Abstract
Rad9 is required for the activation of DNA damage checkpoint pathways in budding yeast. Rad9 is phosphorylated after DNA damage in a Mec1- and Tel1-dependent manner and subsequently interacts with Rad53. This Rad9-Rad53 interaction has been suggested to trigger the activation and phosphorylation of Rad53. Here we show that Mec1 controls the Rad9 accumulation at double-strand breaks (DSBs). Rad9 was phosphorylated after DSB induction and associated with DSBs. However, its phosphorylation and association with DSBs were significantly decreased in cells carrying a mec1Δ or kinase-negative mec1 mutation. Mec1 phosphorylated the S/TQ motifs of Rad9 in vitro, the same motifs that are phosphorylated after DNA damage in vivo. In addition, multiple mutations in the Rad9 S/TQ motifs resulted in its defective association with DSBs. Phosphorylation of Rad9 was partially defective in cells carrying a weak mec1 allele (mec1-81), whereas its association with DSBs occurred efficiently in the mec1-81 mutants, as found in wild-type cells. However, the Rad9-Rad53 interaction after DSB induction was significantly decreased in mec1-81 mutants, as it was in mec1Δ mutants. Deletion mutation in RAD53 did not affect the association of Rad9 with DSBs. Our results suggest that Mec1 promotes association of Rad9 with sites of DNA damage, thereby leading to full phosphorylation of Rad9 and its interaction with Rad53.
The integrity of genomic information is critical to the survival and propagation of all cellular organisms. Environmental stresses and normal cellular processes can cause DNA damage that compromises genomic stability. To ensure the proper response to DNA damage, cells use a set of surveillance mechanisms termed checkpoint controls. Checkpoint machinery monitors genomic integrity and activates a variety of DNA damage responses, including cell cycle arrest and alterations in gene expression (6).
DNA damage checkpoint pathways transmit signals through evolutionarily conserved kinases (39). These kinases include the family of high-molecular-weight protein kinases, i.e., ATM (mammals), ATR (mammals), MEC1 (the budding yeast Saccharomyces cerevisiae), TEL1 (budding yeast), and rad3+ (the fission yeast Schizosaccharomyces pombe) (1). Each of these genes falls into two family groups based on homology; ATM is related most closely to TEL1, while ATR is more closely related to MEC1 and rad3+. These large protein kinases regulate the activation of two downstream protein kinases, represented by mammalian Chk1 and Chk2 (1, 39). Chk2 contains a forkhead-associated (FHA) domain, and Chk2 homologs are encoded by RAD53 and cds1+ in budding yeast and fission yeast, respectively (39). Kinases related to mammalian Chk1 have been identified and called Chk1 in budding yeast and fission yeast (39).
In budding yeast, Mec1 plays a critical role in the DNA damage checkpoint controls throughout the cell cycle (14), whereas Tel1 plays a minor role (17, 20, 28). Mec1 physically interacts with Ddc2 (also called Lcd1 and Pie1), a protein that exhibits limited homology to the fission yeast Rad26 and mammalian ATRIP proteins (2, 5, 21, 24, 35). Mec1 and Ddc2 function as a complex and localize to sites of DNA damage, suggesting that the Mec1-Ddc2 complex interacts with aberrant DNA structures or the DNA repair apparatus after DNA damage (13, 16, 25). Recent evidence supports the model in which Ddc2 binds to replication protein A (RPA)-coated single-stranded DNA (ssDNA) and thereby the Mec1-Ddc2 complex localizes to sites of DNA damage (40). Mec1 regulates the phosphorylation and activation of the Rad53 and Chk1 protein kinases (27, 28, 31). Rad53 plays a critical role in DNA damage checkpoints throughout the cell cycle (14), whereas Chk1 acts in the G2/M-phase cell cycle arrest in response to DNA damage (27). Thus, the Mec1 and Rad53 kinases constitute a central DNA damage checkpoint pathway.
Phosphorylation and activation of Rad53 is also controlled by DDC1, MEC3, RAD17, and RAD24 (14). Genetic evidence has suggested that RAD17, RAD24, MEC3, and DDC1 operate in the same pathway (14). DDC1, MEC3, and RAD17 are homologs of the mammalian RAD9, HUS1, and RAD1 genes, respectively, each of which encodes a protein structurally related to PCNA (39). Consistently, Ddc1, Mec3, and Rad17 form a PCNA-related complex and function as a complex (12). RAD24 is related to the RAD17 gene in mammals and encodes a protein structurally similar to the subunits of replication factor C (RFC) (39). Rad24 interacts with the four small RFC subunits, Rfc2, Rfc3, Rfc4, and Rfc5, to form an RFC-related complex (10, 18). The Rad24 complex regulates the recruitment of the Ddc1-Mec3-Rad17 complex to sites of DNA damage (13, 16).
RAD9 is the prototype DNA damage checkpoint gene (36) and is required for the phosphorylation and activation of Chk1 and Rad53 (9, 27, 29). Rad9 is related to Crb2 in fission yeast, although no clear mammalian homolog has been identified (39). Rad9 is phosphorylated after DNA damage in a manner dependent on Mec1 and Tel1 (7, 34). Members of the ATM and ATR kinase families phosphorylate serine or threonine in S/TQ motifs (1). Consistently, multiple S/TQ motifs within Rad9 are phosphorylated in response to DNA damage (29). Moreover, multiple mutations in the Rad9 S/TQ motifs cause a defect in the activation and phosphorylation of Rad53 after DNA damage (29). In addition to Mec1 and Tel1, the Rad17 and Rad24 complexes contribute to Rad9 phosphorylation after DNA damage (7, 34). Phosphorylated Rad9 interacts physically with Rad53 in vivo (7, 32, 34). Rad53 contains two FHA domains, which have the ability to bind to specific phosphopeptides. Both of the Rad53 FHA domains can interact with Rad9 phosphopeptides (4, 29), and mutations of conserved amino acids in the second FHA domain abolish the DNA damage-induced Rad53 phosphorylation (32). Furthermore, phosphorylated Rad9 facilitates the activation and phosphorylation of Rad53 in vitro (9). These data have supported the model in which phosphorylated Rad9 acts as machinery to interact with the Rad53 FHA domain and activate the Rad53 kinase. Thus, phosphorylation of Rad9 after DNA damage is a crucial step in the transduction of the checkpoint signal. Because Mec1 localizes to sites of DNA damage, it is possible that Mec1 phosphorylates Rad9 at DNA damage sites. Previously, Rad9 was shown to form foci in the presence of DNA damage (16) and associate with replication origins when DNA replication and damage checkpoints have deteriorated (11). However, whether Rad9 is recruited to sites of DNA damage has not been clearly demonstrated.
