Abstract
Most A/T specific heterocyclic diamidine derivatives need at least four A/T base pairs for tight binding to the DNA minor groove. Addition of a GC base pair to A/T sequences typically causes a large decrease in binding constant. The ability to target biologically important sequences of DNA could be significantly increased if compounds that could recognize A/T sites with an intervening GC base pair could be designed. The kinetoplast DNA sequence of parasitic microorganisms, for example, contains numerous three A/T binding sites that are separated by a single G. A series of compounds were prepared to target the AAAGTTT sequence as a model system for discovery of “G-jumpers”. The new synthetic compounds have two aromatic-amidine groups for A/T recognition, and these are connected through an oxy-methylene linker to cross the GC. CD experiments indicated a minor groove binding mode, as expected, for these compounds. Tmax, surface plasmon resonance, and isothermal titration calorimetry experiments revealed 1:1 binding to the AAAGTTT sequence with an affinity that depends on compound structure. Benzimidazole derivatives gave the strongest binding and had generally good solution properties. The binding affinities to the classical AATT sequence were similar to that for AAAGTTT for these extended compounds, but binding was weaker to the AAAGCTTT sequence with two intervening GC base pairs. Binding to both AAAGTTT and AATT was enthalpy driven for strong binding benzimidazole derivatives.
Many cationic-heterocyclic compounds selectively target A/T sequences by binding in the DNA minor groove. With the exception of polyamides, targeting GC and mixed AT/GC sequences has been difficult with this class of compounds. Minor groove binding diamidines are AT specific minor groove binding agents, such as DB75 and pentamidine (Figure 1), that have good uptake into many cells with relatively low toxicity (1–3). DB289, an orally active prodrug of DB75, which has reached phase III clinical trials against sleeping sickness, is active against other microbial parasites (1,4–6). Pentamidine has been available for many years and is widely used against trypanosomiasis, leishmaniasis and P. carinii pneumonia (4, 7–9). In a recent study of diamidine minor groove binders, it was shown that stacked minor groove dimer formation of DB293 in an ATGA binding site of the Pit-1 and Brn-3 promoter sequences inhibits binding of these two transcription factors to the major groove of DNA through structural changes in the minor groove (10). Cooperative stacked dimer formation in a single binding site is one of the methods to target DNA sequences with high specificity, but so far there is no general way to target mixed sequences with heterocyclic dications (11).
Figure 1.
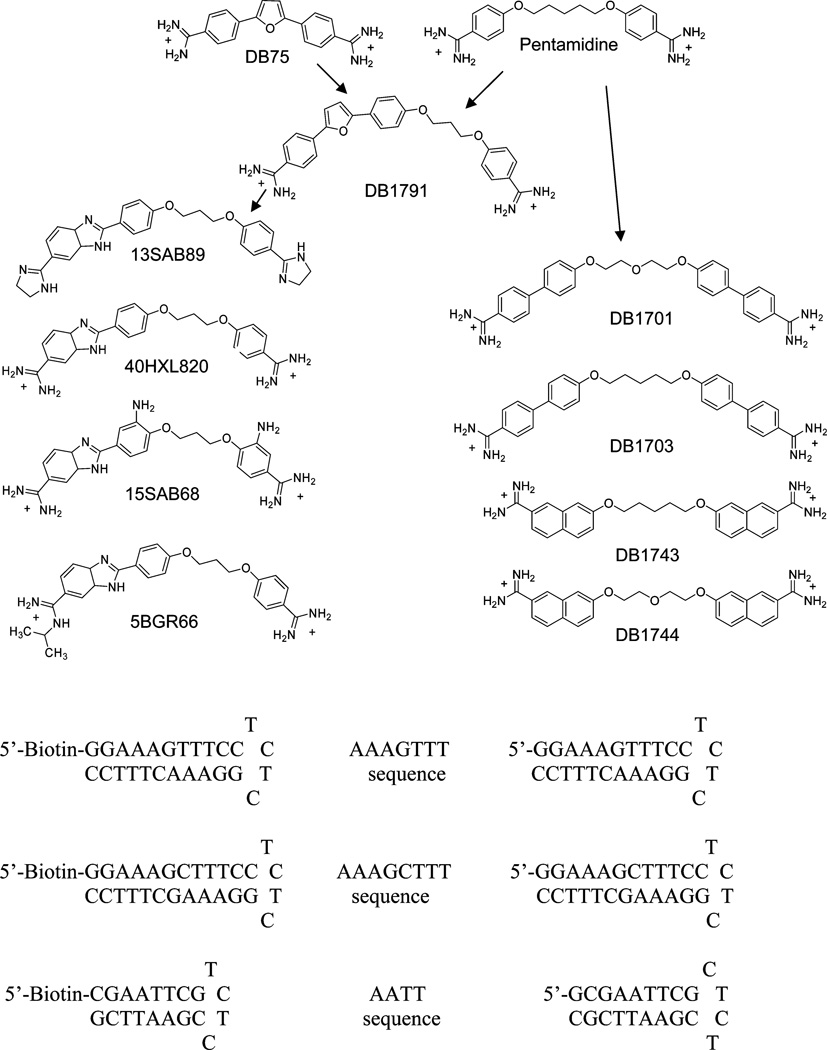
Compound structures and DNA sequences.
Kinetoplast minicircle mitochondrial DNAs (kDNA)1 in disease-causing kinetoplastid parasites have numerous A/T binding sites in thousands of repeated minicircles (5, 12, 13). Compounds that could bind cooperatively to such repeated sequences that are separated by one or more GC base pairs could have very high selectivity for targeting parasites. Cooperativity in adjacent binding sites in regions with A/T sequences, such as kDNA is, however, difficult to achieve with currently available compounds. Although some compounds show strong cooperativity in binding to two adjacent binding sites in short duplex DNAs (14), the cooperativity decreases in less dynamic, hairpin DNAs, which resemble longer sequences (15).
Adjacent sites in kDNA might also be selectively targeted by compounds with linkers that could cross intervening GC base pairs in the minor groove. The kDNA minicircles have numerous sequences with three consecutive A/T bases separated by a single GC base pair (16). Compounds that could bind to such sequences, as well as to the classical binding sites with four or more AT base pairs, would have significantly enhanced selectivity for kDNA. Thus, the goal of this work is to design compounds that are able to selectively target sequences, such as AAAGTTT, and bind tightly to two A/T binding sites of 5–6 base pairs total while crossing 1–2 intervening GC base pairs. The AAAGTTT and AAAGCTTT sequences have been chosen as model systems (Figure 1) and the classical AATT site along with a single AAA site were used for comparison. To test which types of structures could bind to the target sequences, compounds that are similar to the biologically active anti-parasitic diamidines, DB75 and pentamidine (Figure 1) were initially designed and synthesized. This design concept includes two connected aromatic-amidine moieties that are specific for AT base pair sequences. Since the amino group of G in a GC base pair is in the minor groove, the AT specific groups were connected with oxy-alkyl linkers. The concept is to place oxygens in the linker to provide H-bond acceptors for interaction with the G-amino group. The interactions of the compounds with the different DNA model systems have been investigated by several different biophysical methods, and compounds that bind strongly to the AAAGTTT sequence have been identified.
MATERIALS AND METHODS
Materials and Buffers
The synthesis of 40HXL820 has been published (17); the synthesis of 5BGR66, 15SAB68, and 13SAB89 will be published elsewhere; and the synthesis of all other compounds is described below. DNA sequences (Figure 1) were purchased from Integrated DNA Technologies. MES 10 buffer containing 0.01 M [2-(N-morpholino) ethanesulfonic acid] (MES), 0.001 M EDTA, and 0.1 M NaCl with a pH of 6.25 adjusted with 1 M NaOH solution was used for Tmax, ITC, and CD experiments. Tris buffer containing 0.01 M hydroxymethyl aminomethane hydrochloride (Tris), 0.1 M NaCl, and 0.001 EDTA with a pH of 7.0 adjusted with 1 M NaOH solution was used for SPR experiments, since better sensorgrams were obtained with that buffer.
Synthesis of 2-(4-(3-(4-Amidinophenoxy)propoxy)phenyl)-5-(4-amidinophenyl)furan Dihydrochloride (DB1791)
Anhydrous K2CO3 (20.7 g, 0.15 mol) was added to a stirred solution of 4-cyanophenol (11.9 g, 0.1 mol) in anhydrous acetonitrile (120 mL), after 30 min 1,3-dibromopropane (60.6 g, 0.3 mol) was added, and the reaction mixture was heated at reflux for 24 h (TLC monitored). The solvent was removed under reduced pressure; treated with water; extracted with 100 mL of EtOAc; washed with 5% NaOH, brine, and water; dried over anhydrous MgSO4; filtered; and concentrated under reduced pressure. The residue was chromatographed over silica gel, eluting with hexane to hexane:EtOAc (9:1), which yielded white crystalline solid 3-(4-cyanophenoxy)-1-bromopropane, 16.5 g (68%): mp 40 °C; 1H NMR (DMSO-d6) 7.77 (d, 2H, J = 8.4 Hz), 7.14 (d, 2H, J = 8.4 Hz), 4.20 (t, 2H, J = 6 Hz), 3.67 (t, 2H, J = 6 Hz), 2.29 (quintet, 2H, J = 6 Hz); 13C NMR (DMSO-d6) 162.8, 134.7, 119.7, 116.3, 103.7, 66.58, 57.9, 32.5; MS m/e 240 (M+). Anal. Calcd for C10H10BrNO: C, 50.02; H, 4.19; N; 5.83. Found: C, 50.23; H, 3.97; N, 5.72.
