Summary
Biotechnical production processes often operate with plasmid‐based expression systems in well‐established prokaryotic and eukaryotic hosts such as Escherichia coli or Saccharomyces cerevisiae, respectively. Genetically engineered organisms produce important chemicals, biopolymers, biofuels and high‐value proteins like insulin. In those bioprocesses plasmids in recombinant hosts have an essential impact on productivity. Plasmid‐free cells lead to losses in the entire product recovery and decrease the profitability of the whole process. Use of antibiotics in industrial fermentations is not an applicable option to maintain plasmid stability. Especially in pharmaceutical or GMP‐based fermentation processes, deployed antibiotics must be inactivated and removed. Several plasmid addiction systems (PAS) were described in the literature. However, not every system has reached a full applicable state. This review compares most known addiction systems and is focusing on biotechnical applications.
Introduction
Fermentations of microorganisms in biotechnological production processes often depend on foreign genetic information located on plasmids, e.g. the production of human insulin (Johnson, 1983). Further industrial relevant plasmid‐based processes for the production of, for example, amylases, lipases, proteases, vitamins or antibiotics were described (Vary et al., 2007). Plasmids are separate genetic elements and autonomously replicated from the chromosomes. Furthermore, several fields of research utilize plasmids as important and essential tools. The success of plasmid‐based microbial production systems significantly depends on plasmid stability, copy number and choice of suitable promotors. Plasmids are frequently used to introduce the formation of additional proteins or even for the establishment of complete metabolic pathways. Actual experiments in the fields of gene therapy and genetic vaccination using naked plasmid DNA as therapeutic vectors were successful. For these reasons plasmids became an important medical and economical product (Friehs, 2004). The easy transfer into a new host and the occurrence in multiple copies in the cells provides advantages. Frequently used cloning and expression plasmids harbour antibiotic resistance genes and require the addition of antibiotics to the cultivation medium for plasmid maintenance. Whereas this procedure is feasible at the laboratory scale, it is not applicable at large‐scale cultivations in industry due to the high costs and due to ecological constraints. Moreover, plasmid instability can even occur despite the addition of antibiotic during cultivation (Zabriskie and Arcuri, 1986). Chromosomal gene integration is another strategy to stabilize foreign genes; it is mostly associated with single‐copy insertions which often cause lower levels of recombinant protein or negative polar effects in contrast to the use of (multi‐copy) plasmids.
Most naturally occurring plasmids as well as most constructed plasmids do not encode information required for viability or survival of the host cell. They often provide advantages to the host under specific environmental conditions such as utilizing certain chemical compounds or providing resistances towards adverse substances (Nordstrom and Austin, 1989). Plasmids may exert a metabolic burden to the host in comparison to plasmid‐free cells (Zielenkiewicz and Cegłowski, 2001). Diverse mechanisms have been evolved to ensure stable maintenance of plasmids in cells (Nordstrom and Austin, 1989): (i) site‐specific recombination systems function as plasmid maintenance systems for most high‐copy plasmids (Alonso et al., 1996; Grindley et al., 2006), (ii) active partition systems, essential for distribution of plasmids with moderate or low copy number, consisting of two proteins and a centromere‐like site actively segregating plasmids to progeny cells during cell division (Gerdes et al., 2000; Funnell and Slavcev, 2004), and (iii) plasmid addiction systems (PAS) preventing the survival of plasmid‐free cells due to selective killing (Zielenkiewicz and Cegłowski, 2001;Gerdes et al., 2005).
The present review is divided into three major parts. The first section ‘Variants of PAS and their functioning’ is a synopsis of current PAS. It depicts several examples and classifies the PAS related to their functionality. The following section ‘PAS and their applications’ focuses on applied PAS with biotechnical background. Here a couple of approved examples are displayed and discussed in more detail. Finally, the section ‘Conclusion and outlook: applications and perspectives of PAS in biotechnology’ summarizes the entire topic and provides comments on possible future developments of PAS.
Variants of PAS and their functioning
This section provides an overview of three major groups of PAS according to their principle of function: (i) toxin/antitoxin (TA)‐based systems, (ii) metabolism‐based systems and (iii) operator repressor titration (ORT) systems (Fig. 1). It must be emphasized that most of the presented PAS occur naturally (see the host organisms in Tables 1–3) and were modified for special applications or adapted to extend the range of hosts. Exceptions occur in the metabolism‐based and ORT systems which are completely synthetic engineered PAS.
Figure 1.
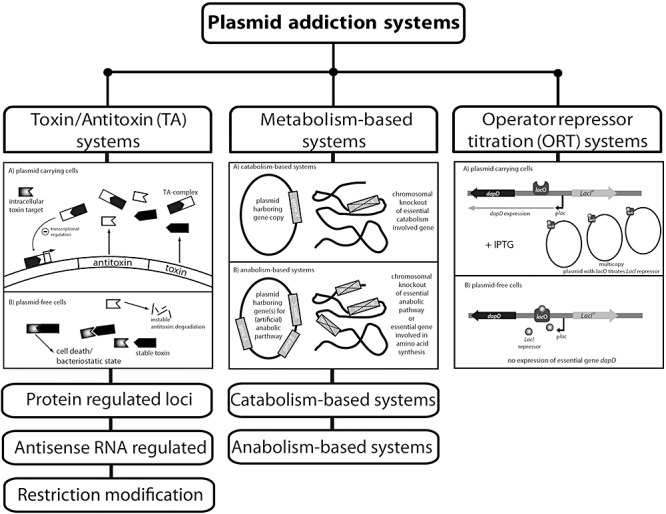
Overview and classification of the most common plasmid addiction systems (PAS) depending on their functionality and principle of mechanism. Toxin/antitoxin (TA) systems contain three separate subgroups with protein regulated loci, antisense RNA‐regulated mechanism or specific restriction modification. The general function of this group is illustrated above by the mechanism of TA systems. Metabolism‐based systems are divided into catabolism and anabolism‐based systems depending on their target pathway and complementation principle. The operator repressor titration (ORT) system is emphasized as a stand‐alone group.
Table 1.
Overview of toxin/antitoxin‐based addiction systems that were described in literature.
| Name of locus | Toxin/antitoxin | Organism | References |
|---|---|---|---|
| hok/sok | Hok/Sok | E. coli | Gerdes et al. (1986a,b); Gerdes (1988); Thisted and Gerdes (1992); Pedersen and Gerdes (1999); Kobayashi (2004) |
| par | RNA I/RNA II | E. faecalis | Weaver and Tritle (1994); Weaver et al. (1996; 2004); Greenfield et al. (2000) |
| ldrD–rdlD | LdrD/RdlD | E. coli | Kawano et al. (2002) |
| ratA–txpA | RatA/TxpA | B. subtilis | Silvaggi et al. (2005) |
| symER | SymR/SymE | E. coli | Kawano et al. (2007) |
Table 3.
Overview of restriction–modification (RM) systems.
| Name of locus | Toxin/antitoxin | Organism | References |
|---|---|---|---|
| ecoRI | EcoRI/M.EcoRI | E. coli | Hedgpeth et al. (1972); Kulakauskas et al. (1995) |
| ecoRII | EcoRII/M.EcoRII | E. coli | Som et al. (1987); Som and Friedman (1993); Karyagina et al. (1997); Takahashi et al. (2002); Ohno et al. (2008) |
| ecoRV | EcoRV/M.EcoRV | E. coli | Nakayama and Kobayashi (1998) |
| ssoII | SsoII/M.SsoII | Shigella sonnei | Karyagina et al. (1997); Kobayashi (2004) |
| paeR71 | PaeR71/M.PaeR71 | P. aeroginosa | Wilson and Murray (1991); Kusano et al. (1995); Kobayashi (2004) |
| pvuII | PvuII/M.PvuII | P. vulgaris | Tao et al. (1991); Cheng et al. (1994); Calvin‐Koons and Blumenthal (1995); Gong et al. (1997) |
| bsp6I | Bsp6I/M.Bsp6I | Bacillus sp. RFL6I | Kulakauskas et al. (1995); Lubys and Janulaitis (1995) |
| haeII | HaeII/M.HaeII | Haemophilus aegyptius | Tu et al. (1976) |
| dcm–vsr | None/Dcm | E. coli | Lieb (1991); Palmer and Marinus (1994); Takahashi et al. (2002); Bunting et al. (2003) |
Table displays the host origin, involved toxin/antitoxin as described in literature.
TA‐based systems
The principle of TA systems is the counteraction of two proteins from which one is a stable toxin and the other is an unstable antitoxin. Mainly, three different groups of TA systems can be distinguished: (i) antisense RNA‐regulated loci, (ii) protein‐regulated loci and (iii) restriction modification systems (Fig. 1).
In TA systems the antitoxin is usually encoded upstream of the toxin gene (Fig. 1). By binding to each other, the antidote neutralizes the toxin and prevents the organism from cell death. In the case of plasmid loss, the durable toxin is still active, whereas the unstable antidote is degraded rapidly. Thus, the persistent toxin exerts its toxic effect and leads to cell death of the segregant. This mechanism favours stable maintenance of plasmids. The TA systems of antisense RNA‐regulated loci have a toxin and antitoxin as antagonistic elements partially on the RNA level. Those mRNA‐sequences bind to their target and induce cell death. The antidote is either RNA or a protein, and it inhibits the translation of the toxin RNA and therefore neutralizes toxic effects (Jensen and Gerdes, 1995). In the subgroup of TA systems based on protein‐regulated loci, the two antagonists are proteins. The group of so‐called restriction modification systems differs from the two other groups in that the toxin is a restriction enzyme which cleaves the host DNA at special nucleotide sequences and thus kills the cell. The corresponding antitoxin is a modification enzyme that methylates the target sequence thereby protecting the DNA from cleavage (Kobayashi, 2004).
In 2007, a web‐based tool was developed, ‘Rapid Automated Scan for Toxins and Antitoxins in Bacteria’ (‘RASTA‐Bacteria’, accessible at http://genoweb.univ‐rennes1.fr/duals/RASTA‐Bacteria), to improve and relieve the identification of putative TA loci in prokaryotic genomes (Sevin and Barloy‐Hubler, 2007). The software ‘RASTA‐Bacteria’ annotates toxin and antitoxin genes based on general characteristics: (i) TA systems consist of, at least, two genes, (ii) with a size of a few hundreds nucleotides each and (iii) 1–20 overlapping nucleotides in general, (iv) with the antitoxin usually being located upstream of the toxin and (v) with the antitoxin gene shorter than the toxin gene. Although restriction–modification systems belong also to TA systems, they were excluded from the study as well as the ω‐ε‐ζ TA family with its three components due to their specific characteristics.
‘RASTA‐Bacteria’ confirmed all, except two, TA loci that were previously predicted by Pandey and Gerdes (2005), whereby the exceptions were not detected because of their low homology or low confidence score. ‘RASTA‐Bacteria’ was able to identify new loci in all studied genomes except in the obligate intracellular organisms without TA systems and in Bacillus spp., which harbour a single TA locus that was known before. Among the 532 TA loci predicted in this study, the major family was vapBC comprising 37% of the members, whereas five other families (mazEF, relBE, higBA, hipBA, parDE) each comprised only about 10% of the loci. In contrast, phd/doc and ccdAB were less frequently detected (3% and 0.009%, respectively); the other TA loci remained unclassified. ‘RASTA‐Bacteria’ was also able to analyse eukaryotic genomes, although it was not designed for that task. However, further improvements are necessary, since not all TA families have been included, yet, and the predicted TA loci have to be validated in detailed characterization studies. The currently known TA systems are summarized in Table 1.
Antisense RNA‐regulated systems. These systems consist of a toxin and antitoxin; both are in most cases RNA molecules. The toxin is encoded by a stable mRNA which upon expression forms a protein that may kill the cell if it is present in high concentrations. In antisense RNA‐regulated systems, a small, unstable transcript that functions as antidote binds to the toxin‐encoding RNA and inhibits its translation (Gerdes et al., 1997).
The most intensively studied example is the hok/sok system from Escherichia coli (Kobayashi, 2004). This system stabilizes plasmid R1 in E. coli and is encoded in the parB region. The hok/sok locus of plasmid R1 from E. coli consists of three genes and is one of the best‐characterized antisense RNA‐regulated TA loci (Gerdes et al., 1997). The hok gene encodes a toxic protein that leads to an irreversible damage of the cell membrane potential (Gerdes et al., 1986a,b). As a result the cell is rapidly killed and shows a characteristic ‘ghost morphology’. Expression and translation of hok requires the mok gene; the translation of the latter is blocked by the sok reading frame encoding an unstable antisense RNA (Thisted and Gerdes, 1992). Sok‐RNA binds to hok mRNA, and the resulting hok mRNA:Sok‐RNA duplex is cleaved by RNase III (Gerdes et al., 1992).
Another system based on the unique par locus, which was found in Enterococcus faecalis, is so far the only post‐segregational‐killing system detected in Gram‐positive bacteria. The par locus consists of two genes which encode the 210‐nucleotide antisense RNA, RNA II and the 63‐nucleotide toxin‐encoding RNA I (fst) (Greenfield et al., 2000). Because their sequences are complementary to each other, RNA II is capable of regulating the transcription of RNA I and suppresses fst translation (Weaver et al., 1996). The par toxin, Fst, is activated by removal of RNA II when the plasmid is lost (Weaver et al., 2004). Figure 2 describes the par system as a representative of the antisense RNA‐regulated systems.
Figure 2.
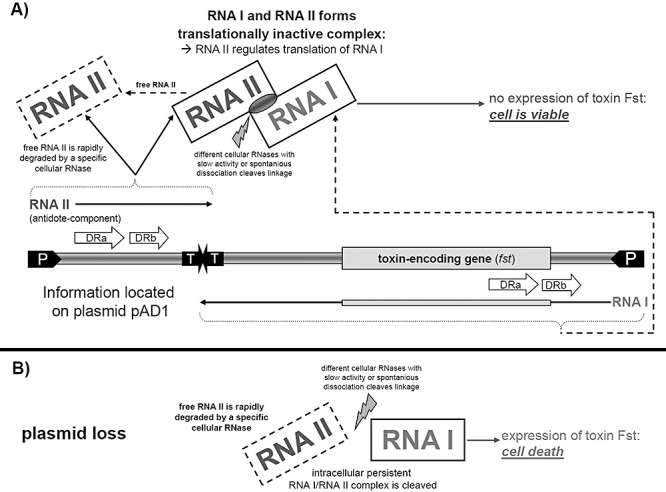
Principle and scheme of an antisense RNA‐regulated toxin–antitoxin plasmid addiction system based on the par locus of Enterococcus faecalis. The natural occurring par locus contains the RNA I and RNA II encoding sequence in series on plasmid pAD1. The complete in vivo regulation and function of the native par locus is not known in detail, yet. Plasmid loss leads to cell death because of the persistence of the RNA I–RNA II complex and subsequent toxin expression after removal and degradation of the RNA II molecule (Weaver et al., 2004). A. Plasmid carrying cells encode the RNA II fragment which acts as the antitoxin being an antisense RNA that inhibits translation of the toxin message by forming a complex with the chromosomally encoded RNA I. Downstream of the promoter (P) both sequences encoding RNA I and RNA II transcripts contain direct repeats (DRa; DRb) with a bidirectional rho‐independent transcription terminator (TT). They provide the complementary regions required for interaction of RNA I and RNA II, because transcription occurs in opposite directions across DRa and DRb. While RNA I and RNA II form a translationally inactive complex, the toxic gene product Fst does not appear, and the cells are viable. The RNA I/RNA II complex is cleaved either by an RNase or by spontaneous dissociation of the two components resulting in the release of RNA I. Since there is a counteraction by the acquisition of the RNA I/RNA II complex, cells still hold a viable state. B. In the case of plasmid loss the open reading frame encoding the toxin on the persistent RNA I is translated by the ribosomes and the toxin Fst leads to cell death by disrupting the cell membrane.
