Summary
MLL fusion proteins in leukemia induce aberrant transcriptional elongation and associated chromatin perturbations, however the upstream signaling pathways and activators that recruit or retain MLL oncoproteins at initiated promoters are unknown. Through functional and comparative genomic studies, we identified an essential role for NF-κB signaling in MLL leukemia. Suppression of NF-κB led to robust anti-leukemia effects that phenocopied loss of functional MLL oncoprotein or associated epigenetic cofactors. The NF-κB subunit RELA occupies promoter regions of crucial MLL target genes and sustains the MLL-dependent leukemia stem cell program. IKK/NF-κB signaling is required for wild-type and fusion MLL protein retention and maintenance of associated histone modifications providing a molecular rationale for enhanced efficacy in therapeutic targeting of this pathway in MLL leukemias.
Keywords: MLL, NF-κB, HOX, MEIS, leukemia
Introduction
MLL is an epigenetic regulator that loses its inherent histone methyltransferase activity and acquires abnormal functionalities due to fusions with various partner proteins in leukemias. Although most MLL fusion proteins are strong transactivators that induce inappropriate expression of target genes, they do not typically display features of classical activators that recruit RNA polymerase II. Rather, substantial evidence shows that many MLL fusion proteins assemble into multi-protein complexes involved in transcriptional elongation and associated chromatin modifications. Epigenetic factors known to physically interact with MLL oncoproteins include lens epithelium-derived growth factor (LEDGF) (Yokoyama and Cleary, 2008), histone methyltransferase DOT1L (Okada et al., 2005), chromobox homolog 8 (CBX8) (Tan et al., 2011), histone demethylase KDM1A (Harris et al., 2012), and bromodomain-containing 4 (BRD4) (Zuber et al., 2011b). Chemical inhibitors that target some of these accessory factors validate their crucial roles in leukemia pathogenesis and offer promising therapeutic strategies to block inappropriate expression of key subordinate genes such as HOXA9 and MEIS1 that suppress differentiation and induce aberrant self-renewal capabilities in progenitors critical for leukemogenic potential (Wong et al., 2007).
The DOT1L histone methyltransferase is implicated to play a central role physically interacting with several MLL fusion partners including AF10, AF9, and ENL (Okada et al., 2005; Zhang et al., 2006; Bitoun et al., 2007). Aberrant levels of its H3K79 methylation mark, which denotes recently elongated genes, are a key feature of MLL primary target genes (Krivtsov et al., 2008; Bernt et al., 2011), suggesting it might contribute a common mechanism of oncogenic activation in MLL leukemias. The elongation factor P-TEFb is also implicated to serve a central role. Fusion partners that account for most MLL leukemias assemble into higher-order protein complexes that contain P-TEFb (Lin et al., 2010; Yokoyama et al., 2010), and are aberrantly tethered or indirectly recruited to MLL target genes (Yokoyama et al., 2010). P-TEFb phosphorylates the C-terminus of stalled RNA polymerase II as well as factors (NELF, DSIF) that otherwise pause the polymerase after promoter initiation (Fujinaga et al., 2004; Yamada et al., 2006). The role of MLL fusion associated factors such as P-TEFb in elongation of initiated transcripts strongly suggests that MLL oncoproteins must function in partnership with more conventional transcriptional activators. However, little is known about the upstream signaling pathways or activators that may recruit, retain or cooperate with MLL oncoproteins at subordinate genes.
These studies were undertaken to identify requisite signaling pathways that sustain the transcriptional roles of MLL oncoproteins in leukemia pathogenesis.
Results
Functional and comparative genomics implicate the NF-κB signaling pathway in MLL leukemia pathogenesis
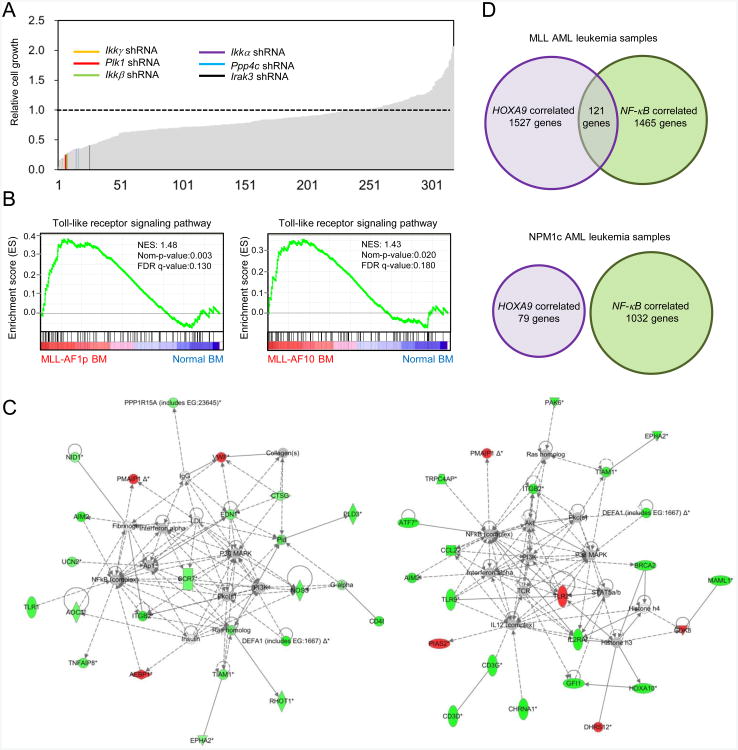
A functional genomic screen was conducted to identify essential signaling pathways in MLL leukemia cells. A lentiviral-based shRNA knockdown approach was applied on a gene-per-gene basis using a library of 321 shRNAs that target 211 phosphoregulators (Table S1) with potential roles in stem cell biology (Lee et al., 2012). An initial screen and subsequent confirmatory rounds of analysis quantified the effects of specific shRNA knockdown on growth of myeloid progenitors transformed by MLL oncogenes (Figure S1A). This identified 33 candidate kinases or phosphatases whose knockdown resulted in 50% or greater impairment (Figure S1B). Among the high-scoring candidates, several have implicated roles in mediating NF-κB signaling, including the IKK complex (IKKα, IKKβ, and IKKγ) (Karin and Ben-neriah, 2000), polo-like kinase 1 (PLK1) (Lin et al., 2011), protein phosphatase 4 catalytic subunit (PPP4C) (Yeh et al., 2004), and IL-1R-associated kinase 3 (IRAK3) (Wesche et al., 1999) (Figure 1A), suggesting that the NF-κB pathway may contribute to MLL leukemias.
Figure 1. Enrichment of NF- κB signaling in MLL leukemia cells.
(A) Single cell-based shRNA screening was used to identify the effects of kinase andphosphatase knockdowns on the growth of mouse myeloid cells transduced by MLL-AF9. Combined results from two independent replicates are expressed as the relativecell number compared to cells transduced with control shRNA. Potential candidatesassociated with NF-κB signaling are indicated.
(B) GSEA analyses demonstrate that expression of genes associated with the Toll-likereceptor signaling pathway is enriched in leukemic BM cells from mice with AMLinduced by MLL-AF1p (left) and MLL-AF10 (right) compared to normal BM. Thenormalized enrichment scores (NES) are based on analysis of a public dataset (GSE13796).
(C) Aberrantly methylated NF-κB gene networks in two MLL-associated epigeneticallydefined human AML clusters. Genes with DNA hypomethylation compared with normalCD34+ cells are shown in green, whereas hypermethylated genes appear in red.
(D) Comparison of HOXA9-correlated and NF-κB-correlated (correlation > 0.5) gene expression in human MLL acute myeloid leukemias showed 121 unique genes whose expression overlapped (p = 1×10-16, calculated using R package: SAGx_1.32.0 under R version 2.15.3). A similar analysis in NPM1c acute myeloid leukemias showed no overlap.
See also Figure S1 and Tables S1 and S2.
