Abstract
A three layer (trilayer) polymeric micelle system based on the self-association of the triblock polymer poly(ethylene glycol)-b-poly{N-[N-(2-aminoethyl)-2-aminoethyl] aspartamide}-b-poly(ε-caprolactone) (PEG-b-PAsp(DET)-b-PCL) has been synthesized and investigated for combination delivery of rapamycin (RAP) and siRNA targeting Y-box binding protein-1 (siYB-1). The trilayer micelle is composed of (a) a hydrophilic poly(ethylene glycol) (PEG) block constituting the outer layer to improve pharmacokinetics, (b) an intermediate compartment composed of the cationic poly{2-[(2-aminoethyl)amino] ethyl aspartamide} (PAsp(DET)) segment for interacting with siYB-1, and (c) an inner hydrophobic poly(ε-caprolactone) (PCL) compartment for encapsulation of RAP. A major advantage of this system is biocompatibility since PEG and PCL are both approved by the FDA, and PAsp(DET) is a non-toxic pH responsive cationic poly(amino acid)-based polymer. In this study, it has been shown that PCL can encapsulate RAP with high loading efficiencies, and PAsp(DET) can successfully interact with siRNA for efficient transfection/knockdown with negligible cytotoxicity. The enhanced therapeutic efficacy of RAP/ siYB-1 micelles was demonstrated in cell cultures and in a PC3 xenograft nude mouse model of human prostate cancer. Herein, we demonstrate that trilayer micelles are a promising approach to improve the simultaneous delivery of combination siRNA/drug therapies.
Keywords: Micelles, Combination therapy, Animals, siRNA, YB-1, Rapamycin
1. Introduction
Rapamycin (RAP) (914 g/mol) was originally used as an antifungal agent [1], and later developed as an approved immuno-suppressant and potential anticancer drug [2–5]. Preclinical data with RAP revealed inhibition of tumor growth in a number of cell lines, including lung [2], cervix [4], colon [6], prostate [2,7], and breast [8] cell carcinomas with a typical IC50 < 50 nm. The anticancer mechanism of the drug involves blocking vascular endothelial growth factor (VEGF) production, stimulating endothelial cells [9,10] and inducing cell cycle arrest at the transition from G1-S phase [11,12]. In addition, RAP is the most commonly used chemical to induce autophagy, a lysosome-based pathway that is responsible for the degradation and recycling of proteins and intracellular components for maintaining cellular homeostasis [13]. Unfortunately, autophagy protects some cancer cells by blocking the apoptotic pathway [14], and further suggests that RAP as a cancer therapeutic be administered in combination with other drugs. In addition, the systemic delivery of RAP constitutes a major challenge in the field of cancer therapy due to the drug’s poor solubility in water (2.6 mg/ml) [15], low bioavailability, and dose-limiting toxicity [16]. To date, only one successful formulation of RAP into poly(ethylene glycol)-b-poly(ε-caprolactone) (PEG-b-PCL) micelles has been reported [17].
Small interfering RNA (siRNA) targeting Y-box binding protein-1 (siYB-1) has attracted great interest as a therapeutic agent because of its ability to efficiently knockdown (kd) genes associated with cancer cell proliferation and multiple drug resistance (MDR) [18]. YB-1 is an oncogenic transcription/translation factor and regulates DNA- and mRNA-dependent events in eukaryotic cells [19–21]. YB-1 is also a pronounced marker of tumors because of its overexpression in cancer cells [22–25]. Moreover, YB-1 translocation from the cytoplasm to the nucleus stimulates transcription of a number of genes encoding protective proteins, including those responsible for MDR [26–28]. As a result, the inhibition of YB-1 expression has been found to sensitize cancer cells to chemother-apeutic drugs such as doxorubicin (DOX) [29] or paclitaxel (PTX) [30]. Likewise, we have found that the combination of siYB-1 with RAP greatly enhanced sensitivity of human PC3 prostate cancer cells to RAP.
