Abstract
Cap'n'collar (CNC) family basic leucine zipper transcription factors play crucial roles in the regulation of mammalian gene expression and development. To determine the in vivo function of the CNC protein Nrf3 (NF-E2-related factor 3), we generated mice deficient in this transcription factor. We performed targeted disruption of two Nrf3 exons coding for CNC homology, basic DNA-binding, and leucine zipper dimerization domains. Nrf3 null mice developed normally and revealed no obvious phenotypic differences compared to wild-type animals. Nrf3−/− mice were fertile, and gross anatomy as well as behavior appeared normal. The mice showed normal age progression and did not show any apparent additional phenotype during their life span. We observed no differences in various blood parameters and chemistry values. We infected wild-type and Nrf3−/− mice with acute lymphocytic choriomeningitis virus and found no differences in these animals with respect to their number of virus-specific CD8 and CD4 T cells as well as their B-lymphocyte response. To determine whether the mild phenotype of Nrf3 null animals is due to functional redundancy, we generated mice deficient in multiple CNC factors. Contrary to our expectations, an absence of Nrf3 does not seem to cause additional lethality in compound Nrf3−/−/Nrf2−/− and Nrf3−/−/p45−/− mice. We hypothesize that the role of Nrf3 in vivo may become apparent only after appropriate challenge to the mice.
Basic leucine zipper factors of the cap'n'collar (CNC) family regulate gene expression, tissue differentiation, and development in a variety of organisms. A unique region, the CNC domain, located amino terminal to the basic DNA-binding region, defines this group of proteins (4, 7, 43). The functional role of the CNC domain remains unknown (3). The CNC family includes the Drosophila CNC, Caenorhabditis elegans Skn-1, and vertebrate p45 NF-E2, Nrf1, Nrf2, Nrf3, Bach1, and Bach2 proteins (4, 7, 9, 11, 12, 30, 32, 37, 43, 44, 48, 52). The roles of some of the mammalian CNC factors have been extensively analyzed. NF-E2 is a heterodimer composed of a p45 protein, a CNC family member, and a small Maf protein (4, 5, 48). The expression of the p45 NF-E2 subunit is restricted to hematopoietic progenitor, erythroid, megakaryocytic, and mast cells (4). Homozygous p45 null mice suffer from a mild anemia and a profound decrease in platelets as a result of a block in the biogenesis of platelets (56, 57). This leads to the death of most animals from hemorrhage. Homozygous Nrf1 null mice die in utero. One laboratory reported that the absence of Nrf1 in a mixed 129/C57BL/6 strain results in a non-cell autonomous defect in definitive erythropoiesis which leads to death at mid- to late gestation (14). Others reported the death at embryonic day 7.5 due to abnormal mesoderm formation in an outbred Black Swiss background (24). When those investigators backcrossed the mutation in the C57BL/6J inbred mouse strain, homozygous mutant mouse embryos had no mesoderm block but did have a severe fetal anemia due to a defect in definitive hematopoiesis, which resolved at embryonic day 18.5 (39). Nrf1 appears to be critical for the redox balance during development (18). Targeted disruption of the Nrf2 locus showed that this protein is dispensable for mouse development (16). Compound p45−/−/Nrf2−/− mice do not exhibit defects beyond those seen with the loss of p45 alone, suggesting that Nrf2 is not the compensating factor in the mild erythrocyte phenotype of p45 null mice (34, 38). A combined absence of Nrf1 and Nrf2 results in early embryonic lethality and severe oxidative stress (36). In the adult organism, Nrf2 appears to participate in the response to oxidative stress (29, 35, 61). Several reports showed that Nrf2 plays an important role in the expression of detoxification and antioxidant enzymes (2, 13, 15, 25, 29, 54, 60, 62).
The CNC factors have been shown to form heterodimers with the small Maf factors, members of the Maf (proto-)oncogene family of bZIP transcription factors (6, 46). The founding member of the Maf family, the v-Maf oncogene, was isolated as the transforming agent of an avian retrovirus (50). Related Maf proteins can be divided into two subgroups: the small Mafs, including MafF, MafG and MafK factors, and large Mafs, including c-Maf, MafA, MafB (Kreisler), L-Maf, and Nrl proteins (6, 46). Small Mafs are widely expressed, but levels vary considerably (6). Small Maf homodimers that function as transcriptional repressors have been reported (22, 28, 49). In contrast, when small Mafs associate with the CNC factor p45 NF-E2 or Nrf2, the heterodimer serves as an activator of gene transcription (6, 25, 33, 62).
To better understand the possible in vivo functions of the recently identified Nrf3 protein, we isolated a mouse genomic clone comprising three exons coding for Nrf3. We performed genetic mapping and fluorescence in situ hybridization (FISH) analysis localizing the Nrf3 gene to mouse chromosome 6. Gene targeting of the mouse Nrf3 locus yielded healthy and fertile mice. Compound Nrf3/Nrf2- and Nrf3/p45-deficient animals were also viable. We speculate that a physiological role for Nrf3 may be uncovered by exposing the mice to challenging conditions.
MATERIALS AND METHODS
Isolation of a genomic clone.
We used a human NRF3 cDNA probe generated by random priming of a 950-bp BglII fragment, corresponding to amino acids 171 to 487 of human NRF3 (V. Blank, unpublished data) to screen a mouse 129SvJ genomic library (Stratagene) at reduced stringency. We isolated a 15-kb mouse genomic clone containing three exons of the mouse Nrf3 gene that were identified by DNA sequencing.
FISH analysis. (i) Chromosomal slide preparation.
Lymphocytes were isolated from mouse spleen and cultured at 37°C in RPMI 1640 medium supplemented with 15% fetal calf serum, 3 μg of concanavalin A/ml, 10 μg of lipopolysaccharide/ml, and 50 μM β-mercaptoethanol. After 44 h, the cultured lymphocytes were treated with 0.18 mg of bromodeoxyuridine/ml for an additional 14 h. The synchronized cells were washed and recultured at 37°C and treated with bromodeoxyuridine for 4 h in minimum essential medium alpha with thymidine (2.5 μg/ml). Chromosome slides were prepared by a conventional method used for human chromosome preparation (hypotonic treatment, fixation, and air drying).