PANTHER analysis showed the enrichment of identified candidate kinases/phosphatases in the Toll-like receptor signaling pathway (data not shown). Consistent with this, expression of transcripts encoding proteins of Toll-like signaling pathways (KEGG pathway: HSA04620) that mediate p105-dependent NF-κB activation (Table S2) are enriched in MLL-AF1p and MLL-AF10 leukemia cells compared to normal bone marrow cells (Figure 1B) (enrichment also observed in MLL-AF9 leukemia cells but did not achieve statistical significance; result not shown), further suggesting the importance of NF-κB signaling in mouse models of MLL leukemia.
Network analysis of global DNA methylation profiles in human MLL-associated acute myeloid leukemia (AML) also implicates the NF-κB pathway. AMLs with MLL chromosomal translocations cluster in two of 16 distinct subgroups defined by DNA methylation profiles (Figueroa et al., 2010), and are distinguished by hypomethylation of genes that organize into NF-κB networks (Figure 1C) consistent with a role for NF-κB signaling in sustaining MLL leukemia. Furthermore, gene expression analysis in the same AML cohort identified 121 genes whose expression correlated with both HOXA9 and NF-κB expression in MLL-associated AML whereas no overlapping genes were observed in NPM1c mutant AML, another genetic subtype associated with aberrant HOXA9 expression (Figure 1D). Taken together, these findings suggest a potential pathogenic role of the NF-κB signaling pathway in MLL-associated leukemia.
MLL leukemia cells are preferentially dependent on IKK/NF-κB signaling
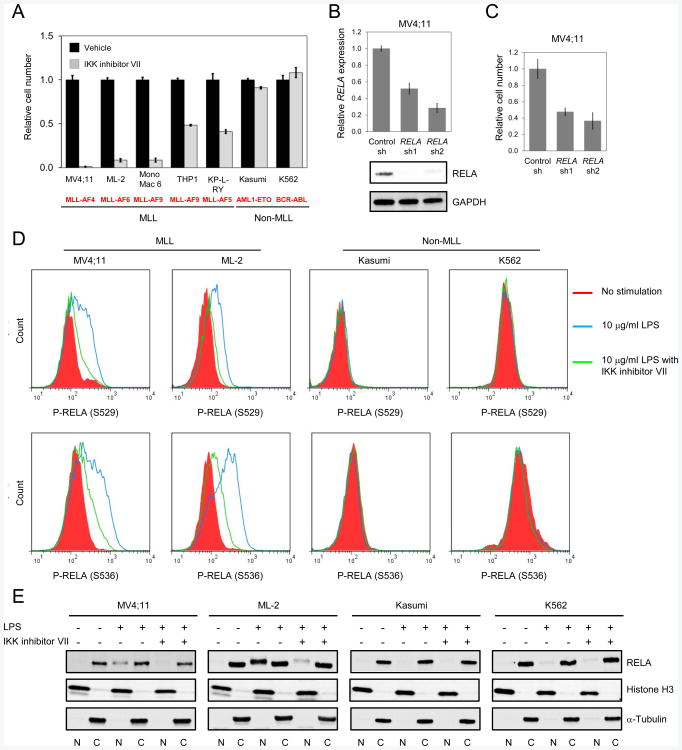
Pharmacologic inhibitors of the IKK complex, a major upstream regulator of NF-κB signaling, were assessed for their effects on the growth of human cell lines representative of different genetic subtypes of AML. Cell lines with MLL aberrations displayed enhanced sensitivity to three different IKK inhibitors compared to non-MLL leukemia cells (Figures 2A and S2A-C). Furthermore, knockdown of the NF-κB subunit RELA (p65), a transcriptional effector of the pathway, showed marked impairment of cell growth that correlated with the extent of knockdown in the MLL leukemia cell lines MV4;11 (Figures 2B and 2C) and ML-2, but not in non-MLL cells K562 (Figure S2D).
Figure 2. Differential sensitivity of human MLL leukemia cells to IKK/NF- κB signaling.
(A) The growth of human myeloid leukemia cell lines was assessed after 4 days culture in the absence or presence of 2 μM IKK inhibitor VII. Results are expressed as the relative cell number compared to vehicle treated cells.
(B and C) MV4;11 human leukemia cells were transduced with lentiviral vectors expressing the indicated shRNAs. Relative RELA transcript and protein levels were measured by qRT-PCR and western blot analysis, respectively (B). Cell numbers (C) were enumerated at 3 days, and expressed relative to cells transduced with control shRNA.
(D) Human myeloid leukemia cell lines were serum starved (0.1% FBS) overnight, then stimulated with 10 μg/ml LPS for 30 min with or without IKK inhibitor VII pretreatment, and analyzed by phospho-flow cytometry with antibodies specific to p65 RELA phosphorylated at S529 or S536. Representative results are shown from three independent experiments.
(E) Human leukemia cells were treated as (D). Nuclear (N) and cytoplasmic (C) fractions were collected and detected by antibodies specific to RELA, histone H3 (nuclear control), and α-Tubulin (cytoplasmic control). All error bars represent SD of triplicate analyses. See also Figure S2.
Consistent with observations that phosphorylation of RELA modulates NF-κB transcriptional activity, LPS stimulation induced substantial RELA phosphorylation (Figure 2D) and nuclear localization (Figure 2E) in MLL leukemia cell lines compared to non-MLL cell lines, and was blocked by IKK inhibition. These results demonstrate that human MLL leukemia cells are highly responsive to NF-κB upstream activation, and are dependent on pathway signaling for sustained in vitro growth.
RELA is required for initiation and establishment of MLL-mediated oncogenesis
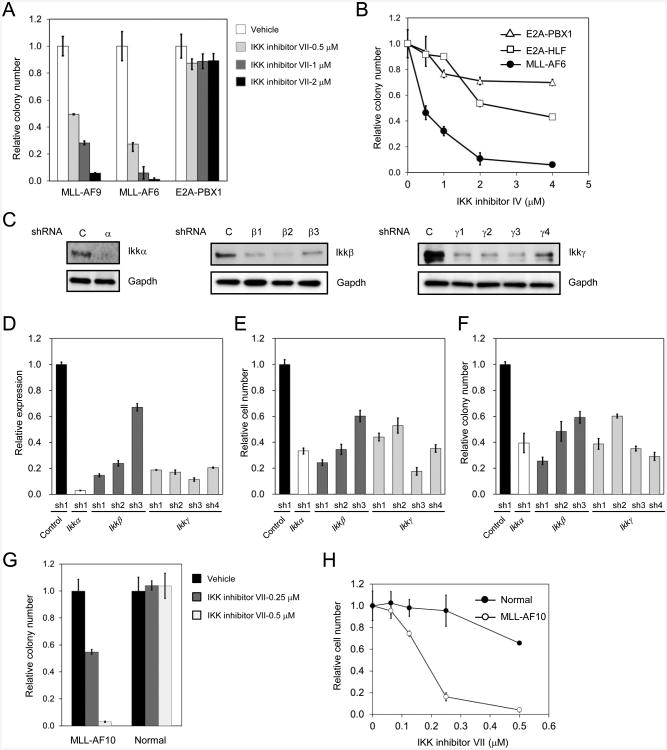
A murine transformation model was employed to further investigate the role of NF-κB in MLL leukemogenesis. Primary mouse myeloid progenitors transduced with MLL oncogenes display enhanced self-renewal in vitro and develop AML in vivo that accurately models the features of human MLL leukemia. A selective requirement for NF-κB signaling in this model was evidenced by sensitivity to IKK inhibitors, which substantially reduced clonogenic potentials of myeloid progenitors transduced by MLL oncogenes (MLL-AF6 and MLL-AF9), as compared to progenitors transduced by other fusion oncogenes (E2A-PBX1 or E2A-HLF) (Figures 3A and 3B). Similarly, genetic reduction of Ikkα, Ikkβ, or Ikkγ levels by shRNA-mediated knockdown (Figures 3C and 3D) resulted in decreased cell growth and clonogenicity in MLL oncogene transduced cells (Figures 3E and 3F) and AML cells (Figures S3A and S3B) but not in E2A-PBX1 transduced cells and normal hematopoietic progenitors (Figure S3C). Growth and clonogenicity suppression correlated with Ikkβ expression knockdown level, and Ikkα and Ikkγ inhibition to a lesser extent. Differential effects of IKK inhibition on colony formation and cell growth were also observed in MLL leukemia cells compared with normal hematopoietic progenitors (Figures 3G and 3H).