To a well-stirred solution of 4-methoxyphenylboronic acid (1.8 g, 0.012 mol) and 2-bromo-5-(4-cyanophenyl)furan (2.48 g, 0.01 mol) in 30 mL of toluene under N2 was added a solution of 2 M Na2CO3 (12 mL). After stirring the solution for 15 min, Pd(PPh3)4 (0.46 g, 4 mol %) was added. The reaction mixture was allowed to reflux under N2 for 10–12 h (TLC monitored). The reaction mixture was diluted with water, extracted with CH2Cl2, washed with 10% NH4OH and water, dried, and concentrated under reduced pressure. The residue was chromatographed over silica gel eluting with hexane to EtOAc:hexane (9:1) to yield yellow crystalline solid 2-(4-cyanophenyl)-5-(4-methoxyphenyl)furan, 2.1 g (76%): mp 299–300 °C; 1H NMR (DMSO-d6) 7.99 (d, 2H, J = 9 Hz), 7.89 (d, 2H, J = 9 Hz), 7.81 (d, 2H, J = 9 Hz), 7.34 (d, 1H, J = 3.6 Hz), 7.05 (d, 2H, J = 9 Hz), 7.03 (d, 1H, J = 3.6 Hz), 3.83 (s, 3H); 13C NMR (CDCl3) 159.7, 150.6, 134.6, 132.6, 132.0, 128.6, 127.7, 125.6, 123.6, 114.3, 114.4, 110.6, 106.2, 55.4; MS m/e 275 (M+ + 1). Anal. Calcd for C18H13NO2: C, 78.53; H, 4.76; N, 5.09. Found: C, 78.41; H, 4.69; N, 5.26.
A solution of BBr3 (5 g, 0.02 mol) in 20 mL of CH2Cl2 was added dropwise to an ice-cold stirred solution of 2-(4-cyanophenyl)-5-(4-methoxyphenyl)furan (2.75 g, 0.01 mol) in CH2Cl2 (100 mL), and the resulting solution was stirred for 12 h (TLC followed), cooled, poured into an ice–water mixture, extracted twice with 100 mL of CH2Cl2, washed with water and brine water, dried over anhydrous Na2SO4, and concentrated under reduced pressure to yield a yellow solid 2-(4-cyanophenyl)-5-(4-hydroxyphenyl)furan, 2.1 g (81%): mp >300 °C; 1H NMR (DMSO-d6) 9.64 (br, 1H), 7.91 (d, 2H, J = 8.4 Hz), 7.82 (d, 2H, J = 8.4 Hz), 7.66 (d,2H, J = 8.4 Hz), 7.23 (d, 1H, J = 3.2 Hz), 6.87 (d, 2H, J = 8.4 Hz), 6.86 (d, 1H, J = 3.2 Hz); 13C NMR (DMSO-d6) 158.3, 155.6, 150.3, 134.7, 133.3, 126.1, 124.0, 121.6, 119.4, 116.3, 112.1, 109.3, 106.6; MS m/e 261 (M+). Anal. Calcd for C17H11NO2: C, 78.18; H, 4.24; N, 5.36. Found: C, 77.91; H, 4.39; N, 5.26.
A mixture of 2-(4-cyanophenyl)-5-(4-hydroxyphenyl)furan (1.04 g, 0.004 mol), 3-(4-cyanophenoxy)-1-bromopropane (0.96 g, 0.004 mol), and anhydrous K2CO3 (1.43 g, 0.006 mol) in 50 mL of anhydrous CH3CN was heated at reflux under N2 for 12 h (TLC followed); solvent was removed under reduced pressure, and 50 mL of water was added to the residue; and the solvent was filtered, the solid was suspended into 20 mL of water and 10 mL of 5% NaOH, and the suspension was stirred for 10 min, filtered, washed with water, and dried. The solid was crystallized from hot EtOAc:hexane, yellow solid 2-(4-(3-(4-cyanophenoxy)propoxy) phenyl)-5-(4-cyanophenyl)furan, 1.40 g (83%): mp 159–60 °C; 1H NMR (CDCl3) 7.79 (d, 2H, J = 8.4 Hz), 7.69 (d, 2H, J = 9 Hz), 7.67 (d, 2H, J = 9 Hz), 7.61 (d, 2H, J = 8.4 Hz), 6.99 (d, 2H, J = 8.8 Hz), 6.98 (d, 2H, J = 8.8 Hz), 6.90, (d, 1H, J = 3.6 Hz), 6.67 (d, 1H, J = 3.6), 4.26 (t, 2H, J = 6 Hz), 4.23 (q, 2H, J = 6 Hz), 2.34 (t, 2H, J = 6 Hz); 13C NMR (CDCl3): 162.1, 158.8, 155.1, 150.7, 134.6, 134.0, 132.6, 125.6, 123.6, 123.4, 119.2, 119.1, 115.2, 114.9, 110.6, 109.8, 106.3, 104.1, 64.8, 64.2, 29.1; MS m/e 420 (M+). Anal. Calcd for C27H20N2O3: C, 77.13; H, 4.79; N. 6.66. Found: C, 77.36; H, 4.71; N, 6.79.
Five milliliters of 1 M LiN(TMS)2 in THF (0.84 g, 0.005 mol) was added under N2 to a solution of the above bisnitrile (0.42 g, 0.001 mol) in 10 mL of THF at 0 °C, and the resulting solution was stirred at room temperature for 12 h, cooled to 0 °C, and acidified with ethanol saturated with HCl gas; after 4 h of stirring, the solvent was removed under reduced pressure, the residue was triturated with dry ether and filtered, the solid was dissolved in ice-water and basified with 2 N NaOH to pH 10, and the precipitated yellow solid was filtered, washed with cold water, and dried. The solid was suspended in 10 mL of absolute EtOH, and the suspension was treated with 1 mL of HCl-saturated EtOH and filtered; after removing solvent, the residue was stirred with anhydrous ether for 30 min, filtered, and washed with dry ether to yield 0.46 g (84%) of the bisamidine as a yellow salt: mp >300 °C; 1H NMR (DMSO-d6) 9.46 (br, 2H), 9.45 (br, 2H), 9.38 (br, 2H), 9.33 (br, 2H), 7.99 (d, 2H, J = 8.4 Hz), 7.97 (d, 2H, J = 8.4 Hz), 7.79 (d, 2H, J = 8.4 Hz), 7.50 (d, 2H, J = 8.4 Hz), 7.48 (d, 2H, J = 8.4 Hz), 7.30 (d, 1H, J = 3.2 Hz), 7.06 (d, 2H, J = 8.4 Hz), 6.98 (d, 1H, J = 3.2 Hz), 4.30 (t, 2H, J = 6 Hz), 4.22 (t, 2H, J = 6 Hz), 2.24 (quintet, 2H, J = 6 Hz); 13C NMR (DMSO-d6) 166.1, 165.6, 159.1, 154.9, 153.4, 150.9, 135.4, 130.7, 129.7, 129.4, 126.1, 123.7, 123.3, 120.7, 115.6, 114.5, 112.0, 107.5, 65.5, 65.0, 29.1; MS m/e 455 (M+ + 1). Anal. Calcd for C27H26N4O3 • 2HCl • 1.5H2O: C, 58.48; H, 5.63; N, 10.14. Found: C, 58.51; H, 5.31; N, 10.34.
Synthesis of 1,5-Bis(4′-amidino-1,1′-biphenyl-4-yloxy)-3-oxapentane Dihydrochloride (DB1701)
Anhydrous K2CO3 (2.07 g, 0.015 mol) was added to a solution of 4′-hydroxybiphenyl-4-carbonitrile (1.95 g, 0.01 mol) and 1,5-dibromo-3-oxa-pentane (1.16 g, 0.005 mol) in 50 mL of acetonitrile. The solution was heated at reflux for 12–15 h (TLC followed); solvent was removed under reduced pressure; water was added; and the solid was filtered, washed with 2 N NaOH and water, dried, and recrystallized from hot EtOAc–hexane to yield white solid 1,5-bis(4′-cyano-1,1′-biphenyl-4-yloxy)-3-oxapentane, 1.68 g (82%): mp 149–50 °C; 1H NMR (DMSO-d6) 7.82 (d, 4H, J = 8.7 Hz), 7.78 (d, 4H, J = 8.7 Hz), 7.66 (d, 4H, J = 8.7 Hz), 7.06 (d, 4H, J = 8.7 Hz), 4.21–4.18 (m, 4H), 3.87–3.83 (m,4H); 13C NMR (DMSO-d6) 159.1, 144.1, 132.8, 130.4, 128.3, 126.9, 119.0, 115.1, 109.1, 69.0, 67.3; MS m/e 460 (M+). Anal. Calcd for C30H24N2O3: C, 78.24; H, 5.25; N, 6.08. Found: C, 78.25; H, 5.22; N, 6.02.
Following the above procedure for DB1791, the bisnitrile (0.46 g, 0.001 mol) and 5 mL of 1 M LiN(TMS)2 in THF (0.84 g, 0.005 mol) yielded 0.48 g (82%) of the title bisamidine as a white hydrochloride salt: mp 120–1 °C; 1H NMR (DMSO-d6) 9.32 (br, 4H), 9.2 (vbr, 4H), 7.93 (d, 4H, J = 7.8 Hz), 7.87 (d, 4H, J = 7.8 Hz), 7.72 (d, 4H, J = 7.8 Hz), 7.09 (d, 4H, J = 7.8 Hz), 4.21 (t, 4H, J = 4.8 Hz), 3.88 (t, 4H, J = 4.8 Hz); 13C NMR (DMSO-d6) 162.5, 159.1, 144.9, 130.6, 128.8, 128.3, 126.3, 125.8, 115.2, 69.1, 67.4; MS m/e 495 (M+ + 1). Anal. Calcd for C30H30N4O3 • 2HCl • H2O: C, 61.53; H, 5.85; N, 9.56. Found: C, 61.678; H, 5.77; N, 9.38.
