Summary
Two continuous‐flow bench‐scale bioreactor systems populated by mixed communities of acidophilic sulfate‐reducing bacteria were constructed and tested for their abilities to promote the selective precipitation of transition metals (as sulfides) present in synthetic mine waters, using glycerol as electron donor. The objective with the first system (selective precipitation of copper from acidic mine water containing a variety of soluble metals) was achieved by maintaining a bioreactor pH of ∼2.2–2.5. The second system was fed with acidic (pH 2.5) synthetic mine water containing 3 mM of both zinc and ferrous iron, and varying concentrations (0.5–30 mM) of aluminium. Selective precipitation of zinc sulfide was possible by operating the bioreactor at pH 4.0 and supplementing the synthetic mine water with 4 mM glycerol. Analysis of the microbial populations in the bioreactors showed that they changed with varying operational parameters, and novel acidophilic bacteria (including one sulfidogen) were isolated from the bioreactors. The acidophilic sulfidogenic bioreactors provided ‘proof of principle’ that segregation of metals present in mine waters is possible using simple online systems within which controlled pH conditions are maintained. The modular units are versatile and robust, and involve minimum engineering complexity.
Introduction
Waters draining abandoned metal mines and mine waste repositories are characteristically acidic (sometimes extremely so) and enriched with dissolved transition metals and aluminium (Nordstrom, 2000). The physicochemical characteristics of mine‐impacted waters (MIWs) vary from location to location, as these are dictated by a number of geochemical, climatic, hydrological and other factors. Microbially enhanced oxidative dissolution of sulfide minerals is a prime cause of water pollution associated with metal mines (Johnson and Hallberg, 2003). Bacteria such as Acidithiobacillus spp. and Leptospirillum spp. are well known for their abilities to use reduced chemicals (ferrous iron and/or reduced sulfur) as sources of energy, and to use the energy released from these reactions to fix carbon dioxide and thereby produce new biomass. These autotrophic bacteria have minimal nutritional requirements, and their abilities to tolerate elevated concentrations of dissolved metals in acidic solutions enables them to exploit the seemingly hostile environments that characterize mine spoils, mineral tailings and MIWs. Acidity derives from the oxidation of the reduced sulfur moiety in sulfide minerals, and also the hydrolysis of ferric iron in the case of iron‐containing minerals, such as the most ubiquitous of all sulfides, pyrite (FeS2). Equation 1 depicts the complete oxidation of the most abundant copper sulfide mineral in the lithosphere, chalcopyrite:
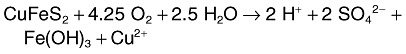 |
1 |
The low pH of the leach liquors produced allows metals, such as copper and zinc, which are released from the oxidative dissolution process to remain in solution. Aluminium does not occur as a sulfide mineral, but many aluminosilicates are susceptible to acid dissolution and, as a results concentrations of this metal are also usually much higher in MIWs than in non‐impacted (circum neutral‐pH) streams.
The severe impact that MIWs can have on the local and wider environment means that control of their formation or, if this is not pragmatic, remediation of waters draining metal mines is generally regarded as a priority issue for regulatory authorities. The most widely used approach for remediating MIWs is to aerate (to oxidize ferrous iron to ferric) and add an alkalizing chemical (such as CaO) in order to raise water pH and to precipitate metals as hydroxides and carbonates. Aggregation and thickening of the metal hydroxide flocs produces a sludge which typically contains ∼30% solids in the ‘high‐density sludge’ application. However, this active chemical process has numerous drawbacks, including operating and reagent costs, and the need to dispose of the polymetallic sludge generated in designated landfill sites. In addition, potentially useful and valuable metal resources are not recovered in chemical remediation of MIWs. Compost bioreactors (sometimes called ‘constructed anaerobic wetlands’) that use microbial reductive processes to immobilize metals in MIWs also suffer from a number of drawbacks (Johnson and Hallberg, 2002) These reactors are fuelled by bulky organic materials (usually a mixture of straw or sawdust, and animal manure) which require periodic replacement, and again metals are not recovered but are ‘locked up’ within the spent compost, which is therefore usually categorized as a toxic waste.
An alternative approach for remediating MIWs, which, like compost bioreactors, harnesses the abilities of microorganisms to generate alkalinity and to immobilize metals, is referred to generically as ‘active biological treatment’. In such systems, microorganisms that catalyse redox transformations of iron or sulfur are maintained in reactors where conditions can be optimized for their activities and, like active chemical treatment, this approach requires continuous inputs of reagents and more intensive management. Two distinct variants have been demonstrated as pilot‐scale or full‐scale systems. One uses acidophilic bacteria to oxidize ferrous iron and thereby facilitate iron removal from MIWs by hydrolysis and precipitation of the ferric iron produced. A pilot‐scale operation of this kind has been operating at Nochten, in east Germany, for over 3 years, removing iron from contaminated groundwater and producing schwertmannite [Fe8O8(OH)6SO4] as a by‐product (Heinzel et al., 2009). Other metals often present in MIWs are, however, more effectively removed as sulfides than as hydroxide or carbonate phases. Since metal sulfides have different solubilities, they can be selectively precipitated by controlling solution pH, which will determine the concentration of the reactant, S2− (Steudel, 2000). Sulfidogenic bacteria generate hydrogen sulfide primarily by using either sulfate or elemental sulfur as an electron acceptor, and an organic (e.g. ethanol) or inorganic (e.g. hydrogen) electron donor. Since MIWs usually contain elevated concentrations of sulfate (which, like many of the metals present, derives from the oxidation of sulfide minerals; Eq. 1) using sulfate‐reducing bacteria (SRB) for mine water amelioration is particularly pertinent. A drawback to this strategy has been, however, that characterized species of SRB are highly sensitive to even mild acidity, and do not grow at pH < 5.5 (Koschorreck, 2008). This characteristic has required SRB used for remediation of MIWs to be maintained in reactors where direct contact with the acidic wastewaters is avoided, which has implications for the design and operating costs of engineered systems.
There have been some reports of novel species of SRB that are acid‐tolerant or acidophilic. Kimura and colleagues (2006) reported that a mixed culture of a SRB isolated from a geothermal site on the Caribbean island of Montserrat (‘Desulfosporosinus acidophilus’ M1) and an acidophilic heterotroph (Acidocella sp. PFBC) grew in a synthetic liquid media maintained at pH 3.8–4.2, where it promoted the selective precipitation of zinc sulfide. Senko and colleagues (2009) also isolated an acid‐tolerant Desulfosporosinus sp. (from sediment of MIW from a coal mine) that reduced sulfate, iron(III), manganese(IV) and uranium(VI) at pH 4.2. Rowe and colleagues (2007) reported that SRB other than Desulfosporosinus spp. were responsible for precipitating copper (as CuS) in a microbial ‘mat’ found in a stream draining an abandoned copper mine in south‐west Spain.
Here we describe the commissioning and performances of continuous‐flow bioreactors containing mixed communities of acidophilic SRB and other bacteria, designed to remove selectively copper and zinc (as sulfides) from MIWs.