The E. coli ldrD–rdlD locus in is a third example of an antisense RNA‐regulated locus. Here, the ldrD gene encodes a peptide that kills the host cell if overexpressed. The antitoxin, RdlD, functions as a trans‐acting regulator of ldrD translation (Kawano et al., 2002).
The ratA–txpA system occurs in Bacillus subtilis. The accumulation of the toxin, a peptide encoded by the txpA gene, is blocked by the antitoxin, an antisense RNA encoded by ratA gene (Silvaggi et al., 2005).
The last example of antisense RNA‐regulated addiction systems is the symER locus, where the toxin SymR is repressed by the RNA‐molecule SymE (Kawano et al., 2007). SymE synthesis is not only inhibited at the level of translation, but also at the level of transcription or protein by the LexA repressor and Lon protease. An important difference to all other TA systems is that the toxin is not rapidly degraded. One part of the two molecules that might be attacked by the Lon protease is the toxin, and not the antitoxin (Kawano et al., 2007). This system has not yet been analysed in more detail.
Protein‐regulated systems. In protein‐regulated systems (PRS) two proteins are encoded by two adjacent coexpressed genes encoding the stable toxin and the metastable antitoxin. In most PRS the antitoxin gene is located upstream of the toxin gene (Jensen and Gerdes, 1995). Protein‐encoded addiction systems are mostly organized in operons and are autoregulated with the antitoxin or the TA complex repressing the promoter of this operon (Tam and Kline, 1989a,b, see Fig. 1). The accumulation of the toxic protein in plasmid‐free cells will lead to cell death. There are at present several examples known which all function according to the same principle (see Table 2). Whereas the functions and genetic structures of most PRS are similar, they differ slightly in their sequence. In their location PRS may partially chromosally or completely extrachromosomally located. Further variables are the target of the toxin and the mechanism by which the antitoxin is degraded (Engelberg‐Kulka and Glaser, 1999). Whereas the role of plasmid located PRS as plasmid‐stabilization systems was demonstrated by several studies, the biological role of chromosomally located TA loci is subject of debate. Hitherto the function of chromosomal located systems is not completely investigated. Magnuson (2007) discussed several possible functions ranging from ‘genomic junk’ over ‘programmed cell arrest’ to ‘growth control’. Others proposed that chromosomal PRS induce programmed cell death (Engelberg‐Kulka et al., 2005; Kolodkin‐Gal and Engelberg‐Kulka, 2006), while Gerdes and others suggested that these PAS only inhibit cell growth (Pedersen et al., 2002; Buts et al., 2005; Gerdes et al., 2005). These contrary views probably reflect differences in the activities of TA‐encoded toxins. For example, cell death caused by mazEF in E. coli has been observed as a response to stress conditions (Sat et al., 2001; 2003; Hazan et al., 2004; Kolodkin‐Gal and Engelberg‐Kulka, 2006), whereas induction of relBE (and in some cases mazEF) did not affect cell viability (Christensen and Gerdes, 2003; Christensen et al., 2003). An outstanding example is the MqsR/MqsA system which regulates more than its own locus and is implicated in the role of cell‐persister formation (Kim and Wood, 2009;Kim et al., 2009). Furthermore, some PRS are associated to biofilm formation (Kim et al., 2009; Kolodkin‐Gal et al., 2009). A comprehensive overview of PRS with continuative literature is given in Table 2.
Table 2.
Overview of addiction systems with protein‐regulated loci displaying the host origin, involved toxin/antitoxin as described in literature.
Restriction/modification‐based systems. In restriction/modification (RM)‐based systems a restriction endonuclease functions as the toxin whereas a modification enzyme serves as the antitoxin. Therefore, it typically consists of two genes. One gene encodes a DNA endonuclease (ENase). The other gene encodes a DNA methyltransferase (MTase) which prevents DNA cleavage by transferring a methyl group specifically to a base and inhibits cleavage at the respective site (Handa and Kobayashi, 1999; Handa et al., 2000; Ichige and Kobayashi, 2005). RM addiction systems act as plasmid‐stabilization systems (Kulakauskas et al., 1995; Nakayama and Kobayashi, 1998) in that way that the MTase‐antitoxin protects the cellular target of the ENase‐toxin. (Kobayashi, 2004). This variant of addiction system was not investigated in detail, yet. It seems to be different to the two other TA addiction systems described above regarding the regulatory mechanism of post‐segregational killing: the toxic effect is conferred by the different stabilities of the two involved protein components in the PAS and in antisense RNA systems, whereas in the RM systems, the dilution of both proteins by cell growth activates the lethal activity of the restriction enzyme (Ichige and Kobayashi, 2005). Upon plasmid loss and further cell growth, the ENase‐toxin and MTase‐antitoxin are both ‘diluted’ and are proteolytically degraded in the daughter cells. Furthermore, the amount of the MTase target is simultaneously increasing. The intracellular concentration of the RM enzyme decreases then to a level at which not enough methylase activity remains to modify all DNA sites. Consequently, non‐methylated sites appear and DNA cleavage might lead to cell death (Kulakauskas et al., 1995). RM systems have been reviewed in detail (Kobayashi, 2004). A scheme and a short overview are given in Fig. 3 and Table 3.
Figure 3.
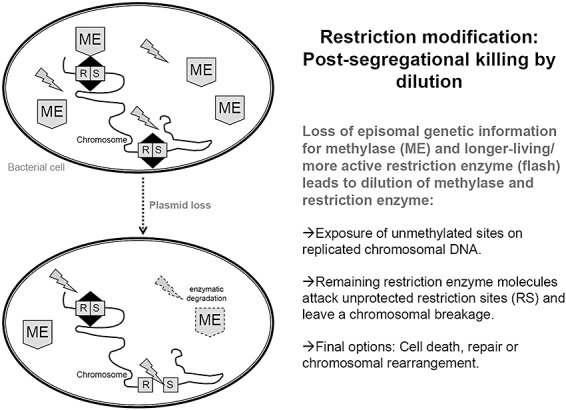
Scheme of the restriction modification‐based post‐segregational killing principle by dilution of episomal coded genetic information for methylase and restriction enzyme. Due to the reduced activity or proteolytic degradation of the specific DNA protecting methylase (ME), the persistent or more active restriction enzyme (flash) cleaves the chromosomal DNA at its unmethylated restriction sites (RS), finally leading to extensive chromosomal degradation.
Metabolism‐based systems
Metabolism‐based addiction systems can be divided into catabolism‐ and anabolism‐based systems.
Catabolism‐based systems. Catabolism‐based systems (CBS) utilize the necessity of enzymes that degrade an essential carbon and energy source for the microorganism. If the gene for an essential enzyme of the catabolism of such a compound is inactivated, the cells are impaired to use it for growth. However, episomal expression of the respective gene on a plasmid restores the catabolic pathway. Ralstonia eutropha H16 is a Gram‐negative and facultative chemolithoautotrophic bacterium capable of using gluconate or fructose as carbon and energy sources for growth. Both compounds are catabolized via the Entner–Doudoroff pathway (Gottschalk et al., 1964; Blackkolb and Schlegel, 1968). The key intermediate of this pathway 2‐keto‐3‐deoxy‐6‐phosphogluconate (KDPG) is converted to glyceraldehyde‐3‐phosphate and pyruvate by KDPG‐aldolase (Kovachevich and Wood, 1955). This key enzyme is encoded by eda in R. eutropha H16, which is located on chromosome 2 (Pohlmann et al., 2006). A plasmid containing eda was transferred into the KDPG aldolase‐negative mutant R. eutropha H16‐PHB−4Δeda to restore KDPG‐aldolase activity, thereby yielding a first example for a CBS. In addition to eda, the plasmid contained the cyanophycin synthetase gene (cphA) thereby allowing improved production of cyanophycin (Voß and Steinbüchel, 2006)
Anabolism‐based system. Anabolism‐based addiction systems (ABS) depend on at least one gene for an essential anabolic pathway. If such a gene is inactivated and if a copy of the intact gene is localized on a plasmid, that is important for a biotechnological process, the production of the respective compound becomes dependant on the presence of the plasmid.
Chromosomally encoded auxotrophic markers like URA or LEU2, which are complemented by an intact plasmid‐encoded gene, are used in the genetics of yeasts and other eukaryotes since many years for selection of plasmids (Strausberg and Strausberg, 2001). One example is the URA3 gene, encoding orotidine‐5′‐phosphate decarboxylase. Ura3 represents an essential enzyme in pyrimidine biosynthesis in Saccharomyces cerevisiae; its gene is often used on plasmids for complementation of auxotrophic yeast strains (Hensing et al., 1995). However, such systems were scarcely employed for production processes in biotechnology. Unsatisfying results were obtained when leucine auxotrophy in a xylitol production strain of S. cerevisiae was complemented. Due to the release of leucine from plasmid‐containing cells also cells without plasmids containing the LEU2 auxotrophic complementation selection marker grew (Meinander and Hahn‐Hägerdal, 1997). On the other hand, successful complementation of auxotrophy for uracil and proline combined with expression of heterologous proteins was reported in Pseudomonas fluorescens (Schneider et al., 2005).
Recently, a new anabolic addiction system was developed that relies on the isoprenoid biosynthesis pathway (Kroll et al., 2009). In this ABS the native MEP pathway in E. coli was disrupted through deletion of the ispH gene, thereby disenabling the cells to catalyse the final step of the MEP pathway yielding IPP or DMAPP (Rohdich et al., 2002). Deletion of this gene is lethal, due to the absence of essential precursors for isoprenoid biosynthesis (McAteer et al., 2001). A suppression of this auxotrophic mutation by IPP uptake from the medium is not possible because of the phosphorylated nature of the compound. A synthetic plasmid, which harbours the relevant genes of an artificial IPP‐producing mevalonate (MVA) pathway, was transferred to E. coli before generating the ΔispH knockout, thereby enabling synthesis of isoprenoids via the foreign pathway in the ispH deletion mutant. This system is independent of minimal media and the use of defined carbon sources. The applicability of this addiction system was shown by improved production of cyanophycin in E. coli (Kroll et al., 2009).
ORT‐based systems
To avoid plasmid maintenance through selectable marker genes, Williams and colleagues (1998) developed a novel addiction system depending on plasmid‐borne ORT utilizing the lac repressor. They integrated a kanamycin resistance gene into the chromosome of E. coli and placed it under control of the operator–promoter region of the lac operon, resulting in a conditionally essential chromosomal gene under kanamycin selection. The lac repressor protein (LacI) binds to the lac operator (lacO) and therefore represses the expression of the kanamycin resistance gene. When IPTG is available as an inducer, LacI is titrated by lacO, and the E. coli cells were able to grow in media containing kanamycin due to expression of the corresponding resistance gene. The same result was obtained when high‐copy‐number plasmids harbouring lacO were used for ORT. In this case the plasmid was stably maintained over 72 generations (Williams et al., 1998). In a subsequent study, ORT by using high‐copy‐number plasmids was reported in E. coli strains with the essential chromosomal dapD gene under control of the lac operator–promoter region (Cranenburgh et al., 2001; Hanak et al., 2003). DapD is involved in lysine biosynthesis and provides diaminopimelate (DAP) for cross‐linking of peptidoglycan chains (Schleifer and Kandler, 1972; Richaud et al., 1984). Alterations in lacO sequences and plasmid copy number were also studied in E. coli with LacI‐repressed dapD gene (Cranenburgh et al., 2004). Figure 1 illustrates the previously described ORT system using the regulation of dapD expression. The lacO of the wild‐type E. coli lac operon or its palindromic sequence provided sufficient levels of ORT, and lower‐copy‐number plasmids derived from pBR322, with 39–55 copies per cell (Müller‐Hill et al., 1968), were shown to be as effective in ORT as high‐copy‐number plasmids.
PAS and their applications
Naturally occurring PAS can be considered as regulatory elements in prokaryotic or eukaryotic cells, which control by different mechanisms such as plasmid stability or the adjustment of the transcriptional rate. Hitherto, the precise functions of naturally occurring addiction systems are not fully understood and different hypothesis exist although the main role of these addiction systems is to maintain plasmid stability in daughter cells. Since the discovery of TA systems in the 1980s and due to an increasing interest in antibiotic‐independent strategies for stable plasmid maintenance in biotechnology, great efforts have been made to further develop existing TA systems and to increase our knowledge about their characteristics and functionality. The gained knowledge has been used for the implementation of TA systems in a large variety of applications (see Tables 4 and 5). The following section describes the applications of biological containment systems for genetically engineered microorganisms (GEMs) in bioremediation processes, the delivery of antigens to the human immune system and the production of compounds based on enhanced plasmid stability.
Table 4.
TA systems used for bacterial containment and in bioremediation.
| Application | TA system | Killing induction | References |
|---|---|---|---|
| Model containment system | hok | Tryptophan starvation | Molin et al. (1987) |
| hok | Time‐dependent | Molin et al. (1987; 1993) | |
| hok | Lactose or IPTG | Bej et al. (1988) | |
| hok/sok | Phosphate starvation | Schweder et al. (1992) | |
| sacBa | Sucrose | Recorbet et al. (1993) | |
| gef | Time‐dependent | Klemm et al. (1995) | |
| colE3/immE3, ecoRI | Lactose or IPTG | Torres et al. (2003) | |
| Bioremediation | gef | Absence of degradable compounds | Contreras et al. (1991); Jensen et al. (1993); Ronchel et al. (1995) |
| colE3a | Lactose or IPTG | Munthali et al. (1996a,b) | |
| stva | Absence of degradable compounds | Szafranski et al. (1997); Kaplan et al. (1999) | |
| asda, gef | Absence of degradable compounds | Ronchel and Ramos (2001) |
TA‐less killing function.
All listed model containment systems have been constructed in E. coli and the active containment systems were designed for the use of P. putida in bioremediation.
Table 5.
TA systems used in a variety of biotechnical applications.
| Application | Organism | TA system | Killing induction | References |
|---|---|---|---|---|
| Live vector vaccines | S. typhi | hok/sok | Plasmid loss | Galen et al. (1999) |
| S. typhimurium | dapDa,b | Absence of lactose or IPTG | Garmory et al. (2005) | |
| Phage therapy | E. coli | gef, chpBK | Lactose or IPTG | Westwater et al. (2003) |
| E. coli | bglII | Lactose or IPTG | Hagens and Bläsi (2003) | |
| P. aeruginosa | bglII | Lactose or IPTG | Hagens et al. (2004) | |
| Vectors | E. coli | ccdB | Insert‐less MCS | Bernard et al. (1994) |
| C. neoformans | ccdB | Lactose or IPTG | Mondon et al. (2000) | |
| F. tularensis ssp. tularensis | ORF4‐ORF5 | Plasmid loss | LoVullo et al. (2006) | |
| E. coli | ccdAB | Plasmid loss | Wegerer et al. (2008) | |
| Anti‐antitoxin PNAs | E. coli | hok/sok | Antitoxin sequestration | Faridani et al. (2006) |
| Gene silencing | E. coli | RNA I/RNA II | – | Pfaffenzeller et al. (2006a,b) |
| RASTA‐Bacteriab | – | – | – | Sevin and Barloy‐Hubler (2007) |
TA‐less killing function.
Web‐based annotation tool.
Bacterial containment systems were listed in Table 4.