Synthesis of 1,5-Bis(4′-amidino-1,1′-biphenyl-4-yloxy)pentane Dihydrochloride (DB1703)
A mixture of 4′-hydroxybiphenyl-4-carbonitrile (1.95 g, 0.01 mol), 1,5-dibromopentane (1.15 g, 0.005 mol), and 2.07 g (0.015 mol) of anhydrous K2CO3 in anhydrous CH3CN was heated at reflux for 12–18 h (TLC followed); after removal of solvent and treatment with water, the separated solid was filtered, washed with water, and crystallized from EtOAc:hexane as white solid 1,5-bis(4′-cyano-1,1′-biphenyl-4-yl-oxy) pentane, 1.6 g (79%): mp 127–8 °C; 1H NMR (CDCl3): 7.71 (d, 4H, J = 8.4 Hz), 7.66 (d, 4H, J = 8.4 Hz), 7.56 (d, 4H, J = 8.8 Hz), 7.03 (d, 4H, J = 8.8 Hz), 4.09 (t, 4H, J = 6.8 Hz), 1.93 (quintet, 4H, J = 6.8 Hz), 1.74 (sept, 2H, J = 6.8 Hz); 13C NMR (CDCl3) 159.7, 154.2, 132.6, 131.4, 128.4, 127.1, 119.1, 115.1, 110.1, 67.9, 29.0, 22.8; MS m/e 458 (M+). Anal. Calcd for C31H26N2O2: C, 81.19; H, 5.72; N, 6.11. Found: C, 80.98; H, 5.62; N, 6.02.
Following the above procedure for DB1791, the dinitrile (0.458 g, 0.001 mol) and 5 mL of 1 M LiN(TMS)2 in THF (0.84 g, 0.005 mol) yielded 0.46 g (79%) the hydrochloride salt of the title bisamidine as a white solid: mp 300 °C; 1H NMR (DMSO-d6/D2O) 7.82(d, 4H, J = 8.4 Hz), 7.79 (d, 4H, J = 8.4 Hz), 7.66 (d, 4H, J = 9 Hz), 7.04 (d, 4H, J = 9 Hz), 4.07 (t, 4H, J = 6.6 Hz), 1.85–1.79 (m, 4H), 1.67–1.55 (m, 2H); 13C NMR (DMSO-d6) 165.2, 159.3, 144.9, 130.3, 128.8, 128.3, 126.3, 125.7, 115.1, 67.5, 28.4, 22.2; MS m/e 493 (M+ + 1); Anal. Calcd for C31H32N4O2 • 2HCl • H2O: C, 63.80; H, 6.21; N, 9.60. Found: C, 63.86; H, 6.09; N, 9.40.
Synthesis of 1,5-Bis(7-amidinonaphthalen-2-yloxy)pentane Dihydrochloride (DB1743)
A mixture of 7-cyano-2-naphthol (1.68 g, 0.01 mol), 1,5-dibromopentane (1.15 g, 0.005 mol), and 2.07 g (0.015 mol) of anhydrous K2CO3 in anhydrous CH3CN was heated at reflux for 12–18 h (TLC followed); after removing solvent and treating with water, the separated solid was filtered, washed with water, and crystallized from EtOAc:hexane as white solid 1,5-bis(7-cyanonaphthalen-2-yloxy)pentane, 1.6 g (79%): mp 259–60 °C; 1H NMR (DMSO-d6) 8.39 (brs, 2H), 8.06 (d, 2H, J = 8 Hz), 7.96 (d, 2H, J = 8.8 Hz), 7.61 (d, 2H, J = 8 Hz), 7.48 (brs, 2H), 7.38 (d, 2H, J = 8.8 Hz), 4.70 (t, 4H, J = 4.6 Hz), 2.00 (q, 4H, J = 4.6 Hz), 1.68 (q, 2H, J = 4.6 Hz); 13C NMR (DMSO-d6) 157.7, 133.4, 132.7, 129.8, 129.6, 129.1, 124.0, 122.1, 119.3, 108.8, 107.1, 67.7, 28.3, 25.3; MS m/e 406 (M+). Anal. Calcd for C27H22N2O2: C, 79.78; H, 5.45; N, 6.89. Found: C, 80.04; H, 5.54; N, 6.95.
Using the procedure described for DB1791, the above bisnitrile (0.406 g, 0.001 mol) and 5 mL of 1 M LiN(TMS)2 in THF (0.84 g, 0.005 mol) yielded 0.4 g (74%) of the title bisamidine as a white hydrochloride salt: mp 217–18 °C; 1H NMR (DMSO-d6) 9.50 (br, 4H), 9.43 (br, 4H), 8.42 (d, 2H, J = 8 Hz), 8.02 (d, 2H, J = 8.4 Hz), 7.95 (d, 2H, J = 9 Hz), 7.70 (dd, 2H, J = 8.4 Hz, J = 1.8 Hz), 7.44 (dd, 2H, J = 2.4 Hz), 7.35 (dd, 2H, J = 9 Hz, J = 2.4 Hz), 4.18 (t, 4H, J = 6.6 Hz), 1.90 (quintet, 4H, J = 6.6 Hz), 1.70 (quintet, 2H, J = 6.6 Hz); 13C NMR (DMSO-d6) 167.0, 158.4, 134.0, 131.3, 130.1, 129.1, 128.8, 126.5, 122.5, 121.9, 108.8, 68.7, 29.1, 23.0; MS m/e 441 (M+ + 1). Anal. Calcd for C27H28N4O2 • 2HCl • 1.75H2O: C, 59.50; H, 6.19; N, 10.28. Found: C, 59.66; H, 6.19; N, 10.17.
Synthesis of 1,5-Bis(7-amidino-2-naphthalenoxy)-3-oxapentane Dihydrochloride (DB1744)
Anhydrous K2CO3 (2.07 g, 0.015 mol) was added to a solution of 6-cyano-2-naphthol (1.69 g, 0.01 mol) and 1,5-dibromo-3-oxa-pentane (1.16 g, 0.005 mol) in 50 mL of acetonitrile. The solution was heated at reflux for 12–15 h (TLC followed); solvent was removed; the residue was treated with water; and the solid was filtered, washed with 2 N NaOH and water, dried, and recrystallized from hot EtOAc–hexane to yield 1,5-bis-(7-cyano-2-naphthalenoxy)-3-oxa-pentane as a white solid, 1.68 g (82%): mp 216–17 °C; 1H NMR (DMSO-d6) 8.30 (d, 2H, J = 1.8 Hz), 7.97 (d, 2H, J = 8.4 Hz), 7.90 (d, 2H, J = 9 Hz), 7.55 (dd, 2H, J = 1.8 Hz, J = 8.4 Hz), 7.43 (d, 2H, J = 2.4 Hz), 7.33 (dd, 2H, J = 2.4 Hz, J = 9 Hz), 4.23 (m, 4H), 3.83 (m, 4H); 13C NMR (DMSO-d6) 157.5, 133.4, 132.7, 129.9, 129.6, 129.1, 124.1, 122.0, 199.4, 108.8, 107.2, 68.8, 67.6; MS m/e 408 (M+). Anal. Calcd for C26H20N2O3: C, 76.45; H, 4.93; N, 6.86. Found: C, 76.22; H, 4.86; N, 6.78.
Following the procedure for DB1791, the above bisdinitrile (0.408 g, 0.001 mol) and 5 mL of 1 M LiN(TMS)2 in THF (0.84 g, 0.005 mol) yielded 0.42 g (77%) of white hydrochloride salt of the title bisamidine: mp 240–2 °C; 1H NMR (DMSO-d6) 9.42 (br, 4H), 9.29 (br, 4H), 8.35 (d, 2H, J = 1.5 Hz), 8.03 (d, 2H, J = 8.4 Hz), 7.95 (d, 2H, = 9 Hz), 7.67 (dd, 2H, J = 1.5 Hz, J = 8.4 Hz), 7.46 (d, 2H, J = 2.1 Hz), 7.36 (dd, 2H, J = 2.1 Hz, J = 2.9 Hz), 4.31 (dd, 4H, J = 6 Hz, J = 4.8 Hz), 3.95 (dd, 4H, J = 6 Hz, J = 4.8 Hz); 13C NMR (DMSO-d6) 166.0, 157.2, 132.9, 130.4, 129.2, 128.1, 127.7, 125.6, 121.4, 121.0, 107.9, 68.8, 67.5; MS m/e 438 (M+ + 1). Anal. Calcd for C26H26N4O3 • 2HCl • 1.5H2O: C, 57.56; H, 5.76; N, 10.23. Found: C, 57.72; H, 5.62; N, 10.17.
Thermal Melting (Tmax)
DNA thermal melting experiments were conducted using Cary 300 BIO UV–vis at a wave-length of 260 nm. The absorbance of samples containing free DNA as a reference and different ratios of compounds in MES 10 buffer using 1 cm quartz cuvettes were monitored as a function of temperature. The temperature was monitored by a thermistor inserted in a reference cuvette and was increased by a rate of 0.5 °C/min. The thermal melting point was calculated from a plot of dA260/dT vs temperature, and the maximum point was taken as Tmax.
The concentration of DNA was determined from absorbance by using the nearest neighbor extinction coefficient (18) and adjusted to 2 × 10−6 M. The ratio of compound to DNA was set to 0.5:1, 1:1, 1.5:1, and 2:1 to look at the melting point changes vs ratio and to make sure all the binding sites were filled by compounds.