As such, multifunctional nanoparticles (NP) are highly desirable for the delivery of effective drug combinations and the only means to ensure that both drugs will simultaneously reach cancer cells. In drug delivery, chemical and/or physical optimizations are typically required of the vehicle to efficiently deliver any single one therapeutic agent. Obviously, co-delivery of two or more variety of drugs (e.g.siRNA/drug or drug/drug) requires compartmentalization in the design of the NP to satisfy the different chemical properties of each drug. To date, there have been few drug delivery systems reported in the literature for the co-delivery of both drug and gene. The need for drug compartmentalization was demonstrated with cationic core–shell NP encapsulating PTX in the core and condensing either plasmid DNA (pDNA) or siRNA on the cationic surface [31]. In another drug compartmentalization system, NP were formed by conjugating DOX to poly(lactic-co-glycolide) (PLGA) polymers, and these in turn were entrapped within liposomes made of PEG-block-distearoylphosphatidylethanolamine (PEG-b-DSPE), phosphatidylcholine and cholesterol for optimal loading of combretastatin within the outer lipid bilayer (combretastatin is an anti-angiogenesis drug) [32]. Still, another design utilized NP made from PEG-b-PLGA polymers decorated with nucleic acid ligands (aptamers) to encapsulate docetaxel in the core and intercalate DOX within the aptamers at the surface [33]. Although promising, some of these systems rely on complicated NP assemblies that are ultimately inherently too complex to scale up.
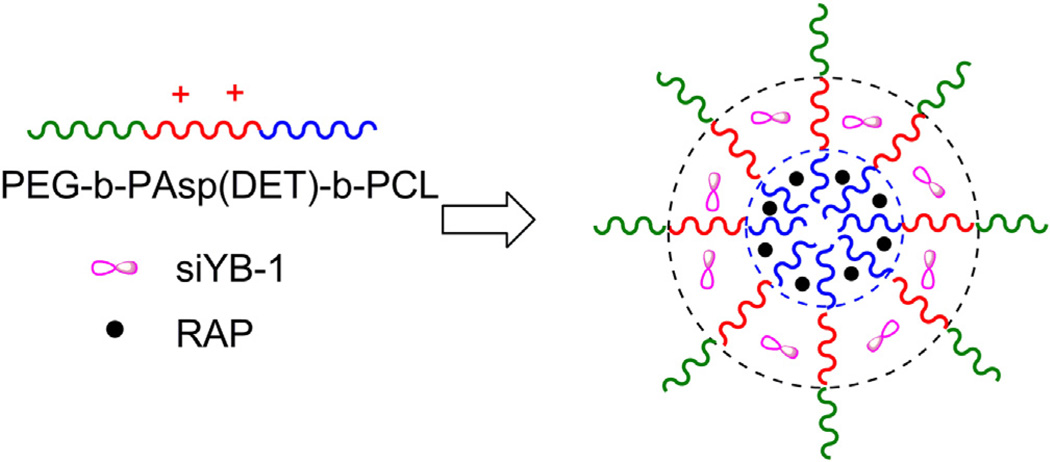
In the present study, we report on the synthesis and characterization of three layer micelles formed from the triblock polymer PEG-b-PAsp(DET)-b-PCL for delivery of RAP and siYB-1. Each drug is encapsulated within a different compartment of the micelle (Fig.1), and the design successfully incorporates biocompatibility, simultaneous loading of RAP and siYB-1 at high loading capacities, and relies on self-association of polymer chains to form NP. More specifically, the trilayer micelle is composed of (a) a hydrophilic PEG block constituting the outer layer to improve pharmacokinetics, (b) an intermediate compartment composed of the cationic PAsp(DET) segment for interacting with siYB-1, and (c) an inner hydrophobic PCL compartment for encapsulation of RAP. In addition, we also report on the therapeutic efficacy of combination RAP/siYB-1 micelles in cultured cells and in a PC3 xenograft mouse model of human prostate cancer.
Fig. 1.
The trilayer micelle is composed of (a) a hydrophilic PEG block constituting the outer layer to improve pharmacokinetics, (b) an intermediate compartment composed of the cationic PAsp(DET) segment for interacting with siYB-1, and (c) an inner hydrophobic PCL compartment for encapsulation of RAP.
