Abstract
Glutaredoxins are small redox proteins able to reduce disulfides and mixed disulfides between GSH and proteins. Synechocystis sp. PCC 6803 contains three genes coding for glutaredoxins: ssr2061 (grxA) and slr1562 (grxB) code for dithiolic glutaredoxins while slr1846 (grxC) codes for a monothiolic glutaredoxin. We have analyzed the expression of these glutaredoxins in response to different stresses, such as high light, H2O2 and heat shock. Analysis of the mRNA levels showed that grxA is only induced by heat while grxC is repressed by heat shock and is induced by high light and H2O2. In contrast, grxB expression was maintained almost constant under all conditions. Analysis of GrxA and GrxC protein levels by western blot showed that GrxA increases in response to high light, heat or H2O2 while GrxC is only induced by high light and H2O2, in accordance with its mRNA levels. In addition, we have also generated mutants that have interrupted one, two, or three glutaredoxin genes. These mutants were viable and did not show any different phenotype from the WT under standard growth conditions. Nevertheless, analysis of these mutants under several stress conditions revealed that single grxA mutants grow slower after H2O2, heat and high light treatments, while mutants in grxB are indistinguishable from WT. grxC mutants were hypersensitive to treatments with H2O2, heat, high light and metals. A double grxAgrxC mutant was found to be even more sensitive to H2O2 than each corresponding single mutants. Surprisingly a mutation in grxB suppressed totally or partially the phenotypes of grxA and grxC mutants except the H2O2 sensitivity of the grxC mutant. This suggests that grxA and grxC participate in independent pathways while grxA and grxB participate in a common pathway for H2O2 resistance. The data presented here show that glutaredoxins are essential for stress adaptation in cyanobacteria, although their targets and mechanism of action remain unidentified.
Keywords: glutaredoxin, stress, redox regulation, cyanobacteria, high light, heat shock, oxidative stress, metal resistance
Introduction
Glutaredoxins are small redox proteins that were first discovered as alternatives to thioredoxin as electron donors for ribonucleotide reductase (Holmgren, 1976). Glutaredoxin structures are closely related to those of thioredoxins, the so-called thioredoxin fold, and they also catalyze disulfide reduction through a dithiol mechanism, as thioredoxins, though some glutaredoxins use a monothiol mechanism (Fernandes and Holmgren, 2004; Meyer et al., 2009). Furthermore, glutaredoxins are able to catalyze protein glutathionylation/deglutathionylation, a function initially ascribed for these enzymes (Rouhier et al., 2008; Meyer et al., 2009), but recent data suggest that thioredoxins could also contribute to deglutathionylating reaction in yeast and plants (Greetham et al., 2010; Bedhomme et al., 2012). Glutaredoxins generally use the GSH/glutathione reductase system for reduction, although some of them can also accept electrons directly from thioredoxin reductases or can be reduced by thioredoxins or other glutaredoxins (Zaffagnini et al., 2008; Couturier et al., 2009a; Meyer et al., 2009). Glutaredoxins can function as alternative electron donors for ribonucleotide reductase in E. coli (but not in yeast), in sulfate assimilation, as electron donors for 3′-phosphoadenosine 5′-phosphosulfate synthase or for methionine sulfoxide reductases (Meyer et al., 2009; Hanschmann et al., 2013; Lillig and Berndt, 2013; Toledano et al., 2013). Moreover, some glutaredoxins present peroxidase activity and/or are able to reduce some peroxiredoxins thus contributing to resistance to oxidative stress (Rouhier et al., 2001, 2002; Finkemeier et al., 2005; Hanschmann et al., 2010; Pedrajas et al., 2010). In addition, it has also been shown that some glutaredoxins are able to bind Fe-S clusters and are involved in some aspects of Fe-S cluster biogenesis and its regulation (Muhlenhoff et al., 2010; Rouhier, 2010; Couturier et al., 2011; Kumar et al., 2011; Li and Outten, 2012; Boutigny et al., 2013).
Six different classes of glutaredoxins have been described in photosynthetic organisms (Couturier et al., 2009a), where class I and II correspond to classical dithiolic and monothiolic glutaredoxins, respectively, that are present in all organisms. Class IV is restricted to photosynthetic eukaryotes and class III is specific to land plants. On the other hand class VI seems to be restricted to cyanobacteria and class V is only present in cyanobacteria and some proteobacteria (Benyamina et al., 2013). In photosynthetic eukaryotes the repertoire of glutaredoxin proteins is larger than in other organisms, which suggests that they could play critical roles regulating processes related to photosynthesis (Rouhier et al., 2008; Couturier et al., 2009a). The exact roles of glutaredoxins in photosynthetic organisms remain unclear mainly because several paralogs of each have been identified in many plant genomes and only a few mutants are available (Couturier et al., 2009a; Meyer et al., 2009, 2012). In Arabidopsis only a few mutants in glutaredoxin genes have been analyzed. GrxS14 and GrxS17 class II glutaredoxins have been shown to be essential for oxidative stress tolerance and heat tolerance, respectively, while class III glutaredoxins have been shown to be essential for flower development and pathogen resistance (Li et al., 2009; Murmu et al., 2010; Zander et al., 2011). In contrast, several of these proteins have been characterized biochemically indicating that glutaredoxins could play multiple roles, including functions as electron donors, deglutathionylation of target proteins or as Fe-S cluster sensors (Rouhier et al., 2001; Gelhaye et al., 2003; Rouhier et al., 2003, 2005; Feng et al., 2006; Vieira Dos Santos et al., 2007; Bandyopadhyay et al., 2008; Zaffagnini et al., 2008; Couturier et al., 2009b, 2011; Gao et al., 2011; Bedhomme et al., 2012).
In cyanobacteria much less is known about glutaredoxins and their functions. All cyanobacteria contain at least two different glutaredoxins: one from the classical dithiolic subgroup (class I) and one from the monothiolic subgroup (class II) (Couturier et al., 2009a). Moreover, some cyanobacteria contain genes encoding for additional dithiolic glutaredoxins (class I) or for proteins from classes V and VI. Synechocystis sp. PCC 6803 (hereafter Synechocystis) contains 3 genes coding for glutaredoxins: ssr2061 (grxA also known as grx2) and slr1562 (grxB also known as grx1) code for dithiolic glutaredoxins that belong to class I (Couturier et al., 2009a), while slr1846 (grxC also known as grx3) codes for a monothiolic glutaredoxin that belongs to class II (Couturier et al., 2009a). The Synechocystis glutaredoxins have been implicated in responses to metals and metalloids (Lopez-Maury et al., 2009; Marteyn et al., 2009; Kim et al., 2012; Marteyn et al., 2013) and oxidative stress (Li et al., 2007; Marteyn et al., 2009). Moreover, putative targets of GrxA have been identified using a monocysteinic glutaredoxin mutant (Li et al., 2007). GrxC has been shown to be a dimer and to bind a 2Fe-2S cluster that is ligated by one cysteine from each subunit of the dimer and two molecules of GSH (Picciocchi et al., 2007; Iwema et al., 2009). In contrast, the in vivo functions of GrxC have not been explored in cyanobacteria.
Here we have characterized mutant strains lacking one, two, or all glutaredoxin genes, in all possible combinations, in response to several stresses in Synechocystis. These have showed that GrxA and GrxC play a prominent role in protecting cells from stress. Analysis of double and triple mutants showed a complex pattern of genetic interactions, suggesting that they can be involved in different pathways which are all interconnected. Furthermore, expression of grxA and grxC genes was induced by some of these stresses, while the grxB gene was expressed constitutively under all conditions analyzed. GrxA and GrxC proteins levels did not always correspond to their mRNA levels under these conditions suggesting that expression of glutaredoxins could be controlled also post-transcriptionally.
Materials and methods
Strains and culture conditions
Synechocystis cells were grown photoautotrophically on BG11C (Rippka et al., 1979) at 30°C under continuous illumination (50 μE·m−2·s−1) and bubbled with a stream of 1% (v/v) CO2 in air. For plate cultures, the medium was supplemented with 1% (wt/vol) agar. Kanamycin, chloramphenicol and spectinomycin were added to a final concentration of 50 μg mL−1, 20 μg mL−1, and 5 μg mL−1, respectively. BG11C medium was supplemented with different concentrations of CuSO4, NiSO4, CdCl2, Na2SeO4, and Na2SeO3 when indicated. Experiments were performed using cultures from the mid-logarithmic phase (3–5 μg chlorophyll mL−1) cultivated without antibiotics. For high light conditions, cultures were illuminated with white light at an intensity of 500 μE·m−2·s−1, and the temperature was kept at 30°C by applying a 5-cm-thick water filter. For heat shock conditions, cells were grown in a water bath at 42°C. For oxidative stress conditions, 1 mM hydrogen peroxide was added. Synechocystis strains and their relevant genotypes are described in Table 1.
Table 1.