Results
The long‐term performances of the two 2.3 l working volume sulfidogenic bioreactors were assessed by operating them in parallel as anaerobic continuous‐flow systems for over 300 days. During this time, flow rates of both systems fluctuated depending on several factors, including the pH differential between the influent liquor and that set for the bioreactor, and rates of bacterial sulfidogenesis which were also affected by pH. In general, pH values measured in the lower biofilm layers in the reactors were similar to those of the liquid phases above the bead beds, although they tended to be lower (∼0.2–1.0 pH unit) when flow rates were relatively fast and when the pH of the influent liquors were lowered.
Bioreactor I
The major objective with bioreactor I was to determine conditions which allowed the selective precipitation of copper sulfide from feed liquors that also contained soluble zinc and ferrous iron. The mean flow rate with this bioreactor was 49 (± 27, standard deviation) ml h−1, corresponding to a mean dilution rate of 0.021 h−1, with maximum and minimum rates of 135 and 20 ml h−1 (Fig. 1). All of the copper present in the influent liquor was precipitated in the bioreactor for the greater part of the experiment (Fig. 2). At times when soluble copper was detected in the bioreactor liquor, this was due to short‐term perturbations in the system (reflux of copper sulfate from the gas trap into the reactor, due to low nitrogen pressure) and, on each occasion, the bioreactor recovered rapidly. In contrast, none of iron in the influent liquor was retained in the bioreactor. Concentrations of iron were invariably greater than the 1 mM present in the feed water, due to: (i) dissolution of small amounts of FeS deposited during the initial commissioning of the bioreactor, and (ii) microbially and acid‐enhanced corrosion of some of the stainless steel components of the bioreactor (Dinh et al., 2004). Since separation of copper and iron as solid and liquid phases was readily achieved in bioreactor I, the main challenge was to establish conditions where copper would precipitate (as a sulfide) but zinc would be retained in solution. When the bioreactor pH was set at 3.6, > 99% of the zinc in the influent liquor was precipitated, along with the copper, within the bioreactor (Fig. 1). By progressively lowering both the bioreactor pH and the concentration of the electron donor (glycerol) in the influent liquor, it was possible to retain increasing amounts of zinc in solution. Between days 147 and 236, the bioreactor was maintained at pH 2.4, and the amount of zinc precipitated stayed reasonably stable at 47 ± 16% (23 sampling time points) even though influent glycerol concentrations was lowered from 4 to 1.5 mM during this time. However, by decreasing the bioreactor pH still further (ultimately to pH 2.2) and the influent glycerol concentration to 0.7 mM, it was possible precipitate only 8 ± 2% (nine samples) of zinc within the bioreactor, while maintaining > 99% removal of copper from solution. Analysis of the solid residue that accumulated in bioreactor I confirmed that it was predominantly copper sulfide.
Figure 1.
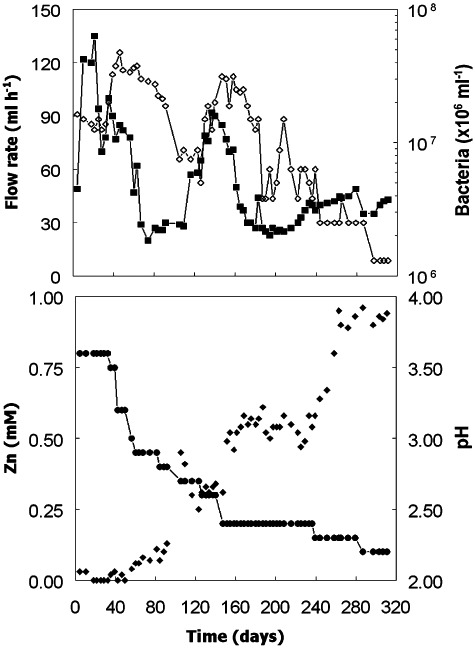
Changes in flow rates ( ), pH (●), concentrations of soluble zinc (◆) and numbers of planktonic‐phase bacteria (◊) in bioreactor I during the first phase of operation. The feed liquor contained 1 mM copper, zinc and ferrous iron and its pH was progressively lowered from 2.5 to 2.1 on day 21.
), pH (●), concentrations of soluble zinc (◆) and numbers of planktonic‐phase bacteria (◊) in bioreactor I during the first phase of operation. The feed liquor contained 1 mM copper, zinc and ferrous iron and its pH was progressively lowered from 2.5 to 2.1 on day 21.
Figure 2.
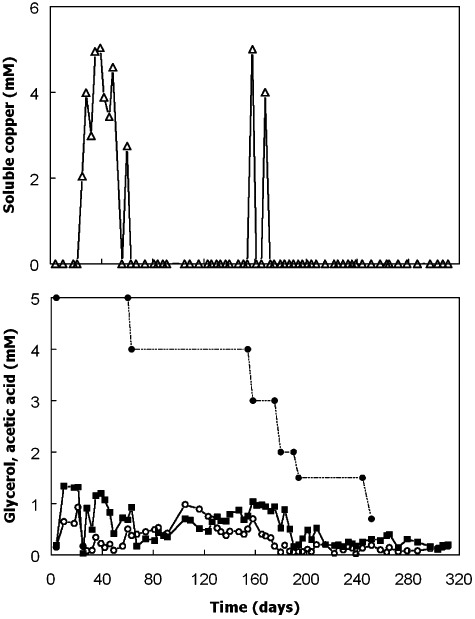
Changes in concentrations of soluble copper (▵), glycerol (○) and acetic acid ( ) in bioreactor I during the first phase of operation. Concentrations of glycerol in the feed liquor are also shown (●). The feed liquor contained 1 mM copper, zinc and ferrous iron and its pH was progressively lowered from 2.5 to 2.1 on day 21.
) in bioreactor I during the first phase of operation. Concentrations of glycerol in the feed liquor are also shown (●). The feed liquor contained 1 mM copper, zinc and ferrous iron and its pH was progressively lowered from 2.5 to 2.1 on day 21.
Bacterial numbers in the upper liquid phase in bioreactor I varied between 1.3 × 106 and 4.75 × 107 ml−1 (Fig. 1) and were more strongly correlated with influent glycerol concentrations (r = 0.77) than with bioreactor pH (r = 0.56; Fig. S1). Most of the glycerol added to the bioreactor was oxidized completely to carbon dioxide; although acetic acid was detected in all samples analysed, it was always present in low concentrations, and corresponded to a mean of ∼26% of the glycerol that was oxidized throughout the 311 trial period.
Bioreactor I was subsequently operated (for 56 days) with a feed liquor similar in composition to water draining the abandoned Mynydd Parys copper mine, with the bioreactor pH maintained at 2.6. All of the copper, but none of the ferrous iron, aluminium or manganese, were precipitated within the bioreactor under these conditions (Fig. 3). As before, some precipitation of zinc (sulfide) occurred (∼75% of that in the influent) at pH 2.6 when the glycerol concentration in the feed liquor was 3 mM. Decreasing the glycerol concentration to 0.7 mM lowered the amount of zinc precipitated to about 30% of that in the synthetic mine water, and this was depressed further (to ∼25%) when the yeast extract content of the influent was lowered from 0.01% to 0.005%. Numbers of bacteria in the upper liquid phase of the bioreactor decreased significantly as a result of lowering the concentrations of both glycerol and yeast extract (Fig. 3). Concentrations of acetic acid corresponded to a mean of ∼9.5% of the glycerol that was oxidized during this phase of operation.