(ii) Probe labeling and in situ hybridization.
The DNA probe was biotinylated with dATP by using a Gibco BRL BioNick labeling kit (15°C, 1 h) (26). The procedure for FISH detection was performed as described previously (26, 27). Briefly, slides were baked at 55°C for 1 h. After RNase A treatment, the slides were denatured in 70% formamide in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 2 min at 70°C followed by dehydration with ethanol. Probes were denatured at 75°C for 5 min in a hybridization mixture consisting of 50% formamide and 10% dextran sulfate and prehybridized for 15 min at 37°C. Probes were loaded on the denatured slides. After overnight hybridization, slides were washed, detected, and amplified by using a published method (26); FISH signals and the DAPI (4′,6′-diamidino-2-phenylindole) banding pattern were recorded separately. Images were captured and combined with a charge-coupled device camera, and the assignment of the FISH mapping data with chromosomal bands was achieved by superimposing FISH signals with DAPI-banded chromosomes (27).
(iii) Analysis.
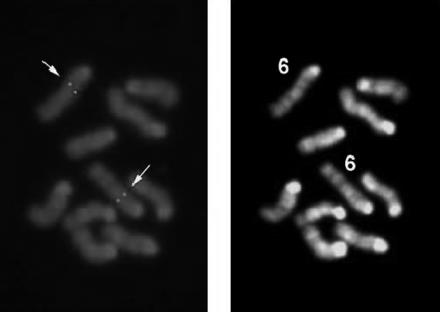
Under the conditions used, the detection efficiency was 72% for this probe (among 100 checked mitotic figures, 72 showed hybridization signals on one pair of chromosomes). Since DAPI banding was performed to identify the specific chromosome, the assignment between signals from the probe and mouse chromosome 6 was obtained. The detailed position was further determined based on the summary from 10 photos.
Generation of mice lacking Nrf3.
We used the isolated Nrf3 genomic clone to generate 7.5- and 3.2-kb fragments to use as 5′ and 3′ segments flanking two exons of the Nrf3 gene. These were cloned into the pTKLNCL targeting vector (a kind gift from Richard Mortensen). The resulting plasmid was linearized at a NotI site, chaperone oligonucleotide linkers were ligated onto the sticky ends to form hairpins (47), and the modified plasmid was electroporated into mouse TC1 embryonic stem (ES) cells (a kind gift from Phil Leder) (21). Subsequently, we grew the cells in the presence of G418 and ganciclovir and selected drug-resistant clones. Using Southern blot analysis, we identified clones that had undergone homologous recombination. All clones with a normal 40XY karyotype were injected into C57BL/6J blastocysts to generate chimeric offspring. High-level (up to 90%) chimeric mice were bred to C57BL/6J and 129S6 females to obtain heterozygous offspring. Nrf3 null mice were obtained by further breeding of the Nrf3+/− animals. Mice were kept in sterile microisolators at all times.
RNA preparation and Northern blot hybridization analyses.
Mouse tissues were isolated and immediately frozen in liquid nitrogen and stored at −80°C until RNA isolation. Total RNA was isolated by using the Trizol reagent according to the manufacturer's instructions (Invitrogen). The RNA concentration was determined by measuring the absorbance at a wavelength of 260 nm. For Northern blotting, 10 μg of each RNA sample was then separated by formaldehyde-agarose gel electrophoresis, transferred to a nylon membrane (Amersham), and subjected to hybridization as previously described, with minor modifications (20). The Nrf3 probe was generated by random priming of a 572-bp SacII/PstI fragment (corresponding to amino acids 131 to 321 of mouse Nrf3) by using [32P]dCTP (Amersham) and the Klenow fragment of DNA polymerase I according to standard methods (Roche). Membranes were stripped in 0.1% sodium dodecyl sulfate at 95°C.
Phenotypical characterization.
Hematological parameters were measured with the ADVIA 120 hematology analyzer (Bayer Diagnostics, Tarryton, N.Y.) and analyzed with a software package specific to mice. Biochemical parameters were measured by use of standard biochemical assays on a Hitachi 917 analyzer (Roche Diagnostics, Indianapolis, Ind.).
Viral infections.
All mouse infections were done by intraperitoneal injection of 2 × 105 PFU of the Armstrong CA 1371 strain of lymphocytic choriomeningitis virus (LCMV). Stocks of the Armstrong CA 1371 strain of LCMV were grown and quantified as previously described (2). Mice were sacrificed on days 8 and 30 post-LCMV Armstrong infection.
Flow cytometry and intracellular staining for IFN-γ.
Intracellular staining for gamma interferon (IFN-γ) was used to quantify virus-specific T cells as previously described (3). Briefly, 106 splenocytes were cultured in vitro with or without peptide stimulation in the presence of brefeldin A (Golgistop) (BD Pharmingen, La Jolla, Calif.) for 5 h, followed by surface and intracellular stain. CD8 T cells were stimulated with the dominant major histocompatibility complex (MHC) class I-restricted LCMV peptide NP396-404 (0.2 μg/ml), and the CD4 T cells were stimulated with the MHC class II-restricted LCMV peptide GP61-80 (2 μg/ml). The CD8 (clone 53-6.7), CD4 (clone RM4-5), and IFN-γ (clone XMG1.2) antibodies were purchased from BD Pharmingen.
Quantification of LCMV-specific ASC.
LCMV-specific antibody secreting cells (ASC) were quantified by using a previously described enzyme-linked immunospot assay method (4). Lysate from LCMV-infected BHK cells was used as antigen. Biotinylated goat anti-mouse immunoglobulin G [IgG](γ) antibodies were used as detecting antibodies (Caltag Laboratories, San Francisco, Calif.). Horseradish peroxidase-conjugated Avidin D was purchased from Vector Laboratories (Burlingame, Calif.).
Generation of compound CNC transcription factor null mice.
