Abstract
Screening the Saccharomyces cerevisiae disruptome, profiling transcripts, and determining changes in protein expression have identified an important new role for the high-osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) pathway in the regulation of adaptation to citric acid stress. Deletion of HOG1, SSK1, PBS2, PTC2, PTP2, and PTP3 resulted in sensitivity to citric acid. Furthermore, citric acid resulted in the dual phosphorylation, and thus activation, of Hog1p. Despite minor activation of glycerol biosynthesis, the inhibitory effect of citric acid was not due to an osmotic shock. HOG1 negatively regulated the expression of a number of proteins in response to citric acid stress, including Bmh1p. Evidence suggests that BMH1 is induced by citric acid to counteract the effect of amino acid starvation. In addition, deletion of BMH2 rendered cells sensitive to citric acid. Deletion of the transcription factor MSN4, which is known to be regulated by Bmh1p and Hog1p, had a similar effect. HOG1 was also required for citric acid-induced up-regulation of Ssa1p and Eno2p. To counteract the cation chelating activity of citric acid, the plasma membrane Ca2+ channel, CCH1, and a functional vacuolar membrane H+-ATPase were found to be essential for optimal adaptation. Also, the transcriptional regulator CYC8, which mediates glucose derepression, was required for adaptation to citric acid to allow cells to metabolize excess citrate via the tricarboxylic acid (TCA) cycle. Supporting this, Mdh1p and Idh1p, both TCA cycle enzymes, were up-regulated in response to citric acid.
In nature, acidity, or low pH, is an environmental stress to which microorganisms are routinely exposed. However, acid stress is not simply the toxic effect of a high hydrogen ion concentration but is also dependent on the chemical nature of the acid to which the organism is exposed. In reality, there is no single type of acid stress but a host of different inhibitory effects depending on the nature of the acid(s) present, for example, weak acids versus strong acids or organic acids versus inorganic acids. It is well-known that different weak organic acids have hugely different inhibitory effects on microorganisms even when applied at identical pH values (32). Thus, we could hypothesize that due to the chemical diversity of acid stress, it is unlikely that an organism could develop a single general response to acid stress in the same fashion as, for example, the heat shock response to heat stress. In truth, whatever the acid present, an organism still has to adapt to growth in the presence of a high external hydrogen ion concentration. Thus, this component of acid stress is not unique, and in Saccharomyces cerevisiae, there is evidence of a general response to combat this effect (40). However, other inhibitory effects of acids that are entirely dependent on the unique chemistry of each counter ion must be considered. This implies that any microorganism exposed to the acid may induce a unique stress response specific for that particular ion. In fact, available data support this hypothesis. For example, there has been much research on the effects of the weak organic acid food preservatives, sorbic and benzoic acid, on S. cerevisiae. These studies have indicated the importance of the Pdr12 ATP binding cassette (ABC) transporter in the development of resistance to some weak acids (26). In addition, sorbic acid has been shown to up-regulate a number of stress proteins and chaperones, including Hsp26p, Hsp42p, and Hsp70p isoforms and many oxidative stress defense enzymes (7).
The recently identified transcription factor War1p has been implicated in activating PDR12 through an uncharacterized signal transduction event that detects weak organic acid stress (22). This work also found that PDR12 induction requires a unique cis-acting weak acid response element (WARE) in the PDR12 promoter. In contrast, the transcriptional response to acetic acid, another weak organic acid, is not well characterized, but it is known that resistance is mediated by a different plasma membrane transporter of the major facilitator superfamily, AZR1 (39). Thus, these studies have demonstrated that S. cerevisiae has evolved dedicated stress responses to counteract the inhibitory effects of different organic acids.
In this study, we wished to characterize the response of S. cerevisiae to citric acid, which is an organic acid ubiquitous in nature and has not been studied in terms of cellular resistance mechanisms. Citric acid is an intermediary metabolite of the tricarboxylic acid (TCA) cycle and is a key component of normal respiratory metabolism in S. cerevisiae. There are four genes listed in the SGD database (http://www.yeastgenome.org/) under the gene ontology heading of citrate metabolism; ACO1, CIT1, CIT2, and CIT3. Deletion of either ACO1 (encoding aconitate hydratase) or CIT1 (encoding citrate synthase) results in a growth defect in the presence of a nonfermentable (respiratory) carbon source, indicating the importance of citric acid metabolism to the generation of energy via oxidative phosphorylation (37).
Citric acid is a sharp-tasting, water-soluble organic acid that occurs naturally in the juice of lemons and other sour fruits at concentrations typically between 10 and 40 mM. Yeasts are known to occur naturally on the surfaces of such fruits, so it is not unreasonable to propose that yeasts may have evolved to adapt to this form of acid stress. Typical yeast species found in citrus juices include Candida parapsilosis, Candida stellata, Saccharomyces cerevisiae, Torulaspora delbrueckii, and Zygosaccharomyces rouxii (15).
Citric acid is a tri-protic acid with three carboxylic acid groups with pKa values of 3.13, 4.76, and 6.4, respectively. Citric acid and its salts are commonly used in the food and beverage industry as organic acidulants and pH control agents. In manufactured foods and beverages, citric acid is most often used as a flavor adjunct to improve taste, but it is also used as a pH control agent and preservative to prevent microbial growth, often in conjunction with other weak acid preservatives, such as sorbic or benzoic acid, and as a chelating agent to prevent food spoilage. Despite hygienic manufacturing and the use of organic acid preservatives, yeast and mold spoilage still results in huge economic losses of foods and beverages. It has been estimated that food losses to microbial growth in the United Kingdom alone exceeds £100 million each year and that up to 25% of all food crops are lost in the developing world due to spoilage (11).
Yeast spoilage often occurs because the organism is able to adapt and grow under extreme conditions such as low pH in the presence of organic acids. Therefore, fundamental understanding of the molecular mechanisms employed by the yeast that allow it to grow at low pH in the presence of citric acid is also of applied interest.
The combination of whole-genome transcript analysis and proteomics has established the capability to investigate the mode of action of antimicrobial agents at the molecular level in detail. The rationale for this approach is that sublethal exposure to an antimicrobial agent will alter the homeostasis of an organism such that a transcriptional response will be initiated to restore homeostasis in addition to the induction of any resistance mechanisms. The resulting gene, or protein, expression profile serves as a signature of the inhibitor used, and in the case of antimicrobials whose modes of action are unknown, incriminates various genes involved with the inhibited cellular process(es). Thus, functional genomic techniques are being used increasingly to investigate and determine the mode(s) of action of antimicrobials (for example, the effect of isoniazid on Mycobacterium tuberculosis [42] and effects of antifungals, such as fluconazole and ketoconazole, on S. cerevisiae [3]).
In this study, we screened the S. cerevisiae genome deletion set to identify genes mediating resistance to citric acid stress. We also studied differential protein expression and changes in the transcriptome induced by exposure to citric acid stress to identify key components of the molecular response of S. cerevisiae to this compound. Our results have shown that the mitogen-activated protein kinase (MAPK) high-osmolarity glycerol (HOG) pathway regulates resistance to citric acid stress. Thus, we present evidence for an important new role for the HOG pathway in yeast.
MATERIALS AND METHODS
Yeast strains.
The strain of S. cerevisiae used in this study was BY4741, a derivative of S288C with the genotype MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 obtained from Research Genetics (Huntsville, Ala.). All gene deletions in this study were from the Research Genetics BY4741 MATa haploid genome deletion set, which contains 4,847 yeast strains with all nonessential open reading frames (ORFs) disrupted by the KanMX cassette (http://www-sequence.stanford.edu/group/yeast_deletion_project/PCR_strategy.html). All key deletion strains were verified by PCR.
Media and growth assays.
Yeast cells were grown either in malt extract (ME) medium (0.6% malt extract, 0.6% dextrose, 0.18% maltose, 0.12% yeast extract) or in defined minimal medium (0.67% BDH defined medium without amino acids but with 2% glucose) supplemented with appropriate amino acids and bases (20 mg ml−1). Citric acid was added to media from a stock solution of 3.5 M, and the pH was adjusted to 3.5 using 10 M NaOH.
A growth test was developed to screen the entire S. cerevisiae genome gene deletion set for sensitivity or resistance to citric acid. First, using a 48-pin replica plater, individual yeast mutant strains were inoculated from the strain collection stored in 96-well plates into sterile 96-well plates containing fresh ME broth. After overnight incubation at 30°C, growth was monitored by optical density, and approximately 5 μl of the individual cultures was spotted, at an optical density at 600 nm (OD600) of 1.0, using a 48-pin replica plater, onto ME agar medium containing either 0 or 400 mM citric acid, pH 3.5. Colony growth was inspected after 48 h of incubation at 30°C.
More-detailed sensitivity assays were performed by spotting 10-fold serial dilutions of an exponentially growing culture (OD600 of 0.1) onto ME agar plates with increasing concentrations of citric acid (0 to 400 mM), pH 3.5. Colony growth was inspected as described above.
To accurately determine citric acid sensitivity by calculating changes in growth rate, shaking flask experiments were performed. The yeast cells were grown at 30°C in ME medium, pH 3.5, with or without 150 or 300 mM citric acid with shaking at 200 rpm.
Proteome analysis.
Protein was extracted from duplicate mid-exponential-phase cultures at an OD600 of 0.35 (200 ml) grown in ME medium either in the presence or absence of 300 mM citric acid, pH 3.5. Cells were pelleted and washed twice in ice-cold, sterile double-distilled water. Cell pellets were resuspended in 1.5 ml of lysis buffer {8 M urea, 2 M thio-urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM dithiothreitol, 40 mM Tris base, 1% Pharmalyte 3-10 (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom), fungal protease inhibitor cocktail (Sigma)}, followed by the addition of an equal volume of 425- to 600-μm-diameter glass beads (Sigma). Cells were disrupted using a mini-bead beater (Biospec, Bartlesville, Okla.) five times for 1 min each time, with 1 min between bead beating on ice. Glass beads were allowed to settle before the lysate was removed and transferred to a 1.5-ml microcentrifuge tube. Cell debris was removed by centrifugation, and the resulting soluble protein preparation was stored at −80°C.