Circular Dichroism (CD)
The experiments were done in a 1 cm quartz cuvette on a Jasco J-810 spectrometer with MES 10 buffer at 25 °C. The wavelength region from 230 to 450 nm was used to cover both DNA and compound spectral regions. The desired ratios were obtained by adding compounds to the cell containing constant amounts of DNA.
Surface Plasmon Resonance (SPR)
SPR experiments were conducted as previously described (19) using a BIAcore 2000 biosensor system. A four channel streptavidin-coated chip (BIAcore SA) was used to manually immobilize biotin-labeled DNA hairpins on three channels with the first channel left blank as a reference. The steady-state experiments were conducted by injecting different concentrations of compounds at 50 µL/min in Tris buffer containing 0.005% surfactant P-20. Corrected sensorgrams at each concentration were obtained by subtraction of the signal from the blank channel. The maximum response unit (RUmax) was calculated from DNA molecular weight, compound molecular weight, and the amount of DNA that was immobilized on each channel (19, 20). The number of binding sites as well as binding constants were calculated from fitting the RU vs free compound concentration in a two-site or one-site (K2 = 0) model:
RU = RUmax(K1Cfree+2K1K2Cfree2)/(1+K1Cfree+K1K2Cfree2)
where K1 and K2 are macroscopic equilibrium constants and Cfree is the compound concentration in the injecting buffer (20).
Isothermal Titration Calorimetry (ITC)
ITC experiments were performed using a MicroCal VP-ITC (MicroCal Inc., Northampton, MA) controlled with a computer equipped with VP-2000 viewer instrument software. The data were analyzed with Origin 5.0 software. In ITC experiments, 10 µL increments of compound in MES 10 buffer at 0.1 mM concentration were injected into the sample cell, which was filled with 0.01 mM AAAGTTT sequence. Injections were done every 300 s for a total of 30 injections with the exception of the first injection with 2 µL volume. The duration of each injection was set to 20 s with 290 rpm. The observed heat for each peak was measured by integration of the peak area with respect to time. A blank titration was conducted with the same conditions with injection of compound to the sample cell-containing buffer. The corrected interaction heat for each injection was calculated by subtraction of a blank titration from the DNA titrations. The binding enthalpy was determined by fitting the data in the 0–0.5, relatively linear, molar-ratio range to obtain the binding enthalpy for the strong binding site. The complexity of the ITC curves at higher ratios (secondary binding and an ITC dip near a ratio of 1.0) prevented us from obtaining unique fits to the results at higher ratios.
RESULTS
Compound Design
Since DB75 and pentamidine have both been used in successful clinical treatment of kinetoplastid parasitic diseases (1, 4, 5), the initial compound design was based on the use of similar units to design potential agents for binding to AT sites that are separated by a GC base pair as in kDNA (Figure 1). The idea was to use the linker of pentamidine to cross the GC base pair and to use aromatic systems larger than the pentamidine phenylamidines to bind to the two A/T sites. In the simplest extended compounds, the phenyls of pentamidine were replaced with biphenyl and naphthyl groups. In more extended compounds, one of the phenyls was extended with the phenyl-furan aromatic system of DB75 (DB1791). An even more extended derivative was prepared with both phenyls replaced with the DB75 system, but the compound had poor solubility. In a final set of derivatives, one phenyl of pentamidine was extended with a benzimidazole group. Benzimidazoles, for example with Hoechst33258, typically bind very strongly to A/T DNA sequences and that was the rationale behind this design (21, 22). The amidine substitutes in Figure 1 are protonated under the experimental conditions of the experiments reported here, and all compounds are dications. The unsubstituted benzimidazole pKa is near 5.5, while benzimidazoles substituted in the 5 or 6 position with electron withdrawal groups such as a protonated amidine have much lower pKa values (23). The benzimidazole substitutes in Figure 1 are thus neutral under experimental conditions.
Thermal Melting
ΔTmax (Tmax of complex − Tmax of DNA) measurements are a useful way to qualitatively rank compounds based on their relative binding affinity (24). The DB75 and benzimidazole analogs of pentamidine had the largest Tmax increase (9–11 °C, Table 1), and their thermal melting profiles with the AAAGTTT hairpin sequence are shown in Figure 2. Replacement of the three-methylene linker in the DB75 analog (DB1791) with a five-methylene linker reduced the ΔTmax from 9 to 5 °C (not shown). In an attempt to improve binding affinity, amidine, isopropyl amidine, and imidazoline cationic groups were used in the benzimidazole series but the ΔTmax values were similar for all modified cationic groups. Binding of the biphenyl and naphthyl compounds to the AAAGTTT sequence gave smaller ΔTmax values (Table 1). Since the binding of these compounds to the AATT sequence was also weak, the Tmax experiments with other sequences were not done.
Table 1.
Tmax Results of Compounds with AAAGTTT, AAAGCTTT, and AATT Sequencesa
| AAAGTTT | AAAGCTTT | AATT | AAA | |
|---|---|---|---|---|
| Tmax | 69.5 | 72.5 | 71.0 | 72.0 |
| 5BGR66 | 10.5 | 6.0 | 11.0 | 5.0 |
| 15SAB68 | 11.0 | 6.5 | 11.0 | 4.5 |
| 13SAB89 | 10.0 | 5.5 | 9.0 | 4.0 |
| DB1791 | 9.0 | 6.0 | 5.5 | 1.0 |
| DB1701 | 4.0 | 1.5 | ||
| DB1703 | 6.0 | 1.5 | ||
| DB1744 | 4.0 | 2.5 | ||
| DB1743 | 6.0 | 2.0 |
The first row is the DNA melting point while the following rows are the ΔTmax (Tmaxcomplex − TmaxDNA) values for the complexes. The experiments were done in MES 10 buffer. The ratio of compound to DNA is 2:1.
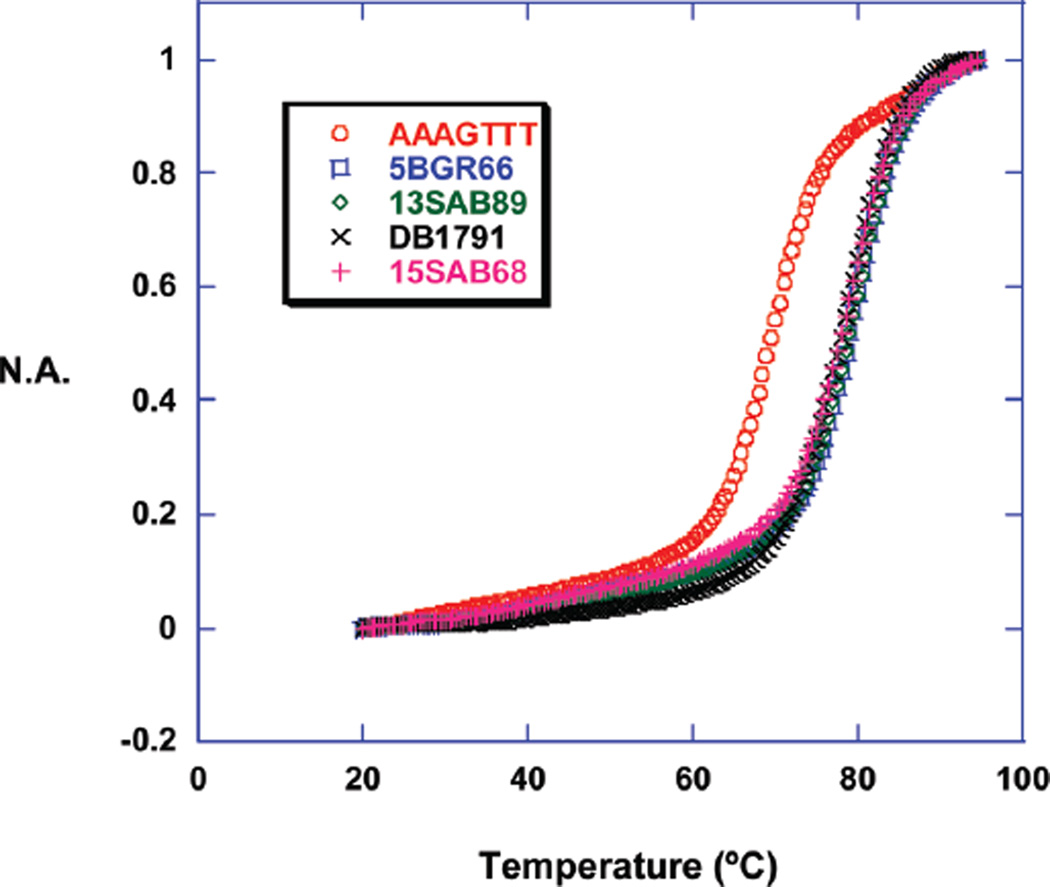
Figure 2.
Normalized absorbance (N.A.) vs temperature of compound-AAAGTTT complexes.
The melting point experiments show that compounds containing a phenyl-furan or a benzimidazole group have good affinity for the AAAGTTT sequence. To compare the effect of the length of DNA binding sequences, the benzimidazole containing compounds and DB1791 were evaluated with the AAAGCTTT, AATT, and AAA hairpins (Figure 1). The ΔTmax values decreased on increasing the DNA GC length in AAAGCTTT, but it was similar for the AATT and AAAGTTT sequences (Table 1). The low ΔTmax values for the single AAA sequence indicate that three consecutive A/T base pairs are not enough for binding of these compounds. The Tmax of AAAGTTT with DB75, a very well studied diamidine compound, increased by 5 °C, which shows that three A/T base pairs are not enough for strong binding of this compound and that it does not likely bind in a cooperative manner. The Tmax of AAAGTTT with pentamidine, another well studied minor groove binder, increased by 3 °C, in agreement with its expected weaker binding.
