escherichia coli RNA polymerase (8, 9), about 8 bp for RNA polymerase II (10), and about 8 bp for T7 RNA polymerase (11–13). RNA polymerases can accommodate only a 2–3-nucleotide transcript in their open complex (14), and to continue beyond that requires further melting of downstream DNA while maintaining promoter contacts, and by keeping the upstream edge of the bubble constant, the bubble expands. Not surprisingly, then, RNA polymerases maintain their initial promoter contacts through the synthesis of RNA up to about the length of the ultimate RNA-DNA hybrid in stable elongation complexes. This “initially transcribing” phase of transcription, in which promoter contacts are maintained as the bubble expands, is characteristically unstable, leading to the premature release of short abortive RNAs, with the complex recycling to repeat the de novo initiation event.
The instability of initially transcribing complexes might arise from several energetic factors. The requirement of melting downstream duplex (bubble expansion) to synthesize RNA beyond about 2–3 bases could impede forward progression, whereas premature reannealing of the already melted DNA strands (partial or complete bubble collapse) would competitively displace the nascent RNA (15). The drawing in of downstream DNA into the active site could lead to energetically unfavorable compaction or steric “scrunching” of the DNA template strand in the active site cavity. This could present an energetic barrier that similarly opposes forward progression and/or destabilizes the nascent transcript (14, 16, 17). Finally, we now understand that as the RNA-DNA hybrid grows, it pushes against elements of the protein involved in promoter binding, and this steric clash could impede forward progression and/or destabilize the hybrid (6, 14, 18–20). All of these potential stresses fall broadly under the category of a “stressed intermediate,” and it has been assumed that this stress leads to abortive dissociation of nascent RNA (5).
It is important to note that although abortive propensity is very sequence-dependent, all RNA polymerases display abortive cycling. Thus, it must reflect a fundamental property of the transcription mechanism. The well studied RNA polymerases fall into two distinct classes as follows: the evolutionarily related “multisubunit” bacterial and eukaryotic enzyme family, and the “single subunit” family represented by bacteriophage T7, mitochondrial, and chloroplast RNA polymerases. All RNA polymerases are characterized not only by a relatively unstable (abortive) initiation phase, but this phase is also followed by a significant structural rearrangement associated with the transition to stable elongation. Although the structural details of initiation complexes from single subunit and multisubunit RNA polymerases are known (14, 21–23), the energetic and mechanistic details of these structural changes and of abortive cycling are yet to be completely understood.
Our recent studies with T7 RNA polymerase have established in that system that neither downstream bubble collapse nor DNA scrunching stress contributes substantially to instability in initially transcribing complexes (7). Structures of T7 RNA polymerase complexes halted at positions +7 and +8 show retention of promoter contacts accompanied by a significant rotation of the N-terminal platform (24). It is expected that as the RNA-DNA hybrid grows, it will start “pushing” on the N-terminal platform driving the latter to rotate. In the multisubunit bacterial RNA polymerase, the RNA-DNA hybrid is thought to similarly “push” on the σ3.2 linker, displacing the linker from the RNA exit channel (18, 19). This likely has the same effect as in T7 RNA polymerase; hybrid push drives a structural change in the protein that drives a weakening of promoter contacts, ultimately triggering promoter release and the opening or formation of the RNA exit channel. Necessarily, however, the protein must push back on the hybrid, which could destabilize the hybrid and lead directly to abortive loss of the RNA. Our recent studies with a T7 RNA polymerase mutant support this model (25).
We now extend these studies to the initially transcribing complexes of σ70-E. coli RNA polymerase transcribing on variants of the T5 N25 promoter (26, 27). Specifically, we have studied the role of scrunching and of downstream DNA bubble collapse in initially transcribing complexes by designing DNA constructs with a gap or nick to reduce scrunching stress, and also by introducing mismatch sequences at the downstream end of a halted transcription bubble to prevent downstream DNA bubble collapse. Note that this work addresses only the conventional abortive cycling that is thought to occur before normal promoter release; it does not address “very long” abortive products seen on some constructs, which are thought to arise via a different mechanism (28).
In E. coli RNA polymerase, it is thought that abortive complexes are backtracked complexes (29). In an initially transcribing complex, the upstream end of the transcription bubble is held in position by the interactions of σ with promoter DNA, as well as with the core polymerase. Backtracking involves displacement of the 3′ end of the RNA from the template, necessarily stopping addition of the next nucleotide. Unlike backtracking during elongation, it also leads to a net shortening of the RNA-DNA hybrid (because the 5′ end of the RNA is not yet displaced from the hybrid, there is no upstream single-stranded RNA to reanneal in compensation). Both of these effects would be expected to kinetically favor the abortive dissociation of the nascent RNA. Extrusion of the 3′ end of the RNA into the secondary channel also makes the RNA accessible to the GreB cleavage factor (29, 30). Thus, in this work, we use GreB cleavage as a marker of backtracking-induced abortive complexes. We have also studied the effects of a mutant σ70 on backtracked complexes formed on engineered DNA constructs.
The results below argue that in the multisubunit RNA polymerases, steric compaction of DNA strands plays no role, and downstream bubble collapse plays only a minor role, as driving forces in abortive cycling. The results instead support a model in which “hybrid-push” is the primary energetic contributor to abortive cycling. Together, these and recent results suggest that despite very different structural details, all RNA polymerases share common mechanistic fundamentals.
MATERIALS AND METHODS
Enzymes and Proteins
E. coli RNA polymerase holoenzyme and core enzyme were purchased commercially from Epicentre (an Illumina company). The mutant σ70 and GreB proteins were generously provided by Lilian Hsu.
DNA Oligonucleotides
DNA oligonucleotides were purchased commercially from Eurofins MWG Operon. All DNA strands used in this study extend from position −60 upstream to +35 downstream, except the fork junction construct that extends from −12 to +35 on the nontemplate strand and from −4 to +35 on the template strand. Except where indicated, the sequence of the promoter is that of the T5 N25 promoter (31).
Transcription Assays
Transcription assays were carried out in a total volume of 10 μl at 37 °C for 20 min. Unless otherwise indicated, DNA and enzyme concentrations were 100 nm each in transcription buffer (50 mm Tris-HCl, 200 mm KCl, 10 mm MgCl2, 10 mm β-mercaptoethanol, and 10 μg/ml acetylated BSA). Transcription was initiated with a final concentration of 100 μm of ATP and UTP each, to allow halting at position +8. RNA was 5′-labeled with [γ-32P]ATP or internally labeled with [γ-32P]ATP or [α-32P]UTP, as indicated. Transcription was quenched by addition of an equal volume of formamide stop solution (95% formamide, 40 mm EDTA, 0.01% bromphenol blue and 0.01% xylene cyanol). The RNA products were resolved on 20% polyacrylamide gels containing 7 m urea and visualized with a Fuji phosphorimager.