To ensure that equal concentrations of protein were loaded onto two-dimensional gels, the samples were prerun on one-dimensional sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gels minigels (Bio-Rad Criterion) and visualized by staining with colloidal Coomassie blue R-250 (Bio-Rad). The concentrations of proteins were determined empirically and adjusted accordingly. Comparative two-dimensional electrophoresis was performed using a MultiphorII apparatus for isoelectric focusing (IEF) (Amersham Pharmacia Biotech) as previously described (29) and a Hoefer-DALT apparatus (Amersham Pharmacia Biotech) for the second dimension. Immobilized pH gradient 18-cm strips, pH 4.5 to 5.5, pH 4 to 7, and pH 6 to 11 (Amersham Pharmacia Biotech), were used for IEF. SDS-10% polyacrylamide gels (22 cm by 25 cm) (10% Duracryl [Genomic Solutions, Ann Arbor, Mich.[, 0.4 M Tris [pH 8.8], 0.1% SDS, 0.1% ammonium persulfate, 0.01% N,N,N′,N′-tetramethylethylenediamine [TEMED]) were used for the second dimension. Protein samples were first run on analytical gels (with approximately 0.1 mg of total protein) and were visualized using a silver stain kit (Genomic Solutions). Silver-stained gels were scanned using an Image Scanner II (Amersham Pharmacia Biotech). Protein spots showing obvious and reproducible changes in expression (present on both gel sets from duplicate experiments) were catalogued. Subsequently, identical protein samples were run on preparative gels (with approximately 1 mg of total protein) and stained with Sypro ruby stain as described by the manufacturer (Genomic Solutions). The Sypro ruby gels were scanned using an FLA-5000 scanner (Fuji Photo Film Europe GmbH) at an excitation wavelength of 473 nm and emission of 618 nm. The protein spots with altered expression catalogued previously were excised from these gels for identification by peptide mass fingerprinting.
Images were exported to Microsoft PhotoDraw 2000 version 2.0 for annotation and presentation. The isoelectric point and molecular weight of the proteins of interest were calculated from protein migration as previously described (5).
Mass spectrometry.
Protein spots of interest were excised from the gels and digested in the gel with trypsin (Promega) using an Investigator Progest digestion robot (Genomic Solutions). Half the sample was desalted and concentrated using a micro C18 column (0.2 μl of ZipTip) (Millipore, Gloucestershire, United Kingdom) according to the manufacturer's instructions. The peptides were eluted directly from the tip onto the target in 1.5 μl of α-cyano-4-hydroxycinnamic acid (saturated stock prepared in acetonitrile-0.2% trifluoroacetic acid [50:50] and then diluted 1/5 in acetonitrile-0.2% trifluoroacetic acid [60:40]). Spectra were obtained on a Micromass TofSpec 2E instrument (Micromass, Manchester, United Kingdom), equipped with a 337-nm-wavelength laser and operated in reflectron mode. The data were calibrated using the tryptic peptides of β-galactosidase (Sigma) and lock mass corrected using a Glu-Fibrinopeptide B spike.
Monoisotopic peptide masses were selected using BioLynx ProteinProbe (Micromass) and submitted for peptide mass matching against the National Center for Biotechnology Information (NCBI) database using the Mascot search engine (Matrix Science).
RNA isolation, cDNA synthesis, miniarray hybridization, and analysis.
Total RNA was isolated from mid-exponential-phase (OD600 of 0.3) control and yeast cells exposed to 300 mM citric acid (pH 3.5) for 20 min using the RNA minikit (Qiagen). RNA was used as a template for cDNA synthesis using Moloney murine leukemia virus reverse transcriptase in the STRIP-EZ RT StripAble cDNA probe synthesis and removal kit (Ambion). The resulting cDNA probe is stable under hybridization conditions but can be readily cleaved after detection by a reagent in the probe degradation buffer. Unincorporated nucleotides were separated from the 33P-labeled cDNA probe by passage through a Sephadex G-50 column. More than 98% of S. cerevisiae ORFs were contained on two separate Yeast GeneFilters (Research Genetics), which were simultaneously, but separately, hybridized with one of two probes (made from RNA isolated from control cells and cells exposed to citric acid, respectively) as described previously (7). Data were collected using the phosphorimaging function of a FLA-5000 scanner (Fuji Film). The membranes were stripped using the STRIPeasy kit (Ambion) per the manufacturer's instructions. These membranes were then rehybridized with freshly made probes (as described above) from three identical, replicate cultures, resulting in three independent hybridizations. The hybridization data were analyzed using Pathways software (Research Genetics). Expression ratios are shown for genes that on average changed more than 1.5-fold over the three replicate hybridization experiments. To allow for comparison, the filter images were normalized by the Pathways software using all data points on the array. This resulted in expression ratios of 1 for genes unlikely to be affected by citric acid stress (e.g., ACT1, encoding actin).
Measurement of osmolarity.
Osmolarities were determined in suitably diluted samples using a fully calibrated Westcor 5100c vapor pressure osmometer according to the manufacturer's instructions (Westcor, Inc., Logan, Utah). The osmolarities of ME medium (pH 3.5) and ME medium containing citric acid, NaCl, KCl, and sorbitol were determined. Standard curves were produced in ME medium (pH 3.5) with sorbitol, KCl, and NaCl, using a concentration range of 50 mM to 1 M.
Determination of intracellular glycerol.
Cultures of wild-type S. cerevisiae BY4741 MATa in ME medium (pH 3.5) were incubated at 30°C with shaking at 200 rpm in the presence of 0, 150, or 300 mM citric acid, 230 or 460 mM sorbitol, 100 or 210 mM KCl, or 125 or 250 mM NaCl. A 1-ml aliquot was removed and incubated for 10 min at 100°C, and cell debris was pelleted at 13,000 rpm (Eppendorf Mini-Spin) for 2 min. Glycerol concentrations were determined using the glycerol determination kit (Boehringer Mannheim) per the manufacturer's instructions.
Complementation of Δhog1 and overexpression of HOG1.
HOG1 and its promoter, 600 bp upstream of the ATG codon, were PCR amplified from S. cerevisiae BY4741 genomic DNA, isolated using a genomic DNA extraction kit (Qiagen). The resulting PCR product was cloned into the multiple cloning site of pRS313, a single-copy HIS3 vector and pRS413, a multicopy HIS3 vector (35). Δhog1 and BY4741 strains were transformed with these plasmids using the high-efficiency lithium acetate method (10).
Western blotting of Hog1p and Hog1p phosphorylation.
Isolation of total soluble protein and subsequent Western blotting of Hog1p and phosphorylated Hog1p were performed as previously described (44). Briefly, total soluble protein was isolated from S. cerevisiae BY4741, Δhog1, Δssk1, and Δpbs2 strains grown in ME medium (pH 3.5) after a 10-min exposure to either 300 mM citric acid or 0.4 M NaCl. Equal concentrations of protein (15 μg) were loaded and separated by SDS-polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membranes. Dual phosphorylation of Hog1p was observed using a dually phosphorylated (Thr174 and Tyr176) p38 antibody (New England Biolabs). Hog1p was detected using an anti-C-terminal Hog1p antibody (Yc20; Santa Cruz Biotechnology). Antibody binding was visualized using a Supersignal Westfemto maximum sensitivity kit (Pierce) after binding of a horseradish peroxidase-conjugated secondary antibody (Sigma).
RESULTS
Citric acid inhibits growth of S. cerevisiae.
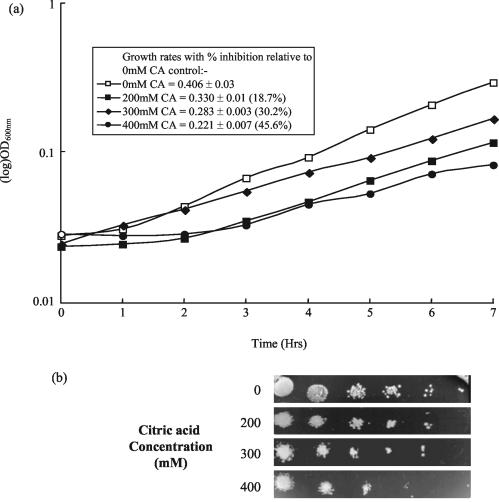
Growth of S. cerevisiae BY4741 was inhibited in the presence of increasing concentrations of citric acid in both ME broth (pH 3.5) (Fig. 1a) and on ME agar (pH 3.5) (Fig. 1b). Growth rate was reduced from a value of 0.406 in the control culture to 0.221 (45.6% inhibition) in the presence of 400 mM citric acid (Fig. 1a). Exposure to 300 mM citric acid resulted in a 30.2% reduction in the growth rate.
FIG. 1.
Growth of S. cerevisiae BY4741 is inhibited in the presence of citric acid. (a) Cultures of S. cerevisiae BY4741 were grown to late exponential phase in ME medium alone (pH 3.5) and in the presence of 200, 300, and 400 mM citric acid (CA). (b) Serial dilutions of mid-exponential-phase cultures of S. cerevisiae BY4741 were spotted onto ME agar plates at pH 3.5 with and without the presence of 200, 300, and 400 mM citric acid. Plates were incubated for 48 h at 30°C before the plates were photographed.
Screening the yeast disruptome identified many genes that confer resistance to citric acid stress.
To identify key genes involved in mediating resistance to citric acid stress, we conducted a phenotypic screen of all the 4,847 nonessential gene deletions in S. cerevisiae BY4741 MATa. Yeast deletion strains were spotted onto plates in the presence or absence of 400 mM citric acid. Of the 4,847 nonessential gene deletions tested; 69 (1.4%) showed sensitivity to citric acid compared to the parent (Table 1). No citric acid-resistant phenotypes were detected. Of the 69 citric acid-sensitive gene deletions, 10 were of particular interest because they encode known regulatory proteins that could be involved in signaling an adaptive response to citric acid stress. The 10 gene deletions of interest are RCS1 (iron-regulated transcriptional repressor), MSN4 (transcriptional activator in response to stress), HOG1 (MAPK), BUB1 (serine/threonine protein kinase), CKA1 (casein kinase II), PTK2 (protein kinase involved in polyamine uptake); BCK1 (MEKK), SSK1 (two-component signal transducer), TPD3 (serine/threonine protein phosphatase 2A), and RRD1 (strong similarity to human phospho-tyrosyl phosphatase activator).
TABLE 1.