Containment systems for GEMs
Pollution of the environment with persistent, toxic and xenobiotic chemicals is of global concern and requires remediation of these sites (Paul et al., 2005). Bioremediation relies on the degradative pathways for various organic substrates as sources of carbon and energy, which have evolved in microorganisms over the time, and which may be more eco‐friendly and cost‐effective, in comparison with physicochemical processes (Pieper and Reineke, 2000; Cases and de Lorenzo, 2005). For an effective taxonomic affiliation of microorganisms involved in bioremediation processes, polyphasic approaches as a combination of more traditional phenotypic and physiological approaches with modern molecular biology techniques are particularly successful because only a community of microorganisms provides a sufficient metabolic diversity for disposal of these anthropogenic pollutions (Pieper and Reineke, 2000). In addition, physicochemical and also special biological remediation processes are frequently used for the removal of chemicals which resist biodegradation (Dua et al., 2002).
Based on the multitude of sequenced genomes, gene clusters encoding degradative pathways for various aromatic and other hazardous organic compounds were cloned and characterized (Sangodkar et al., 1989; Samanta et al., 2002). Several GEMs with novel or improved degradative properties were constructed, and their potentials as biocatalysts were investigated (Pieper and Reineke, 2000; Furukawa, 2003; Lovley, 2003). Owing to security concerns and fears of non‐predictable horizontal gene transfer and uncontrolled proliferation when GEMs would be released to the environment, legal requirements restrict possible applications of GEMs in bioremediation, although they were already successfully investigated under laboratory conditions (Diamand, 1999; Paul et al., 2005). Therefore, bacterial containment systems were developed by combining degradative pathways for organic compounds mainly with TA systems for the use of GEMs in bioremediation (see Table 4). These systems aim at killing the cells by toxin expression, for example by induction through amino acid starvation when they escape the designated environment. TA‐absent containment systems using GEMs expressing bacterial haemoglobin to degrade aromatic compounds under hypoxic conditions have been reviewed elsewhere (Urgun‐Demirtas et al., 2006). Table 4 summarizes containment systems and gives cross references to the relevant literature.
Model containment systems for controlled killing of bacteria. The first model containment system was designed in E. coli with the hok gene (see above), encoding the Hok toxin gene in fusion with the promoter of the tryptophan operon (Molin et al., 1987). Under controlled conditions such as in fermentation processes with sufficient supply of tryptophan in the medium, the hok gene is not expressed due to an inactivated tryptophan promoter. Inadvertent escape to the environment with tryptophan starvation or genetic rearrangement of the containment system led to expression of the toxin and therefore to cell death. Molin and colleagues suggested that time‐dependent toxin induction regardless of growth rate would be more efficient than toxin expression depending on cell division, since the latter is unpredictable in nature (Molin et al., 1987; 1993). Therefore, they placed a hok gene under control of the invertible fimA promoter periodically expressing type I fimbriae in E. coli. The plasmid‐located fim gene cluster provides antagonistically acting fimB and fimE gene products which are involved in the regulation of the trans activated fimA promoter. Because FimB and FimE act as simple activator/repressor proteins, FimB mediates an active promoter configuration resulting in fimA transcription (Molin et al., 1987). The investigated mutant E. coli MC1000 harboured either one or three additional different compatible plasmids with different copy numbers in addition to the above described plasmid carrying the fim cluster. In addition, plasmids containing the genes fimB alone, fimB in combination with fimE or only fimE were also investigated. Finally, if the positive regulator FimB is present in excess, the cells are killed due to Hok toxin expression. In a slowly growing population such as occurring in the environment, stochastic dying of a cell fraction would lead to a gradual reduction of viable cell counts over time. However, in a fast‐growing population such time‐dependent killing would be insignificant with regard to the growth rate, thus permitting the controlled containment of bacteria under certain environmental conditions. These results were confirmed in a subsequently performed experiment that used the gef gene, a hok homologue, under control of the fimA promoter in E. coli (Klemm et al., 1995).
A related suicide vector‐based hok expressing model containment system with additionally mediated carbenicillin resistance was under control of the lac promoter (Bej et al., 1988). Cultivation upon induction with IPTG under antibiotic selection led to mutational resistance against hok expression, whereas induction under non‐selective conditions resulted in a toxin‐mediated killing of 90–99% of the cell population. Surviving cells, grown under the latter condition, had developed a resistance against the still active hok gene with reduced energy‐generating membrane functions and exponential growth. There may be also the following explanations (Bej et al., 1988): a mutation in the lacI gene occurred, which prevented hok induction, or in the cells carrying the suicide vector making them resistant to Hok polypeptide. Thus and due to the results obtained during cultivation in presence of carbenicillin, the authors concluded that their system was not elaborated enough to ensure the fail‐safe containment of GEMs.
Additionally, plasmid maintenance by the complete parB locus, encoding the hok/sok TA system fused to the phosphate‐repressed promoter of the alkaline phosphatase gene (PphoA), was investigated in E. coli (Schweder et al., 1992). This model containment system provided hok‐mediated cell killing due to phosphate limitation, which often occurs as a growth‐limiting factor in the environment. Phosphate is an essential constituent in media, thus it ensures plasmid maintenance under these controlled conditions. Analyses of non‐selective and glucose‐limited cultivations of strains harbouring a plasmid with hok/sok or a PphoA‐hok/sok construct revealed no plasmid‐free cells after 50 generations or only 3% plasmid loss between the 50th and 60th generation. When grown in phosphate‐limited medium, cell killing was observed after 4 h of cultivation but about 5% of the population still remained viable after 8 h. Because of the more effective hok‐based model containment systems Schweder and colleagues suggested that the integration of the complete parB locus and resulting hok mRNA:Sok‐RNA interactions might have been unfavourable for the efficacy of cell killing.
Development of functional bacterial containment systems applicable in bioremediation.Contreras and colleagues (1991) constructed the first active containment system in Pseudomonas putida to degrade a variety of substituted benzoates by the use of two plasmid‐borne elements. Enhanced substrate range and a 50‐fold increased affinity for 3‐methylbenzoate as effector were achieved in the substitution mutant XylSR45T. The gef gene, a hok homologue, was placed under control of a LacI‐repressed promoter and the corresponding lacI gene under the influence of a XylS‐regulated promoter. This provided a suicide function resulting in cell killing through Gef toxin activity in the absence of substituted benzoates as XylS effectors. That means no active form of XylS was formed allowing lacI expression and therefore no gef (toxin) expression downstream of a Ptac promoter. However, as observed in previous studies (Molin et al., 1987; Bej et al., 1988), few cells became resistant for uninvestigated reasons.
To study factors that limit the killing efficiency of containment systems, plasmids harbouring the relF gene, homologous to hok and gef, fused with the lac promoter were used in E. coli (Knudsen and Karlström, 1991). The factors were determined to be the mutation frequency of the toxin gene with 10−6 per cell per generation, preventing cells from being killed, and a basal toxin expression, which results in a reduced growth under controlled conditions, but provides a selective growth advantage to cells with mutational resistance towards toxin activity. A containment system with a second copy of the relF‐encoding plasmid led to a reduced mutation rate of < 5 × 10−9 per cell and generation. In a subsequent study, a plasmid harbouring two relF genes, which were arranged to prevent inactivation by deletion or recombination and insertional inactivation, resulted in a mutation rate of 10−8 per cell and generation (Knudsen et al., 1995).
The active containment system of Contreras and colleagues (1991) was altered in a way that a transposase‐less transposon bearing the gef gene in fusion with a LacI‐repressed promoter for random integration into the P. putida chromosome, and the XylSR45T mutant with p‐ethylbenzoate as additional effector molecule were used (Jensen et al., 1993). Toxin resistance occurred with a frequency between 10−5 and 10−6 per cell per generation and 10−8 in cells harbouring two copies of the suicide function respectively. A P. putida strain with the chromosomally integrated transposon was used for alkylbenzoate degradation and its applicability for bioremediation in alkylbenzoates‐containing soils was demonstrated (Ronchel et al., 1995).
Additional containment systems have been reported: a sucrose‐sensitive E. coli strain harbouring a chromosomally integrated sacB gene encoding levansucrase, which converts sucrose to levan that causes cell lysis (Recorbet et al., 1993), or the degradation of polychlorinated biphenyls by a P. putida strain that is killed due to colicin E3 RNase activity (Munthali et al., 1996a,b).
Further improvements were achieved with a novel concept in P. putida based on the stv gene encoding streptavidin (Szafranski et al., 1997; Kaplan et al., 1999), which interacts with biotin and thus inhibits biotin‐depending enzymes. The constructed P. putida strain was used for bioremediation of aromatic hydrocarbons and in the absence of the effector 3‐methylbenzoate, a reduction of at least 90% of the population was detected with a mutation rate from 10−7 to 10−8 per cell and generation.
Ronchel and Ramos (2001) designed a novel bacterial suicide containment system ensuring controlled killing without the risk of mutational inactivation. The Pm promoter was fused to the essential asd gene, which is therefore expressed in the presence of substituted benzoates as effector molecules. The asd gene encodes the aspartate‐semialdehyde dehydrogenase which is essential for biosynthesis of methionine, threonine and isoleucine, lysine and DAP as a constituent of peptidoglycan layers (Truffa‐Bachi and Cohen, 1968). The additional integration of a containment system reported previously (Ronchel et al., 1995, see above) resulted in an undetectable mutation rate (< 10−9 per cell per generation), and the respective GEM population of P. putida was eliminated in less than 25 days in soil experiments.
An alternative dual model containment system consists of two TA loci, the colicin E3‐immunity E3 system and the EcoRI restriction–modification system, with plasmid‐borne toxins and both antitoxins stably integrated into the E. coli chromosome (Torres et al., 2003). The so‐called ‘containment efficacy’ of this dual containment system was experimentally determined and higher than the ones of single containment systems, but lower than the expected values of theoretical calculations (Knudsen and Karlström, 1991). Mutated clones that escaped the expected killing effect showed deletions or insertions in one or both lethal genes. However, horizontal gene transfer in this dual system was effectively decreased and the presented system provides a valuable strategy to increase containment.
Pandey and colleagues (2005) conceptualized a novel active containment system with improved plasmid maintenance and regulated survivability in the environment. Two different TA systems would be necessary, one with plasmid‐borne toxin and chromosomally integrated antitoxin, the other in reverse arrangement. Both toxin genes would be constitutively expressed, and the antitoxins would be under control of promoters of catabolic operons such as Pm. In the absence of effector molecules which induce antitoxin expression, the cells would be killed due to toxin activity. Thus, survival upon escaping from a defined environment would be prevented. Horizontal transfer of the plasmid would result in toxin‐mediated cell killing since the respective antitoxin would be lacking in the recipient cell.
Use of TA systems in antimicrobial treatments. Several applications have been developed, which use TA systems for stable plasmid maintenance and killing of progeny cells without plasmids, in antimicrobial treatments. Among these applications are expression plasmids for the delivery of heterologous antigens to the human immune system in attenuated Salmonella spp. live vector vaccine strains that were shown to be effective in treatments of Salmonella infections. To treat infections of E. coli or Pseudomonas aeruginosa, other approaches were based on genetically engineered phages, which inject DNA inter alia thereby killing the bacteria. Specific peptide nucleic acids (PNAs) were constructed as a novel class of antimicrobial compounds to sequester antitoxin‐RNA and to enable toxin activity in E. coli. TA system‐dependent shuttle vectors have been also designed as genetic tools for the highly pathogenic bacterium Francisella tularensis (see below).
Plasmids expressing heterologous antigens for vaccine therapies in Salmonella spp.
Expression of immunogenic and protective antigens for delivery to the human immune system from plasmids in attenuated Salmonella spp. live vector vaccine strains are one possibility in vaccine development (Galen et al., 1999; Garmory et al., 2002). Sufficient expression level of antigens and stable plasmid maintenance define the efficacy of live vector vaccine strains. A pre‐clinical model of murine intranasal immunization was established for Salmonella typhi CVD 908‐based live vector vaccines in mice (Galen et al., 1997) and various heterologous antigens have been successfully expressed within this strain (Barry et al., 1996). Although an addiction system for plasmid maintenance based on an essential gene was designed in attenuated Salmonella typhimurium strains (Nakayama et al., 1988) and expressed successfully heterologous antigens in mice (Karem et al., 1995; Srinivasan et al., 1995; Covone et al., 1998), stabilization of plasmids with this system in S. typhi vaccine strains failed, yet (Tacket et al., 1997). Nakayama and colleagues (1988) constructed a S. typhimurium mutant lacking the asd gene. Mutants without a plasmid‐borne asd gene or cells grown in absence of DAP were killed.
To alternatively enhance plasmid maintenance and stability in S. typhi, two sets of novel non‐catalytic multicopy expression plasmids were developed that harboured the hok/sok antisense RNA‐regulated TA locus (see section above) and one or both plasmid partition functions encoded by par (Wahle and Kornberg, 1988) and parA (Dam and Gerdes, 1994) to avoid random segregation (Galen et al., 1999). The sets contained either the oriE1 or the ori15A origin of replication expected to result in approximately 60 or 15 copies per cell, respectively, and a derivative of the osmolarity‐inducible ompC promoter cassette (PompC1) from E. coli was used (Norioka et al., 1986). The plasmids also harboured the gfpuv gene encoding a UV‐optimized variant of the green fluorescent protein (GFPuv) as a test antigen. Plasmid maintenance was measured by quantification of GFPuv‐mediated fluorescence using flow cytometry in the absence of antibiotics. These experiments revealed that plasmids containing the higher‐copy‐number oriE1 origin of replication were very unstable and even when cultivated in the presence of antibiotic selection, 50–62% of the cell population did not express measurable amounts of GFPuv any longer, depending on the plasmid's maintenance functions. Examinations revealed that non‐fluorescing bacteria were viable and sensitive to antibiotic due to plasmid loss.
Consistent with other reports, these results indicate that, in contrast to plasmids containing the oriE1 replicon, the hok/sok TA system improves the stability of plasmids with ori15A origin of replication (Gerdes, 1988), although it does not ensure killing of all plasmid‐free cells (Wu and Wood, 1994). To enhance plasmid stability, hok/sok was combined with the parDE and pnd loci; however, long‐time cultivation in the absence of an antibiotic did not result in complete plasmid stability (Pecota et al., 1997). According to recent studies (Stephens et al., 2006), this result gives rise to doubts about strategies using higher‐copy‐number plasmids to express heterologous antigens in S. typhi as the metabolic burden promotes plasmid loss. Galen and colleagues (1999) suggested that the design of S. typhi live vector vaccines strains with stabilized low‐copy‐number plasmids expressing heterologous antigens to be a promising strategy in vaccine development. These plasmids should be able to express high levels of heterologous antigens in response to an environmental signal that occurs in vivo at the destination area of immunological induction which the vaccine organism has to reach.
Maintenance of high‐copy‐number plasmids in the absence of selectable marker genes was achieved by using ORT to successfully express the Yersinia pestis F1 antigen in an attenuated S. typhimurium strain (Garmory et al., 2005). The lac repressor gene was chromosomally integrated to ensure effective repression of the essential chromosomal dapD gene, which was placed under control of the lac operator–promoter region. Despite this, a basal expression of dapD in the absence of IPTG was detected; however, growth was only possible in liquid media. Cultivation of S. typhimurium for 5 days demonstrated ORT‐mediated stable plasmid maintenance without plasmid loss. In vivo experiments in mice revealed recombinant bacteria, although significantly lower levels were particularly measured due to the metabolic burden of harbouring high‐copy‐number plasmids. To provide immunization against plague in mice, the kanamycin resistance gene of a plasmid expressing Y. pestis F1 antigen (Titball et al., 1997) was exchanged for the palindromic lac operator, and no plasmid loss was detected in S. typhimurium over 5 days. This was the first time that a single dose of S. typhimurium‐based vaccine was shown to protect against plague. A comprehensive overview of the mentioned systems for vaccines therapies is provided in Table 3.