Strains used in this work.
| Strain | genotype | ORF mutated | Antibiotic resistance | References |
|---|---|---|---|---|
| WT | Synechocystis sp. PCC 6803 | Lab stock | ||
| SGRXA | grxA::C.C1 | ssr2061 | Cm | Lopez-Maury et al., 2009 |
| SGRXB | grxB::SpΩ | slr1562 | Sp | Lopez-Maury et al., 2009 |
| SGRXC | grxC::C.K1 | slr1846 | Km | Lopez-Maury et al., 2009 |
| SGRXAB | grxA::C.C1 grxB::SpΩ | ssr2061 slr1562 | Cm Sp | Lopez-Maury et al., 2009 |
| SGRXAC | grxA::C.C1 grxC::C.K1 | ssr2061 slr1846 | Cm Km | Lopez-Maury et al., 2009 |
| SGRXBC | grxB::SpΩ grxC::C.K1 | slr1562 slr1846 | Sp Km | Lopez-Maury et al., 2009 |
| SGRXABC | grxA::C.C1 grxB::SpΩ grxC::C.K1 | ssr2061, slr1562 slr1846 | Cm Sp Km | Lopez-Maury et al., 2009 |
RNA isolation and northern blot analysis
Total RNA was isolated from 30 mL samples of Synechocystis cultures in the mid-exponential growth phase (3–5 μg chlorophyll mL−1). Extractions were performed by vortexing cells in the presence of phenol-chloroform and acid-washed baked glass beads (0.25–0.3 mm diameter) as previously described (Garcia-Dominguez and Florencio, 1997). Five microgram of total RNA was loaded per lane and electrophoresed in 1.2% agarose denaturing formaldehyde gels (Sambrook et al., 1989) and transferred to nylon membranes (Hybond N-Plus; Amersham). All probes were synthesized by PCR and oligonucleotide pairs used are described in Table 2 and were 32P-labeled with a random-primer kit (Amersham Biosciences) using α-[32P] dCTP (3000 Ci/mmol). Prehybridization, hybridization, and washes were in accordance with Amersham instruction manuals. All filters were stripped and re-hybridized with the constitutively expressed rnpB gene from Synechocystis as loading control (Vioque, 1992). Hybridization signals were quantified with a Cyclone Phosphor System (Packard).
Table 2.
Oligonucleotides used in this work.
| Oligonucleotide | Sequence |
|---|---|
| GrxA250F | GGCTGTCTCGGCAAAAATTG |
| GrxA250R | GGTCCAACTTGCCTGCACCATC |
| GrxB331F | GGCTAATTTGTTCAACTGGC |
| GrxB331R | CTAGGCTGGGTTAGGAGGAG |
| GrxC295F | GCAAGAATTGATCAGTTGGTC |
| GrxC295R | GCCACTTCTAACATTTCCTGC |
| isiAF | CATAGGTCTCGGGTGGAC |
| isiAR | TAAAGCTGATGGCTAATG |
| pgr5_F | GGAGTCACTCATATGTTCGCCC |
| pgr5_R | CTCAGTTTCTCGAGAATTATTG |
| hspA_F | CCACACATCAGGAGTTAACAT |
| hspA_R | TTGATCATCTAGGGTCAGGAGC |
Anti-Grx antibody production, western blotting and preparation of crude extracts from Synechocystis cells
Anti-GrxA and anti-GrxC antisera were obtained according to standard immunization protocols by injecting GrxA and GrxC purified proteins in rabbits. For analysis of GrxA and GrxC protein levels in Synechocystis cells grown under different conditions, crude extracts were prepared using glass beads in 50 mM Tris HCl pH 8.0, 50 mM NaCl. Protein concentration in cell-free extracts or purified protein preparations was determined by the method of Bradford, using ovalbumin as a standard and the specified amounts of proteins were resolved using SDS-PAGE gels. Proteins were thereafter electroblotted onto nitrocellulose membranes that were blocked in PBS containing 0.1% tween 20 and 5% of skimmed milk and incubated with anti-GrxA (1:1000) or anti-GrxC (1:3000). Anti-GSI (1:50.000) was used as loading control.
Results
Glutaredoxins are not essential in Synechocystis
We have previously generated Synechocystis mutants lacking each or all of the glutaredoxin genes but their phenotypes were not analyzed in detail (Lopez-Maury et al., 2009). All mutant strains were viable in all possible combinations and were fully segregated when grown on solid media. Here, we have further analyzed their phenotypes in our standard growth conditions (BG11C, 30°C, 1% CO2 bubbled in air, 50 μE·m−2·s−1) during the growth curve. Growth of the mutants was analyzed in cultures inoculated at OD750nm = 0.1 (which is equivalent to 0.5 μg chl mL−1) and growth was monitored following both chlorophyll content and OD750nm for 5 days when it leveled off around 30 μg chl mL−1 (Figure 1A) and an OD750nm = 6 (Figure S1). None of the strains presented a substantial reduced growth rate during the lag, exponential, linear or stationary phases of the growth curve. We have also studied expression of the glutaredoxin genes during the growth curve. Signals for all three mRNAs could be detected during all growth phases although grxC was induced slightly (2 to 3-fold) during the early exponential growth phase and both grxA and grxB remained almost constant during all phases (Figures 1B,C). Furthermore, we have also analyzed GrxA and GrxC protein levels during the growth phases, using specific antibodies raised against the corresponding proteins, and the proteins levels did not change substantially (Figure 1D). We have further analyzed expression of the three glutaredoxins genes during the growth curve in single mutants lacking one of the other genes in order to study if there were any compensatory effects in their expression. None of the mutant strains presented changes in mRNA levels of the remaining glutaredoxins when compared to the WT strain (Figures 2A–C, S2). We have also analyzed protein levels in exponentially growing cells (3–5 μg chl ml−1) of single mutant strains although neither GrxA or GrxC protein levels changed (Figure 2D). These data suggest that glutaredoxins play a less prominent role than thioredoxins under non-stress conditions and contrast with data from yeast or E. coli in which at least one glutaredoxin is essential for growth (Draculic et al., 2000; Fernandes and Holmgren, 2004; Ortenberg et al., 2004; Buchanan and Balmer, 2005; Toledano et al., 2013).
Figure 1.
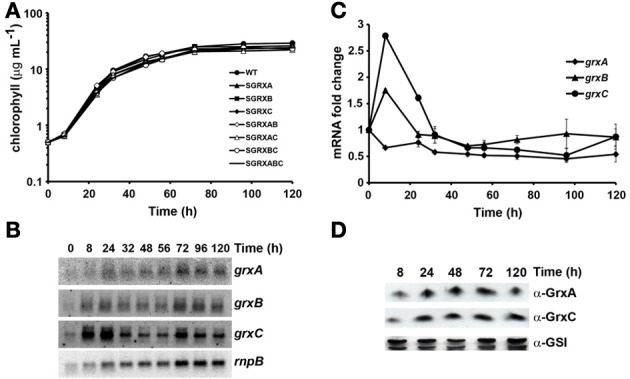
Glutaredoxins are not essential under standard growth conditions. (A) Semi-logarithmic representation of growth of Synechocystis glutaredoxin mutants strains under standard conditions. WT (•), SGRXA (▲), SGRXB (■), SGRXC (♦), SGRXAB (◊), SGRXAC (Δ), SGRXBC (○), and SGRXABC (−) strains were inoculated at 0.5 μg chlorophyll mL−1 and growth was monitored by measuring chlorophyll content. (B) Northern blot analysis of grxA, grxB, and grxC expression along the growth curve. Total RNA was isolated from WT cells grown in BG11C at the indicated times. The filter was hybridized with grxA, grxB, and grxC probes and subsequently stripped and re-hybridized with an rnpB probe as a control. (C) Quantification of relative mRNA levels of grxA, grxB, and grxC during the growth curve. Radioactive signals were quantified and normalized to the rnpB signal. Plots of relative mRNA levels vs. time were drawn; data represent average of 3 independent experiments and error bars represent SE. grxA (♦), grxB (▲), and grxC (•). (D) Western blot analysis of GrxA and GrxC levels during the growth curve. WT cells were grown in BG11C medium and cells were harvested at the indicated times. Fifteen microgram of total protein from soluble extracts were separated by 15% SDS-PAGE and subjected to western blot to detect GrxA, GrxC, and GSI.
Figure 2.
