Abstract
The flavonoids FAA and DMXAA showed impressive activity against solid tumors in mice, but failed clinical trials. They act on a previously unknown molecular target(s) to trigger cytokine release from leukocytes, which causes tumor-specific vascular damage and other anti-tumor effects. We show that DMXAA is a competitive agonist ligand for mouse STING (stimulator of interferon genes), a receptor for the bacterial PAMP cyclic-di-GMP (c-di-GMP) and an endogenous second messenger cyclic-GMP-AMP. In our structure-activity relationship studies, STING binding affinity and pathway activation activity of four flavonoids correlated with activity in a mouse tumor model measured previously. We propose that STING agonist activity accounts for the anti-tumor effects of FAA and DMXAA in mice. Importantly, DMXAA does not bind to human STING, which may account for its lack of efficacy or mechanism-related toxicity in man. We propose that STING is a druggable target for a novel innate immune activation mechanism of chemotherapy.
Introduction
Most human tumors contain large numbers of innate immune cells that are thought to support tumor growth and angiogenesis.1, 2 Converting these cells to an anti-tumor phenotype holds promise as an approach to chemotherapy that would complement cytotoxic, hormonal and targeted drugs. The anti-cancer flavonoids flavone acetic acid (FAA) and 5,6– dimethylxanthenone-4-acetic acid (DMXAA) are thought to damage tumors by activating innate immune cells, at least in mice, but their clinical development has been hindered by lack of knowledge of their molecule target(s). FAA was discovered by screening natural flavonoids on mouse solid tumor models. It was shown to act by triggering secretion of inflammatory cytokines from innate cells that damage tumors by multiple effector mechanisms3. DMXAA was developed as a more potent analog, and shown to act primarily by inducing tumor-specific acute vascular damage, again downstream of cytokine secretion.4-6 Both drugs were effective for treatment of syngeneic and human xenograft solid tumors in mice, with a single dose sometimes curative.5 Both drugs failed in clinical trials, though DMXAA went as far as phase III.7-9
An important clue to the molecular target(s) of DMXAA is its ability to activate the TBK1-IRF3 pathway in macrophages.10 This pathway activates expression of cytokine genes, notably interferon-β (IFN-β), in response to pathogen-associated molecular patterns (PAMPs), including viral DNA and cyclic-dinucleotides (see diagram in Supplementary Figure 1A). As IFN-β knockout mice showed compromised DMXAA efficacy, the TBK1-IRF3 pathway is considered to play a key role in the anti-cancer effects of DMXAA. How DMXAA activates TBK1-IRF3 pathway to induce IFN-β secretion remained unclear. In this study we showed that DMXAA is a competitive (with c-di-GMP) agonist of mouse STING (mSTING), but shows no binding to human STING (hSTING). We propose that STING agonist activity accounts for the anti-tumor effects of FAA and DMXAA in mice, and lack of this activity on hSTING explains lack of efficacy or mechanism related toxicity in man.
Results and Discussion
DMXAA activates the TBK1-IRF3 pathway in a STING dependent manner
We hypothesized that DMXAA might bind to the TBK1-IRF3 activating protein STING, because it contains a small molecule binding site for cyclic dinucleotide PAMPs11,12 (Supplementary Figure 1A). We first confirmed that DMXAA induces IFN-β secretion in the mouse macrophage cell line Raw264.713, and confirmed that this induction depends on TBK1 kinase using a small molecule inhibitor14 (Supplementary Figure 1D). Immunoblotting with an antibody to phospho-Ser 172 provided a convenient readout of TBK1 activation.15 TBK1 in Raw264.7 cells became phosphorylated in response to DMXAA in a dose- and time-dependent manner (Supplementary Figure 1B). We then used an IRF3-driven luciferase reporter cell line to confirm that DMXAA induces IRF3 activation and nuclear translocation (Supplementary Figure 1C). To test if mSTING is required for activation of TBK1 by DMXAA we performed shRNA knockdown studies in another mouse cell line, L929, which was easier to transfect and also has a functional STING-TBK1-IRF3 pathway.16 When mSTING was successfully knocked down using shRNA oligo 1, TBK1 phosphorylation in response to DMXAA was reduced (Figure 1A upper panel). Unsuccessful knockdown using oligo 2 had no effect, providing a control for the effects of transfection reagents. A side note is that DMXAA and c-di-GMP triggered STING activation resulted in a shift in its gel mobility, presumably due to the change of its phosphorylation states, a phenomenon that has been previously reported.16 Consistent with the TBK1 phosphorylation studies, IFN-α/β induction by DMXAA was also reduced only when mSTING was successfully knocked down (Figure 1A lower panel). These results demonstrate that mSTING is required for the DMXAA activated TBK1-IRF3-IFN-α/β axis.
