Abstract
Spatiotemporal control of gene expression is central to animal development. Core promoters represent a previously unanticipated regulatory level by interacting with cis-regulatory elements and transcription initiation in different physiological and developmental contexts. Here, we provide a first and comprehensive description of the core promoter repertoire and its dynamic use during the development of a vertebrate embryo. By using cap analysis of gene expression (CAGE), we mapped transcription initiation events at single nucleotide resolution across 12 stages of zebrafish development. These CAGE-based transcriptome maps reveal genome-wide rules of core promoter usage, structure, and dynamics, key to understanding the control of gene regulation during vertebrate ontogeny. They revealed the existence of multiple classes of pervasive intra- and intergenic post-transcriptionally processed RNA products and their developmental dynamics. Among these RNAs, we report splice donor site-associated intronic RNA (sRNA) to be specific to genes of the splicing machinery. For the identification of conserved features, we compared the zebrafish data sets to the first CAGE promoter map of Tetraodon and the existing human CAGE data. We show that a number of features, such as promoter type, newly discovered promoter properties such as a specialized purine-rich initiator motif, as well as sRNAs and the genes in which they are detected, are conserved in mammalian and Tetraodon CAGE-defined promoter maps. The zebrafish developmental promoterome represents a powerful resource for studying developmental gene regulation and revealing promoter features shared across vertebrates.
Precise spatial and temporal control of the transcription of protein-coding and noncoding genes is a fundamental process underlying development and differentiation of multicellular organisms. The core promoter, which is a relatively short stretch of sequence around the transcription start site (TSS), contains regulatory information for the recruitment of general transcription initiation factors (GTFs), necessary for the formation of pre-initiation complex. Recent evidence points at the core promoter as an important determinant of developmental transcription control. This is based on two advances: (1) the discovery of a variety of GTF proteins and complexes that may replace TFIID or its subunits during various stages of development; (2) the recognition of a previously unanticipated diversity of core promoter types and features, which suggests differential core promoter usage by subsets of genes (for reviews, see Müller et al. 2007; Goodrich and Tjian 2010; Juven-Gershon and Kadonaga 2010; Ohler and Wassarman 2010; Lenhard et al. 2012). The diversity of core promoters may reflect alternative integration points for developmental signals and plays a role in differential transcription regulation (D'Alessio et al. 2009; Müller et al. 2010).
Cap analysis of gene expression (CAGE) has given rise to an improved annotation and description of core promoters on a genomic scale (Kodzius et al. 2006), revealing intricate details about TSS usage and dynamics at single nucleotide resolution (Carninci et al. 2006). It has revealed that most promoters lack a TATA-box, which was previously considered as the seeding element for transcription initiation. Despite a number of alternative core promoter motifs (Juven-Gershon et al. 2008), a global code for core promoters is still elusive. Additionally, the organismal and developmental roles of the diversity of core promoters and associated motifs are not yet understood in the complexity of a vertebrate animal. CAGE technology provides the opportunity to classify noncoding RNAs generated by post-transcriptional processing in human and other genomes (Kapranov et al. 2007; Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009; Hoskins et al. 2011). However, the existence and biological relevance of these noncoding RNAs have not yet been demonstrated in vivo.
Despite progress in our understanding of promoters, we lack genome-scale data of core promoter usage and the dynamics of it under changing conditions in a developing vertebrate embryo. The early ontogeny of the zebrafish, like other anamniotes, is characterized by a dramatic transition with global changes in transcriptional activities during the mid-blastula transition (MBT) (Kane and Kimmel 1993; Schier 2007). Before the MBT, a pluripotent cell mass evolves from the fertilized egg without transcriptional activity. The transcriptome at this time reflects the transcription program acting in the oocyte of the mother. During MBT, activation of the zygotic genome occurs in parallel with maternal mRNA degradation (Mathavan et al. 2005), providing the necessary transcriptome changes for specification and determination of cell fates during differentiation. Post-translational modification of histones has been shown to be predictive for core promoter regions (Wardle et al. 2006) and has been suggested to play a role in promoter regulation in anamniote development (Akkers et al. 2009; Vastenhouw et al. 2010; Lindeman et al. 2011). Accurate promoter prediction based on mapping of TSSs during development is needed to decipher the complex interplay between DNA sequence determinants for transcription initiation and epigenetic regulation on core promoters. The lack of precise TSS data so far has restricted the study of developmental regulatory mechanisms of transcription initiation in vertebrates due to the unreliable TSS position detection based on cDNA/EST and RNA-seq data and scarcity of available data sets.
Here we have set out to generate the first global description of TSS usage during key stages of vertebrate embryonic development at single nucleotide resolution. We have coupled CAGE maps to protein-coding and noncoding transcripts by RNA sequencing and to post-translational histone modifications associated with promoters (H3K4me3) by ChIP sequencing. These data sets provide a quantitative description of TSS usage on a genome scale. We have chosen critical phases of vertebrate ontogeny, including the maternal to zygotic transition at MBT and the subsequent stages of differentiation leading to formation of the body plan and organ systems. We reveal an extraordinary dynamic in promoter usage that takes place during development of the vertebrate embryo. We show that the onset of transcription and subsequent differentiation is characterized by the developmentally regulated appearance of gen(om)e-wide capped 5′-ends of RNAs in many genes, and describe an entire hitherto unknown layer of RNA species overlapping known genes with specific signatures occurring in exons, introns and 3′-UTRs of specific sets of developmentally active genes. We uncover evolutionarily conserved features of core promoters, which include a novel vertebrate specific initiator sequence shared by a subset of membrane/transport-associated genes in human, showing that our zebrafish data set has the potential to reveal promoter features shared by all vertebrates.
Results
Genome-wide detection of 5′-ends of capped transcripts during zebrafish ontogeny
In order to map promoter usage during vertebrate development, we identified TSSs by CAGE analysis of zebrafish RNA samples collected from 12 developmental stages. A total of over 83 million reads were generated by Illumina sequencing, resulting in 3.7–8.2 million reads mapped to the zebrafish genome per stage (Supplemental Table 1). The CAGE signal mapped to the genome revealed the detailed developmental dynamics of individual core promoter usage. As an illustrative example, Figure 1A–C shows a genome browser view of the promoter-associated data sets for the ncalda gene. H3K4me3 histone modification marks analyzed by ChIP-seq offer further support for the promoter regions (Fig. 1A).
Figure 1.
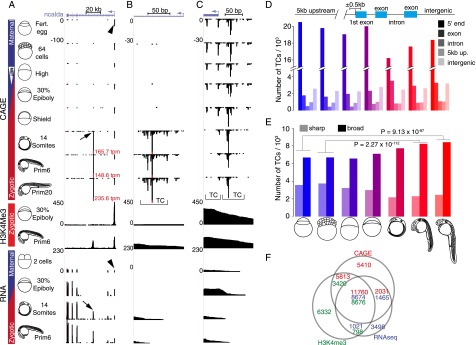
Mapping of transcription initiation in zebrafish embryo development. (A–C) Genome browser view of ncalda gene with CAGE-seq, ChIP-seq, and RNA-seq tracks from selected developmental stages. Schematic representation of developmental stages is on the left. Vertical bar with blue (maternal) and red (zygotic) bars indicates transcriptional activity of the genome. White arrowhead indicates the onset of zygotic transcription at the mid-blastula transition (MBT). Vertical scales on the left of tracks are tpm values and fixed within experiments. Height of the CTSS bars is proportional to the number of CAGE tags aligned to that position. Transcript clusters (TC) of varying width are labeled with brackets. (A) Full-length transcripts of ncalda indicating two promoter regions (arrow and arrowhead) were detected by CAGE and verified by H3K4me3 peaks and RNA-seq data. (Fert) Fertilized. (B) High-resolution mapping of zygotically active novel alternative TSS (arrow in A) of ncalda gene. (C) High-resolution mapping of continuously active Ensembl annotated TSS (arrowhead in A) of ncalda gene. (D, top) Schematic of gene structures for analysis of distribution of TCs. (Bottom) Number of TCs overlapping with the annotated segments of the genome is shown at the developmental stages indicated by schematics. Colors from blue to red indicate transition from maternal to zygotic transcriptomes. (E) Distribution of sharp and broad TCs at selected developmental stages. Shades of color indicate gene segment, blue to red transition indicates maternal to zygotic transition of transcriptome. P-values of one-tailed Fisher's exact test for selected comparisons are denoted above the bars. (F) Intersection of Ensembl gene 5′-ends detected by CAGE (>1 tpm, shown in red), RNA-seq (>1 rpkm, shown in blue), and H3K4me3 peaks (in green) at prim-6 stage.
