Abstract
Background
The genetic basis of the autoimmune disease type 1 diabetes (T1D) has now been largely determined, so now we can compare these findings with emerging genetic knowledge of disorders and phenotypes that have been negatively or positively associated with T1D historically. Here, we assessed the role in T1D of variants previously reported to be associated with atopic diseases and epithelial barrier function, profilaggrin (), and those that affect the expression levels of the proinflammatory cytokines tumour necrosis factor (TNF)-α, interleukin (IL)-1β, interferon (IFN)γ and IL-18.
Methods
We genotyped single nucleotide polymorphisms (SNPs): − 105/rs28665122 in SELS or SEPS1 (selenoprotein), three single nucleotide polymorphisms in IL18 (−105/rs360717, + 183/rs5744292 and + 1467/rs574456) and R501X/rs61816761 in FLG, the major locus associated with atopic dermatitis and predisposing to asthma, in a minimum of 6743 T1D cases and 7864 controls.
Results
No evidence of T1D association was found for any of the SNPs we genotyped at FLG, SELS or IL18 (p ≥ 0.03), nor with haplotypes of IL18 (p = 0.82). Review of previous T1D genome-wide association results revealed that four (human leucocyte antigen (HLA), gasdermin B/ORM1 (Saccharomyces cerevisiae)-like/gasdermin B/, GSDMB/ORMDL3/GSDMA and IL2RB) of ten loci recently reported to be associated with asthma were associated with T1D (p≤0.005).
Conclusions
These results show that there are shared genetic associations for atopy-related traits and T1D, and this might help in the future to understand the mechanisms, pathways and environmental factors that underpin the rapid rise in incidence of both disorders in children. Copyright © 2011 John Wiley & Sons, Ltd.
Keywords: autoimmune diabetes, atopy, asthma, cytokines, proinflammation
Introduction
Type 1 diabetes (T1D) is a multifactorial autoimmune disease which results from cell-mediated immune destruction of pancreatic β cells, a complex process controlled by many genes, in particular, the human leucocyte antigen (HLA) class II and class I genes 1. Considerable overlap in the map locations of susceptibility loci in T1D, celiac disease, Graves' disease and rheumatoid arthritis has been reported (http://www.t1dbase.org) indicating shared aetiologies 1, 2. However, T1D incidence has increased steadily over the last few decades, especially in children under 5 years of age, and is predicted to double again in this age group by 2020 3, 4. T1D shares this childhood pandemic with asthma, allergy and atopic illnesses, suggesting perhaps common environmental factors and susceptibility alleles 1, 3, 4. Atopy is the development of adverse hypersensitivity immune reactions against environmental antigens, usually associated with immunoglobulin E, and includes atopic dermatitis, asthma, allergic rhinitis, allergic conjunctivitis and food allergy 5. Stene and Joner found an inverse association between atopic dermatitis and T1D 6, supporting the EURODIAB substudy which found a reduced risk of T1D in children exhibiting atopic eczema 7. However, another group found that asthma was more common in children with T1D than those without in a Finnish birth register study, suggesting that Th1 and Th2 immune-mediated disorders can coexist 8.
Among the variants studied in atopic diseases, loss-of-function null mutations in the FLG gene are the most widely replicated and are the largest genetic risk factors for atopic dermatitis and also increase risk of asthma and peanut allergy 9–11. The two most common null FLG variants in the UK, R501X and 2282del4, are rare (<3%) and show high penetrance for atopic dermatitis in heterozygous and compound heterozygous genotypes. FLG encodes a precursor protein, profilaggrin, which is a constituent of the cornified envelope of the skin. Filaggrin plays a vital role in maintaining hydration levels in the epidermis and preventing the entry of potentially harmful chemical and biological antigens which can elicit immune responses. Therefore, FLG null mutations may increase skin permeability to molecules or pathogens which could influence T1D development.
Proinflammatory cytokines, such as TNF-α, IL-1β, IFN-γ, IL-1α, IL-6 and IL-18, have been implicated in the pathogenesis of T1D 12. Selenoprotein S (SELS) is involved in the retro-translocation of misfolded proteins from the endoplasmic reticulum to the cytosol in response to endoplasmic reticulum stress and inflammation, leading to the activation of transcription factor NF-κB, which in turn activates a number of genes including IL1B, TNF and IL6. Selenoprotein S is encoded by SEPS1 (also known as SEPS1 or VIMP), on chromosome 15q26.3 13. SELS has a single nucleotide polymorphism (SNP) in the promoter region located in a putative endoplasmic reticulum stress-response element. The minor A allele of SNP rs28665122 G > A is associated with decreased SELS expression and increased plasma levels of the proinflammatory cytokines IL-1β, TNF-α and IL-6 13.