Figure 3. Inhibition of IKK signaling suppresses cell growth and colony formation of mouse MLL leukemia cells.
(A and B) Mouse myeloid progenitors transformed by the indicated oncogenes (bone marrow c-kit+ cells transduced with MLL oncogene) were plated in methylcellulose medium with different concentrations of IKK inhibitor VII (A) or IV (B). Colonies were enumerated after 5 days and expressed relative to cells treated with vehicle (DMSO) alone.
(C and D) Mouse myeloid progenitors transformed by MLL-AF9 were stably transduced with lentiviral vectors expressing control shRNA or shRNAs targeting Ikkα, Ikkβ or Ikkγ. Protein (C) and relative mRNA (D) levels were determined by western blot analysis and qRT-PCR, respectively.
(E and F) Mouse MLL-AF9 transduced cells were treated as in panels C and D. Viable cell numbers at day 2 (E) and colony numbers at day 5 (F) were enumerated and expressed relative to the number obtained with control shRNA transduced cells.
(G) Mouse MLL-AF10 leukemia cells and bone marrow c-kit+ cells (normal hematopoietic progenitors) were plated in methylcellulose medium with different concentrations of IKK inhibitor VII. Colonies were enumerated after 5 days and expressed relative to cells treated with vehicle (DMSO) alone.
(H) The growth of mouse MLL-AF10 leukemia cells and c-kit+ cells was assessed after 2 days culture in the absence or presence of the indicated concentrations of IKK inhibitor VII. Results are expressed as relative cell number compared to vehicle treated cells. All error bars represent SD of triplicate analyses. See also Figure S3.
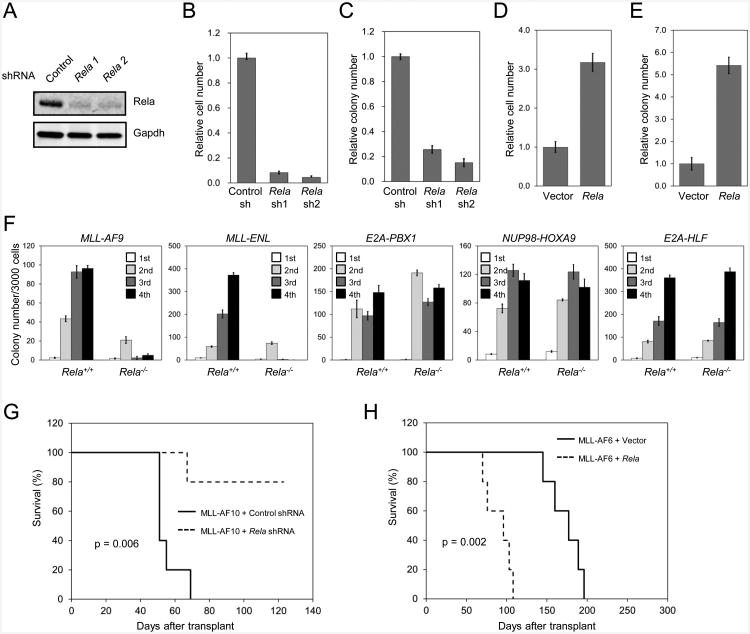
Rela knockdown also induced a pronounced reduction of cell growth and clonogenicity in MLL transformed and leukemia cells but not normal hematopoietic progenitors (Figures 4A-C, S4A-C). Conversely, forced Rela expression increased cell growth and clonogenicity in MLL oncogene transduced cells (Figures 4D and E). To further study the Rela requirement, myeloid progenitors were isolated from fetal livers of Rela+/+; or Rela-/- mouse embryos at embryonic day 13.5 (E13.5), transduced with MLL oncogenes (MLL-AF9 or MLL-ENL) or oncogenes involved in other genetic subtypes of acute leukemia (E2A-PBX1, NUP98-HOXA9 or E2A-HLF), and serially plated in methylcellulose medium to assess self-renewal. Rela-/- cells were unable to sustain colonies induced by MLL oncogenes beyond the second plating yet were fully capable of continuous replating induced by non-MLL oncogenes (Figure 4F), demonstrating a specific requirement for NF-κB at early stages of MLL-mediated oncogenic transformation.
Figure 4. RELA is required for MLL leukemia development.
(A-C) Mouse MLL-AF9 cells were transduced with lentiviral vectors expressing control or Rela shRNAs. Protein levels of Rela were detected by western blot analysis (A). Cell numbers (B) and colony numbers (C) were enumerated after 2 and 5 days, respectively, and expressed relative to the numbers obtained with control shRNA transduced cells.
(D and E) Mouse MLL-AF9 cells were transduced with Rela over-expression or control vectors. Cell numbers (D) and colony numbers (E) were enumerated after 3 and 5 days, respectively, and expressed relative to the numbers obtained with control vector transduced cells.
(F) Hematopoietic progenitors obtained from mouse fetal livers (E13.5) of wild-type (Rela+/+) or Rela knockout (Rela-/-) embryos were transduced with the indicated oncogenes and used for serial myeloid replating assays. Representative results from two independent replicates through four rounds of serial methylcellulose culture are shown as colony number per 3000 cells.
(G) Survival curves are shown for cohorts of mice transplanted with mouse MLL-AF10 leukemia cells (5 × 105) transduced with control or Rela shRNAs (n = 5 each cohort). Acute leukemia was confirmed by peripheral blood leukocyte count and necropsy. Log-rank Test was used for statistical analysis (p = 0.006).
(H) Survival curves are shown for mice transplanted with mouse MLL-AF6 transformed cells (1 × 106) co-transduced with Rela or control vectors (n = 5 each cohort). Log-rank Test was used for statistical analysis (p = 0.002).
All error bars represent SD of triplicate analyses. See also Figure S4.
To assess if Rela is required to establish MLL leukemia in vivo, MLL-AF10 AML and MLL-AF9 transformed cells were transduced with lentiviral constructs expressing control or Rela shRNAs, and transplanted into syngeneic recipients. Rela knockdown resulted in reduced leukemia penetrance and increased survival (Figures 4G and S4D). Conversely, hyper-expression of Rela in MLL-AF6 transduced cells substantially shortened leukemia latency and survival of transplanted mice (Figure 4H), suggesting that Rela serves a critical role in leukemia development. Our results using different MLL fusions further demonstrate a requirement for NF-κB in the broad context of MLL leukemia.
The NF-κB pathway maintains proliferation, survival and differentiation arrest of MLL leukemia cells
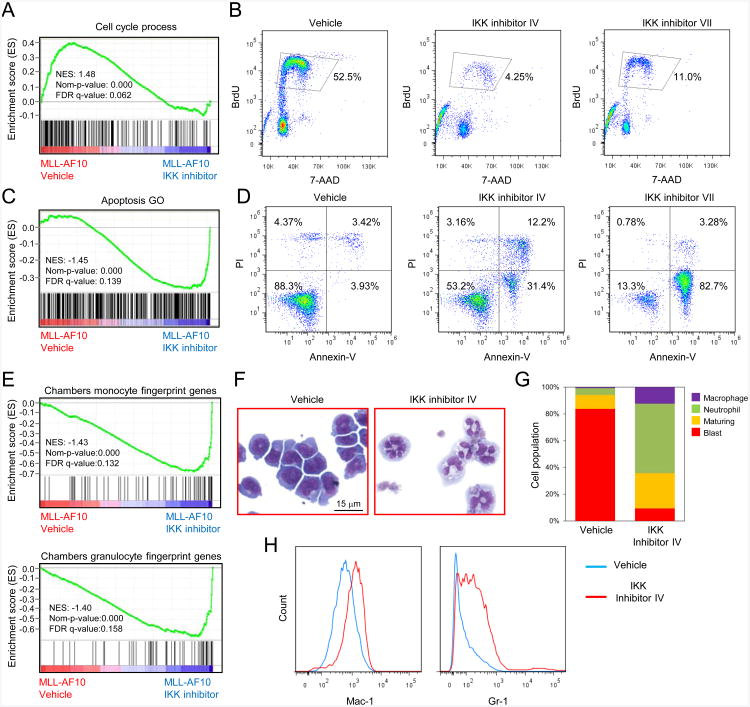
Gene expression profiling indicated that genes with decreased expression in MLL leukemia cells treated with IKK inhibitors for 24 hr were significantly enriched for cell cycle genes (GO: 0022402) (Subramanian et al., 2005), whereas up-regulated genes were enriched for apoptosis (GO: 0006915) and monocyte/granulocyte signature genes (Chambers et al., 2007) (Figures 5A, 5C, 5E, and S5A and Table S3).
