Abstract
Lysine acetylation modulates the activities of nonhistone regulatory proteins and plays a critical role in the regulation of cellular gene transcription. In this study, we showed that the transcriptional coactivator p300 acetylated β-catenin at lysine 345, located in arm repeat 6, in vitro and in vivo. Acetylation of this residue increased the affinity of β-catenin for Tcf4, and the cellular Tcf4-bound pool of β-catenin was significantly enriched in acetylated form. We demonstrated that the acetyltransferase activity of p300 was required for efficient activation of transcription mediated by β-catenin/Tcf4 and that the cooperation between p300 and β-catenin was severely reduced by the K345R mutation, implying that acetylation of β-catenin plays a part in the coactivation of β-catenin by p300. Interestingly, acetylation of β-catenin had opposite, negative effects on the binding of β-catenin to the androgen receptor. Our data suggest that acetylation of β-catenin in the arm 6 domain regulates β-catenin transcriptional activity by differentially modulating its affinity for Tcf4 and the androgen receptor. Thus, our results describe a new mechanism by which p300 might regulate β-catenin transcriptional activity.
β-Catenin was originally described as a component of cell-cell adhesion complexes, where it binds to E-cadherin. More recently, β-catenin was shown to be a key effector of the Wnt signaling pathway, which plays a pivotal role in growth and cell fate at early and late developmental stages (reviewed in references 37, 38, and 49). In the absence of Wnt signals, the cytosolic pool of β-catenin is maintained at a low level by targeted degradation in a multiprotein complex including the suppressor adenomatous polyposis coli (APC), Axin, glycogen synthase kinase 3, and casein kinase I α (16, 30, 41, 52, 53). Wnt activation abrogates the degradation of β-catenin and induces its accumulation and translocation into the nucleus, where it binds one of the four members of the T-cell factor/lymphoid enhancer factor (Tcf/Lef) family and activates transcription of target genes (4, 23). Growing evidence has associated Wnt signaling with tumor development. Constitutive Wnt signaling in cancer cells results mainly from genetic defects in the N-terminal region of the β-catenin gene itself or in the APC or Axin gene, which induce in all cases the stabilization and nuclear translocation of β-catenin (reviewed in reference 38).
Although it is well established that the formation of nuclear β-catenin/Tcf complexes plays a pivotal role in the activation of Wnt target genes, the fine mechanisms of transcriptional activation and regulation are still under investigation (5, 17). In the absence of β-catenin, the Tcf/Lef transcription factors act as transcriptional repressors by recruiting proteins such as Groucho/TLE, CtBP, and histone deacetylase (6-9, 28, 40). Upon Wnt activation, the binding of β-catenin to Tcf generates a bipartite transcription factor, in which Tcf provides the DNA binding domain and the C terminus of β-catenin provides the transactivation domain, therefore inducing a transcriptional switch. Recent physical and biochemical studies of the β-catenin-Tcf interaction have provided detailed information on the mode of β-catenin recognition by Tcf. Binding regions have been mapped to the N-terminal domain of Tcf/Lef and armadillo (arm) repeats 3 to 8 of β-catenin, with critical hot spots within repeat 8 (46). The crystal structure of β-catenin/Tcf complexes further revealed that the core arm repeat domain of β-catenin forms a superhelix of helices, providing a long, positively charged groove that engages the negatively charged β-catenin binding domain of Tcf (13, 14, 39). These studies outlined the importance of two critical lysine residues of β-catenin, K312 and K435, called the charged buttons, located in arm repeats 5 and 8.
Different aspects of the regulation of Tcf-dependent transcription by β-catenin have been unraveled. β-Catenin might recruit the basal transcription machinery via its interaction with the TATA-binding protein and Pontin 52 (TIP 49) (3, 18). β-Catenin has also been shown to interact with cellular factors essential for its transcriptional activity, such as pygopus and Lgs/BCl9, or with proteins involved in histone modification and chromatin remodeling, such as CBP/p300 and Brahma/Brg-1 (2, 20, 25, 33, 36, 43, 44). A crucial role for CBP/p300 in β-catenin/Tcf activity has been demonstrated during Xenopus embryogenesis and β-catenin-associated transformation (43, 44).
The mechanism by which CBP/p300 stimulate transcription is likely multifactorial (reviewed in references 12 and 27). CBP/p300 can contribute to the formation of a multiprotein activation complex bridging various factors to the general transcription machinery. In addition, CBP/p300 possess intrinsic histone acetyltransferase (HAT) activity, and histone acetylation regulates promoter activity by relieving chromatin-dependent repression. More recently, CBP/p300 have been shown to acetylate a growing number of nonhistone proteins, notably transcription factors such as p53, E2F, HMG I(Y), HNF-4, and human immunodeficiency virus Tat (15, 22, 31, 34, 42). Acetylation of these factors may affect different biological functions, including DNA binding affinity, transcriptional activity, stability, and subcellular localization. Recent studies have reported acetylation of β-catenin at lysine 49 and acetylation of the Caenorhabditis elegans Tcf/Lef homolog POP-1 at three neighboring residues (K185, 187, and 188), suggesting that acetylation might also be involved in the transcriptional activity of β-catenin-Tcf complexes (11, 50).
In this study, we showed that β-catenin is acetylated in vivo and in vitro by p300. We found that lysine K345, located in arm repeat 6, is an acetyl acceptor and that acetylation of K345 increases the affinity of β-catenin for Tcf4. We further demonstrated that acetylation of K345 specifically enhanced the coactivator function of β-catenin in a Tcf-dependent manner. Our data thus identify a new mechanism implicated in the regulation of β-catenin transcriptional activity and in the activation of Wnt-responsive genes.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa, 293, and SW480 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Cells were transiently transfected with β-catenin, p300, and Tcf4 vectors as indicated in the figure legends with the Lipofectamine reagent (Invitrogen). Total amounts of transfected DNA were kept constant by addition of empty pcDNA3 vector. All transfection experiments were repeated at least three times in duplicate. For luciferase assays, cells were lysed and assayed for luciferase activity 48 h posttransfection. Because p300 was found to activate transcription of the thymidine kinase-β-galactosidase plasmid used to normalize luciferase activity for transfection efficiency, it could not be used for normalization; the results were confirmed by multiple independent assays.
Antibodies.
Monoclonal antibodies against β-catenin were purchased from Transduction Laboratories. Antihemagglutinin (anti-HA), anti-Axin, and anti-APC antibodies were from Babco, Santa Cruz, and Oncogene Research Products, respectively. Monoclonal anti-acetyl-lysine antibodies were from Cell Signaling Technology and Upstate Biotechnology.
Plasmids.