The frequencies, at which individual nucleotide positions were mapped by 5′-end CAGE, were measured as tag per million (tpm) and called CAGE transcript start sites (CTSSs) (Carninci et al. 2006). The CTSS distribution revealed consistent and reproducible patterns over neighboring stages (Fig. 1B,C). The CTSS positions with adjacent CTSS falling within 20 bp were clustered into transcript clusters (TCs) of varying width (brackets in Fig. 1B,C), which aid in determining the TSS distribution within a promoter (Carninci et al. 2006). The developmental dynamics indicate maternal- and zygotic-specific changes in TC positions during ontogeny. TCs in early (pre-MBT) stages indicate maternal gene products inherited from the oocyte (arrowhead in Fig. 1A). A previously unannotated major TC downstream from the annotated promoter is up-regulated after the shield stage, indicating a zygote-specific alternative promoter also confirmed by RNA-seq (arrows in Fig. 1A). The reproducibility of CAGE, both in terms of extent and complexity of initiation site usage, was verified by biological replicates of the prim-6 stage (Supplemental Fig. 1A,B). Furthermore, to account for the number of CTSSs and TCs in a manner robust to sequencing depth across samples, we determined their number at different thresholds (Supplemental Fig. 1C; Supplemental Table 2). The change in the number of CTSSs and TCs that account for varying percentage of total CAGE tags was consistent between adjacent stages (Supplemental Fig. 1D,E), and showed that a relatively small proportion of CTSSs accounts for the majority of the signal and suggests a low level of noise. The quantitative nature of the CAGE-based prediction of promoter activity is demonstrated via the correlation between the developmental dynamics of promoter usage and temporal dynamics of gene activity measured by RNA-seq. (Supplemental Fig. 1F; Supplemental Table 3).
To investigate the properties of CAGE tags associated with different parts of annotated genes, we segmented Ensembl gene transcripts (Flicek et al. 2013) and their genomic vicinity into gene-associated regions (Fig. 1D). The overall majority of CAGE tags were detected at gene 5′-ends within 500 bp from the annotated start sites (Fig. 1D). The width of zebrafish TC mapping to the vicinity of known gene 5′-ends followed the mammalian dichotomy of sharp (or focused) and broad (or diffuse) promoters (Fig. 1E; Supplemental Fig. 2A,B; Lenhard et al. 2012). There was a significant decrease in the number of sharp TCs from the maternal to zygotic stages while the usage of broad TCs increased after the start of zygotic genome activation and peaked at organogenesis (Fig. 1E). The distribution of the number of TCs within single promoter regions (within 500 bp of 5′-ends of Ensembl and novel RNA-seq transcripts) revealed widespread usage of multiple TCs particularly prevalent in maternal stages, followed by noticeable reduction during zygotic stages (Supplemental Fig. 2C).
CAGE, RNA-seq, and H3K4me3 ChIP-seq reliably detect promoters of coding and noncoding RNAs during development
To estimate the extent of the current RefSeq and Ensembl transcript models (including non-embryonic transcripts) (Howe et al. 2013) detected by CAGE, we compared TCs around 5′-ends of genes. At stringent threshold (≥1 tpm), ∼70% and 52% of gene models (RefSeq and Ensembl, respectively) are detected during early embryonic stages (Supplemental Table 4). At lower TC thresholds (>0.5 tpm), the coverage of Ensembl transcripts is similar to that detected by RNA-seq (Pauli et al. 2012). To demonstrate the concordance of CAGE TCs, RNA-seq 5′-ends, and H3K4me3 peaks (Supplemental Table 5), we calculated their intersection at the prim-6 stage (Fig. 1F). Taken together, CAGE-seq supports the bulk of transcripts detectable in embryogenesis, improves precision of gene 5′-end detection at nucleotide resolution (Fig. 1; Supplemental Fig. 3A), and provides direct insight into the developmental dynamics of TSSs usage.
We detected 926 out of the 1133 lncRNAs reported (Pauli et al. 2012) of which 625 showed evidence for transcription in at least two or more consecutive stages. In addition, CAGE analysis revealed transcriptional initiation events corresponding to numerous previously unannotated noncoding transcripts, including antisense noncoding RNA products (Supplemental Fig. 3D). In a number of cases, CAGE proved to be more sensitive than RNA-seq, robustly detecting 459 novel promoters of intergenic transcripts without RNA-seq evidence (Supplemental Table 6), of which 327 were supported by H3K4me3 enrichment, suggesting the detection of promoters of novel genes (Supplemental Fig. 3B,C).
Identification and developmental dynamics of alternative promoters
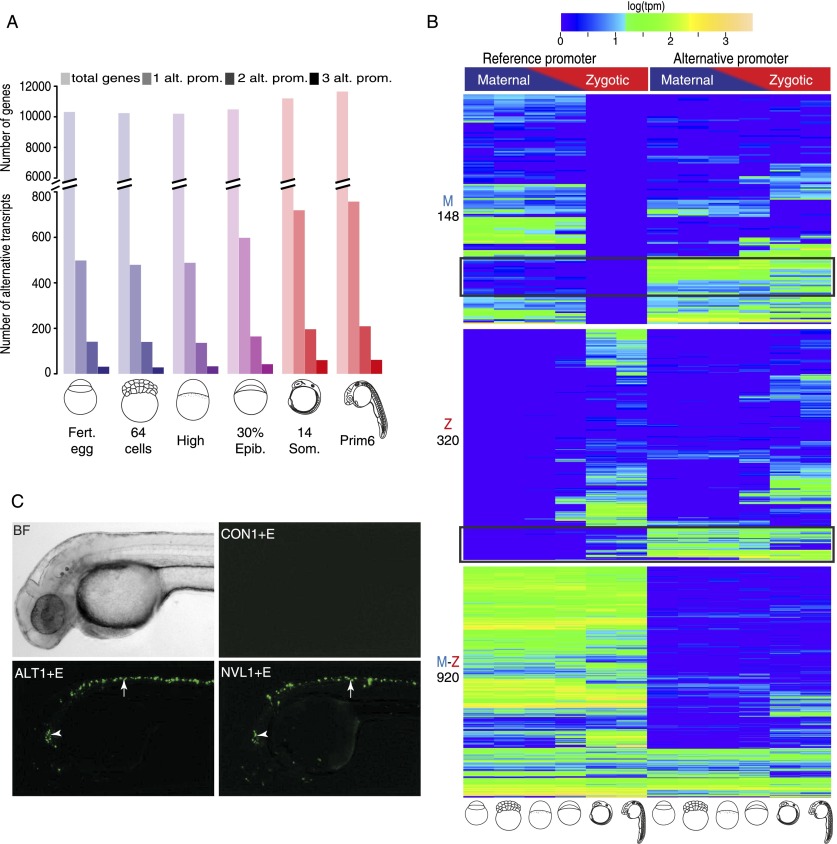
Vertebrate genes often possess multiple promoters, which result in distinct transcripts with potentially different function (Davuluri et al. 2008). To identify alternative promoters of genes, we isolated Ensembl and RNA-seq transcripts (Pauli et al. 2012) with different 5′-ends (see Methods) and compared them with CAGE-seq supported gene 5′-ends. We identified 1612 genes with at least one alternative promoter (Fig. 2A; Supplemental Table 7) of which 586 promoters were novel. The dynamics of alternative promoter usage during development was quantified by measuring tpm values of TCs (Fig. 2A,B). This analysis indicates complex and often independent regulatory patterns between alternative promoters during early development (Fig. 2B). Notably, we identified a set of genes, where the promoter regions discovered by CAGE are preferentially used over the previously annotated gene 5′-ends (black rectangles in Fig 2B). Alternative promoters carry both broad and sharp TCs independently (Supplemental Fig. 4A).