In this study, we found that Rad9 associates with double-strand break (DSB) lesions and that its association requires Mec1 kinase activity. We found that Rad9 associated with DSBs in wild-type cells but not in cells carrying mec1Δ or kinase-defective mec1 mutations. Mec1 phosphorylated serine or threonine of the Rad9 S/TQ motifs in vitro, residues that are required for DNA damage-induced phosphorylation of Rad9 in vivo. Accordingly, mutations in the S/TQ motifs caused a defect in the association of Rad9 with DSBs. Rad9 associated with DSBs in cells carrying a weak mec1 mutation as efficiently as in wild-type cells, although phosphorylation of Rad9 was partially decreased in the mec1 mutants. Our results suggest that Mec1 promotes association of Rad9 with sites of DNA damage and then fully phosphorylates Rad9 at the damage sites.
MATERIALS AND METHODS
Plasmids.
To construct amino-terminally hemagglutinin (HA)-tagged RAD9, the 5′ noncoding and amino-terminal regions of the RAD9 gene were amplified by PCR with two primer sets to fuse the HA epitope. The YIp-RAD9-HA plasmid was constructed by a three-part ligation of the SalI-MluI-treated 5′ noncoding fragment and the SpeI-MluI-treated amino-terminal fragment with SalI-SpeI-linearized YIplac128 (8). The SalI-PshAI fragment of YIp-RAD9-HA and the PshAI-PstI fragment of pRS316-rad97XA (29) were cloned into YIplac204 (8), generating YIp-RAD97XA-HA. The pGST-RAD9(S/TQ) and pGST-RAD9(AQ) plasmids were constructed as follows. The S/TQ cluster region (amino acids 361 to 656 of Rad9) was amplified by PCR with the wild-type RAD9 gene and pRS316-rad97XA as templates. The resulting PCR products were treated with BamHI and XhoI and cloned into pGEX6P-1 (Amersham Pharmacia Biotech). YIpT-RAD53-HA was constructed from YCpT-RAD53-HA (13) by BglII digestion, blunting with Klenow fragment, and subsequent self-ligation. YCp-RAD53-myc was constructed from YCp-RAD53-HA (30) after replacement of a DNA fragment encoding the HA epitope with one encoding the myc epitope. The YCp-based pGAL-HO plasmid was previously described (13).
Strains.
The RAD9-HA::TRP1 strains were obtained by transforming YIp-RAD9-HA after treatment with PshAI and replacing the LEU2 marker with TRP1. The RAD53-HA::TRP1 strains were obtained by transforming YIpT-RAD53-HA after treatment with SacI. The rad97XA-HA::TRP1 strains were constructed by transformation of YIp-RAD97XA-HA after treatment with SnaBI. The mec1-KN-HA strain was generated by transformation with PCR products amplified with pWJ1077 (23) in combination with YEp-MEC1-KN-HA (35) as previously described (23). Construction of cells carrying MATa-inc and ADH4cs::HIS2 was described previously (19). The mec1-81-HA strains were constructed with YIp-MEC1-HA as previously described (35). The mec1-81, rad17Δ::LEU2, rad24Δ::LEU2, rad53Δ::LEU2, sml1Δ::LEU2, and tel1Δ::KanMX mutations were described elsewhere (20, 30, 35). All of the strains used in this study are isogenic to KSC006 (30) and are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Genotypea |
|---|---|
| KSC1520 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 |
| KSC1623 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 sml1Δ::LEU2 |
| KSC1535 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 mec1Δ::LEU2 sml1Δ::LEU2 |
| KSC1624 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 mec1-KN::KanMX sml1Δ::LEU2 |
| KSC1625 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 tel1Δ::KanMX sml1Δ::LEU2 |
| KSC1689 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 rad53Δ::LEU2 sml1Δ::LEU2 |
| KSC1713 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 mec1-81 sml1Δ::LEU2 |
| KSC1527 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 rad17Δ::LEU2 |
| KSC1532 | MATa-inc RAD9-HA::TRP1 ADH4cs::HIS2 rad24Δ::LEU2 |
| KSC1626 | MATa-inc rad97xA-HA::TRP1 ADH4cs::HIS2 |
| KSC1628 | MATa-inc RAD53-HA::TRP1 ADH4cs::HIS2 sml1Δ::LEU2 |
| KSC1629 | MATa-inc RAD53-HA::TRP1 ADH4cs::HIS2 mec1Δ::LEU2 sml1Δ::LEU2 |
| KSC1630 | MATa-inc RAD53-HA::TRP1 ADH4cs::HIS2 mec1-81 sml1Δ::LEU2 |
| KSC1333 | MATα MEC-HA::URA3 sml1Δ::LEU2 |
| KSC1627 | MATα mec1-81-HA::URA3 sml1Δ::LEU2 |
| KSC1752 | MATα mec1-KN-HA::URA3 sml1Δ::LEU2 |
All of the the strains are isogenic to KSC006 (30).
Immunoblotting and immunoprecipitation.
Immunoblotting was performed as previously described (30), with the following modification. Cells were suspended in 10 mM Tris Cl (pH 8)-1 mM EDTA and incubated on ice after addition of 7/100 volumes of 5 M NaOH and 2-mercaptoethanol each for 4 min. Ice-cold trichloroacetic acid was subsequently added to a final concentration of 1%, and the mixture was further kept on ice for 10 min. Cells were then centrifuged and washed twice with ice-cold acetone. After drying, cells were suspended with sample buffer containing 1 mM phenylmethylsulfonyl fluoride and physically disrupted. Yeast cells were precultured in medium containing 2% sucrose and then arrested by incubation with 15 μg of nocodazole per ml for 120 min. To induce HO expression, galactose was added to the culture to a final concentration of 2%. Immunoprecipitation and cellular fractionation were done as described previously (35).
Measurement of DSB processing.
Yeast cells were precultured in medium containing 2% sucrose and then arrested with 15 μg of nocodazole per ml for 120 min at G2/M. To induce HO expression, galactose was added to a final concentration of 2%. Purified DNA was fixed to a positively charged nylon membrane and probed with sequences complementary to DNA strands near the HO restriction site at the ADH4 locus as previously described (19). Cleavage at the ADH4 locus was confirmed by Southern blot analysis (13).
In vitro kinase assay.
In vitro kinase assay of Mec1 was done as described previously (20, 30). Kinase assay for Mec1 was performed in 40 μl of kinase buffer by the addition of 10 mCi of [γ-32P]ATP (3,000 Ci/mmol; Amersham Pharmacia Biotech), substrate [2 μg of glutathione S-transferase (GST)-RAD9(S/TQ) or GST-RAD9(AQ) and 0.6 μg of PHAS-1], and ATP to 100 μM.
Chromatin immunoprecipitation assay.