Circular Dichroism
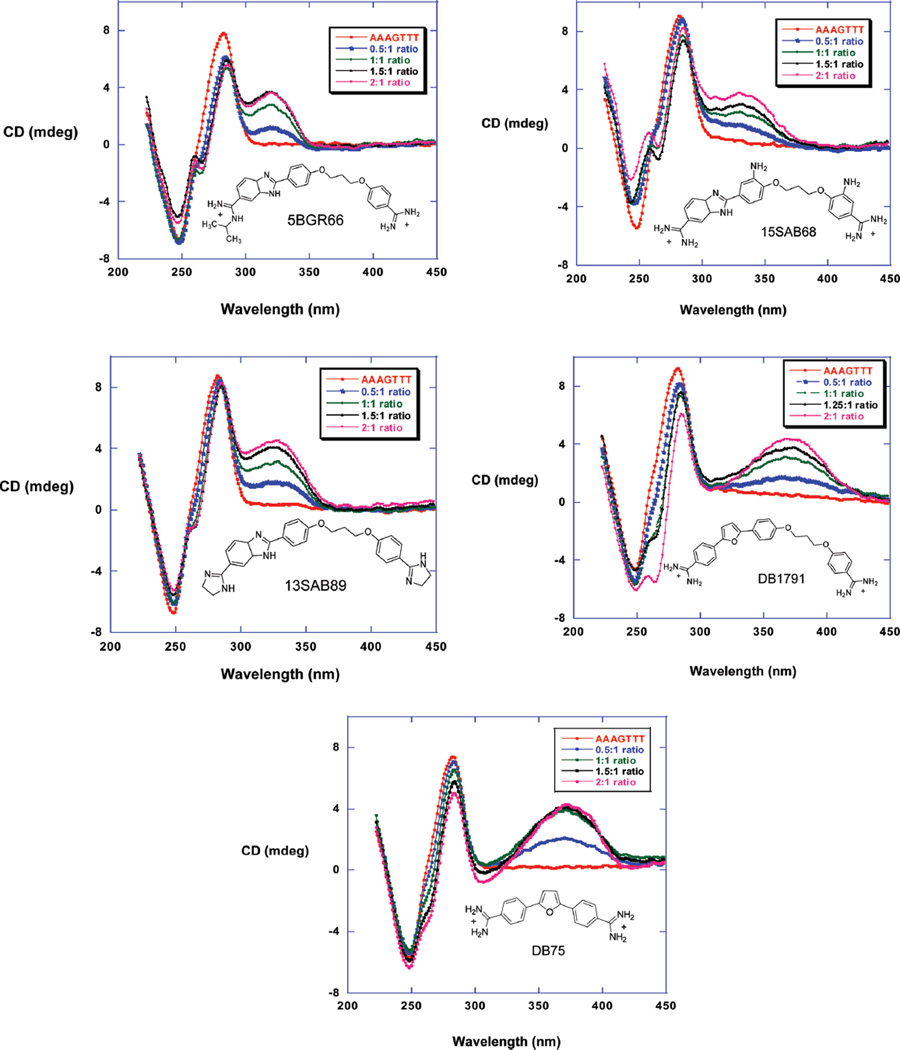
CD experiments are useful to identify the binding mode of minor groove binding compounds(25,26). The unbound compounds of Figure 1 do not show any CD signals; however, a strong and positive signal is induced upon binding to AAAGTTT (Figure 3). The CD signal changes decrease slightly after a 1:1 ratio indicating that the strong primary binding is followed by weak secondary binding in good agreement with SPR results (below). The induced CD signal of DB75 with the AAAGTTT sequence (Figure 3) is similar to that for the longer compounds in agreement with a minor groove binding mode (27, 28).
Figure 3.
CD spectra of 5BGR66, 13SAB89, 15SAB68, DB1791, and DB75 titrated with the AAAGTTT sequence. The added ratios of compounds to the DNA from bottom to top are 0.5, 1, 1.5, and 2. The experiments were done in MES 10 buffer at 25 °C.
Surface Plasmon Resonance
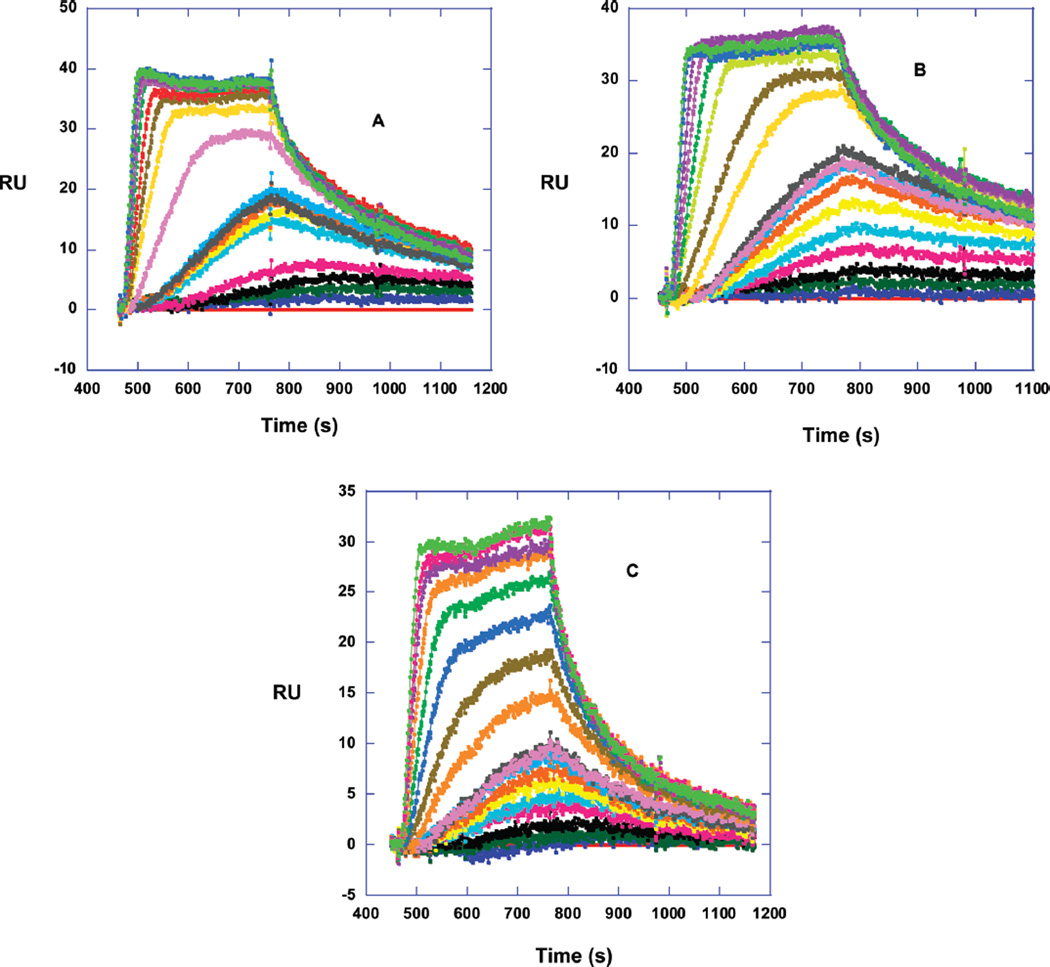
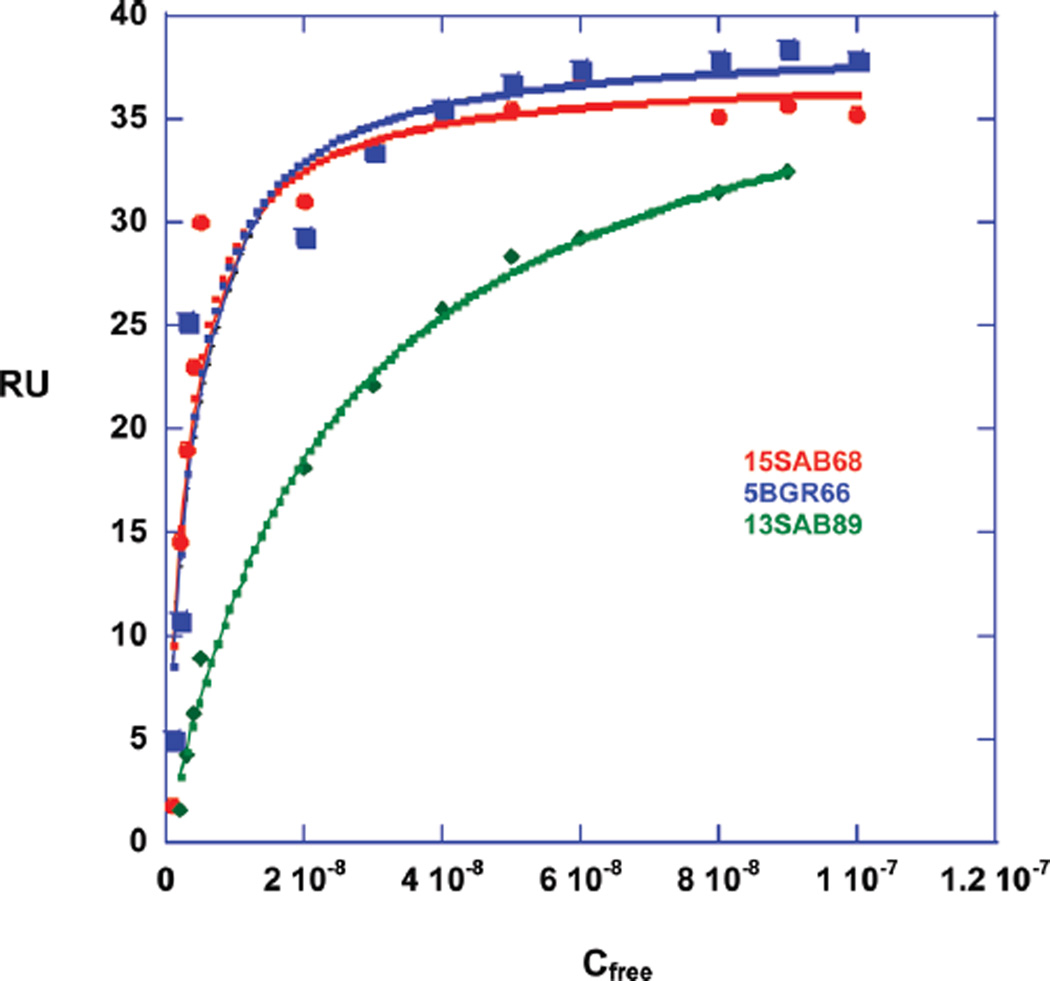
Since SPR is sensitive to the mass bound to immobilized DNA and does not need any label or radioactivity, it provides an excellent method to quantitatively investigate small molecule–DNA interactions (19, 29, 30). The method is able to quantitatively determine the kinetics, stoichiometry, binding constant, and the cooperativity of small molecule–nucleic acid interactions. The benzimidazole compounds, which showed the best interaction with AAAGTTT in Tmax and CD experiments, were titrated against three immobilized DNAs on a four-channel sensor chip. The AAAGTTT sequence was the primary target of interest, and the AATT and AAAGCTTT sequences were chosen as references to compare the effect of the classical AATT and the AAA/TTT sites with two intervening base pairs in comparison to a one base pair gap in the AAAGTTT sequence. The SPR sensorgrams of the benzimidazole 5BGR66 (Figure 4A) show slow but increasing kinetics of association from 1 to 10 nM concentration. At the highest concentrations, the association is fast enough for the sensorgrams to reach a steady state. From 1 to 10 nM concentration of free compounds, the RUs at the steady-state were predicted from fitting the curves to a 1:1 global kinetics model that includes a mass transfer term (19, 29, 30). After 10 nM, the steady state regions were used directly. The other benzimidazoles have the same behavior as 5BGR66 (Figure 4B,4C). The binding constants were determined by plotting the steady-state RU values as a function of the compound concentration in the flow solution, the free compound concentration, as described in Materials and Methods. The plot shows saturation of DNA at approximately 10−7 M concentration of compound (Figure 5) and that the interaction is not cooperative. The fitting of RU vs Cfree shows very strong binding to the AAAGTTT sequence with one strong binding site and weaker nonspecific binding with a binding constant that is around 100 times less than the primary binding (Knonspecific ≈ 105 M−1). The nonspecific binding is typical of organic dications at high ratios to DNA (31). The results are in good agreement with Tmax and CD experiments.
Figure 4.
SPR sensorgrams for 5BGR66 (A), 15SAB68 (B), and 13SAB89 (C) binding to AAAGTTT. The concentrations start from 1 to 100 nM from bottom to top. The experiments were done in Tris buffer at 25 °C.
