Summary
An intracellular approach for monitoring protein production in Staphylococcus aureus is described. mCherry, fused to the dodecapeptide Tip, was capable of inducing tetracycline repressor (TetR). Time‐ and concentration‐dependent production of mCherry could be correlated to TetR‐controlled GFPmut2 activity. This approach can potentially be extended to native S. aureus proteins.
Staphylococcus aureus is a pathogenic bacterium causative of numerous human diseases. A profound understanding of S. aureus biology is imperative for finding cures against infections caused by strains recalcitrant to antibiotics. Changes in the transcriptome and proteome patterns have been elucidated for many S. aureus strains and mutants (see Nagarajan and Elasri, 2007; François et al., 2010; Hecker et al., 2010 for recent overviews); however, analysing intracellular protein production of S. aureus, especially during infection, remains challenging. Translational fusions of genes of interest to easily detectable and quantifiable reporters, such as lacZ or gfp, result in a considerable increase of the deduced proteins in molecular mass, which may not only render them less abundant in the cell but potentially also less soluble or impeded in localization or activity (Cubitt et al., 1995; Lesley et al., 2002). In contrast, proteins bearing short peptide appendices, exemplified by the hexa‐histidine‐ or the nine‐amino‐acid encompassing strep‐tag (Hochuli et al., 1988; Schmidt and Skerra, 1994), typically used to facilitate purification, generally provide high chances of retained activity. The dodecapeptide Tip (TetR‐inducing peptide) has been identified as an alternative inducer of tetracycline (Tc) repressor TetR (Klotzsche et al., 2005), an extensively characterized and widely applied regulator in bacteria (Bertram and Hillen, 2008). Notably, in contrast to TetR's conventional low‐molecular‐weight effectors Tc or anhydrotetracycline (ATc) (Degenkolb et al., 1991), TetR(B) from transposon Tn10 is susceptible to Tip, which is not the case for the frequently used TetR(BD) hybrid (Schnappinger et al., 1998; Schubert et al., 2001). This specific interaction and the chemical nature of Tip can be exploited in the context of a genetic read‐out system, which monitors the specific production of a target protein tagged with Tip. To date, the expression of at least six full‐length or truncated genes translationally fused to Tip could be correlated to an increase of lacZ activity subject to TetR control (Klotzsche et al., 2005; Schlicht et al., 2006). Since the Tip system has only been applied in Escherichia coli and Salmonella so far (C. Berens, pers. comm.), we decided to adapt Tip‐dependent TetR induction to S. aureus, a Gram‐positive bacterium of broad general and clinical interest, in a proof‐of‐concept study.
To set‐up strains capable of responding to Tip, a tetR(B) expression cassette was inserted into the chromosomal lip locus of S. aureus SA113 (wt) and its derivative RAB171 (harbouring a TetR‐controlled Pxyl/tet‐gfpmut2 fusion). To this end, tetR(BD) was exchanged against tetR(B) in a pRAB2‐derived vector, which was subsequently integrated as described (Stary et al., 2010). Oligonucleotides, plasmids and bacterial strains are listed in Tables 1 and 2; further details are available on request. Epifluorescence microscopy (DM 5500 B microscope and DFC 360 FX camera, Leica) confirmed that cells of the RAB171‐derived strain RAB201B (Pxyl/tet‐gfpmut2, tetR(B)), but not those of RAB200B (tetR(B)), devoid of a TetR controlled reporter gene, exhibited bright green fluorescence when cultured with a final concentration of 0.4 µM ATc (data not shown). Thus, TetR(B) (henceforth referred to as TetR for convenience) can functionally regulate gene expression in this genomic architecture, as shown for TetR(BD) before (Stary et al., 2010). Subsequent excision of the aphAIII resistance markers of RAB200B and RAB201B by Cre recombinase (Leibig et al., 2008) yielded kanamycin‐sensitive strains RAB210B and RAB211B respectively (Fig. 1A). As a prototype carrier for Tip, the monomeric protein mCherry was chosen (Shaner et al., 2004), which is an established red fluorescent reporter in S. aureus (Malone et al., 2009; Pereira et al., 2010). Intracellular detection and quantification does not require substrates or co‐factors and its structural properties, including solvent exposed N‐ and C‐termini (Shu et al., 2006), make it very suitable for protein fusions. In analogy to comparable fluorescent protein encoding genes, successfully adapted to the codon usage of S. aureus (Sastalla et al., 2009; Paprotka et al., 2010), a new mcherry allele termed gp‐mcherry (Gram‐positive adapted mCherry) was designed (DNA 2.0, Menlo Park, CA, USA). Furnished with the ribosomal binding site of sod (Franke et al., 2007), it was cloned into the xylose inducible vector pKX15. Cloning of further synthetic DNA fragments (Entelechon, Regensburg) into