Anti‐Sok PNA oligomers in E. coli (Faridani et al., 2006)
Faridani and colleagues (2006) developed a method to sequester Sok‐RNA competitively to inhibit hok mRNA:Sok‐RNA interactions in E. coli. They designed PNA oligomers that were complementary to the 5′ end of Sok‐RNA to prevent Sok‐mediated degradation of hok mRNA; this led to cell killing through synthesis of Hok toxin. To enhance uptake in E. coli, the PNA oligomers were attached to the synthetic peptide (KFF)3K that increases the permeability of the outer membrane (Vaara and Porro, 1996). Already 10 µM of one of the anti‐Sok PNAs prevented growth of E. coli cells for as long as 20 h, whereas a lower concentration or other PNAs exerted only a transitive growth arrest. Surprisingly, the inhibitory effect of the global transcription inhibitor rifampicin on viable cell counts was significantly lower than that of the PNA oligomers even at a 20‐fold higher concentration. Thus, anti‐Sok PNAs are more bactericidal to E. coli containing a hok/sok TA system than rifampicin. After treatment of an E. coli culture harbouring hok/sok with either anti‐Sok PNA or rifampicin, dead ‘ghost’ cells occurred and mature hok mRNA was accumulated. In contrast to antibiotics, which are rapidly exported from cells, there is evidence that PNA oligomers accumulate in E. coli (Good et al., 2001).
Peptide nucleic acids are potent competitive inhibitors of toxin mRNA:antitoxin‐RNA interactions. To enhance their bactericidal activity for clinical applications, their cell influx and toxicity must be further enhanced. Antisense agents targeting essential genes are effective against E. coli infections in mice (Geller et al., 2005; Tan et al., 2005). Faridani and colleagues (2006) proposed that chromosomal antitoxin‐RNAs may be sequestered by the cells as a resistance mechanism against PNAs. Mutations that do not inactivate the toxin, could lead to cell death if only the formation of toxin mRNA:antitoxin‐RNA is prevented. PNA oligomers may therefore present a new class of effective antimicrobials for treatment of bacterial infections.
Novel shuttle vectors for use in F. tularensis ssp. tularensis (LoVullo et al., 2006)
The Gram‐negative bacterium F. tularensis is one of the most infectious pathogens known and causes the zoonotic disease tularaemia, which is rarely fatal when treated with appropriate antibiotics, even in its more serious pneumonic and typhoidal forms, but infected persons are often severely debilitated (Titball et al., 2007). Biosafety‐level three conditions are necessary for manipulation of F. tularensis (Dennis et al., 2001), and most of the genetic tools such as E. coli–Francisella shuttle vectors, derived from the cryptic Francisella‐plasmid pFNL10 (Norqvist et al., 1996), have been developed for the moderate pathogenic subspecies novicida and a holarctica‐derived attenuated live vaccine strain (LVS, see Table 5). Recently, genetic tools including shuttle vectors, transposons and allelic replacement strategies have been exhaustively reviewed (Frank and Zahrt, 2007). A challenge in the development of genetic tools for F. tularensis is its resistance to β‐lactam antibiotics and the restricted use of other antibiotics that are administered in tularaemia treatments (Baker et al., 1985).
In need to develop genetic tools and to test marker genes for the most pathogenic F. tularensis ssp. tularensis, LoVullo and colleagues (2006) constructed novel E. coli–Francisella shuttle vectors, which were derived from the cryptic Francisella‐plasmid pFNL10 (Pomerantsev et al., 2001). In a future perspective, an attenuated F. tularensis ssp. tularensis vaccine strain might be more effective in providing protective antigens for delivery to the human immune system than the existing vaccine strains derived from other F. tularensis subspecies (Conlan and Oyston, 2007). A spontaneous mutant of a specific F. tularensis ssp. tularensis strain was recently discovered and shown to be both safer and more protective against F. tularensis infections in mice than the holarctica‐derived LVS (Twine et al., 2005).
Further applications based on TA systems
Besides the broad applications in bioremediation and antimicrobial treatments, a few other usages of TA systems were reported. Among them are cloning vectors that harbour a toxin gene as a positive selection marker. Plasmids with ColE1‐derived origin of replication were successfully shown to silence a chromosomal target gene due to toxin mRNA:antitoxin‐RNA interactions. To relieve and improve the identification of novel TA systems, a web‐based tool for identifying TA loci in prokaryotes has been developed (Sevin and Barloy‐Hubler, 2007).
Incorporation of TA systems into cloning vectors. Two novel positive selection vectors derived from the high‐copy‐number vectors pUC18/19 have been constructed (Bernard et al., 1994). The 3.0 kb vectors harboured both an ampicillin resistance gene and the ccdB toxin gene with incorporated MCS differently orientated in both vectors, under control of the lac promoter. The cytotoxic active gene ccdB was found on the ccd (control of cell death) locus on the F plasmid and kills the hosts by interfering with the DNA–gyrase cleavable complex (Bernard, 1995). To avoid killing of vector‐harbouring cells, maintenance in DNA gyrase mutants suppressing the toxin activity or hosts expressing lac repressor was necessary. By insertion of DNA fragments of at least 150 bp into the MCS, positive selection upon IPTG induction was possible due to disruption of the ccdB gene and subsequent occurrence of viable colonies. In a further study, the ampicillin resistance gene was replaced by the kanamycin or chloramphenicol resistance gene (Bernard, 1995).
A novel ccdB‐based shuttle vector for transformation of the basidiomycetous yeast Cryptococcus neoformans was designed that combined positive selection in bacteria with functions for enhanced episomal maintenance in C. neoformans (Mondon et al., 2000). This approach was used to force plasmid stability with a multitude of positive influences from known elements on plasmid stability in yeasts. Autonomously replicating plasmids have been shown in several yeasts to be lost during mitosis under non‐selective conditions. Therefore, the ccdB toxin gene, telomere sequences, the STAB fragment enhancing stable plasmid maintenance (Varma and Kwon‐Chung, 1998) and the URA5 gene as a selectable marker on media that contain 5‐fluoroorotic acid (Edman and Kwon‐Chung, 1990) were assembled to the novel vector. The above mentioned STAB‐DNA sequence itself resulted in an enhanced episomal stability in more than 60% of the transformants. Furthermore, plasmid‐borne prototrophic stability in these transformants was enhanced up to 80% (up from 0 to 5% regarding to the control experiment). Although vector‐mediated CcdB activity was shown to inhibit growth of E. coli, no such effect was detected with C. neoformans.
Maintenance of ColE1 plasmids due to gene silencing based on RNA interactions. Plasmids derived from vectors with a ColE1‐type origin of replication offer the possibility to silence engineered target genes by toxin mRNA:antitoxin‐RNA interactions (Pfaffenzeller et al., 2006a,b). The replicational regulatory network of these plasmids consists of RNA II that functions as a primer by hybridizing to a region in the origin of replication and RNA I, inhibiting this hybridization by prior binding to RNA II (Brenner and Tomizawa, 1991). Pfaffenzeller and colleagues (2006a) integrated the green fluorescent protein (GFP) chromosomally into E. coli and obtained sequences of different length complementary to RNA I near to the ribosomal binding site. As a result the use of a ColE1‐type plasmid should lead to translation inhibition by interactions of RNA I with the mRNA of the target gene. Investigations on GFP expression in presence and absence of a ColE1‐type plasmid showed no difference for the control strain without RNA I‐complementary sequence. A decrease in the GFP expression of 36% was observed for the strain with entire RNA I‐complementary sequence in the presence of plasmid‐encoded RNA II. However, a strain that included only a partially complementary sequence showed a significantly lower GFP expression without ColE1‐type plasmid, whereas almost no expression was detected in the presence of the plasmid. These results indicate that not only sequence length but also structural stability were important for the silencing of the target gene. Finally fermentation experiments confirmed the result that a partial complementary sequence to RNA I ensures almost entire gene silencing over 22 h.
Integration of the ccdAB TA systems into rhamnose‐inducible expression vector. Expression systems for plasmid‐encoded heterologous genes need special requirements: (i) they have to ensure high yields of enzymes and proteins (ii) in a cost‐effective production (iii) at high plasmid stability (iv) while the expression has to be tightly controllable (Stumpp et al., 2000; Wegerer et al., 2008). A versatile expression vector is pJOE3075, which contains the genes for l‐rhamnose uptake and catabolism under control of their native promoter (rhaPBAD) as a positive regulator (Stumpp et al., 2000). The completely sequenced expression vector pJOE4056.2, a derivative of pJOE3075, harbours the enhanced green fluorescent protein (eGFP) gene under control of the rhaPBAD promoter (Wegerer et al., 2008). To optimize pJOE4056.2, a stem loop structure was initially inserted into the transcription initiation region of the rhaPBAD promoter resulting in a fourfold increased eGFP production (Wegerer et al., 2008). However, consequences on plasmid maintenance were not detected. Subsequently, combinations of the genetic modules rop, ccdAB and cer with regard to their advantages for the above mentioned qualities of plasmid‐encoded expression systems were investigated. The genetic modules have different functions: rop is involved in the maintenance of constant but low copy numbers for plasmids (Cesareni et al., 1982), ccdAB encodes a TA system, with a toxin activity leading to inhibition of DNA gyrase and therefore to cell killing, and cer does not only participate in the resolution of plasmid multimers (Summers and Sherratt, 1984) but also contains a genetic element the transcript of which retards cell division in the presence of plasmid multimers and functions in plasmid maintenance (Patient and Summers, 1993; Balding et al., 2006). Deletion of rop led to a more than threefold increase in plasmid DNA without a simultaneous rise in eGFP production due to the high content of eGFP already available. The inadvertent insertion of a cer gene tandem had a tremendous stabilizing effect because more than 90% of the cells retained the plasmid under antibiotic selection, whereas over 50% of the population lost a cer‐less plasmid. Excision of one cer gene did not affect plasmid maintenance but increased the yield of eGFP. Surprisingly, the integration of ccdAB resulted in an additional plasmid loss; Wegerer and colleagues (2008) ascribed this observation to the fact that two independently positive effects in combination might present a disadvantage. Since expression vectors with rhaPBAD promoter are already efficient, tightly regulated and well balanced, further improvements might be difficult to establish and unfavourable. Therefore, the novel constructed plasmid pWA21 containing the stem loop, a single cer gene and the rop gene offered both high expression levels and an accurate plasmid stability.
Applications in enzyme production
The applicability of complemented P. fluorescens mutants with uracil and proline auxotrophy was analysed during production of α‐amylase and nitrilase (Schneider et al., 2005). Two plasmids were used: one plasmid encoding the respective enzyme under control of a LacI‐repressible promoter and tetracycline resistance, while the other plasmid harboured lacI and the kanamycin resistance gene (kamR). In a first step, the tetracycline resistance gene was replaced by pyrF, and subsequently (kamR) was exchanged by proC. Fermentation experiments at the 20 l scale over 75 h showed no difference in enzyme activity and optical density between antibiotic‐containing plasmids, those encoding PyrF and kanamycin resistance and plasmids that complemented both auxotrophic mutations. Thus, plasmid loss led to auxotrophy for uracil and proline and plasmid‐free progeny cells were killed. Plasmid maintenance was measured with two different methods and revealed that 94–97% of the population retained the plasmids even after 62 generations. The detection of a slower growth rate of auxotrophic cells compared with complemented cells even on medium supplemented with uracil and praline was cited as reason for the small fraction of plasmid‐free segregants.
Production of the biopolymer cyanophycin
Cyanophycin (CGP) is a polymer of biotechnological interest (Mooibroek et al., 2007), which consists of a polyaspartic acid backbone linked with arginine moieties (Simon, 1976). Different approaches to increase the production of CGP in R. eutropha H16 without the addition of antibiotics to stabilize plasmid maintenance were investigated. Strategies based on different plasmid types harbouring the cyanophycin synthetase gene of Synechocystis sp. PCC6308 or a chromosomal integration failed and did not enhance the production of CGP in R. eutropha H16 yielding only very low CGP contents. Therefore, in an alternative attempt to enhance the CGP content of the cells, the eda‐dependent CAS was constructed (Voß and Steinbüchel, 2006). Flasks experiments revealed high CGP contents up to 40.0% (w/w) of cell dry weight without CGP precursor substrates. The CGP contents obtained in the absence of plasmid‐stabilizing antibiotics were only slightly lower. Fed‐batch fermentations at the 30 l and 500 l scale confirmed the results of the flasks experiments. Even in the absence of antibiotics, only 7% of the cells had lost the plasmid because the eda gene is essential for growth of R. eutropha H16 on gluconate or fructose.
The recently developed novel branch of ABS by Kroll and colleagues in 2009 revealed nearly full plasmid stability and represents a novel PAS without a possible bypass/loophole. This system was used to produce CGP and aims at providing precursors for isoprenoid biosynthesis as the target anabolic pathway. The use of this class of ABS with its independence of using defined carbon sources or cultivation media like mineral media may be very interesting for industrial fermentations. It provides also the possibility to use cheap agricultural commodities such as corn steep liquor or melasse as cultivation media.
Conclusion and outlook: applications and perspectives of PAS in biotechnology
Recombinant microorganisms are frequently used for the production of proteins, enzymes, organic compounds – e.g. amino acids, sugars, vitamins, alkanes and their various derivatives, antibodies and drugs in large‐scale fermentation processes. These processes are often based on heterologous expression of plasmid‐encoded genes and the systems have to meet the following quality criteria: (i) the respective products must be obtained with high yields, (ii) production must be cost‐effective, (iii) plasmid stability must be high, while (iv) expression has to be certainly and tightly controllable (Jana and Deb, 2005; Sørensen and Mortensen, 2005;Wendisch et al., 2006; Wegerer et al., 2008). The host of choice for production of many recombinant proteins is in particular the Gram‐negative bacterium E. coli for several reasons: (i) its economical and fast high‐density cultivation possibilities (ii) combined with its simplicity, (iii) the extensive biochemical and genetic knowledge, and (iv) the large variety of available biotechnological tools for high‐level expression of a multitude of recombinant proteins (Jana and Deb, 2005; Sørensen and Mortensen, 2005). Other organisms such as Corynebacterium glutamicum or the yeast Pichia pastoris are used for the industrial production of amino acids or therapeutic glycoproteins with fully humanized N‐glycosylation structures respectively (Macauley‐Patrick et al., 2005; Wildt and Gerngross, 2005; Wendisch et al., 2006). The importance of biotechnical processes in industrial production is steadily increasing, due to rising oil prices and the demand for ecologically feasible production (Wendisch et al., 2006). As most established systems for heterologous productions are plasmid‐based, they often require antibiotics for plasmid selection. This is not only cost‐intensive but also entails potential risks to the environment from the antibiotics in industrial waste as well as from the spreading of the resistance genes when cells would escape inadvertently (Hägg et al., 2004). Furthermore, the use of antibiotics in pharmaceutical production is limited since recombinant therapeutics require a contamination‐free product or their use is prohibited for fear of emerging antibiotic resistances when they can be used in antimicrobial treatments (LoVullo et al., 2006). Thus, there is a strong need for alternative systems that maintain plasmids highly stable and antibiotic‐free.
