Significance
Transformation allows naturally competent bacteria to take up DNA from the environment and integrate the DNA into the chromosome by recombination. In Gram-negative bacteria, the DNA-uptake machinery shuttles the incoming DNA across the outer membrane, the periplasmic space, and the inner membrane. This study investigates the DNA-uptake complex of the human pathogen Vibrio cholerae, using a cellular biology-based approach. We visualized different components of this multicomponent complex, including a type IV pilus appendage, determined their (co)localization within the bacterial cell, and conducted an analysis of competence-gene mutants. We conclude that the uptake of DNA occurs via (at least) a two-step process.
Abstract
Natural competence for transformation is a mode of horizontal gene transfer that is commonly used by bacteria to take up DNA from their environment. As part of this developmental program, so-called competence genes, which encode the components of a DNA-uptake machinery, are expressed. Several models have been proposed for the DNA-uptake complexes of competent bacteria, and most include a type IV (pseudo)pilus as a core component. However, cell-biology–based approaches to visualizing competence proteins have so far been restricted to Gram-positive bacteria. Here, we report the visualization of a competence-induced pilus in the Gram-negative bacterium Vibrio cholerae. We show that piliated cells mostly contain a single pilus that is not biased toward a polar localization and that this pilus colocalizes with the outer membrane secretin PilQ. PilQ, on the other hand, forms several foci around the cell and occasionally colocalizes with the dynamic cytoplasmic-traffic ATPase PilB, which is required for pilus extension. We also determined the minimum competence regulon of V. cholerae, which includes at least 19 genes. Bacteria with mutations in those genes were characterized with respect to the presence of surface-exposed pili, DNA uptake, and natural transformability. Based on these phenotypes, we propose that DNA uptake in naturally competent V. cholerae cells occurs in at least two steps: a pilus-dependent translocation of the incoming DNA across the outer membrane and a pilus-independent shuttling of the DNA through the periplasm and into the cytoplasm.
Natural competence for genetic transformation is one of three modes of horizontal gene transfer (HGT) in prokaryotes and is often tightly regulated (1–3). Large pieces of DNA containing a series of genes can be transferred by natural transformation without the need for direct interaction with other microbes or mobile genetic elements. This process can foster rapid evolution, and HGT is known to be involved in the spread of antibiotic resistance, adaptation to new environmental niches, and the emergence of new pathogens.
Many bacterial species are able to enter a state of natural competence, including the human pathogen Vibrio cholerae. In this bacterium, competence is induced upon growth on chitinous surfaces (3, 4), the natural habitat of V. cholerae (5). Although we have gained a reasonably clear understanding of the regulatory network driving competence induction in this organism (for a review, see ref. 3), almost nothing is known about its DNA-uptake machinery. Indeed, the sophisticated DNA-uptake complexes used by naturally competent bacteria during transformation are still poorly characterized (6), especially in Gram-negative bacteria in which the transforming DNA (tDNA) must cross two membranes and the periplasmic space (including the peptidoglycan layer) to enter the cytoplasm and recombine with the chromosome (the latter step is not required if the tDNA consists of plasmid DNA). Interestingly, the majority of competence-protein localization studies using cellular microbiology approaches are based on studies performed with the Gram-positive bacterium Bacillus subtilis. For B. subtilis, a multicomponent protein machine may be responsible for DNA uptake (1, 7, 8), as many transformation proteins colocalize to the pole(s) of the cell (9–12). Furthermore, using single-molecule experiments with laser tweezers, Hahn et al. showed that DNA binding and uptake also occur preferentially at the cell pole (9). It is unknown whether a polar localization pattern of the DNA-uptake machinery is universal for all naturally competent bacteria and essential for its functionality. We addressed this question and demonstrate that upon competence induction, V. cholerae cells produce a type IV pilus (Tfp)-like appendage that extends beyond the outer membrane. We also visualized other components of the DNA-uptake complex, using fluorescently labeled fusion proteins, and showed that those components and the pilus are not strictly associated with the cell poles of V. cholerae. Furthermore, we identified a minimal set of competence genes required for efficient transformation of V. cholerae. We show that most gene products within this competence regulon contribute to DNA uptake and efficient transformation, even though the Tfp-related competence proteins are not entirely essential for transformation. These data provide unique insight into the function of the competence proteins with respect to DNA transfer across the outer membrane, the periplasm, or the inner membrane and suggest an at least two-step DNA-uptake process in the Gram-negative bacterium V. cholerae.
Results
Identification of Components of the DNA-Uptake Complex in Naturally Competent V. cholerae Cells.
Although the regulation of natural competence differs widely between Gram-negative bacteria (3, 13), the core components of the DNA-uptake machinery are often conserved, and the uptake process might be close to universal; however, major knowledge gaps still exist with respect to the mechanistic aspects of DNA uptake, as also noted recently by Rosemary Redfield and coworkers [“our knowledge of the proteins responsible for DNA uptake and transformation is piecemeal” (ref. 14, p. 5245)]. Thus, the aim of this study was to better understand DNA uptake in naturally competent V. cholerae. First, we examined genes (including neighboring genes in the same operon) that were significantly and reproducibly up-regulated upon growth of the bacterium on chitin (4, 15) or upon chitin-independent competence induction (through expression of the transformation regulator gene tfoX) (4, 16, 17) and excluded those genes with a potential function in chitin colonization and degradation or in a general stress response (including protein-folding chaperones). Next, we screened for genes with homologs in other naturally transformable bacteria (Table S1) and demonstrated that all these competence genes were also strictly conserved in other naturally transformable species of the genus Vibrio (3, 18) (Table S1). The majority of this gene set was predicted to encode components of a Tfp, as previously suggested (15). We also identified a cluster of genes (VC0857–0861) encoding (among other products) putative type IV pilins, which we included in further analysis even though no clear homologs were identified in naturally competent bacteria such as Neisseria gonorrheae, Haemophilus influenzae, or B. subtilis (Table S1). On the basis of the data provided below, we define this set of 19 genes (Table S1) as the minimum (but most likely still incomplete) competence regulon of V. cholerae.