the obtained vector pRAB32‐gp gave rise to plasmids pRAB32‐ntgp and pRAB32‐ctgp, encoding mCherry N‐ or C‐terminally linked to Tip respectively (Fig. 1B). RAB210B and RAB211B cells carrying either one of the three pRAB32 plasmids (Fig. 1A) were cultivated in rich basic medium (BM) (Bera et al., 2005) without glucose and analysed for xylose‐dependent red fluorescence using three different methods. In all strains induced by adding xylose to a final concentration of 0.5% (w/v), cells glowed strongly red, indicating high levels of functional mCherry irrespective of any Tip‐appendix. In contrast, weak but clearly discernible green fluorescence was observed only in induced RAB211B (pRAB32‐ntgp) cells, as depicted in Fig. 2A. The same reporter strains were subjected to flow cytometry (FACSCalibur, BD) and accordingly, a shift towards red fluorescence by about one order of magnitude was observed in all populations of induced cultures. Again, gfpmut2 expression appeared to be solely triggered in the strain disposing of N‐terminally Tip‐tagged mCherry (Fig. 2B). In order to quantify red and green fluorescence, cells harvested at different time‐points during exponential growth phase were analysed using a microplate reader (Infinite M200 Pro, Tecan). Activity profiles of mCherry and GFPmut2 expressed from the reporter strains were subsequently monitored in a time‐resolved and xylose‐dependent manner. As depicted in Fig. 3A, red fluorescence increased steadily over time in xylose‐induced cells, albeit somewhat less pronounced in case of C‐Tip mCherry. This might be due to improper folding or lower protein amounts, one or both of which might also be causative for the inefficiency of C‐Tip mCherry to induce GFPmut2 production. This also agrees with previous results, in which C‐terminally Tip‐tagged TrxA had proven to be a less effective inducer of TetR than N‐tip TrxA in E. coli (Klotzsche et al., 2005). The observed GFPmut2 activity was also consistent with observations from microscopy and flow cytometry, because only cells expressing N‐terminally tagged mCherry, fluoresced both red and green (Fig. 3B). To provide a control for full, rapid and direct induction of TetR, RAB211B was induced by 0.4 µM ATc. In comparison, green fluorescence due to Tip‐mediated induction in RAB211B (pRAB32‐ntgp) was delayed by ∼ 200 min. This might in part be attributed to 15 min maturation time for mCherry (Shaner et al., 2004), but most likely primarily reflects the time span between xylose induction and the production of sufficient amounts of N‐Tip mCherry to efficiently induce TetR. Induced RAB211B (pRAB32‐ntgp) cells reached ∼ 30% of the fluorescence intensity determined for RAB211B with ATc. Indeed, binding constants of Tip to TetR had been established to be about 2–3 orders of magnitude lower than for Tc effectors (Degenkolb et al., 1991; Klotzsche et al., 2005). As indicated by Figs 2 and 3A, the induced reporter strain appears to produce high amounts of mCherry when cultured with 0.5% xylose. According to a study by Zhang and colleagues (2000), a rough estimation indicates the presence of mCherry in a range of ∼ 104 proteins per cell in this state. Reducing the inducer concentrations for RAB211B (pRAB32‐ntgp) resulted in stepwise decreased red and green fluorescence (Fig. 4). Thus, the system appears to be capable of graded responses to different amounts of the Tip‐tagged target protein. Strains with enhanced sensitivity might be required to detect the presence of native S. aureus proteins that are only weakly or moderately produced. Increasing the inducer to TetR ratio is one promising way, since strains with less TetR, controlling the Pxyl/tet promoter upstream of gfpmut2, putatively respond better to lower levels of Tip‐tagged proteins (Klotzsche et al., 2005). To this end, S. aureus strains bearing autoregulated tetR, which generally ensures a balanced level of repressor molecules, might be exploited (Gründling and Schneewind, 2007), optionally combined with improved TetR/Tip pairs (Klotzsche et al., 2005; Klotzsche et al., 2007; Daam et al., 2008). Taken together, proof‐of‐concept for a functional Tip‐tagging architecture in S. aureus was achieved using mCherry as a carrier protein. Based upon our observations that N‐tip mCherry both glowed red and moonlighted as an inducer for TetR, it appears feasible to apply this approach to expression profiling of S. aureus encoded proteins. To this end, tip could be either fused to genes of interest on plasmids for chromosomal integration (Brückner, 1997; Arnaud et al., 2004; Bae and Schneewind, 2006), or it could be randomly attached to chromosomal genes via an integrative element in a transposon‐like fashion, as demonstrated for E. coli before (Schlicht et al., 2006). The Tip‐tagging technique might be particularly useful for in vivo grown S. aureus cells, e.g. in infection models, in which standard proteomic techniques might not be applicable.