2. Materials and methods
2.1. Materials
2.1.1. Chemicals and biologicals
l-aspartic acid β-benzyl ester, triphosgene, diethylenetriamine (DET), benzene, anhydrous dichloromethane (DCM), hexane and N,N-dimethylformamide (DMF) were purchased from Sigma–Aldrich (Milwaukee, WI). Methoxy-ω-amino PEG (PEG5k−NH2, Mn = 5 KDa, Mw/Mn = 1.04) was purchased from Polymer Science. Dialysis tubing (MWCO = 3500) was obtained from SpectraPor. The monomer -caprolactone (Alfa Aesar) was distilled over CaH2 and used fresh. The catalyst trimethylaluminum (AlMe3) was purchased from TCI (Portland, OR). The anticancer drug RAP was purchased from LC systems. For gel retardation studies, plasmid DNA (pDNA) coding for luciferase (pGL4) contained 5320 base pairs (Promega). For drug release studies, total RNA (tRNA) was isolated from mouse placenta by using a Qiagen RNA purification kit. For kd studies, siRNA against luciferase (siLuc) was obtained from Promega (Madison, WI). For cytotoxicity studies, siYB-1 (sequence was previously reported [30]) was obtained from IDT (sense: 5′-UUUGCUG-GUAAUUGCGUGGdTdT-3′; antisense: 5′-CCACGCAAUUACCAGCAAAdTdT-3′).
2.1.2. Cell culture
Dulbecco’s Modified Eagle’s Medium (DMEM), phosphate buffered saline (PBS), fetal bovine serum (FBS), trypsin-EDTA (0.05% trypsin, 0.48 mM EDTA in HBSS) and penicillin-streptomycin were purchased from Cellgro (MediaTech, Herndon, VA). LNCap and PC3 are human prostate cancer cells obtained from ATCC and cultured according to ATCC protocols. LNCap cells were transduced with lentiviral vectors per standard protocols to generate a stable cell line expressing luciferase (LNCap-Luc).
2.1.3. Knockdown and cytotoxicity assays
For kd assays, the Luciferase Assay System was purchased from Promega (Madison, WI). Luminescence was measured with a single tube luminometer (Berthold Technologies). Protein content was obtained using the DC protein assay kit from BioRad (Hercules, CA), absorbance was measured with the Spectramax 190 microplate reader (Molecular Devices, Sunnyvale, CA), and results were fit to a known protein calibration curve. Cytotoxicity was assessed through the resazurin dye.
2.1.4. Animals
All aspects of the animal studies were performed in accordance with the guidelines defined by the Animal Research Committee of the University of Wisconsin. Nude male mice (7–8 week old) were obtained from Jackson Laboratory and divided into five groups (5 mice per group) for xenograft tumor studies. General anesthesia to animals was induced with 1.5% isoflurane/oxygen. Tumor volume (volume = 0.5 × l × w2) and animal body weights (bw) were monitored daily for 10 days.
2.1.5. Instruments
1H NMR spectra were obtained with a Varian Unity-Inova 400 MHz NMR spectrometer (Palo Alto, CA), with temperature regulation at 80 °C or otherwise as indicated. Chemical shifts are reported in ppm, with respect to the deuterated solvent used. All polymers were characterized by 1H NMR and gel permeation chromatography (GPC) (Shodex LF-804 GPC column) at every step of the synthesis.
2.1.6. Statistical analyses
Results were presented as mean ± SEM, n = 3–6. To compare between data sets, Graphpad Prism 5 Software was used to perform one way analysis of variance (ANOVA). A p < 0.01 and p < 0.001 were considered significant and denoted by * or ** respectively to indicate statistical differences.
2.2. Methods
2.2.1. Synthesis of poly(ethylene glycol)-block-poly(β-benzyl l-aspartate) or PEG-b-PBLA
The monomer β-benzyl-l-aspartate N-carboxy-anhydride (BLA-NCA) was prepared using the Fuchs-Farthing method, through the cyclization of β-benzyl-l-aspartic acid with triphosgene, followed with purification by crystallization in dry THF/hexane. Next, diblock polymer PEG-b-PBLA was prepared by the ring-opening polymerization of BLA-NCA initiated by the terminal −NH2 of PEG5k−NH2 (MW 5 kDa) as previously reported [34]. Typically, BLA-NCA (349 mg, 1.4 mmol) was dissolved in 1 ml DMF and 5 ml DCM, and added into PEG5k−NH2 (200 mg, 0.04 mmol) dissolved in 5 ml DCM. The reaction was stirred at 35 °C under argon for 24 h. The mixture was then precipitated in diethyl ether three times, and freeze-dried with benzene to yield a solid white powder. The degree of polymerization (DP) and size distribution of the polymer were checked by 1H NMR and GPC.