In transcription assays to observe backtracking of RNA polymerase, a 10-fold excess of GreB was added to the enzyme-DNA complex before initiating transcription with nucleoside triphosphates. The RNA products were internally labeled in these reactions to be able to resolve GreB-cleaved RNA products.
Transcription initiation from fork junction DNA constructs showed misinitiation in the presence of all four NTPs. To restrict initiation to the +1 position, the dinucleotide pApU was included, as indicated, to a concentration of 500 μm (32).
Quantification
Turnover numbers were calculated by dividing the concentration of the halted 8-mer RNA by the limiting concentration of enzyme-DNA complex (100 nm). Note that if the concentration of active enzyme is lower than this, the reported turnover numbers will represent lower limits, but comparisons between constructs will remain unchanged. Percent fall off calculations were carried out by calculating the ratio of the amount of RNA released of a given length to the total amount of RNA of that length and longer (3).
RESULTS
A full understanding of transcription regulatory events such as promoter proximal pausing and abortive cycling requires an understanding of the energetics (or more appropriately, the relative kinetics of competing processes) of initially transcribing complexes. What factors do or do not contribute to the stability of halted complexes or of the tendency to release abortive products during initial transcription? Factors that have been implicated in the past include the steric scrunching of template and/or nontemplate strands of the DNA, the overall energetics of bubble expansion, and the local expansion/collapse of the edges of the melted bubble (7, 17, 33). In this study, we tested each of these as potential contributors to abortive cycling.
Removal of Downstream Bubble Collapse by Local Mismatching
Previous studies in the T7 RNA polymerase system have shown that artificially halted initially transcribing complexes are destabilized by collapse of the DNA from the downstream end of the melted bubble (33). More specifically, mismatching of the DNA at the downstream edge of the bubble to prevent collapse stabilizes halted complexes against dissociation of the RNA. However, during full run synthesis in the presence of all four nucleoside triphosphates (NTPs), similar mismatching at corresponding sites around abortive positions leads to only small changes in abortive fall off.
In this study, we have characterized transcription from similar DNA constructs in the E. coli RNA polymerase system. Introducing mismatched sequences at the downstream end of a halted transcription complex bubble prevents the reannealing of the downstream DNA but should not alter the steric stress of scrunched template or nontemplate DNA in the transcription bubble. The results shown in Fig. 1A demonstrate clearly that in the E. coli system mismatching of the bubble in the initially transcribed region also has only small and nonsystematic effects on abortive propensities during full run synthesis (in the presence of all four NTPs).
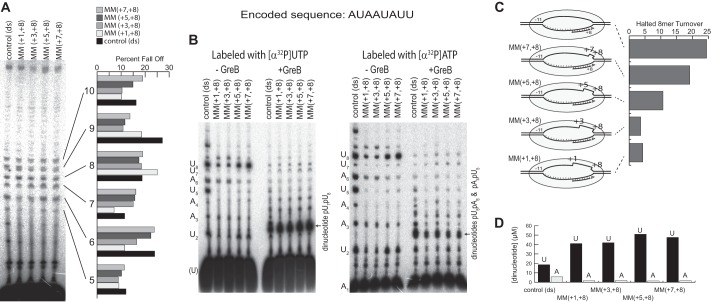
FIGURE 1.
Removal of downstream bubble collapse in complexes halted at position +8. A, runoff transcription on control and MM(+1,+8) DNA, showing quantification of abortive percent fall off profiles. B, transcription (−GreB) in complexes halted at position +8 (by inclusion of only ATP and UTP in the reaction). To detect backtracking, parallel reactions were carried out in the presence of 1 μm cleavage factor GreB. To assign GreB cleavage products, each set of reactions was run in parallel with either [α-32P]UTP versus [α-32P]ATP as the label. C, schematics showing the DNA constructs used, noting regions of DNA mismatch in a halted complex and showing quantification of halted 8-mer turnover on each construct, from the data in B. D, quantification of differential labeling ([α-32P]UTP versus [α-32P]ATP) of the dinucleotide GreB cleavage product confirms that it is primarily the pUpU predicted from backtracking of the halted 8-mer, with some pApU and pUpA from backtracked abortive complexes at positions +5 and +6, respectively.
In artificially halted complexes (in this case, by including only ATP and UTP to force a halt at position +8), stability (as measured by turnover) is increased by increasing extents of mismatching within the bubble (as quantified in Fig. 1C). Transcription on constructs mismatched at the downstream edge of the bubble shows reduced turnover compared with the fully complementary DNA (double strand control). The DNA construct with 8 bases mismatched from position +1 to +8 shows ∼5-fold lower turnover than the control, and mismatching 6 bases from position +3 to +8 yields similarly reduced turnover. Mismatching 4 bases from position +5 to +8 reduces turnover, but to a lesser extent, whereas mismatching only 2 bases from position +7 to +8 yields a still smaller effect.
Note that halted transcription on the fully double-stranded construct produces a 5-mer transcript (Fig. 1B, −GreB) that is not observed in runoff transcription (Fig. 1A) nor on the mismatched constructs. That it is not observed in runoff transcription is consistent with its assignment as a halted (at position +8) product deriving from misinitiation at position +4. That it is dramatically reduced in halted transcription from the mismatched constructs suggests that mismatching in the +1 to +8 region reduces this proposed misinitiation (the basis for this is not clear).
Although in halted transcription, the turnover number for the two base-mismatched construct MM(+7,+8) is reduced only about 20% from that of the control DNA, the amount of abortives (in particular, the 6-mer) is strikingly low. This suggests that the halted complex is stabilized more than the 20% reduction would predict, because a complex that aborts less should be more efficient at synthesis of the 8-mer halted RNA.
To confirm that the reduced accumulation of halted 8-mer RNA on mismatched constructs arises from a failure of the 8-mer product to turn over, rather than from a failure to initiate, we carried out a parallel set of reactions, but in the presence of the cleavage factor GreB. This factor stimulates cleavage of backtracked RNAs that are expected to arise from abortive or halted complexes (34). In this case, (only) the halted 8-mer RNA, which has the sequence AUAAUAUU, should yield the primary dinucleotide cleavage product pUpU, silent in the [α-32P]ATP, while the two highest aborting complexes at positions +5 and +6 should yield the cleavage products pApU and pUpA, respectively, and will be labeled by both [α-32P]ATP and [α-32P]UTP.