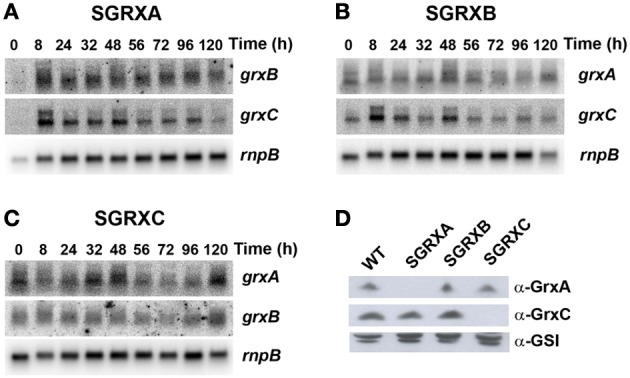
Glutaredoxin gene expression is not altered in single glutaredoxin mutants. (A) Northern blot analysis of grxB and grxC expression during different stages of the growth curve in SGRXA. Total RNA was isolated from SGRXA cells grown in BG11C at the indicated times. The filter was hybridized with grxB and grxC probes and subsequently stripped and re-hybridized with an rnpB probe as a control. (B) Northern blot analysis of grxA and grxC expression during different states of the growth curve in SGRXB. Total RNA was isolated from SGRXB cells grown in BG11C at the indicated times. The filter was hybridized with a grxA and grxC probes and subsequently stripped and re-hybridized with an rnpB probe as a control. (C) Northern blot analysis of grxA and grxB expression during different states of the growth curve in SGRXC. Total RNA was isolated from SGRXC cells grown in BG11C at the indicated times. The filter was hybridized with a grxA and grxB probes and subsequently stripped and re-hybridized with an rnpB probe as a control. (D) Western blot analysis of GrxA and GrxC levels in glutaredoxin mutants. WT, SGRXA, SGRXB, and SGRXC cells were grown in BG11C medium and cells were harvested at exponential phase (3–5 μg chl mL−1). Fifteen microgram of total protein from soluble extracts were separated by 15% SDS-PAGE and subjected to western blot to detect GrxA, GrxC, and GSI.
Responses to different stress conditions
In order to further characterize the glutaredoxin mutant strains we have analyzed both the growth and expression of glutaredoxins in response to different stress treatments, namely high light intensity, heat shock, hydrogen peroxide stress, and metal stress.
High light intensity
In order to study the effect of high light treatment we transferred exponentially growing cells (3–5 μg chl mL−1) from our standard light intensity (50 μE·m−2·s−1) to 500 μE·m−2·s−1 and followed glutaredoxin gene expression in response to this change in light intensity. This light intensity was chosen because it did not induce photoinhibition (Figure 3) and in fact WT cells grew faster under this illumination, but was high enough to reveal a phenotype in light sensitive mutants (Perez-Perez et al., 2009a). First, we analyzed expression of glutaredoxin genes in response to this treatment; as a marker gene we have used pgr5, the mRNA of which has been described to be induced by high light treatment (Allakhverdiev et al., 2002). pgr5 was transiently induced in our experiment, with a peak induction at 15 min (Figures 3A, S3A). After the shift to high light, grxC mRNA levels increased linearly during the 5 h of the treatment, while levels of grxA and grxB did not significantly change during the treatment (Figures 3A,B). In contrast, both GrxA and GrxC protein levels increased after this treatment (Figure 3C).
Figure 3.
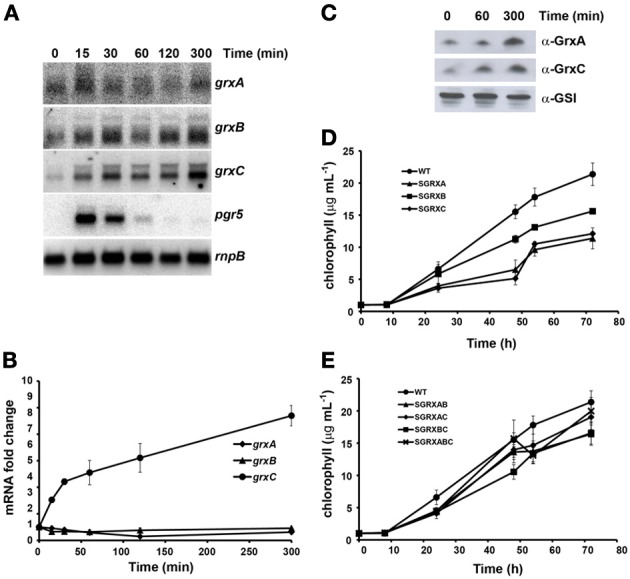
Effect of high light on glutaredoxin gene expression and growth. (A) Northern blot analysis of grxA, grxB, grxC, and pgr5 expression in response to a shift from 50 to 500 μE·m−2·s−1 light intensity. Total RNA was isolated from exponentially growing WT cells at the indicated times after the shift. The filter was hybridized with grxA, grxB, grxC, and pgr5 probes and subsequently stripped and re-hybridized with an rnpB probe as a control. (B) Quantification of relative mRNA levels of grxA, grxB, and grxC in response to a shift from 50 to 500 μE·m™·s−1 light intensity. Radioactive signals were quantified and normalized to the rnpB signal. Plots of relative mRNA levels vs. time were drawn; data represent average of 3 independent experiments and error bars represent SE. grxA (♦), grxB (▲), and grxC (•). (C) Western blot analysis of GrxA and GrxC levels in response to a shift from 50 to 500 μE·m−2·s−1. WT cells were grown in BG11C medium and samples were collected at the indicated times after the shift. 15 μg of total protein from soluble extracts were separated by 15% SDS-PAGE and subjected to western blot to detect GrxA, GrxC, and GSI. (D) Growth of glutaredoxin single mutants strains after a shift to high light. WT (•), SGRXA (▲), SGRXB (■), SGRXC (♦) were grown until the exponential phase, diluted to 1 μg chlorophyll mL−1 and shifted to high light intensity. Growth was monitored by measuring chlorophyll content. (E) Growth of glutaredoxin double and triple mutants strains after a shift to high light. WT (•), SGRXAB (▲), SGRXAC (♦), SGRXBC (■), and SGRXABC (x) were grown until the exponential phase, diluted to 1 μg chlorophyll mL−1 and shifted to high light intensity. Growth was monitored by measuring chlorophyll content.
In order to clarify the roles of the three different glutaredoxins we analyzed growth of the different mutants after a shift from normal to high light. To this end, exponentially growing cells (3–5 μg chl mL−1) were diluted to 1 μg chl mL−1, shifted to 500 μE·m−2·s−1 and growth of the strains was monitored measuring chlorophyll content (Figures 3D,E) and OD750nm for 72 h (Figure S3). Growth of the SGRXA (grxA) and the SGRXC (grxC) mutant strains was markedly retarded after the shift to high light while that of SGRXB (grxB) was only slightly reduced (Figures 3D, S3). Double and triple mutant strains behaved more similarly to SGRXB and WT strains (Figure 3E). These results suggest that both GrxA and GrxC play a role in adaptation to high light and that grxB mutation was able to suppress mutations in both grxA and grxC.
Heat shock
We have performed experiments by shifting cells grown at 30–42°C, that is, a non-lethal heat shock treatment. As for the experiments involving high light treatment, we analyzed expression of glutaredoxins both at mRNA and protein levels. As a control we analyzed expression of the hspA mRNA, which was induced rapidly after the shift from 30 to 42°C and returned to almost initial levels after 5 h as previously described (Figures 4A, S4; Fang and Barnum, 2004; Suzuki et al., 2005; Tuominen et al., 2006). grxA mRNA levels decreased slightly during the first hour after the shift to 42°C and increased afterwards reaching more than 2 fold induction after 5 h of the treatment. In contrast grxC mRNA decreased 2–3 times and remained at this level along the course of the experiment, while grxB mRNA did not change significantly during this treatment (Figure 4B). In accordance with mRNA levels GrxA protein levels increased appreciably after 5 h (Figure 4C). However, GrxC protein levels remained constant despite the fact its mRNA levels were repressed. These results suggest that either GrxC was stabilized and/or that grxC mRNA translation was enhanced after heat shock.
Figure 4.