A chromatin immunoprecipitation assay was performed essentially as previously described (19). Yeast cells were precultured in medium containing 2% sucrose and then arrested with 15 μg of nocodazole per ml for 120 min. To induce HO expression, galactose was added to a final concentration of 2%. Immunoprecipitation of cross-linked DNA was performed after sonication with anti-HA (12CA5) monoclonal antibodies. DNA was also purified from the whole-cell extract (designated “input”). PCR was performed under nonsaturating conditions, in which the rate of PCR amplification is proportional to the substrate concentration and cycling. The sequences of the primers for the HO1 set at the ADH4 locus are 5′-TCTATTAATGAGCCGAGACCGGTA-3′ and 5′-CGCATGTGAATGACACACGAAAGT-3′, those for the HO2 set at the ADH4 locus are 5′-CATTATTCTCGGAAGTAGAGTCGA-3′ and 5′-TTCGCGAGAAGAAGGTACATGATC-3′, and those for the SMC2 locus are 5′-AAGAGAAACTTTAGTCAAAACATGGG-3′ and 5′-CCATCACATTATACTAACTACGG-3′.
RESULTS
Phosphorylation of Rad9 in response to the HO-induced DSB.
To determine whether localization of Rad9 is regulated by DNA damage, we examined the association of Rad9 with regions near DSBs generated by the site-specific HO endonuclease. For this purpose, we used strains in which the MAT locus has been replaced with MATa-inc, containing a mutated HO cleavage site, and a separate cleavage site is introduced into the ADH4 locus (19) (Fig. 1A). In this background, HO induces a single DSB at the ADH4 locus, which in turn activates the Rad53 checkpoint pathway at the G2/M phase (22, 33). Rad9 is phosphorylated following DNA damage, as shown by decreased electrophoretic mobility, and this phosphorylation of Rad9 is involved in activation and phosphorylation of Rad53 (7, 29, 34). We then examined the Rad9 phosphorylation status in response to the HO-induced DSB with cells expressing an HA-tagged Rad9 protein (Fig. 1B). Cells carrying the GAL-HO plasmid were grown initially in sucrose to repress HO expression and then transferred to medium containing nocodazole to arrest at G2/M. After arrest, galactose was added to induce HO expression. Cells were collected at various times and subjected to immunoblotting analysis. Rad9 became phosphorylated 2 h after HO expression, as evidenced by the appearance of modified forms.
FIG. 1.
Phosphorylation of Rad9 and its association with HO-induced DSBs. (A) Schematic of the HO cleavage site at the ADH4 locus (ADH4cs). An HO cleavage site, labeled HIS2, was introduced at the ADH4 locus of chromosome (Chr.) VII. The primer pairs were designed to amplify regions 1 and 2 kb from the HO cleavage site. An arrow represents the telomere. The black and gray bars indicate the probes used to examine the rate of degradation of the DSB ends (see Fig. 8). (B) Rad9 phosphorylation after HO expression. Cells expressing Rad9-HA (KSC1520) were transformed with pGAL-HO. Transformed cells were grown in sucrose to repress HO expression and synchronized at G2/M phase with nocodazole. After arrest, the culture was incubated with galactose to induce HO expression. Cells were collected at the indicated times and then subjected to immunoblotting analysis. (C) Association of Rad9 with sites near the HO-induced DSB. Cells expressing Rad9-HA (KSC1520) were transformed with pGAL-HO. Transformed cells were grown as described for panel B to induce HO expression. Cells were harvested at the indicated times and subjected to chromatin immunoprecipitation (IP). PCR was performed with the primers for the ADH4 locus and the control SMC2 locus. PCR products of the respective input extracts are shown in parallel.
Association of Rad9 with the HO-induced DSB.
We next investigated whether Rad9 associates with the HO-induced DSB (Fig. 1C). Cells expressing HA-tagged Rad9 were transformed with the GAL-HO plasmid and grown as described above to induce HO expression. Cells were collected at various times, and extracts prepared after formaldehyde cross-linking were sonicated and subjected to immunoprecipitation with anti-HA antibodies. Coprecipitated DNA was extracted and amplified by PCR with either primer sets (HO1 and HO2) corresponding to regions near the HO restriction site on the ADH4 locus on chromosome VII or primers for the SMC2 locus containing no cleavage site on chromosome VI. PCRs with the primer pairs HO1 and HO2 amplify regions 1.0 and 2.0 kb distant from the cleavage site, respectively (Fig. 1A). PCR amplification with the HO1 and HO2 primer sets was detected in cells carrying the GAL-HO plasmid after incubation with galactose (Fig. 1C). In contrast, there was no increase in the PCR product amplified from the SMC2 locus after incubation with galactose (Fig. 1C). No PCR amplification was observed in untagged cells or in cells lacking a HO cleavage site at the ADH4 locus (data not shown). These results indicate that Rad9 associates with sites near the HO-induced DSB.
Effect of the mec1Δ or tel1Δ mutation on association of Rad9 with DSBs.
DNA damage-induced phosphorylation of Rad9 is dependent on Mec1 and Tel1 (7, 34). We therefore tested whether HO-induced Rad9 phosphorylation requires Mec1 and/or Tel1 in G2/M-arrested cells (Fig. 2A). Because the lethality of the mec1 disruption is suppressed by sml1 mutation (38), we examined the effect of the mec1Δ mutation in an sml1Δ background. Rad9 was phosphorylated in tel1Δ mutants similar to wild-type cells after HO expression, but phosphorylation was not detected in mec1Δ mutants. Treatment with 4-nitroquinoline-1-oxide is known to induce phosphorylation of Rad9 in a process dependent on Tel1, and partial phosphorylation of Rad9 is observed in mec1Δ cells (7). Consistently, Rad9 was partially phosphorylated in the same mec1Δ mutant strain after treatment with 4-nitroquinoline-1-oxide at G2/M (data not shown). Thus, Mec1 is essential for the HO-induced phosphorylation of Rad9, whereas Tel1 has little involvement.
FIG. 2.