Toxin/antitoxin systems as the major PAS group can be further divided into three subtypes: (i) antisense RNA‐regulated loci, whose antisense RNA‐antitoxin binds to toxin mRNA and thereby inhibits toxin translation (Gerdes et al., 1997), (ii) restriction modification systems with a restriction endonuclease cleaving foreign DNA and a methyltransferase that prevents host DNA from being cleaved due to methylation (Kobayashi, 2004), and (iii) the classical proteic TA systems, which consists of seven TA gene families (Pandey and Gerdes, 2005), whose toxin and antitoxin are both actively present as proteins (Engelberg‐Kulka and Glaser, 1999; Gerdes et al., 2005). TA systems were also discovered to be encoded chromosomally (Masuda et al., 1993; Aizenman et al., 1996; Pedersen and Gerdes, 1999), but their physiological role remains under speculative debate (Magnuson, 2007). The prevalent hypothesis is that they are involved in general environmental stress responses, e.g. to amino acid starvation, by induction of a bacteriostatic state and thus regulating the synthesis of macromolecules, such as DNA or proteins, depending on external nutrient supply (Gerdes et al., 2005; Kolodkin‐Gal and Engelberg‐Kulka, 2006). However, recent studies contradict this hypothesis by demonstrating that chromosomally encoded TA systems did not lead to selective advantages under stressful conditions (Szekeres et al., 2007; Tsilibaris et al., 2007; Saavedra de Bast et al., 2008).
Since the discovery of TA systems as natively occurring PAS in 1983 by Ogura and Hiraga, great efforts have been undertaken to identify additional TA systems and to enhance our knowledge on their characteristics and functionality (Gerdes et al., 2005; Pandey and Gerdes, 2005). The results have been used for implementation of TA systems in a large variety of applications. A major field of application is the construction of containment systems as artificial PAS for GEMs that are used in bioremediation (Lovley, 2003; Paul et al., 2005). These GEMs, mostly strains of E. coli or P. putida, are only viable in a specific environment for the desired task, e.g. the degradation of aromatic, phenolic or other organic compounds (Pieper and Reineke, 2000). Inadvertent escape or horizontal gene transfer is prevented as this induces cell killing mediated by inhibited expression of essential genes. This containment principle can also be employed on GEMs used for biochemical, biotechnological or pharmaceutical processes in industrial production (Molin et al., 1987; Schweder et al., 1992). The use of TA systems for maintenance of plasmids delivering heterologous antigens to the human immune system enables the necessarily antibiotic‐free production and represents a second large area of application. For example, expression plasmids in attenuated Salmonella spp. live vector vaccine strains were shown to be effective in treatments of bacterial infections (Galen et al., 1999; Garmory et al., 2005). Infections of E. coli or P. aeruginosa were treated with genetically engineered phages, which inject, inter alia, DNA‐encoded toxins that lead to bacterial cell death (Westwater et al., 2003; Hagens et al., 2004).
Finally, the following PAS should be mentioned: (i) TA systems as the major group of reported PAS, (ii) metabolism‐based PAS including the hitherto only catabolism‐based plasmid addiction system (CBS) (Voß and Steinbüchel, 2006), (iii) an outstanding ABS utilizes the biosynthesis pathway of isoprenoids (Kroll et al., 2009), (iv) complementation of auxotrophic mutations, e.g. for uracil and proline combined with expression of heterologous proteins in P. fluorescens, was reported (Schneider et al., 2005), and (v) ORT is based on essential chromosomal genes under control of the lac operator–promoter region, resulting in repression by binding of the lac repressor protein (Williams et al., 1998). Expression is only enabled in presence of plasmids that also harbour a lac operator–promoter region to titrate the repressor protein away from the chromosomal gene.
The increasing importance of the biotech industry with its interest in stable and recombinant production of proteins and chemicals especially based on renewable resources will be a key factor to predict PAS an important significance for the future. Nevertheless, not every PAS has reached an industrial applicable status. This might be the problem of an insufficient stabilization during the cultivation process or due to the difficult to integrate the system in existing production processes. Chromosomal integration of genes instead of using plasmid‐based systems as a simple solution is not obvious in every case. Important factors like polar effects or limited gene copy numbers are just two disadvantages of this alternative. The development of further applications for addiction systems will be helpful to overcome current problems in biotechnology or pharmaceutical industry. For example, the field of cancer treatment could be supported by the use of special addiction modules. Toxin molecules like RelE or Kid/Kis induce or trigger apoptosis in human cells (Kristoffersen et al., 2000; de la Cueva‐Mendez et al., 2003). As an example, this selective killing mechanism of eukaryotic (cancer) cells could be a future development based on the knowledge of naturally occurred addiction systems.
References
- Afif H., Allali N., Couturier M., Melderen L.V. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison‐antidote system. Mol Microbiol. 2001;41:73–82. doi: 10.1046/j.1365-2958.2001.02492.x. [DOI] [PubMed] [Google Scholar]
- Agarwal S., Agarwal S., Bhatnagar R. Identification and characterization of a novel toxin–antitoxin module from Bacillus anthracis. FEBS Lett. 2007;581:1727–1734. doi: 10.1016/j.febslet.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Aizenman E., Engelberg‐Kulka H., Glaser G. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine 3′,5′‐bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.C., Ayora S., Canosa I., Weise F., Rojo F. Site‐specific recombination in gram‐positive theta‐replicating plasmids. FEMS Microbiol Lett. 1996;142:1–10. doi: 10.1111/j.1574-6968.1996.tb08399.x. [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Aravind L. New connections in the prokaryotic toxin–antitoxin network: relationship with the eukaryotic nonsense‐mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.N., Hollis D.G., Thornsberry C. Antimicrobial susceptibility testing of Francisella tularensis with a modified mueller‐hinton broth. J Clin Microbiol. 1985;22:212–215. doi: 10.1128/jcm.22.2.212-215.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding C., Blaby I., Summers D. A mutational analysis of the ColE1‐encoded cell cycle regulator rcd confirms its role in plasmid stability. Plasmid. 2006;56:68–73. doi: 10.1016/j.plasmid.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Barry E.M., Gomez‐Duarte O., Chatfield S., Rappuoli R., Pizza M., Losonsky G. Expression and immunogenicity of pertussis toxin S1 subunit‐tetanus toxin fragment C fusions in Salmonella typhi vaccine strain CVD 908. Infect Immun. 1996;64:4172–4181. doi: 10.1128/iai.64.10.4172-4181.1996. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech F.W., Jørgensen S.T., Diderichsen B., Karlström O.H. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 1985;4:1059–1066. doi: 10.1002/j.1460-2075.1985.tb03739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej A.K., Perlin M.H., Atlas R.M. Model suicide vector for containment of genetically engineered microorganisms. Appl Environ Microbiol. 1988;54:2472–2477. doi: 10.1128/aem.54.10.2472-2477.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P. New ccdB positive‐selection cloning vectors with kanamycin or chloramphenicol selectable markers. Gene. 1995;162:159–160. doi: 10.1016/0378-1119(95)00314-v. [DOI] [PubMed] [Google Scholar]
- Bernard P., Gabant P., Bahassi E.M., Couturier M. Positive‐selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- Black D.S., Kelly A.J., Mardis M.J., Moyed H.S. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1991;173:5732–5739. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.S., Irwin B., Moyed H.S. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J Bacteriol. 1994;176:4081–4091. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackkolb F., Schlegel H.G. Katabolische Repression und Enzymhemmung durch molekularen Wasserstoff bei Hydrogenomonas. Arch Mikrobiol. 1968;62:129–143. [PubMed] [Google Scholar]
- Bodogai M., Ferenczi S., Bashtovyy D., Miclea P., Papp P., Dusha I. The ntrPR operon of Sinorhizobium meliloti is organized and functions as a toxin–antitoxin module. Mol Plant Microbe Interact. 2006;19:811–822. doi: 10.1094/MPMI-19-0811. [DOI] [PubMed] [Google Scholar]
- Bravo A., De Torrontegui G., Díaz R. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol Gen Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- Brenner M., Tomizawa J. Quantitation of ColE1‐encoded replication elements. Proc Natl Acad Sci USA. 1991;88:405–409. doi: 10.1073/pnas.88.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.L., Grigoriu S., Kim Y., Arruda J.M., Davenport A., Wood T.K. Three dimensional structure of the MqsR:MqsA complex: a novel TA pair comprised of a toxin homologous to RelE and an antitoxin with unique properties. PLoS Pathog. 2009;5:e1000706. doi: 10.1371/journal.ppat.1000706. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde P.P., Davis B.M., Yuan J., Waldor M.K. Characterization of a higBA toxin–antitoxin locus in Vibrio cholerae. J Bacteriol. 2007;189:491–500. doi: 10.1128/JB.00909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting K.A., Roe S.M., Headley A., Brown T., Savva R., Pearl L.H. Crystal structure of the Escherichia coli dcm very‐short‐patch DNA repair endonuclease bound to its reaction product‐site in a DNA superhelix. Nucleic Acids Res. 2003;31:1633–1639. doi: 10.1093/nar/gkg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buts L., Lah J., Dao‐Thi M., Wyns L., Loris R. Toxin–antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci. 2005;30:672–679. doi: 10.1016/j.tibs.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Calvin‐Koons M.D., Blumenthal R.M. Characterization of pPvu1, the autonomous plasmid from Proteus vulgaris that carries the genes of the PvuII restriction–modification system. Gene. 1995;157:73–79. doi: 10.1016/0378-1119(94)00618-3. [DOI] [PubMed] [Google Scholar]
- Camacho A.G., Misselwitz R., Behlke J., Ayora S., Welfle K., Meinhart A. In vitro and in vivo stability of the ε2ζ2 protein complex of the broad host‐range Streptococcus pyogenes pSM19035 addiction system. Biol Chem. 2002;383:1701–1713. doi: 10.1515/BC.2002.191. et al. [DOI] [PubMed] [Google Scholar]
- Cases I., De Lorenzo V. Genetically modified organisms for the environment: stories of success and failure and what we have learned from them. Int Microbiol. 2005;8:213–222. [PubMed] [Google Scholar]
- Cegłowski P., Boitsov A., Chai S., Alonso J.C. Analysis of the stabilization system of pSM19035‐derived plasmid pBT233 in Bacillus subtilis. Gene. 1993a;136:1–12. doi: 10.1016/0378-1119(93)90441-5. [DOI] [PubMed] [Google Scholar]
- Cegłowski P., Boitsov A., Karamyan N., Chai S., Alonso J.C. Characterization of the effectors required for stable inheritance of Streptococcus pyogenes pSM19035‐derived plasmids in Bacillus subtilis. Mol Gen Genet. 1993b;241:579–585. doi: 10.1007/BF00279900. [DOI] [PubMed] [Google Scholar]
- Cesareni G., Muesing M.A., Polisky B. Control of ColE1 DNA replication: the rop gene product negatively affects transcription from the replication primer promoter. Proc Natl Acad Sci USA. 1982;79:6313–6317. doi: 10.1073/pnas.79.20.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Balendiran K., Schildkraut I., Anderson J.E. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny I., Gazit E. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J Biol Chem. 2004;279:8252–8261. doi: 10.1074/jbc.M308263200. [DOI] [PubMed] [Google Scholar]
- Christensen S.K., Gerdes K. RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48:1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- Christensen S.K., Mikkelsen M., Pedersen K., Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci USA. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.K., Pedersen K., Hansen F.G., Gerdes K. Toxin–antitoxin loci as stress‐response‐elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J Mol Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- Christensen‐Dalsgaard M., Gerdes K. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol Microbiol. 2006;62:397–411. doi: 10.1111/j.1365-2958.2006.05385.x. [DOI] [PubMed] [Google Scholar]
- Clissold P.M., Ponting C.P. PIN domains in nonsense‐mediated mRNA decay and RNAi. Curr Biol. 2000;10:R888–R890. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- Conlan J.W., Oyston P.C.F. Vaccines against Francisella tularensis. Ann N Y Acad Sci. 2007;1105:325–350. doi: 10.1196/annals.1409.012. [DOI] [PubMed] [Google Scholar]
- Contreras A., Molin S., Ramos J. Conditional‐suicide containment system for bacteria which mineralize aromatics. Appl Environ Microbiol. 1991;57:1504–1508. doi: 10.1128/aem.57.5.1504-1508.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covone M.G., Brocchi M., Palla E., Da Silveira W.D., Rappuoli R., Galeotti C.L. Levels of expression and immunogenicity of attenuated Salmonella enterica serovar typhimurium strains expressing Escherichia coli mutant heat‐labile enterotoxin. Infect Immun. 1998;66:224–231. doi: 10.1128/iai.66.1.224-231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranenburgh R.M., Hanak J.A., Williams S.G., Sherratt D.J. Escherichia coli strains that allow antibiotic‐free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 2001;29:E26. doi: 10.1093/nar/29.5.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranenburgh R.M., Lewis K.S., Hanak J.A.J. Effect of plasmid copy number and lac operator sequence on antibiotic‐free plasmid selection by operator‐repressor titration in Escherichia coli. J Mol Microbiol Biotechnol. 2004;7:197–203. doi: 10.1159/000079828. [DOI] [PubMed] [Google Scholar]
- De La Cueva‐Mendez G., Mills A.D., Clay‐Farrace L., Diaz‐Orejas R., Laskey R.A. Regulatable killing of eukaryotic cells by the prokaryotic proteins Kid and Kis. EMBO J. 2003;22:246–251. doi: 10.1093/emboj/cdg026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daines D.A., Wu M.H., Yuan S.Y. VapC‐1 of nontypeable haemophilus influenzae is a ribonuclease. J Bacteriol. 2007;189:5041–5048. doi: 10.1128/JB.00290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam M., Gerdes K. Partitioning of plasmid R1: ten direct repeats flanking the parA promoter constitute a centromere‐like partition site parC, that expresses incompatibility. J Mol Biol. 1994;236:1289–1298. doi: 10.1016/0022-2836(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Davis T.L., Helinski D.R., Roberts R.C. Transcription and autoregulation of the stabilizing functions of broad‐host‐range plasmid RK2 in Escherichia coliAgrobacterium tumefaciens and Pseudomonas aeruginosa. Mol Microbiol. 1992;6:1981–1994. doi: 10.1111/j.1365-2958.1992.tb01371.x. [DOI] [PubMed] [Google Scholar]
- Dennis D.T., Inglesby T.V., Henderson D.A., Bartlett J.G., Ascher M.S., Eitzen E. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. et al. [DOI] [PubMed] [Google Scholar]
- Diamand E. Genetically modified organisms and monitoring. J Environ Monit. 1999;1:108N–110N. doi: 10.1039/a908767b. [DOI] [PubMed] [Google Scholar]