IL-18 induces the production of IFNγ, TNF-α and IL-1. It has been implicated in the pathogenesis of several immune disorders including juvenile idiopathic arthritis and Crohn's disease 14, and serum IL-18 levels are higher in newly diagnosed T1D cases compared to controls 15. Recent studies have highlighted the importance of IL18 haplotypic effects on IL18 expression: IL18 haplotypes carrying the C allele at − 105/rs360717 (5′ untranslated region) and G allele at + 183/rs5744292 (3′ untranslated region) are associated with a decrease in IL-18 at both the mRNA and protein level 16. Another allele associated with lower IL-18 serum levels is the C allele of the IL18 intronic SNP rs5744256 (T > C), p = 1 × 10−5 17. A recent genome-wide association study (GWAS) found two SNPs, rs2115763 and rs1834481, independently and convincingly associated with IL-18 levels 18. These SNPs are in linkage disequilibrium with rs5744256, r2 = 0.2, D′ = 1 and r2 = D′ = 1, respectively, in individuals of European ancestry in the Centre D'Etude du Polymorphisme Humain DNA samples. However, these SNPs have yet to be directly tested for T1D association. The recent asthma GWAS identified several regions including an association within the IL18R1 gene 19.
In this study, we investigated whether SNPs in SELS, IL18 and FLG were associated with T1D. We also investigated whether there was any overlap between the genetic regions associated with asthma 19 and T1D 20, 21.
Methods
Subjects and genotyping
Genotyping was performed on DNA samples from a minimum of 6743 British childhood-onset T1D cases and 7864 British controls, all of whom were of self-reported white ethnicity 22. Samples were genotyped for rs28665122, rs360717, rs5744292, rs5744256 and rs61816761 using TaqMan® allele discrimination assays developed by Assay-By-Design (Applied Biosystems, Warrington, UK), following the manufacturer's protocol, as described previously 22. The appropriate ethics committees approved the collection of all DNA samples, and written consent was obtained from all individuals, or parents of individuals who were too young to consent.
(Applied Biosystems, Warrington, UK), following the manufacturer's protocol, as described previously 22. The appropriate ethics committees approved the collection of all DNA samples, and written consent was obtained from all individuals, or parents of individuals who were too young to consent.
Candidate gene association tests
Statistical analyses were performed using STATA version 10 (http://www.stata.com). Association was tested, and odds ratios with 95% confidence intervals calculated, by logistic regression models. Disease status was treated as the outcome variable in the logistic model and the allele/genotype of the test SNP used as independent variable(s). Cases and controls were stratified within the regression models into 12 broad regions of Great Britain to account for variation in disease incidence and allele/genotype frequency across the country 23, 24. The appropriateness of the multiplicative allelic effects model assumption was tested using a likelihood ratio test. Haplotypes were generated using SNPHAP (www-gene.cimr.cam.ac.uk/clayton/software) and tested for association using a logistic regression model. Disease status was used as the dependant variable and counts of haplotypes weighed by the posterior probabilities as independent variables. Geographical region was included as strata. Robust variance estimates were used and a multiplicative effects model assumed. Age-at-diagnosis effects were tested in cases, in a linear regression model with age-at-diagnosis as dependent variable and SNP genotype as independent variable.
We had > 90% power to detect an effect with an odds ratio ≥ 1.12 for an α of 10−7 for rs28665122 (SELS) and > 80% power to detect a similar effect for the IL18 SNPs. FLG null alleles are rare (frequency < 3%), and, hence even with our large sample size the study was underpowered.
Results
We genotyped the most common FLG null mutation R501X/rs61816761 in 7688 T1D cases and 9354 controls but no association was found (p = 0.82; Table 1). As the effect of null alleles in FLG is semi-dominant, in addition to genotyping R501X/rs6181671, we genotyped the second most common null mutation in this gene, 2282del4, which has a reported minor allele frequency (MAF) of 0.01–0.02 9 in 384 individuals (a random selection of cases and controls), and only one heterozygous individual was identified. Owing to the low MAF of these FLG null mutations and their semi-dominant mode of inheritance, over 11,000 cases and the same number of controls would be required to have 80% power to detect an association with an odds ratio ≥ 1.2 between FLG variants and T1D at α = 0.05. Therefore, no further samples were genotyped.