Figure 2.
Identification and developmental dynamics of alternative initiation sites. (A) Frequency and developmental dynamics of alternative promoters. Colors reflect maternal to zygotic transition as in Figure 1. Genes with up to three alternative promoters are plotted (see Supplemental Table 7 for details). Shades indicate alternative promoter numbers, color transition indicates maternal to zygotic transition of transcriptome. (B) Clustering of three sets of genes based on their reference promoter activity. Annotated reference promoters (as assigned by Ensembl 71) are on the left and alternative promoters on the right. Genes are clustered in three groups according to the reference promoter being active during maternal (M), zygotic (Z), or maternal and zygotic (M-Z) stages. Total number of genes in each group is indicated in the left. Black rectangles indicate genes where the previously unannotated alternative promoter's activity is preferential over that of the annotated reference promoter. (C) Fluorescent Venus reporter activity driven by alternative (ALT1) and novel (NVL1) core promoters attached to a neural specific enhancer (E) in transgenic embryos. Control (CON) indicates a random DNA fragment replacing a promoter. Maximum projections of embryos overlaid from a single injection experiment are shown (see details in Supplemental Table 8). Bright-field (BF) image of a single zebrafish embryo is shown for reference. (Arrowhead) Cerebellum; (arrow) spinal cord activity.
To validate the predicted promoter function of TSS regions detected by CAGE, we tested the transcription initiating potential of a set of alternative promoters and a set of previously unannotated 5′-ends of genes by transgenic reporter assays in embryos. We cloned five alternative promoters and five novel promoter regions in reporter construct (Supplemental Table 8). Nine out of 10 predicted core promoters activated neural-specific reporter expression in transient transgenic zebrafish, when linked to the heterologous neural specific islet1 zCREST2 enhancer (Uemura et al. 2005; Gehrig et al. 2009), while control fragments from random loci remained silent in this assay (Fig. 2C; Supplemental Fig. 4B,C). These results support CAGE as a detection tool for functional promoters in zebrafish.
Evolutionary conservation of sequence characteristics suggest functional components of promoters
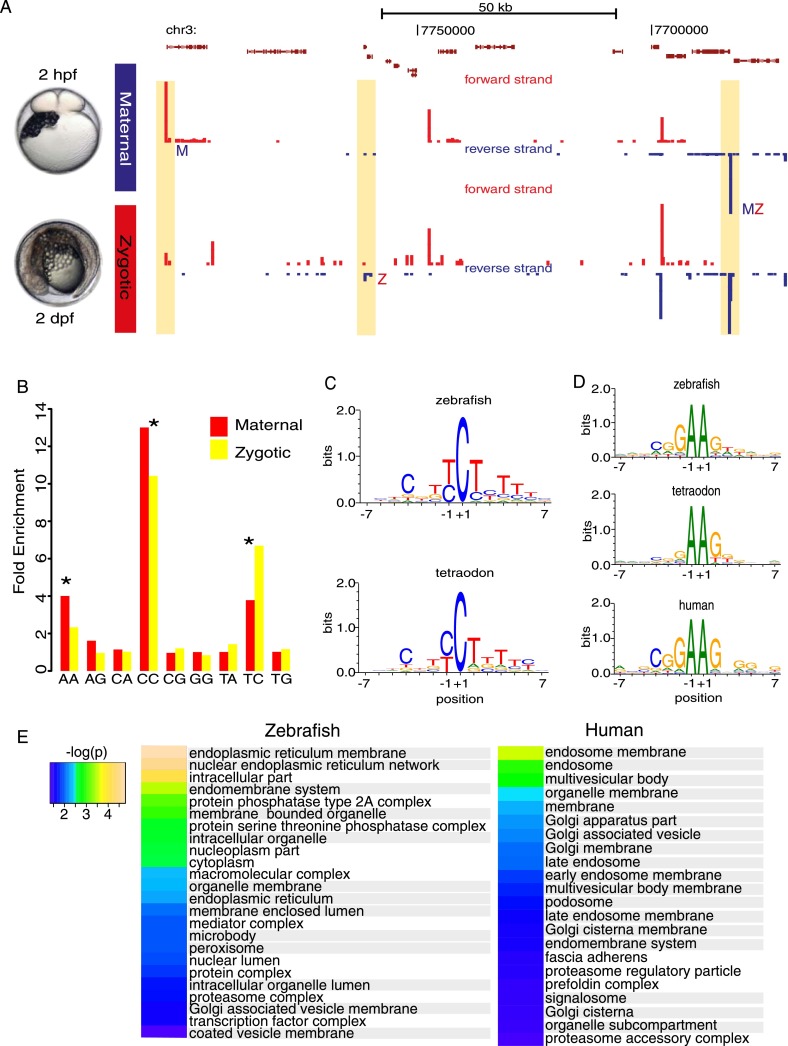
Most vertebrate core promoters are characterized by the relative scarcity of promoter motifs (Lenhard et al. 2012). To detect core promoter-associated sequences and to improve the prediction of functional core promoter features, we used a comparative genomic strategy. We sought evolutionarily conserved features of promoters by comparing zebrafish data to that of the small genome species Tetraodon nigroviridis (spotted green pufferfish) (Jaillon et al. 2004). We bred Tetraodon in the laboratory (Watson et al. 2009; A Zaucker, T Bodur, J Gehrig, Y Hadzhiev, F Loosli, H Roest Crollius, C Watson, F Müller, in prep.) and carried out CAGE analysis of its promoterome. Example of the Tetraodon CAGE-seq data with overview of the results is shown in Figure 3A and Supplemental Table 1.
Figure 3.
Sequence characteristics of developmentally regulated core promoters are evolutionarily conserved. (A) A genome browser view with annotated Tetraodon genes (top of the panel) along with CAGE-seq tracks from two developmental (maternal and zygotic) stages. In yellow boxes, core promoter regions of annotated genes expressed specifically at maternal (M), zygotic (Z), or maternal and zygotic stages (MZ). CTSSs in red and blue indicate sense and antisense direction, respectively. (B) Correlation of dinucleotides of CTSSs (−1,+1) between zebrafish and Tetraodon orthologs represented as fold enrichments vs. expected by chance. Asterisks denote significant correlations (P ≤ 0.05). Only dinucleotides, which occur at TSSs, are shown. (C,D) Sequence logos and their information content of initiator motifs for selected dinucleotides: (C) CC/TC and (D) AA dinucleotides. Human genes with “AA” initiation motifs were plotted from ENCODE cell lines (see Methods). (E) Enriched GO terms of genes with AA initiator. Identical terms are highlighted in gray. Heat map represents the –log (P-values) of enriched GO terms.
We exploited the high resolution of TSSs detection by CAGE-seq and searched for known TSS-associated motifs (TATA-box, GC-box, CAAT-box, Inr, DPE, DRE, MTE), (Juven-Gershon et al. 2008) and novel core promoter motifs by k-mer enrichment analysis (Frith et al. 2008). However, no constrained motifs were found enriched at specific positions in relation to TSSs by scoring matrices from JASPAR (Portales-Casamar et al. 2010), except for SP1 and TATA-box motifs (data not shown). Both motifs were significantly enriched in Tetraodon orthologs of zebrafish genes, suggesting evolutionarily conserved mechanisms for gene-specific transcription initiation among teleosts.