Screening the yeast disruptome identified genes required for optimal growth of S. cerevisiae in the presence of 400 mM citric acid (pH 3.5)a
| Functional category and ORF | Gene | Characteristic or description |
|---|---|---|
| Metabolism | ||
| Lipid, fatty acid, and isoprenoid metabolism | ||
| YER019W | ISC1 | Weakly similar to human and mouse neutral sphingomyelinase |
| YMR207c | HFA1 | Strongly similar to acetyl coenzyme A carboxylase |
| YMR202w | ERG2 | C8 sterol isomerase |
| YLR056w | ERG3 | C5 sterol desaturase |
| YGL012w | ERG4 | Sterol C24 reductase |
| Carbohydrate metabolism | ||
| YGL156w | AMS1 | α-Mannosidase |
| YLR377c | FBP1 | Fructose-1,6-bisphosphatase |
| YBR126C | TPS1 | α,α-Trehalose-phosphate synthase |
| Nucleotide metabolism | ||
| YMR300c | ADE4 | Amidophosphoribosyltransferase |
| Amino acid metabolism | ||
| YGL154c | LYS5 | l-Aminoadipate-semialdehyde dehydrogenase, small subunit |
| YEL046c | GLY1 | l-Threonine aldolase, low-specific |
| YGL148w | ARO2 | Chorismate synthase |
| YHR025w | THR1 | Homoserine kinase |
| YCR053w | THR4 | Threonine synthase (o-p-homoserine p-lyase) |
| Metabolism of energy reserves | ||
| YBL058w | SHP1 | Potential regulatory subunit for Glc7p |
| Energy generation activities | ||
| YOL079w | Similar to NADH dehydrogenases | |
| Regulation of interaction with cellular environment | ||
| Ion homeostasis | ||
| YGR217W | CCH1 | Calcium channel protein |
| YHR060w | VMA22 | Vacuolar ATPase assembly protein |
| YGR020c | VMA7 | H+-ATPase V1 domain 14-kDa subunit, vacuolar |
| YBR127C | VMA2 | H+-ATPase V1 domain 60-kDa subunit, vacuolar |
| YKL080w | VMA5 | H+-ATPase V1 domain 42-kDa subunit, vacuolar |
| YEL051w | VMA8 | H+-ATPase V1 domain 32-kDa subunit, vacuolar |
| YEL027w | CUP5 | H+-ATPase V0 domain 17-kDa subunit, vacuolar |
| YAL026c | DRS2 | P-type amino-phospholipid-ATPase |
| YLR447c | VMA6 | H+-ATPase V0 domain 36-kDa subunit, vacuolar |
| YDL185w | TFP1 | H+-ATPase V1 domain 69-kDa catalytic subunit, vacuolar |
| YHR026w | PPA1 | H+-ATPase 23-kDa subunit, vacuolar |
| Cellular transport and transport mechanisms | ||
| YDR320c | SWA2 | Clathrin-binding protein required for normal clathrin function |
| Control cellular organization | ||
| Cell wall | ||
| YCL007c | CWH36 | Affects the mannoprotein layer of the cell wall |
| YLR390w-a | CCW14 | Secretory stress response protein 1 |
| YGR036c | CAX4 | Required for full levels of dolichol-linked oligosaccharides in the endoplasmic reticulum |
| YJL095W | BCK1 | Cell wall integrity pathway, nutrient sensing, and growth control |
| YDR323c | PEP7 | Vacuolar segregation protein |
| YJR059W | PTK2 | Involved in polyamine uptake |
| Cell fate | ||
| YNL084c | END3 | Required for endocytosis and cytoskeletal organization |
| YGR167w | CLC1 | Clathrin light chain |
| YDR388w | RVS167 | Reduced viability upon starvation protein |
| YAL016w | TPD3 | Ser/Thr protein phosphatase 2A, regulatory chain A |
| YDR099W | BMH2 | Suppressor of clathrin deficiency |
| Transcription | ||
| YBR112c | CYC8 | General repressor of transcription |
| YPL129w | ANC1 | TFIIF subunit (transcription initiation factor), 30 kDa |
| YOR295w | UAF30 | Subunit of RNA polymerase I transcription factor |
| YOR290c | SNF2 | Component of SWI/SNF global transcription activator complex |
| YMR228w | MTF1 | RNA polymerase-specific factor, mitochondrial |
| YKL062w | MSN4 | Transcriptional activator |
| YGL071W | RCS1 | Activates transcription in response to iron deprivation |
| Protein synthesis | ||
| YBR191w | RPL21A | Ribosomal protein L21.e |
| YNL073w | MSK1 | Lysyl-tRNA synthetase, mitochondrial |
| YOL023w | IFM1 | Translation initiation factor 2, mitochondrial |
| Cellular communication/signal transduction pathway | ||
| YMR016c | SOK2 | Regulatory protein in the protein kinase A signal Transduction pathway |
| YLR113w | HOG1 | Ser/Thr protein kinase of MAPK family |
| YLR006c | SSK1 | Two-component signal transducer |
| Protein fate | ||
| YLR148w | PEP3 | Vacuolar membrane protein |
| YPL045w | VPS16 | Vacuolar sorting protein |
| YLR396c | VPS33 | Vacuolar sorting protein |
| YCR069w | SCC3 | Peptidyl-prolyl cis-trans isomerase precursor |
| YML078w | CPR3 | Cyclophilin (peptidylprolyl isomerase) |
| YMR150C | IMP1 | Protease, mitochondrial |
| Cell rescue, defense, and virulence | ||
| Detoxification | ||
| YJR104c | SOD1 | Copper-zinc superoxide dismutase |
| Cell cycle and DNA processing | ||
| YIL153w | RRD1 | Similar to human and rabbit phosphotyrosyl phosphatase activators and YPL152w |
| YGR188c | BUB1 | Ser/Thr protein kinase |
| YIL035c | CKA1 | Casein kinase II, catalytic alpha chain |
| YPL239W | YAR1 | Ankyrin repeat-containing protein |
| YJR144w | MGM101 | Mitochondrial genome maintenance protein |
| Unknown function | ||
| YMR216c | SKY1 | Similar to Schizosaccharomyces pombe dsk1, human SRPK1, and other protein kinases |
| YDL204W | Similar to hypothetical protein YDR233c | |
| YLR326w | ||
| YOR331c | Questionable ORF | |
| YLR334c | Questionable ORF | |
| YOR161C | Similarity to Caenorhabditis elegans cosmid F35C8 | |
| YKL169c | ||
| YBR101c | Weakly similar to S. pombe hypothetical protein SPBC3B9.01 | |
| YLR346c | Weakly similar to YGR035c | |
| YHR198c | ||
| YKL102c | ||
| YEL045c | Weakly similar to cytochrome c oxidase III of Trypanosoma brucei kinetoplast | |
| YLR047c | Similar to hypothetical protein YGL160w | |
| YPL144w | Hypothetical protein |
Ten of the 69 citric acid-sensitive gene deletions were of particular interest, because the genes encode known regulatory proteins that could be involved in signalling an adaptive response to citric acid stress. The 10 genes are shown in bold type.
Using the Gene Ontology (GO) database (http://fatigo.bioinfo.cnio.es/), we analyzed the functional categories of citric acid-sensitive gene deletions. Of the total of 69 genes, 56 had GO at level 3, biological process, which resulted in the following GO profile: 55.1% metabolism, 46.4% cell growth and/or maintenance, 17.4% homeostasis, 11.6% response to external stimulus, 10.1% response to stress, 4.3% cell communication, 2.9% reproduction, 1.45% morphogenesis, 1.45% growth, and 15.94% GO terms present at other levels.
Growth in the presence of citric acid induces changes in protein expression.
We measured changes in the S. cerevisiae proteome occurring in response to growth in the presence of 300 mM citric acid (pH 3.5) over the pH separation ranges of 4.5 to 5.5, 4.0 to 7.0, and 6.0 to 11.0. These proteins were subsequently identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry of the peptides produced by in-gel digestion with trypsin, followed by database searching using the peptide masses derived from each trypsinized protein (see Table S1 in the supplemental material).
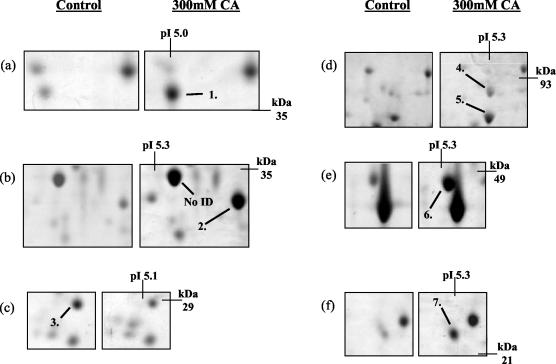
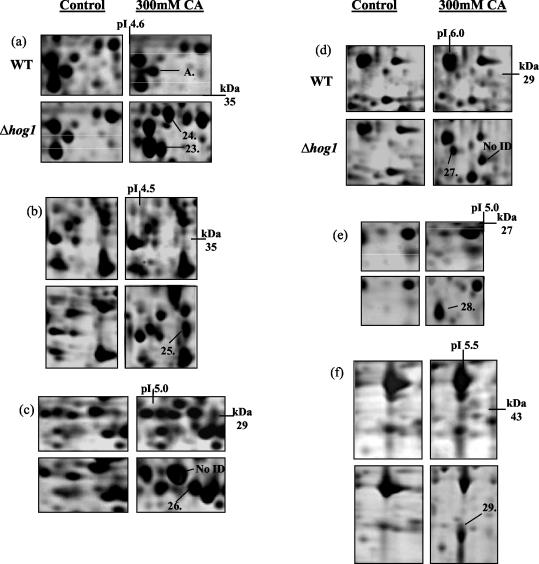
Over the pH range 4.5 to 5.5, we reproducibly detected the up-regulation of six new proteins and the down-regulation of one protein due to the presence of 300 mM citric acid, pH 3.5 (Fig. 2). The up-regulated proteins were identified as putative adenosine kinase (Ado1) (protein 1), dl-glycerol-3-phosphate (Gpp2) (protein 2), imidazole glycerol phosphate synthase (His7) (protein 4), stress-induced protein (Sti1) (protein 5), carbamyl phosphate synthase (Ura2) (protein 6), and pyruvate decarboxylase isozyme 1 (Pdc1) (protein 7) (Fig. 2a, b, d, e, and f). The down-regulated protein was identified as an hypothetical ORF (Ylr301w) (protein 3 in Fig. 2c).
FIG. 2.
Identification of changes in protein expression induced in S. cerevisiae BY4741 due to growth in the presence of 300 mM citric acid, pH 3.5. Total soluble proteins were separated by IEF over the pH range 4.5 to 5.5. Proteins were prepared from control cells (grown in ME medium [pH 3.5]) and cells grown in ME medium (pH 3.5) plus 300 mM citric acid (CA). The positions of Ado1p (protein 1), Gpp2p (protein 2), hypothetical ORF Ylr301wp (protein 3), His7p (protein 4), Sti1p (protein 5), Ura2p (protein 6), and Pdc1p (protein 7) are indicated. Molecular masses (in kilodaltons) are shown on the y axis, and pI values are shown on the x axis. A representative result from two replicate experiments is shown. No ID, no protein identification.
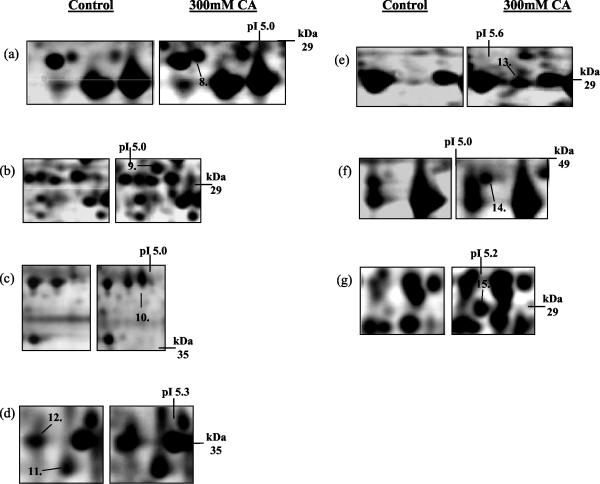
Similarly, over the pH ranges 4 to 7 and 6 to 11, we detected the up-regulation of eight (Fig. 3) and seven proteins (Fig. 4), respectively. The up-regulated proteins on the pH 4 to 7 gels were identified as heat shock protein 70 isoform (Ssa1) (protein 8), vacuolar ATPase V1 domain subunit E (Vma4) (protein 9), 40S ribosomal protein S0-A (Rps0ap) (protein 10), dl-glycerol-3-phosphatase (Gpp1) (protein 11), inorganic pyrophosphatase (Ipp1) (protein 12), enolase 2 (Eno2) (protein 13), heat shock protein 70 isoform (Ssb2) (protein 14), and heat shock protein 26 (Hsp26) (protein 15) (Fig. 3a, b, c, d, e, and f).