The results presented in Fig. 1B show that in the presence of GreB, substantial amounts of dinucleotide are observed for complexes halted at position +8. Differential labeling with [α-32P]ATP and [α-32P]UTP shows that the product is primarily the predicted pUpU product (all other potential products would contain A), as quantified in Fig. 1D. The accumulated quantity of this product is higher in the mismatched constructs than in the double-stranded control, consistent with a higher occupancy (increased stability) of the halted complex in the downstream-mismatched constructs. The high levels of GreB-mediated cleavage argue that mismatching at the downstream edge of the bubble does not reduce backtracking (required for cleavage). In other words, collapse of the downstream edge of the bubble does not drive backtracking.
Introduction of Nicks or Gaps to Reduce Steric Scrunching
An alternative source of potentially destabilizing energetics during initial transcription comes from the fact that forward progression of RNA polymerase draws DNA into the active site. Thus, abortive instability might arise from the potential stress caused by compaction of DNA strands (“steric scrunching”) in the active site channel as the polymerase translocates from position +3 or +4 to the point of promoter release and the transition to elongation (14, 16, 17).
Scrunching of a strand of DNA into a restricted volume would be subject to the energetic constraints imposed by the DNA backbone. Introduction of a nick in the DNA (by assembling the strand from two pieces) should loosen some of those constraints, allowing more conformational freedom in the backbone. Introduction of a nick also removes a negatively charged phosphate (the oligonucleotides have 3′- and 5′-hydroxyls), removing volume and reducing potential phosphate charge-charge repulsion. Nicking DNA should also reduce the energetics of bubble collapse, because the cooperativity of collapse is reduced. Indeed, nicking within the initially melted region of the promoter has been shown to significantly enhance open complex formation (35). Gapping the DNA removes entire nucleotides and so should more dramatically reduce any unfavorable energetics of steric scrunching.
The results presented in Fig. 2C demonstrate that simple nicking of the DNA, while otherwise retaining the base pairing that drives collapse, yields overall turnover of a complex halted at position +8 comparable with that of native double-stranded DNA. Similarly, gapping the DNA by removing nontemplate nucleotides −2 and −1 in the gap(−3/+1) construct also yields native halted complex turnover. This is consistent with a model in which collapse of the downstream edge of the bubble (beginning at or near position +8) contributes to instability in initially transcribing complexes halted at position +8, because all of these constructs retained downstream collapse. To confirm this result, the downstream strand was altered to reintroduce a mismatch from position +1 to +8, or from position +5 to +8, while retaining the nick between nontemplate bases −1 and +1. As expected, in both cases, turnover decreased dramatically, as seen with the mismatched but fully contiguous constructs. Fig. 2D shows the runoff product from transcription on the DNA constructs used in Fig. 2C, in the presence of all four NTPs.
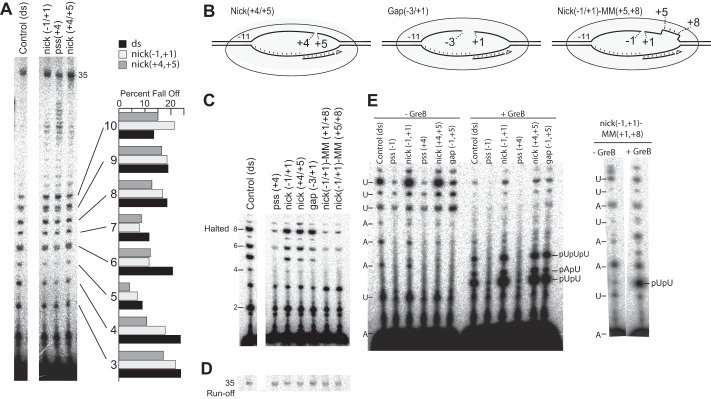
FIGURE 2.
Nicking or gapping the DNA does not stabilize initially transcribing complexes. A, runoff transcription (35-mer) on sample nicked and mismatched constructs (with all four NTPs). B, schematics illustrating nick/gap/mismatch nomenclature. C, transcription halted at +8 on various DNA constructs. D, runoff (35-mer) transcription on the constructs from C. E, GreB susceptibility of various nicked, gapped, and/or mismatched DNA constructs halted at position +8. The sequences of the templates used are shown alongside the gels. In all experiments, RNA was labeled with [α-32P]ATP.
We have included in Fig. 2C the results with a construct, pss(+4), analogous to a construct studied with T7 RNA polymerase, in which the nontemplate strand is removed completely downstream of position +4. Although T7 RNA polymerase initiates well with a downstream single-stranded template (36, 37), E. coli RNA polymerase has been shown to require duplex DNA downstream for proper initiation (38). The results of Fig. 2C appear to confirm this observation, as very little transcript is produced in halted transcription. Although the pss(+4) construct does show significant runoff product (Fig. 2A), this result is likely misleading. Although the results of Fig. 2C confirm that initiation on a construct that is single-stranded downstream of the start site is poor, after an initial single round of full-length synthesis on a partially single-stranded template, the construct will no longer be partially single-stranded, as the product RNA should stably take the place of the missing nontemplate strand (either remaining as a persistent hybrid or reannealing after synthesis). Subsequent promoter binding events will now occur on a gap(+4,+5) construct, with a downstream RNA-DNA duplex. Because the latter prefers an A-form structure, this suggests that the downstream β-clamp can interact with either a B- or A-form duplex. Finally, note that although scrunching strain is presumably reduced in the nicked DNA constructs, the abortive profiles of runoff transcription reactions in Fig. 2A are comparable with that of the fully double-stranded and complementary control DNA, suggesting that neither bubble collapse nor nontemplate strand scrunching is the primary driving force in abortive cycling.
Downstream Bubble Collapse and Nontemplate Scrunching Play Little Role in Backtracking of Initially Transcribing Complexes
As noted above, a correlation has been established previously that positions (beyond about +5) with high abortive probability are also subjected to backtracking, as evidenced by GreB-mediated cleavage and rescue (39). Backtracking extrudes the 3′ end of the RNA into the secondary channel, rendering it susceptible to GreB-mediated cleavage. This yields short (primarily dinucleotide) cleavage products corresponding to the 3′ end of the transcript (40). Cleavage brings the RNA-DNA hybrid back to the active site register, allowing continued extension of the RNA (or potentially, a second round of GreB cleavage).
When RNA polymerase backtracks in the elongation phase, the transcription bubble moves upstream, with simultaneous reannealing of downstream DNA and re-melting of upstream DNA (41–43). Total hybrid length is maintained, because displacement of the 3′ end of the RNA is balanced by reannealing of RNA at the 5′ end. However, in the initially transcribing phase, backtracking is expected to yield reannealing of only the downstream DNA, as the upstream end of the transcription bubble is held in position by the strong interactions between the σ2.4 domain and the −10 region of the promoter. Additionally, because there is no single-stranded RNA at the 5′ end to reanneal, displacement of the 3′ end of the RNA yields a net shortening of the RNA-DNA hybrid. Thus, backtracking during initial transcription would be expected to destabilize hybrids.