Figure 5.
RU values vs free compound concentration. The RUs from 1 to 10 nM were calculated from kinetic fitting with the mass transfer model. The RUs from 10 to 100 nM were measured from steady-state fitting. The data were fitted in a one-site model for 15SAB68 (red) and 5BGR66 (blue) and a two-site model for 13SAB89 (green) with one strong and one nonspecific binding. The nonspecific binding is approximately 100 times weaker than the primary binding.
Binding of the benzimidazoles to AATT is similar to the binding to AAAGTTT while binding to AAAGCTTT is reduced (Table 3). DB75, which is a well-studied diamidine compound that binds to four consecutive A/T base pairs, was used as reference. It binds strongly to the AATT sequence with K = 9.3 × 106, which is in good agreement with previous studies (32). Its binding to AAAGTTT (K = 2.00 × 106) and AAAGCTTT (K = 1.45 × 106) is reduced, as expected for three base pair A/T sequences. The results for 40HXL820 and DB1791 complexes could not be used due to unknown erratic SPR responses, perhaps due to aggregation or sensorchip surface nonsubtractable adsorption.
Table 3.
Gibbs Energy Values for Compounds Binding to Different DNA Sequencesa
| 5BGR66 | 15SAB68 | 13SAB89 | |
|---|---|---|---|
| AAAGTTT | −11.5 | −11.6 | −10.4 |
| AAAGCTTT | −10.4 | −10.1 | −9.4 |
| AATT | −11.2 | −11.0 | −10.6 |
The experiments are done at 25 °C and in Tris buffer. The errors are 0.3 kcal/mol.
Isothermal Calorimetry
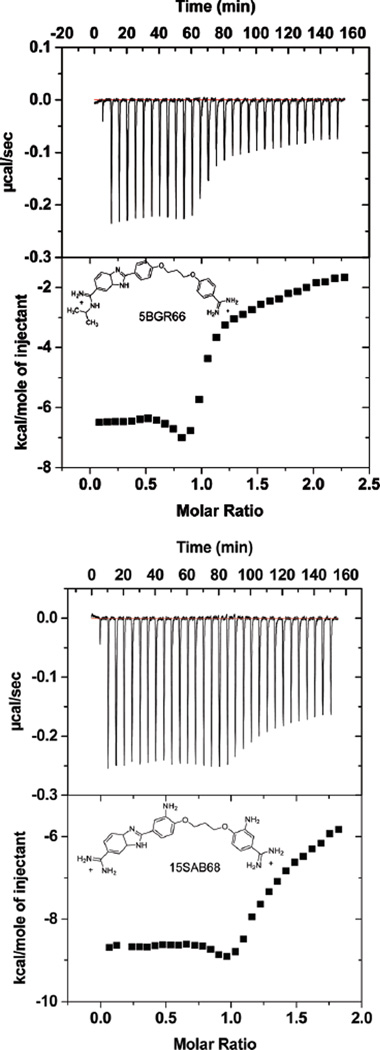
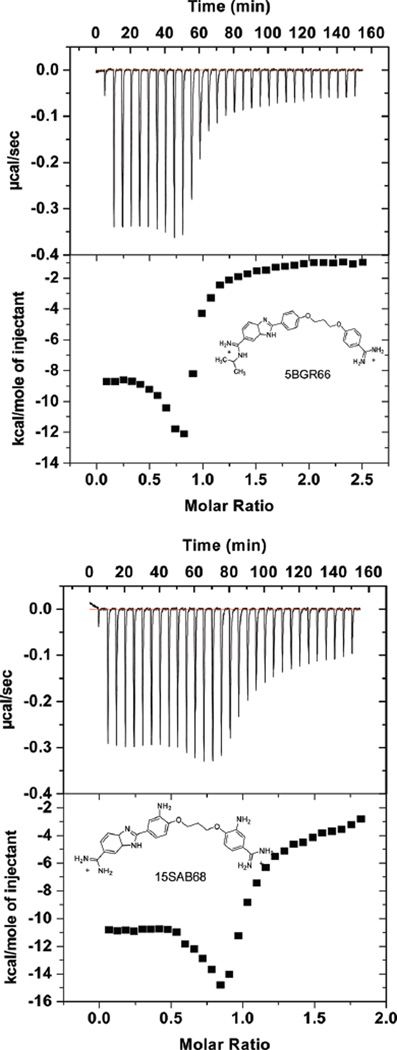
ITC experiments to determine the binding enthalpy were conducted with the benzimidazole compounds and the AAAGTTT and the AATT hairpin sequences. With the SPR results for ΔG°, the ITC ΔH allows calculation of full thermodynamic profiles for binding. The entropy was calculated from ΔG° = ΔH − TΔS (33–35). Figure 6 shows plots of heat vs molar ratio for injecting compound into a DNA solution and subtracting the heat of dilution for the compound. The data were fitted only in the molar ratio range 0–0.5 to obtain ΔH1 for the strong binding sites (Table 2). An ITC dip near a ratio of 1.0 and weaker, nonspecific binding at higher ratios complicate fitting. We have not attempted to assume a model and fit the high-ratio part of the ITC curve (36–39). The titration of 5BGR66 and 15SAB68 with the AATT sequence had the same pattern as the AAAGTTT sequence, but the heats of complex formation are somewhat more favorable than with AAAGTTT (Figure 7).
Figure 6.
ITC curves for binding of 5BGR66 (top) and 15SAB68 (bottom) to the AAAGTTT sequence. In each graph, the top plot is corrected experimental data and the bottom is the molar heat vs molar ratio of compound to the DNA. The experiments were done in MES 10 buffer at 25 °C.
Table 2.
Thermodynamic Profile of Compounds Titrated with AAAGTTT Sequencea
| Ka (M−1) | ΔG (kcal/mol) | ΔH (kcal/mol) | TΔS (kcal/mol) | |
|---|---|---|---|---|
| 5BGR66 | 2.8 × 108 | −11.5 | −6.6 | 4.9 |
| 15SAB68 | 3.4 × 108 | −11.6 | −8.7 | 2.9 |
| 13SAB89 | 4.5 × 107 | −10.4 | −1.4 | 9.0 |
The SPR experiments were done at 25 °C and in Tris buffer, while the ITC experiments were conducted in MES 10 buffer. The Ka values and thus ΔG values are measured by SPR; the ΔH values are measured by ITC. The experimental errors for ITC and SPR experiments are 0.1 and 0.3 kcal/mol, respectively.