Association of Rad9 with the HO-induced DSB in mec1 and tel1Δ mutant cells. (A and B) Effects of mec1Δ, tel1Δ, and mec1-KN mutations on HO-induced phosphorylation of Rad9 and its association with the DSB. Wild-type (WT) (KSC1623) cells and mec1Δ (KSC1535), mec1-KN (KSC1624), and tel1Δ (KSC1625) mutant cells expressing Rad9-HA were transformed with pGAL-HO. Transformed cells were grown in sucrose to repress HO expression and synchronized at G2/M phase with nocodazole. After arrest, the culture was incubated with galactose for 3 h to induce HO expression (+), while part of the culture was maintained in sucrose to repress HO expression (−). Cells were then subjected to immunoblotting analysis (A) or chromatin immunoprecipitation (IP) assay (B). (C) Intracellular distribution of Rad9 in wild-type and mec1Δ mutant cells. Wild-type (KSC1623) and mec1Δ mutant (KSC1535) cells expressing Rad9-HA were harvested and spheroplasted. Spheroplasts were homogenized to prepare whole-cell extract (W) and then separated into cytoplasmic (C) and nuclear (N) fractions. Aliquots were analyzed on immunoblots with anti-HA, anti-glucose-6-phosphate dehydrogenase (G6PD), and anti-nuclear pore complex (NPC) antibodies. (D) Extended kinetics analysis of the association of Rad9 with the DSB. Wild-type (KSC1623) and mec1Δ mutant (KSC1535) cells expressing Rad9-HA were transformed with pGAL-HO and then subjected to chromatin immunoprecipitation assay as described for Fig. 1C.
We next examined the association of Rad9 with the HO-induced DSB in mec1Δ and tel1Δ mutants (Fig. 2B). Association of Rad9 with the DSB was significantly decreased in mec1Δ mutants compared to that in wild-type cells, whereas no apparent defect was detected in tel1Δ mutant cells. The decreased association of Rad9 with DSBs in mec1Δ mutants would be explained if the Rad9 protein localized to the nucleus and its cellular localization might be affected by the mec1Δ mutation. To exclude this possibility, we examined the intracellular distribution of Rad9 in wild-type and mec1Δ cells (Fig. 2C). Whole-cell extracts were fractionated into two separate cytoplasmic and nuclear fractions. Equal-volume aliquots of these fractions and whole-cell extract were analyzed on immunoblots to detect Rad9-HA. As a control for the fractionation, we assessed each fraction for the presence of cytoplasmic glucose-6-phosphate dehydrogenase and a nuclear pore complex protein. Most Rad9 proteins were separated in the nuclear fractions in wild-type cells, consistent with the previous finding that Rad9 is predominantly localized to the nucleus (16). Rad9 was fractionated to nuclei in mec1Δ mutants as well. Thus, the mec1Δ mutation does not affect the cellular localization of Rad9.
To address whether Mec1 kinase activity is important, we examined the Rad9 association in cells carrying a kinase-negative mec1 allele (mec1-KN mutants) (35). We found that phosphorylation of Rad9 and its association with DSBs were not detectable in mec1-KN mutants (Fig. 2A and B). Altogether, these results suggest that Mec1 kinase activity is essential for HO-induced Rad9 phosphorylation and association of Rad9 with the HO-induced DSB.
Rad9 is homologous to Crb2 of fission yeast, whereas Mec1 is closely related to fission yeast Rad3. Recently, Du et al. (3) showed that accumulation of Crb2 at sites corresponding to DSBs can be separated into two steps, i.e., initial recruitment and continued retention, and that Rad3 controls the retention of Crb2 at DSBs. We therefore addressed whether association of Rad9 with DSBs might be regulated in a similar manner. If Mec1 controlled the retention of Rad9, transient association of Rad9 with DSBs could be detected in mec1Δ mutants while Rad9 recruitment is in progress. Alternatively, if Mec1 controlled the recruitment of Rad9, no Rad9 association should be detected in mec1Δ mutants. To define the Rad9 recruitment process, we extended the time course analysis examining the association of Rad9 after HO expression. Rad9 association reached a plateau at 3 h after HO expression (Fig. 2D), suggesting that Rad9 is actively recruited to the DSB within 3 h after HO expression. In contrast, no apparent association was detected in mec1Δ mutants during the time course (Fig. 2D). Southern blot analysis indicated that the cleavage at the ADH4 locus occurred within 1 h after HO expression in both wild-type and mec1Δ mutant cells (data not shown). These results suggest that Mec1 is required for Rad9 recruitment to DSBs. However, it remains possible that Mec1 controls Rad9 retention because our assay might not be sensitive enough to detect transient association of Rad9 with DSBs in mec1Δ mutant cells.
Effects of mutations in S/TQ motifs on the association of Rad9 with DSBs.
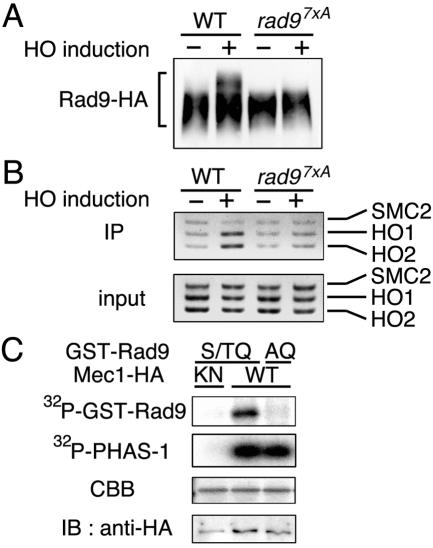
Rad9 contains a total of 14 S/TQ motifs, which are consensus sites of phosphorylation by the ATM and ATR family proteins (1). Recent evidence demonstrated that seven serine or threonine residues (Thr390, Thr398, T410, T420, S435, T457, and T603) within the S/TQ motifs are redundantly required for DNA damage-induced phosphorylation (29). The Rad97XA mutant protein, in which these seven serine and threonine residues are all replaced with alanine, is very poorly phosphorylated after DNA damage and is defective in checkpoint activation.
To further address the possibility that phosphorylation of Rad9 is required for its association with the HO-induced DSB, we examined whether the rad97XA mutation affects HO-induced phosphorylation and association of Rad9 with the DSB (Fig. 3). We found that, in contrast to wild-type Rad9, Rad97XA did not become phosphorylated after HO expression (Fig. 3A). Moreover, association of Rad97XA with the HO-induced DSB was significantly decreased (Fig. 3B). Rad97XA was fractionated into the nuclear fraction similarly to wild-type Rad9 (data not shown). These results indicate that the rad97XA mutation causes a defect in the association of Rad9 with the HO-induced DSB.
FIG. 3.
Effects of mutations in the Rad9 S/TQ motifs on the association with DSBs and in vitro phosphorylation by Mec1. (A) Phosphorylation of the Rad97XA mutant protein after HO expression. RAD9-HA (KSC1520) and rad97xA-HA (KSC1626) cells were transformed with pGAL-HO and analyzed as described for Fig. 2A. WT, wild type. (B) Association of the Rad97xA mutant protein with the HO-induced DSB. RAD9-HA (KSC1520) and rad97xA-HA (KSC1626) cells carrying pGAL-HO were analyzed as described for Fig. 2B. IP, immunoprecipitate. (C) In vitro phosphorylation of the Rad9 S/TQ motifs by Mec1. Extracts were prepared from MEC1-HA (KSC1333) and mec1-KN-HA (KSC1752) cells and subjected to immunoprecipitation with anti-HA-antibodies. The immunoprecipitated Mec1 proteins were assayed for kinase activity with mixtures of PHAS-1 with GST-RAD9(S/TQ) or GST-RAD9(AQ) as substrates. The top two autoradiograms show 32P incorporation into the GST-RAD9 fusion proteins or PHAS-1. In the third autoradiogram, Coomassie brilliant blue (CBB) staining shows the amounts of the GST-RAD9 fusion proteins. In the bottom autoradiogram, immunoblotting (IB) with anti-HA antibodies indicates the amount of the Mec1 protein used for the kinase assay.