Figure 5. IKK inhibition decreases proliferation, increases apoptosis and induces differentiation of MLL leukemia cells.
(A) GSEA plot shows downregulation of cell cycle process related genes in mouse MLL-AF10 leukemia cells treated for 24 hr with IKK inhibitor versus vehicle treated cells.
(B) Mouse MLL-AF10 leukemia cells were cultured in the presence of 2 μM IKK inhibitor IV or 0.5 μM IKK inhibitor VII for 2 days, and BrdU incorporation was quantified by flow cytometry analysis.
(C) GSEA plot shows upregulation of apoptosis related genes in mouse MLL-AF10leukemia cells treated for 24 hr with IKK inhibitor versus vehicle treated cells.
(D) Mouse MLL-AF10 leukemia cells were cultured in the presence of 1 μM IKK inhibitor IV or 0.5 μM IKK inhibitor VII for 3 days. The annexin-V positive and PI negative populations constitute early apoptotic cells.
(E) GSEA plots show upregulation of monocyte or granulocyte fingerprint genes in mouse MLL-AF10 leukemia cells treated for 24 hr with IKK inhibitor versus vehicle treated cells.
(F) Light microscopy of May-Grunwald/Giemsa-stained mouse MLL-AF10 leukemia cells after 2 days of IKK inhibitor IV treatment (2 μM).
(G) Quantification of leukemia cell populations with indicated morphological features after 2 days of IKK inhibitor IV treatment (2 μM).
(H) Flow cytometry analysis of Mac-1 and Gr-1 surface expression by mouse MLL-AF10 leukemia cells after 2 days of 2 μM IKK inhibitor IV treatment.
See also Figure S5 and Table S3.
Consistent with gene expression profiles, FACS analysis demonstrated a marked reduction of proliferation following 2 day IKK inhibitor treatment of mouse (Figures 5B and S5B) and human (Figures S5C and S5D) MLL leukemia cells, whereas non-MLL leukemia cells (K562) were minimally affected (Figures S5C and S5D). Annexin-V staining confirmed the presence of apoptosis at 3 days (Figure 5D). Notably, the differentiation block of MLL leukemia cells was reversed by IKK inhibitor IV treatment (2 nM), which induced morphological maturation (Figures 5F and 5G) associated with increased expression of myeloid differentiation antigens Mac-1 and Gr-1 (Figure 5H) within 2 days. Similar results were found by treating with IKK inhibitor VII (data not shown), different concentration of IKK inhibitor IV (data not shown), or in MLL-AF9 transduced cells (Figures S5E and S5F). These results phenocopy the loss of functional MLL fusion protein complex and indicate that IKK/NF-κB signaling serves a crucial role in supporting proliferation, sustaining survival, and arresting differentiation of MLL transformed and leukemia cells.
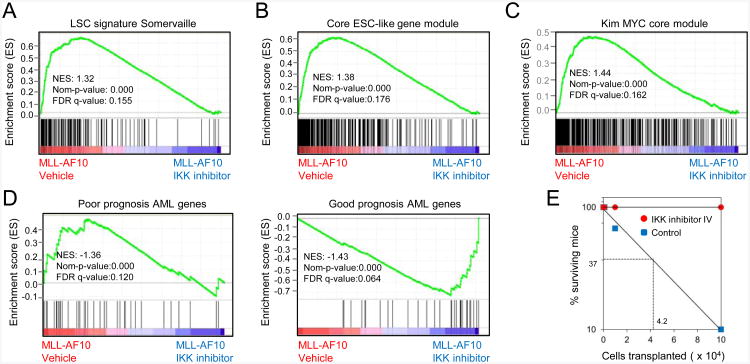
IKK/NF-κB signaling sustains LSC potential
In murine AML induced by MLL-AF9, expression of NF-κB target genes is enriched in leukemic granulocyte/macrophage progenitors (L-GMPs), which have features of LSCs, compared with normal GMPs (Figure S6A and Table S4) (Chen et al 2008) consistent with a possible role of the NF-κB pathway in LSCs. In IKK inhibitor-treated mouse and human leukemia cells genes showing decreased expression were significantly enriched for a transcriptional signature previously shown to distinguish LSCs from non-self-renewing leukemia cells and involved in LSC maintenance (Somervaille et al., 2009) (Figures 6A and S6B and Table S4). A transcriptional program that MLL LSCs share with embryonic stem cells (ESCs) (Somervaille et al., 2009) was also down-regulated in treated cells (Figures 6B and S6C and Table S4). The ESC-like program overlaps with a transcriptional program controlled by the MYC oncoprotein (Kim et al., 2010), and MYC core module genes (the set of genes bound by the combination of MYC, MAX, MYCN, DMAP1, E2F1, E2F4, and ZFX in murine embryonic stem cells) (Kim et al., 2010) were also down-regulated in IKK inhibitor-treated cells (Figure 6C and Table S4). Since poor prognosis in a diverse set of human malignancies is associated with expression of an ESC-like program (Wong et al., 2007; Ben-Porath et al., 2008; Yagi et al., 2003), its down-regulation by IKK inhibition has potential therapeutic implications. Indeed, a gene set associated with poor prognosis in pediatric AML was down-regulated in IKK inhibitor-treated cells concomitant with up-regulation of a good prognosis gene set (Figures 6D and S6D and Table S4).
Figure 6. IKK inhibition reduces the LSC population.
(A-C) GSEA plots show downregulation of MLL LSC maintenance signature genes (A), core ESC-like gene module (B), and MYC core module genes (C) in mouse MLL-AF10 leukemia cells treated for 24 hr with IKK inhibitor versus vehicle treated cells.
(D) GSEA plots show downregulation of poor prognosis AML genes and upregulation of good prognosis AML genes in mouse MLL-AF10 leukemia cells treated with IKK inhibitor versus vehicle treated cells.
(E) Limit-dilution analyses show the estimated cell number of transplanted mouse MLL-AF10 leukemia cells required to initiate AML in sublethally irradiated recipient mice (n = 3 for each cell dose). MLL-AF10 leukemia cells were pretreated with IKK inhibitor IV (2 μM) or control vehicle for 2 days. Viable cells used for transplantation were confirmed by Trypan Blue staining. Mice were followed for 190 days, with the longest disease latencies being 84 days.
See also Figure S6 and Table S4.
Functional studies confirmed that IKK inhibition negatively impacts LSCs. Pretreatment of MLL-AF10 and MLL-AF9 AML cells with IKK inhibitor for 2 days not only significantly reduced cell growth (Figure S6E) but resulted in a substantial reduction in colony forming cells (CFCs) in the remaining viable cell population (Figure S6F), which correlate with LSCs (Somervaille and Cleary, 2006). Limit-dilution secondary transplantation of inhibitor-pretreated AML cells showed at least a 10-fold reduction in LSC frequency (Figure 6E). Thus, the NF-κB pathway is critically required for maintenance of LSCs in MLL leukemia.