Figure 1. Identification of a molecular target of DMXAA.
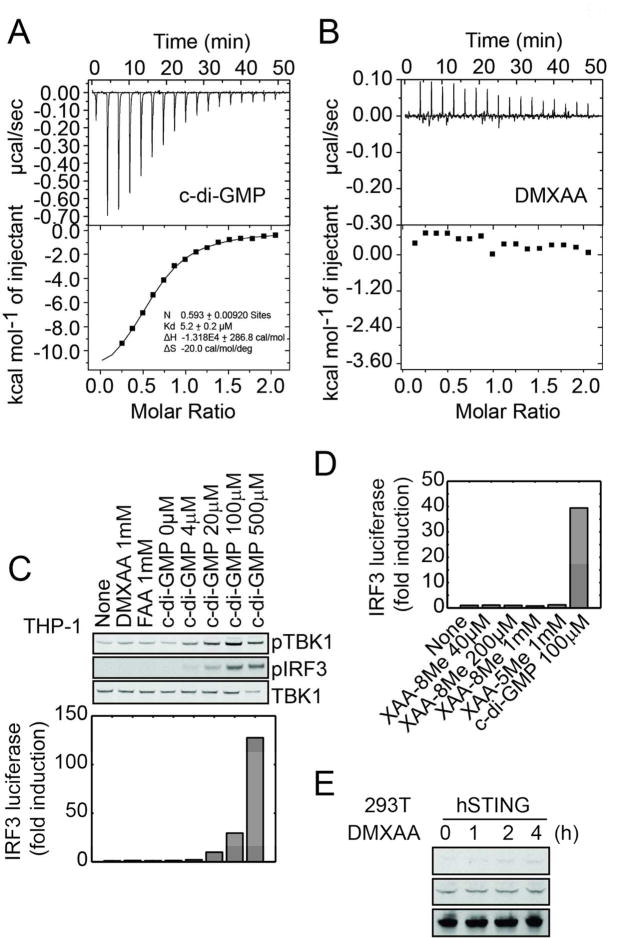
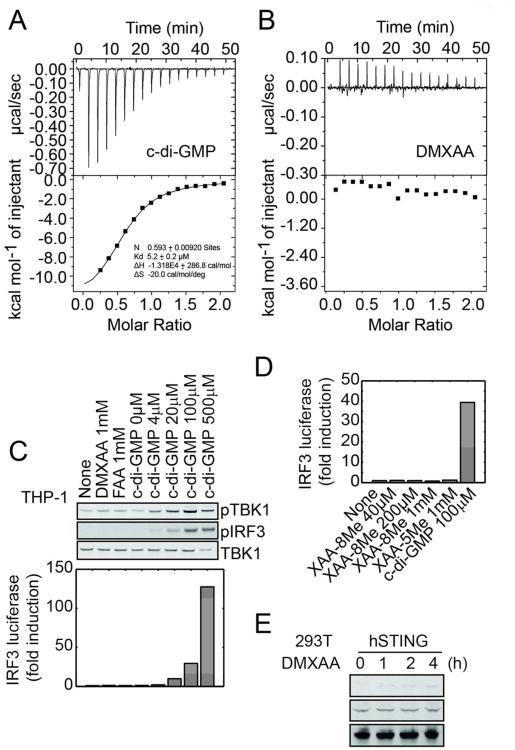
(A) mSTING knockdown studies in L929 cells using lentiviral delivered shRNAs. Cells were treated with 500 μM DMXAA for an hour or 7.5 μM c-di-GMP for 3 hours. C-di-GMP was delivered by digitonin permeabilization. TBK1 activity was measured by monitoring phospho-serine 172 levels (upper panel) and IFN-β was measured using a B16 IFN-α/β reporter cell line (lower panel). Error bars represent mean ± s. d. (n=3) An asterisks (*) indicate p <0.05 and double asterisks indicate p < 0.005 (t test). (B) Overexpress studies of mSTING in 293T cells. Cells were transfected with c-di-GMP (2 μg/ml) or treated with DMXAA (500 μM). (C) Isothermal titration calorimetry measurement of mSTING-CTD interacting with c-di-GMP or with (D) DMXAA.
To test if forcing STING expression in a non-responding cell line is sufficient to confer DMXAA responsiveness, we transfected human 293T cells, which express TBK1, but do not respond to c-di-GMP. Overexpression of full-length mSTING made these cells responsive to both c-di-GMP and DMXAA (Figure 1B). Gel mobility shift of activated STING was less noticeable in this case, perhaps due to signal saturation when STING is overexpressed. Together, both knockdown and ectopic expression experiments support a role of STING in the DMXAA response.