To test whether the scrunching of the nontemplate DNA strand drives polymerase backtracking during initial transcription, we probed the above complexes for susceptibility to GreB cleavage. Complexes halted (and backtracked) at position +8 are expected to yield the primary GreB cleavage product pUpU. As shown in Fig. 2E, complexes that yield relatively unstable halted complexes at +8 (extensive turnover in −GreB measurements) also yield extensive cleavage products in the presence of GreB. Partially single-stranded controls, pss(−1) and pss(+4) show little accumulation of GreB cleavage products, most likely because they are expected (and observed) to initiate very poorly (38).
Thus, as before, in the nicked and gapped constructs, restoration of downstream bubble collapse, even in the absence of steric strand scrunching, restores native instability. Appearance of the GreB cleavage product pUpU on the nick(−1,+1)−MM(+1,+8) DNA shows that backtracking occurs independently of DNA scrunching and downstream bubble collapse.
Evidence That “Hybrid Push” Plays a Role in the Backtracking of Initially Transcribing Complexes
In the bacterial RNA polymerase, the growing RNA-DNA hybrid is thought to push against the σ3.2 “linker” subdomain, which connects promoter binding regions σ2 and σ4, and which itself makes weak contacts with the core RNA polymerase (19). The σ3.2 linker sits in the RNA exit channel so that it is displaced by the growing hybrid, weakening promoter interactions and allowing the transition to elongation (19, 20, 44, 45). Murakami et al. (19) have proposed that competition between this linker and the growing hybrid could lead to abortive instability.
Indeed, a point mutation, S506F, in the σ3.2 linker region of σ70, which weakens its interaction with the core enzyme as well as directs the linker away from the active site, leads to reduced abortive cycling (18, 19, 44, 46). The hybrid push model (19, 25) would predict that this mutation should also lead to decreased backtracking and increased stability of the complex, as the linker pushes back less against the nascent hybrid.
The results presented in Fig. 3, A and C (quantified in B and D), demonstrate that on double-stranded DNA with two different initial sequences, the S506F mutant holoenzyme halted at position +8 turns over about 2–3-fold less than does the wild type complex, suggesting that the halted complex at position +8 is more stable in the mutant. Although the mutant has been previously reported to produce ∼30% less full-length product (18), the data in Fig. 3E show that both wild type and mutant holoenzyme yield comparable amounts of full-length runoff product, suggesting that initiation is native-like in the mutant.
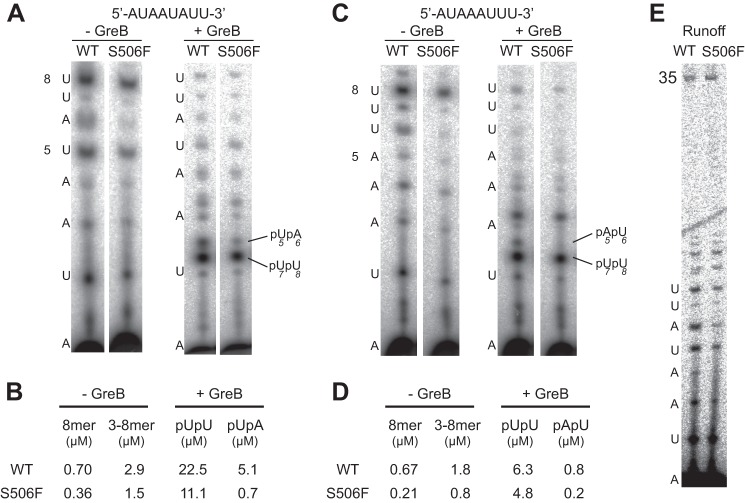
FIGURE 3.
σ70 S506F low-aborting mutant yields less back tracking in a complex halted at +8. Transcription is carried out in the absence (left) and presence (right) of GreB with wild type and S506F mutant on double-stranded DNA with initial sequence AUAAUAUU (A) and initial sequence AUAAAUUU (the radioactive label is [α-32P]UTP) (B). C and D, quantification of cleaved dinucleotides (pUpU and pUpA) from wild type and mutant complex are shown in μm from A and B, respectively. E, runoff transcription with WT and mutant complex shows comparable full-length products on DNA with the initial sequence AUAAUAUU.
As above, the nature of halted initially transcribing complexes in the S506F mutant can be probed by addition of GreB. The results presented in Fig. 3 show that the accumulation of the pUpU GreB cleavage product associated with complexes halted at position +8 is 2–3 times lower in the mutant than in the wild type complex. If the 8-mer halted complex turns over less (accumulates more at steady state) in the mutant, the lower amounts of cleavage product must arise from lower rates of RNA cleavage mediated by GreB. The most likely explanation for this observation is that such complexes backtrack less in the mutant than in the wild type protein, a result predicted by the model in which the σ3.2 linker “pushes back” less in the mutant than in the wild type.
Note that the primary product of backtracking on these two sequences is the dinucleotide pUpU. In most cases, transcription resumes efficiently following GreB cleavage, rescuing abortive cycling. In principle, however, after this cleavage, a second backtracking event would allow GreB cleavage to produce the dinucleotide pUpA for the sequence in Fig. 3A and pApU for the sequence in Fig. 3C. The results suggest this and show that alternative dinucleotides (that each contain A and U) accumulate at rates 4–7-fold lower in the S506F mutant. If this interpretation is correct, the results suggest that this extensive backtracking is driven much less in the reduced-abortive S506F mutant, further supporting the role of hybrid push in backtracking and dissociation of abortive RNAs.
Holoenzyme Abortively Cycles in the Absence of Scrunching
To probe more directly whether pushing of the σ3.2 linker on the growing RNA-DNA hybrid facilitates abortive release of initially transcribing RNA in the native enzyme, we characterized transcription from fork junction DNA constructs, which allow initiation in the complete absence of σ (38, 47). In these constructs, the nontemplate strand of the DNA extends from upstream position −12 to downstream position +35, whereas the template strand extends from only −4 upstream to +35 downstream. In addition, the bases in the two strands from positions −4 to −1 (only) are designed to be noncomplementary, as shown in Fig. 4A. The four unpaired bases at the 3′ end of the template strand mimic the downstream end of an open complex and, together with clamp interactions with the downstream duplex, allow the initiation of transcription (48).