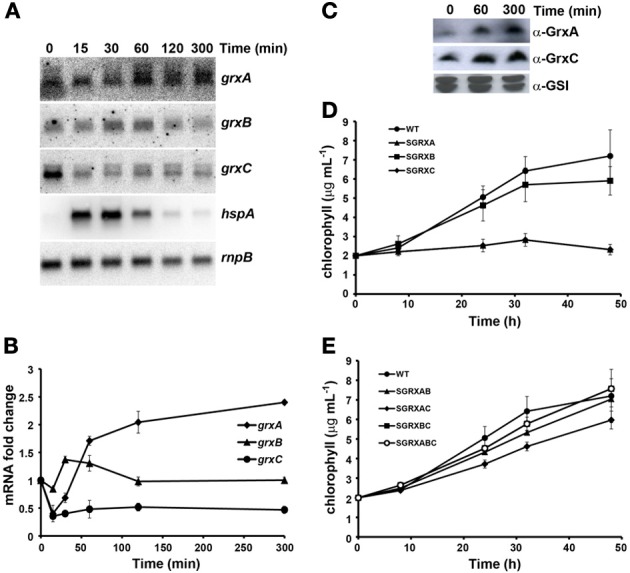
Effect of heat shock on glutaredoxin gene expression and growth. (A) Northern blot analysis of grxA, grxB, grxC, and hspA expression in response to heat shock. Total RNA was isolated from exponentially growing WT cells at the indicated times after the shift from 30 to 42°C. The filter was hybridized with grxA, grxB, grxC, and hspA probes and subsequently stripped and re-hybridized with an rnpB probe as a control. (B) Quantification of relative mRNA levels of grxA, grxB, and grxC in to response to heat shock. Radioactive signals were quantified and normalized to the rnpB signal. Plots of relative mRNA levels vs. time were drawn; data represent average of 3 independent experiments and error bars represent SE. grxA (♦), grxB (▲), and grxC (•). (C) Western blot analysis of GrxA and GrxC levels in response to heat shock. WT cells grown in BG11C and samples were collected at the indicated times after a shift from 30 to 42°C. Fifteen microgram of total protein from soluble extracts were separated by 15% SDS-PAGE and subjected to western blot to detect GrxA, GrxC, and GSI. (D) Growth of glutaredoxin single mutants strains after heat shock. WT (•), SGRXA (▲), SGRXB (■), and SGRXC (♦) were grown until the exponential phase, diluted to 2 μg chlorophyll mL−1 and shifted from 30 to 42°C. Growth was monitored by measuring chlorophyll content. (E) Growth of glutaredoxin double and triple mutant strains after heat shock. WT (•), SGRXAB (▲), SGRXAC (♦), SGRXBC (■), and SGRXABC (○) were grown until the exponential phase, diluted to 2 μg chlorophyll mL−1 and shifted from 30 to 42°C. Growth was monitored by measuring chlorophyll content.
To analyze the growth of the different strains exponentially growing cells (3–5 μg chl mL−1) were diluted to 2 μg chl mL−1, shifted from 30 to 42°C and growth of the strains was monitored measuring chlorophyll content (Figures 4D,E) and OD750nm (Figures S4B,C). Analysis of mutant strains at elevated temperatures revealed that most strains showed some degree of growth inhibition. Strikingly, the SGRXA and SGRXC were completely unable to grow under this condition (Figure 4D). Surprisingly, SGRXABC, SGRXAB, SGRXBC strains, and to a lesser extent SGRXAC strain, were less sensitive to heat stress than SGRXA and SGRXC, suggesting again that mutations in grxB could partially suppress grxA and grxC mutations and that grxA and grxC mutation suppress partially each other. All the above results suggest that both GrxA and GrxC could play a very important role in protecting cells from heat shock.
Hydrogen peroxide stress
We have analyzed the effect of addition of 1 mM of hydrogen peroxide (H2O2) both at mRNA and protein levels and on growth of the mutant strains. As a marker gene we used the isiAB operon, which has been reported to be induced after this treatment (Li et al., 2004a; Kanesaki et al., 2007), with a peak at 30 min in our experiments (Figures 5A, S5). grxC mRNA was induced transiently with a peak at 1h after the treatment and decreased afterwards. In contrast, both grxA and grxB mRNA levels remained nearly constant during the time course (Figures 5A,B). However, both GrxA and GrxC protein levels increased during the experiment with maximum levels reached after 5 h of treatment (Figure 5C). These results suggest that GrxA is stabilized in response to oxidative stress or, alternatively, that its translation is increased, while GrxC accumulation followed its mRNA induction pattern but with a delay in protein accumulation.
Figure 5.
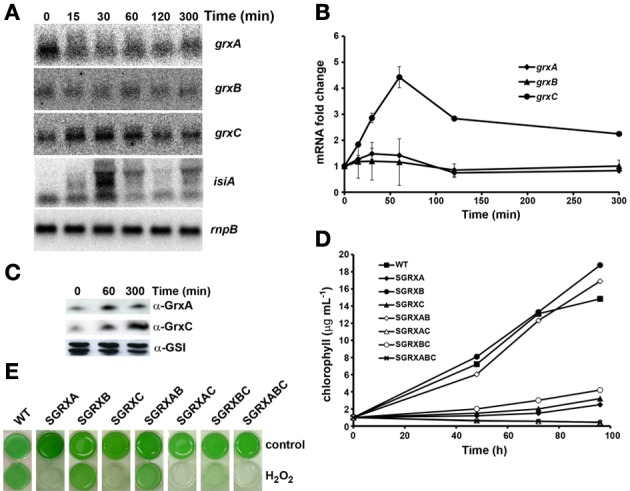
Effect of H2O2 addition on glutaredoxin gene expression and growth. (A) Northern blot analysis of grxA, grxB, grxC, and isiA expression in response to H2O2 addition. Total RNA was isolated from WT cells grown in BG11C at the indicated times after a 1 mM H2O2 addition. The filter was hybridized with grxA, grxB, grxC, and isiA probes and subsequently stripped and re-hybridized with an rnpB probe as a control. (B) Quantification of relative mRNA levels of grxA, grxB, and grxC in response to H2O2 addition. Radioactive signals were quantified and normalized to the rnpB signal. Plots of relative mRNA levels vs. time were drawn; data represent average of 3 independent experiments and error bars represent SE. grxA (♦), grxB (▲), and grxC (•). (C) Western blot analysis of GrxA and GrxC levels in response to hydrogen peroxide. WT cells grown in BG11C and samples were collected at the indicated times after 1mM H2O2 addition. Fifteen microgram of total protein from soluble extracts were separated by 15% SDS-PAGE and subjected to western blot to detect GrxA, GrxC, and GSI. (D) Growth of glutaredoxin mutants strains after H2O2 addition. WT (■), SGRXA (♦), SGRXB (•), SGRXC(▲), SGRXAB (◊), SGRXAC (Δ), SGRXBC (○), and SGRXABC (x) were grown until the exponential phase, diluted to 1 μg chlorophyll mL−1 and 1 mM H2O2 was added. Growth was monitored by measuring chlorophyll content. (E) Photograph of glutaredoxin mutants grown in liquid BG11C or BG11C supplemented with 1 mM H2O2 for 5 days.
In order to study the sensitivity of the different strains, cells were grown until they reached the exponential growth phase (3–5 μg chl mL−1), then cultures were diluted to 1 μg chl mL−1 and 1 mM of H2O2 was added to the cultures. Growth was monitored for 4 days by measuring chlorophyll content (Figures 5D,E). Both SGRXA and SGRXC mutant strains presented a clear sensitivity to the H2O2 treatment and were unable to grow under this condition (Figures 5D,E). However, in contrast to high light and heat shock treatments, after addition of H2O2 the SGRXAC double mutant strain presented an enhanced sensitivity, suggesting that GrxA and GrxC participate in parallel pathways that cooperate in response to H2O2 (Figures 5D,E). Furthermore, mutation of grxB partially suppresses the sensitivity of grxA mutants (SGRXAB was less sensitive than SGRXA) but had almost no effect on grxC mutation (compare SGRXC and SGRXBC which were almost identical). The triple mutant SGRXABC showed a degree of sensitivity comparable to that of SGRXAC, which suggested that the grxB mutation was not able to suppress the stress sensitivity of the SGRXAC strain.
Metal stress
We have previously shown that GrxA was essential for arsenate resistance as mutants lacking grxA were sensitive to arsenate in the media and GrxA was the best electron donor in vitro for the main arsenate reductase from Synechocystis (Lopez-Maury et al., 2009). Furthermore, GrxA (Grx2) was suggested to be involved in selenate resistance (Marteyn et al., 2009) and recently GrxB has been implicated in mercury and uranium resistance (Marteyn et al., 2013). In order to further analyze the sensitivity of glutaredoxin mutants to different metals, we have analyzed the growth of all mutants in the presence of selenate, selenite, cadmium, copper, and nickel. All mutant strains, except SGRXC, grew as well as the WT in the presence of 2 μM of Cd, 3 μM of Cu, 5 μM of Ni and 30 μM Na2SeO3 or 30 μM Na2SeO4 (Figure 6). The SGRXC strain presented hypersensitivity to nickel and copper, and to a lesser extent to Cd (Figure 6A), while the double mutants SGRXAC and SGRXBC (lacking grxA or grxB in addition to grxC) and triple mutants (lacking all glutaredoxin genes) strains behaved like the WT. This suggests that compensatory mechanisms were activated in double and triple mutant strains that protect against the toxic effect of these metals. However, although these mutations were able to suppress grxC mutation, they did not enhance resistance in a WT background (Figure 6A).
Figure 6.