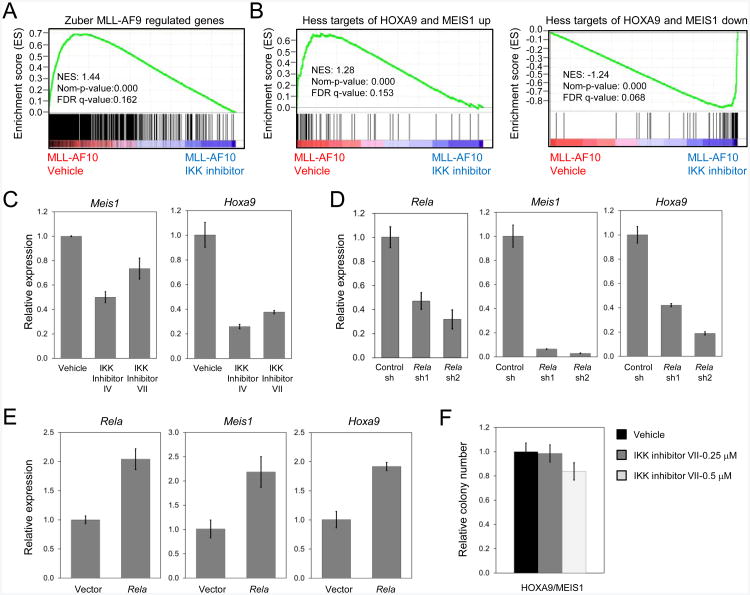
RELA controls essential gene expression programs in MLL leukemia by direct regulation of MEIS1 and HOXA9
Genes showing decreased expression in MLL leukemia cells treated with IKK inhibitors were significantly enriched for genes previously shown to be regulated by the MLL oncoprotein (gene sets down-regulated upon MLL-AF9 withdrawal in murine MLL-AF9;NrasG12D AML cells) (Zuber et al., 2011a) or for MLL-ENL direct targets (Wang et al., 2011) (Figures 7A and S7A and Table S5) (other published direct target gene sets were also down-regulated in IKK inhibitor-treated cells but the enrichment p-value did not achieve statistical significance). Transcripts for primary MLL target genes Hoxa9 and Meis1 were significantly reduced in IKK inhibitor-treated leukemia cells (Figure 7C), which were enriched for HOXA9 and MEIS1 down-regulated gene sets (Hess et al., 2006) (Figures 7B and S7B and Table S5). Rela(RELA) knockdown in MLL-transduced mouse progenitors or human leukemia cell lines also markedly reduced Hoxa9(HOXA9) and Meis1(MEIS1) transcript levels (Figures 7D, S7C, and S7D), whereas forced Rela expression substantially increased their levels (Figure 7E). These results indicated that NF-κB upregulates HOXA9/MEIS1 expression. Consistent with this suggestion, myeloid progenitors transformed by forced expression of Hoxa9 and Meis1 to bypass the MLL oncoprotein were relatively resistant to IKK inhibitor VII (Figures 3G and 7F).
Figure 7. IKK/NF- κB signaling regulates Meis1 and Hoxa9gene expression.
(A) GSEA plot shows downregulation of MLL-AF9 regulated genes in mouse MLL-AF10leukemia cells treated with IKK inhibitor versus vehicle treated cells.
(B) GSEA plots show downregulation of HOXA9 and MEIS1 upregulated target genes (left) and upregulation of HOXA9 and MEIS1 downregulated target genes (right) in mouse MLL-AF10 leukemia cells treated with IKK inhibitor versus vehicle treated cells.
(C) Mouse MLL-AF10 leukemia cells were cultured in the absence or presence of IKK inhibitors (IV or VII) for 2 days. Meis1 or Hoxa9 transcripts were quantified by qRT-PCR, and expressed relative to vehicle treated cells.
(D) Mouse MLL-AF9 transformed cells transduced with lentiviral vectors expressing control or Rela shRNAs were assessed for Rela, Meis1 or Hoxa9 transcript levels by qRT-PCR. Results are displayed relative to control shRNA-transduced cells.
(E) Mouse MLL-AF9 transformed cells transduced with retroviral vector expressing Rela or control vector were assessed for Rela, Meis1 or Hoxa9 transcript levels by qRT-PCR. Results are displayed relative to control vector-transduced cells.
(F) Mouse HOXA9/MEIS1 leukemia cells were plated in methylcellulose medium with different concentrations of IKK inhibitor VII. Colonies were enumerated after 5 days and expressed relative to cells treated with vehicle (DMSO) alone. All error bars represent SD of triplicate analyses.
See also Figure S7 and Table S5.
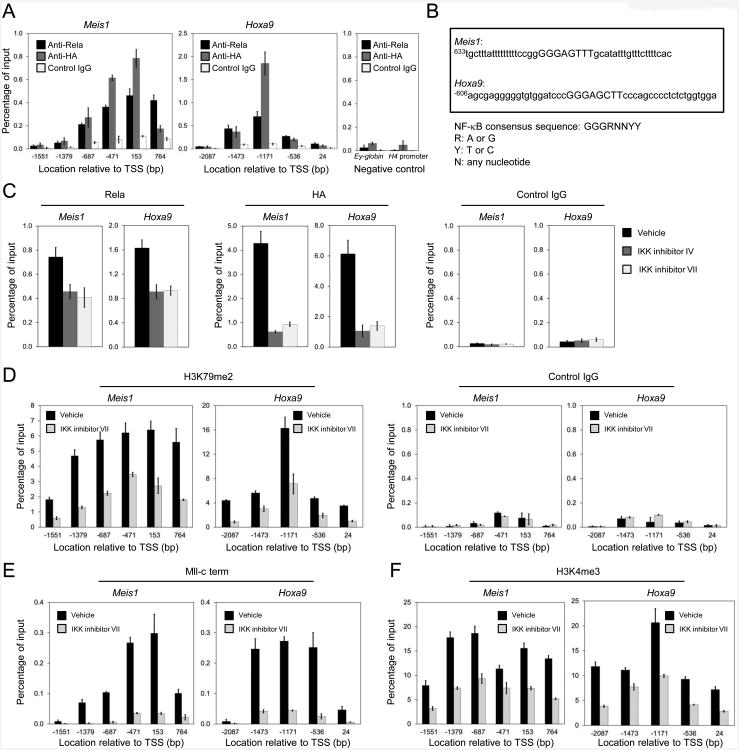
Promoter occupancy and epigenetic roles of MLL proteins are dependent on IKK/NF-κB signaling
In support of a direct role for NF-κB in Hoxa9 and Meis1 transcriptional regulation, chromatin immunoprecipitation (ChIP) demonstrated Rela occupancy that peaked within specific regions of the Hoxa9 and Meis1 promoters in proximity of consensus NF-κB binding sites (Figures 8A and 8B) in mouse AML cells. The Rela occupancy profile was similar to that of MLL-AF10 detected with anti-HA antibody specific for HA-tagged MLL-AF10 in AML cells (Figure 8A). The observed Rela occupancy was reduced by treatment with IKK inhibitors and therefore dependent on IKK/NF-κB signaling (Figure 8C). Notably, IKK inhibition also substantially reduced promoter occupancy of MLL-AF10 (Figure 8C) indicating that chromatin association of the MLL oncoprotein was highly dependent on IKK/NF-κB signaling. The marked reduction of promoter-associated MLL-AF10 occurred despite stable cellular MLL-AF10 protein levels in treated cells (Figure S8). IKK inhibition also reduced histone H3K79 dimethylation (Figure 8D) consistent with reduced occupancy of MLL-AF10, which interacts with the DOT1L H3K79 histone methyltransferase. Aberrant recruitment of the latter promotes broad spreading of the H3K79 dimethyl mark in MLL target genes characteristic of “epigenetic lesions” (Guenther et al., 2008). IKK inhibitor treatment also substantially reduced promoter occupancy of wild-type MLL (Figure 8E) and the level of H3K4 trimethylation (Figure 8F) on the Meis1 and Hoxa9 genes. Thus, NF-κB signaling is specifically required for promoter association of MLL proteins and maintenance of epigenetic marks necessary for transcription of key MLL target genes in leukemia cells.
Figure 8. Promoter occupancies of MLL wild-type and oncoprotein are dependent on IKK/NF- κB signaling.