Table 1.
Association of FLG single nucleotide polymorphism, R501X/rs61816761 (C > T), in 7688 type 1 diabetic cases and 9354 controls
| Minor allele/genotype | Controls n (frequency) | Cases n (frequency) | Odds ratio (95% confidence interval) | p-value |
|---|---|---|---|---|
| T | 452 (0.02) | 368 (0.02) | 1.01 (0.88–1.17) | 0.82 |
| C/C | 8907 (0.95) | 7323 (0.95) | 1.00 (reference) | — |
| T/C | 442 (0.05) | 362 (0.05) | 1.02 (0.88–1.18) | — |
| T/T | 5 (0.00) | 3 (0.00) | 0.88 (0.20–3.81) | — |
The p-value reported is for the multiplicative allelic effects model (which has an appropriate assumption).
n, number of chromosomes with allele T or number of individuals carrying the listed genotype.
The SNP -105/rs28665122 within SELS is the most associated SNP with proinflammatory cytokine levels 13, but was not associated with T1D risk (p = 0.08; Table 2). The three SNPs associated with IL-18 levels, − 105/rs5744292, + 1467/rs5744256 and + 183/rs360717, were also not associated with T1D (p ≥ 0.03; Table 3), nor were the IL18 haplotypes defined by the two SNPs − 105/rs5744292 and + 183/rs360717 (p = 0.82; Table 4), which have been shown to be associated with decreased IL-18 levels 16, 25.
Table 2.
Association of SELS single nucleotide polymorphism, -105/rs28665122 (G > A), in 8063 type 1 diabetic cases and 10,320 controls
| Allele or genotype | Controls n (frequency) | Cases n (frequency) | Odds ratio (95% confidence interval) | p-value |
|---|---|---|---|---|
| A | 2715 (0.17) | 2020 (0.15) | 0.94 (0.88–1.01) | 0.08 |
| G/G | 7792 (0.76) | 6160 (0.76) | 1.00 (reference) | — |
| G/A | 2341 (0.23) | 1786 (0.22) | 1.02 (0.89–1.18) | — |
| A/A | 187 (0.02) | 117 (0.02) | 0.89 (0.21–1.26) | — |
The p-value reported is for the multiplicative allelic effects model (which fitted the data).
n, number of chromosomes with allele A or number of individuals carrying the listed genotype.
Table 3.
Association of IL18 variants with type 1 diabetes
| n | n (frequency) | ||||||
|---|---|---|---|---|---|---|---|
| SNP | Controls | Cases | Minor allele | Controls | Cases | Odds ratio (95% | p-value |
| genotype | confidence interval) | ||||||
| − 105/rs5744292 | 7864 | 6743 | C | 3932 (0.25) | 3405 (0.25) | 1.01 (0.96–1.07) | 0.60 |
| T/T | 4392 (0.56) | 3765 (0.56) | 1.00 (reference) | — | |||
| T/C | 3012 (0.38) | 2551 (0.38) | 0.99 (0.92–1.06) | — | |||
| C/C | 460 (0.06) | 427 (0.06) | 1.09 (0.94–1.25) | — | |||
| + 1467/rs5744256 | 7904 | 6755 | C | 4112 (0.26) | 3581 (0.27) | 1.02 (0.97–1.08) | 0.39 |
| T/T | 4313 (0.55) | 3667 (0.54) | 1.00 (reference) | — | |||
| T/C | 3070 (0.39) | 2595 (0.38) | 0.99 (0.93–1.06) | — | |||
| C/C | 521 (0.07) | 493 (0.07) | 1.11 (0.97–1.27) | — | |||
| + 183/rs360717 | 9093 | 7370 | A | 4900 (0.27) | 3994 (0.27) | 1.01 (0.96–1.06) | — |
| G/G | 4820 (0.53) | 3945 (0.54) | 1.00 (reference) | 0.03 | |||
| G/A | 3646 (0.40) | 2856 (0.39) | 0.95 (0.89–1.01) | — | |||
| A/A | 627 (0.07) | 569 (0.08) | 1.12 (0.99–1.26) | — | |||
p-values are reported for the multiplicative allelic effects model at rs5744292 and rs5744256 as it was found to be an appropriate approximation, whereas the genotype effects model which makes no assumption about the mode of inheritance was required for rs360717.