Mixed C57BL/6J-129S6 background Nrf3−/− animals were first crossed to Nrf2+/− mice (>90% C57BL/6J generated through a backcross of 129X1 mice to C57BL/6J mice), a kind gift from Paul Ney, St. Jude Children's Hospital, Memphis, Tenn. Double heterozygote Nrf3+/−/Nrf2+/− mice were then used to obtain compound Nrf3−/−/Nrf2−/− animals. To generate Nrf3−/−/p45−/− mice, offspring with an Nrf3−/−/p45+/− genotype on a C57BL/6J-129S6 background were mated (57).
RESULTS
Chromosomal mapping of the Nrf3 gene.
The identification of the Nfe2l3 gene encoding Nrf3, a new member of CNC family, has been described (32). Nrf3 dimerizes with the small bZIP Maf protein MafK, and the resulting heterodimers bind to NF-E2/MARE (Maf recognition element)-type DNA-binding sites present in the regulatory enhancer and promoter regions of numerous genes (6, 32). To determine the chromosomal localization and to perform gene targeting of the mouse Nrf3 (Nfe2l3) locus, we isolated a 15-kb genomic fragment. The clone comprised three exons of the mouse Nrf3 gene, which were identified by DNA sequencing. We performed FISH analysis using the 15-kb genomic fragment as a probe and unequivocally mapped the mouse Nrf3 gene to chromosome 6, region B2-B3, a region syntenic to that of its human homolog (26) (Fig. 1). We confirmed this result by performing genetic mapping using interspecific backcross panels from the Jackson Laboratory (53; data not shown). Our mapping results confirm unpublished radiation hybrid localization data (Jackson Laboratory, MGI:1339958). As has been reported for other CNC transcription factor loci (10, 32, 40, 41, 63), the mouse Nrf3 gene colocalizes with members of the Hox and collagen gene families.
FIG. 1.
Example of FISH mapping results for Nrf3. The left panel shows the FISH signals on the mouse chromosome, and the right panel shows the same mitotic figure stained with DAPI to identify mouse chromosome 6.
Gene targeting of the mouse Nrf3 locus.
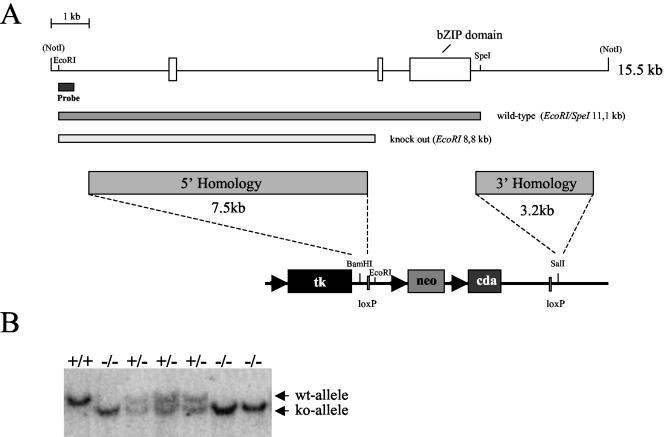
To perform targeted disruption of the mouse Nrf3 locus, we constructed a targeting vector replacing two exons of the gene by vector sequences (Fig. 2A). This replacement results in an aberrant Nrf3 transcript lacking the regions coding for the CNC homology, the basic DNA-binding, and the leucine zipper dimerization domains. The absence of the DNA-binding and dimerization domains should yield a nonfunctional Nrf3. Following electroporation of the targeting construct into mouse TC1 ES cells, we obtained 3 out of 103 correctly targeted clones. The microinjection of two of these clones into mouse blastocysts resulted in high-level chimeric (>80 to 90%) offspring. Germ line transmission of the Nrf3 knockout allele has been achieved with mice generated from one of the original ES cell clones, and the resulting Nrf3+/− mice were bred to obtain Nrf3−/− offspring (Fig. 2B). The genotypes of the offspring corresponded well to the expected Mendelian ratio of 25% homozygous wild-type, 50% heterozygous, and 25% homozygous knockout mice, suggesting that Nrf3 null mice developed normally and reached adulthood.
FIG. 2.
Generation of Nrf3-deficient mice. (A) Replacement of two exons of the Nrf3 gene, comprising the sequences coding for the bZIP domain, by the neomycin-cytosine deaminase cassette of the targeting vector. Nrf3 gene exons are represented by open boxes, and the exon encoding the Nrf3 bZIP domain is shown. The targeting vector contains 7.5 and 3.2 kb of Nrf3 gene homologous genomic sequences on the 5′ and 3′ sides of the neomycin-cytosine deaminase cassette, respectively. The probe used for Southern analysis and the expected fragments from EcoRI/SpeI digests of wild-type and knockout genomic DNA are indicated. tk, thymidine kinase; neo, neomycin; cda, cytosine deaminase. (B) Generation of Nrf3−/− mice. Southern blot analysis of Nrf3+/− mouse matings yielding offspring with homozygous wild-type (wt) and heterozygous and homozygous mutant (null) genotypes. ko, knockout.
Nrf3 gene expression pattern in wild-type and knockout mice.
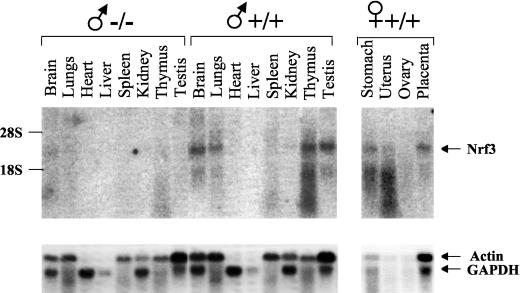
It has been shown that human NRF3 is highly expressed in the placenta and Burkitt’s lymphoma cell lines and at lower levels in the heart, brain, lung, kidney, leukocytes, colon, small intestine, thymus, and spleen (32). We performed Northern blot analysis of mouse tissues (Fig. 3). Murine Nrf3 mRNA is expressed in a wide variety of tissues, although at different levels. The levels of expression are high in the brain, thymus, testis, and placenta, and medium levels are observed in the uterus, stomach, and lung. Transcript levels are low in the kidney, and minimal or no expression is found in the heart, liver, spleen, and ovary. We hypothesize that Nrf3 may perform multiple roles in different tissues in the mouse. Interestingly, the Nrf3 gene tissue expression pattern in the mouse differs slightly from that reported in humans (32), indicating that Nrf3 might have distinct functions in both mammalian species. To confirm the absence of functional Nrf3 transcripts in gene-targeted mice, we performed Northern analysis comparing Nrf3-targeted and wild-type animals. As shown in Fig. 3, transcripts are not present in Nrf3−/− mice.