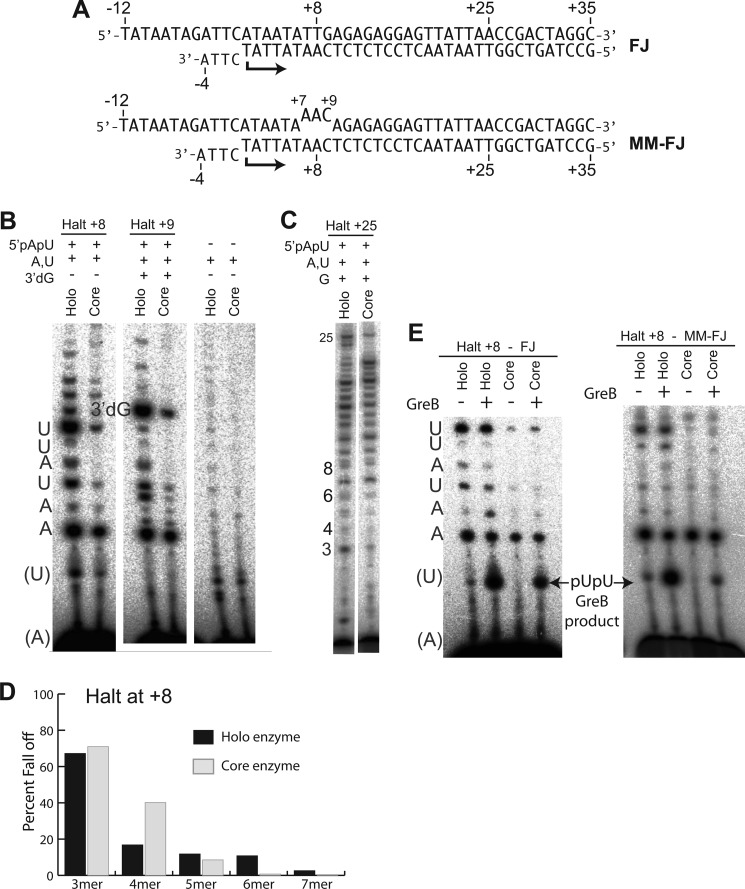
FIGURE 4.
Test of the hybrid push model, absence of σ. A, fork junction constructs used as DNA templates, natively matched (FJ) or mismatched (MM-FJ) at positions +7 to +9. B, halted 8-mer and 9-mer transcription initiated with pApU, using holoenzyme and core enzyme on fork junction DNA (FJ). C, as in B but halting at position +25. D, percent fall off numbers calculated from B show no abortives at positions +6 and +7 with core polymerase. E, GreB cleavage of halted 8-mer transcription on fork junction (FJ) and mismatched fork junction (FJ-MM) DNA, comparing holoenzyme and core enzyme. In all cases, productive transcription was initiated with pApU (as noted) and labeling was with [α-32P]ATP.
This design of the fork junction allows us to carry out de novo initiation of transcription in the presence and absence of σ. The results presented in Fig. 4 show that initiation by holoenzyme (holo) produces normal levels of abortive and full-length products, despite the complete absence of DNA upstream of position −4 on the template strand and the mismatching of bases from positions −4 through −1. Thus, all of the traditional elements of scrunching and upstream bubble collapse are lacking, yet abortives are produced in the presence of σ.
Absence of σ Stabilizes the Initially Transcribing Complex
Although the holoenzyme aborts and turns over normally on the fork junction construct, the results presented in Fig. 4B show that initiation by the core enzyme (completely lacking σ) leads to a much more stably halted complex at position +8. That the low amount of accumulating 8-mer is due to increased stability of the complex, rather than poor initiation, is confirmed by the fact that transcription to position +25 (elongation phase) as shown in Fig. 4C is similar in both the core and holoenzymes.
Critically, the absence of σ in the core complex also leads to a drastic reduction in abortive products longer than 4–5 bases (Fig. 4, B and D). This result confirms the role of σ in abortively destabilizing RNA in initially transcribing complexes beyond 4–5 bases (19, 46). It has been noted that very short hybrids (less than 4–5 bp here) would be expected to be intrinsically unstable and to push less against the protein; thus, it is expected that the absence of σ might not significantly reduce very short abortives (18). The details of this balance may be polymerase-specific (49).
Our model suggests that backtracking during initial transcription is a result of the pushing back of the σ3.2 linker against the hybrid. In agreement with this model, the results in Fig. 4E reveal a large reduction in GreB cleavage products in transcription complexes halted at position +8 in the absence of σ. The residual amounts of dinucleotides remaining are likely due to induced backtracking of stably halted 8-mer complexes by spontaneous downstream bubble collapse. To test this proposal, we introduced mismatch sequences at the halt site to prevent downstream bubble collapse and its induced backtracking. As predicted, GreB cleavage products from core polymerase on the fork junction are reduced still further. Together, these results argue for a very significant role for the σ subunit (specifically region 3.2) in driving abortive cycling, with a small contribution from downstream bubble collapse.
DISCUSSION
RNA polymerases must establish strong protein-promoter DNA contacts to achieve regulated initiation of transcription, but they must also release those contacts to progress to stable elongation. A wealth of recent structural data provides a framework for understanding initially transcribing RNA polymerases. It is clear that promoter contacts are retained during initial transcription, as downstream DNA is melted and drawn in, and the bubble expands, leading to scrunching of the DNA (16, 17). It is also clear that on promoter release, the upstream edge of the bubble collapses to return the elongation complex bubble to a smaller size. It is logical to assume that accumulated energetic stress is used to couple the energy of phosphoryl transfer to driving timed promoter release.
In principle, this stress can come from several different sources. First, the progressive increase in size of the melted bubble surely requires energy. Second, the drawn in template and nontemplate strands may undergo steric crowding or distortion. Third, the growing hybrid is blocked by an element of the protein and must “push” that element aside (21, 24). Ascertaining the relative roles of these processes in initial transcription requires direct tests, by the introduction of controlled perturbations into the system. In this study, we extend our earlier studies in the T7 RNA polymerase single subunit model system to the structurally unrelated multisubunit RNA polymerases, specifically, to the multisubunit model E. coli RNA polymerase.
Role of Bubble Collapse
We have previously presented evidence that in T7 RNA polymerase, reannealing of the DNA bubble at its downstream end contributes to instability of halted complexes but does not drive the release of abortive products during normal transcription (7, 33). The data presented in Fig. 1 demonstrate that in the multisubunit E. coli RNA polymerase, removing this driving force by the introduction of mismatches similarly increases the stability of halted complexes but does not systematically reduce abortive probabilities during readthrough transcription. The results also argue that collapse from the upstream end of the bubble similarly does not drive abortive cycling. The results from mismatched DNA constructs are supported by the observation in Fig. 2A that runoff transcription on partially single-stranded DNA, which as noted above may yield a hybrid-gapped construct, similarly shows no systematic reduction in abortive cycling. Finally, in transcription from downstream fork junction constructs, shown in Fig. 4B, the lack of complementary upstream DNA precludes any contribution from upstream bubble collapse, yet abortive products persist.