Construction of Competence-Gene Mutants and Assessment of Their Phenotypes.
To test for the involvement of all identified gene products in natural transformation, we deleted each gene from the wild-type strain of V. cholerae (strain and plasmid lists in Table S2 and S3, respectively). A deletion of the gene encoding the prepilin peptidase PilD led to a significant growth defect, as previously noted (19). For this reason, the mutant was not considered further. No other knockout strains were affected in growth, but they were severely or completely impaired in natural transformability under chitin-inducing competence conditions (Table S4). We also deleted the second copy of pilT (VC0463, pilU) (20), with no effect on natural transformation (Table S4).
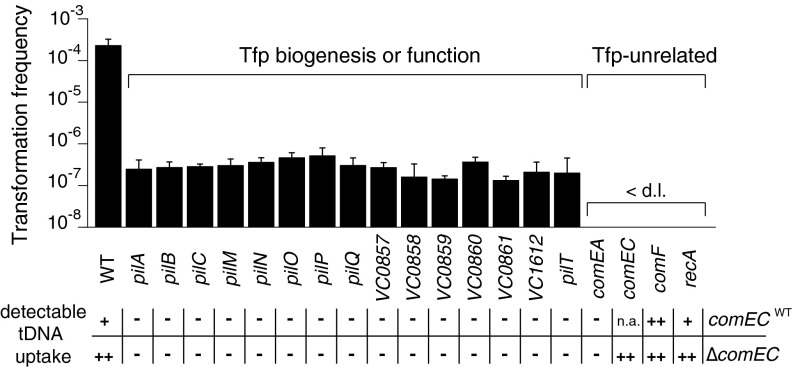
As we recently demonstrated, only a subpopulation of bacteria induces the expression of competence genes upon growth on chitin surfaces, which is most likely a reflection of the heterogeneous conditions surrounding the chitin surface (17). We thus decided to study the role of the individual components of the transformation machinery via our recently established chitin-independent transformation assay (17, 21). As for chitin-mediated induction (Table S4), all strains were transformation impaired in this assay; however, low numbers of transformants were reproducibly observed in all strains lacking Tfp-related components, which was not the case for strains lacking comEA, comEC, comF, and recA (Fig. 1). Importantly, complementation assays in which the deleted genes were encoded on a plasmid and driven by the PBAD promoter rescued the natural transformability to statistically significant levels, even though the stoichiometry of the components is expected to be impaired under these commonly used trans-complementation conditions (Table S5). Furthermore, we observed severe toxicity when two genes (pilP and pilQ) were expressed in Escherichia coli, which is consistent with the lower complementation efficiency observed in V. cholerae (Table S5).
Fig. 1.
Natural transformation and DNA uptake in strains lacking individual competence proteins. The natural transformability of a wild-type strain and derivatives lacking individual competence genes was tested in a chitin-independent assay (17) (all strains contained TntfoX). Shown are the average transformation frequencies of at least three independent biological replicates. Error bars indicate SD. All mutants were significantly impaired in natural transformation (P < 0.02). < d.l., below detection limit. The average detection limit of nontransformable strains was 7.1 × 10−8 (±3.3 × 10−8). Uptake of DNA was tested by detection of internalized tDNA in a whole-cell duplex-PCR assay (16); the results are indicated below the graph (row comECWT). The DNA-uptake assay was also performed in double-knockout strains from which comEC was concomitantly absent (row ΔcomEC). Labels: −, no tDNA detectable; +/++, tDNA detected by PCR; n.a., not applicable. Band intensities were judged relative to that of the wild-type strain (Fig. S1).
Distinguishing Between DNA Transport Across the Outer Membrane and Shuttling of the DNA Across the Inner Membrane.
After we identified this set of competence genes, we aimed to test their role in DNA transport across the two membranes. Indeed, defects in transport across either of the two membranes would affect natural transformability; however, a recently developed DNA-uptake assay (16) allowed us to distinguish these two processes via the accumulation of tDNA within the periplasm in mutants defective for transport across the inner membrane. As indicated in Fig. 1, we detected tDNA only in strains lacking comEC, comF, or recA and not in Tfp-related mutants or a comEA− strain. We then deleted the inner membrane-channel–encoding gene comEC in all mutant backgrounds and tested the double-knockout strains for DNA uptake to classify the competence proteins according to their function upstream or downstream of ComEC. The concomitant absence of comEC did not result in accumulation of tDNA in any of the Tfp-related mutants (or in the comEA mutant), suggesting that the proteins act upstream of ComEC (Fig. 1). An increase in accumulated tDNA was observed in cells lacking recA (Fig. 1 and Fig. S1), confirming that RecA acts in the cytoplasm (and thus downstream of ComEC). The amount of detectable tDNA did not increase in a comF mutant following codeletion of comEC (Fig. 1 and Fig. S1), suggesting that the proteins function along the same pathway (or step) to mediate DNA translocation across the inner membrane.
Visualization of the Competence-Related Tfp of V. cholerae.