The β-catenin expression vector pCMV-T41Aβ-catenin, carrying a Myc-tagged dominant stable β-catenin mutated at residue 41 (threonine to alanine), has been described previously (47). Mutations of lysine residue K345 and/or K49 to R or A were generated in this construct with the QuickChange XL site-directed mutagenesis kit (Stratagene) and verified by sequencing. Glutathione S-transferase (GST)-arm 1-12 and deletion mutants were generated by cloning PCR-amplified fragments of the β-catenin armadillo domain in pGEX-5X-1 (Amersham Biosciences). GST-arm 1-12 K354R and GST-arm 1-12 K345R were generated by site-directed mutagenesis (Stratagene). The HA-Tcf 4 construct and reporter plasmid pTOP-FLASH were kindly provided by H. Clevers and have been described previously (23). The androgen receptor (AR) expression vector was provided by G. Castona, and expression vectors for wild-type p300 and the p300 ΔHAT mutant deleted of amino acids 1413 to 1721 were provided by Y. Nakatami. GST p300 HAT (a gift of S. Emiliani) was generated by cloning a PCR-amplified fragment of p300 (positions 1195 to 1810) into the pGEX vector.
Recombinant proteins and in vitro binding assays.
GST-p300 HAT and GST-arm proteins were expressed in Escherichia coli BL21 and purified by glutathione-Sepharose affinity (Sigma) according to standard protocols. Tcf4 and AR proteins were in vitro translated in the presence of [35S]methionine with the TNT-coupled reticulocyte lysate system (Promega). For the in vitro binding assay and competition experiments, 35S-labeled Tcf4 or AR was mixed with GST or GST-arm 1-12 bound to Sepharose beads, in the absence or presence of competitor peptide (337LWTTSRVLKVLSVCSSN353), acetylated (Ac-arm 6 peptide) or not (arm 6 peptide) at K345 for 2 h at 4°C in binding buffer (20 mM HEPES [pH 7.9], 200 mM NaCl, 1 mM MgCl2, 0.5% NP-40, 1 mM dithiothreitol, and 1 mg of bovine serum albumin/ml). The beads were then washed six times with binding buffer, and bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Acetyltransferase assays.
For in vivo acetylation analysis, 293 cells were washed in phosphate-buffered saline at 24 h posttransfection, and incubated in methionine- and cysteine-free Dulbecco's modified Eagle's medium supplemented with 2% fetal bovine serum for 1 h at 37°C. Cells were then metabolically labeled with [3H]sodium acetate at 1 mCi/ml for 1 h at 37°C. Whole-cell extracts were prepared in lysis buffer (300 mM NaCl, 50 mM Tris [pH 7.5], 0.5% Triton X-100, 1 mM dithiothreitol) and protease inhibitor cocktail (Life Technologies), immunoprecipitated with monoclonal β-catenin antibody, and analyzed on SDS-8% PAGE. Gels were fixed in 30% methanol and 10% acetic acid, soaked in Amplify (Amersham), dried, and exposed to X-ray film.
In vitro acetylation assays were performed with a GST fusion protein containing the catalytic domain of p300 or with PCAF or p300 proteins produced and purified from a baculovirus overexpression system (35); 1 μg of GST-arm protein was incubated with PCAF or GST-p300 at 30°C for 1 h in 20 μl of reaction buffer (50 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol, 0.1 mM EDTA, 50 mM KCl, 5% glycerol, 10 μM sodium butyrate, protease inhibitor cocktail) and [14C]acetyl-coenzyme A (1 μCi/reaction; Amersham). Proteins were resolved on SDS-PAGE, and gels were treated as described above.
Immunoprecipitation and Western blotting.
Cells were lysed either in buffer A (150 mM NaCl, 50 mM Tris-HCl [pH 7.4], 0.5% [vol/vol] Nonidet P-40, and protease inhibitor cocktail) or in buffer B (300 mM NaCl, 50 mM Tris [pH 7.5], 0.5% Triton X-100, and protease inhibitor cocktail) as indicated and quickly frozen on dry ice. Lysates were thawed, centrifuged at 14,000 rpm for 10 min, and cleared by incubation with 30 ml of a protein A-protein G mixture. Antibodies were incubated with 50 μl of 50% (vol/vol) protein A-protein G-Sepharose for 2 h and added to the lysates. After overnight incubation, the beads were extensively washed with the same buffer, and bound proteins were resolved by SDS-PAGE.
For Western blot analysis, proteins were transferred to a nitrocellulose membrane and probed with the indicated antibody for 1 h. Reactive proteins were developed with secondary antibodies conjugated to alkaline phosphatase and visualized with chemiluminescence according to the manufacturer's protocol (Tropix). For quantitative measurements, gels were analyzed with a video acquisition system (Intelligent Dark Box II; Fuji) and Image Gauge software (Fuji).
EMSA.
For the electrophoretic mobility shift assay (EMSA), in vitro-translated Tcf 4 was incubated with GST-arm 1-12 in binding buffer (20 mM HEPES, pH 7.9, 75 mM NaCl, 1 mM dithiothreitol, 2 mM MgCl2, 10% glycerol) with 1 μg of poly(dI · dC) for 10 min at room temperature. Then [α32P]dCTP-labeled Tcf probe (105 cpm) was added, and the mixture was incubated for an additional 20 min at room temperature. The DNA probes used were complementary pairs of synthetic oligonucleotides with overhangs containing the consensus Tcf site (Tcf1, 5′-AGCTGGTAAGATCAAAGGG-3′, and Tcf2, 5′-TGCCGCCCTTTGATCTTACC-3′; the Tcf site is italic). For competition assays, increasing amounts of peptide were incubated for 5 min at room temperature with in vitro-translated Tcf4 prior to addition of arm 1-12. DNA-protein complexes were separated on a 4% acrylamide gels. Gels were dried and either exposed to X-ray films or visualized and quantified with a PhosphorImager (Storm Imaging System; Molecular Dynamics) as indicated in the figure legends.
RESULTS
β-Catenin is acetylated by p300 on lysine 345.
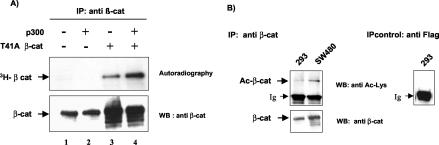
It has been shown that the CBP/p300 acetyltransferases can bind β-catenin and activate Tcf-dependent gene expression (20, 33, 43, 44). Because a growing number of nonhistone regulatory proteins are covalently modified by acetylation (reviewed in references 1 and 24), we investigated whether β-catenin is acetylated in vivo. To this end, we used either 293 cells transfected with a stable dominant form of β-catenin (T41A-β-catenin) or SW480 cells in which wild-type β-catenin is constitutively active owing to defective APC. 293 cells were transfected with T41A-β-catenin alone or in combination with p300, and metabolically labeled with 3H-labeled sodium acetate. After immunoprecipitation of β-catenin with a specific monoclonal antibody, proteins were separated on SDS-polyacrylamide gels. Acetylation of T41A-β-catenin was detected by the incorporation of radioactive acetate, and overexpression of p300 increased the amount of the acetylated form but not the total amount of β-catenin protein (Fig. 1A, lanes 3 to 4). Although endogenous β-catenin was efficiently immunoprecipitated in nontransfected 293 cells, the acetylated form was almost undetectable after autoradiography (Fig. 1A, lanes 1 to 2). By contrast, in SW480 colon carcinoma cells, immunoprecipitation of endogenous wild-type β-catenin followed by Western blot analysis with anti-acetyl-lysine antibodies confirmed that endogenous β-catenin was acetylated (Fig. 1B). Moreover, a faint signal corresponding to endogenous acetylated β-catenin was also detected in 293 cells with anti-acetyl-lysine antibodies (Fig. 1B), suggesting that anti-acetyl-lysine Western blotting might be more sensitive for detecting β-catenin acetylation than in vivo labeling.