Next, we searched for evolutionarily conserved transcription initiation sites using dinucleotide frequency analysis in all possible combinations at the −1,+1 nucleotides of TSSs in pairs of zebrafish and Tetraodon orthologs. The enrichment analysis revealed three sets of conserved dinucleotides, suggesting biological significance (Fig. 3B). Two dinucleotides, CC and TC, are part of a broader TC/CT motif (Fig. 3C), similar to the TCT initiator, specific to highly expressed protein translation-associated genes (Parry et al. 2010). Indeed, the genes that carry these motifs are also protein translation-associated genes in zebrafish (Supplemental Fig. 5A).
The third example for enriched dinucleotides contains AA at the TSSs, representing a noncanonical initiation signal, which is identified as a part of a novel initiator motif, GAAG (Fig. 3D). The GAAG motif is found in a small subset of genes (557 in zebrafish and 150 in Tetraodon) (Supplemental Table 9); although genes with AA initiator sequence can have an alternative canonical initiator in the promoter region, the AA initiator is dominant (Supplemental Fig. 5B). A search for the motif in the CAGE data from ENCODE cell lines (The ENCODE Project Consortium 2011; Djebali et al. 2012; Harrow et al. 2012) revealed its existence in human (152 genes, Fig. 3D) and suggests that this novel initiator is universal across vertebrates. Gene Ontology (GO) analysis of genes with the GAAG motif revealed an association with vesicles, vesicle transport, and membrane-associated proteins, both in fish and human (Fig. 3E). Thus, the GAAG motif is a novel initiator, used by a small orthologous set of genes suggesting an evolutionarily conserved, noncanonical transcription initiation mechanism during vertebrate development.
Pervasive nonconventional exonic CAGE tags suggest post-transcriptionally processed RNAs
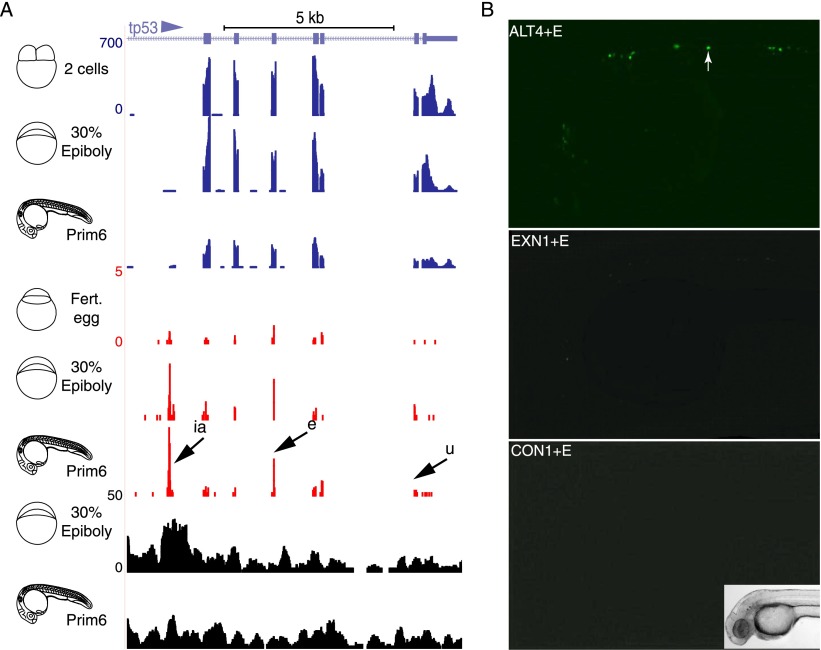
While the large majority of CAGE tags were found at promoters, pervasive CAGE signals were also detected in introns, exons and 3′-UTR sequences (Fig. 1D; Supplemental Fig. 2C), similar to that shown in mammalian cells (Carninci et al. 2006). CAGE tags revealed the developmentally regulated production of RNAs at intragenic sites, including coding exons and 3′-UTR sequences (Fig. 4A; Supplemental Tables 10, 11). Previously, exonic RNAs were suggested to be of post-transcriptional origin (Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009). To test this, CAGE tags, which did not map to the genome, were aligned to zebrafish cDNA. Up to 15% of the unmapped CAGE tags aligned to available cDNAs spanning exon–exon junctions, suggesting that these tags are from spliced RNAs (Supplemental Fig. 6A). To independently verify the existence of the observed intragenic RNAs, we compared their CAGE tags with a zebrafish small RNA library (Wei et al. 2012) and identified significant overlap (58%, Fisher test: P < 2.2 × 10−16) in the same exons.
Figure 4.
Exonic CAGE tags during development. (A) A genome browser view of the intragenic region of the tp53 gene. Arrow with “ia” indicates the intronic alternative promoter of the delta117p53 variant (Chen and Peng 2009; Chen et al. 2009). Arrows labeled “e” and “u” indicate RNA start sites of exonic and 3′-UTR regions, respectively. (B) Lack of fluorescent Venus reporter activity in maximum projection overlays of 36 hpf embryos injected with an exonic CAGE marked candidate promoter region (EXN1) as compared with an active core promoter (ALT4) and a negative control (CON1) linked to a neural specific enhancer (E) (details in Supplemental Table 8). Insert of a bright-field image of an embryo is shown as reference for view of fluorescence image.
We next monitored the developmental dynamics of exon-associated CAGE tags. Exonic TCs appear before zygotic transcription initiation at MBT, which together with the spliced RNA alignment analysis argues further against de novo transcriptional activity in their production (Fig. 4A; Supplemental Fig. 6B). Exonic tags appear to correlate mostly with gene 5′-end expression (Supplemental Fig. 6C). While the biological significance of intragenic RNA products remains unknown, it is notable that GO analysis suggests that exonic tags are enriched in genes associated with translation and cellular metabolism (Supplemental Fig. 6D,E). Furthermore, post-transcriptional cleavage of lncRNAs, such as MALAT1 (Mercer et al. 2010; Ulitsky et al. 2011; Pauli et al. 2012) described for both human and zebrafish, is detected by exonic CAGE tags (data not shown).
We hypothesized that the initiation sites in exons detected by CAGE do not reflect transcriptional promoter activity. To test this we selected five exonic CAGE start site regions (Supplemental Table 8) and tested them similarly to predicted gene 5′-end promoters in transgenic reporter assays. None of the five regions activated significant reporter activity and were comparable to three random control genomic regions without CAGE tags (Fig. 4B; Supplemental Fig. 4C). These results support the notion that exonic CAGE tag-defined RNAs are of post-transcriptional origin and not initiated from intragenic promoters.