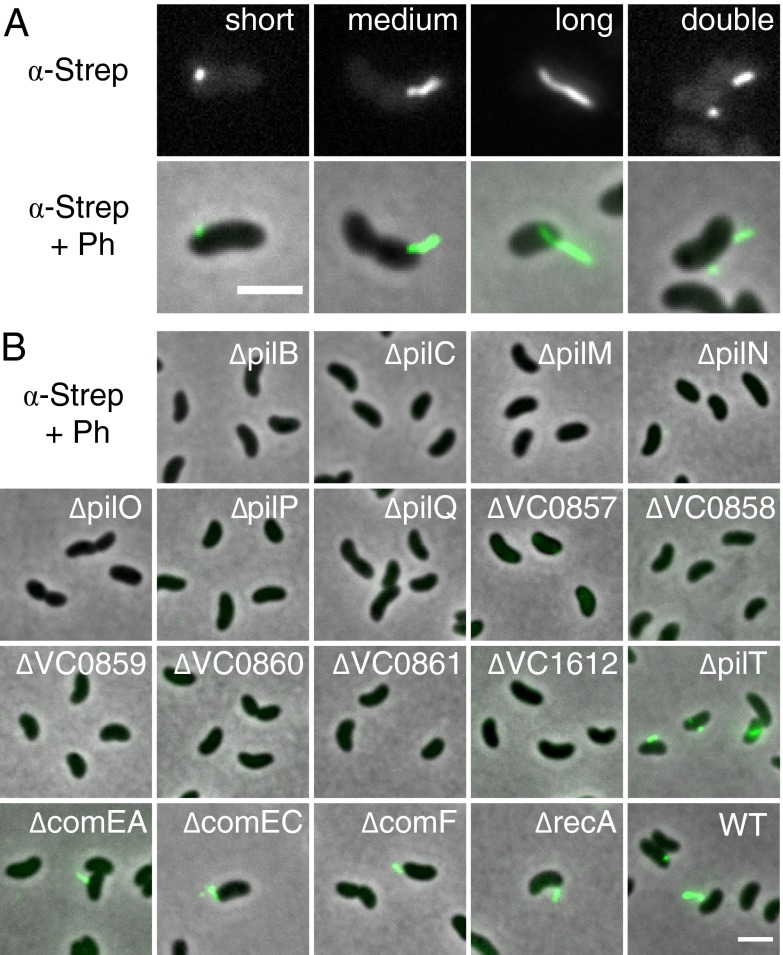
As described above, a majority of competence proteins show homology to Tfp components; however, no such pilus could be visualized in an earlier study on V. cholerae (4). The existence of a shorter competence pseudopilus, which would not reach far beyond the outer membrane, was therefore suggested (4). We tested this idea by constructing a functional, tagged (Strep-tag II) version of the major pilin subunit (PilA). To ensure expression from the indigenous pilA promoter, we inserted the pilA-strep allele onto the chromosome of V. cholerae, thereby replacing the wild-type copy of pilA. We subjected the resulting strain to chitin-independent competence induction (17) and determined the earliest point at which the bacteria are fully transformable (Fig. S2). Then, the bacteria were treated with a fluorescently labeled antibody against PilA-Strep, which enabled us to visualize competence-induced pili of different lengths (Fig. 2A). In 95% of piliated cells, we observed only one pilus per cell (quantified in Table S6). We also observed many free-floating pili, suggesting that shearing occurred rapidly due to the fragility of the pilus. Irrespective of size, pili were observed at cell poles (∼40% of cells), as well as at the 1/4 or 3/4 and central positions (Fig. S3A). Furthermore, as pilus-expressing cells maintained their polar flagellum (as demonstrated by costaining of both appendages), we could show that the pilus was able to localize to either of the two poles (e.g., the old or the new pole) (22) (Fig. S3).
Fig. 2.
Visualization of the competence-induced pilus. The major pilus subunit PilA was fused to an affinity tag, and the construct (pilA-strep) was brought onto the chromosome, replacing the wild-type copy of pilA. Pili were visualized by immunofluorescence microscopy, using an Oyster488 conjugated anti–Strep-tag antibody. (A) Diverse polar and nonpolar pili structures were observed. Depicted are fluorescence images (Upper) and overlays with the phase contrast images (Lower). (B) The majority of competence-gene mutants are no longer piliated. Shown are merged images (fluorescence in green plus phase contrast). Strain details are described in Table S2 (all contained TntfoX). (Scale bars, 2 μm.)
Contribution of Other Competence Proteins to the Production of Detectable Tfp.
To obtain a first insight into which other competence proteins might be involved in Tfp biogenesis or stabilization, we transferred the pilA-strep allele into all knockout strains described above (replacing the wild-type copy of pilA) and tested the respective strains for surface-exposed pili. All mutants lacking Tfp-related genes, including the newly identified gene cluster (VC0857–0861; Table S1), were nonpiliated (Fig. 2B). In contrast, mutants that were nontransformable in the assay described above (comEA, comEC, comF, and recA; Fig. 1) still displayed pili. A mutant carrying a deletion of pilT, which potentially encodes a retraction ATPase, still possessed pili as well (Fig. 2B).
Localization of Other Competence Proteins Within Competent V. cholerae Cells.