DMXAA binds to mSTING directly
To test if mSTING binds DMXAA we expressed and purified the C-terminal ligand-binding domain (CTD, amino acids 139-378). This domain was shown to form a stable dimer that binds c-di-GMP at the dimer interface.17 We confirmed its binding to c-di-GMP using isothermal titration calorimetry (ITC), finding that one c-di-GMP molecule binds to one mSTING dimer (N = 0.57) with a dissociation constant (Kd) of ∼110 nM. Using the same assay, DMXAA bound to mSTING with a similar affinity (Kd ∼ 130 nM). The shape of the binding curve and the apparent stoichiometry, N = 0.97, suggested that two DMXAA molecules bind cooperatively to one mSTING dimer.
DMXAA competes with c-di-GMP for binding to mSTING binding
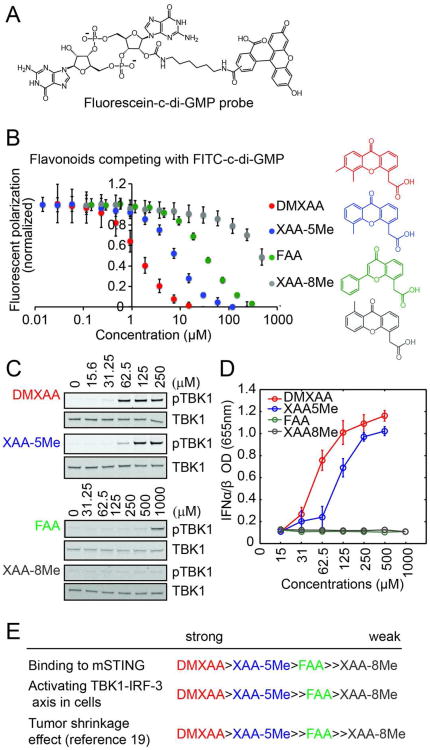
To provide additional support for DMXAA binding to STING, and test competition with the natural ligand c-di-GMP, we developed a fluorescence polarization (FP) assay. C-di-GMP derivatized on one of the ribose hydroxyl groups was used as the probe (Figure 2A). Titration of mSTING CTD into this probe caused a concentration-dependent increase in polarization anisotropy indicative of probe binding (Supplementary Figure 2A). Unlabeled c-di-GMP competed out this signal (Supplementary Figure 2B). DMXAA also competed in this assay, with an apparent IC50 similar to that of c-di-GMP (Supplementary Figure 2C). The presence of detergent did not affect the apparent IC50 of DMXAA suggesting that, despite its hydrophobicity, DMXAA does not form aggregates at the concentrations tested (Supplementary Figure 2D). Together, our data show that DMXAA is an agonist ligand for mSTING that mimics bacteria-derived cyclic di-nucleotide PAMPs11 and the host second messenger cyclic-GMP-AMP12.
Figure 2. Structural function relationship studies with anti-cancer flavonoids.
(A) Chemical structure of fluorescein-labeled c-di-GMP as the probe used in the fluorescence polarization assay. (B) Competition studies of four flavonoid analogs with the fluorescent probe. (C) Induction of TBK1 phosphorylation by flavonoids in Raw264.7 cells. Cells were treated with each drug for 2 hours. (D) Induction of IFN-α/β by anti-cancer flavonoids. Cells were treated with each drug for 4 hours. Error bars represent mean ± s. d. (n=3) (E) Relative activities of four flavonoids in binding to mSTING, in activating the TBK1-IRF3 axis in cells, and in mouse tumor models.
Structure activity relationship studies of four flavonoids
We next measured structure-activity relationships across four flavanoids that had previously been compared for their activity in mice (Figure 2). We measured binding to the CTD of mSTING using the in vitro FP assay (Figure 2B), TBK1 phosphorylation in Raw264.7 cells (Figure 2C) and IFN-α/β production in Raw264.7 cells using a B16 IFN-α/β reporter cell line (Figure 2D). Rank order of potency in these assays was compared to each other and to rank order for inducing hemorrhagic necrosis in the mouse tumor model as previously reported by the Baguley group.18, 19 The rank order of activity was the same in all assays (Figure 2E). These data are consistent with STING being the relevant target of DMXAA, though they do not conclusively prove this point, nor rule out additional targets.