Intronic CAGE tags exhibit functional subclasses and are developmentally regulated
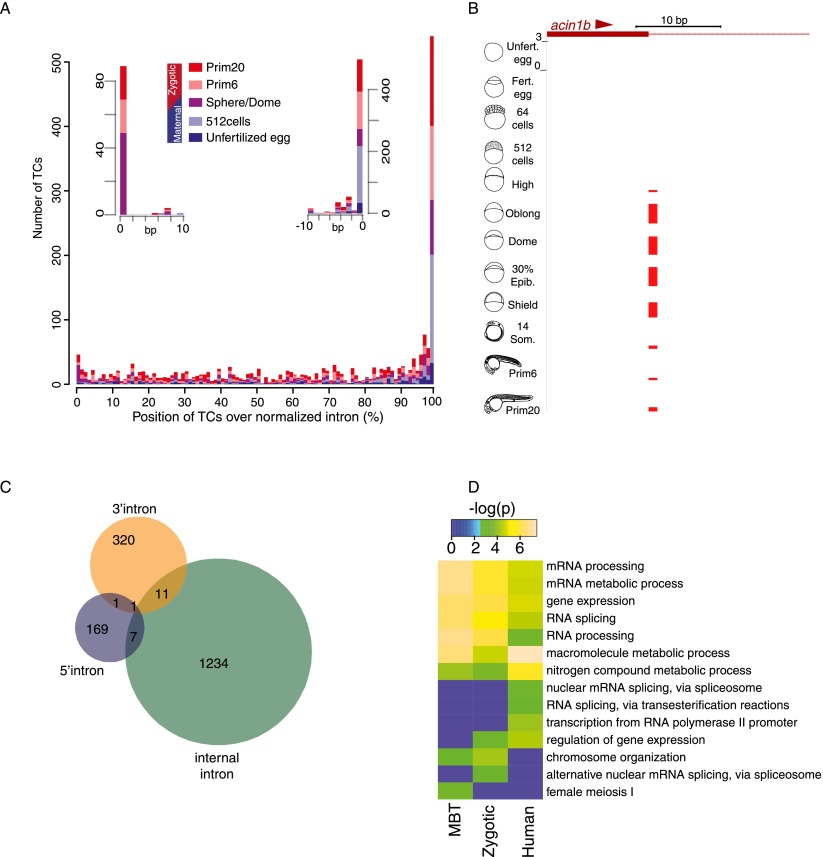
Next we addressed the dynamics and distribution of intronic CAGE tags that were not assigned to intronic alternative promoters based on transcript models from Ensembl or RNA-seq. These tags may represent novel unannotated promoters but could also reflect processed RNAs. Such processed RNAs may include splice site-associated RNAs (sRNAs), which have recently been described in HeLa cells (Valen et al. 2011). To assess the existence and dynamics of such sRNAs during zebrafish development, we aligned intronic CAGE tags within introns. The positional distribution of intronic tags revealed 5′- and 3′-end-specific intronic TCs (Fig. 5A). An example of a developmentally regulated intron 5′-associated RNA start site is demonstrated in Figure 5B. The intron 5′-end associated RNAs are exclusively produced in zygotically active stages (Fig. 5A,B) consistent with splicing-associated activity and in contrast with other intronic RNAs, which are present throughout development (Fig. 5A). Consistent with their distinct developmental dynamics 5′, 3′, and intra-intronic RNAs are produced mostly in nonoverlapping sets of genes (Fig. 5C). Intra-intronic tags show both positive and negative correlation with gene 5′-end and exonic activity (Supplemental Fig. 7A,B), suggesting that many of these intronic RNA products are not constitutive degradation products of mRNAs and are independent of exonic RNAs.
Figure 5.
Distribution and developmental regulation of intronic CAGE tags. (A) Distribution of all intronic TCs aggregated and aligned in windows of 1% length of a normalized intron. TCs in specific stages are shown in colors as indicated. Insets show aggregates of CAGE tags aligned at single bases, up to 10 bases from either side of intron ends. (B) A genome browser view of splice donor site of the acin1b gene and associated intronic 5′-end CAGE tags. (C) Venn diagram of intersection and number of genes with various types of intronic TCs. (D) Enriched GO terms of genes with intron 5′-end CTSSs in zebrafish and human. The heat map represents the –log (P-values) of significantly enriched GO terms. (MBT) Mid-blastula transition.
Interestingly, intron 5′-end RNAs, unlike other intronic CAGE tagged RNAs, are detected mainly in genes, which themselves encode splicing-associated proteins (Fig. 5D; Supplemental Fig 7C,D; Supplemental Table 12). A similar association of intron 5′-end RNAs with splicing-related genes was also observed in human cells (Fig. 5D). Thus, our data together demonstrate that intron 5′-end specific RNAs are evolutionarily conserved and are property of splicing-associated genes.
Intragenic CAGE tags do not carry core promoter features
Several lines of evidence so far argue for exonic and intronic tags representing nonconventional RNA initiation sites and are likely generated by post-transcriptional mechanisms. To test this hypothesis further we asked whether the 5′-end sequence environment of intragenic CTSSs resemble that of known transcriptional initiation sites of conventional core promoters. Core promoters are characterized by the initiator sequence (YYA[+1]NWYY) (Bucher 1990) or, more generally, YR(+1) consensus previously established in mouse and human (Carninci et al. 2006). Dinucleotide frequency analysis demonstrated strikingly different characteristics of start sites at gene 5′-ends from intragenic sites (Fig. 6A). Exonic tags (both spliced and unspliced) are marked primarily with G stretches at their 5′-end, previously associated only with 3′-UTR CAGE signal (Carninci et al. 2006). This signal was also detected in exonic tags on the MALAT1 lincRNA, in both zebrafish and human (Supplemental Fig. 8A). The exonic start signal is clearly different from the functional initiator sequence observed at the gene 5′-end of promoters (Fig. 6A,B), and shows mild nucleotide preference, with similarity to exon start sites in human (Fig. 6B). The prevalence of a G base at the start of exon tags is not due to a potential experimental G bias introduced by the CAGE method, which is compensated by removal of mismatching Gs at the 5′-end of all tags (Supplemental Fig. 8B; Supplemental Table 13). Thus, the dinucleotide frequency patterns suggest a fundamentally different way for the production of exonic and promoter transcripts. Furthermore, exonic and intronic CAGE tags lack the promoter-associated post-translational histone modification mark H3K4me3 (Bernstein et al. 2005), which is in contrast to the sequence regions around gene 5′-end associated TSSs (Supplemental Fig. 8C).
Figure 6.
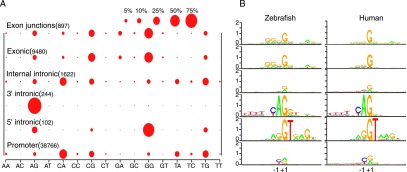
Intragenic CAGE tags do not carry core promoter features. (A) Dinucleotide frequency analysis of dominant CTSSs (−1,+1 bp) of gene 5′-end promoter and of intragenic RNA products. Relative abundance of dinucleotides is shown in bubbles of varying size. Number of TCs analyzed from 12 stages are indicated in brackets (repeat incidences in multiple stages are not included). (B) Sequence logos and their information content of dominant CTSSs (−1,+1 bp) at gene 5′-end and intragenic sites in zebrafish and human.
These data, together with the independent developmental dynamics, cDNA alignment, and reporter gene functional assays, argue that exon and intron 5′- and 3′-end associated RNAs are biochemically independent products from full-length RNAs. Based on the sequence and histone modification pattern analysis, intragenic CAGE-detected start sites do not reflect canonical core promoters and are strong candidates for RNAs generated by post-transcriptional processing.
Discussion
In this study we have provided the first quantitative mapping of the developmental dynamics of TSSs usage at single nucleotide resolution on a genome scale for both protein-coding and noncoding genes during vertebrate embryo development. CAGE analysis has revealed developmental TSS usage at a previously unparalleled resolution, leading to the description of a large number of alternative promoters as well as the detection of positionally constrained sequence features of developmentally active promoters including a novel initiator sequence. We have shown that the transition from maternal to zygotic genome activity including the onset of zygotic transcription is characterized by the appearance of a previously unappreciated and pervasive production of intragenic processed RNAs overlapping known genes and having specific intronic and exonic signatures. We presented several lines of evidence showing that many intragenic CTSSs represent several classes of cleaved RNAs generated by post-transcriptional processing, and some of which are characteristic to subsets of genes acting in concordance during development. Among other assays, we carried out transgenic reporter assays which—keeping in mind the limits of the small number of examples tested—argue for the non-promoter nature of exonic RNA start regions. It is yet unclear if these RNAs carry a conventional cap structure, but it is worth mentioning that recapping may occur downstream from mRNA production and may also occur in the cytoplasm (Otsuka et al. 2009). Intriguingly, intron 5′-end RNAs were found in RNA processing and particularly in RNA splicing-associated genes, which suggests the existence of a mechanism which links a specific splicing activity with the regulation of expression of the splicing machinery. While the size and role of the intragenic RNA products and the mechanism of their generation remain unknown, their abundance and increased intron coverage at subsets of developmentally regulated genes indicate a possibly important regulatory function. In particular, the biological association with splicing genes is conserved between fish and human and suggests a fundamental property of vertebrates.