Figure 3.
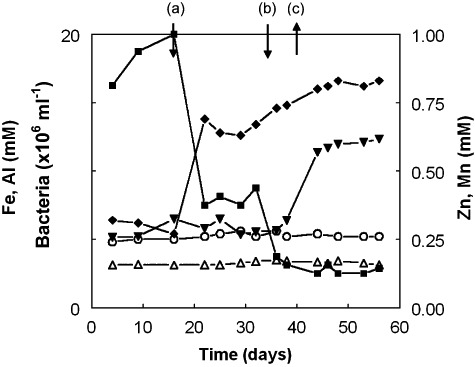
Changes in concentrations of soluble zinc (◆), iron (▾), aluminium (▵) and manganese (○), and numbers of planktonic‐phase bacteria ( ) in bioreactor I during the second phase of operation. The feed liquor was a synthetic mine water based on the chemical composition of a stream draining the abandoned Mynydd Parys copper mine in north Wales. The arrows indicate the points at which: (a) the glycerol concentration in the feed liquor was lowered from 3 mM to 0.7 mM; (b) the yeast extract concentration was lowered from 0.01% to 0.005% (w/v); (c) the ferrous iron was increased from 5 mM to 10 mM.
) in bioreactor I during the second phase of operation. The feed liquor was a synthetic mine water based on the chemical composition of a stream draining the abandoned Mynydd Parys copper mine in north Wales. The arrows indicate the points at which: (a) the glycerol concentration in the feed liquor was lowered from 3 mM to 0.7 mM; (b) the yeast extract concentration was lowered from 0.01% to 0.005% (w/v); (c) the ferrous iron was increased from 5 mM to 10 mM.
Bioreactor II
Flow rates into and out of bioreactor II were comparable with those of bioreactor I, with a mean value and standard deviation of 57 ± 18 ml h−1 (corresponding to a dilution rate of 0.025 h−1) and maximum and minimum values of 105 and 17 ml h−1 respectively (Fig. 4) from day 0 to day 335. For most of this time, ferrous iron concentrations in the bioreactor were slightly greater than the 3 mM present in the influent liquor (3.2 ± 0.23 mM between days 88 and 335; n = 49 sampling points; Fig. 4), although between days 0 and 84, the mean ferrous iron concentration in the effluent liquor was slightly less than that in the influent (2.8 ± 0.41; n = 18 sampling points), possibly due to the higher pH (4.9 to 4.8) at which the bioreactor was maintained up to day 84 than afterwards (pH 4.5 to 4.0). Concentrations of zinc were below levels of detection in most of the samples analysed (Fig. 4), although significant concentrations of soluble zinc (27% of that in the influent) were measured in the bioreactor shortly after the aluminium content of the feed liquor was increased from 16 to 30 mM. On the four occasions when aluminium concentrations in the bioreactor liquor were determined, these were found to be very similar to those present in the feed used at those times (Fig. 5A) confirming that very little of the aluminium was precipitated within the bioreactor. Analysis of the solid residue that accumulated in bioreactor II confirmed that it was predominantly zinc sulfide.
Figure 4.
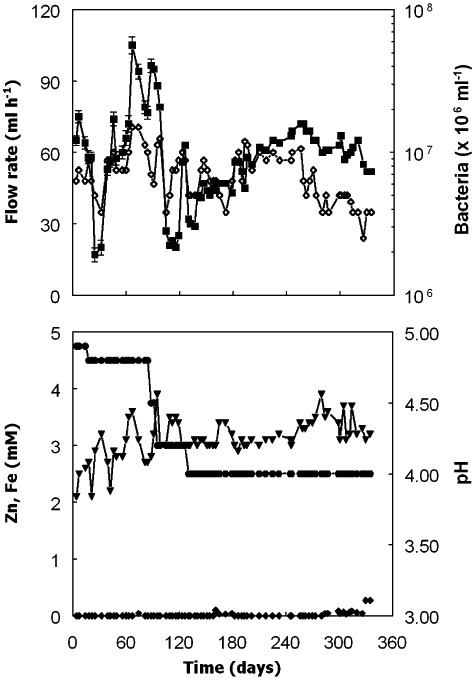
Changes in flow rates ( ), pH (●), concentrations of soluble zinc (◆) and soluble iron (▾), and numbers of planktonic‐phase bacteria (◊) in bioreactor II. The composition of the feed liquor was based on that of an acidic (pH 2.5) stream draining the abandoned Cwm Rheidol mine in mid‐Wales, and contained 3 mM ferrous iron and 3 mM zinc and varying concentrations of aluminium, as major soluble metals.
), pH (●), concentrations of soluble zinc (◆) and soluble iron (▾), and numbers of planktonic‐phase bacteria (◊) in bioreactor II. The composition of the feed liquor was based on that of an acidic (pH 2.5) stream draining the abandoned Cwm Rheidol mine in mid‐Wales, and contained 3 mM ferrous iron and 3 mM zinc and varying concentrations of aluminium, as major soluble metals.
Figure 5.
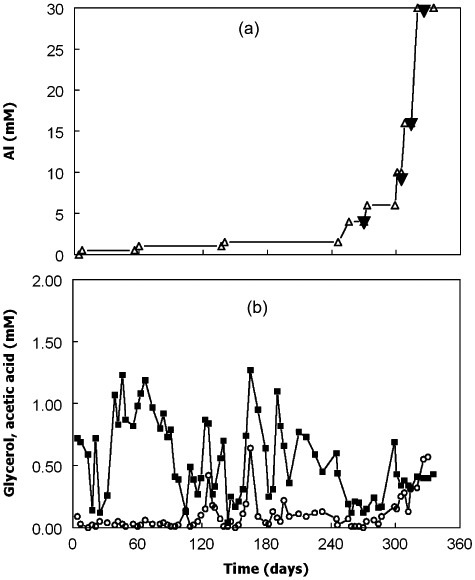
(A) Concentrations of aluminium of the feed liquor (▵) and those determined in bioreactor liquor on four sampling occasions (▴); (B) concentrations of glycerol (○) and acetic acid ( ) in the bioreactor. Concentrations of glycerol in the feed liquor were maintained at 4 mM throughout the experiment.
) in the bioreactor. Concentrations of glycerol in the feed liquor were maintained at 4 mM throughout the experiment.
Bacterial numbers in bioreactor II declined from a mean of 8.1 ± 2.6 × 106 ml−1 from days 0 to 259, when the maximum aluminium concentration in the feed liquor was 1.5 mM, to 4.7 ± 1.2 × 106 ml−1 from days 259 to 335, when aluminium concentrations were increased to up to 30 mM (Fig. 4). Analysis of glycerol in the bioreactor indicated that effective oxidation (98%, mean figure; n = 64 sampling points) of glycerol was occurring from day 0 to day 300 but that this declined to a mean of 92% (n = 8 sampling points) when the aluminium concentration was increased from 6 to 30 mM (Fig. 5B). Acetic acid was detected in the bioreactor throughout the experiment (Fig. 5B), but generally in small concentrations (0.04–1.27 mM; n = 73 sampling points), corresponded to a mean of ∼13% of the glycerol that was oxidized throughout the 335‐day experiment.