FIG. 3.
Nrf3 transcript analysis of wild-type and gene-targeted mice. Northern blot analysis showing Nrf3 mRNA levels in various tissues in wild-type and Nrf3 null mice was done. Ten micrograms of total RNA was used per lane. The same blot was probed with actin- and GAPDH (glyceraldehyde-3-phosphate dehydrogenase gene)-specific control probes.
Phenotypic analysis of Nrf3-deficient mice.
We performed a phenotypic analysis of the mice deficient for Nrf3. The general behavior including feeding and mating of Nrf3 null mice does not seem to be altered under nonchallenging conditions compared to that of their wild-type littermates. These mice also have a normal life span and do not develop any observable abnormalities. We analyzed Nrf3−/− animals with respect to their gross anatomy and multiple blood parameters, including red blood cell numbers, white blood cell numbers, and erythrocyte cellular indices (mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, and mean cell hemoglobin concentration) (data not shown). In addition, we examined several biochemical parameters in plasma (glucose, blood urea nitrogen, cholesterol, triglyceride, aspartate transaminase, alanine transaminase, lactate dehydrogenase, iron, total bilirubin, and direct bilirubin) (data not shown). No differences between Nrf3 null mice and wild-type littermates were found. As it is possible that the loss of Nrf3 function may be compensated for by the overexpression of other CNC transcription factors, we analyzed the expression levels of other family members in the Nrf3−/− mice. We found that the transcript levels of the closely related family members p45, Nrf1, and Nrf2 appear not to be altered in the Nrf3 null mice in the tissues analyzed in Fig. 3 (data not shown).
Response of Nrf3 null mice to viral infection.
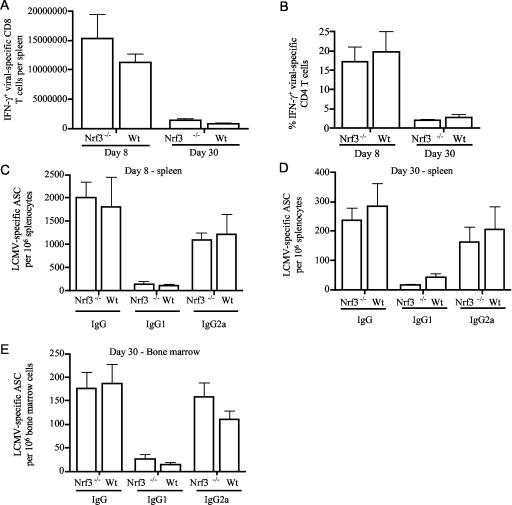
The murine Nrf3 gene is expressed at higher levels in the murine thymus, testis, placenta, lung, and brain. Since the animals are housed in a pathogen-free environment, a role for Nrf3 in thymic function and immune defense mechanisms may have remained undetected. Thus, as Nrf3 is highly expressed in the thymus, we wanted to determine if Nrf3 plays a role in the adaptive immune response. To determine this, we infected Nrf3−/− and control mice with acute LCMV. The CD8, CD4, and B-cell responses to acute LCMV infection have been well characterized (1, 51, 58). The peak CD8, CD4, and B-cell response to LCMV occurs on day 8 postinfection. As shown in Fig. 4, we did not detect any significant differences between the Nrf3−/− and control mice at day 8. The numbers of virus-specific CD8 (Fig. 4A) and CD4 (Fig. 4B) T cells were comparable between the Nrf3−/−and control mice. We also found no difference in the B-cell response to LCMV. The amounts of plasma or ASC secreting LCMV-specific IgG, IgG2a, the dominant isotype, or IgG1 (Fig. 4C) were equivalent. To determine whether the Nrf3 deficiency had any effect on T-cell memory development, we sacrificed mice 30 days post-LCMV infection. As shown in Fig. 4, both the control and Nrf3−/− mice had comparable numbers of virus-specific CD8 (Fig. 4A) and CD4 (Fig. 4B) T cells at this later time point. The numbers of LCMV-specific ASC in both the spleen and bone marrow were also comparable at day 30 postinfection (Fig. 4D and E). Our data suggest that Nrf3 null mice do not show major differences in their viral response. In summary, our phenotypic analysis of Nrf3 null mice suggested functional redundancy among CNC family proteins.
FIG. 4.
Response of wild-type and Nrf3 null mice to LCMV infection. Nrf3−/− mice and control mice have comparable T- and B-cell responses to LCMV. IFN-γ production by virus-specific CD8 (A) and CD4 (B) T cells was determined on days 8 and 30 postinfection. Spleen cells from day 8 or day 30 infected mice were stimulated in vitro for 5 h with either MHC class I-restricted epitope NP396-404 (A) or MHC class II-restricted epitope GP61-80 (B). The number of LCMV-specific ASC was determined at day 8 (C) and day 30 (D) in the spleen and on day 30 in the bone marrow (E). LCMV-specific IgG, IgG2a, and IgG1 was determined by an enzyme-linked immunospot assay. The data shown represent a mean of four mice per group. Wt, wild type.
Generation of compound Nrf3−/−/Nrf2−/− and Nrf3−/−/p45−/− mice.