Rad9 phosphorylation after DNA damage is defective in mec1Δ mutants, suggesting that Mec1 might directly phosphorylate the Rad9 S/TQ motifs in vitro. To test this, we made two GST-Rad9 fusion proteins: GST-RAD9(S/TQ), which contains the seven S/TQ motifs, and GST-RAD9(AQ), which possesses the same region but replaces serine or threonine in the motifs with alanine. PHAS-1 was used as a control substrate (15, 20) and incubated together with GST-RAD9(S/TQ) or GST-RAD9(AQ). Extracts were prepared from cells expressing the wild-type Mec1-HA or kinase-negative Mec1-KN-HA protein and immunoprecipitated with anti-HA antibodies. Immunoprecipitates were then subjected to a kinase assay. We found that GST-RAD9(S/TQ) and PHAS-1 were phosphorylated in immunoprecipitates containing Mec1-HA but not in those containing Mec1-KN-HA (Fig. 3C). In contrast, no specific phosphorylation of GST-RAD9(AQ) was observed with Mec1-HA (Fig. 3C). Thus, Mec1 phosphorylates serine and threonine residues of the Rad9 S/TQ motifs in vitro. Together, these data support a model in which Mec1 phosphorylates the S/TQ motifs and then facilitates the association of Rad9 with DSBs.
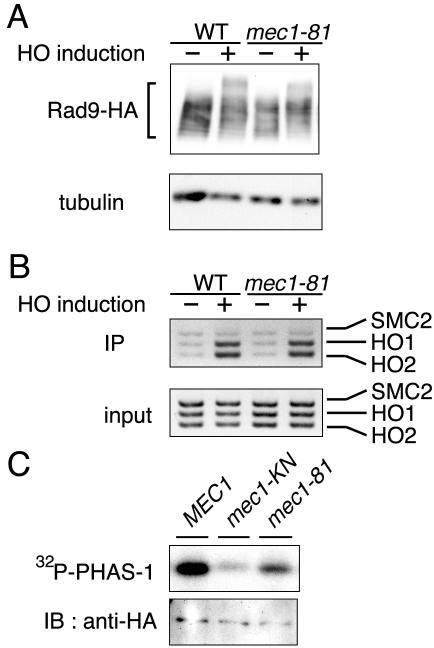
Effect of the mec1-81 mutation on the phosphorylation of Rad9 and its association with DSBs.
Since Mec1 controlled the phosphorylation of Rad9 and its association with DSBs, it is possible that the association of Rad9 with DSBs is coupled with its phosphorylation status. If this were the case, both phosphorylation of Rad9 and its association would be reduced in cells carrying weak mec1 mutation alleles. To test this possibility, we examined the phosphorylation of Rad9 and its association with DSBs in mec1-81 mutant cells (20) (Fig. 4A and B). The Rad9 phosphorylation level in mec1-81 mutants was low compared with that in wild-type cells, and fully modified Rad9 was not detected in the mec1-81 mutants (Fig. 4A). However, no significant defect in the association of Rad9 with DSBs was detected in mec1-81 mutants (Fig. 4B).
FIG. 4.
Effect of the mec1-81 mutation on the association of Rad9 with DSBs and Mec1 kinase activity. (A) Phosphorylation of Rad9 in mec1-81 mutants after HO expression. Wild-type (WT) (KSC1623) and mec1-81 mutant (KSC1713) cells expressing Rad9-HA were transformed with pGAL-HO. Cells were then subjected to immunoblotting analysis as described for Fig. 2A. Tubulin was detected as a loading control. (B) Association of Rad9 with the HO-induced DSB in mec1-81 mutants. Wild-type (KSC1623) and mec1-81 mutant (KSC1713) cells expressing Rad9-HA were transformed with pGAL-HO. Cells were then subjected to chromatin immunoprecipitation (IP) assay as described for Fig. 2B. (C) Phosphorylation of PHAS-1 by the Mec1-81 mutant protein in vitro. Extracts were prepared from MEC1-HA (KSC1333), mec1-81-HA (KSC1627), and mec1-KN-HA (KSC1752) cells and subjected to immunoprecipitation with anti-HA antibodies. The immunoprecipitated Mec1 proteins were assayed for kinase activity with PHAS-1 as a substrate. The autoradiogram shows 32P incorporation into PHAS-1 (top), and immunoblotting (IB) with anti-HA antibodies indicates the amount of Mec1 protein used for the kinase assay (bottom).
To understand the effect of mec1-81 mutations, we examined the kinase activity of the Mec1-81 mutant protein in vitro (Fig. 4C). Extracts were prepared from cells expressing the wild-type Mec1-HA, Mec1-81-HA, or kinase-negative Mec1-KN-HA protein and immunoprecipitated with anti-HA antibodies. Immunoprecipitates were then subjected to kinase assay with PHAS-1 as a substrate. PHAS-1 phosphorylation was reduced in the immunoprecipitates containing Mec1-81-HA compared with those containing Mec1-HA. Thus, although Mec1 kinase activity is essential for association of Rad9 with DSBs, its association does not appear to require full kinase activity of Mec1.
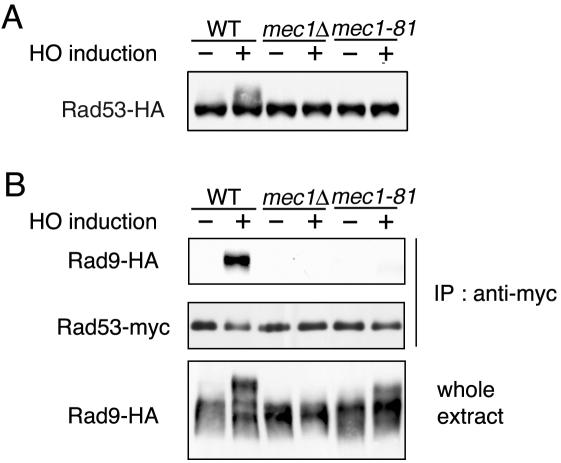
Relationships among Mec1-dependent phosphorylation, DSB binding, and interaction with Rad53.