Table 1.
Oligonucleotides used in this study.
| Primer | Sequence (5′→3′) | Characteristics and comments |
|---|---|---|
| GPMCHE‐F | GGATCCGAATTCTTAGGAGGATGATTATTTATGGTGAGCAAGGGCGAG | EcoRI, sod ribosomal binding site; cloning of gp‐mcherry into pJL71 |
| GPMCHE‐R | GGATCCGGCGCGCCTTACTTGTACAGCTCGTCCATGC | AscI, cloning of gp‐mcherry into pJL71 |
| gpmCh_fw_BamHI | GATCGGATCCAGGAGGATGATTATTTATGGT | BamHI, cloning of gp‐mcherry into pRAB32‐gp |
| gpmCh_rev_XmaI | CTAGCCCGGGTTACTTGTACAGCTCGTC | XmaI, cloning of gp‐mcherry into pRAB32‐gp |
Endonuclease restriction sites are given in boldface, ribosomal binding sites are italicized and start codons are underlined.
Table 2.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristic(s) | References and comments |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F‐(ϕ80dΔlacZ M15)Δ(lacZYA‐argF) U169hsdR17(r‐M) recA1 endA1 relA1 deoR)λ‐, phoA supE44thi‐1, gyrA96 | Hanahan (1983) |
| S. aureus | ||
| RAB171 | SA113 fdh::Pxyl/tet‐gfpmut2 | Stary et al. (2010) |
| RAB200B | SA113 lip::Pt17‐tetR(B), lox66‐aphAIII‐lox71 | This study |
| RAB201B | SA113 lip::Pt17‐tetR(B), lox72 | This study |
| RAB210B | SA113 lip::Pt17‐tetR(B), lox66‐aphAIII‐lox71, fdh::Pxyl/tet‐gfpmut2 | This study |
| RAB211B | SA113 lip::Pt17‐tetR(B), lox72, fdh::Pxyl/tet‐gfpmut2 | This study |
| RN4220 | NCTC8325‐4 derivative, acceptor of foreign DNA | Iordanescu and Surdeanu (1976) |
| SA113 (ATCC 35556) | NCTC8325 derivative, agr‐, 11 bp deletion in rbsU | Iordanescu and Surdeanu (1976) |
| Plasmids | ||
| pJL71 | pCN54 derivative (Charpentier et al., 2004) bearing gp‐mcherry | This study |
| pKX15 | aphAIII, xylR, PxylA, pTX15 derivative (Peschel et al., 1996) | This study, aphAIII cloned from pDG782 (Guérout‐Fleury et al., 1995) via StuI and Acc65I |
| pRAB2‐B | cat, bla, ‘lip’‐Pt17‐tetR(B)‐lox66‐aphAIII‐lox71‐‘lip’ pBT2 derivative (Brückner, 1997) | This study, tetR(B) cloned from pWH1925‐derivative (Scholz et al., 2004) via XbaI and BstEII |
| pRAB32‐gp/‐ntgp/‐ctgp | pKX15 derivative bearing native, N‐terminally or C‐terminally tip‐tagged gp‐mcherry | This study, gp‐mcherry cloned from pJL71, attachment of tip‐tags by commercially purchased DNA fragments (Entelechon, Regensburg) |
Antibiotics were used, where appropriate, at the following final concentrations: ampicillin 100 µg ml−1, chloramphenicol 10 µg ml−1 and kanamycin 15 µg ml−1.
Figure 1.