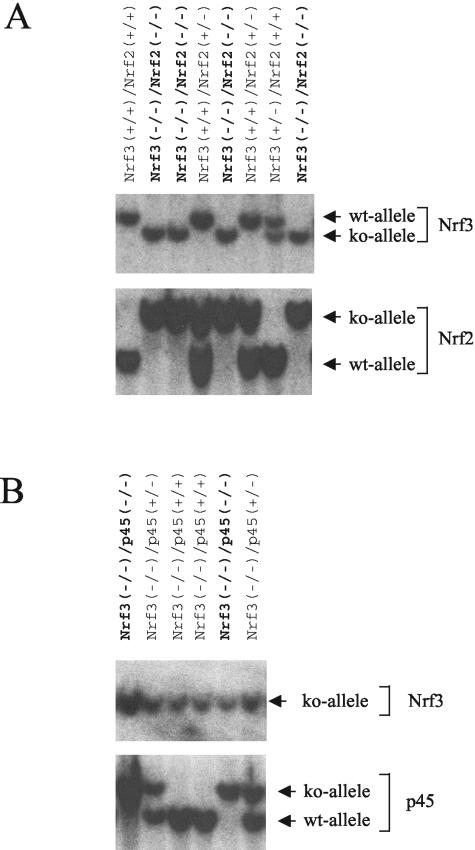
To determine whether the mild phenotype of Nrf3 null animals is due to functional redundancy among CNC transcription factors, we generated compound null animals deficient for Nrf3/Nrf2 and Nrf3/p45. Southern blot analysis confirmed the genotype of the offspring of these matings (Fig. 5) (38, 57). We found that Nrf3−/−/Nrf2−/− mice develop normally and are viable and fertile, as we obtained live compound null animals at a rate at least equal to the expected Mendelian ratio (Table 1). In additional matings, we obtained some Nrf3−/−/p45−/− animals that survived to adulthood (Table 1). We thus conclude that the absence of the Nrf3 gene does not cause additional lethality in these compound mutant mice.
FIG. 5.
Generation of compound Nrf3−/−/Nrf2−/− and Nrf3−/−/p45−/− null mice. Southern blot analyses of Nrf3+/−/Nrf2+/− mice matings (A) and Nrf3−/− and p45+/− mice matings (B) are shown. Double knockout (ko) animals are indicated in boldface type. wt, wild type.
TABLE 1.
Viability of compound Nrf3−/−/Nrf2−/− and Nrf3−/−/p45−/− micec
| Genotype | Alive (%) | Dead (%) | Expected Mendelian ratio (%) |
|---|---|---|---|
| Nrf3+/−/Nrf2+/− matingsa | |||
| Nrf3+/+/Nrf2+/+ | 5.8 | 6.25 | |
| Nrf3+/+/Nrf2+/− | 14.5 | 12.50 | |
| Nrf3+/+/Nrf2−/− | 4.4 | 6.25 | |
| Nrf3+/−/Nrf2+/+ | 14.5 | 12.50 | |
| Nrf3+/−/Nrf2+/− | 21.7 | 25.00 | |
| Nrf3+/−/Nrf2−/− | 5.8 | 12.50 | |
| Nrf3−/−/Nrf2+/+ | 2.9 | 6.25 | |
| Nrf3−/−/Nrf2+/− | 15.9 | 12.50 | |
| Nrf3−/−/Nrf2−/− | 14.5 | 6.25 | |
| Nrf3−/−/p45+/− matingsb | |||
| Nrf3−/−/p45+/+ | 33.3 | 0 | 25.0 |
| Nrf3−/−/p45+/− | 46.0 | 2.3 | 50.0 |
| Nrf3−/−/p45−/− | 16.1 | 2.3 | 25.0 |
Offspring viability of Nrf3+/−/Nrf2+/− matings. A sample of 69 animals (nine litters) is shown.
Offspring viability of Nrf3−/−/p45+/− matings. A sample of 87 animals (12 litters) is shown.
The observed survival of the animals through adulthood, as well as the expected Mendelian ratio of genotypes, is indicated.
DISCUSSION
To further characterize the role of the Nrf3 transcription factor in mammalian gene expression, we isolated a mouse genomic clone comprising three exons of the Nrf3 locus. We used this clone to determine the chromosomal localization of the murine Nrf3 gene by FISH analysis and genetic mapping. As shown in Fig. 1, we unequivocally localized the mouse Nrf3 gene to chromosome 6, region B2-B3, confirming unpublished radiation hybrid data (MGI:1339958). The mouse Nrf3 maps close to the murine Hoxa cluster, a map location syntenic to the region containing the human Nrf3 locus on chromosome 7 p15-7 p14 (bacterial artificial chromosome clone no. RG119C02) (32). This mapping result further strengthens the hypothesis, proposed previously (10, 32), that the genetic loci of p45, Nrf1, Nrf2, and Nrf3 have evolved from a single gene. Mouse and human versions of these genes colocalize to a cluster of genes comprising members of the collagen and Hox gene families (10, 32, 40, 41, 63). The p45 (human, 12q13.1-13.3; mouse, 15), Nrf1 (human, 17q21.3; mouse, 11), Nrf2 (human, 2q31; mouse, 2), and Nrf3 (human, 7p14-15; mouse, 6) genes map to each of the four known hox clusters, hoxC, hoxB, hoxD, and hoxA, respectively. Duplication and subsequent diversification of each family member may allow for the fine-tuning of CNC-dependent transcription in mammals.
To investigate the in vivo role of Nrf3, we performed gene targeting by homologous recombination in the mouse. We found that Nrf3 heterozygote matings generated offspring in the expected Mendelian ratio. Gross anatomy and behavior also appeared normal. It has been shown that p45 NF-E2 plays crucial roles in blood cell function, particularly in platelet biogenesis and erythroid cell-specific gene expression (31, 55-57). However, analysis of common blood parameters and chemistry values of Nrf3−/− mice did not reveal any significant changes. We bred Nrf3 null mice, females and males, and found that both sexes were fertile and that females were able to bear normal-sized litters. Thus, although Nrf3 is highly expressed in the placenta and testis, and at medium levels in the uterus, its function can be substituted in these tissues. It was previously proposed that Nrf3, in conjunction with Nrf2, may have a role in keratinocyte function (8), but we did not observe any skin abnormalities in Nrf3−/− animals. We observed a limited number of animals for up to 22 months, but no abnormalities were noticeable with advancing age. Thus, Nrf3 null mice developed normally and are indistinguishable from their wild-type littermates when held in a sterile environment. It is possible that Nrf3 is absolutely required only in adult mice, and a phenotype may be exposed only under challenging conditions. We speculated that Nrf3 may play a role in the adaptive immune response due to its high expression in the thymus, but viral insult with the LCMV model (1, 51, 58) did not expose differences between wild-type and Nrf3 knockout animals. The mixed C57BL/6J-129S6 genetic background of these mice might contribute to alleviate the phenotype, but Nrf3−/− mice in a pure inbred 129 background still develop and grow normally (A. Derjuga and V. Blank, unpublished observations). In summary, our analysis showed that Nrf3 null mice exhibit no obvious phenotype.