FAA and DMXAA lacked definitive anti-cancer efficacy in man, and their side effects were not obviously suggestive of an interferon-inducing activity.20 We therefore tested if they are ligands for the CTD of hSTING, which shares 89% sequence identity and also forms a stable dimer that binds c-di-GMP.21-25 We first tested for pathway activity in THP-1 cells, a human monocytic leukemia cell line that is known to respond to c-di-GMP.26 As expected, c-di-GMP induced TBK1 phosphorylation and IRF3 activation in THP1 cells (Figure 3C). DMXAA and FAA, however, showed no activity, even at 1 mM concentration. A recent report showed that XAA-8Me increased IL-6, IL-8 and TNFα secretion in human leukocytes27. We tested whether XAA-5Me and XAA-8Me can induce IRF3 driven luciferase production in THP-1 cells and observed no activity even at 1 mM (Figure 3D), suggesting that the reported cytokine release by XAA-8Me occurs through unrelated pathways. Overexpression of full-length hSTING in 293T cells did not render them responsive to DMXAA as assayed by TBK1 by phosphorylation (Figure 3E), unlike overexpression of mSTING (Figure 1B). However, hSTING expression appeared more toxic, and it was difficult to assay c-di-GMP responsiveness, so this assay may not be trustworthy.
Figure 3. Measuring DMXAA's activities in human cells and towards hSTING.
(A) Isothermal titration calorimetry measurement of hSTING-CTD interacting with c-di-GMP or with (B) DMXAA. (C) TBK1 phosphorylation in THP-1 cells (upper panel). Induction of IRF3 driven luciferase (lower panel). Cyclic-di-GMP was delivered using digitonin permeabilization. Cells were incubated with or without c-di-GMP containing permeabilization buffer for 30 min. The buffer was removed by centrifugation and cells were incubated in fresh medium for 4 hours. (D) Induction of IRF3 driven luciferase in THP-1 cells. Cells were treated with each flavonoid for 4 hours in (C) and (D). (E) Overexpress studies of hSTING in 293T cells. Cells were treated with DMXAA (500 μM) for indicated times.
DMXAA does not bind to hSTING
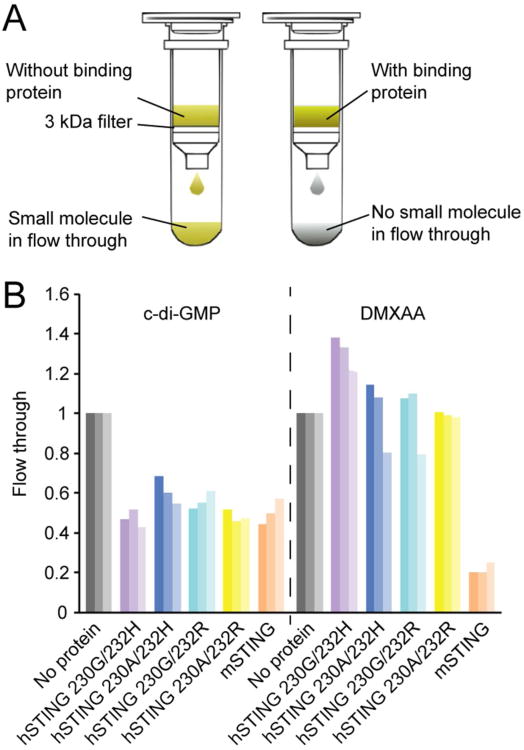
To test binding activity we expressed and purified hSTING CTD (amino acid residues 139-379) and confirmed binding to c-di-GMP using ITC assay (Figure 3A). We observed a Kd of approximately 5.2 μM and a stoichiometry of 0.59, both of which agree well with the reported value.21-24 Using the same assay we detected no interaction of DMXAA with hSTING CTD (Figure 3B). We attempted the FP assay, but found that the fluorescent probe (Figure 2A) showed no detectable change in anisotropy when hSTING was added, suggesting lack of binding (data not shown). To test the negative binding result by ITC more rigorously, we evaluated genetic variants in hSTING. There are four naturally occurring SNPs in the hSTING CTD, 230G/A and 232R/H.28 We expressed hSTING CTD with all four possible combinations and tested c-di-GMP and DMXAA binding using a rapid centrifugal ultrafiltration assay, a modern version of the equilibrium dialysis assay (Figure 4A). All four hSTING variants bound c-di-GMP, but not DMXAA, in this assay (Figure 4B). Together, our data suggest that DMXAA either does not bind hSTING, or binds it too weakly to score in our assays. We estimate the upper bound of detection by ITC to be ∼100 μM, so the affinity of DMXAA for the CTD of hSTING must be at least 1000 fold lower than for mSTING.
Figure 4. Measuring DMXAA binding to hSTING variants.
(A) Scheme of the rapid centrifugal ultrafiltration assay. (B) Binding studies of c-di-GMP and DMXAA with mSTING and hSTING variants. Three bars with the same color, but different shades represent results from three biological repeats using different batches of purified proteins.