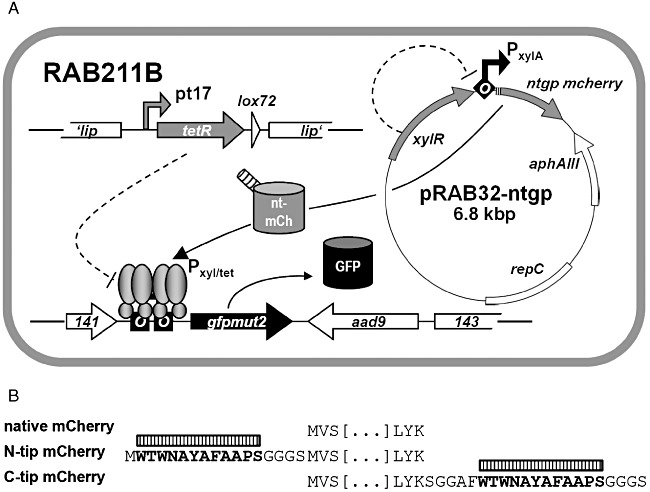
A. Schematic representation of RAB211B (pRAB32‐ntgp). Tip or tip are symbolized by hatched tubes or boxes respectively. Repression is symbolized by dashed lines. Squares or diamonds marked ‘O’ denote TetR‐ or XylR‐binding sites respectively. Promoters are represented by bent arrows (with TetR bound to Pxyl/tet). nt‐mCh: N‐terminally Tip‐tagged mCherry; GFP: GFPmut2. B. Representation of Tip‐tagged or native mCherry proteins' primary structure termini. The Tip dodecapeptide is given in bold and is marked by a hatched box.
Figure 2.
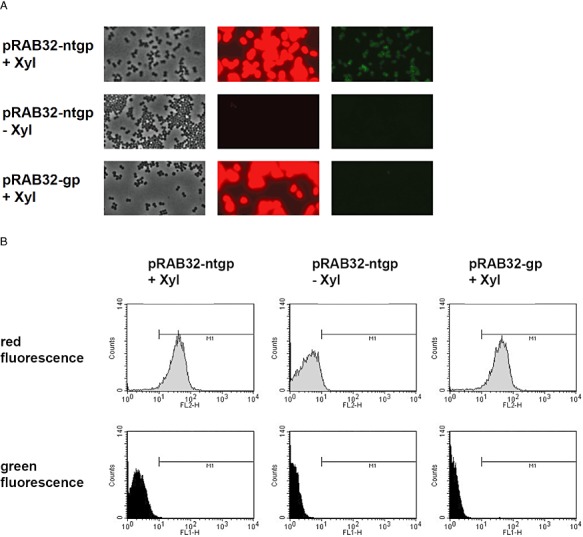
RAB211B cells harbouring vectors and grown under conditions as indicated. A. Microscopic observation of cells 330 min after xylose induction in phase‐contrast (left), using filters for red‐ (middle) or green‐fluorescence (right). B. Flow cytometric analysis of cells carrying pRAB32‐gp or ‐ntgp grown in medium with or without xylose. Upper panels show populations' red fluorescence, lower panels indicate green fluorescence. The M1 threshold was arbitrarily set to 10. It should be noted that GFPmut2 fluorescence is also partially detected in the FL2‐H channel.
Figure 3.
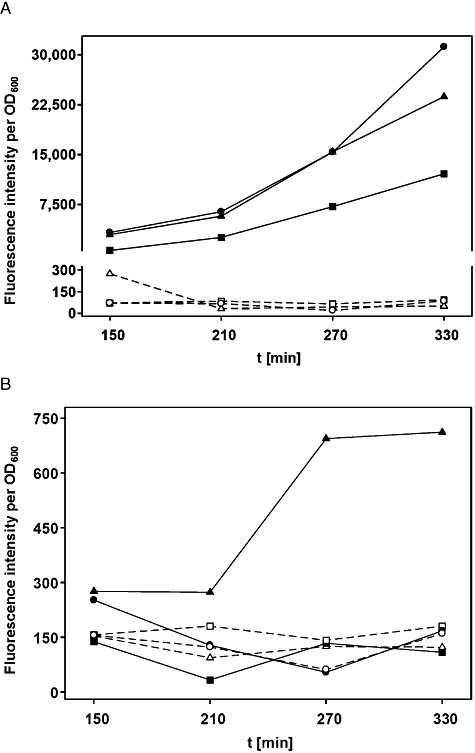
Fluorescence intensities of RAB211B cells bearing pRAB32‐gp (circles), pRAB32‐ntgp (triangles) or pRAB32‐ctgp (squares), (A) red‐, (B) green‐fluorescence. Open symbols (connected by dashed lines) represent growth under non‐induced conditions and filled symbols denote cells induced with xylose. Fluorescence values were correlated to cell densities (OD600).