The mild phenotype of Nrf3 null mice suggested functional redundancy by closely related transcription factors. We speculated that other CNC proteins, because of their structural homologies, similar DNA-binding specificities, and overlapping expression pattern (45), might compensate for the loss of Nrf3. We found that p45 NF-E2, Nrf1, and Nrf2 transcript levels appear to be unchanged in Nrf3 null mice (data not shown), but compensation for the absence of Nrf3 may not necessarily require higher expression levels. Several other CNC family members have been analyzed by gene targeting. Although p45 NF-E2 and Nrf1 null mice show severe phenotypes, Nrf2−/− and Bach1−/− mice develop normally and are fertile (14, 16, 24, 35, 57, 59). Compound Nrf2−/−/p45−/− mice do not exhibit defects beyond those seen with the loss of p45 alone (38). The absence of Bach1 leads to a high level of expression of heme oxygenase 1 (59). Subsequent analyses of Nrf2-deficient mice showed that this protein is important for liver function in various responses to stress-inducing agents, including antioxidants or drugs such as acetaminophen (13, 17, 19, 23, 29, 36, 42). As Nrf3 is not expressed in the liver (Fig. 3), an analogous function for Nrf3 in the liver is unlikely. However, Nrf3 may be important for similar responses to stress or other insults in other tissues. To test the hypothesis of functional redundancy among CNC proteins, we generated compound Nrf3−/−/Nrf2−/− and Nrf3−/−/p45−/− mice. Unexpectedly, these compound knockout mice are viable (Table 1). We have recently obtained compound Nrf3−/−/Nrf2−/−/p45−/− mice, and preliminary observations suggest that some of the triple-compound knockout mice also survive (Derjuga and Blank, unpublished). In conclusion, the absence of Nrf3-dependent transcription in Nrf2 or p45 animals does not cause added lethality, as would be expected if these factors were functionally redundant. It may be that the functions of Nrf3/Nrf2 or Nrf3/p45 are compensated for by the remaining CNC proteins, Nrf1, Bach1, and Bach2, or by unrelated factors. Thus, the generation of different combinations of CNC transcription factor null animals may be required to obtain insights into Nrf3 function. In addition, future experiments exposing the generated single and compound knockout mice to challenging and stressful conditions may help uncover the role of Nrf3 in vivo.
Acknowledgments
We thank Phil Leder for providing ES cells, Richard Mortensen for providing the targeting vector, and Paul Ney for providing Nrf2 null mice. We thank Tam Thompson and Hong Ye for microinjection of mouse blastocysts (through the Children's Hospital Gene Manipulation Facility, funded by NIHP30-HD 18655), Carlo Brugnara for blood and biochemical parameter analysis, Daniel Martineau for mouse pathology, Benoît Chénais for critical reading of the manuscript, and Joanne Levy, Lynne Montross, Cameron Trenor, Mark Fleming, and Mary Barter for valuable discussions.
T.S.G. is a Leukemia and Lymphoma Society Scholar, and R.A. acknowledges the support of the NIH (grant no. AI30048). N.C.A. is an Associate Investigator of the HHMI and acknowledges the support of the NIH (grant no. HL51057). R.A.S. is a Scholar of the Leukemia and Lymphoma Society and acknowledges the support of the NIH (grant no. HL63143). This work was partly supported by a PSIIRI collaboration grant of the Quebec Ministry of Economic and Regional Development and Research to V.B., R.A., and R.A.S. V.B. is a Chercheur Boursier of the Fonds de la Recherche en Santé du Québec. These studies were funded by grants of the Canadian Institutes of Health Research and the Cancer Research Society Inc. to V.B.