In summary, we found that mSTING expression is necessary for activation of the TBK1-IRF3 pathway by DMXAA in mouse cells, that ectopic expression of mSTING in human cells confers DMXAA responsiveness, and that DMXAA binds to the CTD of mSTING competitively with the natural ligand c-di-GMP. These data show that DMXAA is an agonist ligand for mSTING. Rank orders of four flavonoids in structure-activity studies are consistent with mSTING being the relevant target protein for activation of hemorrhagic necrosis in a syngeneic mouse tumor model. IFN-β receptor knockout reduces the response of mouse tumors to DMXAA,13 consistent with this hypothesis. We have not formally proven that mSTING is the relevant target for tumor responses, and additional targets may exist. Nevertheless, given the broad efficacy of FAA and DMXAA across mouse solid tumor models, and their novel mechanisms of action, which include anti-vascular activity, NK cell activation, and induction of T cell immunity in some models, we suggest that STING is an exciting new candidate target for cancer treatment.
Our cellular activity and biochemical binding data for activity of DMXAA on hSTING were entirely negative, which makes formal conclusions difficult. However, they raise the possibility that lack of potent binding to hSTING was responsible for lack of efficacy and of mechanism-related toxicity of anti-cancer flavonoids in man. We suggest that STING has not yet been properly evaluated as a therapeutic target in man, and that new compounds with proven agonist activity on hSTING are required for this test. DMXAA was reported to show some activity in on the NFkB pathway human leukocyte cell lines29 and some clinical activity in phase II trials20. It could be the case that DMXAA has additional therapy-relevant targets, or that it binds to hSTING with an affinity too low to detect in ITC assays. Differential binding of DMXAA to mSTING and hSTING CTDs is surprising given their high sequence identity and similar folds. The natural ligand c-di-GMP binds in a very different way to the two proteins, as revealed by recent co-crystal structures (summarized in Supplementary Figure 3A). mSTING CTD dimer closes more tightly around c-di-GMP, which may account for significantly higher binding affinity of the mouse homolog (∼50 fold in our assays). Different c-di-GMP binding modes may also explain the failure of the fluorescent derivative (Figure 2A) to bind hSTING. These differences in structure and affinity for a natural ligand suggest evolutionary divergence of STING ligand-binding activity between mice and men, whose biological significance is currently unclear. This difference will be important when evaluating STING as a target for chemotherapy of cancer and infectious diseases.
During the submission process of our manuscript, the Fitzgerald group reported findings similar to ours.30, 31 The two studies are complementary in that different biochemical and biological assays were used to draw similar conclusions. Also very recently, the Hornung group reported another mouse-selective STING agonist called CMA, which was originally developed as an anti-viral drug that promotes interferon release.32 Like DMXAA, CMA also showed impressive results in mice, in the case in anti-viral assays, but failed in men. CMA and DMXAA are structurally similar. A crystal structure with mSTING in the Hornung paper reveals two molecules of CMA binding per mSTING dimmer, consistent with the 2:2 stoichiometry we measured for DMXAA by ITC. Together, the three studies solve the mystery of the flavonoids' target and lack of activity in men, and underscore the untested potential of hSTING agonists as anti-tumor and anti-viral agents.
Methods
Reagents and antibodies
DMXAA was purchased from Wuhan Sunrise Technology. Flavone acetic acid, XAA-5Me, and XAA-8Me were kindly provided by Dr. John Tallarico at Novartis Institutes for Biomedical Research in Cambridge, MA. C-di-GMP and fluorescein-labeled c-di-GMP were purchased from Biolog Life Science Institute. Flavonoids were dissolved in aqueous solution with equimolar NaHCO3 and lyophilized to form the water-soluble sodium salts, which were used in all experiments. Rabbit polyclonal antibodies against TBK, pTBK (S172), pIRF3 (S396), STING were purchased from Cell Signaling Technology. Mouse monoclonal HA (F-7) antibody was purchased from Santa Cruz Biotechnology Inc. Reagents for the Luciferase and SEAP assays were purchased from InvivoGen. The HA-tagged mSTING plasmid was a generous gift from Dr. Russell Vance at University of California-Berkeley.