FIG. 3.
Identification of changes in protein expression induced in S. cerevisiae BY4741 due to growth in the presence of 300 mM citric acid, pH 3.5. Total soluble proteins were separated by IEF over the pH range 4 to 7. Proteins were prepared from control cells (grown in ME medium [pH 3.5]) and cells grown in ME medium (pH 3.5) plus 300 mM citric acid (CA). The positions of Ssa1p (protein 8), Vma4p (protein 9), Rps0ap (protein 10), Gpp1p (protein 11), Ipp1p (protein 12), Eno2p (protein 13), Ssb2p (protein 14), and Hsp26p (protein 15) are indicated. Molecular masses (in kilodaltons) are shown on the y axis, and pI values are shown on the x axis. A representative result of two replicate experiments is shown.
FIG. 4.
Identification of changes in protein expression induced in S. cerevisiae BY4741 due to growth in the presence of 300 mM citric acid, pH 3.5. Total soluble proteins were separated by IEF over the pH range 6 to 11. Proteins were prepared from control cells grown in ME medium [pH 3.5]) and cells grown in ME medium (pH 3.5) plus 300 mM citric acid (CA). The positions of Atp1p (protein 16), Imd3p (protein 17), Ilv5p (protein 18), Mdh1p (protein 19), Sis1p (protein 20), Aro3p (protein 21), and Idh1p (protein 22) are indicated. Molecular masses (in kilodaltons) are shown on the y axis, and pI values are shown on the x axis. A representative result of two replicate experiments is shown.
The up-regulated proteins identified on the pH 6 to 11 gels were mitochondrial F1F0-ATPase alpha subunit (Atp1) (protein 16), IMP dehydrogenase homolog (Imd3) (protein 17), aceto-hydroxyacid reductoisomerase (Ilv5) (protein 18), mitochondrial malate dehydrogenase (Mdh1) (protein 19), Hsp40 family chaperone (Sis1) (protein 20), 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase (Aro3) (protein 21), and isocitrate dehydrogenase 1 alpha-4-beta-4 subunit (Idh1) (protein 22) (Fig. 4).
A number of the up-regulated proteins on the gels were identified as polypeptide fragments (e.g., Pdc1, Ssa1, Eno2, and Ssb2). However, like all the other proteins listed, expression of the proteins represented by these fragments was altered in duplicate experiments. Comparison of the experimental versus predicted molecular weights and pIs of the identified proteins is shown in Table S1 in the supplemental material. The predicted and experimental molecular weights were mostly in agreement, apart from the polypeptide fragments discussed previously. However, the experimental and predicted pIs varied, even in intact proteins, which could be explained by the presence of different posttranslational modifications.
As before, we analyzed the GO database functional categories of the citric acid-induced changes in protein expression. Of the total of 24 proteins with up-regulated expression, 22 had GO at level 3, biological process, which resulted in the following GO profile: 83.3% metabolism, 20.8% cell growth and/or maintenance, 8.3% response to stress, 4.17% response to external stimulus, and 4.17% homeostasis.
Comparison of the above proteins with the genome-wide, citric acid-sensitive, gene deletion screen in Table 1 revealed no correlation between proteins with altered expression due to growth in the presence of citric acid and the citric acid-sensitive gene deletions.
Growth in the presence of citric acid induces changes in gene expression.
The gene expression profile of S. cerevisiae grown in the presence of 300 mM citric acid (pH 3.5) for 20 min was compared to that of cells grown in ME medium (pH 3.5) alone. Genes which showed a greater than 1.5-fold induction due to the presence of citric acid are shown in Table S2 in the supplemental material. The data are listed according to putative functional categories, as described in the MIPS database, and are expressed as the means and standard deviations from three independent experiments.
Of the 6,144 ORFs, 68 (1.1%) showed greater than a 1.5-fold induction (see Table S2 in the supplemental material) and 12 of these genes showed an increase greater than 2.5-fold: glyceraldehyde-3-phosphate dehydrogenase 2 (TDH2), alpha-trehalose-phosphate synthase (TPS1), glycerol-3-phosphate dehydrogenase (GPD1), cell surface glycoprotein (SED1), stress-inducible aldehyde dehydrogenase (ALD3), three cell wall-associated proteins (SPI1, CWP1, and CCW14), and four ORFs of unknown function (YDL204w, YGL037c, YGR086c, and YFR001w).
Of the 68 up-regulated genes, GO database analysis, at level 3 of biological process, revealed 43 genes with the following profile: 44.1% metabolism, 26.5% cell growth and/or maintenance, 13.2% response to stress, 8.8% response to external stimuli, 8.8% cell communication, 4.41% reproduction, 2.94% morphogenesis, 2.94% homeostasis, 2.94% growth, 1.47% death, 1.47% cell death, 1.47% ageing, 1.47% conjugation, and 36.8% with GO terms at other levels.
Furthermore, 54 (0.9%) of the 6,144 ORFs showed a decrease in expression greater than 1.5-fold (see Table S3 in the supplemental material). Of these ORFs, 33 showed repression more than 2.5-fold and were nearly all structural components of the ribosome, apart from pyruvate decarboxylase 1 (PDC1), a putative ATP-dependent RNA helicase (ECM16), and a gene mediating glucose repression (GSF2). GO analysis of these genes, performed exactly as described above, revealed 63.0% metabolism, 11.1% cell growth and/or maintenance, 1.85% homeostasis, 1.85% response to external stimulus, and 11.1% GO terms present at other levels. The majority of genes that were repressed mediate protein synthesis and were largely structural constituents of the ribosome.
Comparison of these genes induced or repressed in response to citric acid with the genome-wide, citric acid-sensitive, gene deletion screen in Table 1 revealed 10 genes whose expression was altered upon exposure to citric acid and whose corresponding gene deletion strain also exhibited sensitivity to citric acid: TPS1 (α, α-trehalose-phosphate synthase), FBP1 (fructose-1,6-bisphosphate), IMP1 (protease, mitochondrial), VMA2 (H+-ATPase V1 domain 60-kDa subunit, vacuolar), BMH2 (suppressor of clathrin deficiency), CCW14 (secretory stress response protein 1), CYC8 (general repressor of transcription), YAR1 (ankyrin repeat-containing protein), and YDL204W and YOR161c both encoding proteins of unknown function. These 10 genes represent 8.2% of the total of 122 genes that displayed altered expression in response to citric acid.
Detailed phenotypic analysis of deletion mutants reveals a role for the MAPK HOG pathway in resistance to citric acid.
Results from the genome-wide screen (sensitivity of HOG1, RRD1, SSK1, and MSN4 deletion strains), protein expression analysis (up-regulation of Gpp1p and Gpp2p), and transcript analysis (induction of GPD1, GPP2, TPS1, and CTT1) clearly suggested a role for the HOG pathway in mediating resistance to citric acid stress.
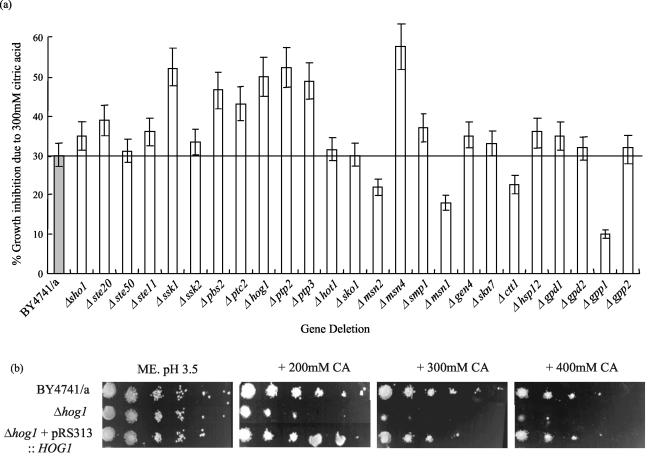
To further investigate the possibility that citric acid resistance may be regulated via the HOG pathway, we investigated the growth sensitivity or resistance of the nonessential gene deletions within the entire HOG pathway, including general stress genes, and genes known to be regulated by HOG1 that are involved in glycerol biosynthesis. This experiment was performed in more detail than the initial genome deletion screen by assaying sensitivity to citric acid using shaking flask cultures and calculating change in growth rate. This was necessary because we have detected discrepancies in the results from sensitivity assays performed on agar compared to liquid culture (for example, Δpbs2, Δptp2 and Δptp3). In some cases, strains show sensitivity in liquid culture but not on agar (see below). Growth of the parent strain and the following HOG pathway gene deletion strains was monitored in ME broth (pH 3.5) with and without 300 mM citric acid: regulatory sensor (Δsho1), upstream control system (Δssk1), upstream kinases (Δste20 and Δste50), MAP kinase kinase kinase (Δste11 and Δssk2), MAP kinase kinase (Δpbs2), MAP kinase (Δhog1), phosphatases (Δptp2 and Δptp3), transcriptional regulators (Δmsn1, Δmsn2, Δmsn4, Δhot, Δsmp1, Δsko1, Δgen4, and Δskn7), and the target genes (Δctt1, Δhsp12, Δgpd1, Δgpd2, Δgpp1, and Δgpp2).
It is clear that important gene deletions within the HOG MAPK pathway induce sensitivity to citric acid (Fig. 5a). In fact, upon exposure to 300 mM citric acid, deletion of the protein kinases SSK1, PBS2, and HOG1 resulted in approximately 20% additional reduction in growth rate relative to the growth rate of the parent (Fig. 5a). Furthermore, deletion of the transcription factor MSN4 gene showed approximately 25% additional inhibition of growth rate and deletion of the protein phosphatases PTC2, PTP2, and PTP3 also showed enhanced sensitivity upon exposure to citric acid. Interestingly, deletion of GPP1 resulted in approximately 20% increase in growth rate, or resistance, to citric acid relative to the parent.
FIG. 5.
Growth inhibition of S. cerevisiae BY4741 MATa (BY4741/a) and single deletion mutants of genes that constitute the HOG pathway or are known to regulate or be regulated by the HOG pathway. (a) Cultures were grown to late exponential phase in ME medium (pH 3.5) in the presence or absence of 300 mM citric acid. For each strain, growth inhibition is expressed as the percent inhibition of growth rate. This was calculated by measuring the growth rate in the presence or absence of 300 mM citric acid and expressing the additional inhibition seen in the citric acid-treated culture relative to the untreated culture as a percentage. Three replicate experiments were performed. The values are shown as the means ± standard deviations (error bars) of the three experiments. (b) Complementation of the Δhog1 citric acid-sensitive phenotype. Growth inhibition of S. cerevisiae BY4741 MATa (BY4741/a), Δhog1 mutant, and Δhog1 mutant carrying pRS313::HOG1 is shown on ME agar (pH 3.5) and ME agar (pH 3.5) plus 200, 300, and 400 mM citric acid (CA). A representative result is shown.