Molecular analysis of bioreactor communities
Bacterial 16S rRNA genes were routinely amplified from both bioreactors, and subjected to semi‐quantitative terminal restriction enzyme fragment length polymorphism (T‐RFLP) analysis to assess changes in microbial community structures with varying operating conditions. All attempts at amplifying archaeal 16S rRNA genes were unsuccessful (in contrast to positive controls), indicating that methanogenic prokaryotes were absent in both bioreactors. Results from T‐RFLP analysis (Fig. 6) show that bacterial populations in both bioreactors changed in response to varying operational parameters. In the case of bioreactor I, the dominant terminal restriction fragment (T‐RF) when the reactor was maintained at pH 3.6 corresponded to a novel Firmicute (coded IR2) isolated from this bioreactor (see below). The same T‐RF was present when the pH of bioreactor I was lowered to pH 2.4 and then to 2.2, but in smaller relative abundance (Fig. 6A). T‐RFs corresponding to sulfidogenic bacteria were 138 nt (which accounted for 54% of the total peak area at pH 2.4, but which was detected in a small proportion at pH 3.6 and not detected at lower pH values than 2.4) and 214 nt (which accounted for, at most, 6% of total peak area) in length (HaeIII digests). The 138 nt corresponded to a novel acidophilic SRB (CEB3) which was also isolated from bioreactor I (see below) while the 214 nt restriction fragment is common to both Desulfosporosinus M1 and ‘Desulfobacillus (Db.) acidavidus’. Other T‐RFs identified were 253 nt (corresponding to Acidithiobacillus ferrooxidans), and 231 nt, which was only founded when the reactor I was maintained at pH 3.6, and corresponded to a Gram‐positive Actinobacterium isolate (IR1) isolated from bioreactor II maintained at pH 4.0 (described below). The relative abundance of the T‐RF corresponding to At. ferrooxidans increased as the pH of bioreactor I was lowered, from 6% of total peak area at pH 3.6 to 20% at pH 2.2 (Fig. 6A).
Figure 6.
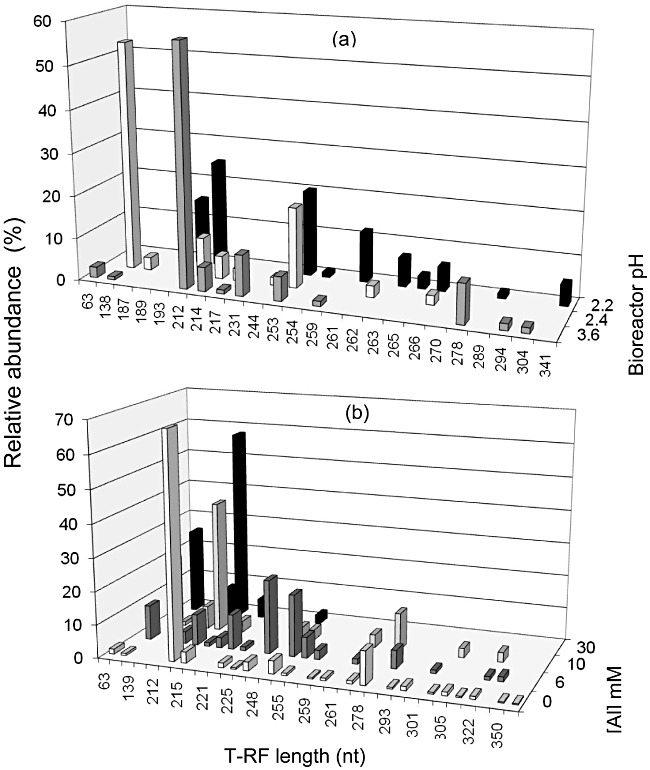
Terminal restriction enzyme fragment length polymorphism (T‐RFLP) analysis of planktonic‐phase bacterial communities: (A) bioreactor I, sampled at different pH values; (B) bioreactor II, sampled at different aluminium concentrations in the feed liquor. Terminal restriction fragments (T‐RFs) are of HaeIII digests of 16S rRNA genes.
In the case of bioreactor II, T‐RFs corresponding to confirmed sulfidogenic bacteria accounted for more of the summated T‐RF peak areas of all four samples analysed than was the case with bioreactor I. The T‐RF corresponding to the novel SRB isolate CEB3 (138 nt) increased progressively in relative peak area from < 1% to 26% as the aluminium concentration in the feed liquor was increased, while that corresponding to Desulfosporosinus M1/‘Db. acidavidus’ (214 nt) was the dominant T‐RF in three of the four samples analysed (Fig. 6B). T‐RFs corresponding to At. ferrooxidans, and to the novel isolates IR1 and IR2, were also found in T‐RFLP profiles of samples from bioreactor II but all three were less abundant than in bioreactor I.
Isolation and phylogenetic analysis of bacteria
The appearance of T‐RFs of sizes that were not registered in the acidophile databank held in the authors' laboratory prompted attempts to isolate the ‘unknown’ bacteria that corresponded to these T‐RFs. Three such isolates were obtained (Table 1). One of these (isolate CEB3) was confirmed to be an acidophilic SRB (sulfide was produced in pH 3.7 growth medium) while sulfidogenesis was not observed with isolates IR1 or IR2. The presence of At. ferrooxidans in the bioreactors inferred from T‐RFLP profiles (T‐RF of 253 nt with HaeIII digests) was confirmed by isolating this facultative anaerobe from the bioreactors on ferrous iron‐containing overlay media (Johnson and Hallberg, 2007).
Table 1.
Some physiological and phylogenetic characteristics of the novel bacteria isolated from bioreactor I and II.
| Isolate | Bioreactor pHa | Major physiological characteristics | Amplification product length (nt) (16S rRNA gene) | GenBank accession No. | T‐RF lengthb (nt) | Nearest relative | Identity (%) |
|---|---|---|---|---|---|---|---|
| CEB3 | 2.4 | Spore‐forming motile rods; obligately anaerobic sulfidogen | 1314 | JF346160 | 138 | Uncultured Desulfitobacterium sp. clone E41 (Winch et al., 2009) | 95.0 |
| IR1 | 4.0 | Rod‐shaped motile cells; spores not observed; facultative anaerobe; non‐sulfidogen | 1303 | JF346161 | 231 | Uncultured Actinobacterium clone (Akob et al., 2008) | 98.0 |
| IR2 | 2.2 | Spore‐forming motile rods; facultative anaerobe; non‐sulfidogen | 1302 | JF346159 | 212 | Uncultured Alicyclobacillus sp. clone (G13_bac) (Winch et al., 2009) | 99.0 |
At the time of isolation.
Digested with the restriction enzyme HaeIII.