- Dua M., Singh A., Sethunathan N., Johri A.K. Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biotechnol. 2002;59:143–152. doi: 10.1007/s00253-002-1024-6. [DOI] [PubMed] [Google Scholar]
- Eberl L., Givskov M., Schwab H. The divergent promoters mediating transcription of the par locus of plasmid RP4 are subject to autoregulation. Mol Microbiol. 1992;6:1969–1979. doi: 10.1111/j.1365-2958.1992.tb01370.x. [DOI] [PubMed] [Google Scholar]
- Edman J.C., Kwon‐Chung K.J. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg‐Kulka H., Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- Engelberg‐Kulka H., Hazan R., Amitai S. mazEF: a chromosomal toxin–antitoxin module that triggers programmed cell death in bacteria. J Cell Sci. 2005;118:4327–4332. doi: 10.1242/jcs.02619. [DOI] [PubMed] [Google Scholar]
- Falla T.J., Chopra I. Joint tolerance to beta‐lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA. Antimicrob Agents Chemother. 1998;42:3282–3284. doi: 10.1128/aac.42.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falla T.J., Chopra I. Stabilization of Rhizobium symbiosis plasmids. Microbiology. 1999;145:515–516. doi: 10.1099/13500872-145-3-515. [DOI] [PubMed] [Google Scholar]
- Faridani O.R., Nikravesh A., Pandey D.P., Gerdes K., Good L. Competitive inhibition of natural antisense Sok–RNA interactions activates Hok‐mediated cell killing in Escherichia coli. Nucleic Acids Res. 2006;34:5915–5922. doi: 10.1093/nar/gkl750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fico S., Mahillon J. TasA–tasB, a new putative toxin–antitoxin (TA) system from Bacillus thuringiensis pGI1 plasmid is a widely distributed composite mazE–doc TA system. BMC Genomics. 2006;7:259. doi: 10.1186/1471-2164-7-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.W., Zahrt T.C. Genetics and genetic manipulation in Francisella tularensis. Ann N Y Acad Sci. 2007;1105:67–97. doi: 10.1196/annals.1409.008. [DOI] [PubMed] [Google Scholar]
- Friehs K. Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol. 2004;86:47–82. doi: 10.1007/b12440. [DOI] [PubMed] [Google Scholar]
- Funnell B.E., Slavcev R.A. Partition systems of bacterial plasmids. In: Funnell B.E., Phillips G.J., editors. ASM Press; 2004. pp. 81–103. [Google Scholar]
- Furukawa K. ‘Super bugs’ for bioremediation. Trends Biotechnol. 2003;21:187–190. doi: 10.1016/S0167-7799(03)00054-4. [DOI] [PubMed] [Google Scholar]
- Galen J.E., Gomez‐Duarte O.G., Losonsky G.A., Halpern J.L., Lauderbaugh C.S., Kaintuck S. A murine model of intranasal immunization to assess the immunogenicity of attenuated Salmonella typhi live vector vaccines in stimulating serum antibody responses to expressed foreign antigens. Vaccine. 1997;15:700–708. doi: 10.1016/s0264-410x(96)00227-7. et al. [DOI] [PubMed] [Google Scholar]
- Galen J.E., Nair J., Wang J.Y., Wasserman S.S., Tanner M.K., Sztein M.B., Levine M.M. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908‐htrA. Infect Immun. 1999;67:6424–6433. doi: 10.1128/iai.67.12.6424-6433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmory H.S., Brown K.A., Titball R.W. Salmonella vaccines for use in humans: present and future perspectives. FEMS Microbiol Rev. 2002;26:339–353. doi: 10.1111/j.1574-6976.2002.tb00619.x. [DOI] [PubMed] [Google Scholar]
- Garmory H.S., Leckenby M.W., Griffin K.F., Elvin S.J., Taylor R.R., Hartley M.G. Antibiotic‐free plasmid stabilization by operator‐repressor titration for vaccine delivery by using live Salmonella enterica serovar typhimurium. Infect Immun. 2005;73:2005–2011. doi: 10.1128/IAI.73.4.2005-2011.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit E., Sauer R.T. The Doc toxin and Phd antidote proteins of the bacteriophage P1 plasmid addiction system form a heterotrimeric complex. J Biol Chem. 1999a;274:16813–16818. doi: 10.1074/jbc.274.24.16813. [DOI] [PubMed] [Google Scholar]
- Gazit E., Sauer R.T. Stability and DNA binding of the Phd protein of the phage P1 plasmid addiction system. J Biol Chem. 1999b;274:2652–2657. doi: 10.1074/jbc.274.5.2652. [DOI] [PubMed] [Google Scholar]
- Geller B.L., Deere J., Tilley L., Iversen P.L. Antisense phosphorodiamidate morpholino oligomer inhibits viability of Escherichia coli in pure culture and in mouse peritonitis. J Antimicrob Chemother. 2005;55:983–988. doi: 10.1093/jac/dki129. [DOI] [PubMed] [Google Scholar]
- Gerdes K. The parBhoksok) locus of plasmid R1: a general purpose plasmid stabilization system. Bio/Technology. 1988;6:1402–1405. [Google Scholar]
- Gerdes K. Toxin–antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid‐free cells. Proc Natl Acad Sci USA. 1986a;10:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Bech F.W., Jørgensen S.T., Løbner‐Olesen A., Rasmussen P.B., Atlung T. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986b:2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Nielsen A., Thorsted P., Wagner E.G. Mechanism of killer gene activation: antisense RNA‐dependent RNase III cleavage ensures rapid turn‐over of the stable Hok, SrnB and PndA effector messenger RNAs. J Mol Biol. 1992;226:637–649. doi: 10.1016/0022-2836(92)90621-p. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Gultyaev A.P., Franch T., Pedersen K., Mikkelsen N.D. Antisense RNA‐regulated programmed cell death. Annu Rev Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Christensen S.K., Løbner‐Olesen A. Prokaryotic toxin–antitoxin stress response loci. Nat Rev Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- Gerlitz M., Hrabak O., Schwab H. Partitioning of broad‐host‐range plasmid RP4 is a complex system involving site‐specific recombination. J Bacteriol. 1990;172:6194–6203. doi: 10.1128/jb.172.11.6194-6203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong W., O'Gara M., Blumenthal R.M., Cheng X. Structure of Pvu II DNA‐(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good L., Awasthi S.K., Dryselius R., Larsson O., Nielsen P.E. Bactericidal antisense effects of peptide–PNA conjugates. Nat Biotechnol. 2001;19:360–364. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- Gotfredsen M., Gerdes K. The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- Gottschalk G., Eberhardt U., Schlegel H.G. Verwertung von Fructose durch Hydrogenomonas H 16 (I.) Arch Mikrobiol. 1964;48:95–108. [PubMed] [Google Scholar]
- Grady R., Hayes F. Axe‐txe, a broad‐spectrum proteic toxin–antitoxin system specified by a multidrug‐resistant, clinical isolate of Enterococcus faecium. Mol Microbiol. 2003;47:1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- Greenfield T.J., Ehli E., Kirshenmann T., Franch T., Gerdes K., Weaver K.E. The antisense RNA of the par locus of pAD1 regulates the expression of a 33‐amino‐acid toxic peptide by an unusual mechanism. Mol Microbiol. 2000;37:652–660. doi: 10.1046/j.1365-2958.2000.02035.x. [DOI] [PubMed] [Google Scholar]
- Grindley N.D., Whiteson K.L., Rice P.A. Mechanisms of site‐specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- Gross M., Marianovsky I., Glaser G. MazG – a regulator of programmed cell death in Escherichia coli. Mol Microbiol. 2006;59:590–601. doi: 10.1111/j.1365-2958.2005.04956.x. [DOI] [PubMed] [Google Scholar]
- Hagens S., Bläsi U. Genetically modified filamentous phage as bactericidal agents: a pilot study. Lett Appl Microbiol. 2003;37:318–323. doi: 10.1046/j.1472-765x.2003.01400.x. [DOI] [PubMed] [Google Scholar]
- Hagens S., Habel A., Von Ahsen U., Von Gabain A., Bläsi U. Therapy of experimental Pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob Agents Chemother. 2004;48:3817–3822. doi: 10.1128/AAC.48.10.3817-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägg P., De Pohl, Wa Johanna, Abdulkarim F., Isaksson L.A. A host/plasmid system that is not dependent on antibiotics and antibiotic resistance genes for stable plasmid maintenance in Escherichia coli. J Biotechnol. 2004;111:17–30. doi: 10.1016/j.jbiotec.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Hanak J., Cranenburgh R., Gorman S. 2003. , and ) Plasmid stabilisation in vivo. Patent application WO 03/097838.
- Handa N., Kobayashi I. Post‐segregational killing by restriction modification gene complexes: observations of individual cell deaths. Biochimie. 1999;81:931–938. doi: 10.1016/s0300-9084(99)00201-1. [DOI] [PubMed] [Google Scholar]
- Handa N., Ichige A., Kusano K., Kobayashi I. Cellular responses to postsegregational killing by restriction–modification genes. J Bacteriol. 2000;182:2218–2229. doi: 10.1128/jb.182.8.2218-2229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R., Sat B., Reches M., Engelberg‐Kulka H. Postsegregational killing mediated by the P1 phage ‘addiction module’phd‐doc requires the Escherichia coli programmed cell death system mazEF. J Bacteriol. 2001;183:2046–2050. doi: 10.1128/JB.183.6.2046-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R., Sat B., Engelberg‐Kulka H. Escherichia coli mazEF‐mediated cell death is triggered by various stressful conditions. J Bacteriol. 2004;186:3663–3669. doi: 10.1128/JB.186.11.3663-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgpeth J., Goodman H.M., Boyer H.W. DNA nucleotide sequence restricted by the RI endonuclease. Proc Natl Acad Sci USA. 1972;69:3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensing M.C., Rouwenhorst R.J., Heijnen J.J., Van Dijken J.P., Pronk J.T. Physiological and technological aspects of large‐scale heterologous‐protein production with yeasts. Antonie Van Leeuwenhoek. 1995;67:261–279. doi: 10.1007/BF00873690. [DOI] [PubMed] [Google Scholar]
- Hopper S., Wilbur J.S., Vasquez B.L., Larson J., Clary S., Mehr I.J. Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infect Immun. 2000;68:896–905. doi: 10.1128/iai.68.2.896-905.2000. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Hoz A.B., Ayora S., Sitkiewicz I., Fernández S., Pankiewicz R., Alonso J.C., Cegłowski P. Plasmid copy‐number control and better‐than‐random segregation genes of pSM19035 share a common regulator. Proc Natl Acad Sci USA. 2000;97:728–733. doi: 10.1073/pnas.97.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Hoz A.B., Pratto F., Misselwitz R., Speck C., Weihofen W., Welfle K. Recognition of DNA by omega protein from the broad‐host range Streptococcus pyogenes plasmid pSM19035: analysis of binding to operator DNA with one to four heptad repeats. Nucleic Acids Res. 2004;32:3136–3147. doi: 10.1093/nar/gkh633. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichige A., Kobayashi I. Stability of EcoRI restriction–modification enzymes in vivo differentiates the EcoRI restriction–modification system from other postsegregational cell killing systems. J Bacteriol. 2005;187:6612–6621. doi: 10.1128/JB.187.19.6612-6621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé A., Ogura T., Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985;163:841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana S., Deb J.K. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl Microbiol Biotechnol. 2005;67:289–298. doi: 10.1007/s00253-004-1814-0. [DOI] [PubMed] [Google Scholar]
- Jensen R.B., Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- Jensen L.B., Ramos J.L., Kaneva Z., Molin S. A substrate‐dependent biological containment system for Pseudomonas putida based on the Escherichia coli gef gene. Appl Environ Microbiol. 1993;59:3713–3717. doi: 10.1128/aem.59.11.3713-3717.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Pogliano J., Helinski D.R., Konieczny I. ParE toxin encoded by the broad‐host‐range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- Johnson I.S. Human insulin from recombinant DNA technology. Science. 1983;219:632–637. doi: 10.1126/science.6337396. [DOI] [PubMed] [Google Scholar]
- Johnson E.P., Strom A.R., Helinski D.R. Plasmid RK2 toxin protein ParE: purification and interaction with the ParD antitoxin protein. J Bacteriol. 1996;178:1420–1429. doi: 10.1128/jb.178.5.1420-1429.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.L., Mello C., Sano T., Cantor C., Smith C. Streptavidin‐based containment systems for genetically engineered microorganisms. Biomol Eng. 1999;16:135–140. doi: 10.1016/s1050-3862(99)00040-6. [DOI] [PubMed] [Google Scholar]
- Karem K.L., Chatfield S., Kuklin N., Rouse B.T. Differential induction of carrier antigen‐specific immunity by Salmonella typhimurium live‐vaccine strains after single mucosal or intravenous immunization of BALB/c mice. Infect Immun. 1995;63:4557–4563. doi: 10.1128/iai.63.12.4557-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karyagina A., Shilov I., Tashlitskii V., Khodoun M., Vasil'ev S., Lau P.C., Nikolskaya I. Specific binding of SsoII DNA methyltransferase to its promoter region provides the regulation of ssoII restriction–modification gene expression. Nucleic Acids Res. 1997;25:2114–2120. doi: 10.1093/nar/25.11.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M.E., Strugnell R.A., Rood J.I. Molecular characterization of a genomic region associated with virulence in Dichelobacter nodosus. Infect Immun. 1992;60:4586–4592. doi: 10.1128/iai.60.11.4586-4592.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M., Oshima T., Kasai H., Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35‐amino‐acid cell‐killing peptide and a cis‐encoded small antisense RNA in Escherichia coli. Mol Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- Kawano M., Aravind L., Storz G. An antisense RNA controls synthesis of an SOS‐induced toxin evolved from an antitoxin. Mol Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo S.K., Loll B., Chan W.T., Shoeman R.L., Ngoo L., Yeo C.C., Meinhart A. Molecular and structural characterization of the PezAT chromosomal toxin–antitoxin system of the human pathogen Streptococcus pneumoniae. J Biol Chem. 2007;282:19606–19618. doi: 10.1074/jbc.M701703200. [DOI] [PubMed] [Google Scholar]
- Kim Y., Wood T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2009;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Wang X., Ma Q., Zhang X.S., Wood T.K. Toxin–antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J Bacteriol. 2009;191:1258–1267. doi: 10.1128/JB.01465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P., Jensen L.B., Molin S. A stochastic killing system for biological containment of Escherichia coli. Appl Environ Microbiol. 1995;61:481–486. doi: 10.1128/aem.61.2.481-486.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S.M., Karlström O.H. Development of efficient suicide mechanisms for biological containment of bacteria. Appl Environ Microbiol. 1991;57:85–92. doi: 10.1128/aem.57.1.85-92.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen S., Saadbye P., Hansen L.H., Collier A., Jacobsen B.L., Schlundt J., Karlström O.H. Development and testing of improved suicide functions for biological containment of bacteria. Appl Environ Microbiol. 1995;61:985–991. doi: 10.1128/aem.61.3.985-991.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I. Genetic addiction: a principle of gene symbiosis in a genome. Plasmid Biol. 2004:105–144. [Google Scholar]
- Kolodkin‐Gal I., Engelberg‐Kulka H. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J Bacteriol. 2006;188:3420–3423. doi: 10.1128/JB.188.9.3420-3423.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin‐Gal I., Hazan R., Gaathon A., Carmeli S., Engelberg‐Kulka H. A linear pentapeptide is a quorum‐sensing factor required for mazEF‐mediated cell death in Escherichia coli. Science. 2007;318:652–655. doi: 10.1126/science.1147248. [DOI] [PubMed] [Google Scholar]