To confirm that deletion of Δhog1 caused sensitivity to citric acid, we performed a complementation experiment by introducing HOG1 into the Δhog1 deletion mutant on a single-copy plasmid (pRS313::HOG1) under the control of its own promoter. Upon transformation of this plasmid into the Δhog1 strain, sensitivity to citric acid was abolished (Fig. 5b). Furthermore, we overexpressed HOG1 by cloning the ORF into the multicopy vector pRS413 and transforming BY4741. Subsequently, transformants were found not to display any significant phenotype upon exposure to citric acid (data not shown).
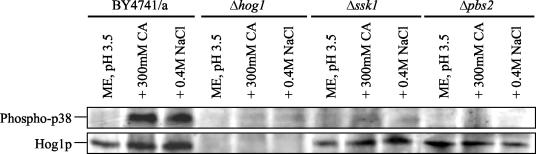
Exposure to citric acid results in the phosphorylation and activation of Hog1p.
Ultimately, stimulation of the HOG pathway results in the activation of the MAP kinase Hog1p via the dual phosphorylation of threonine-174 and tyrosine-176 (33). To determine whether exposure to citric acid resulted in the activation of Hog1p, we used an antibody that specifically detects the dually phosphorylated form of Hog1p, and thus, the active kinase (38). Phosphorylation of Hog1p was determined by Western blot analysis in the parent strain and the Δhog1, Δssk1, and Δpbs2 deletions after a 10-min exposure to 300 mM citric acid. For a positive control, we also determined the degree of phosphorylation of Hog1p after exposure to 0.4 M NaCl, a concentration previously shown to result in Hog1p phosphorylation (17). Protein loading was checked using an antibody against Hog1p, and as expected, no Hog1p was detected in the Δhog1 strain, but Hog1p was present at similar levels in the parent strain and both the Δssk1 and Δpbs2 strains (Fig. 6).
FIG. 6.
Western blot demonstrating the dual phosphorylation of Hog1p upon exposure to citric acid. Soluble protein extracts were prepared from S. cerevisiae BY4741 MATa (BY4741/a) and Δhog1, Δssk1, and Δpbs2 mutants with or without a 10-min exposure to 300 mM citric acid (CA) and 0.4 M NaCl in ME medium (pH 3.5). Dually phosphorylated forms of Hog1p were detected using anti-phospho-p38. To check that equal amounts of protein were loaded in the lanes, Hog1p was detected using a polyclonal anti-C-terminal Hog1p antibody. A representative result is shown.
Hog1p is phosphorylated, and thus activated, upon exposure to either 300 mM citric acid or 0.4 M NaCl (Fig. 6). Upon exposure to citric acid or NaCl, we observed no phosphorylation of Hog1p in either the Δssk1 or Δpbs2 strain. These results confirm that both Ssk1p and Pbs2p are important upstream components of the HOG pathway that are required for optimal activation of Hog1p in response to both citric acid and NaCl stress. This also explains why the Δssk1 and Δpbs2 strains are almost as sensitive to citric acid stress as the Δhog1 strain.
Activation of the HOG pathway by citric acid is not due to osmotic stress.
The HOG pathway is involved in the adaptation of cells to a high-osmolarity environment and is required for the growth of S. cerevisiae in media supplemented with high concentrations of NaCl (0.9 M) or sorbitol (1.5 M) (13). Therefore, we considered the possibility that because relatively high concentrations of citric acid were required to inhibit yeast growth, the observed inhibitory effect of citric acid could be a consequence of osmotic stress. If true, this would also explain our observation that the HOG pathway plays a role in mediating resistance to citric acid.
To investigate this possibility, we measured the osmolarity of 150 and 300 mM citric acid in ME medium (pH 3.5) using a vapor pressure osmometer. The osmolarity, calculated from two independent readings, of ME medium (pH 3.5) was determined to be 83 mmol kg−1 (Table 2). The addition of 150 and 300 mM citric acid increased the osmolarity to 263 and 460 mmol kg−1, respectively. To compare this osmolarity with concentrations of compounds commonly used to study osmotic stress in S. cerevisiae, we generated a standard curve of increasing concentrations of sorbitol, KCl, and NaCl in ME medium (pH 3.5) versus measured osmolarity (data not shown). From this standard curve, the osmolarity of 150 mM citric acid was found to be equivalent to that of 230 mM sorbitol, 100 mM KCl, and 125 mM NaCl, and the osmolarity of 300 mM citric acid was equivalent to that of 460 mM sorbitol, 210 mM KCl, and 250 mM NaCl (Table 2).
TABLE 2.
Effects of equivalent osmolarities of citric acid, sorbitol, KCl, and NaCl on glycerol production and growth rate of S. cerevisiae BY4741 MATa and Δhog1 deletion mutant
| Treatment | Osmolarity (mmol kg−1) | Glycerol level (mmol g of protein−1) after 3 h | % Growth inhibition
|
|
|---|---|---|---|---|
| BY4741 MATa | Δhog1 | |||
| ME medium (pH 3.5) (control) | 83 | 2.9 | -a | - |
| 150 mM citric acid | 263 | 4.3 | 10 | 33 |
| 300 mM citric acid | 460 | 6.8 | 34 | 59 |
| 230 mM sorbitol | 263 | 5.4 | - | - |
| 460 mM sorbitol | 460 | 8.3 | - | 9.3 |
| 100 mM KCl | 263 | 4.5 | - | - |
| 210 mM KCl | 460 | 5.2 | - | 13.2 |
| 125 mM NaCl | 263 | 4.2 | - | - |
| 250 mM NaCl | 460 | 6.3 | - | 14.5 |
-, no effect on growth rate.
Next, we measured the effects of 150 and 300 mM citric acid (pH 3.5) and equivalent osmolarities of sorbitol, KCl, and NaCl on the growth rate of the parent strain and the Δhog1 deletion strain, which is unable to grow in high-osmolarity media. Table 2 shows the percentage growth inhibition calculated by comparing the growth rate in ME medium (pH 3.5) and the growth rate during exposure to equivalent osmolarities of citric acid, sorbitol, NaCl, and KCl.
The growth rate of the parent strain was inhibited by 10 and 34% in the presence of 150 and 300 mM citric acid, respectively, but no growth inhibition was observed upon exposure to equivalent osmolarities of sorbitol, NaCl, or KCl. Thus, we can conclude that the growth inhibitory effect of citric acid at these concentrations is not due to osmotic stress. Importantly, the Δhog1 strain was very sensitive to citric acid, and growth was inhibited by 33 and 59% by 150 and 300 mM citric acid, respectively. Exposure of Δhog1 to an osmolarity of sorbitol, NaCl, or KCl equivalent to that of 150 mM citric acid resulted in no growth inhibition. There was minor inhibition of Δhog1 upon exposure to an osmolarity of sorbitol, NaCl, or KCl equivalent to that of 300 mM citric acid but only between 9 and 14% compared to 59% with 300 mM citric acid (Table 2). Thus, these results clearly show that the principal inhibitory effect of citric acid is not due to osmotic stress.
To test this conclusion further, we determined whether any citric acid-sensitive gene deletions we identified in our genome-wide screen were known to be sensitive to osmotic stress according to the MIPS mutant phenotype catalogue entry osmotic sensitivity. Of the 43 genes known to display osmotic sensitivity, only genes that encode proteins that are components of the HOG pathway also showed sensitivity to citric acid. All the other osmotically sensitive genes, apart from Δvps33, were not sensitive to citric acid, including Δsat4, Δrvs161, Δgpd1, Δppz1, and Δrts1 (Table 1).
Finally, activation of the HOG pathway results in an increase in intracellular glycerol concentration that allows the cells to grow in a high-osmolarity environment. This response is mediated by an increase in the expression of GPD1 (1). Therefore, we also measured the concentrations of intracellular glycerol in cells exposed to 150 and 300 mM citric acid and compared them to those of cells grown in equivalent osmolarities of sorbitol, KCl, and NaCl. Table 2 shows that after a 3-h exposure to 150 and 300 mM citric acid, the intracellular concentration of glycerol does increase from 2.9 to 4.3 and 6.8 mmol g−1 protein, respectively. This correlates with our observation that exposure to citric acid results in increased expression of GPD1. These values were very similar to the intracellular glycerol concentrations measured when S. cerevisiae was exposed to equivalent osmolarities of sorbitol, KCl, and NaCl (Table 2). Therefore, despite our observation that citric acid activates the HOG pathway via a mechanism distinct from osmotic stress, one of the major consequences of HOG pathway activation, namely, production of intracellular glycerol, does occur during citric acid stress.
Protein expression is up- and down-regulated via the HOG pathway on exposure to citric acid.
We have presented evidence that the HOG pathway mediates adaptation to citric acid. To further investigate how Hog1p is regulating the cellular resistance mechanism to citric acid, we studied changes in protein expression occurring in response to citric acid in the Δhog1 deletion strain in the presence and absence of 300 mM citric acid, pH 3.5. Changes in the Δhog1 proteome were then compared with the changes in the proteome already observed for the parent strain in the presence and absence of citric acid.
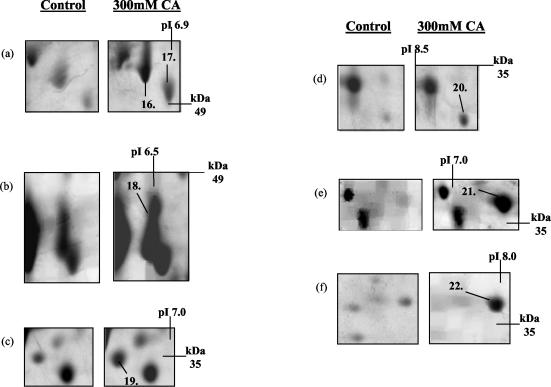
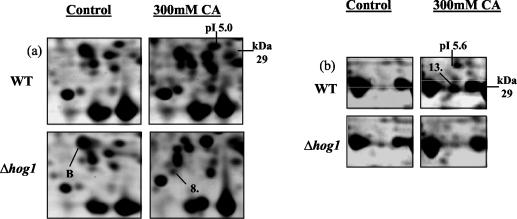
Over the pH range 4 to 7, we reproducibly detected the up-regulation of seven new proteins due to growth in the presence of citric acid; expression of the seven new proteins was dependent on deletion of HOG1 (Fig. 7). These proteins were identified by peptide mass fingerprinting (see Table S4 in the supplemental material). The seven new proteins were brain modulosignalin homologue (Bmh1p) (protein 23), beta subunit of pyruvate dehydrogenase (Pdb1p) (protein 24), dihydroorotate dehydrogenase (Ura1p) (protein 25), fructose bisphosphate aldose (Fba1p) (protein 26), hypothetical protein (Ydr533cp) (protein 27), 6-phosphogluonate dehydrogenase (Gnd1p) (protein 28), and arginase (Car1p) (protein 29) (Fig. 7). We can conclude that HOG1 negatively regulates the expression of these proteins during exposure to citric acid stress.