Discussion
Microbiological sulfate reduction is of fundamental importance in engineered systems, such as compost bioreactors and permeable reactive barriers, which are used to remediate acidic, metal‐rich streams and groundwaters (Johnson and Hallberg, 2005). Biological sulfidogenesis has three features that contribute towards mitigation of MIWs: (i) it is a proton‐consuming reaction, (ii) many transition metals react with the end‐product (sulfide) to form highly insoluble mineral phases and (iii) it lowers the sulfate concentrations of MIWs. Active mine water bioremediation systems that use SRB technology (e.g. Boonstra et al., 1999) can be highly effective, but their widespread application has been limited by construction and operational costs. The prototype system described in the present work minimizes the latter as: (i) it utilizes a single reactor within which both sulfidogenesis and selective metal precipitation can occur, (ii) control of the system is possible by monitoring and maintaining the pH within the reactor, using low‐cost equipment and empirical technology (a pH electrode and meter, and coupled pump), (iii) selective precipitation facilitates recovery of metals (e.g. copper and zinc) that have commercial value, and recycling these can off‐set net costs (Pott and Mattiasson, 2004), (iv) the electron donor used to fuel the process (glycerol) is relatively inexpensive, and (v) the electron acceptor used (sulfate) is present in all MIWs, and therefore no extraneous electron acceptor is required, as in the ‘BioSulphide’ process where elemental sulfur is used as an electron acceptor (Bratty et al., 2006).
The two bioreactors which ran in parallel in the present study, processing different feed liquors, both achieved their key targets of demonstrating selective precipitation of transition metals. In the early stages of operation, with an influent glycerol concentration of 4–5 mM, both zinc and copper were precipitated (as sulfides) within bioreactor I while ferrous iron remained in solution (Eq. 2):
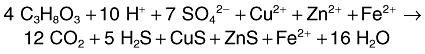 |
2 |
The amount of free hydrogen sulfide produced under such conditions involved a significant consumption of protons, and therefore a relatively large pH increase. To minimize this, and therefore preclude mineralization of ZnS, smaller concentrations of glycerol were required. With a concentration of glycerol in the influent liquor of 0.7 mM, and a bioreactor pH maintained at 2.2, the net reactions that occurred were as depicted in Eq. 3, with only ∼8% of the zinc, > 99.9% of the copper and < 0.1% of the iron in the feed liquor being precipitated:
 |
3 |
In the case of bioreactor II, the feed liquor chemistry was based on water draining an abandoned zinc mine where concentrations of soluble copper were negligible, and the objective here was to produce a ‘clean’ ZnS precipitate. Again, by maintaining an acidic bioreactor pH (though not as low as that of bioreactor I), precipitation of FeS was readily avoided. However, like many other MIWs, the stream draining the Cwm Rheidol mine contains significant amounts of soluble aluminium. Although it does not form a sulfide mineral, aluminium can precipitate as a hydroxide phase (gibbsite) in moderately acidic (pH > 5: Bache, 1986) solutions. Gibbsite forms bulky, gelatinous deposits in such circumstances, and these could cause severe blockage problems in continuous‐flow bioreactors. In the event, segregation of zinc and aluminium was readily achieved by maintaining bioreactor II at pH 4.0–4.5, and supplementing the feed liquor with 4 mM glycerol (Eq. 4):
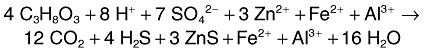 |
4 |
One of the risks with maintaining potentially toxic ions, such as aluminium, in solution is that they may have a negative impact on the SRB that are critical to the remediation process. However, even when the concentration of aluminium in the feed liquor was as high as 30 mM (810 mg l−1; a concentration far greater than that of most MIWs) sulfidogenic activity was maintained in bioreactor II, although increasing concentrations of aluminium caused changes in the bioreactor microbial community, and also resulted in smaller amounts of glycerol being oxidized. Further evidence of the potential of the acidophilic sulfidogenic bioreactors to remediate complex MIWs came from tests with bioreactor I where synthetic Mynydd Parys mine water was used as feed liquor. No precipitation of iron, aluminium or manganese occurred within the reactor, although > 99% of the copper and smaller amounts of zinc were removed.
The key component of the system described is the acidophilic sulfate‐reducing bacterial community. Acidophilic SRB are not well known, although some acid‐tolerant species have been reported (e.g. Johnson, 1995; Hard et al., 1997; Alazard et al., 2010). We included two pure cultures of SRB isolated during earlier studies on the microbial ecology of mine‐impacted environments, in the bioreactor consortium. One of these (Desulfosporosinus M1) was the first acid‐tolerant SRB to be described (Sen and Johnson, 1999) although the proposed species designation (‘Ds. acidophilus’) was also later used for another isolate (Alazard et al., 2010). ‘Desulfobacillus acidavidus’ is a more acidophilic isolate (capable of growth in pure culture in pH 3 liquid media) which was isolated from an anaerobic ‘mat’ community at the Cantareras mine, an enrichment culture of which was also included in the original inoculum used in the pre‐trial bioreactor, and from which the other bacteria detected (and, in some cases, isolated) in the bioreactor liquors would have arisen. A T‐RF restriction fragment that corresponded to both Desulfosporosinus M1 and ‘Db. acidavidus’ dominated the DNA amplified from bioreactor II, but was less abundant in bioreactor I, probably due to the lower pH at which the latter was operated. One of the ‘unknown’ T‐RFs was found to correspond to a novel bacterium (strain CEB3) isolated from bioreactor I, but also detected in bioreactor II. Interestingly, strain CEB3 appeared to be the dominant bacterium present when bioreactor I was operated at pH 2.4, although it was not detected at pH 2.2. It also appeared to became increasingly abundant in bioreactor II when the aluminium concentration in the feed liquor was increased. Preliminary work with a pure culture of isolate CEB3 have confirmed that it is acidophilic, and further characterization of is continuing. The other bacteria detected in bioreactor I, however, are more enigmatic. Sulfidogenesis was confirmed to be a major metabolic process at all pH values at which it was operated, and the stoichiometry of copper and zinc precipitated as sulfides when the feed liquor contained only 0.7 mM glycerol indicated that most of the glycerol oxidized was being coupled to the reduction of sulfate to hydrogen sulfide (Eq. 3). However, on most sampling occasions the dominant T‐RFs obtained of amplified DNA from bioreactor I corresponded to either At. ferrooxidans or a Firmicute related to bacteria of the genus Alicyclobacillus (isolate IR2). Acidithiobacillus ferrooxidans is best known as an iron/sulfur‐oxidizing aerobe, although it can grow anaerobically using ferric iron as electron acceptor, or (in some strains) by sulfur reduction (Pronk et al., 1991; Ohmura et al., 2002). It does not, however, reduce sulfate to hydrogen sulfide, and its presence in both of the sulfidogenic bioreactors was thought due to the fact that the feed liquors were not de‐oxygenated, so At. ferrooxidans was probably using the small amounts of dissolved oxygen present to oxidize ferrous iron to ferric. Oxidized iron would have been rapidly reduced back to ferrous in the prevailing conditions (presence of sulfide, etc.) within the bioreactor. Isolate IR2 was also confirmed to be a facultative anaerobe, although it displayed no propensity to use sulfate as a terminal electron acceptor. This was also the case for other bioreactor isolate (IR1, an Actinobacterium). The roles and significance of isolates IR1 and IR2 in the sulfidogenic bioreactors cannot, at this stage, be ascertained, although they may be involved in syntrophic interactions with the SRB. Kimura and colleagues (2006) reported that Desulfosporosinus M1 and a strain of ‘Acidocella aromatica’ (a non‐sulfidogenic, obligately heterotrophic acidophile) formed a stable syntrophic partnership wherein PFBC metabolized the acetic acid produced by the SRB by its partial oxidation of glycerol, generating molecular hydrogen which was used as a secondary electron donor, by Desulfosporosinus M1.