- Kolodkin‐Gal I., Verdiger R., Shlosberg‐Fedida A., Engelberg‐Kulka H. A differential effect of E. coli toxin–antitoxin systems on cell death in liquid media and biofilm formation. PLoS ONE. 2009;4:e6785. doi: 10.1371/journal.pone.0006785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch S.B., Henderson T.A., Hill T.M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- Kovachevich R., Wood W.A. Carbohydrate metabolism by Pseudomonas fluorescens. IV. Purification and properties of 2‐keto‐3‐deoxy‐6‐phosphogluconate aldolase. J Biol Chem. 1955;213:757–767. [PubMed] [Google Scholar]
- Kristoffersen P., Jensen G.B., Gerdes K., Piskur J. Bacterial toxin–antitoxin gene system as containment control in yeast cells. Appl Environ Microbiol. 2000;66:5524–5526. doi: 10.1128/aem.66.12.5524-5526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll J., Steinle A., Reichelt R., Ewering E., Steinbüchel A. Establishment of a novel anabolism‐based addiction system with an artificially introduced mevalonate pathway: complete stabilization of plasmids as universal application in white biotechnology. Metab Eng. 2009;11:168–177. doi: 10.1016/j.ymben.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Kulakauskas S., Lubys A., Ehrlich S.D. DNA restriction–modification systems mediate plasmid maintenance. J Bacteriol. 1995;177:3451–3454. doi: 10.1128/jb.177.12.3451-3454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K., Naito T., Handa N., Kobayashi I. Restriction–modification systems as genomic parasites in competition for specific sequences. Proc Natl Acad Sci USA. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnherr H., Yarmolinsky M.B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnherr H., Maguin E., Jafri S., Yarmolinsky M.B. Plasmid addiction genes of bacteriophage P1: Doc, which causes cell death on curing of prophage, and Phd, which prevents host death when prophage is retained. J Mol Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- Lieb M. Spontaneous mutation at a 5‐methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genet. 1991;128:23–27. doi: 10.1093/genetics/128.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy V.S., Martín M.T., Camacho A.G., Lurz R., Antelmann H., Hecker M. pSM19035‐encoded zeta toxin induces stasis followed by death in a subpopulation of cells. Microbiology. 2006;152:2365–2379. doi: 10.1099/mic.0.28950-0. et al. [DOI] [PubMed] [Google Scholar]
- Lovley D.R. Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol. 2003;1:35–44. doi: 10.1038/nrmicro731. [DOI] [PubMed] [Google Scholar]
- LoVullo E.D., Sherrill L.A., Perez L.L., Pavelka M.S. Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology. 2006;152:3425–3435. doi: 10.1099/mic.0.29121-0. [DOI] [PubMed] [Google Scholar]
- Lubys A., Janulaitis A. Cloning and analysis of the plasmid‐borne genes encoding the Bsp6I restriction and modification enzymes. Gene. 1995;157:25–29. doi: 10.1016/0378-1119(94)00795-t. [DOI] [PubMed] [Google Scholar]
- McAteer S., Coulson A., McLennan N., Masters M. The lytB gene of Escherichia coli is essential and specifies a product needed for isoprenoid biosynthesis. J Bacteriol. 2001;183:7403–7407. doi: 10.1128/JB.183.24.7403-7407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley‐Patrick S., Fazenda M.L., McNeil B., Harvey L.M. Heterologous protein production using the Pichia pastorisexpression system. Yeast. 2005;22:249–270. doi: 10.1002/yea.1208. [DOI] [PubMed] [Google Scholar]
- Magnuson R.D. Hypothetical functions of toxin–antitoxin systems. J Bacteriol. 2007;189:6089–6092. doi: 10.1128/JB.00958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R., Yarmolinsky M.B. Corepression of the P1 addiction operon by Phd and Doc. J Bacteriol. 1998;180:6342–6351. doi: 10.1128/jb.180.23.6342-6351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson R., Lehnherr H., Mukhopadhyay G., Yarmolinsky M.B. Autoregulation of the plasmid addiction operon of bacteriophage P1. J Biol Chem. 1996;271:18705–18710. doi: 10.1074/jbc.271.31.18705. [DOI] [PubMed] [Google Scholar]
- Makarova K.S., Grishin N.V., Koonin E.V. The HicAB cassette, a putative novel, RNA‐targeting toxin–antitoxin system in archaea and bacteria. Bioinformatics. 2006;22:2581–2584. doi: 10.1093/bioinformatics/btl418. [DOI] [PubMed] [Google Scholar]
- Masuda Y., Miyakawa K., Nishimura Y., Ohtsubo E. chpA and chpBEscherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K., Wilbur J.S., So M., Brennan R.G. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin–antitoxin heterodimers containing pin domains and ribbon–helix–helix motifs. J Biol Chem. 2006;281:37942–37951. doi: 10.1074/jbc.M605198200. [DOI] [PubMed] [Google Scholar]
- Meinander N.Q., Hahn‐Hägerdal B. Fed‐batch xylitol production with two recombinant Saccharomyces cerevisiae strains expressing XYL1 at different levels, using glucose as a cosubstrate: a comparison of production parameters and strain stability. Appl Microbiol. 1997;54:391–399. doi: 10.1002/(SICI)1097-0290(19970520)54:4<391::AID-BIT12>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Meinhart A., Alonso J.C., Sträter N., Saenger W. Crystal structure of the plasmid maintenance system epsilon/zeta: functional mechanism of toxin zeta and inactivation by ε2ζ2 complex formation. Proc Natl Acad Sci USA. 2003;100:1661–1666. doi: 10.1073/pnas.0434325100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger S., Dror I.B., Aizenman E., Schreiber G., Toone M., Friesen J.D. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988;263:15699–15704. et al. [PubMed] [Google Scholar]
- Mhlanga‐Mutangadura T., Morlin G., Smith A.L., Eisenstark A., Golomb M. Evolution of the major pilus gene cluster of Haemophilus influenzae. J Bacteriol. 1998;180:4693–4703. doi: 10.1128/jb.180.17.4693-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Chang Z.T., Horiuchi T. Control of cell division by sex factor F in Escherichia coli. II. Identification of genes for inhibitor protein and trigger protein on the 42.84‐43.6 F segment. J Mol Biol. 1984a;174:627–646. doi: 10.1016/0022-2836(84)90087-1. [DOI] [PubMed] [Google Scholar]
- Miki T., Yoshioka K., Horiuchi T. Control of cell division by sex factor F in Escherichia coli. I. The 42.84–43.6 F segment couples cell division of the host bacteria with replication of plasmid DNA. J Mol Biol. 1984b;174:605–625. doi: 10.1016/0022-2836(84)90086-x. [DOI] [PubMed] [Google Scholar]
- Mittenhuber G. Occurrence of mazEF‐like antitoxin/toxin systems in bacteria. J Mol Microbiol Biotechnol. 1999;1:295–302. [PubMed] [Google Scholar]
- Molin S., Klemm P., Poulsen K., Biehl H., Gerdes K., Andersson P. Conditional suicide system for containment of bacteria and plasmids. Bio/Technology. 1987;5:1315–1318. [Google Scholar]
- Molin S., Boe L., Jensen L.B., Kristensen C.S., Givskov M., Ramos J.L., Bej A.K. Suicidal genetic elements and their use in biological containment of bacteria. Annu Rev Microbiol. 1993;47:139–166. doi: 10.1146/annurev.mi.47.100193.001035. [DOI] [PubMed] [Google Scholar]
- Mondon P., Chang Y.C., Varma A., Kwon‐Chung K.J. A novel episomal shuttle vector for transformation of Cryptococcus neoformans with the ccdB gene as a positive selection marker in bacteria. FEMS Microbiol Lett. 2000;187:41–45. doi: 10.1111/j.1574-6968.2000.tb09134.x. [DOI] [PubMed] [Google Scholar]
- Mooibroek H., Oosterhuis N., Giuseppin M., Toonen M., Franssen H., Scott E. Assessment of technological options and economical feasibility for cyanophycin biopolymer and high‐value amino acid production. Appl Microbiol Biotechnol. 2007;77:257–267. doi: 10.1007/s00253-007-1178-3. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motiejūnaite R., Armalyte J., Markuckas A., Suziedeliene E. Escherichia coli dinJ–yafQ genes act as a toxin–antitoxin module. FEMS Microbiol Lett. 2007;268:112–119. doi: 10.1111/j.1574-6968.2006.00563.x. [DOI] [PubMed] [Google Scholar]
- Moyed H.S., Bertrand K.P. hipA, a newly recognized gene of Escherichia coli K‐12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed H.S., Broderick S.H. Molecular cloning and expression of hipA, a gene of Escherichia coli K‐12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol. 1986;166:399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller‐Hill B., Crapo L., Gilbert W. Mutants that make more lac repressor. Proc Natl Acad Sci USA. 1968;59:1259–1264. doi: 10.1073/pnas.59.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munthali M.T., Timmis K.N., Diaz E. Use of colicin E3 for biological containment of microorganisms. Appl Environ Microbiol. 1996a;62:1805–1807. doi: 10.1128/aem.62.5.1805-1807.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munthali M.T., Timmis K.N., Díaz E. Restricting the dispersal of recombinant DNA: design of a contained biological catalyst. Biotechnology (NY) 1996b;14:189–191. doi: 10.1038/nbt0296-189. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Kelly S.M., Curtiss R., 3rd Construction of an ASD+ expression‐cloning vector: stable maintenance and high level expression of cloned genes in a salmonella vaccine strain. Bio/Technology. 1988;6:693–697. [Google Scholar]
- Nakayama Y., Kobayashi I. Restriction–modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc Natl Acad Sci USA. 1998;95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nariya H., Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Nieto C., Cherny I., Khoo S.K., De Lacoba M.G., Chan W.T., Yeo C.C. The yefM–yoeB toxin–antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation. J Bacteriol. 2007;189:1266–1278. doi: 10.1128/JB.01130-06. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom K., Austin S.J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- Norioka S., Ramakrishnan G., Ikenaka K., Inouye M. Interaction of a transcriptional activator, OmpR, with reciprocally osmoregulated genes, ompF and ompC, of Escherichia coli. J Biol Chem. 1986;261:17113–17119. [PubMed] [Google Scholar]
- Norqvist A., Kuoppa K., Sandström G. Construction of a shuttle vector for use in Francisella tularensis. FEMS Immunol Med Microbiol. 1996;13:257–260. doi: 10.1111/j.1574-695X.1996.tb00248.x. [DOI] [PubMed] [Google Scholar]
- O'Connor E.B., O'Sullivan O., Stanton C., Danielsen M., Simpson P.J., Callanan M.J. pEOC01: a plasmid from Pediococcus acidilactici which encodes an identical streptomycin resistance (aadE) gene to that found in Campylobacter jejuni. Plasmid. 2007;58:115–126. doi: 10.1016/j.plasmid.2007.02.002. et al. [DOI] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini‐F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci USA. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S., Handa N., Watanabe‐Matsui M., Takahashi N., Kobayashi I. Maintenance forced by a restriction–modification system can be modulated by a region in its modification enzyme not essential for methyltransferase activity. J Bacteriol. 2008;190:2039–2049. doi: 10.1128/JB.01319-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oláh B., Kiss E., Györgypál Z., Borzi J., Cinege G., Csanádi G. Mutation in the ntrR gene, a member of the vap gene family, increases the symbiotic efficiency of Sinorhizobium meliloti. Mol Plant Microbe Interact. 2001;14:887–894. doi: 10.1094/MPMI.2001.14.7.887. et al. [DOI] [PubMed] [Google Scholar]
- Palmer B.R., Marinus M.G. The dam and dcm strains of Escherichia coli– a review. Gene. 1994;143:1–12. doi: 10.1016/0378-1119(94)90597-5. [DOI] [PubMed] [Google Scholar]
- Pandey D.P., Gerdes K. Toxin–antitoxin loci are highly abundant in free‐living but lost from host‐associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G., Paul D., Jain R.K. Conceptualizing ‘suicidal genetically engineered microorganisms’ for bioremediation applications. Biochem Biophys Res Commun. 2005;327:637–639. doi: 10.1016/j.bbrc.2004.12.080. [DOI] [PubMed] [Google Scholar]
- Patient M.E., Summers D.K. ColE1 multimer formation triggers inhibition of Escherichia coli cell division. Mol Microbiol. 1993;9:1089–1095. doi: 10.1111/j.1365-2958.1993.tb01238.x. [DOI] [PubMed] [Google Scholar]
- Paul D., Pandey G., Jain R.K. Suicidal genetically engineered microorganisms for bioremediation: need and perspectives. Bioessays. 2005;27:563–573. doi: 10.1002/bies.20220. [DOI] [PubMed] [Google Scholar]
- Pecota D.C., Kim C.S., Wu K., Gerdes K., Wood T.K. Combining the hoksokparDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl Environ Microbiol. 1997;63:1917–1924. doi: 10.1128/aem.63.5.1917-1924.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K., Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- Pedersen K., Christensen S.K., Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- Pfaffenzeller I., Striedner G., Bayer K., Grabherr R. ColE1 derived RNA I as a key molecule in a novel antibiotic free plasmid addiction system. Microb Cell Fact. 2006a;5:P87–P88. [Google Scholar]
- Pfaffenzeller I., Mairhofer J., Striedner G., Bayer K., Grabherr R. Using ColE1‐derived RNA I for suppression of a bacterially encoded gene: implication for a novel plasmid addiction system. Biotechnol J. 2006b;1:675–681. doi: 10.1002/biot.200600017. [DOI] [PubMed] [Google Scholar]
- Pieper D.H., Reineke W. Engineering bacteria for bioremediation. Curr Opin Biotechnol. 2000;11:262–270. doi: 10.1016/s0958-1669(00)00094-x. [DOI] [PubMed] [Google Scholar]
- Pohlmann A., Fricke W.F., Reinecke F., Kusian B., Liesegang H., Cramm R. Genome sequence of the bioplastic‐producing ‘knallgas’ bacterium Ralstonia eutropha H16. Nat Biotechnol. 2006;24:1257–1262. doi: 10.1038/nbt1244. et al. [DOI] [PubMed] [Google Scholar]
- Pomerantsev A.P., Golovliov I.R., Ohara Y., Mokrievich A.N., Obuchi M., Norqvist A. Genetic organization of the Francisella plasmid pFNL10. Plasmid. 2001;46:210–222. doi: 10.1006/plas.2001.1548. et al. [DOI] [PubMed] [Google Scholar]
- Pullinger G.D., Lax A.J. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient‐limited conditions. Mol Microbiol. 1992;6:1631–1643. doi: 10.1111/j.1365-2958.1992.tb00888.x. [DOI] [PubMed] [Google Scholar]
- Recorbet G., Robert C., Givaudan A., Kudla B., Normand P., Faurie G. Conditional suicide system of Escherichia coli released into soil that uses the Bacillus subtilis sacB gene. Appl Environ Microbiol. 1993;59:1361–1366. doi: 10.1128/aem.59.5.1361-1366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richaud C., Richaud F., Martin C., Haziza C., Patte J.C. Regulation of expression and nucleotide sequence of the Escherichia coli dapD gene. J Biol Chem. 1984;259:14824–14828. [PubMed] [Google Scholar]
- Roberts R.C., Helinski D.R. Definition of a minimal plasmid stabilization system from the broad‐host‐range plasmid RK2. J Bacteriol. 1992;174:8119–8132. doi: 10.1128/jb.174.24.8119-8132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R.C., Spangler C., Helinski D.R. Characteristics and significance of DNA binding activity of plasmid stabilization protein ParD from the broad host‐range plasmid RK2. J Biol Chem. 1993;268:27109–27117. [PubMed] [Google Scholar]
- Rohdich F., Hecht S., Gärtner K., Adam P., Krieger C., Amslinger S. Studies on the nonmevalonate terpene biosynthetic pathway: metabolic role of IspH (LytB) protein. Proc Natl Acad Sci USA. 2002;99:1158–1163. doi: 10.1073/pnas.032658999. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchel M.C., Ramos J.L. Dual system to reinforce biological containment of recombinant bacteria designed for rhizoremediation. Appl Environ Microbiol. 2001;67:2649–2656. doi: 10.1128/AEM.67.6.2649-2656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchel M.C., Ramos C., Jensen L.B., Molin S., Ramos J.L. Construction and behavior of biologically contained bacteria for environmental applications in bioremediation. Appl Environ Microbiol. 1995;61:2990–2994. doi: 10.1128/aem.61.8.2990-2994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra De Bast Manuel, Mine N., Van Melderen L. Chromosomal toxin‐antitoxin systems may act as antiaddiction modules. J Bacteriol. 2008;190:4603–4609. doi: 10.1128/JB.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S.K., Singh O.V., Jain R.K. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotechnol. 2002;20:243–248. doi: 10.1016/s0167-7799(02)01943-1. [DOI] [PubMed] [Google Scholar]