2.2.2. Synthesis of the intermediate triblockpoly(ethyleneglycol)-block-poly(β-benzyl l-aspartate)-block-poly(ε-caprolactone) or PEG-b-PBLA-b-PCL
PEG-b-PBLA was used as a macro-initiator for the ring-opening polymerization of -caprolactone. Generally, freshly distilled -caprolactone (166 mg, 1.45 mmol) was added to PEG-b-PBLA (200 mg, 0.016 mmol) dissolved in 10 ml DCM, followed by addition of 6 equivalent AlMe3 (0.096 mmol). The reaction was allowed to proceed for 24 h at 40 °C. The organic solvent and AlMe3 were then removed by rotovap under vacuum. The residues were re-dissolved in DCM, precipitated in diethyl ether, and freeze dried with benzene. The DP for the PCL block was determined by integrating the corresponding proton peak intensities of PCL (−OCH2(CH2)4CO−, δ = 4.0 ppm) to the methylene protons of the PEG chain (−OCH2CH2–, δ = 3.5 ppm) in DMSO-d6 at 80 °C.
2.2.3. Synthesis of the final triblock polymer poly(ethylene glycol)-block-poly{N-[N-(2-aminoethyl)-2-aminoethyl]aspartamide}-block-poly(ε-caprolactone) or PEG-b-PAsp(DET)-b-PCL
DET side chains were substituted into the PBLA segments of PEG-b-PBLA-b-PCL via an aminolysis reaction. Briefly, DET (148.56 mg, 1.44 mmol) was added to PEG-b-PBLA-b-PCL (180 mg, 0.008 mmol) dissolved in 10 ml DMSO, and the reaction was allowed to stir for 24 h at 35 °C. The polymer was dialyzed against ddH2O (SpectraPor, MWCO 3500 Da) for 24 h to remove excess DET and DMSO. The resulting aqueous suspension of polymer was then lyophilized, yielding a solid white powder. To verify there was no chain cleavage of PCL during the reaction, the DP in the final polymer was re-analyzed using 1H NMR, by integrating the proton peak intensity ratio of PCL peaks (−OCH2(CH2)4CO−, δ = 4.0 ppm) to the methylene protons of PEG (−OCH2CH2−, δ = 3.5 ppm) in DMSO-d6 at 80 °C. Because the PAsp(DET) proton peak intensity overlapped with the H2O peak (−NH(CH2)2NH2, δ = 3.1 ppm) we could not use NMR to re-analyze the DP of the PAsp(DET) block, however when the resulting polymer was re-analyzed with GPC there was only one peak which helped support the conclusion that there was no chain cleavage.
2.2.4. Preparation of micelles
Typically, 0.5 mm of triblock polymer PEG-b-PAsp(DET)-b-PCL was dissolved in 1 ml of acetone, then 1 ml of ddH2O was quickly added to the polymer suspension while vigorously stirring. Acetone was removed by evaporation in the hood, followed by centrifugation at 10,000 rpm for 5 min, and passed through a 0.2 µm nylon syringe filter to remove any aggregates that may have formed. RAP-loaded micelles (RAP/NP) and siRNA/RAP-loaded micelles (siRNA/RAP/NP) were prepared in the same manner, by first dissolving RAP with the polymer in acetone and then adding ddH2O with or without siRNA. RAP and siRNA spontaneously load into the micelle during self-association of the polymer chains. Micelles were centrifuged and passed through a nylon filter as before, to remove un-encapsulated drug and/or aggregates.
2.2.5. Characterization of micelles
Particle size and zeta potential of the resulting NP were characterized at 25 °C using a Zeta sizer Nano-ZS (Malvern Instruments, UK) equipped with a He–Ne ion laser (λ = 633 nm). The hydrodynamic diameters of samples were obtained by cumulant method and are reported as Z-average diameters. For zeta measurements, 800 µl of the micelle suspension was transferred to a disposable capillary cell (DTS1061, Malvern Instruments, UK) for analysis.