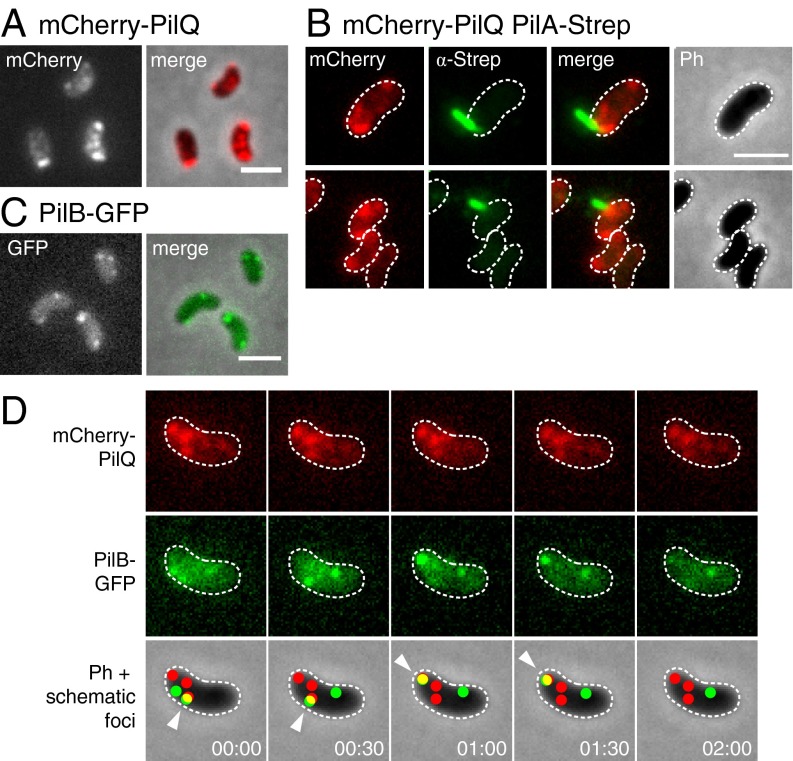
As the Tfp appendage was not strictly located at the pole, we also aimed to localize other components of the DNA-uptake machinery. First, we created a functional translational fusion between the potential outer membrane secretin PilQ and the fluorescent protein mCherry (Tables S2, S3, and S7). Interestingly, we observed that competence-induced cells often contained several PilQ foci (Fig. 3A). Furthermore, colocalization experiments indicated that for the majority (≥76%) of piliated cells (n = 80 cells from three independent biological experiments), the base of the Tfp was located close to one of the PilQ secretin signals, suggesting that both components are indeed part of a larger DNA-uptake complex (Fig. 3B).
Fig. 3.
Cellular localization of specific competence proteins. The wild-type versions of the competence genes pilQ and pilB were replaced by functional translational fusion constructs to visualize their localization. Shown are representative fluorescent images (mCherry, α-Strep, and GFP), phase contrast (Ph) images, and overlays of fluorescent channels/fluorescent and Ph channels (merge). Cells are outlined with dashed lines. V. cholerae strains carrying mCherry-pilQ (A, mCherry N terminally fused to PilQ), mCherry-pilQ and pilA-strep (B), pilB-gfp (C, GFP fused to the C terminus of PilB), and mCherry-pilQ and pilB-gfp (D) were subjected to epifluorescence microscopy (after immunostaining, B). Colocalization of mCherry-PilQ and the pilus was observed in the majority of piliated cells (B). For D, two-color time-lapse microscopy was performed. Representative fluorescent images for mCherry-PilQ (red) and PilB-GFP (green) are indicated. Observed spots for both fluorescent proteins are schematically depicted and projected on the Ph images (Bottom). Colocalization events are indicated by a white arrowhead. The time is given in minutes. (Scale bars, 2 μm.)
Next, we fused the putative pilus-elongating ATPase PilB to the green fluorescent protein (GFP). Again, the fusion protein, which replaced the wild-type copy of PilB, retained its ability to drive natural transformation (Table S7). Like PilQ, PilB did not form a single focus at the pole, but instead formed several distinctive foci throughout the cell (Fig. 3C). In contrast, its putative opponent, the traffic ATPase PilT, showed uniform localization throughout the cytoplasm (with rare formation of distinctive foci; Fig. S4). To test whether the foci of PilB overlap with those of the PilQ secretin, we tested a strain containing both translational fusions (replacing their wild-type counterparts) for cellular colocalization, using time-lapse microscopy. We observed that the PilB foci were often mobile and seemingly traveled from one PilQ focus to the next, resulting in a temporary colocalization of both proteins (Fig. 3D).
Essentiality of ATP Hydrolysis for the Functionality of PilB.
The PilB protein is considered a traffic ATPase (similar to ComGA in B. subtilis and belonging to the family of type II/IV secretion NTPases) and contains several conserved motifs, such as the P-loop–containing Walker A box and an atypical Walker B box (DhhhhGE; h, hydrophobic amino acid) (23). Indeed, the Walker B motif is conserved between PilB of V. cholerae and homologs in other bacteria (Fig. S5A). We therefore decided to test whether ATP hydrolysis is important to PilB’s role in DNA uptake/natural transformation, as for a very different phenotype, the Tfp-dependent gliding motility of Myxococcus xanthus (24). We first cloned a plasmid carrying a pilB with a site-directed mutation (E394A) and tested the variant in a complementation assay with natural transformation as the readout. The single amino acid exchange of the conserved glutamate completely abolished PilB’s functionality (Fig. S5B). Moreover, the PilBE394A variant was unable to restore pilus formation, unlike complementation with wild-type pilB in trans (Fig. S5C).