Table 4.
Association of the two SNP haplotypes, rs5744292 and rs360717, at IL18 in 6123 type 1 diabetes cases and 7321 controls
| n (frequency) | |||||
|---|---|---|---|---|---|
| rs5744292 | rs360717 | Controls | Cases | Odds ratio (95% confidence interval) | p-value |
| T | G | 7032 (0.48) | 5804 (0.47) | 1 (reference) | 0.82 |
| T | A | 3946 (0.27) | 3342 (0.27) | 1.02 (0.96–1.09) | — |
| C | G | 3635 (0.25) | 3076 (0.25) | 1.02 (0.96–1.09) | — |
The C-A rs5744292-rs360717 haplotype had a frequency < 1% and hence is not listed.
The GABRIEL consortium asthma GWAS included 10 365 cases and 16 110 controls recruited from 23 different studies and identified nine regions (ten SNPs) associated with asthma, which are listed in Table S1. Among these regions, ORLMD3/GSDMB was the only non-HLA region shared between childhood-onset asthma and T1D (rs2305480 and rs3894194) 20. Review of the recent GWAS meta-analysis by Barrett et al.
20 revealed that the most atopy-associated SNPs in this region (as reported by several studies), rs2305480 and rs3894194, located in the introns of GSDMB, were also associated with T1D (p = 1.2 × 10−6, 2.7 × 10−4 and 9.3 × 10−7, respectively; Table S1). This was not unexpected as rs2305480 and rs7216389 are in high linkage disequilibrium with rs2290400, the most associated T1D SNP (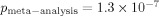 ) 20.
) 20.
Although rs2284033, the asthma-associated SNP at IL2RB, showed suggestive evidence of association with T1D in the Barrett et al. meta-analysis 20 (p = 0.005; Table S1), other SNPs in the region were more convincingly associated with T1D (rs3218253, p = 2.54 × 10−5 21; and rs229541, p = 1.98 × 10−8) 26). However, these T1D-associated SNPs are not in linkage disequilibrium with the asthma-associated SNP rs2284033 (D′ < 0.2) and, therefore, they probably have different effects on the expression of IL2RB, the strongest candidate gene in the region given the importance of the IL-2 pathway in T1D 1.
The GABRIEL consortium also showed that the SNP rs9273349 in the HLA class II region, close to HLA-DRB1, was associated with reduced risk of asthma (odds ratio = 0.85) 19. The SNP had not been typed by any of the GWAS used in the meta-analysis by Barrett et al. 20. However, rs1063355 near HLA-DQB1 which is in linkage disequilibrium with rs9273349 (r2 = 1, D′ = 1 in the Centre D'Etude du Polymorphisme Humain DNA samples was included and so was used as a surrogate in the T1D study. The minor allele at these two SNPs (T) in HLA-DQB1 confers protection in both asthma (odds ratio = 0.85) and T1D (odds ratio = 0.28; 20; Table S1). The minor T allele is in linkage disequilibrium with HLA-DQB1*06, HLA-DQB1*05, HLA-DRB1*15 and HLA-DRB1*01 (r2 = 0.48, 0.28, 0.27 and 0.19, respectively) which are known to confer reduced risk of T1D, specifically the HLA-DRB1*15-HLA-DQB1*06 haplotype (Table S2) 27. We tested for age-at-diagnosis effects at rs1063355 in 3977 T1D cases using the genotypes generated by Barrett et al. 20. The protective genotype at rs1063355 was found to be more common in relatively older onset T1D cases (p = 1.1 × 10−4) with an average age-at-diagnosis of 9.4 years for the cases homozygous for the protective (T) allele and 7.7 years for cases homozygous for the susceptibility allele.
Discussion
We found no evidence for an obvious association between IL18 SNPs and haplotypes and T1D susceptibility despite their known effects on IL-18 serum concentrations. This is consistent with a previous conclusion that the genes with functional variants associated with proinflammatory Th17 pathway-associated molecules and with immune diseases such as Crohn's disease do not alter T1D risk (e.g. CCR6 and IL23R) 1, even though the role of proinflammatory cytokines, IL-1β, TNF-α and IL-17, in pancreatic β cell killing is established. Variants altering their levels are not risk factors for T1D, suggesting that cytokine-killing in T1D is a downstream consequence of the genetically determined autoimmune islet insulitis.