The sulfidogenic bioreactors described here provide ‘proof of principle’ that segregation of metals present in MIWs is possible in online systems by controlling the pH of the reactors. The modular units themselves are versatile and robust, and can be configured to fit into a variety of design options. For example, a single acidic sulfidogenic bioreactor could be used to recover both copper and zinc, selectively, from mine water, by adding sufficient electron donor (glycerol) to cause hydrogen sulfide to be over‐produced in a pH 4, ZnS‐precipitating bioreactor, the excess being delivered in a gas stream to a vessel containing the ‘raw’ mine water, where pH control could be used to facilitate selective mineralization of copper sulfide.
Experimental procedures
Design and commissioning of the sulfidogenic bioreactors
Two sulfidogenic upflow biofilm reactors, based on a system described previously by Jameson and colleagues (2010), were commissioned and assessed as separate units, each for a period of over 300 days. A pre‐trial bioreactor was set up initially, as shown in Fig. 7, in which bacteria were immobilized on a bed (12.5 cm deep) of 1‐ to 2‐mm‐diameter porous glass beads (Poraver Dennert GmbH, Germany). The sterile beads were inoculated with active pure cultures of Desulfosporosinus M1 (Kimura et al., 2006) and ‘Db. acidavidus’ strain CL4 (Jameson et al., 2010), and an enrichment culture of an anaerobic streamer mat from an acidic (pH 2.5) stream draining of an abandoned copper mine in south‐west Spain (Rowe et al., 2007). The bioreactor vessel had a working volume of 2.3 l and was coupled to a FerMac 310/60 unit (Electrolab., UK) which controlled pH, temperature and agitation. Two pH electrodes (Broadley James, UK) were inserted into the bioreactor vessel: one extended into the porous glass bead bed (and was used only to monitor the pH in this part of the reactor) while the other (shorter) electrode extended only in the liquid phase above the biofilm (bead) bed, and was coupled to the acid‐input pump of the control unit. This pump controlled the flow of feed (influent) liquor into the reactor, which was added at a variable rate, required to maintain the pH of the bioreactor liquor at any set value. Since microbial sulfidogenesis is a proton‐consuming reaction, it was essential that the feed liquor was always at a lower pH than that of the bioreactor liquor in order to maintain pH homeostasis. As indicated in Fig. 7, the feed liquor was delivered via an L‐shaped tube which had perforations in its lower length, causing it to migrate through the biofilm bed before it entered the liquid phase above the beads. A drain tube placed above the liquid surface coupled to a second pump on the control unit that was synchronized with the pH pump ensured that the liquid volume within the bioreactor remained constant. The temperature of the bioreactor was set at 30°C, and the vessel contents were stirred gently (at 50 r.p.m.) using a single impellor blade located within the top liquid phase of the bioreactor, in order not to disrupt the bead bed. A continuous stream of nitrogen (∼200 ml min−1) was used to off‐set any oxygen ingress into the bioreactor.
Figure 7.
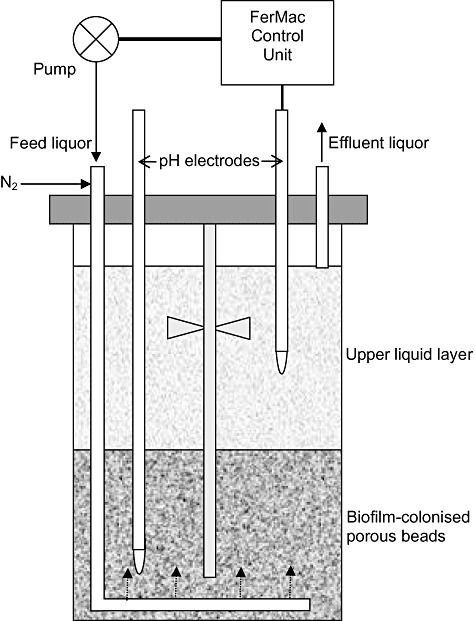
Schematic of the bioreactor vessels used in the present study. The arrows indicated the direction of liquid/gas flow.
The pre‐trial bioreactor was operated at pH varying between 3.5 and 4.5, and was supplied with an influent liquor (pH 2.5) containing 2 mM glycerol (as electron donor), 0.01% (w/v) yeast extract, 2 mM zinc, 1 mM ferrous iron, and basal salts (Wakeman et al., 2008). After 5 months of operation as a continuous‐flow system, the bioreactor was decommissioned, and the biofilm‐containing beads removed and mixed with fresh sterile beads. The bead mixture was split into two equal parts, each of which was placed into a fresh bioreactor vessel, and the two vessels were coupled to two FerMac 310/60 control units (Fig. S2). The two new systems were operated as described for the pre‐trial bioreactor for 1–2 months to allow pre‐conditioning of the units, and then at different operating parameters (described below).
Operation of the sulfidogenic bioreactors
The feed liquor for bioreactor I (Table 2) contained copper, zinc and iron (all at 1 mM), varying concentrations of glycerol (initially 5 mM, decreasing to 0.7 mM), yeast extract (0.01%, w/v) and basal salts. The pH of the feed liquor was set at 2.5 for the day 0–21, and at 2.1 for the next 290 days. In contrast, the bioreactor liquor pH was lowered progressively throughout the experimental period, from a starting value of 3.6 to a final value of 2.2 (Fig. 1). After day 311, the feed liquor for this bioreactor was changed to a synthetic mine water based on the chemistry of a stream draining the abandoned Mynydd Parys copper mine in north Wales (Table 2; Coupland and Johnson, 2004). The main differences between the Mynydd Parys mine water and its synthetic equivalent were the lower pH (2.1, compared with ∼2.5) and more variable ferrous iron concentration (5–10 mM) of the latter. The performance of bioreactor I with a feed liquor of synthetic Mynydd Parys mine water (supplemented with glycerol and yeast extract; Table 1) was tested for 56 days.
Table 2.
Compositions of feed liquors supplied to bioreactors I and II.
| Bioreactor I feed liquor | Bioreactor II feed liquora,b | Synthetic Mynydd Parys mine waterb (bioreactor I) | |
|---|---|---|---|
| pH | 2.5→2.1 | 2.5 | 2.1 |
| Fe2+c | 1.0 | 3.0 | 5.0→10 |
| SO42− | 3.9→6.4 | 12.1→56.4 | 11.7 |
| Cl‐ | 0.27 | Trace | 2.1 |
| H2PO4‐ | 0.15 | Trace | 0.02 |
| Cu2+ | 1.0 | Trace | 0.71 |
| Zn2+ | 1.0 | 3.0 | 1.07 |
| Al3+ | Trace | 0.5→30 | 3.33 |
| Mn2+ | Trace | 0 | 0.22 |
| Na+ | 0.38 | 0.5 | 2.12 |
| NH4+ | 2.72 | Trace | 0.02 |
| K+ | 0.42 | Trace | 0.04 |
| Mg2+ | 0.81 | 2.0 | 0.66 |
| Ca2+ | 0.05 | 0.5 | 1.05 |
| Glycerol | 5.0→0.7 | 4.0 | 3.0→0.7 |
| Yeast extract | 0.01 | 0.01 | 0.01→0.005 |
Based on AMD draining the abandoned Cwm Rheidol zinc mine, mid‐Wales.