- Sangodkar U.M.X., Aldrich T.L., Haugland R.A., Johnson J., Rothmel R.K., Chapman P.J., Chakrabarty A.M. Molecular basis of biodegradation of chloroaromatic compounds. Acta Biotechnol. 1989;9:301–316. [Google Scholar]
- Sat B., Hazan R., Fisher T., Khaner H., Glaser G., Engelberg‐Kulka H. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J Bacteriol. 2001;183:2041–2045. doi: 10.1128/JB.183.6.2041-2045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sat B., Reches M., Engelberg‐Kulka H. The Escherichia coli mazEF suicide module mediates thymineless death. J Bacteriol. 2003;185:1803–1807. doi: 10.1128/JB.185.6.1803-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayeed S., Reaves L., Radnedge L., Austin S. The stability region of the large virulence plasmid of Shigella flexneri encodes an efficient postsegregational killing system. J Bacteriol. 2000;182:2416–2421. doi: 10.1128/jb.182.9.2416-2421.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer K.H., Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O., Schuenemann V.J., Hand N.J., Silhavy T.J., Martin J., Lupas A.N., Djuranovic S. prlF and yhaV encode a new toxin–antitoxin system in Escherichia coli. J Mol Biol. 2007;372:894–905. doi: 10.1016/j.jmb.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J.C., Jenings A.F., Mun D.M., McGovern P.M., Chew L.C. Auxotrophic markers pyrF and proC can replace antibiotic markers on protein production plasmids in high‐cell‐density Pseudomonas fluorescens fermentation. Biotechnol Prog. 2005;21:343–348. doi: 10.1021/bp049696g. [DOI] [PubMed] [Google Scholar]
- Schweder T., Schmidt I., Herrmann H., Neubauer P., Hecker M., Hofmann K. An expression vector system providing plasmid stability and conditional suicide of plasmid‐containing cells. Appl Microbiol Biotechnol. 1992;38:91–93. doi: 10.1007/BF00169425. [DOI] [PubMed] [Google Scholar]
- Sevin E.W., Barloy‐Hubler F. RASTA‐bacteria: a web‐based tool for identifying toxin–antitoxin loci in prokaryotes. Genome Biol. 2007;8:R155. doi: 10.1186/gb-2007-8-8-r155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvaggi J.M., Perkins J.B., Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J Bacteriol. 2005;187:6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R.D. The biosynthesis of multi‐L‐arginyl‐poly(L‐aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim Biophys Acta. 1976;422:407–418. doi: 10.1016/0005-2744(76)90151-0. [DOI] [PubMed] [Google Scholar]
- Smith A.S., Rawlings D.E. Efficiency of the pTF‐FC2 pas poison‐antidote stability system in Escherichia coli is affected by the host strain, and antidote degradation requires the lon protease. J Bacteriol. 1998;180:5458–5462. doi: 10.1128/jb.180.20.5458-5462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobecky P.A., Easter C.L., Bear P.D., Helinski D.R. Characterization of the stable maintenance properties of the par region of broad‐host‐range plasmid RK2. J Bacteriol. 1996;178:2086–2093. doi: 10.1128/jb.178.7.2086-2093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Friedman S. Autogenous regulation of the EcoRII methylase gene at the transcriptional level: effect of 5‐azacytidine. EMBO J. 1993;12:4297–4303. doi: 10.1002/j.1460-2075.1993.tb06114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Bhagwat A.S., Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987;15:313–332. doi: 10.1093/nar/15.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen H.P., Mortensen K.K. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol. 2005;115:113–128. doi: 10.1016/j.jbiotec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Sørvig E., Skaugen M., Naterstad K., Eijsink V.G.H., Axelsson L. Plasmid p256 from Lactobacillus plantarum represents a new type of replicon in lactic acid bacteria, and contains a toxin–antitoxin‐like plasmid maintenance system. Microbiology. 2005;151:421–431. doi: 10.1099/mic.0.27389-0. [DOI] [PubMed] [Google Scholar]
- Srinivasan J., Tinge S., Wright R., Herr J.C., Curtiss R. Oral immunization with attenuated salmonella expressing human sperm antigen induces antibodies in serum and the reproductive tract. Biol Reprod. 1995;53:462–471. doi: 10.1095/biolreprod53.2.462. [DOI] [PubMed] [Google Scholar]
- Stephens J.C., Darsley M.J., Turner A.K. Stabilization of a plasmid coding for a heterologous antigen in Salmonella enterica serotype typhi vaccine strain CVD908‐htrA by using site‐specific recombination. Infect Immun. 2006;74:4383–4386. doi: 10.1128/IAI.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg R.L., Strausberg S.L. Overview of protein expression in Saccharomyces cerevisiae. Curr Protoc Protein Sci. 2001;5:5.6.1–5.6.7. doi: 10.1002/0471140864.ps0506s02. [DOI] [PubMed] [Google Scholar]
- Stumpp T., Wilms B., Altenbuchner J. Ein neues, l‐rhamnose‐induzierbares expressionssystem für Escherichia coli. Biospektrum. 2000;6:33–36. [Google Scholar]
- Summers D.K., Sherratt D.J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984;36:1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- Szafranski P., Mello C.M., Sano T., Smith C.L., Kaplan D.L., Cantor C.R. A new approach for containment of microorganisms: dual control of streptavidin expression by antisense RNA and the T7 transcription system. Proc Natl Acad Sci USA. 1997;94:1059–1063. doi: 10.1073/pnas.94.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres S., Dauti M., Wilde C., Mazel D., Rowe‐Magnus D.A. Chromosomal toxin‐antitoxin loci can diminish large‐scale genome reductions in the absence of selection. Mol Microbiol. 2007;63:1588–1605. doi: 10.1111/j.1365-2958.2007.05613.x. [DOI] [PubMed] [Google Scholar]
- Tacket C.O., Kelly S.M., Schödel F., Losonsky G., Nataro J.P., Edelman R. Safety and immunogenicity in humans of an attenuated Salmonella typhi vaccine vector strain expressing plasmid‐encoded hepatitis B antigens stabilized by the Asd‐balanced lethal vector system. Infect Immun. 1997;65:3381–3385. doi: 10.1128/iai.65.8.3381-3385.1997. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Naito Y., Handa N., Kobayashi I. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction–modification gene complex. J Bacteriol. 2002;184:6100–6108. doi: 10.1128/JB.184.22.6100-6108.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J.E., Kline B.C. Control of the ccd operon in plasmid F. J Bacteriol. 1989a;171:2353–2360. doi: 10.1128/jb.171.5.2353-2360.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J.E., Kline B.C. The F plasmid ccd autorepressor is a complex of CcdA and CcdB proteins. Mol Gen Genet. 1989b;219:26–32. doi: 10.1007/BF00261153. [DOI] [PubMed] [Google Scholar]
- Tan X., Actor J.K., Chen Y. Peptide nucleic acid antisense oligomer as a therapeutic strategy against bacterial infection: proof of principle using mouse intraperitoneal infection. Antimicrob Agents Chemother. 2005;49:3203–3207. doi: 10.1128/AAC.49.8.3203-3207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao T., Bourne J.C., Blumenthal R.M. A family of regulatory genes associated with type II restriction–modification systems. J Bacteriol. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisted T., Gerdes K. Mechanism of post‐segregational killing by the hoksok system of plasmid R1: sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J Mol Biol. 1992;223:41–54. doi: 10.1016/0022-2836(92)90714-u. [DOI] [PubMed] [Google Scholar]
- Tian Q.B., Ohnishi M., Tabuchi A., Terawaki Y. A new plasmid‐encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem Biophys Res Commun. 1996;220:280–284. doi: 10.1006/bbrc.1996.0396. [DOI] [PubMed] [Google Scholar]
- Titball R.W., Howells A.M., Oyston P.C., Williamson E.D. Expression of the Yersinia pestis capsular antigen (F1 antigen) on the surface of an aroA mutant of salmonella typhimurium induces high levels of protection against plague. Infect Immun. 1997;65:1926–1930. doi: 10.1128/iai.65.5.1926-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titball R.W., Sjostedt A., Pavelka M.S., Nano F.E. Biosafety and selectable markers. Ann N Y Acad Sci. 2007;1105:405–417. doi: 10.1196/annals.1409.002. [DOI] [PubMed] [Google Scholar]
- Torres B., Jaenecke S., Timmis K.N., García J.L., Díaz E. A dual lethal system to enhance containment of recombinant micro‐organisms. Microbiology (SGM) 2003;149:3595–3601. doi: 10.1099/mic.0.26618-0. [DOI] [PubMed] [Google Scholar]
- Truffa‐Bachi P., Cohen G.N. Some aspects of amino acid biosynthesis in microorganisms. Annu Rev Biochem. 1968;37:79–108. doi: 10.1146/annurev.bi.37.070168.000455. [DOI] [PubMed] [Google Scholar]
- Tsilibaris V., Maenhaut‐Michel G., Mine N., Melderen L.V. What is the benefit to Escherichia coli of having multiple toxin–antitoxin systems in its genome? J Bacteriol. 2007;189:6101–6108. doi: 10.1128/JB.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S., Ohtsubo E. Autoregulation by cooperative binding of the PemI and PemK proteins to the promoter region of the pem operon. Mol Gen Genet. 1993;237:81–88. doi: 10.1007/BF00282787. [DOI] [PubMed] [Google Scholar]
- Tsuchimoto S., Ohtsubo H., Ohtsubo E. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol. 1988;170:1461–1466. doi: 10.1128/jb.170.4.1461-1466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S., Nishimura Y., Ohtsubo E. The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J Bacteriol. 1992;174:4205–4211. doi: 10.1128/jb.174.13.4205-4211.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C.D., Roychoudhury R., Wu R. Nucleotide recognition sequence at the cleavage site of Haemophilus aegyptius II (HaeII) restriction endonuclease. Biochem Biophys Res Commun. 1976;72:355–362. doi: 10.1016/0006-291x(76)91001-9. [DOI] [PubMed] [Google Scholar]
- Twine S., Byström M., Chen W., Forsman M., Golovliov I., Johansson A. A mutant of Francisella tularensis strain SCHU S4 lacking the ability to express a 58‐kilodalton protein is attenuated for virulence and is an effective live vaccine. Infect Immun. 2005;73:8345–8352. doi: 10.1128/IAI.73.12.8345-8352.2005. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urgun‐Demirtas M., Stark B., Pagilla K. Use of genetically engineered microorganisms (GEMs) for the bioremediation of contaminants. Crit Rev Biotechnol. 2006;26:145–164. doi: 10.1080/07388550600842794. [DOI] [PubMed] [Google Scholar]
- Vaara M., Porro M. Group of peptides that act synergistically with hydrophobic antibiotics against gram‐negative enteric bacteria. Antimicrob Agents Chemother. 1996;40:1801–1805. doi: 10.1128/aac.40.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma A., Kwon‐Chung K.J. Construction of stable episomes in Cryptococcus neoformans. Curr Genet. 1998;34:60–66. doi: 10.1007/s002940050366. [DOI] [PubMed] [Google Scholar]
- Vary P.S., Biedendieck R., Fuerch T., Meinhardt F., Rohde M., Deckwer W.D., Jahn D. Bacillus megaterium‐from simple soil bacterium to industrial protein production host. Appl Microbiol Biotechnol. 2007;76:957–967. doi: 10.1007/s00253-007-1089-3. [DOI] [PubMed] [Google Scholar]
- Voß I., Steinbüchel A. Application of a KDPG‐aldolase gene‐dependent addiction system for enhanced production of cyanophycin in Ralstonia eutropha strain H16. Metab Eng. 2006;8:66–78. doi: 10.1016/j.ymben.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Wahle E., Kornberg A. The partition locus of plasmid pSC101 is a specific binding site for DNA gyrase. EMBO J. 1988;7:1889–1895. doi: 10.1002/j.1460-2075.1988.tb03022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver K.E., Tritle D.J. Identification and characterization of an Enterococcus faecalis plasmid pAD1‐encoded stability determinant which produces two small RNA molecules necessary for its function. Plasmid. 1994;32:168–181. doi: 10.1006/plas.1994.1053. [DOI] [PubMed] [Google Scholar]
- Weaver K.E., Jensen K.D., Colwell A., Sriram S.I. Functional analysis of the Enterococcus faecalis plasmid pAD1‐encoded stability determinant par. Mol Microbiol. 1996;20:53–63. doi: 10.1111/j.1365-2958.1996.tb02488.x. [DOI] [PubMed] [Google Scholar]
- Weaver K.E., Ehli E.A., Nelson J.S., Patel S. Antisense RNA regulation by stable complex formation in the Enterococcus faecalis plasmid pAD1 par addiction system. J Bacteriol. 2004;186:6400–6408. doi: 10.1128/JB.186.19.6400-6408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegerer A., Sun T., Altenbuchner J. Optimization of an E. colil‐rhamnose‐inducible expression vector: test of various genetic module combinations. BMC Biotechnol. 2008;8:2. doi: 10.1186/1472-6750-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendisch V.F., Bott M., Eikmanns B.J. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol. 2006;9:268–274. doi: 10.1016/j.mib.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Westwater C., Kasman L.M., Schofield D.A., Werner P.A., Dolan J.W., Schmidt M.G., Norris J.S. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob Agents Chemother. 2003;47:1301–1307. doi: 10.1128/AAC.47.4.1301-1307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt S., Gerngross T.U. The humanization of N‐glycosylation pathways in yeast. Nat Rev Microbiol. 2005;3:119–128. doi: 10.1038/nrmicro1087. [DOI] [PubMed] [Google Scholar]
- Williams S.G., Cranenburgh R.M., Weiss A.M., Wrighton C.J., Sherratt D.J., Hanak J.A. Repressor titration: a novel system for selection and stable maintenance of recombinant plasmids. Nucleic Acids Res. 1998;26:2120–2124. doi: 10.1093/nar/26.9.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.G., Murray N.E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Wu K., Wood T.K. Evaluation of the hok/sok killer locus for enhanced plasmid stability. Biotechnol Bioeng. 1994;44:912–921. doi: 10.1002/bit.260440807. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Park J.H., Inouye M. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU‐specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284:28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabriskie D.W., Arcuri E.J. Factors influencing productivity of fermentations employing recombinant microorganisms. Enzyme Microb Technol. 1986;8:706–717. [Google Scholar]
- Zhang X.S., Garcia‐Contreras R., Wood T.K. Escherichia coli transcription factor YncC (McbR) regulates colanic acid and biofilm formation by repressing expression of periplasmic protein YbiM (McbA) ISME J. 2008;2:615–631. doi: 10.1038/ismej.2008.24. [DOI] [PubMed] [Google Scholar]
- Zhang J., Zhang Y., Inouye M. Characterization of the interactions within the mazEF addiction module of Escherichia coli. J Biol Chem. 2003;278:32300–32306. doi: 10.1074/jbc.M304767200. [DOI] [PubMed] [Google Scholar]
- Zhao L., Zhang J. Biochemical characterization of a chromosomal toxin–antitoxin system in Mycobacterium tuberculosis. FEBS Lett. 2008;582:710–714. doi: 10.1016/j.febslet.2008.01.045. [DOI] [PubMed] [Google Scholar]
- Zielenkiewicz U., Cegłowski P. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim Pol. 2001;48:1003–1023. [PubMed] [Google Scholar]
- Zielenkiewicz U., Cegłowski P. The toxin‐antitoxin system of the streptococcal plasmid pSM19035. J Bacteriol. 2005;187:6094–6105. doi: 10.1128/JB.187.17.6094-6105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