Hyperphosphorylation of Rad9 leads to its physical interaction with the Rad53 kinase (7, 32, 34). Recent studies have implicated the Rad9-Rad53 complex as machinery that activates Rad53, thereby facilitating Rad53 autophosphorylation (9). Rad53 phosphorylation after DNA damage is decreased in mec1-81 mutants similar to mec1Δ mutants (19). Correspondingly, no apparent Rad53 phosphorylation was detected in mec1-81 or mec1Δ mutants after HO expression (Fig. 5A) (19). To assess the functional link of Rad9 to Rad53 activation, we examined the physical interaction between Rad9 and Rad53 in mec1Δ and mec1-81 mutants after HO expression (Fig. 5B). Cells expressing HA-tagged Rad9 and myc-tagged Rad53 were transformed with the GAL-HO plasmid and grown as described above to induce HO expression. Extracts were prepared from cells and then subjected to immunoprecipitation with anti-myc antibodies. Consistent with the current model in which Rad53 interacts with phosphorylated Rad9, Rad53 was immunoprecipitated with Rad9 in wild-type cells after HO expression, whereas no Rad9-Rad53 interaction was detected in mec1Δ mutants. In contrast, Rad9 was phosphorylated, albeit weakly, in mec1-81 mutants, but the Rad9-Rad53 interaction was significantly reduced in mec1-81 mutants, as observed in mec1Δ mutants.
FIG. 5.
Physical interaction between Rad9 and Rad53 in mec1Δ and mec1-81 mutant cells. (A) Wild-type (WT) (KSC1628) cells and mec1Δ (KSC1629) and mec1-81 (KSC1630) mutant cells expressing Rad53-HA were transformed with pGAL-HO. Cells were then subjected to immunoblotting analysis as described for Fig. 2A. (B) Wild-type (KSC1623) cells and mec1Δ (KSC1535) and mec1-81 (KSC1713) mutant cells expressing Rad9-HA were transformed with YCp-RAD53-myc and pGAL-HO. Transformed cells were grown as described for Fig. 2A to induce HO expression. Extracts were prepared from cells and subjected to immunoprecipitation (IP) with anti-myc antibodies. The extracts and immunoprecipitates were then analyzed on immunoblots with anti-myc and anti-HA antibodies. To detect Rad53-myc proteins, sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis was performed briefly to compact the Rad53 phospho forms.
Since Rad9 functions upstream of Rad53 in the DNA damage checkpoint pathway (9, 29), we examined whether Rad9 associates with the HO-induced DSB in rad53Δ mutants. The effect of the rad53Δ mutation was monitored in an sml1Δ background, because sml1 mutations suppress the lethality of the rad53 disruption (38). Rad9 associated with the HO-induced DSB in rad53Δ mutant cells as efficiently as in wild-type cells (Fig. 6A). Consistently, the rad53Δ mutation did not significantly affect Rad9 phosphorylation (Fig. 6B) (7, 34). Together, these results suggest that the Rad9-Rad53 interaction occurs efficiently after Rad9 associates with DSBs and becomes fully phosphorylated in a Mec1-dependent manner.
FIG. 6.
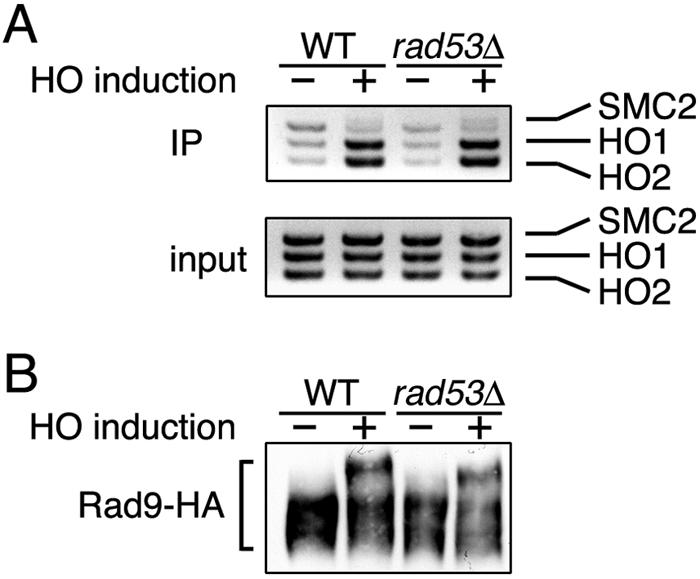
Effect of rad53Δ mutation on association of Rad9 with DSBs. (A) Association of Rad9 with HO-induced DSB in rad53Δ mutants. Wild-type (WT) (KSC1623) and rad53Δ mutant (KSC1689) cells expressing Rad9-HA were transformed with pGAL-HO. Cells were then subjected to a chromatin immunoprecipitation (IP) assay as described for Fig. 2B. (B) Phosphorylation of Rad9 in rad53Δ mutants after HO expression. Wild-type (KSC1623) and rad53Δ mutant (KSC1689) cells expressing Rad9-HA were transformed with pGAL-HO. Cells were then subjected to immunoblotting analysis as described for Fig. 2A.
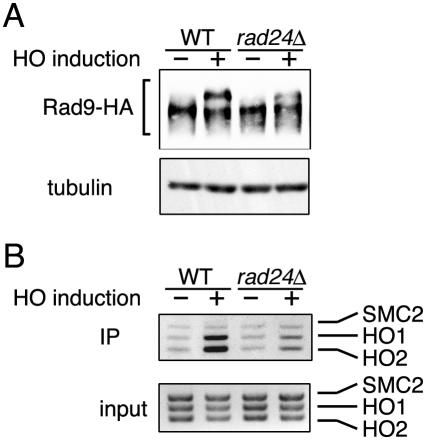
Effect of the rad24Δ mutation on the association of Rad9 with DSBs.
Rad24 forms an RFC-related complex and recruits the Rad17-Mec3-Ddc1 complex to sites of DNA damage. These Rad24, Rad17, Mec3, and Ddc1 proteins are also required for Rad9 phosphorylation after DNA damage. We then examined Rad53 phosphorylation and association with DSBs in rad24Δ mutants after HO expression. Consistent with the previous results (7, 34), the Rad9 phosphorylation level in rad24Δ mutants was partially decreased compared with that in wild-type cells (Fig. 7A). Moreover, association of Rad9 with the HO-induced DSB was lowered in rad24Δ mutants (Fig. 7B). Similar results were obtained with rad17Δ mutants (data not shown). These results indicate that the Rad24 pathway is required for efficient association of Rad9 with the HO-induced DSB.
FIG. 7.