The TSSs data presented here complements mammalian cell culture-based and non-vertebrate animal models such as Drosophila (Ohler et al. 2002; The modENCODE Consortium et al. 2010; Hoskins et al. 2011) and fills the gap by providing the first description of core promoter features during vertebrate development. Several features of promoter diversity showed similarities to that seen in Drosophila and Xenopus sp including sharp and broad peak promoter dynamics (Ni et al. 2010; van Heeringen et al. 2011). However, developmentally active promoters in fish appear to rely much less on positionally constrained motifs than their Drosophila counterparts, suggesting substantially different core promoter regulation mechanisms. Our results demonstrate the global and pervasive changes in promoter utilization during early stages of development when the maternal transcriptome gives way to the activity of the embryonic genome. Nevertheless, the whole embryo analysis in differentiation stages inevitably masks cell and tissue-specific variation in promoter usage and hinders the deciphering of associated promoter codes. The elucidation of developmental promoter codes acting in specific cell types of the embryo will be the challenge of future research.
The comparisons of zebrafish and pufferfish promoterome showed that features such as promoter motif composition are conserved on a per-gene basis, which argues for their functional significance and underscores generality of the detected features in teleost evolution. The comparative approach was taken further with the identification of a novel, evolutionarily conserved initiator characterized by the GAAG motif, which is used by a subset of vesicle and membrane transport-associated genes, demonstrating the utility of genomic analysis of fish models with direct relevance to human transcription regulation.
The resource data generated in this study provides a range of practical applications and benefits and paves the way for functional validation experiments. Widespread occurrence of alternative promoters during development suggests pervasive variability of gene 5′-UTR sequences, with implications for translation start site selection during development. This variability should be taken into account in techniques widely used in disease modeling with zebrafish (Eisen and Smith 2008), such as the design of translation blocking knockdown reagents (e.g., translational start site targeting morpholino antisense oligonucleotides) or the generation of site-specific mutations. Understanding core promoter regulation is central to the informed choice of core promoter for transgene assays designed either to control cell type-specific activities (fluorescence reporter labeling) or to detect and functionally characterize cis-regulatory modules (e.g., enhancer trapping and enhancer tests). Furthermore, the correct identification of core promoters will be critical for finding noncoding mutations that affect development and may lead to phenotypes suitable for disease modeling. The characteristics of core promoters have been proposed to underlie interaction specificity between core promoter and distal acting cis-regulatory modules to secure correct targeting of cognate promoters by enhancers acting over hundreds of kilobases, and core promoters have also been proposed to integrate signaling input, epigenetic, and cell cycle regulation (for review, see Müller et al. 2007).
In conclusion, the high-resolution transcription initiation database for zebrafish and Tetraodon provides the foundation for the comparative analysis of transcription initiation complexes on core promoters during development and the elucidation of developmental codes of transcription initiation in vertebrates. Furthermore, the identification and description of the developmental dynamics of evolutionarily conserved, yet little understood intragenic RNA products will aid in exploiting the zebrafish model in a search for the noncoding regulators of development (for review, see Pauli et al. 2011).
Methods
Collection of RNA and chromatin at embryonic stages in zebrafish
Zebrafish AB* wild-type strains were used. Unfertilized eggs were collected from spawning females. Fertilized eggs and two cells stage were collected from pairwise crosses 10 min after pairing. All other developmental stage samples were collected from several clutches of embryos laid within a 20-min time window and embryos were raised at 28°C in 10% Hanks solution. Embryos were imaged and snap-frozen at the indicated embryonic stages (Kimmel et al. 1995) (for further information on stages used, see Supplemental Table 1). Starting with the 512-cell stage, three further samples were collected 30 min, 60 min, or 90 min after, respectively. Embryos were snap-frozen in liquid nitrogen and stored at −80°C until processing. Repeat samples of prim-6 embryos were collected from different clutches. Batches of embryos (up to 800 embryos each stage) were homogenized in 1 mL TRIzol (Invitrogen) for each 150 embryos using polypropylene pellet pestle (Sigma). Homogenates were merged into a 15-mL falcon tube and RNA was prepared according to manufacturer's instructions. RNA was analyzed on capillary electrophoresis (Bioanalyzer 2100, Agilent).
CAGE library preparation
CAGE library preparation was adapted from Takahashi et al. (2012) and modified to work with Illumina GA IIx sequencers. Five micrograms of total RNA was reverse transcribed with RT random N15 primer (5′-AAGGTCTATCAGCAGNNNNNNNNNNNNNNNC-3′), PrimeScript Reverse Transcriptase in the presence of 0.132 M trehalose and 0.66 M sorbitol. The sample was cap-trapped and a specific linker, containing a 3-bp recognition site and the type III restriction-modification enzyme EcoP15I, (5′-PhosCTGCTGXXXCTGTAGAACTCTGAACCTGTCGGTGG-3′) for both N6 (5′-CCACCGACAGGTTCAGAGTTCTACAGXXXCAGCAGNNNNNNPhos-3′) and GN5 (5′-CCACCGACAGGTTCAGAGTTCTACAGXXXCAGCAGGNNNNNPhos-3′), was ligated to the single-strand cDNA. The priming of the second strand was made with specific primer (5′-BioCCACCGACAGGTTCAGAGTTCTACAG-3′). After second strand synthesis and cleavage with EcoP15I, another linker (1:1 mix of Upper oligonucleotide; 5′-PhosNNTCGTATGCCGTCTTCTGCTTG-3′ and Lower oligonucleotide; 5′-CAAGCAGAAGACGGCATACGA-3′) was ligated. Purified cDNA was amplified with 1 μM each, forward (AATGATACGGCGACCACCGACAGGTTCAGAGTTC) and reverse (CAAGCAGAAGACGGCATACGA) primers with 15 to 18 PCR cycles. PCR products were purified and concentration was adjusted to 10 nM. The CAGE libraries were clustered to GA IIx flowcell at a final concentration of 5 pM, following the Illumina cluster generation protocol kit v.4 and then sequenced with Illumina GA IIx 36 cycles single-read run operation program using specific sequencing primer (CGGCGACCACCGACAGGTTCAGAGTTCTACAG), following the Illumina sequence protocol.
Chromatin immunoprecipitation and sequencing
Approximately 1500 embryos (dome/30% epiboly) or 200 embryos (prim-6) were dechorionated and fixed in 1.85% Formaldehyde in Hanks Media for 20 min at room temperature. Fixation was stopped using 1× Glycine followed by PBS washes (Wardle et al. 2006). ChIP experiments were carried out using the ChIP-IT Express Enzymatic kit (Active Motif) in line with manufacturer's instructions. In brief, embryos were resuspended in lysis buffer, incubated on ice for 20 min, and homogenized using a dounce homogenizer. Nuclei were resuspended in 200 μL digestion buffer. Chromatin was enzymatically sheared for 10 min at 37°C. The reaction was stopped, and 75 μL of sheared chromatin was used for ChIP reactions utilizing 4 μg of anti-H3k4Me3 (Abcam ab8580) or an equivalent volume of water as no antibody control. Samples were incubated overnight at 4°C while rotating. Magnetic beads were washed and decrosslinked for 4 h at 65°C. Samples were proteinase K and RNase A treated and purified using the QIAquick PCR Purification Kit (Qiagen) (dome/30% epiboly) or phenol chloroform extraction (24 hpf). Enrichment of target sequences was determined by qPCR using Power SYBR Green PCR Master Mix (Applied Biosystems). ChIP-seq was performed as described (Soler et al. 2011). In brief, 10 ng of ChIP DNA is end-repaired, ligated to single read adaptors, size selected, and amplified for 18 cycles according to Illumina's ChIP-seq protocol. Cluster generation is performed according to the Illumina Cluster Reagents preparation protocol (http://www.illumina.com). Samples were sequenced for 36 bp on the Illumina GA IIx platform or HiSeq 2000 system. The raw data from the Illumina Genome Analyzer are processed using the IPAR (Integrated Primary Analysis Reporting Software) and the Illumina Genome Analyzer Pipeline (GAP).