Figure 4.
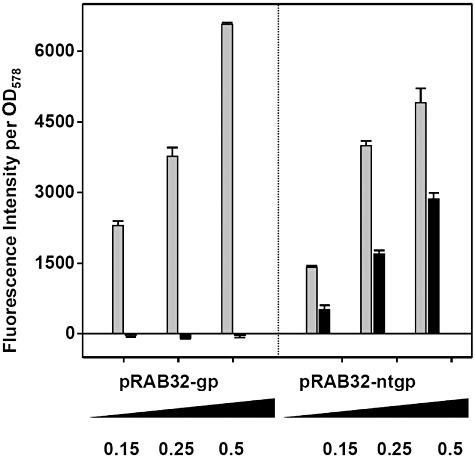
Influence of different xylose concentrations on red‐ (grey bars) and green‐fluorescence (black bars) of RAB211B cells bearing vectors as indicated. Fluorescence values were correlated to cell densities (OD578) and basal fluorescence from non‐induced cells was subtracted from values obtained with 0.15%, 0.25% and 0.5% xylose.
We like to thank Chris Berens for helpful comments, fruitful discussions and critically reading the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft through grants BE4038/1‐1 and BE4038/2‐1 within the DFG priority programme 1316 to Ralph Bertram and by a research fellowship LI1828/2‐1 to Jan Liese.
References
- Arnaud M., Chastanet A., Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low‐GC‐content, gram‐positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T., Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter‐selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Bera A., Herbert S., Jakob A., Vollmer W., Götz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O‐acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- Bertram R., Hillen W. The application of Tet repressor in prokaryotic gene regulation and expression. Microb Biotechnol. 2008;1:2–16. doi: 10.1111/j.1751-7915.2007.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151:1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x. [DOI] [PubMed] [Google Scholar]
- Charpentier E., Anton A.I., Barry P., Alfonso B., Fang Y., Novick R.P. Novel cassette‐based shuttle vector system for gram‐positive bacteria. Appl Environ Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubitt A.B., Heim R., Adams S.R., Boyd A.E., Gross L.A., Tsien R.Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- Daam J., Mehdaoui K., Klotzsche M., Pfleiderer K., Berens C., Hillen W. Functionally important residues of the Tet repressor inducing peptide TIP determined by a complete mutational analysis. Gene. 2008;423:201–206. doi: 10.1016/j.gene.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Degenkolb J., Takahashi M., Ellestad G.A., Hillen W. Structural requirements of tetracycline‐Tet repressor interaction: determination of equilibrium binding constants for tetracycline analogs with the Tet repressor. Antimicrob Agents Chemother. 1991;35:1591–1595. doi: 10.1128/aac.35.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François P., Scherl A., Hochstrasser D., Schrenzel J. Proteomic approaches to study Staphylococcus aureus pathogenesis. J Proteomics. 2010;73:701–708. doi: 10.1016/j.jprot.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Franke G.C., Dobinsky S., Mack D., Wang C.J., Sobottka I., Christner M. Expression and functional characterization of gfpmut3.1 and its unstable variants in Staphylococcus epidermidis. J Microbiol Methods. 2007;71:123–132. doi: 10.1016/j.mimet.2007.08.015. et al. [DOI] [PubMed] [Google Scholar]
- Gründling A., Schneewind O. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J Bacteriol. 2007;189:2521–2530. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout‐Fleury A.M., Shazand K., Frandsen N., Stragier P. Antibiotic‐resistance cassettes for Bacillus subtilis. Gene. 1995;167:335–336. doi: 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hecker M., Becher D., Fuchs S., Engelmann S. A proteomic view of cell physiology and virulence of Staphylococcus aureus. Int J Med Microbiol. 2010;300:76–87. doi: 10.1016/j.ijmm.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Hochuli E., Bannwarth W., Döbeli H., Gentz R., Stüber D. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Nat Biotechnol. 1988;6:1321–1325. [Google Scholar]
- Iordanescu S., Surdeanu M. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J Gen Microbiol. 1976;96:277–281. doi: 10.1099/00221287-96-2-277. [DOI] [PubMed] [Google Scholar]
- Klotzsche M., Berens C., Hillen W. A peptide triggers allostery in tet repressor by binding to a unique site. J Biol Chem. 2005;280:24591–24599. doi: 10.1074/jbc.M501872200. [DOI] [PubMed] [Google Scholar]