REFERENCES
- 1.Ahmed, R., A. Salmi, L. D. Butler, J. M. Chiller, and M. B. Oldstone. 1984. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J. Exp. Med. 160:521-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, J., C. Wicks, D. Stewart, P. Gong, C. Touchard, S. Otterbein, A. M. Choi, M. E. Burow, and J. Tou. 2000. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J. Biol. Chem. 275:27694-27702. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, N. C. 1998. The NF-E2 transcription factor. Int. J. Biochem. Cell Biol. 30:429-432. [DOI] [PubMed] [Google Scholar]
- 4.Andrews, N. C., H. Erdjument-Bromage, M. B. Davidson, P. Tempst, and S. H. Orkin. 1993. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362:722-728. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, N. C., K. J. Kotkow, P. A. Ney, H. Erdjument-Bromage, P. Tempst, and S. H. Orkin. 1993. The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc. Natl. Acad. Sci. USA 90:11488-11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blank, V., and N. C. Andrews. 1997. The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci. 22:437-441. [DOI] [PubMed] [Google Scholar]
- 7.Bowerman, B., B. A. Eaton, and J. R. Priess. 1992. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68:1061-1075. [DOI] [PubMed] [Google Scholar]
- 8.Braun, S., C. Hanselmann, M. G. Gassmann, U. auf dem Keller, C. Born-Berclaz, K. Chan, Y. W. Kan, and S. Werner. 2002. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol. Cell. Biol. 22:5492-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caterina, J. J., D. Donze, C. W. Sun, D. J. Ciavatta, and T. M. Townes. 1994. Cloning and functional characterization of LCR-F1: a bZIP transcription factor that activates erythroid-specific, human globin gene expression. Nucleic Acids Res. 22:2383-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, J. Y., M. C. Cheung, P. Moi, K. Chan, and Y. W. Kan. 1995. Chromosomal localization of the human NF-E2 family of bZIP transcription factors by fluorescence in situ hybridization. Hum. Genet. 95:265-269. [DOI] [PubMed] [Google Scholar]
- 11.Chan, J. Y., X. L. Han, and Y. W. Kan. 1993. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. USA 90:11371-11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, J. Y., X. L. Han, and Y. W. Kan. 1993. Isolation of cDNA encoding the human NF-E2 protein. Proc. Natl. Acad. Sci. USA 90:11366-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan, J. Y., and M. Kwong. 2000. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim. Biophys. Acta 1517:19-26. [DOI] [PubMed] [Google Scholar]
- 14.Chan, J. Y., M. Kwong, R. Lu, J. Chang, B. Wang, T. S. Yen, and Y. W. Kan. 1998. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 17:1779-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, K., and Y. W. Kan. 1999. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. USA 96:12731-12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan, K., R. Lu, J. C. Chang, and Y. W. Kan. 1996. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA 93:13943-13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chanas, S. A., Q. Jiang, M. McMahon, G. K. McWalter, L. I. McLellan, C. R. Elcombe, C. J. Henderson, C. R. Wolf, G. J. Moffat, K. Itoh, M. Yamamoto, and J. D. Hayes. 2002. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 365:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, L., M. Kwong, R. Lu, D. Ginzinger, C. Lee, L. Leung, and J. Y. Chan. 2003. Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell. Biol. 23:4673-4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho, H. Y., A. E. Jedlicka, S. P. Reddy, T. W. Kensler, M. Yamamoto, L. Y. Zhang, and S. R. Kleeberger. 2002. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 26:175-182. [DOI] [PubMed] [Google Scholar]
- 20.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng, C., A. Wynshaw-Boris, F. Zhou, A. Kuo, and P. Leder. 1996. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84:911-921. [DOI] [PubMed] [Google Scholar]
- 22.Dhakshinamoorthy, S., and A. K. Jaiswal. 2000. Small Maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:quinone oxidoreductase1 gene. J. Biol. Chem. 275:40134-40141. [DOI] [PubMed] [Google Scholar]
- 23.Enomoto, A., K. Itoh, E. Nagayoshi, J. Haruta, T. Kimura, T. O'Connor, T. Harada, and M. Yamamoto. 2001. High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59:169-177. [DOI] [PubMed] [Google Scholar]
- 24.Farmer, S. C., C. W. Sun, G. E. Winnier, B. L. Hogan, and T. M. Townes. 1997. The bZIP transcription factor LCR-F1 is essential for mesoderm formation in mouse development. Genes Dev. 11:786-798. [DOI] [PubMed] [Google Scholar]
- 25.Gong, P., B. Hu, D. Stewart, M. Ellerbe, Y. G. Figueroa, V. Blank, B. S. Beckman, and J. Alam. 2001. Cobalt induces heme oxygenase-1 expression by a hypoxia-inducible factor-independent mechanism in Chinese hamster ovary cells: regulation by Nrf2 and MafG transcription factors. J. Biol. Chem. 276:27018-27025. [DOI] [PubMed] [Google Scholar]
- 26.Heng, H. H., J. Squire, and L. C. Tsui. 1992. High-resolution mapping of mammalian genes by in situ hybridization to free chromatin. Proc. Natl. Acad. Sci. USA 89:9509-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heng, H. H., and L. C. Tsui. 1993. Modes of DAPI banding and simultaneous in situ hybridization. Chromosoma 102:325-332. [DOI] [PubMed] [Google Scholar]
- 28.Igarashi, K., K. Kataoka, K. Itoh, N. Hayashi, M. Nishizawa, and M. Yamamoto. 1994. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367:568-572. [DOI] [PubMed] [Google Scholar]
- 29.Itoh, K., T. Chiba, S. Takahashi, T. Ishii, K. Igarashi, Y. Katoh, T. Oyake, N. Hayashi, K. Satoh, I. Hatayama, M. Yamamoto, and Y. Nabeshima. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 236:313-322. [DOI] [PubMed] [Google Scholar]
- 30.Itoh, K., K. Igarashi, N. Hayashi, M. Nishizawa, and M. Yamamoto. 1995. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell. Biol. 15:4184-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, A., E. Ito, T. Toki, K. Kogame, S. Takahashi, K. Igarashi, N. Hayashi, and M. Yamamoto. 1999. Molecular cloning and functional characterization of a new Cap'n'collar family transcription factor Nrf3. J. Biol. Chem. 274:6443-6452. [DOI] [PubMed] [Google Scholar]