Effect of rad24Δ mutation on association of Rad9 with DSBs. (A) Phosphorylation of Rad9 in rad24Δ mutants after HO expression. Wild-type (WT) (KSC1520) and rad24Δ mutant (KSC1532) cells expressing Rad9-HA were transformed with pGAL-HO. Cells were then subjected to immunoblotting analysis as described for Fig. 2A. Tubulin was detected as a control. (B) Association of Rad9 with the HO-induced DSB in rad24Δ mutants. Wild-type (KSC1520) and rad24Δ mutant (KSC1532) cells expressing Rad9-HA were transformed with pGAL-HO and subjected to a chromatin immunoprecipitation (IP) assay as described for Fig. 2B.
Effect of the mec1Δ mutation on accumulation of ssDNA at DSB ends.
Recently, Zhou and Elledge (40) provided evidence that ATRIP or Ddc2 binds to RPA-coated ssDNA, thereby recruiting the ATR-ATRIP or Mec1-Ddc2 complex to sites of DNA damage. DSB ends are degraded primarily by 5′-to-3′ exonuclease activity, producing long 3′-end ssDNA tails (37). As shown above, association of Rad9 with DSBs is significantly decreased in mec1Δ mutants. If Rad9 interacts with ssDNA at DSBs, it is possible that ssDNA accumulation at the DSB end is reduced in mec1Δ mutants compared with that in wild-type cells. We then examined the degradation rate following HO expression from the galactose-inducible GAL10 promoter in G2/M-arrested cells (Fig. 8). Cells carrying the GAL-HO plasmid were grown as described above to induce HO expression. Cells were collected at various times to prepare genomic DNA. Purified DNA was fixed to a membrane and probed with strand-specific sequences near the cleavage site at the ADH4 locus. The 5′-to-3′ degradation occurred in mec1Δ mutants as efficiently as in wild-type cells (Fig. 8A). Moreover, the mec1Δ mutation had little effect on the rate of 3′-to-5′ degradation (Fig. 8B). Thus, the mec1Δ mutation does not impair the DNA degradation at the DSB end, suggesting that ssDNA accumulation by itself does not promote association of Rad9 with DNA lesions.
FIG. 8.
Degradation of HO-induced DSB ends. Wild-type (WT) (KSC1623) and mec1Δ (KSC1535) cells carrying pGAL-HO were grown in sucrose and treated with nocodazole. After arrest at G2/M, the culture was incubated with galactose to induce HO expression. At the indicated time points, aliquots were harvested for DNA preparation. Purified DNAs were fixed to a membrane and probed with 32P-labeled oligonucleotides each complementary to a 5′-to-3′-degrading strand (A) or a 3′-to-5′-degrading strand (B).
DISCUSSION
Rad9 in budding yeast is required for activation of the DNA damage checkpoint pathway. Previous studies have shown that Rad9 is a checkpoint mediator involved in transducing the DNA damage signal. Rad9 is phosphorylated after DNA damage in a Mec1- and Tel1-dependent manner, and phosphorylated Rad9 interacts with Rad53 (7, 9, 32, 34). Furthermore, recent biochemical evidence suggests that the Rad9-Rad53 complex acts as machinery to activate the Rad53 kinase (7, 9, 32, 34). Rad9 forms foci in the presence of DNA damage (16) and associates with replication origins when DNA replication is inhibited in checkpoint-defective cells (11). However, there is no direct evidence that Rad9 associates with regions corresponding to sites of DNA damage. In this study, we showed that Rad9 associates with the HO-induced DSBs and that the association of Rad9 requires Mec1 function. We also showed that although Rad9 is not fully phosphorylated, Rad9 associates efficiently with the DSBs in cells carrying a weak mec1 mutation allele. Our results support the model in which Mec1 promotes the association of Rad9 with DNA lesions and then fully phosphorylates Rad9.
To examine whether Rad9 associates with sites of DNA damage, we used strains carrying a single HO cleavage site, in which the HO-induced DSB is not efficiently repaired by homologous recombination. We found that Rad9 associated with the HO-induced DSB and that its association required Mec1 function. Rad9 contains 14 S/TQ motifs throughout its entire length, motifs that are potentially phosphorylated by ATM and ATR family proteins Mec1 and Tel1. Although any single mutation of the Rad9 S/TQ motifs does not have a significant effect, replacement of the seven S/TQ motifs in the Rad9 central region confers a defect in Rad9 phosphorylation after DNA damage (29). Notably, these multiple replacements in Rad9 caused a defect in its association with DSBs. In addition, Mec1 phosphorylated these Rad9 S/TQ sites in vitro. These results are consistent with the model in which Mec1 phosphorylates S/TQ motifs of Rad9 and promotes its association with sites of DNA damage. However, we cannot exclude the possibility that the replacement-containing Rad97XA mutant protein might be misfolded or debilitated so that it has lost the initial DSB-binding capacity independently of its phosphorylation status (see below). Phosphorylation of the seven S/TQ motifs has been shown to contribute to the interaction with Rad53 (29). Our results here suggest that their phosphorylation is also important for association with sites of DNA damage.
Mec1 promotes the association of Rad9 with DSBs, but Mec1 by itself associates with sites of DNA damage (40). If Mec1 and Rad9 were colocalized, Mec1 could efficiently phosphorylate Rad9. It is thus possible that association of Rad9 with DNA lesions precedes full phosphorylation. Supporting this possibility, we found that Rad9 associated efficiently with DSBs, but its phosphorylation was partially decreased in mec1-81 mutants. We note, however, that the chromatin immunoprecipitation assay might not be as sensitive as the immunoblotting analysis, and thereby a slight decrease in the association of Rad9 could not be detected in mec1-81 mutants. The Mec1-81 mutant protein showed decreased kinase activity in vitro. These results suggest that although all of the S/TQ motifs of Rad9 could be phosphorylated after DNA damage, phosphorylation of some motifs might be sufficient for association of Rad9 with DNA lesions.
Phosphorylated Rad9 interacts with Rad53 through the Rad53 FHA domain (32), and this Rad9-Rad53 interaction is implicated in the activation and phosphorylation of Rad53 (7, 9, 32, 34). Rad9 was phosphorylated after HO expression in wild-type cells but not in mec1Δ mutant cells. Correspondingly, Rad9 interacted with Rad53 in wild-type cells but not in mec1Δ mutant cells. Rad9 is phosphorylated, albeit weakly, after HO expression in mec1-81 mutant cells, but the Rad9-Rad53 interaction was undetectable in these mutant cells. These results suggest that full phosphorylation of Rad9 is important for efficient interaction of Rad9 with Rad53. However, it remained possible that Rad53 binds to specific sets of phosphopeptide sequences of Rad9, and phosphorylation of those sites is defective in mec1-81 mutant cells.