RNA extraction, library preparation, and sequencing
Total RNA was used from four developmental stages (two cells, dome/30% epiboly, 14 somites, and prim-6) and extracted using TRIzol (Invitrogen) according to the manufacturer's instructions and used for subsequent RNA-seq based profiling. The RNA samples were treated with 2U DNase I (Qiagen) per μg RNA sample at 37°C for 10 min. Digested samples were then treated with 20 mg/mL proteinase K (Sigma Aldrich) at 37°C for 45 min. The quality and quantity of total RNA were assessed with the Bioanalyzer 2100 (Agilent). The RNA-seq library was generated following the standard Illumina RNA-seq poly(A)+ protocol and sequenced with 76 bp Paired End reads using an Illumina Genome Analyzer IIx at the Barts and The London Genome Center. SOAPsplice-v1.0 (Huang et al. 2011) was used to align sequences to the zebrafish genome (Zv9/danRer7). A set of custom scripts was used to process SOAPsplice output, and to quantify (as “Rseq-score”) the levels of transcription of annotated zebrafish genes. Briefly, the Rseq-score is a normalized score of total number of RNA-seq reads falling on the first exon divided by the length of the exon. The same approach was tested on second and third exons, to verify if annotation issues could affect the analysis, and the results obtained were very similar, indicating the overall measures of correlation obtained are robust to potential annotation problems.
CAGE mapping and CTSS prediction
The latest build of genome assembly of zebrafish (Zv9) and pufferfish (tetNig2) were downloaded from UCSC Genome Browser (Kuhn et al. 2009). CAGE tags were mapped using Bowtie (Langmead et al. 2009), allowing a maximum of two mismatches and only uniquely mapping tags. Since the CAGE protocol often yields an additional G nucleotide at the 5′-end of the tag, we removed the starting G when mismatching G at the first position and removed tags with an additional mismatch at the second position (affecting 1%–2% of CAGE tags; see Supplemental Table 13). The remaining unique 5′-ends were regarded as CAGE tag-defined transcriptional start sites (CTSSs). The number of CAGE tags mapping to each CTSS across different samples was normalized as in Balwierz et al. (2009) to obtain the normalized number of tags per million (tpm).
CTSS clustering, TCs, and promoter types
Only CTSSs supported by a minimum of 0.5 tpm in at least one stage were used for a stage-specific clustering into transcript clusters (TCs). Neighboring CTSSs were clustered if they were <20 bp apart. To determine the number of CTSSs and TCs with respect to sequencing depth, we sorted the CTSSs (or TCs) based on tpm and counted the minimal number of CTSSs that account for a selected percentage of CAGE tags. Throughout this manuscript we use thresholds of 0.5 tpm when analyzing CTSS and 1 tpm for TCs. To address TC width, we calculated a cumulative distribution of CAGE tags along each TC and determined the position of 10th and 90th percentile. The obtained interquantile range provides a more robust definition of TC width avoiding broadening of cluster at highly expressed clusters. Based on the distribution of the interquantile TC width (between 10th and 90th percentile), we empirically determined a boundary at 10 bp that separates the best sharp from broad TCs. TCs with an interquantile width of <10 bp were classified as sharp, and the rest as broad. For stage specific analysis, CAGE tags (TCs) from 12 developmental stages were classified into three major categories: maternal (0 tpm from shield stage onward), zygotic (0.5 tpm from high stage onward), or transcribed throughout (M-Z: minimum 0.5 tpm values in at least two stages among maternal and zygotic stages).
Genomic location of tags
Current gene model annotations (Ensembl version 71, RefSeq) downloaded from UCSC Genome Browser (Kuhn et al. 2009) and transcript models were built from RNA sequencing data. CAGE tags mapping unambiguously to 5′-UTR, coding exons, introns, 3′-UTR, and promoter regions (±500 bp around annotated TSSs) were classified accordingly. Alternative promoters were defined based on Ensembl and RNA-seq transcript models, by collapsing transcripts with an identical first splice donor site whose 5′-ends were <500 bases apart. If the first base and last base of introns overlapped with intronic TCs, they were subclassified into 5′- and 3′-intronic, and the remaining as intra-intronic. Remaining TCs were classified as novel intergenic transcripts. CAGE tags from the prim-6 stage, which failed to map to the genomic sequence, were mapped to Ensembl cDNA (ver. 65, Zv9) sequences (the longest transcript for each gene) using Bowtie (Langmead et al. 2009). To measure the quantitative nature of CAGE transcript tags, all TCs in the window of ±500 bp of a gene were obtained for each stage and the maximum tpm score was recorded as “cage-score.” Pearson correlation between cage-scores and Rseq-scores was calculated for each of those genes where both cage-score and Rseq-score data were available for all four stages.
GO analysis
GO analyses were performed using the GOstats package from Bioconductor (Falcon and Gentleman 2007). The P-values for the enriched GO terms were corrected by FDR. The –log10 (P-values) were clustered and used to plot the heat maps. Twelve developmental stages were classified into three major categories, maternal (unfertilized egg to 512 cells), MBT (High to Dome/30% Epiboly), and zygotic (shield to Prim20), or transcribed throughout based on tpm values as described for alternative promoters. GO categories with a minimum of five genes and a P-value ≤ 0.05 were considered significantly enriched. All GO-associated statistics are in Supplemental Table 12.
Promoter fragment isolation, reporter constructs, and transgenesis
Test promoter fragments were amplified by PCR from AB* wild-type genomic DNA and cloned into venus fluorescent reporter gene containing vector using a MultiSite Gateway System (Invitrogen) as described in Gehrig et al. (2009). The reporter constructs were verified by sequencing. Genomic coordinates of the cloned fragments are in Supplemental Table 8. Reporter constructs were injected in fertilized zebrafish eggs within 10–25 min after laying with 1–1.2 nL injection solution (20 ng/μL reporter plasmid DNA, 30 ng/μL eCFP mRNA [marker for image processing], and 0.1% Phenol Red). The reporter activity was recorded by automated imaging (Gehrig et al. 2009) and images analyzed with Zebrafish Miner software (Gehrig et al. 2009; M Reischl, A Bartschat, F Eberle, U Liebel, J Gehrig, F Müller, R Mikut, in prep.). The level of reporter expression was measured as pixel intensity value in the corresponding tissue domains (Supplemental Fig. 4C).
Initiator and core promoter features
The analyses of core promoter motifs, initiator usage, and H3K4me3 enrichment were performed using the dominant peak of TCs (tpm ≥ 1). The initiators represent the dominant peak (+1) and the position directly upstream (−1). The R package seqlogo was used to create the sequence logos of the core promoter region (Schneider and Stephens 1990).
Promoter motif analyses and orthologous comparison of promoters
The comparison of all promoter features was performed on orthologous (one to one) gene pairs, on two developmental stages from zebrafish (fertilized egg and prim-6) and pufferfish (fertilized egg and 46 hpf). Only genes with TC (≥5 tpm) were used, where the position of highest tpm value of representative TC was used for TSS definition. These criteria resulted in a final list of 2070 (maternal) and 2700 (zygotic) orthologous gene pairs for comparative analyses. Representative initiator dinucleotide position (−1,+1) was determined based on the +1 position of the dominant CTSS of the representative TC. Promoter motif comparison was performed against common promoter motifs from JASPAR (Portales-Casamar et al. 2010). A region ±150 relative to the dominant CTSS was scanned using the JASPAR TFBS Perl module with the default (80%) threshold. Only promoter motifs detected at a specific position relative to the TSS were used for the comparative analyses. Correlation of promoter types (sharp or broad) of orthologous genes was based on representative TCs. Statistical significance of all orthologous correlations analyses was evaluated by χ2 test for independence (P-values ≤ 0.05). Genes containing the AA-initiator were selected based on the initiator dinucleotide determined by the dominant CTSS of the representative TC (≥5 tpm) at a given stage in fish species. In human, the representative initiator dinucleotide for each gene was determined in AG04450, BJ, H1 hESC, HUVEC, and NHEK cell lines from The ENCODE Project (Djebali et al. 2012; Harrow et al. 2012), using a similar procedure as in fish.