Next, we generated a pilBE394A variant as a translational fusion with GFP, which was used to replace the wild-type copy of pilB on the chromosome. By imaging this strain under competence-inducing conditions, we observed that the PilBE394A protein was not degraded and that its localization pattern was comparable to that of PilB-GFP (Fig. S5D); however, in contrast to that in the wild-type counterpart (Fig. 3C), we never observed any dynamic behavior. The V. cholerae strain carrying the pilBE394A-gfp allele at the native pilB locus was also impaired in natural transformation (to the same level as the pilB-knockout strain; Fig. S5B).
Discussion
A Model for DNA Uptake in Naturally Competent V. cholerae Cells.
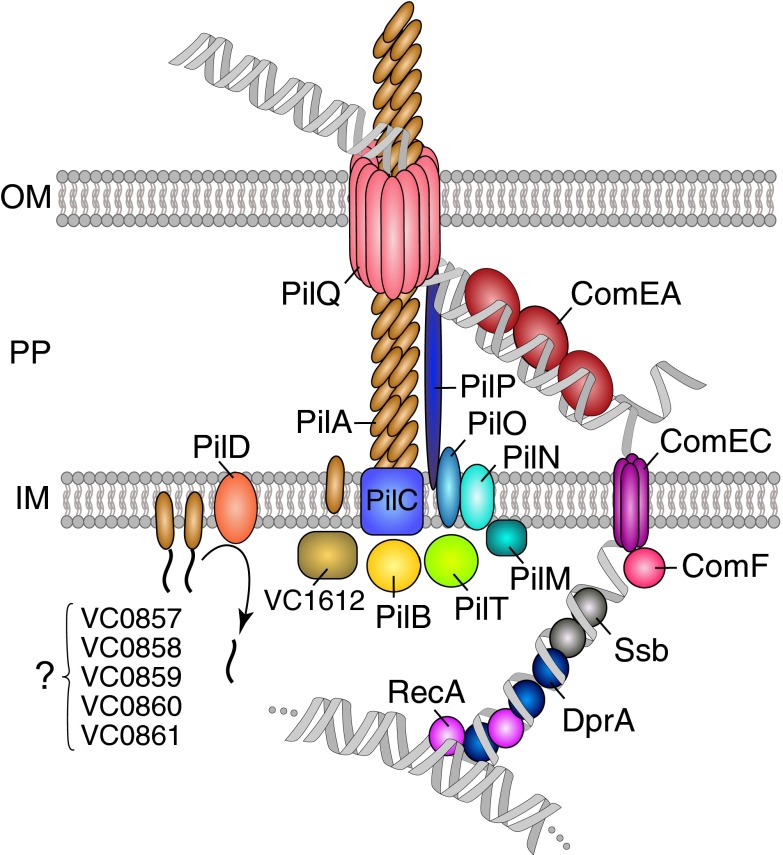
In this paper we propose a unique model of the DNA-uptake machinery of V. cholerae (Fig. 4). The model combines data from this work with those of studies of other transformable bacteria and their Tfp and presents evidence for several important mechanisms that have not been previously addressed experimentally (6–8, 25–31). In the center of the model is a Tfp appendage with PilA as the major pilin subunit (as demonstrated in Fig. 2). Prepilins are processed by the peptidase PilD [as previously demonstrated for another major pilin of V. cholerae, MshA (32), which is not involved in natural transformation] and are incorporated into the growing pilus, which is driven energetically by the traffic ATPase PilB. Indeed, no pili were detectable in a pilB− strain (Fig. 2) or when PilB was unable to hydrolyze ATP (as in the PilBE394A variant; Fig. S5). PilA polymerization might be initiated by the minor pilins encoded by the gene cluster VC0857–0861 as previously suggested for unrelated minor pseudopilins of the Klebsiella oxytoca type II secretion system (33). Indeed, V. cholerae strains lacking genes of the VC0857-0861 cluster were severely impaired in DNA uptake and natural transformation (Fig. 1) and did not produce any detectable surface-exposed pili (Fig. 2).
Fig. 4.
Model of the DNA-uptake machinery of V. cholerae. The proposed model of the DNA-uptake machinery is based on the results provided in this study and on homologies to (predicted) competence proteins from other competent bacteria or other type IV pili-containing organisms. Details are described in the text.
The pilus crosses the outer membrane through the secretin PilQ and pilus retraction is most likely driven by PilT. We showed that both traffic ATPases, PilB and PilT, were required for DNA uptake and efficient transformation of V. cholerae (Fig. 1); however, in contrast to the nonpiliated phenotype of a pilB mutant, the pilT mutant was still piliated. Moreover, although there was no apparent difference in the length of the pili between a wild-type strain containing the pilA-strep construct and the corresponding pilT− strain, we detected significantly more piliated cells for the latter mutant (14% compared with 5.3%; Table S6). Even more strikingly, the percentage of piliated cells containing two or more pili was significantly increased in the pilT− strain (18.7% compared to 3.6% for the WT; Table S6), supporting the hypothesis that PilT plays a role in pilus retraction. A similar increase in multipiliated cells was observed upon overexpression of pilB from a plasmid (Table S6), which is in line with PilB’s putative role as a pilus extension ATPase.