Cell culture, transfection and shRNA infection
Raw264.7, L929 and 293T cells were maintained in DMEM (Cellgro) supplemented with 10% FBS (GIBCO) (v/v) and 1% of penicillin-streptomycin (Cellgro). THP-1 cells were grown in RPMI (Cellgro) supplemented with 10% FBS, 0.1% β-mercaptoethanol (GIBCO) and 1% of penicillin-streptomycin. The HA-tagged mSTING plasmid was a generous gift from Dr. Russell Vance at University of California-Berkeley. Transfection of mSTING in 293T cell line was achieved using Lipofectamine 2000 (Invitrogen). hSTING cDNA plasmid (CCSB-Broad Lentiviral expression human TMEM173, accession number: BC047779, clone ID: ccsBroad304_05465) was purchased from Open BioSystems. Transfection of hSTING in 293T cells was achieved using lentiviral infection followed by puromycin (1 μg mol−1) selection. mSTING targeting shRNA set (GIPZ lentiviral shRNA set) was purchased from Open BioSystems. Lentiviruses carrying mSTING shRNAs were assembled in 293T cells. To knockdown mSTING in L929 cells, cells were infected with the viruses for 12-18 hours and subjected to puromycin (10 μg mol−1) selection for 2 weeks. C-di-GMP is not cell permeable. In order to deliver it into 293T cells, transfecting reagent Lipofectamine 2000 was used. To deliver c-di-GMP into L929, Raw264.7 and THP-1 cells, cells were permeabilized using digitonin permeabilization buffer as previously described (11). In brief, cells were incubated in digitonin permeabilization buffer (50mM HEPES, 100mM KCl, 3mM MgCl2, 0.1mM DTT, 85mM sucrose, 0.2% BSA, 1mM ATP, 0.1mM GTP, 10 μg mol−1digitonin) with or without c-di-GMP for 30 min. Buffer was removed by centrifugation and fresh medium was added for indicated time.
SEAP and luciferase assay
The B16 IFNα/β reporter cell line (Invivogen) was used to measure IFN-α/β levels in Raw264.7 and L929 cells. We followed the manufacturer recommended protocols. Briefly, after drug treatment, conditioned media were collected and added to the B16 reporter cells. After 18 hours, SEAP activity was measured using a Victor plate reader (Perkin Elmer). To measure drug induced IRF3 transcription activity, Raw264.7 cells and THP-1 cells expressing an IRF3 driven luciferease reporter (Invivogen) were used.
Protein purification
DNA sequence encoding mSTING-CTD (139–378) was amplified from HA-tagged mSTING plasmid using Phusion High-Fidelity DNA Polymerase and the primer pair: mSTING-FWD-TATTGAGGCTCACAGAGAACAGATTGGTGGT-ACTCCAGCGGAAGTCTCTGCAGTC And mSTING-REV-GGATCCCCTTCCTGCAGTCACCCGGGCTCGAG- TCAGATGAGGTCAGTGCGGAGTGG
DNA sequence encoding hSTING-CTD (140–379) was amplified from hSTING cDNA clone using primer pair: hSTING-FWD-TATTGAGGCTCACAGAGAACAGATTGGTGGT-GCCCCAGCTGAGATCTCTGCAGTG And hSTING-REV-GGATCCCCTTCCTGCAGTCACCCGGGCTCGAG-TCAAGAGAAATCCGTGCGGAGAGG
The PCR product was inserted into the SapI and XhoI sites of pTB146 (a generous gift from Dr. Thomas Bernhard) using isothermal assembly. His-SUMO tagged mSTING-CTD or hSTING-CTD was expressed in Rosetta™ Competent Cells. Cells were grown in 2XYT media with Ampicilin (100 ng mol−1) and were induced with 0.5 mM IPTG when OD reached 1 and were allowed to grow overnight at 16 °C. Cells were pelleted and resuspended in PBS with 20 mM imidazole and snap frozen in liquid nitrogen. To break cells, two freeze and thaw cycles were preformed followed by sonication. The cell extract was then cleared by ultracentrifugation at 45,000 rpm for 1 h. The cleared supernatant was incubated with Ni-NTA beads (4 mL of beads per liter of bacteria culture). Ni beads were washed with 20 mM imidazole in PBS and protein was eluted with step concentrations of 50 mM to 500 mM imidazole in PBS. Fractions containing his-SUMO-mSTING or hSTING were pooled, concentrated and dialyzed against PBS. The his-SUMO tag was further cleaved overnight at 4 °C using His-tagged Ulp protease (1:100 molar ratio), which was expressed using pTB145 plasmid also from Dr. Thomas Bernhard. The cleaved his-SUMO tag and his-Ulp were removed using Ni-NTA beads to yield mSTING-CTD or hSTING-CTD. The protein was further purified by size-exclusion chromatography in running buffer: 20 mM Tris HCl (pH 8.0),150 mM NaCl, and 5 mM DTT. Fraction containing STING protein was pooled, concentrated, and snap frozen for future use.