FIG. 1.
β-Catenin is acetylated in vivo. (A) 293 cells were transfected with T41A-β-catenin (5 μg) either alone or together with 5 μg of p300 or the empty vector. After metabolic labeling with 3H-labeled sodium acetate, proteins were immunoprecipitated (IP) with β-catenin antibodies and resolved on SDS-PAGE. The gel was then exposed to X-ray film (top panel). Immunoprecipitated proteins were analyzed by Western blotting (WB) with anti-β-catenin antibodies (bottom panel). (B) Endogenous β-catenin was immunoprecipitated from 293 or SW480 cells with anti-β-catenin antibodies. Control immunoprecipitation was performed with an anti-Flag antibody. The samples were resolved by SDS-PAGE, and acetylation was assessed with anti-acetyl-lysine antibodies (upper panel). The amount of β-catenin in the immunoprecipitates was determined with anti-β-catenin antibodies (bottom panel).
We next sought to determine which individual lysine residues of β-catenin were acetylated. For this study, we focused on β-catenin domains involved in its transcriptional activity, particularly on the armadillo repeat domain, composed of 12 copies of a 42-amino-acid sequence (arm repeat), which is involved in the interaction of β-catenin with Tcf/Lef. Bacterially expressed GST-arm 1-12, carrying the full-length armadillo domain, and serial deletion constructs were incubated with purified recombinant p300 HAT in the presence of [14C]acetyl-coenzyme A, and acetylated proteins were detected by autoradiography. Our results show that the armadillo domain of β-catenin was acetylated in vitro by p300 and that acetylation was strongly reduced upon deletion of the arm 6 repeat (Fig. 2A), suggesting that acetylation sites may reside within this domain.
FIG. 2.
β-Catenin is acetylated at lysine residue K345 by p300. (A) Arm domain 6 of β-catenin is acetylated in vitro by p300. (Left panel) Schematic representation of GST-arm repeat constructs used in the in vitro acetylation assay. GST-full-length armadillo construct (GST-arm 1-12) or serial deletion constructs were incubated with GST-p300 recombinant protein and [14C]acetyl-coenzyme A. Reaction products were separated on SDS-10% PAGE. The gel was Coomassie blue-stained (bottom right panel) and then dried and exposed to X-ray film (upper right panel). (B) K345 is an acetyl acceptor in vitro. The amino acid sequence of the arm 6 domain contains two lysines (upper panel). GST-arm 1-12 (wt), GST-arm 1-12 K345R, or GST-arm 1-12 K354R was used for in vitro acetylation assays with GST-p300 recombinant protein. Reaction products were analyzed by SDS-PAGE and autoradiography. (C) In vitro acetylation assays with PCAF recombinant protein revealed acetylated PCAF and histone H3 but no acetylated form of β-catenin. (D) Lysine 345 is acetylated in vivo. HeLa cells were transfected with T41A-β-catenin or T41A-β-catenin K345R. Proteins were immunoprecipitated with β-catenin antibodies and resolved on SDS-PAGE. Blots were hybridized either with anti-acetyl-lysine antibodies or with anti-β-catenin antibodies. WB, Western blotting.
The arm 6 repeat contains two lysine residues (Fig. 2B), and mutant arm 1-12 constructs in which K345 or K354 was changed to arginine were subjected to in vitro acetylation assays. Substitution of K345 but not K354 to arginine eliminated acetylation of arm 1-12 by p300 (Fig. 2B), demonstrating that K345 is the main site of acetylation by p300 within the arm domain. Coomassie staining allowed us to verify that similar amounts of GST fusion proteins were used in the acetylation reactions (Fig. 2B, bottom panel). In contrast, recombinant CBP/p300-associated factor (PCAF) failed to acetylate GST-arm 1-12 in vitro, although it was able to acetylate histone H3 (Fig. 2C), showing that acetylation of β-catenin is achieved specifically by p300.
To confirm that K345 is an acetyl acceptor in vivo, HeLa cells were transfected with either the T41A-β-catenin or T41A-β-catenin K345R expression vector. β-Catenin proteins were immunoprecipitated with specific antibodies, and acetylated β-catenin was detected with anti-acetyl-lysine antibodies. Figure 2D shows that acetylation of the K345R β-catenin mutant was markedly decreased.
Identification of K345 as a residue important for β-catenin/Tcf4 interaction.
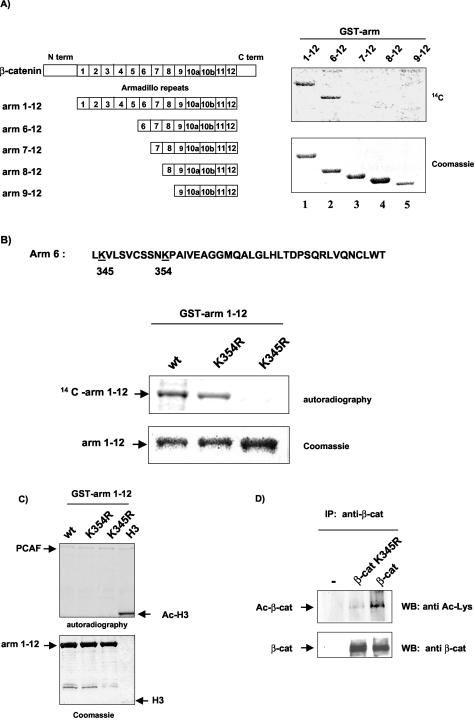
Having identified K345 as an acetyl acceptor, we sought to determine the role of K345 in β-catenin function. Notably, arm 6 is included in the domain involved in β-catenin interaction with Tcf/Lef (46), and this interaction is a critical step for targeting β-catenin to specific promoters and recruitment of transcriptional coactivators. We therefore assessed whether mutation of K345 would impair the β-catenin-Tcf4 interaction. Two β-catenin mutants in which K345 was changed to arginine (T41A-β-catenin K345R) or alanine (T41A-β-catenin K345A) were transfected into HeLa cells, either alone or together with HA-Tcf4. Cell lysates were immunoprecipitated with β-catenin antibodies, followed by immunoblotting analysis with anti-HA antibodies. Figure 3A shows that mutation of K345 to alanine abolished Tcf4 binding, whereas the K345R mutant was still able to bind Tcf4. The loss of binding of the K345A mutant β-catenin to Tcf4 was confirmed by immunoprecipitating the cell lysates with an anti-HA antibody followed by Western blot analysis with anti-β-catenin antibodies (data not shown).