- 33.Kotkow, K. J., and S. H. Orkin. 1995. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol. 15:4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuroha, T., S. Takahashi, T. Komeno, K. Itoh, T. Nagasawa, and M. Yamamoto. 1998. Ablation of Nrf2 function does not increase the erythroid or megakaryocytic cell lineage dysfunction caused by p45 NF-E2 gene disruption. J. Biochem. (Tokyo) 123:376-379. [DOI] [PubMed] [Google Scholar]
- 35.Kwong, M., Y. W. Kan, and J. Y. Chan. 1999. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in γ-gcsL and gss expression in mouse fibroblasts. J. Biol. Chem. 274:37491-37498. [DOI] [PubMed] [Google Scholar]
- 36.Leung, L., M. Kwong, S. Hou, C. Lee, and J. Y. Chan. 2003. Deficiency in Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 278:48021-48029. [DOI] [PubMed]
- 37.Luna, L., O. Johnsen, A. H. Skartlien, F. Pedeutour, C. Turc-Carel, H. Prydz, and A. B. Kolsto. 1994. Molecular cloning of a putative novel human bZIP transcription factor on chromosome 17q22. Genomics 22:553-562. [DOI] [PubMed] [Google Scholar]
- 38.Martin, F., J. M. van Deursen, R. A. Shivdasani, C. W. Jackson, A. G. Troutman, and P. A. Ney. 1998. Erythroid maturation and globin gene expression in mice with combined deficiency of NF-E2 and nrf-2. Blood 91:3459-3466. [PubMed] [Google Scholar]
- 39.Masuoka, H. C., and T. M. Townes. 2002. Targeted disruption of the activating transcription factor 4 gene results in severe fetal anemia in mice. Blood 99:736-745. [DOI] [PubMed] [Google Scholar]
- 40.McKie, J., K. Johnstone, M. G. Mattei, and P. Scambler. 1995. Cloning and mapping of murine Nfe2l1. Genomics 25:716-719. [DOI] [PubMed] [Google Scholar]
- 41.McKie, J., and P. J. Scambler. 1996. The Nfe2l1 gene maps to distal mouse chromosome 11. Mamm. Genome 7:89. [DOI] [PubMed] [Google Scholar]
- 42.McMahon, M., K. Itoh, M. Yamamoto, S. A. Chanas, C. J. Henderson, L. I. McLellan, C. R. Wolf, C. Cavin, and J. D. Hayes. 2001. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 61:3299-3307. [PubMed] [Google Scholar]
- 43.Mohler, J., K. Vani, S. Leung, and A. Epstein. 1991. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech. Dev. 34:3-9. [DOI] [PubMed] [Google Scholar]
- 44.Moi, P., K. Chan, I. Asunis, A. Cao, and Y. W. Kan. 1994. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 91:9926-9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Motohashi, H., T. O'Connor, F. Katsuoka, J. Engel, and M. Yamamoto. 2002. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1-12. [DOI] [PubMed] [Google Scholar]
- 46.Motohashi, H., J. A. Shavit, K. Igarashi, M. Yamamoto, and J. D. Engel. 1997. The world according to Maf. Nucleic Acids Res. 25:2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nehls, M., B. Kyewski, M. Messerle, R. Waldschutz, K. Schuddekopf, A. J. Smith, and T. Boehm. 1996. Two genetically separable steps in the differentiation of thymic epithelium. Science 272:886-889. [DOI] [PubMed] [Google Scholar]
- 48.Ney, P. A., N. C. Andrews, S. M. Jane, B. Safer, M. E. Purucker, S. Weremowicz, C. C. Morton, S. C. Goff, S. H. Orkin, and A. W. Nienhuis. 1993. Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol. Cell. Biol. 13:5604-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen, T., H. C. Huang, and C. B. Pickett. 2000. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J. Biol. Chem. 275:15466-15473. [DOI] [PubMed] [Google Scholar]
- 50.Nishizawa, M., K. Kataoka, N. Goto, K. T. Fujiwara, and S. Kawai. 1989. v-maf, a viral oncogene that encodes a “leucine zipper” motif. Proc. Natl. Acad. Sci. USA 86:7711-7715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oldstone, M. B., R. Ahmed, J. Byrne, M. J. Buchmeier, Y. Riviere, and P. Southern. 1985. Virus and immune responses: lymphocytic choriomeningitis virus as a prototype model of viral pathogenesis. Br. Med. Bull. 41:70-74. [DOI] [PubMed] [Google Scholar]
- 52.Oyake, T., K. Itoh, H. Motohashi, N. Hayashi, H. Hoshino, M. Nishizawa, M. Yamamoto, and K. Igarashi. 1996. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol. 16:6083-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rowe, L. B., J. H. Nadeau, R. Turner, W. N. Frankel, V. A. Letts, J. T. Eppig, M. S. Ko, S. J. Thurston, and E. H. Birkenmeier. 1994. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm. Genome 5:253-274. (Erratum, 5:463.) [DOI] [PubMed] [Google Scholar]
- 54.Satoh, K., K. Itoh, M. Yamamoto, M. Tanaka, M. Hayakari, K. Ookawa, T. Yamazaki, T. Sato, S. Tsuchida, and I. Hatayama. 2002. Nrf2 transactivator-independent GSTP1-1 expression in ‘GSTP1-1 positive' single cells inducible in female mouse liver by DEN: a preneoplastic character of possible initiated cells. Carcinogenesis 23:457-462. [DOI] [PubMed] [Google Scholar]
- 55.Sawado, T., K. Igarashi, and M. Groudine. 2001. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc. Natl. Acad. Sci. USA 98:10226-10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shivdasani, R. A., and S. H. Orkin. 1995. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc. Natl. Acad. Sci. USA 92:8690-8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shivdasani, R. A., M. F. Rosenblatt, D. Zucker-Franklin, C. W. Jackson, P. Hunt, C. J. Saris, and S. H. Orkin. 1995. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell 81:695-704. [DOI] [PubMed] [Google Scholar]
- 58.Slifka, M. K., R. Antia, J. K. Whitmire, and R. Ahmed. 1998. Humoral immunity due to long-lived plasma cells. Immunity 8:363-372. [DOI] [PubMed] [Google Scholar]
- 59.Sun, J., H. Hoshino, K. Takaku, O. Nakajima, A. Muto, H. Suzuki, S. Tashiro, S. Takahashi, S. Shibahara, J. Alam, M. M. Taketo, M. Yamamoto, and K. Igarashi. 2002. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 21:5216-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Venugopal, R., and A. K. Jaiswal. 1996. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-me-diated expression of NAD(P)H:quinone oxidoreductase 1 gene. Proc. Natl. Acad. Sci. USA 93:14960-14965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venugopal, R., and A. K. Jaiswal. 1998. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene 17:3145-3156. [DOI] [PubMed] [Google Scholar]
- 62.Wild, A. C., H. R. Moinova, and R. T. Mulcahy. 1999. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J. Biol. Chem. 274:33627-33636. [DOI] [PubMed] [Google Scholar]
- 63.Yehiely, F., P. Bamborough, M. Da Costa, B. J. Perry, G. Thinakaran, F. E. Cohen, G. A. Carlson, and S. B. Prusiner. 1997. Identification of candidate proteins binding to prion protein. Neurobiol. Dis. 3:339-355. [DOI] [PubMed] [Google Scholar]