(A) ChIP was performed on mouse MLL-AF10 leukemia cells with antibodies against Rela, HA (MLL-AF10), and control IgG. Genomic regions amplified by qPCR are indicated relative to the transcription start site (TSS) on Meis1 and Hoxa9 genes. Ey-globin and H4 promoter primers were used for negative controls.
(B) Promoter sequence of Meis1 and Hoxa9. NF-κB consensus sequence is indicated in capital letters.
(C) Mouse MLL-AF10 leukemia cells were cultured in the absence or presence of IKK inhibitors (IV or VII) for 2 days. ChIP was performed using antibodies against Rela, HA (MLL-AF10), and control IgG.
(D-F) Mouse MLL-AF10 leukemia cells were cultured in the absence or presence of IKK inhibitor VII for 2 days. ChIP was performed using antibodies against H3K79me2 and control IgG (D), Mll-c term (E), and H3K4me3 (F) and qPCR primers amplifying the indicated genomic regions. All error bars represent SD of triplicate analyses.
See also Figure S8.
Discussion
Using pharmacologic, biochemical, genetic and genomic approaches, we demonstrate the requirement of IKK/NF-κB signaling to maintain MLL-mediated transformation in vitro and in vivo, and its important role in promoting leukemia cell proliferation, survival, differentiation arrest, as well as LSC potential. This strong phenocopy with functions of the MLL oncoprotein and associated cofactors reflects that IKK/NF-κB signaling sustains the MLL-dependent LSC gene expression program, and is required for promoter occupancy of the MLL oncoprotein at key target genes HOXA9 and MEIS1. Thus, NF-κB constitutes an upstream signaling pathway to converge on key MLL subordinate genes, in support of a model whereby NF-κB and MLL oncoproteins cooperate to respectively initiate and aberrantly elongate transcription of essential target genes in MLL leukemia pathogenesis.
By employing a non-biased genomic screening approach targeting phosphoregulators, we identified the crucial roles of multiple kinases/phosphatases involved in NF-κB signaling in MLL leukemia cells, including IKKα, IKKβ, IKKγ, PPP4C, IRAK3, and PLK1. In the canonical NF-κB signaling pathway, the IKKα/β/γ complex phosphorylates κBα and triggers its degradation, thereby liberating the NF-κB heterodimer and inducing its nuclear translocation (Karin and Ben-neriah, 2000). PPP4C and IRAK3 also function upstream on the pathway, as their over-expression has been shown to activate NF-κB mediated transcription (Yeh et al., 2004; Wesche et al., 1999). Conversely, PLK1 is downstream since the NF-κB subunit RELA transcriptionally activates its promoter through direct binding (Lin et al., 2011). Inactivation or knockdown of PLK1 showed reduced cell growth in human and mouse MLL cells (data not shown). Notably, PLK1 is also an MLL-AF9 regulated gene (Zuber et al., 2011a) further demonstrating the potential for integrated transcriptional roles of MLL oncoproteins with NF-κB. The identification of multiple direct hits on the NF-κB pathway strongly reinforces its role in MLL leukemia pathogenesis and suggests several alternative strategies to target NF-κB activity in MLL leukemia therapy.
Previous studies have implicated NF-κB in a subset of AML, particularly in chemoresistance and regulation of cell survival (Jiang et al, 2012). FLT3 overexpression and PI3-K signaling contribute to activation of the NF-κB pathway in AML (Takahashi et al, 2005; Birkenkamp et al, 2004), and NF-κB participates in deregulation of the Sp1/NF-κB/HDAC/miR-25b signaling network that drives KIT over-expression in some AMLs (Liu et al, 2010). Furthermore, the AML1-ETO fusion protein lacks the ability of wild type AML1 to attenuate NF-κB resulting in activated NF-κB signaling compared with MLL leukemia cells (Nakagawa et al, 2011). In contrast to the latter, our broader studies using several different experimental systems compared to Nakagawa et al. clearly demonstrate and provide a mechanistic basis for the strong dependence of MLL leukemia cells on NF-κB signaling.
Various signaling pathways and kinases have been implicated in MLL leukemia pathogenesis, including FLT3, which is highly expressed in a subset of MLL-rearranged acute lymphoblastic leukemia (ALL) (Stam et al, 2005), and the AMPK pathway, which directly contributes to the survival of ALL cells with MLL translocations (Accordi et al., 2013). Glycogen synthase kinase 3 (GSK3) supports leukemia cell proliferation and transformation by facilitating the transcriptional activity of HOX proteins (Wang et al., 2008; Wang et al, 2010). However, these studies did not illuminate the upstream signaling pathways or activators that may recruit, retain or cooperate with MLL oncoproteins at subordinate genes.
Our studies demonstrate that IKK/NF-κB signaling impinges on the MLL-dependent transcriptional program and serves a major role in its maintenance and deregulation. MLL target genes previously shown to be regulated by MLL-AF9 are significantly down-regulated in IKK inhibitor treated MLL leukemia cells. This includes Hoxa9 and Meis1, which are essential for leukemia pathogenesis, as well as the transcriptional program subordinate to these key MLL target genes. Thus, in addition to the actions of MLL oncoproteins, NF-κB signaling is also necessary to sustain key MLL target gene expression. Identification of Rela occupancy on Hoxa9 and Meis1 promoters in proximity of NF-κB binding sites (Cartharius et al., 2005) suggests a direct function of NF-κB in their transcriptional regulation.
MLL oncoproteins facilitate aberrant transcription of their target genes by recruitment of elongation factors in conjunction with epigenetic cofactors as opposed to functioning as classical activators to recruit RNA pol II. P-TEFb in particular phosphorylates substrates that otherwise keep RNA pol II paused on primed promoters. Despite extensive implication of MLL oncoproteins in aberrant elongation, the conventional transcriptional activators that may functionally cooperate to recruit or retain MLL fusions at primed promoters have not been defined. Our studies demonstrate that IKK/NF-κB signaling is necessary for promoter occupancy of the MLL oncoprotein and maintenance of histone marks on MLL target genes in leukemia cells. This suggests that NF-κB may initiate the promoter as a pre-requisite for recruitment and/or retention of MLL and associated factors that affect aberrant elongation of the stalled polymerase.
A potential role for NF-κB in recruitment of wild-type MLL to chromatin has recently been reported in other promoter contexts (Wang et al, 2012). Consistent with this, our results also demonstrate the importance of IKK/NF-κB signaling in sustaining H3K4 trimethylation of Hoxa9 and Meis1 genes in AML cells and recruitment of wild-type MLL, whose cooperation with MLL fusions is essential for leukemogenesis (Thiel et al., 2010). Although wild-type MLL has been reported to associate with NF-κB, physical association of RELA or p50 with MLL-fusion proteins was not detected in AML cells by immunoprecipitation (IP) western blot analysis (data not shown) suggesting that NF-κB is not a component of the MLL fusion protein complex and does not directly tether the MLL oncoprotein to chromatin. Although the mechanism underlying their co-dependent functions in MLL leukemia requires further study, one possibility is that establishment of an appropriate chromatin context by NF-κB allows for subsequent binding of the MLL oncoprotein complex given that several of its integral components contain motifs that bind epigenetically modified chromatin or DNA. Alternatively, functional interactions may occur through shared cofactors such as BRD4, a bromodomain protein that positively regulates P-TEFb (Jang et al., 2005) and is present in the MLL oncoprotein higher-order complex. BRD4 not only associates with acetylated histones, but also binds to acetylated NF-κB and coactivates its transcriptional function (Huang et al., 2009). This functional overlap may contribute to the substantial efficacy of therapeutically targeting BRD4 in pre-clinical models of MLL leukemia (Zuber et al., 2011 b).
Small molecule inhibitors targeting the NF-κB pathway are in development, and some are in preclinical testing. CHS828, which impairs LPS induced NF-κB nuclear translocation and transcriptional activation, is currently in phase I/II clinical trial (Hjarnaa et al., 1999). Also in clinical testing is bortezomib, a potent 26S proteasome inhibitor that indirectly inhibits NF-κB activity by preventing κBα proteasomal degradation (Dai et al., 2011). Recent studies, however, indicate that bortezomib can induce NF-κB, rather than inhibit, through calpain (Li et al., 2010) or caspase-independent (Hideshima et al., 2009) mechanisms. Activation of NF-κB by bortezomib has been observed in various tumor types (Hideshima et al., 2009; Li et al., 2010) and normal PBMCs (Hideshima et al., 2009). Other studies suggest that NF-κB inhibition may not be a key mechanism of bortezomib's anti-cancer activity (Chen et al., 2011).