FIG. 3.
K345 of β-catenin is important for β-catenin/Tcf4 binding. (A) HeLa cells were cotransfected with either T41A-β-catenin, T41A-β-catenin K345R, or T41A-β-catenin K345A (5 μg) and with Tcf4-HA expression vector (2.5 μg). Proteins were extracted in lysis buffer A. Following coimmunoprecipitation (IP) with anti-β-catenin antibodies, proteins were resolved on SDS-PAGE, and Tcf4 was detected by Western blotting (WB) with anti-HA antibodies. The amounts of β-catenin in precipitates and Tcf4 in total cell lysates were shown by Western blotting with anti-β-catenin and anti-HA antibodies. (B) HeLa cells were cotransfected with the TOPFLASH reporter (0.1 μg) and T41A-β-catenin or the indicated K345 mutant (0.2 μg) or the empty vector. Luciferase activities were assayed 48 h after transfection. The basal activity of the TOPFLASH plasmid cotransfected with empty vector was arbitrarily set at 1, and other values were calculated accordingly. The data shown are the averages of three separate experiments done in duplicate. (Bottom) Expression levels of the different constructs determined by Western blotting with anti-β-catenin antibodies. (C) HeLa cells were transfected with T41A-β-catenin or the corresponding K345R or K345A mutant. Cellular extracts were prepared in buffer B, and coimmunoprecipitation assays were performed with anti-β-catenin monoclonal antibodies. Immunoprecipitates were resolved on SDS-PAGE, and blots were hybridized with either APC (left) or Axin (right panel) antibodies. Immunoprecipitated β-catenin levels were shown by Western blotting with anti-β-catenin antibodies.
Transfection experiments with the TOPFLASH luciferase reporter containing Tcf/Lef consensus binding sites showed that the K345R mutant activated transcription as efficiently as wild-type β-catenin, while the K345A mutant, which is defective for Tcf4 binding, was less efficient (Fig. 3B). In addition, immunofluorescence studies showed normal nuclear localization of K345A mutant β-catenin, indicating that impaired binding to Tcf4 and transcriptional activity were not due to a defect in nuclear accumulation (data not shown). Collectively, these results indicate that K345 is involved in the binding of β-catenin to Tcf4.
Since β-catenin arm repeats 3 to 8 have been found to be involved in the interaction with other cellular partners such as APC and Axin, we investigated whether mutation of K345 would impair these interactions. HeLa cells were transfected with T41A-β-catenin or T41-β-catenin K345A or K345R, and cell lysates were immunoprecipitated with β-catenin antibodies and analyzed by Western blotting with anti-APC or anti-Axin antibodies. Both wild-type and mutant β-catenins were still able to interact with endogenous APC (Fig. 3C, left panel) and Axin (Fig. 3C, right panel). These results are in agreement with previous data showing that K345A mutant β-catenin was efficiently degraded by ectopic expression of conductin or APC (46).
Acetylation of K345 increases the affinity of β-catenin for Tcf4.
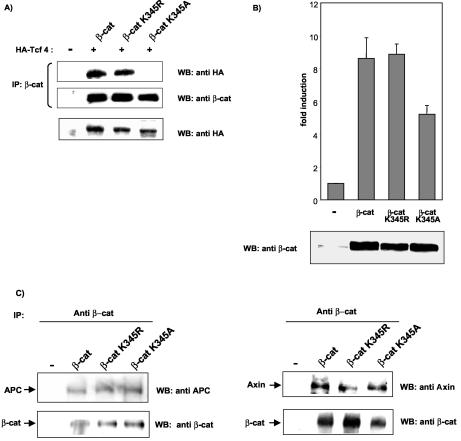
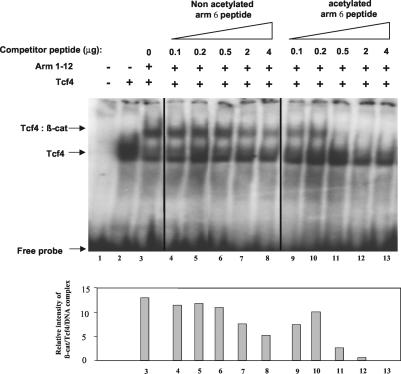
The finding that K345 is important for β-catenin-Tcf4 binding prompted us to examine the functional effect of acetylation on the β-catenin-Tcf4 interaction by EMSA. GST-arm 1-12 or GST-arm 1-12 K345R was incubated in acetylation buffer with recombinant p300 protein in the presence or absence of [14C]acetyl-coenzyme A. Recombinant proteins were either analyzed by SDS-PAGE and autoradiography (Fig. 4A) or incubated with in vitro-translated Tcf4 protein and 32P-labeled synthetic double-stranded DNA containing the consensus Tcf binding motif. EMSA analysis showed that acetylation of arm 1-12 increased its affinity for Tcf4 in a dose-dependent manner (Fig. 4B, compare lanes 3 to 5 with lanes 6 to 8). As previously observed in coimmunoprecipitation assays, arm 1-12 K345R was still able to bind Tcf4, but the binding was not increased when arm 1-12 K345R was incubated with active p300 protein (Fig. 4B, lanes 9 to 14). Moreover, competition experiments with increasing concentrations of a synthetic peptide containing acetylated K345 (ac-arm 6 peptide) or the corresponding nonacetylated peptide (arm 6 peptide) showed that the ac-arm 6 peptide was more effective than its nonacetylated counterpart in competing with the binding of arm 1-12 with Tcf4 (Fig. 5). Taken together, our results demonstrate that acetylation of β-catenin at K345 increases its affinity for Tcf4.
FIG. 4.
Acetylation of K345 increases the affinity of β-catenin for Tcf4. (A) Recombinant GST-arm 1-12 or GST-arm 1-12 K345R or histone H3 was incubated with recombinant p300 protein in the presence or absence of [14C]acetyl-coenzyme A. Acetylated proteins were analyzed by SDS-PAGE and autoradiography. Coomassie blue staining confirmed that equivalent amounts of proteins were loaded. (B) For EMSA, recombinant proteins were eluted from the beads, and identical amounts of proteins were incubated with in vitro-translated Tcf4 and 32P-labeled Tcf probe. Reactions were resolved on 4% acrylamide gels (left panel). The relative intensities of β-catenin/Tcf4/DNA complexes were quantified with a PhosphorImager and are graphically depicted in the right-hand panel.
FIG. 5.
Acetylated arm 6 peptide competes for β-catenin/Tcf binding more efficiently than its nonacetylated counterpart. Acetylated or nonacetylated arm 6 peptide at increasing concentrations was incubated with in vitro-translated Tcf4. GST-arm 1-12 and 32P-labeled Tcf probe were added to the reaction for EMSA analysis, followed by autoradiography. The relative intensities of the β-catenin/Tcf4/DNA complex in the corresponding lanes were quantified by the Syngene Photo Image System (Syngene, Cambridge, United Kingdom) and are shown in bar graphs in the bottom panel.