Halted 8-mer transcription in the presence of GreB shows accumulation of dinucleotides, as expected from backtracked complexes. These dinucleotides are produced primarily from backtracked 8-mer, as ascertained by differential labeling ([α-32P]UTP and [α-32P]ATP) experiments, shown in Fig. 1B. Observation of cleaved dinucleotides from dsDNA complexes, as well as from complexes with mismatched DNA, shows that downstream bubble collapse is not the major contributing factor in backtracking of abortive complexes. The results presented in Fig. 4E suggest only a minor role for downstream bubble collapse in driving backtracking.
Neither Template Nor Nontemplate DNA Steric Scrunching Drives Abortive Cycling
In the initial open complex, the transcription bubble extends from position −11 to position +3, and the nontemplate strand of the upstream end of the bubble interacts with the σ2.4 domain. This strong interaction, along with other structural features of the core enzyme (19–21), keeps the upstream end of the bubble in position during initial promoter-bound transcription (collapse of the upstream end of the bubble on translocation beyond about position +10 facilitates the proper displacement of the 5′ end of the RNA into the RNA exit channel (50, 51)). Hence, as the RNA grows from two nucleotides to eight nucleotides long, the transcription bubble now extends from position −11 to +8. It has been argued that this causes the DNA strands to be compacted (“scrunched”) in the active site channel, another potential source of mechanistically relevant strain (16, 17).
The results of Fig. 2A argue against a role for nontemplate strand scrunching in abortive instability, because complete removal of large regions of the nontemplate strand do not decrease abortive cycling of RNAs up to 8 bases in length. To probe the role of single strand sterics in a more native environment, restoring the potential for downstream collapse, we designed DNA constructs that relieve steric stress either by nicking or gapping the nontemplate strand in the middle of the transcription bubble (the potential for downstream bubble collapse remains, although it could be altered by the increased strand entropy). In this case, the nicks and gaps have no effect on the stability of halted initiation complexes, as shown in Fig. 2C. Thus, the increased stability of halted complexes on partially single-stranded DNA can be assigned to the removal of downstream bubble collapse. Steric constraints within the nontemplate strand play no role in complex instability.
Introducing a nick or a gap in the nontemplate strand also shows backtracking similar to dsDNA, arguing against DNA compaction as a contributing factor toward abortive backtracking. Halted 8-mer transcription in the presence of GreB shows accumulation of pUpU (from backtracked 8-mer RNA) and pUpA (from 6-mer RNA).
Finally, the fork junction constructs described in Fig. 4A completely remove steric constraints in the template strand (binding of the nontemplate strand to the σ2 region might allow retention of some steric constraints in the nontemplate strand). Nevertheless, holoenzyme continues to show significant abortive cycling. Together, the experiments with nicked and gapped DNA and with fork junctions (lacking the template strand) argue strongly against the steric scrunching of individual strands as a major contribution to abortive cycling.
Hybrid Push Drives Backtracking and Abortive Cycling
Crystal structures have revealed that during initial transcription, the σ3.2 linker domain poses a steric obstruction to the nascent RNA-DNA hybrid (19–21). Point mutations (P504L and S506F) in the σ3.2 linker dramatically reduce the amounts of abortive transcripts from a variety of promoters (18, 46, 52), as does a more dramatic C-terminal truncation (20). It has been proposed that the linker serves as a nucleic acid mimic, occupying the RNA exit channel until it is displaced by nascent RNA (19–21). Because it is directly connected to other regions of σ, particularly to region 2, which interacts with the promoter −10 element, displacement of the linker could lead directly to loss of promoter contacts, triggering promoter release (21, 53). To the extent that hybrid growth is driving an unfavorable process (promoter release), that process then necessarily presents a barrier to hybrid growth, destabilizing the RNA in initially transcribing complexes.
The current results show that a single point mutation (S506F) in the σ subunit not only leads to reduced abortives, as previously reported, but also to reduced GreB cleavage products, as presented in Fig. 3, A–C. A reduction in cleavage products indicates less backtracked RNA accessible to GreB cleavage. Transcription profiles with core RNA polymerase on fork junction DNA, as shown in Fig. 4, confirm that the pushing of the σ linker on the nascent RNA-DNA hybrid makes the enzyme backtrack, such that the 3′ end of the RNA is accessible to GreB cleavage. Removal of this push-back (as in the core polymerase, lacking σ) along with removal of downstream bubble collapse (by introduction of a targeted mismatch in DNA) dramatically reduces the accessibility of RNA to GreB cleavage, as revealed in Fig. 4E.
The schematic shown in Fig. 5 summarizes the findings of this study. Upstream bubble collapse and compaction/scrunching of either the single-stranded template or nontemplate strands are not major driving forces for observed instability (abortive cycling) of initially transcribing complexes. Collapse of the bubble from the downstream end of the bubble contributes significantly to instability of halted complexes, but much less so to transcribing complexes, and so it is at most a minor contributor to abortive cycling. By contrast, pushing of the hybrid against a mobile protein element (σ3.2 in the bacterial enzyme) results in substantially increased abortive instability and is the primary energetic contributor to abortive cycling.
FIGURE 5.

Schematic representation of a transcription bubble showing the energetics involved in an initially transcribing complex. Upstream bubble collapse and DNA compaction play no role, although downstream bubble collapse plays a minor role and hybrid-push a major role in abortive cycling.
Mechanism for all RNA Polymerases
The B-finger element in the TFIIB subunit of eukaryotic RNA polymerases, as well as TFB of archaeal RNA polymerase, is thought to play a similar role as the σ3.2 linker in bacterial RNA polymerase (49, 54, 55). The single subunit RNA polymerase family (including T7 RNA polymerase and mitochondrial RNA polymerases) has no structural homology to the multisubunit enzymes. Nevertheless, the rotating N-terminal platform in these enzymes appears to play a role identical in concept (but very different in detail) to the σ3.2 linker, with the same consequence on abortive cycling.
In this general model, hybrid growth drives protein movement, and that movement ultimately drives promoter release. Mutations that lower the initial barrier to protein movement/displacement necessarily stabilize the initially transcribing complexes against abortive dissociation of the nascent RNA. We have recently demonstrated exactly this effect in the T7 RNA polymerase point mutant P266L and have additionally shown that this mutant is delayed (with respect to position/hybrid length) in promoter release (25). Thus, the model in which a protein element pushes back against the growing RNA-DNA hybrid, leading to a direct destabilization of the ternary complex, appears to be applicable to all RNA polymerases.