In contrast, one of the variants more associated with later-onset asthma, rs9273349, was located in the HLA class II region, which is the region in T1D with largest effect on risk. Alleles at this SNP show high linkage disequilibrium with several HLA-DRB1-DQB1 haplotypes, but because the associations and the mechanisms underlying association of the HLA region with asthma are still not clearly defined, in contrast to T1D 27, a biological interpretation of this overlap cannot be made yet. Nevertheless, there is a significant link here, for the first time, between the diseases. Secondly, there is a link between diseases at the chromosome 17q12 and 17q21.1 loci, GSDMB/ORMDL3/ GSDMA (Table S1). The rs12936231 C allele is associated with higher risk of asthma and T1D and with higher expression of GSDMB and ORMDL3 and lower expression of zona pellucida binding protein 2 (ZPBP2) 28. The biological consequences of this co-association remain to be determined.
The increase in the incidence of T1D and atopic illness in the past few decades in children also suggests that this increase is attributable to environmental or lifestyle factors as the genetic pool could not have changed enough to account for such an increase 3, 4. Exposure, or lack of exposure, to certain infectious agents and other homeostatic factors at an early age and the consequences that this could have on the development of the immune system has been an attractive explanation for the rise in incidence, which is supported by animal models of autoimmune disease 1, 5. However, the nature of the immunopathology underlying T1D and atopic illness involves different pathways and there is not much known about the role played by GSDMB/ORMDL3/GSDMA, IL2RB or HLA in atopic disease. Different gene–environmental interactions, with possible involvement of different microorganisms and dietary factors, including the composition of the gut microbiome with diverse effects on immune response and tolerance 29, undoubtedly operate in the aetiology of these two conditions.
Acknowledgments
This work was funded by the Juvenile Diabetes Research Foundation International, the Wellcome Trust and the National Institute for Health Research Cambridge Biomedical Research Centre. The Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (079895). N. Saleh is funded by the Saudi Ministry of Higher Education. C. Wallace is funded by the Wellcome Trust (089989). We thank all study participants and family members. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council (grant G0000934) and the Wellcome Trust (grant 068545/Z/02). We thank The Avon Longitudinal Study of Parents and Children Laboratory in Bristol and the British 1958 Birth Cohort team, including S. Ring, R. Jones, M. Pembrey, W. McArdle, D. Strachan and P. Burton, for preparing and providing the control DNA samples. We acknowledge use of DNA from the UK Blood Services collection of Common Controls (UKBS collection), funded by the Wellcome Trust grant 076113/C/04/Z, by the Wellcome Trust/Juvenile Diabetes Research Foundation grant WT061858 and by the National Institute of Health Research of England. The collection was established as part of the Wellcome Trust Case-Control Consortium. We thank David Dunger, Barry Widmer and the British Society for Paediatric Endocrinology and Diabetes for the TID case collection. We also thank P. Clarke, G. Coleman, S. Duley, D. Harrison, S. Hawkins, T. Mistry and N. Taylor for preparation of DNA samples, E. Adlem for her bioinformatics support and M. Moffatt and W. Cookson for sharing results before publication.
Authors' contribution: N. M. S. performed FLG SNP genotyping, statistical analysis, and drafted the manuscript. S. R. J. genotyped the IL18 SNPs. D. J. S. genotyped the SELS1 SNP. C. W. reviewed the manuscript. J. M. M. H. performed statistical analyses and participated in drafting the manuscript. LB reviewed and edited the manuscript. J. A. T. participated on the conception, design and coordination of the study and wrote the manuscript. H. S. coordinated DNA sample preparation. N. M. W. managed the genotyping data. All authors read and approved the final manuscript.
Conflict of interest
The authors declare that there is no conflict of interest. The manuscript contains original unpublished work, not submitted for publication elsewhere.
Supporting information
Supporting information may be found in the online version of this article.