FIG. 7.
Identification of citric acid-induced proteins whose expression was dependent on deletion of the HOG1 gene. Protein expression in wild-type (WT) S. cerevisiae BY4741 and Δhog1 mutant in the presence or absence of 300 mM citric acid (CA), pH 3.5, is shown. Total soluble proteins were separated by IEF over the pH range 4 to 7. Proteins were prepared from untreated (control) cells (grown in ME medium [pH 3.5]) and cells grown in ME medium (pH 3.5) plus 300 mM citric acid (CA). The positions of Bmh1p (protein 23), Pdb1p (protein 24), Ura1p (protein 25), Fba1p (protein 26), YDR533cp (protein 27), Gnd1p (protein 28), and Car1p (protein 29) are indicated. We also detected a protein whose expression was unaffected by exposure to citric acid alone but whose expression was repressed in the Δhog1 deletion mutant in the presence of citric acid. This protein was identified as Rps0bp (protein A). Molecular masses (in kilodaltons) are shown on the y axis, and pI values are shown on the x axis. A representative result of two replicate experiments is shown.
We also observed that two of the proteins previously identified to be up-regulated upon exposure to citric acid were no longer up-regulated in the Δhog1 deletion strain (Fig. 8). These proteins were identified as heat shock protein 70 isoform (Ssa1p) (protein 8) (Fig. 8a) and enolase 2 (Eno2p) (protein 13) (Fig. 8b) (see Table S1 in the supplemental material). Thus, citric acid-induced expression of these proteins must be under the control of Hog1p.
FIG. 8.
Identification of two citric acid-induced proteins whose expression is dependent on fully functional HOG1. Protein expression in wild-type (WT) S. cerevisiae BY4741 and Δhog1 mutant in the presence or absence of 300 mM citric acid (pH 3.5) is shown. Total soluble proteins were separated by IEF over the pH range 4 to 7. Proteins were prepared from untreated (control) cells (grown in ME medium [pH 3.5]) and cells grown in ME medium (pH 3.5) plus 300 mM citric acid (CA). The positions of Ssa1p (protein 8) and Eno2p (protein 13) are indicated. We also detected a protein whose expression was unaffected by exposure to citric acid alone but whose expression was repressed in the Δhog1 deletion mutant in the presence of citric acid. This protein was identified as Egd2p (protein B). Molecular masses (in kilodaltons) are shown on the y axis, and pI values are shown on the x axis. A representative result of two replicate experiments is shown.
Finally, we detected two proteins whose expression was unaffected by exposure to citric acid alone but whose expression was repressed in the Δhog1 deletion strain only in the presence of citric acid. These proteins were identified as 40S ribosomal protein S0-B (Rps0bp) (protein A) (Fig. 7a) and GAL4 enhancer protein (Egd2p) (protein B) (Fig. 8a). Thus, a functional Hog1p is required to maintain the expression of these two proteins during exposure to citric acid.
None of these Hog1p-regulated proteins correlated with HOG1-regulated genes detected after treatment with 0.7 M NaCl (31). Growth sensitivity assays of the corresponding deletion strains of the above genes did not reveal any sensitivity to citric acid stress, indicating that despite the fact that these proteins are regulated by Hog1p during citric acid stress, they are not themselves essential for optimal adaptation.
DISCUSSION
Taking a functional genomics approach, we have identified and characterized a new and important role for the HOG MAPK pathway in S. cerevisiae, the regulation of an adaptive response to citric acid stress.
Hog1p activation is vital for optimal adaptation to citric acid stress.
Yeast cells respond to environmental stress, such as hyperosmotic, heat, and radiation stress by activating MAPK pathways. The HOG pathway is one such pathway and is essential for the survival of S. cerevisiae in high-osmolarity environments (18). Recent work has indicated that the HOG pathway is also induced by other stress factors, including oxidative stress (36) and heat stress (43).
By screening the S. cerevisiae disruptome, we identified a number of yeast deletions that were hypersensitive to citric acid, including Δmsn4, Δhog1, and Δssk1. Subsequent more-detailed analysis in liquid culture of the sensitivity of deletions in all nonessential genes of the HOG pathway revealed that Δpbs2, Δptc2, Δptp2, and Δptp3 were also sensitive to citric acid (Fig. 9). The citric acid-sensitive phenotype in the Δhog1 deletion strain is specifically due to the loss of HOG1, because we were able to rescue the sensitive phenotype by reinserting HOG1 on a single-copy plasmid. Analysis of protein expression revealed that cells grown in the presence of citric acid up-regulated both isoforms of glycerol-3-phosphatase, Gpp1p and Gpp2p. Finally, study of changes in gene expression induced by citric acid revealed the up-regulation of GPD1 (encoding glycerol-3-phosphate dehydrogenase) and GPP2. The HOG pathway regulates the expression of GPP1, GPP2, and GPD1 (18) (Fig. 9), clearly implying that this pathway plays a role in mediating resistance to citric acid stress.
FIG. 9.
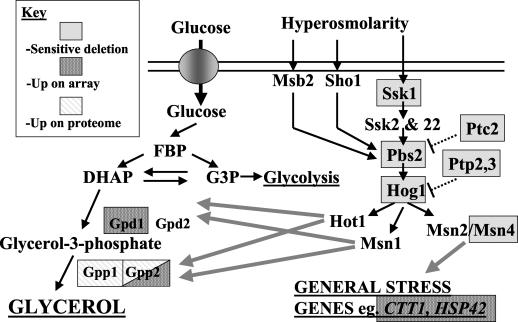
Summary of our findings regarding the role of the HOG pathway in adaptation to citric acid stress in S. cerevisiae. Proteins and genes that are sensitive to the deletion of citric acid or that are up-regulated on the miniarray and proteome are indicated. Abbreviations: FBP, fructose-1,6-biphosphate; DHAP, dihydroxyacetone phosphate; G3P, glucose-3-phosphate.
After this, we studied the effect of citric acid on the phosphorylation state of Hog1p. Activation of Hog1p requires phosphorylation on both Thr174 and Tyr176 (33). Thus, using an antibody that specifically detects Hog1p phosphorylated on both Thr174 and Tyr176, we have shown that citric acid results in the dual phosphorylation, and thus activation, of Hog1p. Indeed, the degree of activation of Hog1p by 300 mM citric acid (pH 3.5) was similar to that induced by exposure to 400 mM NaCl.
The results of previous studies using the inorganic acid HCl to increase the hydrogen ion concentration have suggested that the HOG pathway plays a minor role in regulating gene expression at low pH. Importantly, none of these studies identified HOG1 as essential for growth at these low pH values (20). Schuller et al. (33) showed that a low external pH of 2.8, due to addition of the inorganic acid HCl, resulted in stress response element (STRE) activation in a HOG1-dependent fashion. However, they did not test whether the growth of a Δhog1 mutant was affected at pH 2.8. Furthermore, they were unable to show significant tyrosine phosphorylation of Hog1p upon exposure to pH 2.8 or sorbic acid. Kapteyn et al. (20) noted that reducing culture pH from 5.5 to 3.5, again using the inorganic acid HCl, resulted in major changes in the organization of the S. cerevisiae cell wall. These low pH-induced alterations in yeast cell wall architecture were also found to be dependent on a functional HOG1 gene. However, they did not observe any obvious growth defect when the Δhog1 mutant was grown in media with the pH reduced to 3.5. Thus, they concluded that the Hog1p-dependent changes in cell wall architecture were nonessential under low-pH conditions. Also, we detected no phosphorylation or activation of Hog1p after exposing cells to pH 3.5 alone. Our finding that citric acid-induced activation of Hog1p is necessary for optimal adaptation to this compound is supported by a previous study using mammalian cells (45). In this study, the researchers identified that the mammalian homologue of Hog1p, the p38 protein kinase, is activated at pH 4 and by citric and acetic acid at pH 5.
The fact that inhibition of growth requires exposure to relatively high concentrations of citric acid (150 to 300 mM) led us to explore the possibility that the inhibitory effect of citric acid was simply due to an osmotic shock. However, equivalent osmolarities of sorbitol, KCl, and NaCl to that of 150 mM and 300 mM citric acid (pH 3.5) had little significant inhibitory effect on the growth of either the parent strain or the Δhog1 deletion strain, which is unable to grow in high-osmolarity media. Furthermore, when we tested strains with other gene deletions with known sensitivity to osmotic stress, only the Δvps33 strain and strains with the previously identified HOG pathway genes were sensitive to citric acid. Thus, we can conclude that the inhibitory effect of citric acid is not due to an osmotic stress and that the HOG pathway must be activated by some, as yet unknown, inhibitory effect of this compound.
Comparison of the gene expression profile after exposure to 300 mM citric acid (pH 3.5) with those obtained after exposure to 0.7 M NaCl and 0.95 M sorbitol (31) revealed only 18 genes (of a total of 186 that were up-regulated by both NaCl and sorbitol) that were also up-regulated by citric acid. These included genes involved in glycerol and trehalose metabolism (for example, GPD1, GPP2, TPS1, and TSL1), general stress response (for example, CTT1, HSP42, and DDR48), and genes involved in cell wall organization (for example, SPI1 and CWP1). In terms of down-regulated genes, exposure to NaCl and sorbitol did not induce the drastic down-regulation of protein synthesis that we observed with citric acid stress. In fact, in another gene expression study, Posas et al. (27) detected the up-regulation of 12 ribosomal protein genes due to exposure to 0.4 M NaCl that we identified as being repressed by citric acid. Of 23 proteins that were up-regulated by citric acid, the genes encoding only three proteins were up-regulated by osmotic stress; the three proteins were Gpp1p, Gpp2p, and Hsp26p. Thus, there is little correlation between the expression profiles induced by citric acid and osmotic stress. We found that only the gene deletions on the Sln1p branch of the HOG pathway displayed sensitivity to citric acid. Deletion of any of the nonessential genes in the Sho1 branch had no detrimental effect upon exposure to citric acid. Furthermore, we observed that a Δssk1 mutant, a mutant with a deletion of a protein upstream of Pbs2p (the shared protein where the two pathways converge) in the Sln1p branch of the HOG pathway, was as sensitive to citric acid as Δhog1 mutant. Also, we detected no phosphorylation of Hog1p in the Δssk1 deletion mutant upon exposure to citric acid. Thus, in contrast to osmotic stress, available evidence suggests that adaptation to citric acid occurs mainly via the Sln1 branch of the HOG pathway.