Isothermal titration calorimetry assay
Mouse STING (amino acids 139-378) and hSTING (amino acids 139-344) were dialyzed extensively against running buffer (20 mM Tris, 150 mM NaCl, pH 7.5). DMXAA and c-di-GMP were dissolved in running buffer. Protein concentrations were measured by their absorbance at 280 nm. The concentration of c-di-GMP was decided by its absorbance at 254 nm. DMXAA concentration was decided by weight. Immediately before the titration experiments, both protein and small molecules were centrifuged at 14,000 rpm at 25 °C for 10 min to remove debris and air bubbles. The calorimetric titrations were carried out at 25°C on a microCaliTC200 instrument (GE Healthcare Life Science) with 16 successive injections of 2.4 μL (400 μM) c-di-GMP or DMXAA, spaced 180 s apart, into the sample cell containing 200 μL of 40 μM STING protein. Data analysis was performed using the build-in ORIGIN software. The dissociation constant (Kd), enthalpy change (ΔH), and the stoichiometry (N) were calculated by fitting the thermograms to a model of one set of binding sites. The free energy change (ΔG) and the entropy change (ΔS) were calculated using the equation: .
Centrifugal ultrafiltration assay
Mouse STING (amino acids 139-378) and hSTING (amino acids 140-379) were concentrated to 200 μM to 400 μM and dialyzed extensively against Tris buffer (20 mM Tris, 150 mM NaCl, pH 7.5) to remove imidazole, which interferes with this assay. DMXAA and c-di-GMP were diluted to the same concentration as the protein in Tris-buffer. The small molecules (20 μL) were loaded onto centrifugal devices (NANOSEP 3K OMEGA) with 20 μL of Tris buffer or with 20 μL of STING protein. Flowthrough from a one min spin was read on a nanodrop spectrophotometer (ND-1000). Unbound DMXAA and c-di-GMP were monitored by their UV absorbance at 245 nm and 254 nm respectively. In the case of DMXAA, 150 μL of DMXAA solution was centrifuged through to pre-block the membrane of the centrifugal device.
Fluorescence polarization assay
Fluorescein labeled c-di-GMP mixed with mSTING-CTD with or without the presence of small molecule competitors in PBS in black 384 well plates (10 μL per well). The fluorescence anisotropy was determined on a PerkinElmer EnVisions multi-mode plate reader. The plate can be read immediately after mixing the reagents or after a few hours without change of signals.
Supplementary Material
Acknowledgments
We thank Y. Feng, J. Tallarico at Novartis Institutes for Biomedical Research for providing the flavonoids and helpful discussion. We thank M. Loose for help with protein production, T. Yuan for help with the ITC experiments, and P. Choi for helpful discussions on the manuscript. We thank the Harvard Medical School ICCB screening facility for help with the fluorescence polarization assays. This research was supported by the National Cancer Institute (CA139980) and Novartis Institutes for Biomedical Research. L. Li thanks the Jane Coffin Childs Fund for her postdoctoral fellowship.
Footnotes
Supporting information: Supplementary figures: This material is available free of charge via Internet at http://pubs.acs.org.
References
- 1.Shiao SL, Ganesan AP, Rugo HS, Coussens LM. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev. 2011;25:2559–2572. doi: 10.1101/gad.169029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fridlender ZG, Albelda SM. Tumor-associated neutrophils: friend or foe? Carcinogenesis. 2012;33:949–955. doi: 10.1093/carcin/bgs123. [DOI] [PubMed] [Google Scholar]
- 3.Bibby MC, Phillips RM, Double JA, Pratesi G. Anti-tumour activity of flavone acetic acid (NSC 347512) in mice--influence of immune status. Br J Cancer. 1991;63:57–62. doi: 10.1038/bjc.1991.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woon ST, Reddy CB, Drummond CJ, Schooltink MA, Baguley BC, Kieda C, Ching LM. A comparison of the ability of DMXAA and xanthenone analogues to activate NF-kappaB in murine and human cell lines. Oncol Res. 2005;15:351–364. doi: 10.3727/096504005776449743. [DOI] [PubMed] [Google Scholar]
- 5.Philpott M, Baguley BC, Ching LM. Induction of tumour necrosis factor- alpha by single and repeated doses of the antitumour agent 5,6-dimethylxanthenone-4-acetic acid. Cancer Chemother Pharmacol. 1995;36:143–148. doi: 10.1007/BF00689199. [DOI] [PubMed] [Google Scholar]
- 6.Tozer GM, Kanthou C, Baguley BC. Disrupting tumour blood vessels. Nat Rev Cancer. 2005;5:423–435. doi: 10.1038/nrc1628. [DOI] [PubMed] [Google Scholar]
- 7.Kerr DJ, Kaye SB. Flavone acetic acid--preclinical and clinical activity. Eur J Cancer Clin Oncol. 1989;25:1271–1272. doi: 10.1016/0277-5379(89)90072-2. [DOI] [PubMed] [Google Scholar]