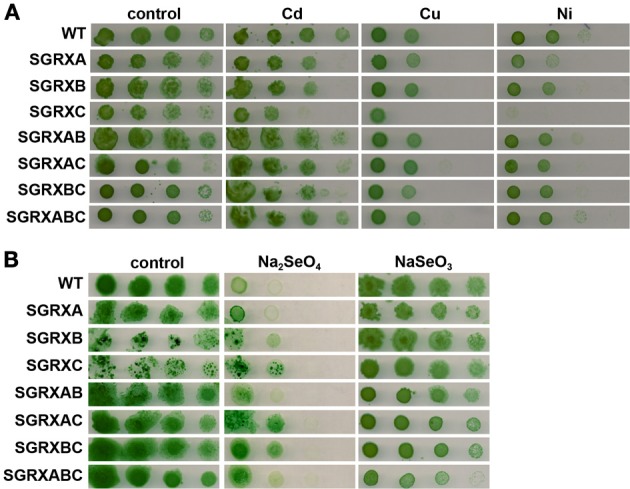
Effect of different metals on glutaredoxin mutants growth. (A) Sensitivity of glutaredoxin mutants to metals. Tolerance of WT, SGRXA, SGRXB, SGRXC, SGRXAB, SGRXAC, SGRXBC, and SGRXABC strains to cadmium, copper, and nickel was examined. Tenfold serial dilutions of a 1 μg chlorophyll mL−1 of exponentially growing cells suspension were spotted onto BG11C supplemented with 2 μM Cd, 3 μM Cu, and 5 μM Ni. Plates were photographed after 5 days of growth. (B) Sensitivity of glutaredoxin mutants to selenium. Tolerance of WT, SGRXA, SGRXB, SGRXC, SGRXAB, SGRXAC, SGRXBC, and SGRXABC strains to selenate (Na2SeO4) or selenite (Na2SeO3) was examined. Ten-fold serial dilutions of a 1 μg chlorophyll mL−1 of exponentially growing cells suspension were spotted onto BG11C supplemented with 30 μM Na2SeO3 or 30 μM Na2SeO4. Plates were photographed after 5 days of growth.
Discussion
Here we have extensively characterized the complete set of glutaredoxins in the cyanobacterium Synechocystis, a model photosynthetic prokaryote. The characterization of the different glutaredoxin mutant strains have revealed that none of the glutaredoxin genes are required under normal growth conditions but that they play an important role in protecting cells from environmental stresses. Our data also indicated that there are complex genetic interactions between glutaredoxins genes and that these interactions are stress dependent. The fact that we have not observed any compensatory effects on expression of glutaredoxin genes or GrxA and GrxC protein levels in mutants lacking other glutaredoxin genes, would suggest that the phenotypes we have observed are most probably linked to downstream processes regulated by glutaredoxins and not to dosage-dependent effects. Furthermore, analysis of the expression of the glutaredoxin genes has shown that these genes were not all under the same regulatory system because their patterns of expression were not coordinated. In fact, the grxB mRNA level did not change significantly under any of the conditions tested, while grxA was only induced by heat shock and grxC was induced after H2O2 and during high light treatment but repressed by heat shock. Although grxC is induced by high light intensities and H2O2, this gene is not under the control of any of the regulatory proteins described to respond to these stress conditions in Synechocystis (Hsiao et al., 2004; Kobayashi et al., 2004; Suzuki et al., 2005; Jantaro et al., 2006; Tuominen et al., 2006; Kanesaki et al., 2007; Tuominen et al., 2008; Horiuchi et al., 2010; Singh et al., 2010; Muramatsu and Hihara, 2012). On the other hand, GrxA protein levels increased under all stress conditions, while the GrxC protein levels were induced only after high light and H2O2, following the changes of its mRNA levels, and was maintained constant after heat shock, despite repression of its mRNA levels. This suggests that in addition to a transcriptional regulation there are also other levels of regulation at the protein level for both glutaredoxins.
Analysis of growth the mutant strains showed that two of the glutaredoxins, GrxA and GrxC, play a prominent role in cell protection against stress, as the single mutants lacking grxA or grxC were sensitive to several stresses. Both GrxA and GrxC were necessary for oxidative stress defense, since mutants lacking one of them were extremely sensitive to the presence of external H2O2 and the mutant lacking both genes showed and additive phenotype (Figure 5). The single mutant lacking grxA (the SGRXA strain) proved to be hypersensitive to H2O2, which is in agreement with phenotypes previously described for this mutant (Li et al., 2007; Marteyn et al., 2009), but the mechanism for this sensitivity and the targets of GrxA are still unknown. GrxA is able to decompose H2O2 in vitro although it is much less efficient than most peroxiredoxins, which are in fact reduced by thioredoxins and not by glutaredoxins in Synechocystis (Perez-Perez et al., 2009b). Therefore, it is unlikely that this activity could account for the protection of cells against H2O2. Besides that, GrxA has been shown to interact with both catalase and peroxiredoxin II (PrxII), although the catalase activity was inhibited by GrxA in Synechocystis extracts (Li et al., 2007) and PrxII eventually did not accept electrons from GrxA (Perez-Perez et al., 2009b). However, it can not be ruled out that glutathionylation is needed for activation and/or protection of these two proteins under oxidative stress conditions. The SGRXA strain was also sensitive to high light and GrxA protein levels increased after this treatment suggesting that GrxA could play a role in high light acclimation. During the high light treatment the excess of light absorbed can generate reactive oxygen species (ROS) leading to oxidative damage and this ROS might affect the growth of the SGRXA strain. Furthermore, the SGRXA strain was also extremely sensitive to heat shock which suggests that GrxA could also play an important role during heat stress. In this regard both GroEL and DnaK1 (two chaperones that are essential for heat adaptation) were also identified as GrxA targets (Li et al., 2007), which suggests that GrxA could be needed for activation or as a helper of both chaperones to function in response to heat shock. Similar phenotypes with respect to oxidative stress sensitivity and heat shock sensitivity have been reported for glutaredoxin mutants in other bacteria (Prinz et al., 1997; Fernandes and Holmgren, 2004; Li et al., 2004b; Benyamina et al., 2013) and in yeast (Luikenhuis et al., 1998; Draculic et al., 2000; Chung et al., 2004), suggesting that dithiolic glutaredoxins could have conserved roles in microorganisms. Finally, we have tested whether SGRXA strains were sensitive to metals and selenium compounds. Any of the tested metals affected significantly the growth of grxA mutant strains (Figure 6), including selenate or selenite which is in disagreement with previous data (Marteyn et al., 2009). Sensitivity to metals is highly influenced by growth conditions, especially light conditions, and this could explain the differences in selenium sensitivity between our results and previously published ones (Marteyn et al., 2009).
The SGRXC strain is unique in that it showed hypersensitivity to all stresses tested. GrxC is a prototypical monothiolic glutaredoxin which has been shown to contain an oxygen labile 2Fe-2S cluster (Picciocchi et al., 2007), and the grxC gene is able to complement the defects of a grx5 yeast mutant (Molina-Navarro et al., 2006). In several organisms it is well-documented that monothiolic glutaredoxins impact Fe-S cluster synthesis and/or assembly although their specific roles are still under discussion. Yeast Grx5 is involved in mitochondrial Fe-S cluster assembly and mutants lacking this protein showed symptoms of oxidative stress under standard growth conditions, accumulation of apoproteins lacking Fe-S clusters in the mitochondria and were unable to grow under aerobic conditions (Rodriguez-Manzaneque et al., 1999, 2002; Belli et al., 2004). A similar phenotype has also been described for a monothiolic glutaredoxin mutant in Sinorhizobium meliloti (Benyamina et al., 2013). Furthermore, in yeast it has been shown that GSH is essential for Fe-S cluster assembly but in this case it affects both cytoplasmic and mitochondrial Fe-S clusters, suggesting a more global impact in Fe-S cluster assembly. This is probably related to its role in binding Fe-S clusters in all monothiolic glutaredoxins in yeast cells (Kumar et al., 2011). In photosynthetic organisms the in vivo role of monothiolic glutaredoxins has been analyzed only in Arabidopsis which has four class II monothiolic glutaredoxins (AtGrxS14–17; Couturier et al., 2009a). Of these AtGrxS14 and AtGrxS16 have been localized to the chloroplast, AtGrxS15 is localized in the mitochondria and chloroplast and AtGrxS17 is a cytosolic protein (Cheng et al., 2006; Bandyopadhyay et al., 2008; Cheng, 2008). It was also shown that both AtGrxS14 and AtGrxS15 mutants are hypersensitive to H2O2 (Cheng et al., 2006; Cheng, 2008), while the AtGrxS17 mutant is sensitive to heat shock and has higher ROS contents under this condition (Cheng et al., 2011). These proteins also present a labile 2Fe-2S cluster (like GrxC) which in vitro could be transferred to Synechocystis ferredoxin (Bandyopadhyay et al., 2008; Liu et al., 2013). Furthermore, these proteins are also able to complement the yeast grx5 mutant strain, like grxC, suggesting that they are functionally conserved (Bandyopadhyay et al., 2008; Liu et al., 2013). Recently it has been shown that the E. coli monothiolic glutaredoxin (GrxD) is able to transfer its Fe-S cluster to ferredoxin (Yeung et al., 2011) but not to MiaB, a radical AdoMet enzyme containing two 4Fe-4S clusters, despite holoGrxD being able to interact with MiaB. This led these authors to propose that GrxD is not involved in Fe-S cluster assembly but in Fe-S cluster repair (Boutigny et al., 2013). This role in repair and/or synthesis of Fe-S cluster could explain SGRXC sensitivity to different stresses, because Fe-S clusters are vulnerable to many stresses, especially H2O2 and metal stress (Imlay, 2006, 2013). In fact destabilization of Fe-S clusters in essential enzymes constitute the main mechanism for metal toxicity in several organisms, including Synechocystis, suggesting that this is a common problem when cells are challenged with an excess of metals (Ranquet et al., 2007; Macomber and Imlay, 2009; Chillappagari et al., 2010; Fantino et al., 2010; Tottey et al., 2012; Xu and Imlay, 2012). The target proteins which require GrxC for Fe-S cluster assembly and/or repair in Synechocystis remain unidentified, but because the SGRXC strain is viable and did not show any growth defects under non-stressed conditions, its targets are unlikely to be essential. Alternatively GrxC could be involved only in Fe-S cluster repair under stress conditions. This is further supported by GrxC induction after several stress treatments, suggesting that cellular requirements for GrxC under these conditions may increase.