Do single subunit RNA polymerases backtrack? From the considerations above, a driving force clearly exists during initial transcription. Although it has never been established that the single subunit T7 RNA polymerase backtracks (there are no known Gre factors to serve as probes), there is the possibility of RNA extruding into the channel through which NTPs enter the active site, just as in the multisubunit enzymes.
Similarities extend beyond simply abortive cycling. In both the bacterial and eukaryotic RNA polymerases, the RNA exit channel exists in the preinitiation complex, but it is not empty. The B-reader/finger of TFIIB and TFB2 (general transcription factors necessary to recruit RNA polymerase II and archaeal RNA polymerase, respectively, to promoter DNA) occupy the RNA exit channel (49, 54, 55). Similarly, the σ3.2 domain occupies the RNA exit channel in bacterial RNA polymerases (19, 20). As noted above, these elements are displaced by the nascent RNA. Interestingly, in eukaryotic polymerase II, recent structural data suggest the hybrid does not experience significant clash until it reaches a much more stable length of about 12–13 bp, consistent with substantially lower abortives in that system (56).
In the single subunit RNA polymerases, the basic story for the transition to elongation is the same, although the details are very different. In these RNA polymerases, the RNA exit channel literally does not exist prior to the large conformational change in the protein that follows promoter release. Thus, the RNA exit channel only forms after (or coincident with) dissociation of the 5′ end of the RNA from the hybrid. In all systems, nature has avoided having an “empty” RNA exit channel during initial transcription.
In summary, it appears that in all promoter-specific DNA-dependent RNA polymerases, nature has coupled the only energy source, nucleotide addition, to driving the timed disruption of promoter contacts. This study provides strong support for a model in which the hybrid is used as a mechanical “rod” to drive disruption of protein-protein and protein-DNA interactions to weaken promoter binding. However, a logical consequence of that use is that the hybrid rod can itself be destabilized either directly and/or by the shortening of the hybrid that correlates with induced backtracking. Thus, abortive cycling is a logical outcome of the need to couple the energy of nucleotide addition to driving the timed release of initial promoter contacts.
Acknowledgment
We thank Lilian Hsu for the gifts of GreB and the S506F mutant of σ70, for many insightful discussions, and for critical readings of the manuscript.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grant 1RO1 GM55002.
REFERENCES
- 1. Carpousis A. J., Gralla J. D. (1980) Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry 19, 3245–3253 [DOI] [PubMed] [Google Scholar]
- 2. Gralla J. D., Carpousis A. J., Stefano J. E. (1980) Productive and abortive initiation of transcription in vitro at the lac UV5 promoter. Biochemistry 19, 5864–5869 [DOI] [PubMed] [Google Scholar]
- 3. Martin C. T., Muller D. K., Coleman J. E. (1988) Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry 27, 3966–3974 [DOI] [PubMed] [Google Scholar]
- 4. Jin D. J., Turnbough C. L., Jr. (1994) An Escherichia coli RNA polymerase defective in transcription due to its overproduction of abortive initiation products. J. Mol. Biol. 236, 72–80 [DOI] [PubMed] [Google Scholar]
- 5. Straney D. C., Crothers D. M. (1987) A stressed intermediate in the formation of stably initiated RNA chains at the Escherichia coli lac UV5 promoter. J. Mol. Biol. 193, 267–278 [DOI] [PubMed] [Google Scholar]
- 6. Kapanidis A. N., Margeat E., Ho S. O., Kortkhonjia E., Weiss S., Ebright R. H. (2006) Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science 314, 1144–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vahia A. V., Martin C. T. (2011) Direct tests of the energetic basis of abortive cycling in transcription. Biochemistry 50, 7015–7022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naryshkin N., Revyakin A., Kim Y., Mekler V., Ebright R. H. (2000) Structural organization of the RNA polymerase-promoter open complex. Cell 101, 601–611 [DOI] [PubMed] [Google Scholar]
- 9. deHaseth P. L., Zupancic M. L., Record M. T., Jr. (1998) RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 180, 3019–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holstege F. C., Fiedler U., Timmers H. T. (1997) Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16, 7468–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ujvári A., Martin C. T. (1996) Thermodynamic and kinetic measurements of promoter binding by T7 RNA polymerase. Biochemistry 35, 14574–14582 [DOI] [PubMed] [Google Scholar]
- 12. Martin C. T., Esposito E. A., Theis K., Gong P. (2005) Structure and function in promoter escape by T7 RNA polymerase. Prog. Nucleic Acid Res. Mol. Biol. 80, 323–347 [DOI] [PubMed] [Google Scholar]
- 13. Cheetham G. M., Jeruzalmi D., Steitz T. A. (1999) Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature 399, 80–83 [DOI] [PubMed] [Google Scholar]
- 14. Cheetham G. M., Steitz T. A. (1999) Structure of a transcribing T7 RNA polymerase initiation complex. Science 286, 2305–2309 [DOI] [PubMed] [Google Scholar]
- 15. Kireeva M. L., Komissarova N., Kashlev M. (2000) Overextended RNA:DNA hybrid as a negative regulator of RNA polymerase II processivity. J. Mol. Biol. 299, 325–335 [DOI] [PubMed] [Google Scholar]
- 16. Cheetham G. M., Jeruzalmi D., Steitz T. A. (1998) Transcription regulation, initiation, and “DNA scrunching” by T7 RNA polymerase. Cold Spring Harbor Symp. Quant. Biol. 63, 263–267 [DOI] [PubMed] [Google Scholar]
- 17. Revyakin A., Liu C., Ebright R. H., Strick T. R. (2006) Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science 314, 1139–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hernandez V. J., Hsu L. M., Cashel M. (1996) Conserved region 3 of Escherichia coli final σ70 is implicated in the process of abortive transcription. J. Biol. Chem. 271, 18775–18779 [DOI] [PubMed] [Google Scholar]
- 19. Murakami K. S., Masuda S., Darst S. A. (2002) Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296, 1280–1284 [DOI] [PubMed] [Google Scholar]
- 20. Vassylyev D. G., Sekine S., Laptenko O., Lee J., Vassylyeva M. N., Borukhov S., Yokoyama S. (2002) Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417, 712–719 [DOI] [PubMed] [Google Scholar]