The RAD24, RAD17, MEC3, and DDC1 genes constitute a DNA damage response pathway. Indeed, Rad24 forms an RFC-related complex and recruits the Rad17-Mec3-Ddc1 complex to sites of DNA damage. These Rad24, Rad17, Mec3, and Ddc1 proteins have been shown to regulate Rad9 phosphorylation after DNA damage. Consistently, Rad9 phosphorylation after HO expression was partially decreased in rad24Δ mutants. Although the mec1-81 mutation did partially decrease Rad9 phosphorylation after HO expression, it did not apparently affect the association of Rad9 with DSBs. In contrast, rad24Δ mutation reduced the association of Rad9 with DSBs as well. One likely explanation is that phosphorylated sites on Rad9 are different in mec1-81 and rad24Δ mutants. Although similar partial phosphorylation occurs after HO expression, phosphorylation involved in DSB binding might be decreased in rad24Δ mutants, but not in mec1-81 mutants. Alternatively, the rad24Δ mutation might affect DNA structure or proteins localizing at sites of DNA damage, thereby reducing the association of Rad9 with the damage sites.
Fission yeast checkpoint protein Crb2 is related to budding yeast Rad9. Recently, Du et al. (3) demonstrated that Crb2 localizes to foci that represent sites of DSBs. In fission yeast, Rad3 is structurally and functionally similar to budding yeast Mec1. Crb2 foci are induced after gamma irradiation in cells lacking Rad3, but the foci disappear immediately in the mutant cells. Thus, Rad3 is required not for the initial recruitment of Crb2 to sites of DNA damage but for its persistent localization. Since Crb2 and Rad9 are related each other, it seems likely that these proteins are regulated similarly. In this scenario, Rad9 could be recruited to DNA lesions independently of Mec1, and after Mec1-dependent phosphorylation, Rad9 would associate stably with the DNA lesions. However, our results suggest that Mec1 may control the recruitment of Rad9 to DSBs. One possible explanation is that recruitment of Rad9 is entirely coupled to its retention at DSBs in budding yeast. Similar to Rad9, Crb2 is phosphorylated after DNA damage, and its damage-induced phosphorylation is dependent on Rad3 (26). Conversely, our results imply that phosphorylation of Crb2 is required for its retention at DNA lesions.
Recent evidence suggests that checkpoint proteins have the ability to bind to ssDNA, thereby associating with DNA lesions (40). However, ssDNA accumulation was little affected in mec1Δ mutants, which exhibited a defect in the association of Rad9 with DSBs. Thus, ssDNA alone does not appear to promote association of Rad9 with DNA lesions, although ssDNA might be involved with the association.
In summary, we showed that Rad9 associates with sites of DNA damage and that its association is controlled by Mec1. However, it remains undetermined what DNA structure or protein connects Rad9 to sites of DNA damage. Moreover, it is unclear how phosphorylated Rad9 at DNA lesions mediates signals to downstream targets. Further experiments will be aimed at understanding the biochemical properties of Rad9 at sites of DNA damage.
Acknowledgments
We thank A. Shinohara, D. Stern, and M. Schwartz for materials; S. Ando and Y. Hirano for plasmid construction; and H. Araki, C. Newlon, and K. Shirahige for discussion.
This work was supported by grants-in-aid from the Ministry of Education and Science of Japan.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 3.Du, L. L., T. M. Nakamura, B. A. Moser, and P. Russell. 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durocher, D., J. Henckel, A. R. Fersht, and S. P. Jackson. 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4:387-394. [DOI] [PubMed] [Google Scholar]
- 5.Edwards, R. J., N. J. Bentley, and A. M. Carr. 1999. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat. Cell Biol. 1:393-398. [DOI] [PubMed] [Google Scholar]
- 6.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 7.Emili, A. 1998. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2:183-189. [DOI] [PubMed] [Google Scholar]
- 8.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 10.Green, C. M., H. Erdjument-Bromage, P. Tempst, and N. F. Lowndes. 2000. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 10:39-42. [DOI] [PubMed] [Google Scholar]
- 11.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1073-1083. [DOI] [PubMed] [Google Scholar]
- 12.Kondo, T., K. Matsumoto, and K. Sugimoto. 1999. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol. Cell. Biol. 19:1136-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 5543:867-870. [DOI] [PubMed] [Google Scholar]
- 14.Longhese, M. P., M. Foiani, M. Muzi-Falconi, G. Lucchini, and P. Plevani. 1998. DNA damage checkpoint in budding yeast. EMBO J. 17:5525-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallory, J. C., and T. D. Petes. 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97:13749-13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 21:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrow, D. M., D. A. Tagle, Y. Shiloh, F. S. Collins, and P. Hieter. 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82:831-840. [DOI] [PubMed] [Google Scholar]
- 18.Naiki, T., T. Shimomura, T. Kondo, K. Matsumoto, and K. Sugimoto. 2000. Rfc5, in cooperation with Rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:5888-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakada, D., K. Matsumoto, and K. Sugimoto. 2003. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 17:1957-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakada, D., T. Shimomura, K. Matsumoto, and K. Sugimoto. 2003. The ATM-related Tel1 protein of Saccharomyces cerevisiae controls a checkpoint response following phleomycin treatment. Nucleic Acids Res. 31:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paciotti, V., M. Clerici, G. Lucchini, and M. P. Longhese. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14:2046-2059. [PMC free article] [PubMed] [Google Scholar]
- 22.Pellicioli, A., S. E. Lee, C. Lucca, M. Foiani, and J. E. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7:293-300. [DOI] [PubMed] [Google Scholar]
- 23.Reid, R. J., I. Sunjevaric, M. Keddache, and R. Rothstein. 2002. Efficient PCR-based gene disruption in Saccharomyces strains using intergenic primers. Yeast 19:319-328. [DOI] [PubMed] [Google Scholar]
- 24.Rouse, J., and S. P. Jackson. 2000. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J. 19:5793-5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rouse, J., and S. P. Jackson. 2002. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol. Cell 9:857-869. [DOI] [PubMed] [Google Scholar]
- 26.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 30.Sugimoto, K., S. Ando, T. Shimomura, and K. Matsumoto. 1997. Rfc5, replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol. Cell. Biol. 17:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 32.Sun, Z., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272-274. [DOI] [PubMed] [Google Scholar]
- 33.Toczyski, D. P., D. J. Galgoczy, and L. H. Hartwell. 1997. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell 90:1097-1106. [DOI] [PubMed] [Google Scholar]
- 34.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wakayama, T., T. Kondo, S. Ando, K. Matsumoto, and K. Sugimoto. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinert, T. A., and L. H. Hartwell. 1988. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241:317-322. [DOI] [PubMed] [Google Scholar]
- 37.White, C. I., and J. E. Haber. 1990. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 9:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, X., E. G. D. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pool. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 39.Zhou, B.-B. S., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 40.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]