We suggest that once short stretches of the tDNA have crossed the outer membrane, either by pilus retraction or by pilus-related opening of the secretin pore, the DNA is further pulled into the periplasm through the binding of ComEA. Indeed, V. cholerae strains devoid of ComEA were nontransformable and did not accumulate detectable levels of tDNA in their periplasm (Fig. S1), whereas deletion of comE1 (comEA homolog) in H. influenzae had only a negligible effect. However, the authors of that study suggested that a paralog encoded elsewhere in the H. influenzae chromosome might compensate for the absence of ComE1. Next in our model (Fig. 4) ComEA shuttles the tDNA through the periplasm and toward the inner membrane aqueous channel ComEC (34), which, potentially in concert with ComF, transports a single DNA strand into the cytoplasm. ComF is a homolog of ComFC from B. subtilis (Table S1); however, disruption of comFC by integration of an erythromycin-resistance cassette resulted in only a weak transformation phenotype in B. subtilis (fivefold lower transformation frequency) compared with a WT strain (35). The cassette was, however, integrated close to the 3′ end of comFC and might have resulted in residual gene-product activity. We deleted all genes of interest rather than create insertional mutants, which resulted in a nontransformable comF-minus strain (Fig. 1), even though the cells were fully piliated (Fig. 2) and able to take up DNA into the periplasm (Fig. 1 and Fig. S1). As tDNA did not further accumulate in the comF-comEC double mutant as in a recA-comEC–negative strain (Fig. 1 and Fig. S1), we suggest that ComF, with the membrane channel protein ComEC, participates in DNA translocation across the inner membrane (Fig. 4). Once inside the cytoplasm, Ssb and DprA bind the ssDNA and facilitate loading of RecA (36), which ultimately catalyzes recombination with the bacterial chromosome.
Our study provides evidence that the localization of the DNA-uptake machinery of V. cholerae is not biased toward the cell pole, as in B. subtilis (9, 10). Indeed, neither the pilus appendage nor the secretin or traffic ATPases localized solely to the pole (Figs. 2 and 3). Whereas we detected only one pilus in the majority of piliated cells (Table S6), several clusters of the outer membrane secretin PilQ and the traffic ATPase PilB were observed in competence-induced cells (Fig. 3). Interestingly, a recent study on the type II secretion system of V. cholerae also showed discrete fluorescent foci along the cell periphery for different components of this macromolecular complex (37). This finding is in contrast to the polar localization pattern described earlier (22), which most likely resulted from an overexpression artifact (37). Indeed, Lybarger et al. concluded that “chromosomal, intraoperon expression conditions are optimal for determining the intracellular locations of fusion proteins” (ref. 37, p. 3149), and such conditions were used throughout this study.
Using time-lapse microscopy, we observed that the PilB clusters were often dynamic, which was not observed for clusters formed by PilQ or by the PilBE394A variant (Fig. S5). On the basis of this observation, it is tempting to speculate that the membrane-bound components of the competence-induced pili, apart from the pilins, are preassembled at different locations around the cell periphery or the pole (as observed for PilQ) and that the temporary interaction with the ATP-hydrolyzing PilB protein initiates pilin polymerization and thus production of Tfp appendages. Interestingly, PilB of M. xanthus primarily localizes to and oscillates between the cell poles, which is not the case for the ATPase-defective PilBE391A variant (38), indicating that PilB dynamics might be widespread among competent and noncompetent bacteria; however, in contrast to the findings by Bulyha et al. (38) in M. xanthus, we did not observe any major accumulations of the retraction ATPase PilT in V. cholerae (Fig. S4).
While the current study was ongoing, Sinha et al. reported on the natural transformation and DNA uptake phenotypes of mutants lacking competence genes in naturally transformable H. influenzae (14). The authors concluded that 17 genes were absolutely required for transformation, of which 14 were presumably involved in the assembly and function of the Tfp part of the DNA-uptake machinery. Whereas the chitin-dependent transformation assays of the competence-gene mutants investigated in our study supported these results for V. cholerae (Table S4), we observed a slight but meaningful difference after chitin-independent transformation. Under the latter conditions, rare transformants were reproducibly detected for all mutants lacking competence proteins involved in Tfp synthesis and function, whereas we never observed rare transformants for mutants lacking Tfp-independent competence genes (such as comEA, comEC, and comF; Fig. 1). These diverging results between chitin-dependent and chitin-independent transformation first appeared contradictory; however, upon closer inspection of the data, we hypothesized that the rare transformants reproducibly observed under chitin-independent conditions might have been under the limit of detection of the chitin-dependent approach (Table S4). Notably, the detection limit of this assay was calculated on the basis of the total number of colony-forming units (cfu) and was therefore based on the assumption that each cell entered the competence state when grown on a chitin surface. However, we have previously shown that due to the heterogeneous environment around the chitin surface, only a part of the population enters the competence state (17). The “real” detection limit based on the number of competent cells is therefore most likely higher, rendering rare transformants undetectable. To test this assumption, we repeated the chitin-dependent transformation assay for three Tfp-related mutants (ΔpilA, ΔpilB, and ΔpilQ) and three Tfp-unrelated mutants (ΔcomEC, ΔcomEA, and ΔcomF); however, rather than directly plate the bacteria on selective medium after their detachment from the chitin surfaces, we first enriched the culture for several hours in rich medium (without selection pressure). Interestingly, under those conditions, we reproducibly detected rare transformants for the Tfp-related mutants but never for the Tfp-unrelated mutants, strongly supporting the data from the chitin-independent assay (Fig. S6).