References
- 1.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32(4):457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Smyth DJ, Plagnol V, Walker NM, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patterson CC, Dahlquist GG, Gyurus E, Green A, Soltesz G. Incidence trends for childhood type 1 diabetes in Europe during 1989– 2003 and predicted new cases 2005-20: a multicentre prospective registration study. Lancet. 2009;373(9680):2027–2033. doi: 10.1016/S0140-6736(09)60568-7. [DOI] [PubMed] [Google Scholar]
- 4.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10(12):861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 5.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010;160(1):1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stene LC, Joner G. Atopic disorders and risk of childhood-onset type 1 diabetes in individuals. Clin Exp Allergy. 2004;34(2):201–206. doi: 10.1111/j.1365-2222.2004.01864.x. [DOI] [PubMed] [Google Scholar]
- 7.Group TESS. Decreased prevalence of atopic diseases in children with diabetes. J Pediatr. 2000;137(4):470–474. doi: 10.1067/mpd.2000.109109. [DOI] [PubMed] [Google Scholar]
- 8.Kero J, Gissler M, Hemminki E, Isolauri E. Could TH1 and TH2 diseases coexist? Evaluation of asthma incidence in children with coeliac disease, type 1 diabetes, or rheumatoid arthritis: a register study. J Allergy Clin Immunol. 2001;108(5):781–783. doi: 10.1067/mai.2001.119557. [DOI] [PubMed] [Google Scholar]
- 9.Palmer CN, Ismail T, Lee SP, et al. Filaggrin null mutations are associated with increased asthma severity in children and young adults. J Allergy Clin Immunol. 2007;120(1):64–68. doi: 10.1016/j.jaci.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Barker JN, Palmer CN, Zhao Y, et al. Null mutations in the filaggrin gene (FLG) determine major susceptibility to early-onset atopic dermatitis that persists into adulthood. J Invest Dermatol. 2007;127(3):564–567. doi: 10.1038/sj.jid.5700587. [DOI] [PubMed] [Google Scholar]
- 11.Brown SJ, Asai Y, Cordell HJ, et al. Loss-of-function variants in the filaggrin gene are a significant risk factor for peanut allergy. J Allergy Clin Immunol. 2010;127(3):661–667. doi: 10.1016/j.jaci.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grunnet LG, Aikin R, Tonnesen MF, et al. Proinflammatory cytokines activate the intrinsic apoptotic pathway in beta-cells. Diabetes. 2009;58(8):1807–1815. doi: 10.2337/db08-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran JE, Jowett JB, Elliott KS, et al. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet. 2005;37(11):1234–1241. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- 14.Sugiura T, Maeno N, Kawaguchi Y, et al. A promoter haplotype of the interleukin-18 gene is associated with juvenile idiopathic arthritis in the Japanese population. Arthritis Res Ther. 2006;8(3):R60. doi: 10.1186/ar1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katakami N, Kaneto H, Matsuhisa M, et al. Serum interleukin-18 levels are increased and closely associated with various soluble adhesion molecule levels in type 1 diabetic patients. Diabetes Care. 2007;30(1):159–161. doi: 10.2337/dc06-1768. [DOI] [PubMed] [Google Scholar]
- 16.Barbaux S, Poirier O, Godefroy T, et al. Differential haplotypic expression of the interleukin-18 gene. Eur J Hum Genet. 2007;15(8):856–863. doi: 10.1038/sj.ejhg.5201842. [DOI] [PubMed] [Google Scholar]
- 17.Frayling TM, Rafiq S, Murray A, et al. An interleukin-18 polymorphism is associated with reduced serum concentrations and better physical functioning in older people. J Gerontol A Biol Sci Med Sci. 2007;62(1):73–78. doi: 10.1093/gerona/62.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He M, Cornelis MC, Kraft P, et al. Genome-wide association study identifies variants at the IL18-BCO2 locus associated with interleukin-18 levels. Arterioscler Thromb Vasc Biol. 2010;30(4):885–890. doi: 10.1161/ATVBAHA.109.199422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace C, Smyth DJ, Maisuria-Armer M, Walker N, Todd JA, Clayton DG. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat Genet. 2010;42(1):68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Todd JA, Walker N, Cooper J, et al. Robust associations of four chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clayton DG, Walker NM, Smyth DJ, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37(11):1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 24.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–687. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiret L, Godefroy T, Lubos E, et al. Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation. 2005;112(5):643–650. doi: 10.1161/CIRCULATIONAHA.104.519702. [DOI] [PubMed] [Google Scholar]
- 26.Cooper JD, Smyth DJ, Smiles AM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40(12):1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nejentsev S, Howson JMM, Walker NM, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450(7171):887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verlaan DJ, Berlivet S, Hunninghake GM, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85(3):377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12(1):5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