Data access
Raw sequencing data have been deposited in the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRA055273. Data tracks (bigWig and bigBed files) containing CAGE transcription start sites as well as H3K4me3 and RNA-seq coverage are available for download at http://zeprome.genereg.net/downloads/danRer7/ and http://zeprome.genereg.net/downloads/tetNig2/. All tracks can be visualized in the form of annotated custom tracks in the UCSC Genome Browser using the following URLs: http://zeprome.genereg.net/downloads/danRer7/Zebrafish_tracks.txt and http://zeprome.genereg.net/downloads/tetNig2/Tetraodon_tracks.txt.
Acknowledgments
We thank the following funding agencies for their support: EUTRACC EU Framework 6 IP project (to F.M., U.S.); EU Framework 7 projects ZF-Health (to F.M., B.L., U.S.); Dopaminet (to F.M., E.S., P.C.); MEXT Grant to RIKEN CLST and OSC; and ENCODE U54 to P.C. We thank Turker Bodur and Andreas Zaucker for pufferfish breeding, Markus Reischl for Zebrafish miner software, Urban Liebel, Ravi Peravali for help with automated microscopy, and James Bull for advice on statistical analyses. We thank Aditi Kanhere and Laszlo Tora for critically reading the manuscript. C.N., Y.H., C.P., V.H., N.L., H.T., A.M.M.S., Y.S., R.F.A., S.A., J.G., A.A., C.E.M.K., A.A.J.v.S., W.F.J.v.I., O.A., S.R., C.W., U.S., P.C., B.L., and F.M. are members of ZEPROME, the Zebrafish Transcriptome and Promoterome Consortium.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.153692.112.
References
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ 2009. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell 17: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balwierz PJ, Carninci P, Daub CO, Kawai J, Hayashizaki Y, Van Belle W, Beisel C, van Nimwegen E 2009. Methods for analyzing deep sequencing expression data: Constructing the human and mouse promoterome with deepCAGE data. Genome Biol 10: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bucher P 1990. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol 212: 563–578 [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Chen J, Peng J 2009. p53 Isoform Δ113p53 in zebrafish. Zebrafish 6: 389–395 [DOI] [PubMed] [Google Scholar]
- Chen J, Ng SM, Chang C, Zhang Z, Bourdon JC, Lane DP, Peng J 2009. p53 isoform Δ113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev 23: 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessio JA, Wright KJ, Tjian R 2009. Shifting players and paradigms in cell-specific transcription. Mol Cell 36: 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH 2008. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet 24: 167–177 [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JS, Smith JC 2008. Controlling morpholino experiments: Don't stop making antisense. Development 135: 1735–1743 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258 [DOI] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. 2013. Ensembl 2013. Nucleic Acids Res 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A 2008. A code for transcription initiation in mammalian genomes. Genome Res 18: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig J, Reischl M, Kalmar E, Ferg M, Hadzhiev Y, Zaucker A, Song C, Schindler S, Liebel U, Muller F 2009. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat Methods 6: 911–916 [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Tjian R 2010. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat Rev Genet 11: 549–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Landolin JM, Brown JB, Sandler JE, Takahashi H, Lassmann T, Yu C, Booth BW, Zhang D, Wan KH, et al. 2011. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res 21: 182–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhang J, Li R, Zhang W, He Z, Lam TW, Peng Z, Yiu SM 2011. SOAPsplice: Genome-wide ab initio detection of splice junctions from RNA-seq data. Front Genet 2: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957 [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga JT 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 339: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT 2008. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol 20: 253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB 1993. The zebrafish midblastula transition. Development 119: 447–456 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, et al. 2006. CAGE: Cap analysis of gene expression. Nat Methods 3: 211–222 [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Karolchik Dl, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. 2009. The UCSC Genome Browser Database: Update 2009. Nucleic Acids Res 37: D755–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhard B, Sandelin A, Carninci P 2012. Metazoan promoters: Emerging characteristics and insights into transcriptional regulation. Nat Rev Genet 13: 233–245 [DOI] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Ostrup O, Winata C, Mathavan S, Müller F, Alestrom P, et al. 2011. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell 21: 993–1004 [DOI] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H, et al. 2005. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet 1: 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, Askarian-Amiri ME, Gardiner BB, Goodall GJ, Grimmond SM, et al. 2010. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res 20: 1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Consortium, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F, Demeny MA, Tora L 2007. New problems in RNA polymerase II transcription initiation: Matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem 282: 14685–14689 [DOI] [PubMed] [Google Scholar]
- Müller F, Zaucker A, Tora L 2010. Developmental regulation of transcription initiation: More than just changing the actors. Curr Opin Genet Dev 20: 533–540 [DOI] [PubMed] [Google Scholar]
- Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J 2010. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods 7: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Wassarman DA 2010. Promoting developmental transcription. Development 137: 15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao GC, Niemann H, Rubin GM 2002. Computational analysis of core promoters in the Drosophila genome. Genome Biol 3: research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR 2009. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol 29: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry TJ, Theisen JW, Hsu JY, Wang YL, Corcoran DL, Eustice M, Ohler U, Kadonaga JT 2010. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev 24: 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF 2011. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12: 136–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. 2012. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22: 577–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A 2010. JASPAR 2010: The greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38: D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF 2007. The maternal-zygotic transition: Death and birth of RNAs. Science 316: 406–407 [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM 1990. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, Boer E, Bryne JC, Thongjuea S, Rijkers E, Demmers J, Ijcken W, Grosveld F 2011. A systems approach to analyze transcription factors in mammalian cells. Methods 53: 151–162 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Lassmann T, Murata M, Carninci P 2012. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat Protoc 7: 542–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura O, Okada Y, Ando H, Guedj M, Higashijima S, Shimazaki T, Chino N, Okano H, Okamoto H 2005. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev Biol 278: 587–606 [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valen E, Preker P, Andersen PR, Zhao X, Chen Y, Ender C, Dueck A, Meister G, Sandelin A, Jensen TH 2011. Biogenic mechanisms and utilization of small RNAs derived from human protein-coding genes. Nat Struct Mol Biol 18: 1075–1082 [DOI] [PubMed] [Google Scholar]
- van Heeringen SJ, Akhtar W, Jacobi UG, Akkers RC, Suzuki Y, Veenstra GJ 2011. Nucleotide composition-linked divergence of vertebrate core promoter architecture. Genome Res 21: 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF 2010. Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464: 922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardle FC, Odom DT, Bell GW, Yuan B, Danford TW, Wiellette EL, Herbolsheimer E, Sive HL, Young RA, Smith JC 2006. Zebrafish promoter microarrays identify actively transcribed embryonic genes. Genome Biol 7: R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CA, Hill JE, Graves JS, Wood AL, Kilgore KH 2009. Use of a novel induced spawning technique for the first reported captive spawning of Tetraodon nigroviridis. Mar Genomics 2: 143–146 [DOI] [PubMed] [Google Scholar]
- Wei C, Salichos L, Wittgrove CM, Rokas A, Patton JG 2012. Transcriptome-wide analysis of small RNA expression in early zebrafish development. RNA 18: 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]