Data kindly supplied by Dr Hugh Potter, Environment Agency, UK.
At point of discharge.
All concentrations shown are mM, except yeast extract (w/v), and arrows indicate changes in concentrations during the course of the experiment.
The composition of the feed liquor for bioreactor II was based on mine water draining an abandoned zinc mine (Cwm Rheidol) in mid‐Wales (Table 2; Edwards and Potter, 2007) which contained zinc, iron and aluminium as major soluble metals. During the course of the experiment, aluminium concentrations in the feed liquor increased from 0.5 to 30 mM, while concentrations of other components of the influent solution remained unchanged. The glycerol and yeast extract concentrations in the feed liquor for bioreactor II were maintained at 4 mM and 0.01%, respectively, and the pH at 2.5. The bioreactor pH (upper liquid phase) was set at 4.9 initially and was lowered, in stages, to 4.0 during the course of the 335‐day experiment.
Physicochemical analysis
Concentrations of transition metals, acetate and glycerol were determined by ion chromatography (Wakeman et al., 2008; Ňancucheo and Johnson, 2010). Soluble transition metal were determined using a Dionex‐320 ion chromatograph fitted with an IonPAC® CS5A column and an AD absorbance detector. Acetate concentrations were measured using a Dionex IC25 ion chromatograph with an Ion Pac AS‐11 column equipped with a conductivity detector. Glycerol was determined using a Dionex ICS 3000 ion chromatography system fitted with a Carbo Pac MA1 column and ED amperometric detector. Aluminium was determined by atomic absorption spectrophotometry using a Varian SpectrAA 220 FS. Metal sulfide precipitates taken from both bioreactors at the end of the experiment were characterized by X‐ray diffraction (XRD) analysis. Solid materials were collected by centrifugation, dried in a vacuum desiccator for 48 h and then ground to a powder. Samples were analysed using a Philips PW3040/60 X'Pert PRO and the data analysed using the PANalytical search‐match program ‘Highscore’.
Microbiological and molecular analysis of bioreactor microbial populations
DNA was routinely extracted from liquid samples taken from both bioreactors using Ultraclean Soil DNA extraction kits (MoBio, CA, USA). The extracted DNA (typically 1 µl) served as template for the amplification of the 16S rRNA genes by PCR as described previously (Okibe et al., 2003). The primers used for amplification for bacterial 16S rRNA were 27F (Lane, 1991) and 1387R (Marchesi et al., 1998). Archaeal 16S rRNA genes were amplified using the primers 20F (Orphan et al., 2000) and 1392R (Lane, 1991), with DNA from the euryarcheote Ferroplasma acidiphilum acting as a positive control. Bacterial communities were analysed by T‐RFLP as described previously by Hallberg and colleagues (2006). The labelled DNA that was amplified by PCR in three independent reactions was digested with the restriction enzymes MspI, Cfol and HaeIII. Assignment of T‐RFs to known acidophilic was accomplished by comparison with a database of previously identified microbes detected in acidic environments (Rowe et al., 2007). T‐RFs of bacteria isolated from the bioreactors were determined using the same protocols. Bacteria in liquid samples were enumerated using a Helber counting chamber marked with Thoma ruling (Hawksley, UK), and viewed with a Leitz Labolux phase‐contrast microscope at ×400 magnification.
Isolation and identification of bacteria from the sulfidogenic bioreactors
Bacteria were isolated from the bioreactors at different stages of operation, by streaking liquid samples onto an overlay medium (Johnson and Hallberg, 2007) supplemented with 4 mM glycerol, 0.02% (w/v) yeast extract, 7 mM zinc and 0.5 mM ferrous iron (pH ∼3.7). Plates were incubated in an anaerobic atmosphere (using the AnaeroGen™ system; Oxoid, UK) at 30°C and removed periodically for visual examination. Representative colonies were subcultured on the same solid medium incubated both anaerobically and aerobically (to test for aerobic growth) and also in a liquid medium equivalent of this medium. Phylogenetic analysis of three isolates obtained was carried out using the method described by Rowe and colleagues (2007).
Acknowledgments
We are grateful to Dr Kevin Hallberg for his help with the molecular work described in this article. Ivan Ňancucheo is grateful to Mecesup Programme of the Chilean Government, and Barrie Johnson is grateful to the Royal Society for the provision of an Industrial Fellowship.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Comparison of correlations between numbers of planktonic‐phase bacteria in bioreactor I during the first phase of operation with (A) bioreactor pH and (B) glycerol concentration in the feed liquor.
Bioreactor II culture vessel and FerMac control unit. Inset: image of Poraver beads at the outset of the experiment, showing dark‐coloured colonized beads mixed with new (white‐coloured) non‐colonized beads.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Akob D.M., Mills H.J., Gihring T.M., Kerkhof L., Stucki J.W., Anastacío A.S. Functional diversity and electron donor dependence of microbial populations capable of U(VI) reduction in radionuclide‐contaminated subsurface sediments. Appl Environ Microbiol. 2008;74:3159–3170. doi: 10.1128/AEM.02881-07. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazard D., Joseph M., Battaglia‐Brunet F., Cayol J.‐L., Ollivier B. Desulfosporosinus acidophilus sp. nov.: a moderately acidophilic sulfate‐reducing bacterium isolated from acid mining drainage sediments. Extremophiles. 2010;14:305–312. doi: 10.1007/s00792-010-0309-4. [DOI] [PubMed] [Google Scholar]
- Bache B.W. Aluminium mobilization in soils and waters. J Geol Soc. 1986;143:699–706. [Google Scholar]
- Boonstra J., van Lier R., Janssen G., Dijkman H., Buisman C.J.N. Biological treatment of acid mine drainage. In: Amils R., Ballester A., editors. Elsevier; 1999. pp. 559–567. [Google Scholar]
- Bratty M., Lawrence R.W., Kratochvil D., Marchant P.B. 2006. pp. 271–281. , and ) Applications of biological H2S production from elemental sulfur in the treatment of heavy metal pollution including acid rock drainage. In Proceedings of the 7th International Symposium of Acid Rock Drainage (ICARD). St. Louis, MO, pp.