The phosphatases Ptp2p, Ptp3p, Ptc1p, Ptc2p, and Ptc3p all affect the level and duration of Hog1p phosphorylation (reviewed in reference 17). Overexpression of any of these proteins suppresses the lethality caused by overactivation of the HOG pathway (18). Mutants lacking both Ptc1p and Ptp2p are inviable, but this lethality is suppressed by additional deletion of HOG1. No other combination of deletion mutations of the two Ptp phosphatases and the three Ptc phosphatases results in this lethality (41). Single deletion PTP2, PTP3, and PTC2 mutants are each sensitive to citric acid in liquid culture. This may indicate that the activity of all these phosphatases is required to down-regulate the HOG pathway to prevent the growth defect induced by hyperactivation of Hog1p after stimulation by citric acid.
Exposure to citric acid up-regulates many stress response genes and proteins.
Induction of the HOG pathway results in the activation of a set of transcription factors, whose main roles are to activate stress response genes and genes involved in glycerol biosynthesis. Many stress-inducible genes are regulated by STREs via the transcription factors Msn2p and Msn4p (23), and it is well-known that upon osmotic shock, Msn2p- or Msn4p-dependent genes require the HOG pathway for induction (33). Of all the known HOG-dependent transcription factors, only deletion of MSN4 resulted in sensitivity to citric acid. This result was surprising, as Msn4p is thought to be functionally redundant with Msn2p, deletion of which resulted in no apparent phenotype upon exposure to citric acid. An important role for Msn4p in mediating the cellular response to citric acid was supported by our observation that a number of genes known to be controlled by Msn2p and Msn4p were up-regulated, including CTT1, ALD3, PNC1, DDR48, and YDL204w.
Citric acid resulted in the up-regulation of proteins and transcripts that have been previously implicated in various stress responses in S. cerevisiae. These included members of the Hsp70 family, Ssa1p and Ssb2p, Hsp26p, Sti1p, DDR48, CTT1, and members of the Hsp40 family of chaperones, Sis1p and HSP42. The increased expression of these proteins or genes during exposure to citric acid could be due to increased damage and aggregation of intracellular proteins or an inhibitory effect of citric acid on the protein synthetic machinery.
The expression of enzymes within the glycolytic and connecting pathways is known to change in response to many stress conditions (9). This altered expression enables the cell to produce stress protectants, compatible solutes, and reserves, such as glycerol and trehalose (17). Expression of both GPD1 and GPP2 is stimulated by nearly all stress factors (6), including exposure to 0.5 M NaCl (a concentration with an osmolarity twice that of 300 mM citric acid) (30). The osmotic stress-induced up-regulation of these genes is known to be dependent on Hog1p (reviewed in reference 17). Deletion of GPD1 alone (and deletion of both GPP1 and GPP2) results in hypersensitivity to osmotic stress (1).
Exposure to citric acid also resulted in the up-regulation of genes and proteins involved in glycerol biosynthesis (GPD1, GPP1, and GPP2) and trehalose biosynthesis (UGP1, TPS1, and TSL1). We also found that the Δtps1 strain was sensitive to citric acid. In support of these observations, we also measured an increase in intracellular glycerol after 3 h of exposure to 150 and 300 mM citric acid. Despite this, deletion of Δgpd1, Δgpd2, Δgpp1, or Δgpp2 did not result in any increased sensitivity to 300 mM citric acid. The Δgpd1 mutant is known to be sensitive to osmomolarity (1), but we observed no sensitivity of this strain to citric acid, further emphasizing that the inhibitory effect of citric acid is not due to osmotic stress.
The expression of both GPD1 and GPP2 is known to be highly dependent on the HOG pathway, via activation of the transcription factor Hot1p, and to a lesser extent Msn1p, but not Msn4p (17). However, single deletion Δhot1 and Δmsn1 mutants showed no sensitivity to citric acid, suggesting that Hot1p may not be as important in mediating the enhanced production of glycerol in response to citric acid stress as it is in the response to osmotic stress. We are currently generating double deletion mutants in key glycerol biosynthesis genes to determine the relative importance of enhanced glycerol production to citric acid tolerance.
Genes regulating glucose derepression are crucial for adaptation to citric acid.
Exposure to citric acid resulted in the up-regulation of two enzymes involved in the TCA cycle, Mdh1p (mitochondrial malate dehydrogenase) and Idh1p (isocitrate dehydrogenase 1 alpha-4-beta-4 subunit). Idh1p is a subunit of an enzyme involved in converting isocitrate into α-ketoglutarate, and Mdh1p converts malate into oxaloacetate. The up-regulation of these enzymes indicates that the cells may be attempting to remove excess citric acid by increasing the rate of metabolism through the TCA cycle, despite the fact the cells are growing on glucose and must be glucose repressed. Supporting this hypothesis, we measured the down-regulation of GSF2, encoding a protein involved in the efficient export of the glucose transporter Hxt1p (34). We believe that this will have the effect of decreasing glucose transport and thus encourage the utilization of the excess citric acid as an energy source. Furthermore, we observed that deletion of the transcriptional repressor CYC8 gene resulted in sensitivity to citric acid stress and that the CYC8 transcript was up-regulated upon exposure to citric acid. Cyc8p is required for derepression of isocitrate lyase (Icl1p), an enzyme that reversibly converts isocitrate to glyoxylate, and is essential for growth on the nonfermentable carbon sources ethanol and acetate. Thus, the activity of CYC8 may be inducing derepression in the presence of citric acid, allowing the cell to metabolize excess citrate. Significantly, it has been postulated that activation of Hog1p results in the phosphorylation of the Sko1p transcription factor, causing the Cyc8/Tup1p transcriptional repressor complex to dissociate, thus allowing the derepression and expression of target genes, such as ICL1 (28). We also identified that deletion of SNF2, a component of the SWI/SNF transcription activator complex, was sensitive to citric acid. Deletion of SNF2 renders cells unable to grow on a nonfermentable carbon source (37). Again, this could prevent cells from metabolizing and removing the excess citrate, thus accounting for the observed growth inhibition.
A functional vacuolar membrane H+-ATPase and plasma membrane calcium channel are crucial for adaptation to citric acid.
The importance of ion homeostasis during citric acid stress is clearly indicated by our results. Citric acid-sensitive deletion mutants included a Δcch1 mutant (CCH1 encodes a plasma membrane Ca2+ channel), in addition to many VMA gene products that encode the constituent subunits of the vacuolar membrane H+-ATPase, which is responsible for acidification of the yeast vacuole (19). In addition, we found that Vma4p (H+-ATPase V1 domain S0-A) was up-regulated in response to citric acid stress. However, only one vacuolar H+-ATPase transcript, VMA2, was induced by citric acid. The vacuole contains a range of proteases and degradative enzymes and has an important role in amino acid, calcium, and intracellular pH (pHi) homeostasis (21). Thus, exposure to citric acid at pH 3.5 may have the effect of reducing pHi, such that activity of the vacuolar H+-ATPase is crucial to allow the cell to maintain pHi homeostasis. Furthermore, in yeast cells, the vacuole contains the major cytosolic Ca2+ pool (8), sequestered via a low-affinity Ca2+/H+ antiporter that utilizes the proton motive force generated by the vacuolar H+-ATPase (25). Therefore, if the activity of the vacuolar H+-ATPase is reduced, then there is no detectable Ca2+ uptake into the vacuole (2). Also, Ca2+ influx into the cell from the vacuole is drastically reduced in Δcch1 cells or cells treated with the chelating agents EGTA or 1,2-bis-(o-aminophenoxy)ethane-N,N,N′,′-tetraacetic acid (tetrapotassium salt) (BAPTA) (24). The inhibitory effect of citric acid on Clostridium botulinum has been reported to be due to citric acid chelating divalent metal ions, particularly calcium (12). Therefore, citric acid may be chelating Ca2+ from the medium, possibly resulting in Ca2+ ion depletion. In fact, we have preliminary data (not shown) demonstrating that the inhibitory effect of citric acid is reduced in the presence of CaCl2. Thus, we propose that disruption of calcium homeostasis may be a principal inhibitory effect of citric acid stress. The chelating activity of citric acid for other metal ions in the media may also be indicated by the sensitivity of the Δrcs1 mutant. RCS1 encodes a transcription factor known to activate transcription in response to iron deprivation. Thus, it may be required for growth in the presence of citric acid if iron levels in the media are drastically reduced via chelation.
HOG1 regulates protein expression in response to citric acid stress.
We identified that several strains with mutations in genes encoding amino acid biosynthesis, Δlys5, Δgly1, Δaro2, Δthr1, and Δthr4 mutants, were sensitive to citric acid. In addition, His7p was up-regulated upon exposure to citric acid. Other weak acids are known to inhibit the uptake of aromatic amino acids, resulting in auxotrophic growth requirements for these amino acids (4). Thus, exposure to citric acid may also be inhibiting the uptake of amino acids.
Amino acid limitation activates two protein kinases, Tor1p and Tor2p, that control many cellular processes, including translation, in response to amino acid availability. In part, this is mediated by controlling the cytosolic localization of transcriptional regulators, including Msn2p and Msn4p. This control is achieved via the 14-3-3 proteins Bmh1p and Bmh2p (reviewed in reference 17). Our results indicate that both the BMH1 and BMH2 transcripts are up-regulated on exposure to citric acid, perhaps in response to amino acid starvation. We also identified that deletion of BMH2 results in sensitivity to citric acid. Importantly, Bmh1p was expressed upon exposure to citric acid, but only in the Δhog1 mutant. This indicates that although transcription of BMH1 is increased on exposure to citric acid, the actual expression of Bmh1p is controlled, or negatively regulated, by Hog1p. Currently, we are testing the hypothesis that this represents a precise feedback mechanism for controlling gene expression via Msn4p during citric acid stress.
During citric acid stress, Hog1p negatively regulates the expression of Pdb1p, Ura1p, Fba1, Ydr533cp, Gnd1p, and Car1p. Thus, it is clear that Hog1p regulates the expression of a broad range of previously unknown proteins with a diverse range of different functions within the cell. However, these Hog1p-regulated proteins are not essential for optimal adaptation to citric acid, because none of the corresponding deletion strains were sensitive to citric acid (results not shown).
Hog1p also positively regulated the expression of proteins in response to citric acid. The citric acid-induced expression of the HSP70 family member Ssa1p and the essential glycolytic protein Eno2p was not detected in the Δhog1 deletion strain under these conditions.
Transcript analysis revealed that citric acid resulted in the repression of a large number of ribosomal genes, a response that is typical after exposure to many stress factors and reflects the slowdown in cell proliferation (16). Interestingly, we identified that the expression of Rps0bp, a 40S ribosomal protein, was repressed in the presence of citric acid in the Δhog1 deletion mutant. Thus, Hog1p may also play a role in posttranscriptional regulation of protein expression. Similarly, Egd2p, a protein that may act as a transcription factor exerting a negative effect on the expression of several genes that are transcribed by RNA polymerase II, was also repressed in the presence of citric acid in the Δhog1 deletion mutant.
In conclusion, this study has clearly shown that Hog1p regulates the expression of a variety of previously unknown proteins with different biological roles in response to citric acid stress.
Changes in gene or protein expression are not essential for growth in the presence of citric acid.