Although those rare transformants might be negligible in a global picture of the DNA-uptake machinery, the difference between the appearance of rare transformants in the Tfp-related mutants and their absence in the Tfp-unrelated mutants is still meaningful, as it provides insight into the mechanistic aspects of the DNA uptake. Indeed, on the basis of these data, we conclude that DNA uptake must occur in at least two independent steps, whereby the second step (mediated by ComEA, ComEC, and ComF) cannot be circumvented. Our data therefore present a unique experimental indication of a two-step (at least) DNA uptake process in an organism containing a competence-induced Tfp, thereby confirming earlier hypotheses (28, 39). Such a distinction between the two groups of competence genes is also reflected by their regulation. We have previously shown that only a minority of (tested) competence genes, namely comEC and comEA, were coregulated by quorum sensing via the transcriptional factor QstR, whereas the Tfp-related competence genes, such as pilA, pilB, and pilQ, were not dependent on quorum sensing (16, 17, 21, 40). However, expression of all tested competence genes was driven by the transformation regulator TfoX and subject to catabolite repression (4, 17, 41).
We suggest that the Tfp part of the DNA-uptake machinery is primarily involved in tDNA translocation across the outer membrane and that the tDNA can occasionally also enter the periplasm in a competence Tfp-independent manner. One of the two other type IV pili encoded on the V. cholerae chromosome, toxin coregulated pilus (TCP) or the mannose-sensitive hemagglutinin (MSH) pilus, may partially compensate for the absence of the competence-related Tfp. To test this hypothesis, we created double- and triple-knockout strains lacking pilA (encoding the major pilin of the competence Tfp) and tcpA (encoding the major pilin subunit of TCP); lacking pilA and mshA (encoding the major pilin subunit of MSH); or missing pilA, mshA, and the whole Vibrio pathogenicity island 1 (VPI-1, which carries all tcp genes). We tested those mutants for rare transformants, but the resulting transformation frequencies (TF) did not differ significantly between those strains (ΔpilA, TF = 1.1 × 10−7 ± 4.4 × 10−8; ΔpilAΔtcpA, TF = 2.6 × 10−7 ± 2.5 × 10−7; ΔpilAΔmshA, TF = 3.1 × 10−7 ± 2.9 × 10−7; ΔpilAΔmshAΔVPI-1, TF = 5.8 × 10−8 ± 6.4 × 10−8), suggesting that neither the TCP nor the MSH pilus contribute to the low transformation frequencies observed in the competence Tfp mutants. Thus, we cannot explain how tDNA enters the periplasm in the absence of the Tfp part of the DNA-uptake machinery.
In summary, our data show that DNA uptake in V. cholerae occurs as (at least) a two-step process. Indeed, the DNA-uptake complex includes a Tfp-like component that is primarily involved in shuttling tDNA across the outer membrane, whereas the competence proteins ComEA, ComEC, and ComF are involved in tDNA transfer downstream. As those two classes of competence genes are also differently regulated, we speculate that the competence-induced Tfp might fulfill a secondary role apart from DNA uptake. Indeed, on the basis of a fitness disadvantage observed for a pilA− strain on chitin surfaces, it was previously suggested that the Tfp contributes to chitin colonization (15), although these fitness experiments were performed in a quorum-sensing–defective strain of V. cholerae that is not naturally transformable. Interestingly, a secondary role for this Tfp would also support a recent hypothesis: Bakkali suggested that DNA uptake might be a side effect of Tfp-mediated bacterial adhesion and twitching motility (42); however, tDNA transfer through the periplasm and across the inner membrane would still require the presence of ComEA, ComEC, and ComF.
Materials and Methods
Strains, Plasmids, and Growth Conditions.
All V. cholerae strains and plasmids used in this study were derivatives of the El Tor strain A1552 (43) and are described in Tables S2 and S3. Unless otherwise stated the bacterial cells were propagated in Luria–Bertani medium (LB) in a shaking incubator at 30 °C.
Natural Transformation and DNA Uptake Assays.
Bacteria were tested for chitin-dependent and chitin-independent natural transformation, as described elsewhere (17, 44, 45). Statistical analysis was conducted in R software (46). Differences in transformation frequencies were considered significant when P values of Welch’s t tests on log-transformed data were below 0.05 (*) or 0.01 (**). DNA uptake was tested using a previously developed whole-cell duplex-PCR assay (16) with minor modifications (SI Materials and Methods).
Epifluorescence Microscopy Experimentation.
Strains carrying chromosomally encoded fluorescent fusion constructs were grown aerobically in LB supplemented with 0.02% l-arabinose at 30 °C for 6–8 h. Samples were washed once in PBS buffer before imaging. Pili were visualized by immunofluorescence microscopy, in which the antibody targeted the tagged version of the major pilin PilA. Details of the epifluorescence microscopy, preparation of bacteria, and the immunofluorescence protocol are provided in SI Materials and Methods.
Other Methods.
Information on design of strains and plasmids, recombinant DNA techniques, and the details of the microscopy approaches and image analysis are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Sandrine Borgeaud and Clémentine Thévoz for technical assistance. This work was supported by the Swiss National Science Foundation Grants 31003A_127029 and 31003A_143356 (to M.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315647110/-/DCSupplemental.