Acetylated β-catenin is preferentially associated with Tcf4 in vivo.
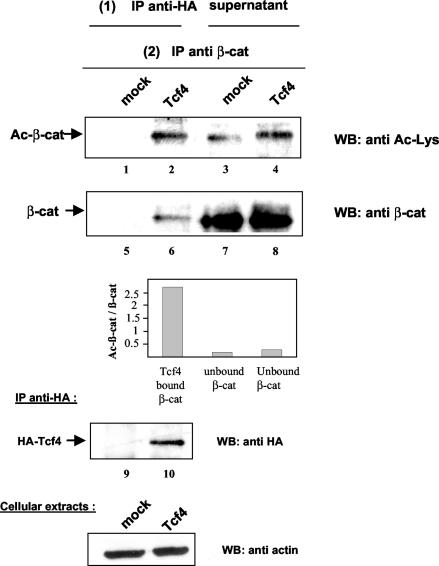
To determine whether acetylation of β-catenin at K345 increases its affinity for Tcf4 in vivo, we investigated whether the pool of acetylated β-catenin was preferentially associated with Tcf4 compared with nonacetylated β-catenin. 293 cells were transfected with an HA-Tcf4 expression vector or the empty vector, and immunoprecipitation was first carried out with an anti-HA antibody. Bound β-catenin was then eluted from the beads in 1% SDS. In a second step, both the eluate and the supernatant obtained from the first immunoprecipitation were immunoprecipitated with β-catenin-specific antibodies. Western blot analysis with anti-acetyl-lysine antibodies or anti-β-catenin antibodies and quantification with a video acquisition system (Fuji) showed that the ratio of acetylated β-catenin to total β-catenin was increased by 10-fold in the Tcf4-bound fraction compared to the results with the unbound fraction (Fig. 6). These data clearly indicate that acetylated β-catenin is preferentially found in Tcf-containing complexes in vivo and lend support to the notion that coactivation of β-catenin/Tcf complexes by p300 is mediated in part by the acetylation of β-catenin.
FIG. 6.
Acetylated β-catenin associates preferentially with Tcf4. 293 cells were transfected with 5 μg of HA-Tcf4 or empty vector. Proteins were extracted in lysis buffer B, and in a first step, Tcf4 and bound proteins were coimmunoprecipitated with anti-HA antibodies. Both the immunoprecipitated (IP) fraction and the supernatant were recovered and used for a second round of immunoprecipitation with anti-β-catenin antibodies. Proteins were resolved on SDS-PAGE and analyzed by Western blotting (WB) with anti-acetyl-lysine antibodies (lanes 1 to 4) or anti-β-catenin antibodies (lanes 5 to 8). The ratios of acetylated β-catenin to total β-catenin were measured with Image Gauge software (Fuji) and are presented in graphic form. Tcf4 levels in the immunoprecipitates were determined by anti-HA Western blotting (lanes 9 and 10). Analysis of cell lysates by Western blotting with antiactin antibodies confirmed that equivalent amounts of proteins were used for immunoprecipitation assays (bottom).
HAT activity of p300 stimulates β-catenin transactivation potential.
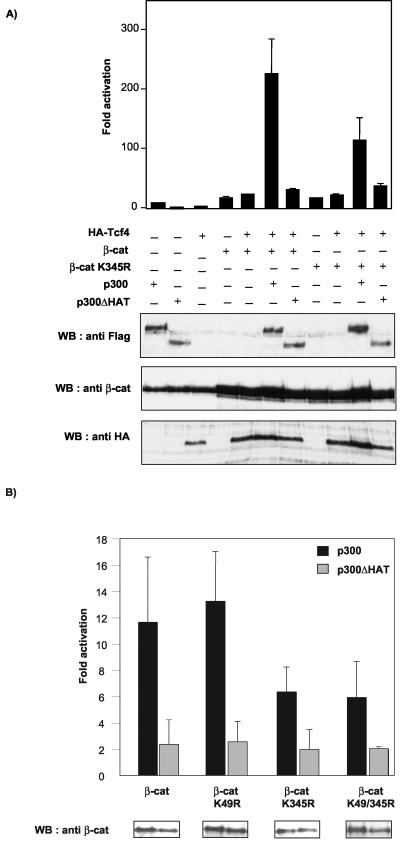
Since acetylation was found to increase the affinity of β-catenin for Tcf4, we next investigated whether acetylation of β-catenin by p300 regulates its transcriptional activity. 293 cells were transfected with the TOPFLASH luciferase reporter alone or with HA-Tcf4, T41A-β-catenin, T41A-β-catenin K345R, p300, or p300 ΔHAT, alone or in combination. As reported previously (20, 43, 44), p300 significantly enhanced β-catenin transcriptional activity (10-fold increase over that with β-catenin/Tcf4) (Fig. 7A). The acetyltransferase activity of p300 contributed to this activation because cotransfection with p300 ΔHAT had little effect on β-catenin/Tcf4 activity. Moreover, the T41A-β-catenin K345R mutant was less efficiently coactivated by p300 (fivefold increase in T41A-β-catenin K345R activity) and p300 ΔHAT could no longer activate this mutant. To rule out a nonspecific effect of p300, we checked the expression levels of β-catenin and Tcf4 in the presence of p300 and observed similar levels (Fig. 7A, bottom panels). We also verified that p300 and the p300 ΔHAT mutant were expressed at comparable levels. These results suggest that coactivation of β-catenin/Tcf by p300 is mediated, at least in part, by acetylation of the β-catenin K345 residue.
FIG. 7.
Cooperation between p300 and β-catenin is severely reduced by K345R mutation. (A) 293 cells were cotransfected with 0.1 μg of TOPFLASH reporter plasmid and either empty pCDNA3 vector or expression vectors for HA-Tcf4 (0.1 μg), T41A-β-catenin (0.25 μg), T41A-β-catenin K345R (0.25 mg), p300 (1.5 μg), and/or p300 ΔHAT (1.5 μg) as indicated. Luciferase activities were determined 48 h posttransfection. The basal TOPFLASH activity was set at 1, and data shown are the averages of three independent experiments performed in duplicate. WB, Western blotting. (Bottom panel) The expression levels of β-catenin, Tcf4, p300, and p300 ΔHAT were revealed by Western blotting with anti-β-catenin, anti-HA, and anti-Flag antibodies, respectively. (B) 293 cells were transfected with TOPFLASH and HA-Tcf4 (0.1 μg) and with either empty pCDNA3 vector or expression vectors for T41A-β-catenin or the indicated lysine mutants (0.25 μg), in combination or not with p300 or p300 ΔHAT (1.5 μg) as indicated. For each β-catenin mutant, TOPFLASH activity in cells cotransfectedwith Tcf4 and empty vector was arbitrarily set at 1. Data shown represent the activation by p300 or p300 ΔHAT for each β-catenin mutant. Values are the averages of three independent experiments performed in duplicate. The expression levels of the different β-catenin constructs in the presence of the coactivator are shown in the insets (bottom).