Overall, we found little correlation between the genes with measured changes in the levels of transcript or protein expressed and the gene deletions that displayed a citric acid-sensitive phenotype. Poor correlation between the levels of transcripts and levels of protein expressed has been observed and discussed previously (7, 14) and will not be discussed further here. After exposing the yeast cells to citric acid, we identified 69 citric acid-sensitive deletions compared to 68 transcripts up-regulated and 54 transcripts down-regulated and 22 proteins up-regulated. Only 9 of 68 genes (13%) whose expression was induced displayed a citric acid-sensitive phenotype when the corresponding gene was deleted. Thus, the majority of genes and proteins whose expression is altered due to exposure to citric acid are not essential for optimal adaptation to this stress. Instead, the activity of existing proteins is essential for adaptation.
Despite this lack of correlation, comparison of the GO ontology profiles in each case revealed that the genes, proteins, and deletions all came from categories with similar biological functions and in similar proportions.
In summary, modern functional genomics tools now make truly global analyses of cellular responses to various factors now possible in S. cerevisiae. Using these approaches, we found that the HOG pathway regulates a unique stress response in S. cerevisiae upon exposure to citric acid. Citric acid activates the Hog1p MAPK pathway, which subsequently regulates the expression of a number of proteins. We found that citric acid does not induce an osmotic stress but does induce a general stress response and glycerol biosynthesis. To counteract the inhibitory effects of citric acid, a number of cellular proteins play a crucial role, including transcription factors mediating glucose derepression, enzymes involved in amino acid biosynthesis, calcium transport channels, and the vacuolar membrane H+-ATPase. In the future, we plan to address the intriguing question of how citric acid stress is physically sensed by the HOG pathway.
Supplementary Material
Acknowledgments
We gratefully acknowledge the financial support of the BBSRC (grant 49/D15224) for this work.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albertyn, J., S. Hohmann, J. M. Thevelein, and B. A. Prior. 1994. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14:4135-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anraku, Y., R. Hirata, Y. Wada, and Y. Ohya. 1992. Molecular genetics of the yeast vacuolar H+-ATPase. J. Exp. Biol. 172:67-81. [DOI] [PubMed] [Google Scholar]
- 3.Bammert, G. F., and J. M. Fostel. 2000. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob. Agents Chemother. 44:1255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, B. E., D. Rossington, M. Mollapour, Y. Mamnun, K. Kuchler, and P. W. Piper. 2003. Weak organic acid stress inhibits aromatic amino acid uptake by yeast, causing a strong influence of amino acid auxotrophies on the phenotypes of membrane transporter mutants. Eur. J. Biochem. 270:3189-3195. [DOI] [PubMed] [Google Scholar]
- 5.Boucherie, H., G. Dujardin, M. Kermorgant, C. Monribot, and P. Slonimski. 1995. Two-dimensional protein map of Saccharomyces cerevisiae: construction of a gene-protein index. Yeast 11:601-613. [DOI] [PubMed] [Google Scholar]
- 6.Causton, H. C., B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander, and R. A. Young. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12:323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Nobel, H., L. Lawrie, S. Brul, F. Klis, M. Davis, H. Alloush, and P. Coote. 2001. Parallel and comparative analysis of the proteome and transcriptome of sorbic acid-stressed Saccharomyces cerevisiae. Yeast 18:1413-1428. [DOI] [PubMed] [Google Scholar]
- 8.Eilam, Y., H. Lavi, and N. Grossowicz. 1985. Mechanism of stimulation of Ca2+ uptake by miconazole and ethidium bromide in yeasts: role of vacuoles in Ca2+ detoxification. Microbios 44:51-66. [PubMed] [Google Scholar]
- 9.Gasch, A. P., and M. Werner-Washburne. 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2:181-192. [DOI] [PubMed] [Google Scholar]
- 10.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 11.Gould, G. W. 1989. Mechanisms of activation of food preservation procedures. Elsevier Sciences Publishers Ltd., London, England.
- 12.Graham, A. F., and B. M. Lund. 1986. The effect of citric acid on growth of proteolytic strains of Clostridium botulinum. J. Appl. Bacteriol. 61:39-49. [DOI] [PubMed] [Google Scholar]
- 13.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gygi, S. P., Y. Rochon, B. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatcher, W. S., M. E. Parish, J. L. Weihe, D. F. Splittstoesser, and B. B. Woodward. 2000. Fruit beverages, p. 565-568. In F. P. Downes and K. Ito (ed.), Methods for microbial examination of food. American Public Health Association, Washington, D.C.
- 16.Herruer, M. H., W. H. Mager, H. A. Raue, P. Vreken, E. Wilms, and R. J. Planta. 1988. Mild temperature shock affects transcription of yeast ribosomal protein genes as well as the stability of their mRNAs. Nucleic Acids Res. 16:7917-7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacoby, T., H. Flanagan, A. Faykin, A. G. Seto, C. Mattison, and I. Ota. 1997. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J. Biol. Chem. 272:17749-17755. [DOI] [PubMed] [Google Scholar]
- 19.Kane, P. M., M. C. Kuehn, I. Howald-Stevenson, and T. H. Stevens. 1992. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H+-ATPase. J. Biol. Chem. 267:447-454. [PubMed] [Google Scholar]
- 20.Kapteyn, J. C., B. ter Riet, E. Vink, S. Blad, H. De Nobel, H. Van Den Ende, and F. M. Klis. 2001. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39:469-479. [DOI] [PubMed] [Google Scholar]
- 21.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kren, A., Y. M. Mamnun, B. E. Bauer, C. Schuller, H. Wolfger, K. Hatzixanthis, M. Mollapour, C. Gregori, P. Piper, and K. Kuchler. 2003. War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol. Cell. Biol. 23:1775-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Pastor, M. T., G. Marchler, C. Schuller, A. Marchler-Bauer, H. Ruis, and F. Estruch. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227-2235. [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto, T. K., A. J. Ellsmore, S. G. Cessna, P. S. Low, J. M. Pardo, R. A. Bressan, and P. M. Hasegawa. 2002. An osmotically induced cytosolic Ca2+ transient activates calcineurin signaling to mediate ion homeostasis and salt tolerance of Saccharomyces cerevisiae. J. Biol. Chem. 277:33075-33080. [DOI] [PubMed] [Google Scholar]
- 25.Ohsumi, Y., and Y. Anraku. 1983. Calcium transport driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 258:5614-5617. [PubMed] [Google Scholar]
- 26.Piper, P., Y. Mahe, S. Thompson, R. Pandjaitan, C. Holyoak, R. Egner, M. Muhlbauer, P. Coote, and K. Kuchler. 1998. The Pdr12 ABC transporter is required for the development of weak organic acid resistance in yeast. EMBO J. 17:4257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Posas, F., J. R. Chambers, J. A. Heyman, J. P. Hoeffler, E. de Nadal, and J. Arino. 2000. The transcriptional response of yeast to saline stress. J. Biol. Chem. 275:17249-17255. [DOI] [PubMed] [Google Scholar]
- 28.Proft, M., A. Pascual-Ahuir, E. de Nadal, J. Arino, R. Serrano, and F. Posas. 2001. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J. 20:1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi, S.-Y., A. Moir, and C. D. O'Connor. 1996. Proteome of Salmonella typhimurium SL1344: identification of novel abundant cell envelope proteins and assignment to a two-dimensional reference map. J. Bacteriol. 178:5032-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rep, M., J. Albertyn, J. M. Thevelein, B. A. Prior, and S. Hohmann. 1999. Different signalling pathways contribute to the control of GPD1 gene expression by osmotic stress in Saccharomyces cerevisiae. Microbiology 145:715-727. [DOI] [PubMed] [Google Scholar]
- 31.Rep, M., M. Krantz, J. M. Thevelein, and S. Hohmann. 2000. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275:8290-8300. [DOI] [PubMed] [Google Scholar]
- 32.Salmond, C. V., R. G. Kroll, and I. R. Booth. 1984. The effect of food preservatives on pH homeostasis in Escherichia coli. J. Gen. Microbiol. 130:2845-2850. [DOI] [PubMed] [Google Scholar]
- 33.Schuller, C., J. L. Brewster, M. R. Alexander, M. C. Gustin, and H. Ruis. 1994. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 13:4382-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherwood, P. W., and H. Carlson. 1999. Efficient export of the glucose transporter Hxt1p from the endoplasmic reticulum requires Gsf2p. Proc. Natl. Acad. Sci. USA 96:7415-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh, K. K. 2000. The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic. Biol. Med. 29:1043-1050. [DOI] [PubMed] [Google Scholar]
- 37.Steinmetz, L. M., C. Scharfe, A. M. Deutschbauer, D. Mokranjac, Z. S. Herman, T. Jones, A. M. Chu, G. Giaever, H. Prokisch, P. J. Oefner, and R. W. Davis. 2002. Systematic screen for human disease genes in yeast. Nat. Genet. 31:400-404. [DOI] [PubMed] [Google Scholar]
- 38.Tamas, M. J., M. Rep, J. M. Thevelein, and S. Hohmann. 2000. Stimulation of the yeast high osmolarity glycerol (HOG) pathway: evidence for a signal generated by a change in turgor rather than by water stress. FEBS Lett. 472:159-165. [DOI] [PubMed] [Google Scholar]
- 39.Tenreiro, S., P. C. Rosa, C. A. Viegas, and I. Sa-Correia. 2000. Expression of the AZR1 gene (ORF YGR224w), encoding a plasma membrane transporter of the major facilitator superfamily, is required for adaptation to acetic acid and resistance to azoles in Saccharomyces cerevisiae. Yeast 16:1469-1481. [DOI] [PubMed] [Google Scholar]
- 40.Viegas, C. A., and I. Sa-Correia. 1991. Activation of plasma membrane ATPase of Saccharomyces cerevisiae by octanoic acid. J. Gen. Microbiol. 137:645-651. [DOI] [PubMed] [Google Scholar]
- 41.Warmka, J., J. Hanneman, J. Lee, D. Amin, and I. Ota. 2001. Ptc1, a type 2C Ser/Thr phosphatase, inactivates the HOG pathway by dephosphorylating the mitogen-activated protein kinase Hog1. Mol. Cell. Biol. 21:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, M., J. DeRisi, H.-H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winkler, A., C. Arkind, C. P. Mattison, A. Burkholder, K. Knoche, and I. Ota. 2002. Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress. Eukaryot. Cell 1:163-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wojda, I., R. Alonso-Monge, J. P. Bebelman, W. H. Mager, and M. Siderius. 2003. Response to high osmotic conditions and elevated temperature in Saccharomyces cerevisiae is controlled by intracellular glycerol and involves coordinate activity of MAP kinase pathways. Microbiology 149:1193-1204. [DOI] [PubMed] [Google Scholar]
- 45.Xue, L., and J. M. Lucocq. 1997. Low extracellular pH induces activation of ERK 2, JNK, and p38 in A431 and Swiss 3T3 cells. Biochem. Biophys. Res. Commun. 241:236-242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.