Our studies demonstrate that NF-κB serves a critical role in sustaining MLL LSC potential. The expression level of Rela quantitatively correlates with LSC activity and leukemia latency likely reflecting its molecular function as a transcriptional regulator of MEIS1, which serves as a rate-limiting regulator of LSC potential and leukemia latency (Wong et al., 2007). LSCs constitute a subpopulation of leukemia cells with unlimited self-renewal and whose acquired drug-resistant properties are responsible for relapse, and therefore represent a crucial target for therapeutic intervention. However, targeting LSCs while sparing hematopoietic stem cells (HSCs) is challenging due to similarities in their biological and molecular properties. Constitutive activation of NF-κB signaling has been observed in primitive AML cells but not in normal primitive hematopoietic cells (Guzman, 2001) suggesting the possibility that dependence on NF-κB may distinguish LSCs from HSCs. Consistent with this notion, indirect inhibition of NF-κB by proteosome blockade induces LSC apoptosis while leaving normal HSCs viable (Guzman et al., 2002). Our studies are consistent with these earlier observations, and demonstrate the differential sensitivity of normal progenitors versus MLL LSCs. The LSC transcriptional program is regulated by NF-κB, which is required for promoter occupancy of MLL proteins and maintenance of subordinate histone marks in chromatin of crucial target genes (Hoxa9 and Meis1). This provides a mechanistic basis for enhanced dependence of MLL leukemia on the NF-κB pathway compared with other genetic subtypes of AML and suggests that targeting the pathway may be particularly efficacious in MLL leukemia.
Experimental Procedures
Inhibitors
IKK inhibitors III, IV and VII (401480, 401481 and 401486, respectively, EMD Chemicals) were dissolved in dimethyl sulfoxide (DMSO) and used at the indicated concentrations.
Cellular fractionation and western blot analysis
Cells were washed with PBS and then lysed in hypotonic buffer (10 mM Hepes (pH 7.5), 25 mM KCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM PMSF, and protease inhibitors). Nuclei were separated from cytoplasmic proteins by centrifugation (1,000 × g) and resuspended in cell extraction buffer (50 mM Tris (pH 7.5), 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, 0.1% SDS, 0.5% deoxycholate, 1 mM PMSF and protease inhibitors) for 30 min on ice. Nuclear fraction was separated from insoluble chromatin fraction by centrifugation (17,000 × g). Western blot analysis was performed as previously described (Yokoyama et al., 2004). The blots were reacted with antibodies specific to RELA (ab7970, Abcam), GAPDH (G9545, Sigma), α-Tubulin (A01410, GenScript), HA (ab9110, Abcam), histone H3 (ab1791, Abcam), Ikkα (2682, Cell Signaling), Ikk β (MAB7155, R&D Systems), Ikkγ (ab137363, Abcam), followed by IRDye secondary antibodies (LI-COR Biosciences) or peroxidase-conjugated secondary antibodies. Images were detected by an Odyssey imaging system (Odyssey Fc, LI-COR Biosciences) or ECL system (GE Healthcare).
In vivo leukemogenesis assays
Mouse MLL-AF10 and MLL-AF9 leukemia cells were transduced with control or Rela shRNAs. After puromycin selection, cells (5 × 105) were transplanted intravenously into sublethally irradiated (450 rads) C57BL/6 mice.
Mouse myeloid cells transformed by MLL-AF6 were transduced with empty vector or Rela expression vector. After puromycin selection, cells (1 × 106) were transplanted intravenously into sublethally irradiated (450 rads) C57BL/6 mice.
All experiments on mice in this study were performed with the approval of and in accordance with the Stanford University Administrative Panel on Laboratory Animal Care.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (Yokoyama et al., 2005) using primary antibodies specific for HA tag (ab9110, Abcam), Rela (ab7970, Abcam), Mll-c term (05-765, Millipore), H3K4me3 (ab8580, Abcam), H3K79me2 (ab3594, Abcam), or control IgG antibodies (ab46540, Abcam). Immuno-complexes were precipitated using Protein A Dynabeads (10001D, Invitrogen) or Protein G Dynabeads (10003D, Invitrogen). Quantitative real-time PCR was performed on the precipitated DNA using primers flanking the Hoxa9, Meis1, Ey-globin and H4 promoter sites. The relative values to input were determined using SYBR green.
Phosphoflow analysis
Leukemia cell lines were serum-starved overnight with 0.1% FBS contained RPMI 1640 medium, and stimulated with 10 μg/ml LPS (L3024-5mg, Sigma) for 30 min with or without IKK inhibitor VII (1 μM) pretreatment for 15 min. Cells were subsequently fixed with 1.5% paraformaldehyde and permeabilized with 100% ice-cold methanol as described (Krutzik and Nolan, 2003). Conjugated antibodies to intracellular proteins P-RELA (S529)-Alexa 488 (558421, BD Biosciences) and P-RELA (S536)-PE (5733, Cell Signaling Technology) were used for staining. Flow cytometry data were acquired on a LSR Fortessa using FACS Diva Software (BD Biosciences) and analyzed using FlowJo (TreeStar Software).
Microarray and GSEA analyses
RNA used for microarray analysis was prepared using an RNeasy Mini kit (QIAGEN). Gene 1.0ST arrays were used according to the manufacturer's instructions. Normalizations of CEL file data were performed using dChip 2010 (DNA-Chip Analyzer) software (Li and Wong, 2001). GSEA analyses were performed using GSEA v2.07 software (http://www.broad.mit.edu/gsea) with 1000 data permutations. Enriched gene sets are selected based on statistical significance (FDR q-value <0.25 and normalized p-value <0.05).
Supplementary Material
Highlights.
IKK/NF- κB signaling sustains the MLL leukemia stem cell program.
RELA occupies the HOXA9 and MEIS1 promoters to regulate their expression.
IKK/NF- κB is required for MLL protein retention on crucial target genes.
Epigenetic regulation by MLL oncoproteins depends on NF- κB.
Significance.
MLL is a large multifunctional epigenetic regulator whose transcriptional activity is corrupted by protein fusions with various partner proteins. Extensive studies have characterized the transcriptional and epigenetic perturbations caused by MLL mutations, however little is known about the upstream signaling pathways and factors that may cooperate with MLL oncoproteins to deregulate critical target genes in leukemia pathogenesis. In a forward genetic screen we identified and validated a crucial role for NF-κB in MLL-mediated transformation. IKK/NF-κB signaling is required for MLL oncoprotein chromatin occupancy and epigenetic function within key target genes of the MLL transcriptional program that sustains leukemia stem cell potential. Therefore, targeting the NF-κB pathway may be particularly efficacious in MLL leukemia compared with other genetic subtypes of AML.
Acknowledgments
We thank Maria Ambrus, Cita Nicolas, and Kevin S. Smith for technical assistance, Norm Cyr for graphical assistance, Howard Y. Chang for RELA heterozygous mice, Beverly S. Mitchell for OCI-AML3 cells, Alejandro Sweet-Cordero for comments on bioinformatics analysis, Wendy J. Fantl for help on phosphoflow analysis, and members of the Cleary lab for helpful discussions. We acknowledge support from the Children's Health Initiative of the Packard Foundation and PHS grant CA116606. H.-P.K. was supported by PHS Grants T32-CA09302 and T32-CA09151, awarded by the National Cancer Institute, DHHS and Dean's Postdoctoral Fellowship at the Stanford School of Medicine; J.D.-A. was supported by the German Research Foundation (DFG, ref. DU 1287/2-1); S.H.K.W. was supported by the Alex's Lemonade Stand Foundation for Childhood Cancer; and D.-F.L. was supported by New York Stem Cell Foundation -Druckenmiller Fellowship.