- Coupland K., Johnson D.B. Geochemistry and microbiology of an impounded subterranean acidic water body at Mynydd Parys, Anglesey, Wales. Geobiology. 2004;2:77–86. [Google Scholar]
- Dinh H.T., Kuever J., Mussmann M., Hassel A.W., Stratmann M., Widdel F. Iron corrosion by novel anaerobic microorganisms. Nature. 2004;427:829–832. doi: 10.1038/nature02321. [DOI] [PubMed] [Google Scholar]
- Edwards P., Potter H. The Cwm Rheidol metal mines remediation project – phase I. In: Cidu R., Frau F., editors. International Mine Water Association; 2007. pp. 181–185. [Google Scholar]
- Hallberg K.B., Coupland K., Kimura S., Johnson D.B. Macroscopic ‘acid streamer’ growths in acidic, metal‐rich mine waters in north Wales consist of novel and remarkably simple bacterial communities. Appl Environ Microbiol. 2006;72:2022–2030. doi: 10.1128/AEM.72.3.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hard B.C., Friedrich S., Babel F.W. Bioremediation of acid mine water using facultatively methylotrophic metal‐tolerant sulfate‐reducing bacteria. Microbiol Res. 1997;152:65–73. [Google Scholar]
- Heinzel E., Janneck E., Glombitza F., Schlömann M., Seifert J. Population dynamics of iron‐oxidizing communities in pilot plants for the treatment of acid mine waters. Environ Sci Technol. 2009;43:6138–6144. doi: 10.1021/es900067d. [DOI] [PubMed] [Google Scholar]
- Jameson E., Rowe O.F., Hallberg K.B., Johnson D.B. Sulfidogenesis and selective precipitation of metals at low pH mediated by Acidithiobacillus spp. and acidophilic sulfate reducing bacteria. Hydrometallurgy. 2010;104:488–493. [Google Scholar]
- Johnson D.B. Acidophilic microbial communities: candidates for bioremediation of acidic mine effluents. Int Biodeterior Biodegradation. 1995;35:41–58. [Google Scholar]
- Johnson D.B., Hallberg K.B. Pitfalls of passive mine drainage. Re/Views. Environ Biotechnol. 2002;1:335–343. [Google Scholar]
- Johnson D.B., Hallberg K.B. The microbiology of acidic mine waters. Res Microbiol. 2003;154:466–473. doi: 10.1016/S0923-2508(03)00114-1. [DOI] [PubMed] [Google Scholar]
- Johnson D.B., Hallberg K.B. Acid mine drainage: remediation options. Sci Total Environ. 2005;338:3–14. doi: 10.1016/j.scitotenv.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Johnson D.B., Hallberg K.B. Techniques for detecting and identifying acidophilic mineral‐oxidising microorganisms. In: Rawlings D.E., Johnson D.B., editors. Springer‐Verlag; 2007. pp. 237–262. [Google Scholar]
- Kimura S., Hallberg K.B., Johnson D.B. Sulfidogenesis in low pH (3.8–4.2) media by a mixed population of acidophilic bacteria. Biodegradation. 2006;17:57–65. doi: 10.1007/s10532-005-3050-4. [DOI] [PubMed] [Google Scholar]
- Koschorreck M. Microbial sulphate reduction at a low pH. FEMS Microbiol Ecol. 2008;64:329–342. doi: 10.1111/j.1574-6941.2008.00482.x. [DOI] [PubMed] [Google Scholar]
- Lane D.J. 16/23s rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. John Wiley and Sons; 1991. pp. 113–175. [Google Scholar]
- Marchesi J.R., Sato T., Weightman A.J., Martin T.A., Fry J.C., Hiom S.J., Wade W.G. Design and evaluation of useful bacterium‐specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ňancucheo I., Johnson D.B. Production of glycolic acid by chemolithotrophic iron‐ and sulfur‐oxidizing bacteria and its role in delineating and sustaining acidophilic sulfide mineral‐oxidizing consortia. Appl Environ Microbiol. 2010;76:461–467. doi: 10.1128/AEM.01832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom D.K. Advances in the hydrogeochemistry and microbiology of acid mine waters. Int Geol Rev. 2000;42:499–515. [Google Scholar]
- Ohmura N., Sasaki K., Matsumoto N., Saiki H. Anaerobic respiration using Fe3+, S0 and H2 in the chemolithotrophic bacterium Acidithiobacillus ferrooxidans. J Bacteriol. 2002;184:2081–2087. doi: 10.1128/JB.184.8.2081-2087.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okibe N., Gericke M., Hallberg K.B., Johnson D.B. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred tank bioleaching operation. Appl Environ Microbiol. 2003;69:1936–1943. doi: 10.1128/AEM.69.4.1936-1943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphan V.J., Taylor L.T., Hafenbradl D., Delong E.F. Culture‐dependent and culture‐independent characterization of microbial assemblages associated with high‐temperature petroleum reservoirs. Appl Environ Microbiol. 2000;66:700–711. doi: 10.1128/aem.66.2.700-711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott B.‐M., Mattiasson B. Separation of heavy metals from water solutions at the laboratory scale. Biotechnol Lett. 2004;26:451–456. doi: 10.1023/b:bile.0000018267.09698.cc. [DOI] [PubMed] [Google Scholar]
- Pronk J.T., Liem K., Bos P., Kuenen J.G. Energy transduction by anaerobic ferric iron reduction in Thiobacillus ferrooxidans. Appl Environ Microbiol. 1991;57:2063–2068. doi: 10.1128/aem.57.7.2063-2068.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe O.F., Sánchez‐España J., Hallberg K.B., Johnson D.B. Microbial communities and geochemical dynamics in an extremely acidic, metal‐rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ Microbiol. 2007;9:1761–1771. doi: 10.1111/j.1462-2920.2007.01294.x. [DOI] [PubMed] [Google Scholar]
- Sen A.M., Johnson D.B. Acidophilic sulphate‐reducing bacteria: candidates for bioremediation of acid mine drainage. In: Amils R., Ballester A., editors. Elsevier; 1999. pp. 709–718. [Google Scholar]
- Senko J.M., Zhang G., McDonough J.T., Bruns M.A., Burgos W.D. Metal reduction at low pH by a Desulfosporosinus species: implications for the biological treatment of acidic mine drainage. Geomicrobiol J. 2009;26:71–82. [Google Scholar]
- Steudel R. The chemical sulfur cycle. In: Lens P., Hulshoff Pol L., editors. International Association on Water Quality; 2000. pp. 1–31. [Google Scholar]
- Wakeman K., Auvinen H., Johnson D.B. Microbiological and geochemical dynamics in simulated heap leaching of a polymetallic sulfide ore. Biotechnol Bioeng. 2008;101:739–750. doi: 10.1002/bit.21951. [DOI] [PubMed] [Google Scholar]
- Winch S., Mills H.J., Kostka J.E., Fortin D., Lean D.R.S. Identification of sulfate‐reducing bacteria in methylmercury contaminated mine tailings by analysis of SSU ribosomal RNA genes. FEMS Microbiol Ecol. 2009;68:94–107. doi: 10.1111/j.1574-6941.2009.00658.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of correlations between numbers of planktonic‐phase bacteria in bioreactor I during the first phase of operation with (A) bioreactor pH and (B) glycerol concentration in the feed liquor.
Bioreactor II culture vessel and FerMac control unit. Inset: image of Poraver beads at the outset of the experiment, showing dark‐coloured colonized beads mixed with new (white‐coloured) non‐colonized beads.