β-Catenin has been shown to be acetylated at K49 by p300 (50). It was therefore of interest to determine whether acetylation of K49 could also be involved in the cooperation between p300 and β-catenin. To this end, we cotransfected β-catenin mutants containing either a single mutation at K49 or K345 or the double mutation into 293 cells and tested whether the transcriptional activity of these mutants was affected by p300 in a TOPFLASH reporter assay. Our results show that while the K49R mutant was fully activated by p300 (13.3-fold), activation of the double mutant K49/345R by p300 was significantly reduced (5.9-fold) (Fig. 7B). We verified by Western blotting that all the β-catenin mutants were expressed at similar levels (Fig. 7B, lower panel). Taken together, our data suggest that acetylation of K345 by p300 contributes to a major extent to the increase in β-catenin transcriptional activity by p300. Our results are in agreement with a previous report showing that mutation of β-catenin at K49 does not impair TOPFLASH transactivation or modulate β-catenin's interaction with Tcf (50).
Acetylation of β-catenin K345 does not affect the affinity of β-catenin for the AR.
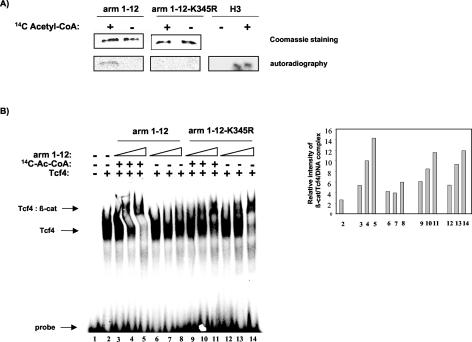
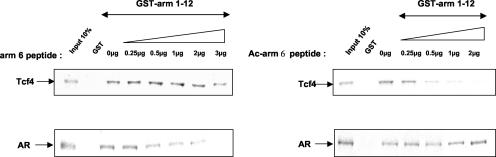
It has been shown recently that β-catenin can interact with the AR and activate transcription in a ligand-dependent fashion (45). Arm repeat 6 of β-catenin was found to be involved in binding with the AR, and the AR was shown to compete with Tcf for β-catenin binding (10, 51). Therefore, we wondered if acetylation of K345 would also increase the binding of β-catenin to the AR. To this end, we performed competition experiments. GST-arm 1-12 was immobilized on agarose beads and incubated with either in vitro-translated Tcf4 (Fig. 8, upper panels) or in vitro-translated AR (Fig. 8, lower panels) in the absence or in the presence of increasing concentrations of nonacetylated or acetylated arm 6 peptide. Consistent with the EMSA results (see Fig. 5), the acetylated arm 6 peptide was a better competitor of β-catenin/Tcf4 binding than the nonacetylated peptide. By contrast, the nonacetylated peptide seemed more efficient in competing the β-catenin and AR binding than the acetylated peptide. Thus, acetylation of K345 might specifically increase the affinity of β-catenin for Tcf4, while acetylation seems to decrease the affinity of β-catenin for the AR.
FIG. 8.
Acetylation of lysine 345 specifically increases the affinity of β-catenin for Tcf4. In vitro-translated Tcf4 (upper panels) and AR (lower panels) were incubated with immobilized GST-arm 1-12 in the presence of increasing amounts of acetylated or nonacetylated arm 6 peptide. Bound proteins were resolved on SDS-PAGE and analyzed by autoradiography.
DISCUSSION
Activation of Wnt target genes by β-catenin/Tcf is tightly controlled by a complex network of regulatory events, including interactions with various cellular partners and different covalent modifications of β-catenin and Tcf4 (17). Notably, the role of CBP and p300 as coactivators of β-catenin is well established, and their recruitment to Tcf-dependent promoters plays a crucial role during development and cell transformation (20, 43). In this study, we have shown that β-catenin is acetylated by p300 in vivo and in vitro and identified β-catenin residue K345 as a target site for acetylation. Substitution of K345 greatly reduced but did not totally eliminate the acetylation signal in vivo. These results are consistent with a recent report showing that CBP acetylates residue K49, located in the N-terminal domain of β-catenin (50). In both studies, PCAF was unable to acetylate β-catenin, indicating that β-catenin acetylation is specifically achieved by CBP/p300. Whether acetylation modulates β-catenin functions is therefore an important issue.
K345 is located in arm repeat 6, a region implicated in β-catenin interaction with a variety of cellular partners. Our mutagenesis studies indicate that K345 is not significantly involved in β-catenin interaction with APC or Axin 1, but it was found to be critical for Tcf4 binding. Interestingly, mutation to alanine abolished the interaction with Tcf4 but mutation to arginine had no effect, probably because the positively charged arginine can maintain the interaction with the negatively charged residues of Tcf4. This interpretation is supported by a recent structural analysis of the β-catenin/Tcf complex, suggesting that charged residues around β-catenin K312, including K345, might interact with the negatively charged region of Tcf4 extending from E23 to E29 and facilitate the initial anchorage of Tcf4 to β-catenin (13). The finding that the β-catenin mutant K345R was still able to bind Tcf4 suggests that acetylation does not serve merely to neutralize a positive charge but may create a novel interface facilitating the binding of β-catenin to Tcf4. Similarly, acetylation has been shown to increase the DNA binding potential of E2F1, although arginine mutants retained the ability to bind DNA (31). The role of K345 in the β-catenin-Tcf4 interaction was further confirmed by the finding that the capacity of the K345A and K345R mutants to bind Tcf4 was strictly correlated with their ability to transactivate the TOPFLASH reporter (Fig. 3B). Reduced transactivation efficiency has also been demonstrated previously for a series of β-catenin mutants defective in Lef-1 binding (46). This effect was not apparently related to abnormal subcellular localization of these β-catenin mutants, since exogenously overexpressed mutants accumulated in the nucleus at the same rate as β-catenin (data not shown). It is also unlikely that substitution of lysine 345 might affect the stability of β-catenin, since it does not impair β-catenin interaction with APC and Axin 1, two major partners within the degradation complex.
An important aspect of our results is that K345 acetylation increases the affinity of β-catenin for Tcf4. This conclusion is supported by several lines of evidence (see Fig. 4 to 8). (i) In the EMSA, p300 increased the binding of the wild-type β-catenin arm domain (arm 1-12) to Tcf4 but had no effect on the corresponding K345R mutant. (ii) In competition assays, a peptide containing acetylated K345 (ac-arm 6 peptide) disturbed this interaction in a dose-dependent manner, more efficiently than the homologous nonacetylated peptide, and (iii) comparable results were also obtained in GST pull-down experiments. (iv) In coimmunoprecipitation assays, the acetylated fraction of endogenous β-catenin was found to associate preferentially with Tcf4. Moreover, the increased affinity of acetylated β-catenin for Tcf4 affected the transcriptional activity of the complex, since cooperation between p300 and β-catenin was significantly reduced upon alteration of lysine 345 of β-catenin to arginine, although this mutant was still able to bind Tcf4. However, the transactivating activity of β-catenin/Tcf complexes involves the recruitment of a large number of cofactors through the β-catenin arm domain (17, 48). Therefore, we cannot completely rule out that acetylation of K345 could also influence the binding of such factors and thereby participate in transcription regulation.