In contrast the SGRXB strain did not display any obvious phenotype, neither were the grxB mRNA levels altered under any of the conditions tested, suggesting that this protein has a minor role and/or a very specialized role. This is supported by the recent data, which showed that grxB mutants were sensitive to mercury and uranium. GrxB regulates the activity of MerA, a mercury reductase, via glutathionylation of one of its active cysteines (Marteyn et al., 2013) and mutation of both merA or grxB confers sensitivity to these compounds. Surprisingly, our results showed that grxB mutation was able to partially suppress most of the phenotypes of grxA mutant strains suggesting that it could be negatively controlling functions activated by GrxA. This is further supported by data showing physical interaction between GrxA and GrxB, which has been observed by two different groups using different experimental approaches (Li et al., 2007; Marteyn et al., 2009). This suggests that these interactions are bona fide interactions and that these proteins could regulate each other's activities. Furthermore, GrxA and GrxB have been proposed to be reduced by Synechocystis NADPH-thioredoxin reductase (encoded by slr0600; NTR), which present homology to bacterial NADPH thioredoxin reductases. Which redoxins are reduced by NTR is controversial because both dithiolic glutaredoxins, GrxA and GrxB, and TrxA have been reported to be reduced by NTR (Li et al., 2007; Hishiya et al., 2008; Marteyn et al., 2009). In addition, a Synechocystis mutant lacking GSH also presented stress sensitivity, although its phenotypes were more severe (Cameron and Pakrasi, 2011) than phenotypes of glutaredoxin mutants described here. This suggests that, as described in yeast (Kumar et al., 2011), GSH could have additional roles in Synechocystis which are not mediated solely by glutaredoxins. One possibility is that the NTR-Grx1/2 or the NTR-TrxA system could work as alternative pathways to reduce GSH in Synechocystis, because this strain lacks a gene for a canonical glutathione reductase (GR) and NADPH dependent GR activity is not detected in extracts (Marteyn et al., 2009; Cameron and Pakrasi, 2011). Similar results have been obtained in yeast in which mutant lacking GR can use both Grx2 and Trx2 to reduce GSH (Morgan et al., 2013). All the above data suggest that there could be a cross-regulation between the thioredoxin and glutaredoxin systems in Synechocystis as it has been shown in other organisms including plants and algae (Gelhaye et al., 2003; Michelet et al., 2005; Rouhier et al., 2008; Zaffagnini et al., 2008; Meyer et al., 2009, 2012; Lillig and Berndt, 2013; Toledano et al., 2013). Finally, analysis of double mutants with grxC shows that grxB was also able to suppress totally or partially the sensitivity phenotype of grxC single mutants to high light, heat shock and metal stress but not to oxidative stress. The relation between these two proteins is completely unknown but is possible that grxB affects GSH metabolism which will probable also alter GrxC function.
In conclusion, we have shown that glutaredoxins are essential for adaptation to several stresses in Synechocystis and that GrxA and GrxC levels increase in response to stress treatments. This induction is mediated mainly by transcriptional induction for GrxC and by posttranscriptional mechanisms in the case of GrxA. Analysis of the different genetic interactions between the glutaredoxin genes suggest the existence of an interconnected genetic network that is perturbed in response to environmental changes. In this regard, it would be interesting to analyze mutants lacking both glutaredoxins and other components of antioxidative system, such as the thioredoxins, as some of the phenotypes we have observed are shared between them.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Ministerio de Economía y Competitividad (MINECO) and the European Regional Fund (FEDER) to Francisco J. Florencio (grant BFU2010–15708, and plan E–CLPN09–001) and by Junta de Andalucía (group BIO–284). Luis López-Maury was recipient of a Junta de Ampliación de Estudios-Doctor contract from Consejo Superior de Investigaciones Científicas (CSIC). We thank Anna Marika Lindhal for critical reading of the manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2013.00428/abstract
References
- Allakhverdiev S. I., Nishiyama Y., Miyairi S., Yamamoto H., Inagaki N., Kanesaki Y., et al. (2002). Salt stress inhibits the repair of photodamaged photosystem II by suppressing the transcription and translation of psbA genes in synechocystis. Plant Physiol. 130, 1443–1453 10.1104/pp.011114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S., Gama F., Molina-Navarro M. M., Gualberto J. M., Claxton R., Naik S. G., et al. (2008). Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 27, 1122–1133 10.1038/emboj.2008.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedhomme M., Adamo M., Marchand C. H., Couturier J., Rouhier N., Lemaire S. D., et al. (2012). Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 445, 337–347 10.1042/BJ20120505 [DOI] [PubMed] [Google Scholar]
- Belli G., Molina M. M., Garcia-Martinez J., Perez-Ortin J. E., Herrero E. (2004). Saccharomyces cerevisiae glutaredoxin 5-deficient cells subjected to continuous oxidizing conditions are affected in the expression of specific sets of genes. J. Biol. Chem. 279, 12386–12395 10.1074/jbc.M311879200 [DOI] [PubMed] [Google Scholar]
- Benyamina S. M., Baldacci-Cresp F., Couturier J., Chibani K., Hopkins J., Bekki A., et al. (2013). Two Sinorhizobium meliloti glutaredoxins regulate iron metabolism and symbiotic bacteroid differentiation. Environ. Microbiol. 15, 795–810 10.1111/j.1462-2920.2012.02835.x [DOI] [PubMed] [Google Scholar]
- Boutigny S., Saini A., Baidoo E. E., Yeung N., Keasling J. D., Butland G. (2013). Physical and functional interactions of a monothiol glutaredoxin and an iron sulfur cluster carrier protein with the sulfur-donating radical S-adenosyl-L-methionine enzyme MiaB. J. Biol. Chem. 288, 14200–14211 10.1074/jbc.M113.460360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan B. B., Balmer Y. (2005). Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56, 187–220 10.1146/annurev.arplant.56.032604.144246 [DOI] [PubMed] [Google Scholar]
- Cameron J. C., Pakrasi H. B. (2011). Glutathione in synechocystis 6803: a closer look into the physiology of a gshB mutant. Plant Signal. Behav. 6, 89–92 10.4161/psb.6.1.14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N. H. (2008). AtGRX4, an Arabidopsis chloroplastic monothiol glutaredoxin, is able to suppress yeast grx5 mutant phenotypes and respond to oxidative stress. FEBS Lett. 582, 848–854 10.1016/j.febslet.2008.02.006 [DOI] [PubMed] [Google Scholar]
- Cheng N. H., Liu J. Z., Brock A., Nelson R. S., Hirschi K. D. (2006). AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J. Biol. Chem. 281, 26280–26288 10.1074/jbc.M601354200 [DOI] [PubMed] [Google Scholar]
- Cheng N. H., Liu J. Z., Liu X., Wu Q., Thompson S. M., Lin J., et al. (2011). Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. J. Biol. Chem. 286, 20398–20406 10.1074/jbc.M110.201707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillappagari S., Seubert A., Trip H., Kuipers O. P., Marahiel M. A., Miethke M. (2010). Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 192, 2512–2524 10.1128/JB.00058-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. H., Kim K. D., Cho Y. J., Roe J. H. (2004). Differential expression and role of two dithiol glutaredoxins Grx1 and Grx2 in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 321, 922–929 10.1016/j.bbrc.2004.07.042 [DOI] [PubMed] [Google Scholar]
- Couturier J., Jacquot J. P., Rouhier N. (2009a). Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell. Mol. Life Sci. 66, 2539–2557 10.1007/s00018-009-0054-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J., Koh C. S., Zaffagnini M., Winger A. M., Gualberto J. M., Corbier C., et al. (2009b). Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site. J. Biol. Chem. 284, 9299–9310 10.1074/jbc.M807998200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J., Stroher E., Albetel A. N., Roret T., Muthuramalingam M., Tarrago L., et al. (2011). Arabidopsis chloroplastic glutaredoxin C5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins. J. Biol. Chem. 286, 27515–27527 10.1074/jbc.M111.228726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draculic T., Dawes I. W., Grant C. M. (2000). A single glutaredoxin or thioredoxin gene is essential for viability in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 36, 1167–1174 10.1046/j.1365-2958.2000.01948.x [DOI] [PubMed] [Google Scholar]