Figure 7.
ITC curves for binding of 5BGR66 (top) and 15SAB68 (bottom) to the AATT sequence. The experimental conditions are the same as Figure 6.
DISCUSSION
AT specific minor groove binding compounds that could recognize A/T sequences of 5–6 base pairs that are separated by 1–2 GC base pairs would significantly increase the selectivity of binding and range of applications of these compounds. Such compounds, for example, would have greater ability to target the kinetoplast minicircle DNA of parasitic microorganisms. Since there are thousands of minicircles in the kinetoplast, this would dramatically increase the ability to selectively target the parasites. Our initial design concept to target such sequences includes two connected aromatic-amidine moieties that are specific for AT base pair sequences. Since the amino group of G in a GC base pair is in the minor groove, the AT specific groups were connected with oxy-alkyl linkers (Figure 1) with oxygens to provide potential H-bond acceptors for the G-amino. Such bifunctional compounds could provide very valuable information for the design of derivatives for other more complex recognition sequences starting with pure AT base pair binding agents.
Conversion of the natural polyamide distamycin to compounds that can also recognize GC base pairs relies on replacing the pyrrole groups of the parent compound with imidazole to interact with the G-amino (40–43). Our design is different in that we want compounds that bind primarily to short, adjacent AT sequences but which can recognize and cross one to two GC base pairs. Diamidines, such as pentamidine and DB75, bind to the minor groove of A/T rich regions of DNA with high specificity and are transported efficiently into cells, such as trypanosomes where they bind to mitochondrial kDNA (5). Thus, aromatic-amidines, based on these compounds, were selected for the A/T recognition units in our design. The parent compounds, such as DB75, have amidines at both ends while the extended compounds have one amidine for each linked component. The X-ray structure of DB75 bound to an AATT sequence shows that the amidine groups can cover almost a full base pair (44), and DB75 needs at least four base pairs for tight binding. Each component of the extended compounds, which have one amidine, can bind to three base pair sites, and connecting them can extend the recognition sequence and increase affinity. The low toxicity with excellent cell uptake of diamidines makes this class of compounds good candidates for therapeutic methods that target DNA sequences that have multiple binding sites for the compounds.
Biophysical experiments were done to investigate the binding of the designed compounds of Figure 1 with the AATT binding site that has been used in a large number of studies as well as the AAAGTTT and AAAGCTTT sites. Three A/T base pairs do not provide a strong binding site for diamidines, and the designed compounds have low affinity for DNA sequences with a single AAA site (Tmax values in Table 1). To obtain strong binding to the sites with three A/T base pairs separated by GC, the new compounds have to cross the GC base pair. Tmax studies indicated that some new compounds could bind strongly to the AAAGTTT sequence but not to AAAGCTTT. The benzimidazole compounds bound well to the classical AATT site, and they thus have a very favorable potential for enhanced binding to complex kDNA sequences. Addition of extra oxygen atoms in the linker did not improve binding relative to the alkyl linkers (compare DB1701–03 to DB1743–44, Table 1), perhaps due to competition with water or incorrect indexing of the oxygen and amino groups. The strongest binding compounds for the AAAGTTT sequence contained more complex heterocycles, phenyl-furan or benzimidazolephenyl, and the benzimidazole compounds were selected for more detailed, comparative studies.
CD experiments were conducted to evaluate the compound binding mode to the mixed AAAGTTT sequence, and the results clearly show that the compounds bind in the minor groove with similar induced CD signals, which are similar to those for classical minor groove binding at A/T sequences. Only minor changes were observed in the DNA CD region. This indicates that the compounds bind to the minor groove in a relatively standard B-form geometry with only minor conformational changes in the A/T binding sites. SPR binding studies provided sensorgrams for the benzimidazole compounds with AAAGTTT that indicate slow dissociation rates (Figure 4). The slow dissociation is consistent with tight binding, and quantitative analysis of the interaction gave equilibrium constants for complex formation at AAAGTTT of 108 M−1 or above (Figure 5 and Table 2). Binding to AATT had similar affinity, but binding to AAAGCTTT was approximately 10-fold lower (Table 3). These results taken together suggest that the compounds can bind strongly in an extended conformation to the two AAA sites in AAAGTTT but can adopt a more folded conformation that binds well to AATT. The extended conformation is, however, not optimized for the two GC base pair intervening sequence in AAAGCTTT.
ITC studies with the two benzimidazole compounds 5BGR66 and 15SAB68 that bind best to AAAGTTT and AATT indicate an enthalpy driven interaction for the initial and most important strong binding site. Although binding to AATT has a low enthalpy for other similar heterocyclic diamidines, binding of the substituted benzimidazoles to both sequences has a favorable enthalpy and low entropy of binding (21, 45). Since the compounds do not have exceptional H-bonding ability, these results are surprising and require more study with additional compounds for detailed explanations.
In summary, the design of oxy-methylene linked aromatic-amidine moieties provided compounds that could bind strongly in the minor groove of short A/T sequences that are separated by a GC base pair. Given the length of the derivatives, it is somewhat surprising that they bind almost as well to the AATT sequence as to AAAGTTT. This may indicate that the compounds can adopt a folded conformation that fits into a four-base pair A/T site and/or they can interact favorably with the GC base pairs at the ends of the AATT sequence. The ability to expand the A/T recognition sequences of classical A/T specific minor groove binding compounds offers exciting possible expansion of the DNA targeting ability of such compounds and could provide new application for cell permeable minor groove binders. The compounds of Figure 1 provide ideas for additional derivatives with different linkers that can target more complex GC base pairs containing DNA sequences. Compounds with a more rigid linker to interact with GC base pairs and inhibit folding will be designed in the future.
Footnotes
This work was supported by grants from NIH AI064200 (to W.D.W. and D.W.B.), the Gates Foundation (to D.W.B., R.R.T., and W.D.W.), and by a Molecular Basis of Disease fellowship (to M.R.). The ITC and SPR instruments were partially purchased with funds from the Georgia Research Alliance.
Abbreviations: Tmax, thermal melting temperature; CD, circular dichroism; SPR, surface plasmon resonance; ITC, isothermal titration calorimetry; MES, 2-(N-morpholino)ethanesulfonic acid; Tris, hydroxymethyl aminomethane hydrochloride; kDNA, kinetoplast DNA.
REFERENCES
- 1.Wilson WD, Nguyen B, Tanious FA, Mathis A, Hall JE, Stephens CE, Boykin DW. Dications that target the DNA minor groove: compound design and preparation, DNA interactions, cellular distribution and biological activity. Curr. Med. Chem. Anti–Cancer Agents. 2005;5:389–408. doi: 10.2174/1568011054222319. [DOI] [PubMed] [Google Scholar]
- 2.Mathis AM, Holman JL, Sturk LM, Ismail MA, Boykin DW, Tidwell RR, Hall JE. Accumulation and intracellular distribution of antitrypanosomal diamidine compounds DB75 and DB820 in African trypanosomes. Antimicrob. Agents Chemother. 2006;50:2185–2191. doi: 10.1128/AAC.00192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathis AM, Bridges AS, Ismail MA, Kumar A, Francesconi I, Anbazhagan M, Hu Q, Tanious FA, Wenzler T, Saulter J, Wilson WD, Brun R, Boykin DW, Tidwell RR, Hall JE. Diphenyl furans and aza analogs: effects of structural modification on in vitro activity, DNA binding, and accumulation and distribution in trypanosomes. Antimicrob. Agents Chemother. 2007;51:2801–2810. doi: 10.1128/AAC.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tidwell RR, Boykin DW. Dicationic DNA minor-groove binders as antimicrobial agents. In: Demeunynck M, Bailly C, Wilson WD, editors. DNA and RNA Binders: From Small Molecules to Drugs. Weinheim, Germany: Wiley-VCH; 2003. pp. 414–460. [Google Scholar]
- 5.Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW. Antiparasitic compounds that target DNA. Biochimie. 2008;90:999–1014. doi: 10.1016/j.biochi.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thuita JK, Karanja SM, Wenzler T, Mdachi RE, Ngotho JM, Kagira JM, Tidwell R, Brun R. Efficacy of the diamidine DB75 and its prodrug DB289, against murine models of human African trypanosomiasis. Acta Trop. 2008;108:6–10. doi: 10.1016/j.actatropica.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Bray PG, Barrett MP, Ward SA, de Koning HP. Pentamidine uptake and resistance in pathogenic protozoa: past, present and future. Trends Parasitol. 2003;19:232–239. doi: 10.1016/s1471-4922(03)00069-2. [DOI] [PubMed] [Google Scholar]