Importantly, we observed that HAT-deficient p300 had little effect on β-catenin-mediated activation of the TOPFLASH reporter (Fig. 7A) and the natural β-catenin-responsive interleukin-8 promoter (data not shown), implying that the acetyltransferase activity of p300 was required for its coactivator function. In addition, while p300 retained some capacity to coactivate the K345R β-catenin mutant, the HAT-deficient p300 was unable to coactivate this mutant. These results strongly suggest that recruitment of p300 by β-catenin on Tcf-dependent promoters fulfills different functions, as previously described in different settings (26). Besides its ability to bridge DNA-associated activators to the basal transcription machinery, p300 might act by altering chromatin structure though intrinsic HAT activity and modulate β-catenin activity though its factor acetyltransferase activity. Such functional duality has been convincingly demonstrated for coactivation of HMGI(Y) and p65 by CBP/p300 or PCAF, because in these cases, the HAT activity stimulates transcription, but the factor acetyltransferase activity could serve the opposite function in turning off transcription (21, 34). Alternatively, as proposed by Miller and Moon (32), overexpressed mutant β-catenin could compete with endogenous β-catenin for binding to the degradation complex, leading to nuclear accumulation of both wild-type and mutant proteins.
Although coactivation of β-catenin by p300 is well established (20, 29, 33, 43, 44), the contribution of HAT and factor acetyltransferase activities has received little attention so far. Recently, Hecht and collaborators reported that coactivation of β-catenin at the siamois promoter is independent of the acetyltransferase activity of CBP/p300 (20). The reasons for the apparent discrepancy between this observation and our present data are unclear, but it might be explained by recent results showing that different Tcf proteins such as Lef1 and Tcf4E could perform specific, nonredundant functions at different natural β-catenin-responsive promoters (19). It will be of interest to determine whether the acetyltransferase activity of p300 is needed for coactivation of β-catenin in different promoter contexts and whether acetylation of β-catenin affects its binding to different members of the Tcf family. Although we have observed already that acetylation of K345 but not that of K49 mediates, at least in part, the transcriptional coactivation of β-catenin/Tcf by p300, further research on the functional effect of lysine 49 acetylation could determine whether it is implicated in modulating the acetylation of lysine 345.
Besides their role in Tcf-dependent transcription, it has been shown that β-catenin and CBP/p300 are transcriptional coactivators of the AR. Chesire and Isaacs (10) put forward the notion of a reciprocal balance between the activation of AR- and Tcf-related transcription by β-catenin, which might be involved in normal prostate development and in prostate tumor progression. In this study, we showed that acetylation of β-catenin at K345 specifically increases the affinity of β-catenin for Tcf4 but seems to decrease the affinity of β-catenin for the AR (Fig. 8). One might thus speculate that this posttranslational modification of β-catenin is involved in differential gene activation through Tcf versus the AR.
In conclusion, the role of p300 in the regulation of Wnt signaling appears to be complex. Thus, acetylation might participate in the activation of specific sets of genes upon Wnt signaling.
Acknowledgments
We thank O. Bischof, K. T. Jeang, J. G. Judde, and R. Kiernan for critical reading of the manuscript. We are grateful to P. Tiollais and A. Dejean for constant interest in this work. We also thank G. Castoria, H. Clevers, S. Emiliani, and Y. Nakatani for kindly providing the constructs used in this study.
This work was supported in part by grant 4395 from the Association pour la Recherche sur le Cancer (ARC). L.L. and Y.W. were funded by an ARC fellowship. C.L. was funded by an ENS fellowship.
REFERENCES
- 1.Bannister, A. J., and E. A. Miska. 2000. Regulation of gene expression by transcription factor acetylation. Cell. Mol. Life Sci. 57:1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, N., A. Hurlstone, H. Musisi, A. Miles, M. Bienz, and H. Clevers. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 20:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, A., S. Chauvet, O. Huber, F. Usseglio, U. Rothbacher, D. Aragnol, R. Kemler, and J. Pradel. 2000. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 19:6121-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens, J., J. P. von Kries, M. Kuhl, L. Bruhn, D. Wedlich, R. Grosschedl, and W. Birchmeier. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382:638-642. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, M., and H. Clevers. 2003. Armadillo/beta-catenin signals in the nucleus-proof beyond a reasonable doubt? Nat. Cell Biol. 5:179-182. [DOI] [PubMed] [Google Scholar]
- 6.Billin, A. N., H. Thirlwell, and D. E. Ayer. 2000. Beta-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol. Cell. Biol. 20:6882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brannon, M., J. D. Brown, R. Bates, D. Kimelman, and R. T. Moon. 1999. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126:3159-3170. [DOI] [PubMed] [Google Scholar]
- 8.Brantjes, H., J. Roose, M. van De Wetering, and H. Clevers. 2001. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 29:1410-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavallo, R. A., R. T. Cox, M. M. Moline, J. Roose, G. A. Polevoy, H. Clevers, M. Peifer, and A. Bejsovec. 1998. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395:604-608. [DOI] [PubMed] [Google Scholar]
- 10.Chesire, D. R., and W. B. Isaacs. 2002. Ligand-dependent inhibition of beta-catenin/TCF signaling by androgen receptor. Oncogene 21:8453-8469. [DOI] [PubMed] [Google Scholar]
- 11.Gay, F., D. Calvo, M. C. Lo, J. Ceron, M. Maduro, R. Lin, and Y. Shi. 2003. Acetylation regulates subcellular localization of the Wnt signaling nuclear effector POP-1. Genes Dev. 17:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giordano, A., and M. L. Avantaggiati. 1999. p300 and CBP: partners for life and death. J. Cell Physiol. 181:218-230. [DOI] [PubMed] [Google Scholar]
- 13.Graham, T. A., D. M. Ferkey, F. Mao, D. Kimelman, and W. Xu. 2001. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat. Struct. Biol. 8:1048-1052. [DOI] [PubMed] [Google Scholar]
- 14.Graham, T. A., C. Weaver, F. Mao, D. Kimelman, and W. Xu. 2000. Crystal structure of a beta-catenin/Tcf complex. Cell 103:885-896. [DOI] [PubMed] [Google Scholar]