- 21. Murakami K. S., Masuda S., Campbell E. A., Muzzin O., Darst S. A. (2002) Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science 296, 1285–1290 [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y., Feng Y., Chatterjee S., Tuske S., Ho M. X., Arnold E., Ebright R. H. (2012) Structural basis of transcription initiation. Science 338, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu X., Bushnell D. A., Silva D. A., Huang X., Kornberg R. D. (2011) Initiation complex structure and promoter proofreading. Science 333, 633–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Durniak K. J., Bailey S., Steitz T. A. (2008) The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science 322, 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramírez-Tapia L. E., Martin C. T. (2012) New insights into the mechanism of initial transcription: the T7 RNA polymerase mutant P266L transitions to elongation at longer RNA lengths than wild type. J. Biol. Chem. 287, 37352–37361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kammerer W., Deuschle U., Gentz R., Bujard H. (1986) Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J. 5, 2995–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hsu L. M., Vo N. V., Kane C. M., Chamberlin M. J. (2003) In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 1. RNA chain initiation, abortive initiation, and promoter escape at three bacteriophage promoters. Biochemistry 42, 3777–3786 [DOI] [PubMed] [Google Scholar]
- 28. Chander M., Austin K. M., Aye-Han N. N., Sircar P., Hsu L. M. (2007) An alternate mechanism of abortive release marked by the formation of very long abortive transcripts. Biochemistry 46, 12687–12699 [DOI] [PubMed] [Google Scholar]
- 29. Hsu L. M., Vo N. V., Chamberlin M. J. (1995) Escherichia coli transcript cleavage factors GreA and GreB stimulate promoter escape and gene expression in vivo and in vitro. Proc. Natl. Acad. Sci. U.S.A. 92, 11588–11592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Opalka N., Chlenov M., Chacon P., Rice W. J., Wriggers W., Darst S. A. (2003) Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114, 335–345 [DOI] [PubMed] [Google Scholar]
- 31. Deuschle U., Kammerer W., Gentz R., Bujard H. (1986) Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures. EMBO J. 5, 2987–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Minkley E. G., Pribnow D. (1973) Transcription of the early region of bacteriophage T7: selective initiation with dinucleotides. J. Mol. Biol. 77, 255–277 [DOI] [PubMed] [Google Scholar]
- 33. Gong P., Martin C. T. (2006) Mechanism of instability in abortive cycling by T7 RNA polymerase. J. Biol. Chem. 281, 23533–23544 [DOI] [PubMed] [Google Scholar]
- 34. Borukhov S., Sagitov V., Goldfarb A. (1993) Transcript cleavage factors from E. coli. Cell 72, 459–466 [DOI] [PubMed] [Google Scholar]
- 35. Li X. Y., McClure W. R. (1998) Stimulation of open complex formation by nicks and apurinic sites suggests a role for nucleation of DNA melting in Escherichia coli promoter function. J. Biol. Chem. 273, 23558–23566 [DOI] [PubMed] [Google Scholar]
- 36. Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maslak M., Martin C. T. (1993) Kinetic analysis of T7 RNA polymerase transcription initiation from promoters containing single-stranded regions. Biochemistry 32, 4281–4285 [DOI] [PubMed] [Google Scholar]
- 38. Mekler V., Minakhin L., Severinov K. (2011) A critical role of downstream RNA polymerase-promoter interactions in the formation of initiation complex. J. Biol. Chem. 286, 22600–22608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hsu L. M., Cobb I. M., Ozmore J. R., Khoo M., Nahm G., Xia L., Bao Y., Ahn C. (2006) Initial transcribed sequence mutations specifically affect promoter escape properties. Biochemistry 45, 8841–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsu L. M. (2009) Monitoring abortive initiation. Methods 47, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Komissarova N., Kashlev M. (1997) RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 272, 15329–15338 [DOI] [PubMed] [Google Scholar]
- 42. Nudler E., Mustaev A., Lukhtanov E., Goldfarb A. (1997) The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89, 33–41 [DOI] [PubMed] [Google Scholar]
- 43. Palangat M., Landick R. (2001) Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J. Mol. Biol. 311, 265–282 [DOI] [PubMed] [Google Scholar]
- 44. Kulbachinskiy A., Mustaev A. (2006) Region 3.2 of the σ subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 281, 18273–18276 [DOI] [PubMed] [Google Scholar]
- 45. Mekler V., Kortkhonjia E., Mukhopadhyay J., Knight J., Revyakin A., Kapanidis A. N., Niu W., Ebright Y. W., Levy R., Ebright R. H. (2002) Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell 108, 599–614 [DOI] [PubMed] [Google Scholar]
- 46. Cashel M., Hsu L. M., Hernandez V. J. (2003) Changes in conserved region 3 of Escherichia coli σ70 reduce abortive transcription and enhance promoter escape. J. Biol. Chem. 278, 5539–5547 [DOI] [PubMed] [Google Scholar]
- 47. Guo Y., Gralla J. D. (1998) Promoter opening via a DNA fork junction binding activity. Proc. Natl. Acad. Sci. U.S.A. 95, 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mekler V., Pavlova O., Severinov K. (2011) Interaction of Escherichia coli RNA polymerase σ70 subunit with promoter elements in the context of free σ70, RNA polymerase holoenzyme, and the β′-σ70 complex. J. Biol. Chem. 286, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Santangelo T. J., Cubonová L., James C. L., Reeve J. N. (2007) TFB1 or TFB2 is sufficient for Thermococcus kodakaraensis viability and for basal transcription in vitro. J. Mol. Biol. 367, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Toulokhonov I., Landick R. (2006) The role of the lid element in transcription by E. coli RNA polymerase. J. Mol. Biol. 361, 644–658 [DOI] [PubMed] [Google Scholar]
- 51. Naryshkina T., Kuznedelov K., Severinov K. (2006) The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J. Mol. Biol. 361, 634–643 [DOI] [PubMed] [Google Scholar]
- 52. Sen R., Nagai H., Hernandez V. J., Shimamoto N. (1998) Reduction in abortive transcription from the λPR promoter by mutations in region 3 of the σ70 subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 273, 9872–9877 [DOI] [PubMed] [Google Scholar]
- 53. Marr M. T., Datwyler S. A., Meares C. F., Roberts J. W. (2001) Restructuring of an RNA polymerase holoenzyme elongation complex by lambdoid phage Q proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 8972–8978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pal M., Ponticelli A. S., Luse D. S. (2005) The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell 19, 101–110 [DOI] [PubMed] [Google Scholar]
- 55. Westover K. D., Bushnell D. A., Kornberg R. D. (2004) Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303, 1014–1016 [DOI] [PubMed] [Google Scholar]
- 56. Sainsbury S., Niesser J., Cramer P. (2013) Structure and function of the initially transcribing RNA polymerase II-TFIIB complex. Nature 493, 437–440 [DOI] [PubMed] [Google Scholar]