- 8.Bouteille B, Oukem O, Bisser S, Dumas M. Treatment perspectives for human African trypanosomiasis. Fundam. Clin. Pharmacol. 2003;17:171–181. doi: 10.1046/j.1472-8206.2003.00167.x. [DOI] [PubMed] [Google Scholar]
- 9.Rosypal AC, Werbovetz KA, Salem M, Stephens CE, Kumar A, Boykin DW, Hall JE, Tidwell RR. Inhibition by dications of in vitro growth of Leishmania major and Leishmania tropica: causative agents of old world cutaneous leishmaniasis. J. Parasitol. 2008;94:743–749. doi: 10.1645/GE-1387.1. [DOI] [PubMed] [Google Scholar]
- 10.Peixoto P, Liu Y, Depauw S, Hildebrand MP, Boykin DW, Bailly C, Wilson WD, David-Cordonnier MH. Direct inhibition of the DNA-binding activity of POU transcription factors Pit-1 and Brn-3 by selective binding of a phenyl-furan-benzimidazole dication. Nucleic Acids Res. 2008;36:3341–3353. doi: 10.1093/nar/gkn208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dervan PB, Doss RM, Marques MA. Programmable DNA binding oligomers for control of transcription. Curr. Med. Chem.: Anti-Cancer Agents. 2005;5:373–387. doi: 10.2174/1568011054222346. [DOI] [PubMed] [Google Scholar]
- 12.Shapiro TA, Englund PT. The structure and replication of kinetoplast DNA. Annu. Rev. Microbiol. 1995;49:117–143. doi: 10.1146/annurev.mi.49.100195.001001. [DOI] [PubMed] [Google Scholar]
- 13.Shlomai J. The structure and replication of kinetoplast DNA. Curr. Mol. Med. 2004;4:623–647. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- 14.Gavathiotis E, Sharman GJ, Searle MS. Sequence-dependent variation in DNA minor groove width dictates orientational preference of Hoechst 33258 in A-tract recognition: solution NMR structure of the 2:1 complex with d(CTTTTGCAAAAG)(2) Nucleic Acids Res. 2000;28:728–735. doi: 10.1093/nar/28.3.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahimian M, Miao Y, Wilson WD. Influence of DNA structure on adjacent site cooperative binding. J. Phys. Chem. B. 2008;112:8770–8778. doi: 10.1021/jp801997v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas S, Martinez LLIT, Westenberger SJ, Sturm NR. A population study of the minicircles in Trypanosoma cruzi: predicting guide RNAs in the absence of empirical RNA editing. BMC Genomics. 2007;8:133. doi: 10.1186/1471-2164-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tidwell RR, Geratz JD, Dann O, Volz G, Zeh D, Loewe H. Diarylamidine derivatives with one or both of the aryl moieties consisting of an indole or indole-like ring. Inhibitors of arginine-specific esteroproteases. J. Med. Chem. 1978;21:613–623. doi: 10.1021/jm00205a005. [DOI] [PubMed] [Google Scholar]
- 18.Cantor CR, Warshaw MM, Shapiro H. Oligo-nucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen B, Tanious FA, Wilson WD. Biosensor-surface plasmon resonance: Quantitative analysis of small molecule-nucleic acid interactions. Methods. 2007;42:150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Davis TM, Wilson WD. Surface plasmon resonance biosensor analysis of RNA-small molecule interactions. Methods Enzymol. 2001;340:22–51. doi: 10.1016/s0076-6879(01)40416-2. [DOI] [PubMed] [Google Scholar]
- 21.Haq I, Ladbury JE, Chowdhry BZ, Jenkins TC, Chaires JB. Specific binding of hoechst 33258 to the d(CGCAAATTTGCG)2 duplex: calorimetric and spectroscopic studies. J. Mol. Biol. 1997;271:244–257. doi: 10.1006/jmbi.1997.1170. [DOI] [PubMed] [Google Scholar]
- 22.Kiser JR, Monk RW, Smalls RL, Petty JT. Hydration changes in the association of Hoechst 33258 with DNA. Biochemistry. 2005;44:16988–16997. doi: 10.1021/bi051769x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapinos LE, Bin Song, Sigel H. Acid-Base and Metal-Ion-Coordinating Properties of Benzimidazole and Derivatives (= 1,3-Dideazapurines) in Aqueous Solution: Interrelation between Complex Stability and Ligand Basicity. Chem.-Eur. J. 1999;5(9):1794–1802. [Google Scholar]
- 24.Wilson WD, Tanious FA, Fernandez-Saiz M, Rigl CT. Evaluation of drug-nucleic acid interactions by thermal melting curves. Methods Mol. Biol. (Totowa, NJ) 1997;90:219–240. doi: 10.1385/0-89603-447-X:219. [DOI] [PubMed] [Google Scholar]
- 25.Lyng R, Rodger A, Norden B. The CD of ligand-DNA systems. 2. Poly(dA-dT) B-DNA. Biopolymers. 1992;32:1201–1214. doi: 10.1002/bip.360320910. [DOI] [PubMed] [Google Scholar]
- 26.Rodger A, Nordén B. Circular Dichroism and Linear Dichroism. New York: Oxford University Press; 1997. [Google Scholar]
- 27.Nguyen B, Tardy C, Bailly C, Colson P, Houssier C, Kumar A, Boykin DW, Wilson WD. Influence of compound structure on affinity, sequence selectivity, and mode of binding to DNA for unfused aromatic dications related to furamidine. Biopolymers. 2002;63:281–297. doi: 10.1002/bip.10073. [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Kumar A, Boykin DW, Wilson WD. Sequence and length dependent thermodynamic differences in heterocyclic diamidine interactions at AT base pairs in the DNA minor groove. Biophys. Chem. 2007;131:1–14. doi: 10.1016/j.bpc.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myszka DG. Kinetic, equilibrium, and thermodynamic analysis of macromolecular interactions with BIACORE. Methods Enzymol. 2000;323:325–340. doi: 10.1016/s0076-6879(00)23372-7. [DOI] [PubMed] [Google Scholar]
- 30.Day YS, Baird CL, Rich RL, Myszka DG. Direct comparison of binding equilibrium, thermodynamic, and rate constants determined by surface- and solution-based biophysical methods. Protein Sci. 2002;11:1017–1025. doi: 10.1110/ps.4330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazur S, Tanious FA, Ding D, Kumar A, Boykin DW, Simpson IJ, Neidle S, Wilson WD. A thermo-dynamic and structural analysis of DNA minor-groove complex formation. J. Mol. Biol. 2000;300:321–337. doi: 10.1006/jmbi.2000.3869. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Kumar A, Boykin DW, Wilson WD. Sequence and length dependent thermodynamic differences in heterocyclic diamidine interactions at AT base pairs in the DNA minor groove. Biophys. Chem. 2007;131:1–14. doi: 10.1016/j.bpc.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladbury JE, Chowdhry BZ. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem. Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- 34.Ladbury JE. Application of isothermal titration calorimetry in the biological sciences: things are heating up. Biotechniques. 2004;37:885–887. doi: 10.2144/04376TE01. [DOI] [PubMed] [Google Scholar]
- 35.Haq I. Thermodynamics of drug-DNA interactions. Arch. Biochem. Biophys. 2002;403:1–15. doi: 10.1016/S0003-9861(02)00202-3. [DOI] [PubMed] [Google Scholar]
- 36.Freyer MW, Buscaglia R, Hollingsworth A, Ramos J, Blynn M, Pratt R, Wilson WD, Lewis EA. Break in the heat capacity change at 303 K for complex binding of netropsin to AATT containing hairpin DNA constructs. Biophys. J. 2007;92:2516–2522. doi: 10.1529/biophysj.106.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freyer MW, Buscaglia R, Nguyen B, Wilson WD, Lewis EA. Binding of netropsin and 4,6-diamidino-2-phenylindole to an A2T2 DNA hairpin: a comparison of biophysical techniques. Anal. Biochem. 2006;355:259–266. doi: 10.1016/j.ab.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 38.Freyer MW, Buscaglia R, Cashman D, Hyslop S, Wilson WD, Chaires JB, Lewis EA. Binding of netropsin to several DNA constructs: Evidence for at least two different 1:1 complexes formed from an -AATT-containing ds-DNA construct and a single minor groove binding ligand. Biophys. Chem. 2006;126:186–196. doi: 10.1016/j.bpc.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Degtyareva NN, Fresia MJ, Petty JT. DNA Conformational Effects on the Interaction of Netropsin with A-tract Sequences. Biochemistry. 2007;46:15136–15143. doi: 10.1021/bi701722x. [DOI] [PubMed] [Google Scholar]
- 40.Dervan PB. Design of sequence-specific DNA-binding molecules. Science. 1986;232:464–471. doi: 10.1126/science.2421408. [DOI] [PubMed] [Google Scholar]
- 41.Lee M, Pon RT, Krowicki K, Lown JW. Structural and dynamic aspects of the sequence specific binding of netropsin and its bis-imidazole analogue on the decadeoxyri-bonucleotide d-[CGCAATTGCG]2. J. Biomol. Struct. Dyn. 1988;5:939–949. doi: 10.1080/07391102.1988.10506436. [DOI] [PubMed] [Google Scholar]
- 42.Lee M, Rhodes AL, Wyatt MD, D’Incalci M, Forrow S, Hartley JA. In vitro cytotoxicity of GC sequence directed alkylating agents related to distamycin. J. Med. Chem. 1993;36:863–870. doi: 10.1021/jm00059a011. [DOI] [PubMed] [Google Scholar]
- 43.Sharma SK, Morrissey AT, Miller GG, Gmeiner WH, Lown JW. Design, synthesis, and intracellular localization of a fluorescently labeled DNA binding polyamide related to the antibiotic distamycin. Bioorg. Med. Chem. Lett. 2001;11:769–772. doi: 10.1016/s0960-894x(01)00064-6. [DOI] [PubMed] [Google Scholar]
- 44.Laughton CA, Tanious F, Nunn CM, Boykin DW, Wilson WD, Neidle S. A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity. Biochemistry. 1996;35:5655–5661. doi: 10.1021/bi952162r. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Kumar A, Boykin DW, Bailly C, Wilson WD. Comparative thermodynamics for monomer and dimer sequence-dependent binding of a heterocyclic dication in the DNA minor groove. J. Mol. Biol. 2002;317:361–374. doi: 10.1006/jmbi.2002.5433. [DOI] [PubMed] [Google Scholar]