- Fang F., Barnum S. R. (2004). Expression of the heat shock gene hsp16.6 and promoter analysis in the cyanobacterium, Synechocystis sp. PCC 6803. Curr. Microbiol. 49, 192–198 10.1007/s00284-004-4340-5 [DOI] [PubMed] [Google Scholar]
- Fantino J. R., Py B., Fontecave M., Barras F. (2010). A genetic analysis of the response of Escherichia coli to cobalt stress. Environ. Microbiol. 12, 2846–2857 10.1111/j.1462-2920.2010.02265.x [DOI] [PubMed] [Google Scholar]
- Feng Y., Zhong N., Rouhier N., Hase T., Kusunoki M., Jacquot J. P., et al. (2006). Structural insight into poplar glutaredoxin C1 with a bridging iron-sulfur cluster at the active site. Biochemistry 45, 7998–8008 10.1021/bi060444t [DOI] [PubMed] [Google Scholar]
- Fernandes A. P., Holmgren A. (2004). Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 6, 63–74 10.1089/152308604771978354 [DOI] [PubMed] [Google Scholar]
- Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L. J., Dietz K. J. (2005). The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 280, 12168–12180 10.1074/jbc.M413189200 [DOI] [PubMed] [Google Scholar]
- Gao X. H., Zaffagnini M., Bedhomme M., Michelet L., Cassier-Chauvat C., Decottignies P., et al. (2011). Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii: kinetics and specificity in deglutathionylation reactions. FEBS Lett. 584, 2242–2248 10.1016/j.febslet.2010.04.034 [DOI] [PubMed] [Google Scholar]
- Garcia-Dominguez M., Florencio F. J. (1997). Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 35, 723–734 10.1023/A:1005846626187 [DOI] [PubMed] [Google Scholar]
- Gelhaye E., Rouhier N., Jacquot J. P. (2003). Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett. 555, 443–448 10.1016/S0014-5793(03)01301-2 [DOI] [PubMed] [Google Scholar]
- Greetham D., Vickerstaff J., Shenton D., Perrone G. G., Dawes I. W., Grant C. M. (2010). Thioredoxins function as deglutathionylase enzymes in the yeast Saccharomyces cerevisiae. BMC Biochem. 11:3 10.1186/1471-2091-11-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanschmann E. M., Godoy J. R., Berndt C., Hudemann C., Lillig C. H. (2013). Thioredoxins, glutaredoxins, and peroxiredoxinsmolecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid Redox Signal 19, 1539–1605 10.1089/ars.2012.4599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanschmann E. M., Lonn M. E., Schutte L. D., Funke M., Godoy J. R., Eitner S., et al. (2010). Both thioredoxin 2 and glutaredoxin 2 contribute to the reduction of the mitochondrial 2-Cys peroxiredoxin Prx3. J. Biol. Chem. 285, 40699–40705 10.1074/jbc.M110.185827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiya S., Hatakeyama W., Mizota Y., Hosoya-Matsuda N., Motohashi K., Ikeuchi M., et al. (2008). Binary reducing equivalent pathways using NADPH-thioredoxin reductase and ferredoxin-thioredoxin reductase in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 49, 11–18 10.1093/pcp/pcm158 [DOI] [PubMed] [Google Scholar]
- Holmgren A. (1976). Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. U.S.A. 73, 2275–2279 10.1073/pnas.73.7.2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi M., Nakamura K., Kojima K., Nishiyama Y., Hatakeyama W., Hisabori T., et al. (2010). The PedR transcriptional regulator interacts with thioredoxin to connect photosynthesis with gene expression in cyanobacteria. Biochem. J. 431, 135–140 10.1042/BJ20100789 [DOI] [PubMed] [Google Scholar]
- Hsiao H. Y., He Q., Van Waasbergen L. G., Grossman A. R. (2004). Control of photosynthetic and high-light-responsive genes by the histidine kinase DspA: negative and positive regulation and interactions between signal transduction pathways. J. Bacteriol. 186, 3882–3888 10.1128/JB.186.12.3882-3888.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A. (2006). Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59, 1073–1082 10.1111/j.1365-2958.2006.05028.x [DOI] [PubMed] [Google Scholar]
- Imlay J. A. (2013). The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11, 443–454 10.1038/nrmicro3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwema T., Picciocchi A., Traore D. A., Ferrer J. L., Chauvat F., Jacquamet L. (2009). Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry 48, 6041–6043 10.1021/bi900440m [DOI] [PubMed] [Google Scholar]
- Jantaro S., Ali Q., Lone S., He Q. (2006). Suppression of the lethality of high light to a quadruple HLI mutant by the inactivation of the regulatory protein PfsR in Synechocystis, PCC 6803. J. Biol. Chem. 281, 30865–30874 10.1074/jbc.M606252200 [DOI] [PubMed] [Google Scholar]
- Kanesaki Y., Yamamoto H., Paithoonrangsarid K., Shoumskaya M., Suzuki I., Hayashi H., et al. (2007). Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J. 49, 313–324 10.1111/j.1365-313X.2006.02959.x [DOI] [PubMed] [Google Scholar]
- Kim S. G., Chung J. S., Sutton R. B., Lee J. S., Lopez-Maury L., Lee S. Y., et al. (2012). Redox, mutagenic and structural studies of the glutaredoxin/arsenate reductase couple from the cyanobacterium Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1824, 392–403 10.1016/j.bbapap.2011.10.012 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Ishizuka T., Katayama M., Kanehisa M., Bhattacharyya-Pakrasi M., Pakrasi H. B., et al. (2004). Response to oxidative stress involves a novel peroxiredoxin gene in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 45, 290–299 10.1093/pcp/pch034 [DOI] [PubMed] [Google Scholar]
- Kumar C., Igbaria A., D'autreaux B., Planson A. G., Junot C., Godat E., et al. (2011). Glutathione revisited: a vital function in iron metabolism and ancillary role in thiol-redox control. EMBO J. 30, 2044–2056 10.1038/emboj.2011.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Outten C. E. (2012). Monothiol CGFS glutaredoxins and BolA-like proteins: [2Fe-2S] binding partners in iron homeostasis. Biochemistry 51, 4377–4389 10.1021/bi300393z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Singh A. K., Mcintyre L. M., Sherman L. A. (2004a). Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 186, 3331–3345 10.1128/JB.186.11.3331-3345.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Hein S., Zou W., Klug G. (2004b). The glutathione-glutaredoxin system in Rhodobacter capsulatus: part of a complex regulatory network controlling defense against oxidative stress. J. Bacteriol. 186, 6800–6808 10.1128/JB.186.20.6800-6808.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Yang Q., Zhang L., Li H., Cui Y., Wu Q. (2007). Identification of novel targets of cyanobacterial glutaredoxin. Arch. Biochem. Biophys. 458, 220–228 10.1016/j.abb.2006.12.010 [DOI] [PubMed] [Google Scholar]
- Li S., Lauri A., Ziemann M., Busch A., Bhave M., Zachgo S. (2009). Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21, 429–441 10.1105/tpc.108.064477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillig C. H., Berndt C. (2013). Glutaredoxins in thiol/disulfide exchange. Antioxid. Redox Signal. 18, 1654–1665 10.1089/ars.2012.5007 [DOI] [PubMed] [Google Scholar]
- Liu X., Liu S., Feng Y., Liu J. Z., Chen Y., Pham K., et al. (2013). Structural insights into the N-terminal GIY-YIG endonuclease activity of Arabidopsis glutaredoxin AtGRXS16 in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 110, 9565–9570 10.1073/pnas.1306899110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Maury L., Sanchez-Riego A. M., Reyes J. C., Florencio F. J. (2009). The glutathione/glutaredoxin system is essential for arsenate reduction in Synechocystis sp. strain PCC 6803. J. Bacteriol. 191, 3534–3543 10.1128/JB.01798-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikenhuis S., Perrone G., Dawes I. W., Grant C. M. (1998). The yeast Saccharomyces cerevisiae contains two glutaredoxin genes that are required for protection against reactive oxygen species. Mol. Biol. Cell 9, 1081–1091 10.1091/mbc.9.5.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L., Imlay J. A. (2009). The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 10.1073/pnas.0812808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteyn B., Domain F., Legrain P., Chauvat F., Cassier-Chauvat C. (2009). The thioredoxin reductase-glutaredoxins-ferredoxin crossroad pathway for selenate tolerance in Synechocystis PCC6803. Mol. Microbiol. 71, 520–532 10.1111/j.1365-2958.2008.06550.x [DOI] [PubMed] [Google Scholar]
- Marteyn B., Sakr S., Farci S., Bedhomme M., Chardonnet S., Decottignies P., et al. (2013). The Synechocystis PCC6803 MerA-like enzyme operates in the reduction of both mercury and uranium under the control of the glutaredoxin 1 enzyme. J. Bacteriol. 195, 4138–4145 10.1128/JB.00272-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y., Belin C., Delorme-Hinoux V., Reichheld J. P., Riondet C. (2012). Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 17, 1124–1160 10.1089/ars.2011.4327 [DOI] [PubMed] [Google Scholar]