- 15.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 16.Hamada, F., Y. Tomoyasu, Y. Takatsu, M. Nakamura, S. Nagai, A. Suzuki, F. Fujita, H. Shibuya, K. Toyoshima, N. Ueno, and T. Akiyama. 1999. Negative regulation of Wingless signaling by D-axin, a Drosophila homolog of axin. Science 283:1739-1742. [DOI] [PubMed] [Google Scholar]
- 17.Hecht, A., and R. Kemler. 2000. Curbing the nuclear activities of beta-catenin. Control over Wnt target gene expression. EMBO Rep. 1:24-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecht, A., C. M. Litterst, O. Huber, and R. Kemler. 1999. Functional characterization of multiple transactivating elements in beta-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem. 274:18017-18025. [DOI] [PubMed] [Google Scholar]
- 19.Hecht, A., and M. P. Stemmler. 2003. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 278:3776-3785. [DOI] [PubMed] [Google Scholar]
- 20.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiernan, R., V. Bres, R. W. Ng, M. P. Coudart, S. El Messaoudi, C. Sardet, D. Y. Jin, S. Emiliani, and M. Benkirane. 2003. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278:2758-2766. [DOI] [PubMed] [Google Scholar]
- 22.Kiernan, R. E., C. Vanhulle, L. Schiltz, E. Adam, H. Xiao, F. Maudoux, C. Calomme, A. Burny, Y. Nakatani, K. T. Jeang, M. Benkirane, and C. Van Lint. 1999. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 18:6106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korinek, V., N. Barker, P. J. Morin, D. van Wichen, R. de Weger, K. W. Kinzler, B. Vogelstein, and H. Clevers. 1997. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science 275:1784-1787. [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramps, T., O. Peter, E. Brunner, D. Nellen, B. Froesch, S. Chatterjee, M. Murone, S. Zullig, and K. Basler. 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 109:47-60. [DOI] [PubMed] [Google Scholar]
- 26.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehrmann, H., L. L. Pritchard, and A. Harel-Bellan. 2002. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv. Cancer Res. 86:41-65. [DOI] [PubMed] [Google Scholar]
- 28.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 95:11590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy, L., C. Neuveut, C. A. Renard, P. Charneau, S. Branchereau, F. Gauthier, J. T. Van Nhieu, D. Cherqui, A. F. Petit-Bertron, D. Mathieu, and M. A. Buendia. 2002. Transcriptional activation of interleukin-8 by beta-catenin-Tcf4. J. Biol. Chem. 277:42386-42393. [DOI] [PubMed] [Google Scholar]
- 30.Liu, C., Y. Li, M. Semenov, C. Han, G. H. Baeg, Y. Tan, Z. Zhang, X. Lin, and X. He. 2002. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108:837-847. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. R., and R. T. Moon. 1997. Analysis of the signaling activities of localization mutants of beta-catenin during axis specification in Xenopus. J. Cell Biol. 139:229-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyagishi, M., R. Fujii, M. Hatta, E. Yoshida, N. Araya, A. Nagafuchi, S. Ishihara, T. Nakajima, and A. Fukamizu. 2000. Regulation of Lef-mediated transcription and p53-dependent pathway by associating beta-catenin with CBP/p300. J. Biol. Chem. 275:35170-35175. [DOI] [PubMed] [Google Scholar]
- 34.Munshi, N., M. Merika, J. Yie, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol. Cell 2:457-467. [DOI] [PubMed] [Google Scholar]
- 35.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 36.Parker, D. S., J. Jemison, and K. M. Cadigan. 2002. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 129:2565-2576. [DOI] [PubMed] [Google Scholar]
- 37.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis-a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 38.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 14:1837-1851. [PubMed] [Google Scholar]
- 39.Poy, F., M. Lepourcelet, R. A. Shivdasani, and M. J. Eck. 2001. Structure of a human Tcf4-beta-catenin complex. Nat. Struct. Biol. 8:1053-1057. [DOI] [PubMed] [Google Scholar]
- 40.Roose, J., M. Molenaar, J. Peterson, J. Hurenkamp, H. Brantjes, P. Moerer, M. van de Wetering, O. Destree, and H. Clevers. 1998. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395:608-612. [DOI] [PubMed] [Google Scholar]
- 41.Rubinfeld, B., B. Souza, I. Albert, O. Müller, S. H. Chamberlain, F. R. Masiarz, S. Munemitsu, and P. Polakis. 1993. Association of the APC gene product with β-catenin. Science 262:1731-1734. [DOI] [PubMed] [Google Scholar]
- 42.Soutoglou, E., N. Katrakili, and I. Talianidis. 2000. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell 5:745-751. [DOI] [PubMed] [Google Scholar]
- 43.Sun, Y., F. T. Kolligs, M. O. Hottiger, R. Mosavin, E. R. Fearon, and G. J. Nabel. 2000. Regulation of beta-catenin transformation by the p300 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 97:12613-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truica, C. I., S. Byers, and E. P. Gelmann. 2000. Beta-catenin affects androgen receptor transcriptional activity and ligand specificity. Cancer Res. 60:4709-4713. [PubMed] [Google Scholar]
- 46.von Kries, J. P., G. Winbeck, C. Asbrand, T. Schwarz-Romond, N. Sochnikova, A. Dell'Oro, J. Behrens, and W. Birchmeier. 2000. Hot spots in beta-catenin for interactions with LEF-1, conductin and APC. Nat. Struct. Biol. 7:800-807. [DOI] [PubMed] [Google Scholar]
- 47.Wei, Y., M. Fabre, S. Branchereau, F. Gauthier, G. Perilongo, and M. A. Buendia. 2000. Activation of beta-catenin in epithelial and mesenchymal hepatoblastomas. Oncogene 19:498-504. [DOI] [PubMed] [Google Scholar]
- 48.Wei, Y., C. A. Renard, C. Labalette, Y. Wu, L. Levy, C. Neuveut, X. Prieur, M. Flajolet, S. Prigent, and M. A. Buendia. 2003. Identification of the LIM protein FHL2 as a coactivator of beta-catenin. J. Biol. Chem. 278:5188-5194. [DOI] [PubMed] [Google Scholar]
- 49.Willert, K., and R. Nusse. 1998. Beta-catenin: a key mediator of Wnt signaling. Curr. Opin. Genet. Dev. 8:95-102. [DOI] [PubMed] [Google Scholar]
- 50.Wolf, D., M. Rodova, E. A. Miska, J. P. Calvet, and T. Kouzarides. 2002. Acetylation of beta-catenin by CREB-binding protein (CBP). J. Biol. Chem. 277:25562-25567. [DOI] [PubMed] [Google Scholar]
- 51.Yang, F., X. Li, M. Sharma, C. Y. Sasaki, D. L. Longo, B. Lim, and Z. Sun. 2002. Linking beta-catenin to androgen-signaling pathway. J. Biol. Chem. 277:11336-11344. [DOI] [PubMed] [Google Scholar]
- 52.Yost, C., M. Torres, J. R. Miller, E. Huang, D. Kimelman, and R. T. Moon. 1996. The axis-inducing activity, stability and subcellular distribution of β-catenin is regulated in. Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 10:1443-1454. [DOI] [PubMed] [Google Scholar]
- 53.Zeng, L., F. Fagotto, T. Zhang, W. Hsu, T. J. Vasicek, W. L. Perry III, J. L. Lee, S. M. Tilghman, B. M. Gumbiner, and F. Costantini. 1997. The mouse-fused locus encodes axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90:181-192. [DOI] [PubMed] [Google Scholar]