References
- 1.Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310(5753):1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claverys JP, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 3.Seitz P, Blokesch M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev. 2013;37(3):336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 4.Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310(5755):1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 5.Lipp EK, Huq A, Colwell RR. Effects of global climate on infectious disease: The cholera model. Clin Microbiol Rev. 2002;15(4):757–770. doi: 10.1128/CMR.15.4.757-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allemand JF, Maier B. Bacterial translocation motors investigated by single molecule techniques. FEMS Microbiol Rev. 2009;33(3):593–610. doi: 10.1111/j.1574-6976.2009.00166.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2(3):241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 8.Burton B, Dubnau D. Membrane-associated DNA transport machines. Cold Spring Harb Perspect Biol. 2010;2(7):a000406. doi: 10.1101/cshperspect.a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell. 2005;122(1):59–71. doi: 10.1016/j.cell.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidane D, Graumann PL. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell. 2005;122(1):73–84. doi: 10.1016/j.cell.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 11.Kramer N, Hahn J, Dubnau D. Multiple interactions among the competence proteins of Bacillus subtilis. Mol Microbiol. 2007;65(2):454–464. doi: 10.1111/j.1365-2958.2007.05799.x. [DOI] [PubMed] [Google Scholar]
- 12.Kaufenstein M, van der Laan M, Graumann PL. The three-layered DNA uptake machinery at the cell pole in competent Bacillus subtilis cells is a stable complex. J Bacteriol. 2011;193(7):1633–1642. doi: 10.1128/JB.01128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58(3):563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha S, Mell JC, Redfield RJ. Seventeen Sxy-dependent cyclic AMP receptor protein site-regulated genes are needed for natural transformation in Haemophilus influenzae. J Bacteriol. 2012;194(19):5245–5254. doi: 10.1128/JB.00671-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meibom KL, et al. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA. 2004;101(8):2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suckow G, Seitz P, Blokesch M. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J Bacteriol. 2011;193(18):4914–4924. doi: 10.1128/JB.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo Scrudato M, Blokesch M. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 2012;8(6):e1002778. doi: 10.1371/journal.pgen.1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Bernardy EE, Hammer BK, Miyashiro T. Competence and natural transformation in vibrios. Mol Microbiol. 2013;89(4):583–595. doi: 10.1111/mmi.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fullner KJ, Mekalanos JJ. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect Immun. 1999;67(3):1393–1404. doi: 10.1128/iai.67.3.1393-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo Scrudato M, Blokesch M. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 2013;41(6):3644–3658. doi: 10.1093/nar/gkt041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott ME, Dossani ZY, Sandkvist M. Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc Natl Acad Sci USA. 2001;98(24):13978–13983. doi: 10.1073/pnas.241411198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filloux A. The underlying mechanisms of type II protein secretion. Biochim Biophys Acta. 2004;1694(1–3):163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Jakovljevic V, Leonardy S, Hoppert M, Søgaard-Andersen L. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J Bacteriol. 2008;190(7):2411–2421. doi: 10.1128/JB.01793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer TF. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene. 1997;192(1):125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 26.Claverys JP, Martin B, Polard P. The genetic transformation machinery: Composition, localization, and mechanism. FEMS Microbiol Rev. 2009;33(3):643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 27.Averhoff B. Shuffling genes around in hot environments: The unique DNA transporter of Thermus thermophilus. FEMS Microbiol Rev. 2009;33(3):611–626. doi: 10.1111/j.1574-6976.2008.00160.x. [DOI] [PubMed] [Google Scholar]
- 28.Krüger NJ, Stingl K. Two steps away from novelty—principles of bacterial DNA uptake. Mol Microbiol. 2011;80(4):860–867. doi: 10.1111/j.1365-2958.2011.07647.x. [DOI] [PubMed] [Google Scholar]
- 29.Burrows LL. Pseudomonas aeruginosa twitching motility: Type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 30.Pelicic V. Type IV pili: E pluribus unum? Mol Microbiol. 2008;68(4):827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- 31.Laurenceau R, et al. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog. 2013;9(6):e1003473. doi: 10.1371/journal.ppat.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marsh JW, Taylor RK. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol Microbiol. 1998;29(6):1481–1492. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 33.Cisneros DA, Bond PJ, Pugsley AP, Campos M, Francetic O. Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J. 2012;31(4):1041–1053. doi: 10.1038/emboj.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Draskovic I, Dubnau D. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: Membrane topology, oligomerization and formation of disulphide bonds. Mol Microbiol. 2005;55(3):881–896. doi: 10.1111/j.1365-2958.2004.04430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Londoño-Vallejo JA, Dubnau D. comF, a Bacillus subtilis late competence locus, encodes a protein similar to ATP-dependent RNA/DNA helicases. Mol Microbiol. 1993;9(1):119–131. doi: 10.1111/j.1365-2958.1993.tb01674.x. [DOI] [PubMed] [Google Scholar]
- 36.Mortier-Barrière I, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130(5):824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 37.Lybarger SR, Johnson TL, Gray MD, Sikora AE, Sandkvist M. Docking and assembly of the type II secretion complex of Vibrio cholerae. J Bacteriol. 2009;191(9):3149–3161. doi: 10.1128/JB.01701-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulyha I, et al. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol Microbiol. 2009;74(3):691–706. doi: 10.1111/j.1365-2958.2009.06891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 40.Blokesch M. A quorum sensing-mediated switch contributes to natural transformation of Vibrio cholerae. Mobile Genet Elements. 2012;2(5):224–227. doi: 10.4161/mge.22284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blokesch M. Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol. 2012;14(8):1898–1912. doi: 10.1111/j.1462-2920.2011.02689.x. [DOI] [PubMed] [Google Scholar]
- 42.Bakkali M. Could DNA uptake be a side effect of bacterial adhesion and twitching motility? Arch Microbiol. 2013;195(4):279–289. doi: 10.1007/s00203-013-0870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yildiz FH, Schoolnik GK. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180(4):773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marvig RL, Blokesch M. Natural transformation of Vibrio cholerae as a tool—optimizing the procedure. BMC Microbiol. 2010;10:155. doi: 10.1186/1471-2180-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Souza Silva O, Blokesch M. Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid. 2010;64(3):186–195. doi: 10.1016/j.plasmid.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 46.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.