Footnotes
Accession Number: Microarray raw data are available for download at Gene Expression Omnibus (http://ncbi.nlm.nih.gov/geo) accession number GSE46252.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Accordi B, Galla L, Milani G, Curtarello M, Serafin V, Lissandron V, Viola G, Te Kronnie G, De Maria R, Petricoin EF, et al. AMPK inhibition enhances apoptosis in MLL-rearranged pediatric B-acute lymphoblastic leukemia cells. Leukemia. 2013;27:1019–1027. doi: 10.1038/leu.2012.338. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1. L Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenkamp KU, Geugien M, Schepers H, Westra J, Lemmink HH, Vellenga E. Constitutive NF-kappaB DNA-binding activity in AML is frequently mediated by a Ras/PI3-K/PKB-dependent pathway. Leukemia. 2004;18:103–112. doi: 10.1038/sj.leu.2403145. [DOI] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff a, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Boles NC, Lin KYK, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant Aa, Sirin O, Weksberg DC, et al. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11:239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Kumar AR, Hudson WA, Li Q, Wu B, Staggs RA, Lund EA, Sam TN, Kersey JH. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13:432–440. doi: 10.1016/j.ccr.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Chen S, Wang L, Pei XY, Kramer LB, Dent P, Grant S. Bortezomib interacts synergistically with belinostat in human acute myeloid leukaemia and acute lymphoblastic leukaemia cells in association with perturbations in NF-κB and Bim. Br J Haematol. 2011;153:222–235. doi: 10.1111/j.1365-2141.2011.08591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, Schifano E, Booth J, van Putten W, Skrabanek L, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman ML, Neerin SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, Luger SM, Jordan CT. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood. 2001;98:2301–2307. doi: 10.1182/blood.v98.8.2301. [DOI] [PubMed] [Google Scholar]
- Guzman ML, Swiderski CF, Howard DS, Grimes Ba, Rossi RM, Szilvassy SJ, Jordan CT. Preferential induction of apoptosis for primary human leukemic stem cells. Proc Natl Acad Sci USA. 2002;99:16220–16225. doi: 10.1073/pnas.252462599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Hess JL, Bittner CB, Zeisig DT, Bach C, Fuchs U, Borkhardt A, Frampton J, Slany RK. c-Myb is an essential downstream target for homeobox-mediated transformation of hematopoietic cells. Blood. 2006;108:297–304. doi: 10.1182/blood-2005-12-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T, Ikeda H, Chauhan D, Okawa Y, Raje N, Podar K, Mitsiades C, Munshi NC, Richardson PG, Carrasco RD, et al. Bortezomib induces canonical nuclear factor-kappaB activation in multiple myeloma cells. Blood. 2009;114:1046–1052. doi: 10.1182/blood-2009-01-199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjarnaa PV, Jonsson E, Latini S, Dhar S, Larsson R, Bramm E, Skov T. CHS 828, a novel pyridyl cyanoguanidine with potent antitumor activity in vitro and in vivo. Cancer Res. 1999;59:5751–5757. [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jiang XJ, Huang KK, Yang M, Qiao L, Wang Q, Ye JY, Zhou HS, Yi ZS, Wu FQ, Wang ZX, et al. Synergistic effect of panobinostat and bortezomib on chemoresistant acute myelogenous leukemia cells via AKT and NF-κB pathways. Cancer Lett. 2012;326:135–142. doi: 10.1016/j.canlet.2012.07.030. [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzik PO, Nolan GP. Intracellular phospho-protein staining techniques for flow cytometry: monitoring single cell signaling events. Cytometry A. 2003;55:61–70. doi: 10.1002/cyto.a.10072. [DOI] [PubMed] [Google Scholar]
- Lavau C, Szilvassy SJ, Slany R, Cleary ML. Immortalization and leukemic transformation of a myelomonocytic precursor by retrovirally transduced HRX-ENL. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Su J, Ang YS, Carvajal-Vergara X, Mulero-Navarro S, Pereira CF, Gingold J, Wang HL, Zhao R, Sevilla A, et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen S, Yue P, Deng X, Lonial S, Khuri FR, Sun SY. Proteasome inhibitor PS-341 (bortezomib) induces calpain-dependent IkappaB(alpha) degradation. J Biol Chem. 2010;285:16096–16104. doi: 10.1074/jbc.M109.072694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Zhang Y, Pan QJ, Yang H, Shi ZZ, Xie ZH, Wang BS, Hao JJ, Zhang TT, Xu X, et al. PLK1 Is transcriptionally activated by NF-κB during cell detachment and enhances anoikis resistance through inhibiting β-catenin degradation in esophageal squamous cell carcinoma. Clin Cancer Res. 2011;17:4285–4295. doi: 10.1158/1078-0432.CCR-10-3236. [DOI] [PubMed] [Google Scholar]
- Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ, Hickey CJ, Yu J, Becker H, Maharry K, et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Shimabe M, Watanabe-Okochi N, Arai S, Yoshimi A, Shinohara A, Nishimoto N, Kataoka K, Sato T, Kumano K, et al. AML1/RUNX1 functions as a cytoplasmic attenuator of NF-κB signaling in the repression of myeloid tumors. Blood. 2011;118:6626–6637. doi: 10.1182/blood-2010-12-326710. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Somervaille TCP, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Somervaille TCP, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, Chang HY, Shurtleff Sa, Downing JR, Cleary ML. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam RW, den Boer ML, Schneider P, Nollau P, Horstmann M, Beverloo HB, van der Voort E, Valsecchi MG, de Lorenzo P, Sallan SE, et al. Targeting FLT3 in primary MLL-gene-rearranged infant acute lymphoblastic leukemia. Blood. 2005;106:2484–2490. doi: 10.1182/blood-2004-09-3667. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Harigae H, Ishii KK, Inomata M, Fujiwara T, Yokoyama H, Ishizawa K, Kameoka J, Licht JD, Sasaki T, et al. Over-expression of Flt3 induces NF-kappaB pathway and increases the expression of IL-6. Leukemia Res. 2005;29:893–899. doi: 10.1016/j.leukres.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20:563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, Zhang H, Liu Z, Ernst P, Koretzky Ga, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QF, Wu G, Mi S, He F, Wu J, Dong J, Luo RT, Mattison R, Kaberlein JJ, Prabhakar S, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117:6895–6905. doi: 10.1182/blood-2010-12-324699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu K, Li S, Liao Y, Du R, Zhang X, Shu HB, Guo AY, Li L, Wu M. MLL1, a H3K4 methyltransferase, regulates the TNFα-stimulated activation of genes downstream of NF-κB. J Cell Sci. 2012;125:4058–4066. doi: 10.1242/jcs.103531. [DOI] [PubMed] [Google Scholar]
- Wang Z, Iwasaki M, Ficara F, Lin C, Matheny C, Wong SHK, Smith KS, Cleary ML. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer Cell. 2010;17:597–608. doi: 10.1016/j.ccr.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TCP, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche H, Gao X, Li X, Kirschning CJ, Stark GR, Cao Z. IRAK-M is a novel member of the Pelle/interleukin-1 receptor-associated kinase (IRAK) family. J Biol Chem. 1999;274:19403–19410. doi: 10.1074/jbc.274.27.19403. [DOI] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TCP, So CWE, So CWE, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Morimoto A, Eguchi M, Hibi S, Sako M, Ishii E, Mizutani S, Imashuku S, Ohki M, Ichikawa H. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849–1856. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Yeh PY, Yeh KH, Chuang SE, Song YC, Cheng AL. Suppression of MEK/ERK signaling pathway enhances cisplatin-induced NF-kappaB activation by protein phosphatase 4-mediated NF-kappaB p65 Thr dephosphorylation. J Biol Chem. 2004;279:26143–26148. doi: 10.1074/jbc.M402362200. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17:198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Somervaille TCP, Smith KS, Rozenblatt-rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J Biol Chem. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Rappaport AR, Luo W, Wang E, Chen C, Vaseva AV, Shi J, Weissmueller S, Fellman C, Taylor MJ, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 2011a;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011b;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.