- 8.Bibby MC, Double JA. Flavone acetic acid--from laboratory to clinic and back. Anticancer Drugs. 1993;4:3–17. doi: 10.1097/00001813-199302000-00001. [DOI] [PubMed] [Google Scholar]
- 9.McKeage MJ, Reck M, Jameson MB, Rosenthal MA, Gibbs D, Mainwaring PN, Freitag L, Sullivan R, Von Pawel J. Phase II study of ASA404 (vadimezan, 5,6-dimethylxanthenone-4-acetic acid/DMXAA) 1800mg/m(2) combined with carboplatin and paclitaxel in previously untreated advanced non-small cell lung cancer. Lung Cancer. 2009;65:192–197. doi: 10.1016/j.lungcan.2009.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, Akira S, Savan R, van Echo D, Fitzgerald KA, Young HA, Ching LM, Vogel SN. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J Exp Med. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts ZJ, Ching LM, Vogel SN. IFN-beta-dependent inhibition of tumor growth by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA) J Interferon Cytokine Res. 2008;28:133–139. doi: 10.1089/jir.2007.0992. [DOI] [PubMed] [Google Scholar]
- 14.Clark K, Plater L, Peggie M, Cohen P. Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J Biol Chem. 2009;284:14136–14146. doi: 10.1074/jbc.M109.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kishore N, Huynh QK, Mathialagan S, Hall T, Rouw S, Creely D, Lange G, Caroll J, Reitz B, Donnelly A, Boddupalli H, Combs RG, Kretzmer K, Tripp CS. IKK-i and TBK-1 are enzymatically distinct from the homologous enzyme IKK-2: comparative analysis of recombinant human IKK-i, TBK-1, and IKK-2. J Biol Chem. 2002;277:13840–13847. doi: 10.1074/jbc.M110474200. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chin KH, Tu ZL, Su YC, Yu YJ, Chen HC, Lo YC, Chen CP, Barber GN, Chuah ML, Liang ZX, Chou SH. Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING. Acta Crystallogr D Biol Crystallogr. 2013;69:352–366. doi: 10.1107/S0907444912047269. [DOI] [PubMed] [Google Scholar]
- 18.Atwell GJ, Rewcastle GW, Baguley BC, Denny WA. Synthesis and anti- tumour activity of topologically-related analogues of flavoneacetic acid. Anticancer Drug Des. 1989;4:161–169. [PubMed] [Google Scholar]
- 19.Rewcastle GW, Atwell GJ, Li ZA, Baguley BC, Denny WA. Potential antitumor agents. 61. Structure-activity relationships for in vivo colon 38 activity among disubstituted 9-oxo-9H-xanthene-4-acetic acids. J Med Chem. 1991;34:217–222. doi: 10.1021/jm00105a034. [DOI] [PubMed] [Google Scholar]
- 20.McKeage MJ, Jameson MB. Comparative outcomes of squamous and non-squamous non-small cell lung cancer (NSCLC) patients in phase II studies of ASA404 (DMXAA) - retrospective analysis of pooled data. J ThoracDis. 2010;2:199–204. doi: 10.3978/j.issn.2072-1439.2010.02.04.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, Gu L. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 23.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, Li Y, Zhang R, Cheng G, Liu ZJ. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 26.Kumagai Y, Matsuo J, Hayakawa Y, Rikihisa Y. Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J Bacteriol. 2010;192:4122–4133. doi: 10.1128/JB.00132-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tijono SM, Guo K, Henare K, Palmer BD, Wang LC, Albelda SM, Ching LM. Identification of human-selective analogues of the vascular-disrupting agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA) Br J Cancer. 2013;108:1306–1315. doi: 10.1038/bjc.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woon ST, Baguley BC, Palmer BD, Fraser JD, Ching LM. Uptake of the antivascular agent 5,6-dimethylxanthenone-4-acetic acid (DMXAA) and activation of NF-kappaB in human tumor cell lines. Oncol Res. 2002;13:95–101. [PubMed] [Google Scholar]
- 30.Prantner D, Perkins DJ, Lai W, Williams MS, Sharma S, Fitzgerald KA, Vogel SN. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) activates stimulator of interferon gene (STING)-dependent innate immune pathways and is regulated by mitochondrial membrane potential. J Biol Chem. 2012;287:39776–39788. doi: 10.1074/jbc.M112.382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conlon J, Burdette DL, Sharma S, Bhat N, Thompson M, Jiang Z, Rathinam VA, Monks B, Jin T, Xiao TS, Vogel SN, Vance RE, Fitzgerald KA. Mouse, but not Human STING, Binds and Signals in Response to the Vascular Disrupting Agent 5,6-Dimethylxanthenone-4-Acetic Acid. J Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species- specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013 doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.