- Meyer Y., Buchanan B. B., Vignols F., Reichheld J. P. (2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43, 335–367 10.1146/annurev-genet-102108-134201 [DOI] [PubMed] [Google Scholar]
- Michelet L., Zaffagnini M., Marchand C., Collin V., Decottignies P., Tsan P., et al. (2005). Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc. Natl. Acad. Sci. U.S.A. 102, 16478–16483 10.1073/pnas.0507498102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Navarro M. M., Casas C., Piedrafita L., Belli G., Herrero E. (2006). Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett. 580, 2273–2280 10.1016/j.febslet.2006.03.037 [DOI] [PubMed] [Google Scholar]
- Morgan B., Ezerina D., Amoako T. N., Riemer J., Seedorf M., Dick T. P. (2013). Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat. Chem. Biol. 9, 119–125 10.1038/nchembio.1142 [DOI] [PubMed] [Google Scholar]
- Muhlenhoff U., Molik S., Godoy J. R., Uzarska M. A., Richter N., Seubert A., et al. (2010). Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12, 373–385 10.1016/j.cmet.2010.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Hihara Y. (2012). Acclimation to high-light conditions in cyanobacteria: from gene expression to physiological responses. J. Plant Res. 125, 11–39 10.1007/s10265-011-0454-6 [DOI] [PubMed] [Google Scholar]
- Murmu J., Bush M. J., Delong C., Li S., Xu M., Khan M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154, 1492–1504 10.1104/pp.110.159111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortenberg R., Gon S., Porat A., Beckwith J. (2004). Interactions of glutaredoxins, ribonucleotide reductase, and components of the DNA replication system of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 7439–7444 10.1073/pnas.0401965101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrajas J. R., Padilla C. A., McDonagh B., Barcena J. A. (2010). Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in Saccharomyces cerevisiae. Antioxid. Redox Signal. 13, 249–258 10.1089/ars.2009.2950 [DOI] [PubMed] [Google Scholar]
- Perez-Perez M. E., Martin-Figueroa E., Florencio F. J. (2009a). Photosynthetic regulation of the cyanobacterium Synechocystis sp. PCC 6803 thioredoxin system and functional analysis of TrxB (Trx x) and TrxQ (Trx y) thioredoxins. Mol. Plant 2, 270–283 10.1093/mp/ssn070 [DOI] [PubMed] [Google Scholar]
- Perez-Perez M. E., Mata-Cabana A., Sanchez-Riego A. M., Lindahl M., Florencio F. J. (2009b). A comprehensive analysis of the peroxiredoxin reduction system in the Cyanobacterium Synechocystis sp. strain PCC 6803 reveals that all five peroxiredoxins are thioredoxin dependent. J. Bacteriol. 191, 7477–7489 10.1128/JB.00831-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciocchi A., Saguez C., Boussac A., Cassier-Chauvat C., Chauvat F. (2007). CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry 46, 15018–15026 10.1021/bi7013272 [DOI] [PubMed] [Google Scholar]
- Prinz W. A., Aslund F., Holmgren A., Beckwith J. (1997). The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272, 15661–15667 10.1074/jbc.272.25.15661 [DOI] [PubMed] [Google Scholar]
- Ranquet C., Ollagnier-De-Choudens S., Loiseau L., Barras F., Fontecave M. (2007). Cobalt stress in Escherichia coli. The effect on the iron-sulfur proteins. J. Biol. Chem. 282, 30442–30451 10.1074/jbc.M702519200 [DOI] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J. B., Herman M., Stanier R. Y. (1979). Generic assigment, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111, 1–61 10.1099/00221287-111-1-1 [DOI] [Google Scholar]
- Rodriguez-Manzaneque M. T., Ros J., Cabiscol E., Sorribas A., Herrero E. (1999). Grx5 glutaredoxin plays a central role in protection against protein oxidative damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 8180–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque M. T., Tamarit J., Belli G., Ros J., Herrero E. (2002). Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13, 1109–1121 10.1091/mbc.01-10-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N. (2010). Plant glutaredoxins: pivotal players in redox biology and iron-sulphur centre assembly. New Phytol. 186, 365–372 10.1111/j.1469-8137.2009.03146.x [DOI] [PubMed] [Google Scholar]
- Rouhier N., Gelhaye E., Jacquot J. P. (2002). Glutaredoxin-dependent peroxiredoxin from poplar: protein-protein interaction and catalytic mechanism. J. Biol. Chem. 277, 13609–13614 10.1074/jbc.M111489200 [DOI] [PubMed] [Google Scholar]
- Rouhier N., Gelhaye E., Sautiere P. E., Brun A., Laurent P., Tagu D., et al. (2001). Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol. 127, 1299–1309 10.1104/pp.010586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N., Lemaire S. D., Jacquot J. P. (2008). The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166 10.1146/annurev.arplant.59.032607.092811 [DOI] [PubMed] [Google Scholar]
- Rouhier N., Villarejo A., Srivastava M., Gelhaye E., Keech O., Droux M., et al. (2005). Identification of plant glutaredoxin targets. Antioxid. Redox Signal. 7, 919–929 10.1089/ars.2005.7.919 [DOI] [PubMed] [Google Scholar]
- Rouhier N., Vlamis-Gardikas A., Lillig C. H., Berndt C., Schwenn J. D., Holmgren A., et al. (2003). Characterization of the redox properties of poplar glutaredoxin. Antioxid. Redox Signal. 5, 15–22 10.1089/152308603321223504 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor. New York, NY: Cold Spring Harbor Laboratory press [Google Scholar]
- Singh A. K., Elvitigala T., Cameron J. C., Ghosh B. K., Bhattacharyya-Pakrasi M., Pakrasi H. B. (2010). Integrative analysis of large scale expression profiles reveals core transcriptional response and coordination between multiple cellular processes in a cyanobacterium. BMC Syst. Biol. 4:105 10.1186/1752-0509-4-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki I., Kanesaki Y., Hayashi H., Hall J. J., Simon W. J., Slabas A. R., et al. (2005). The histidine kinase Hik34 is involved in thermotolerance by regulating the expression of heat shock genes in synechocystis. Plant Physiol. 138, 1409–1421 10.1104/pp.104.059097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledano M. B., Delaunay-Moisan A., Outten C. E., Igbaria A. (2013). Functions and cellular compartmentation of the thioredoxin and glutathione pathways in yeast. Antioxid. Redox Signal. 18, 1699–1711 10.1089/ars.2012.5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S., Patterson C. J., Banci L., Bertini I., Felli I. C., Pavelkova A., et al. (2012). Cyanobacterial metallochaperone inhibits deleterious side reactions of copper. Proc. Natl. Acad. Sci. U.S.A. 109, 95–100 10.1073/pnas.1117515109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen I., Pollari M., Tyystjarvi E., Tyystjarvi T. (2006). The SigB sigma factor mediates high-temperature responses in the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 580, 319–323 10.1016/j.febslet.2005.11.082 [DOI] [PubMed] [Google Scholar]
- Tuominen I., Pollari M., Von Wobeser E. A., Tyystjarvi E., Ibelings B. W., Matthijs H. C., et al. (2008). Sigma factor SigC is required for heat acclimation of the cyanobacterium Synechocystis sp. strain PCC 6803. FEBS Lett. 582, 346–350 10.1016/j.febslet.2007.12.030 [DOI] [PubMed] [Google Scholar]
- Vieira Dos Santos C., Laugier E., Tarrago L., Massot V., Issakidis-Bourguet E., Rouhier N., et al. (2007). Specificity of thioredoxins and glutaredoxins as electron donors to two distinct classes of Arabidopsis plastidial methionine sulfoxide reductases B. FEBS Lett. 581, 4371–4376 10.1016/j.febslet.2007.07.081 [DOI] [PubMed] [Google Scholar]
- Vioque A. (1992). Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20, 6331–6337 10.1093/nar/20.23.6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F. F., Imlay J. A. (2012). Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl. Environ. Microbiol. 78, 3614–3621 10.1128/AEM.07368-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung N., Gold B., Liu N. L., Prathapam R., Sterling H. J., Willams E. R., et al. (2011). The, E. coli monothiol glutaredoxin GrxD forms homodimeric and heterodimeric FeS cluster containing complexes. Biochemistry 50, 8957–8969 10.1021/bi2008883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M., Michelet L., Massot V., Trost P., Lemaire S. D. (2008). Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. J. Biol. Chem. 283, 8868–8876 10.1074/jbc.M709567200 [DOI] [PubMed] [Google Scholar]
- Zander M., Chen S., Imkampe J., Thurow C., Gatz C. (2011). Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol. Plant 5, 831–840 10.1093/mp/ssr113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