- Klotzsche M., Goeke D., Berens C., Hillen W. Efficient and exclusive induction of Tet repressor by the oligopeptide Tip results from co‐variation of their interaction site. Nucleic Acids Res. 2007;35:3945–3952. doi: 10.1093/nar/gkm357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibig M., Krismer B., Kolb M., Friede A., Götz F., Bertram R. Marker removal in staphylococci via Cre recombinase and different lox sites. Appl Environ Microbiol. 2008;74:1316–1323. doi: 10.1128/AEM.02424-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley S.A., Graziano J., Cho C.Y., Knuth M.W., Klock H.E. Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng. 2002;15:153–160. doi: 10.1093/protein/15.2.153. [DOI] [PubMed] [Google Scholar]
- Malone C.L., Boles B.R., Lauderdale K.J., Thoendel M., Kavanaugh J.S., Horswill A.R. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods. 2009;77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan V., Elasri M.O. SAMMD: Staphylococcus aureus microarray meta‐database. BMC Genomics. 2007;8:351. doi: 10.1186/1471-2164-8-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paprotka K., Giese B., Fraunholz M.J. Codon‐improved fluorescent proteins in investigation of Staphylococcus aureus host pathogen interactions. J Microbiol Methods. 2010;83:82–86. doi: 10.1016/j.mimet.2010.07.022. [DOI] [PubMed] [Google Scholar]
- Pereira P.M., Veiga H., Jorge A.M., Pinho M.G. Fluorescent reporters for studies of cellular localization of proteins in Staphylococcus aureus. Appl Environ Microbiol. 2010;76:4346–4353. doi: 10.1128/AEM.00359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A., Ottenwälder B., Götz F. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol Lett. 1996;137:279–284. doi: 10.1111/j.1574-6968.1996.tb08119.x. [DOI] [PubMed] [Google Scholar]
- Sastalla I., Chim K., Cheung G.Y., Pomerantsev A.P., Leppla S.H. Codon‐optimized fluorescent proteins designed for expression in low‐GC gram‐positive bacteria. Appl Environ Microbiol. 2009;75:2099–2110. doi: 10.1128/AEM.02066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicht M., Berens C., Daam J., Hillen W. Random insertion of a TetR‐inducing peptide tag into Escherichia coli proteins allows analysis of protein levels by induction of reporter gene expression. Appl Environ Microbiol. 2006;72:5637–5642. doi: 10.1128/AEM.00739-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T.G., Skerra A. One‐step affinity purification of bacterially produced proteins by means of the ‘Strep tag’ and immobilized recombinant core streptavidin. J Chromatogr A. 1994;676:337–345. doi: 10.1016/0021-9673(94)80434-6. [DOI] [PubMed] [Google Scholar]
- Schnappinger D., Schubert P., Pfleiderer K., Hillen W. Determinants of protein‐protein recognition by four helix bundles: changing the dimerization specificity of Tet repressor. EMBO J. 1998;17:535–543. doi: 10.1093/emboj/17.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz O., Henssler E.M., Bail J., Schubert P., Bogdanska‐Urbaniak J., Sopp S. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol Microbiol. 2004;53:777–789. doi: 10.1111/j.1365-2958.2004.04159.x. et al. [DOI] [PubMed] [Google Scholar]
- Schubert P., Schnappinger D., Pfleiderer K., Hillen W. Identification of a stability determinant on the edge of the Tet repressor four‐helix bundle dimerization motif. Biochemistry. 2001;40:3257–3263. doi: 10.1021/bi001927e. [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shu X., Shaner N.C., Yarbrough C.A., Tsien R.Y., Remington S.J. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- Stary E., Gaupp R., Lechner S., Leibig M., Tichy E., Kolb M., Bertram R. New architectures for Tet‐on and Tet‐off regulation in Staphylococcus aureus. Appl Environ Microbiol. 2010;76:680–687. doi: 10.1128/AEM.02416-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fan F., Palmer L.M., Lonetto M.A., Petit C., Voelker L.L. Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene. 2000;255:297–305. doi: 10.1016/s0378-1119(00)00325-5. et al. [DOI] [PubMed] [Google Scholar]