2.2.6. RAP and siRNA loading into micelles
The concentration of RAP in trilayer micelles was measured by reverse-phase HPLC [17] equipped with a Symmetry Shreld RP18 column from Waters, and monitoring RAP by UV detection at 279 nm (the triblock polymer did not absorb at this wavelength). RAP loading and encapsulation efficiency were calculated using the following equations:
The mobile phase used was a cocktail of 70% acetonitrile, and 30% water containing 0.1% phosphoric acid/1% methanol. The concentration of RAP was increased from 5%, 10%, 25%, to 50% (w/w) while keeping the concentrations of the triblock polymer constant at 0.5 mm and siRNA constant at 39 µg/ml (3 µm). Next, siRNA loading capacity into NP was determined based on a previously published procedure [35]. Briefly, the sample solutions (100 µl) were centrifuged at 15,000× g for 30 min to precipitate the NP and siRNA concentration remaining in the supernatants (50 µl) was determined by measuring absorption at λ = 260 nm using a NanoDrop UV–Vis spectrophotometer. The percentage of siRNA loaded for different RAP concentrations was calculated by subtracting the siRNA concentration in the supernatant from the total concentration.
2.2.7. Complexations studies
For complexations studies with the triblock polymer, 1 µg of pDNA was used instead of siRNA because it was easier to visualize on an agarose gel with ethidium bromide (EtBr). Specifically, control diblock PEG-b-PAsp(DET)/pDNA (no PCL block) or triblock polymer/pDNA complexes were prepared in PBS at various N/P ratios and incubated at RT for 30 min. The complexes were loaded onto a 0.6% agarose gel containing 0.0004% EtBr and ran for 30 min at 100 V, using lithium borate as the running buffer, prior to imaging. Complementing the 2-D gel retardation assay, the minimum amount of polymer needed to form complexes in solution was also investigated through a 3-D EtBr fluorescence assay. Quantitatively, the fluorescence of EtBr is highest when intercalated to pDNA and lowest when un-bound. Upon addition of polymers to pDNA, EtBr molecules get displaced and result in decreased fluorescence measurements characteristic of the system at a specific N/P value.
To investigate further siRNA loading capacity into NP without RAP, siLuc at 0– 50 µg/ml was loaded into trilayer micelles at a constant polymer concentration of 300 µg/ml. We also looked at loading capacity when siLuc was kept constant at 39 µg/ml (3 µm) while polymer concentration was increased above 300 µg/ml, specifically from 355 to 1772 µg/ml (corresponding to N/P 10–50).
2.2.8. RAP release kinetics
For the drug release study, all micelles were prepared at the 5% w/w ratio of RAP to polymer, to maximize RAP encapsulation within the PCL core rather than at the interfaces of micelles as previously done [17]. RAP was loaded into control PEG-b- PCL micelles and was loaded with or without tRNA into PEG-b-PAsp(DET)-b-PCL micelles to compare release kinetics in the absence and presence of nucleic acids. RAP loading levels did not change in the presence of tRNA, as determined by RP-HPLC. The micelle formulations were diluted with respect to RAP concentration to 0.1 mg/ml in ddH2O and loaded into a dialysis cassette. The release study was conducted by dialyzing the cassettes against ddH2O at 37 °C for 7 days. At various time points, 20 µl of suspension was withdrawn from the dialysis cassettes for RAP analysis by RP-HPLC and replaced with 20 µl of ddH2O.
2.2.9. Knockdown studies
Luciferase-expressing LNCaP prostate cancer cells (LNCaP-Luc) were used for kd studies. Generally, the cells were seeded in a 24-well plate at a density of 50,000 cells per well and incubated overnight. Next, triblock PEG-b-PAsp(DET)-b-PCL/siLuc and control diblock PEG-b-PAsp(DET)/siLuc micelles were prepared at N/P 10–50. Briefly, 50 µl of micelles were added into each well containing 450 µl of media; negative control cells were treated with 50 µl of sterile water. The plates were gently shaken before they were returned to the incubator. After 24 h, the luciferase expression level in each well, with respect to total protein concentration, was determined by a luciferase assay system.
2.2.10. Cytotoxicity studies
PC3 cells were seeded in a 96-well plate at 5000 cells per well for 24 h, and then pretreated with 50 nM siYB-1 in trilayer micelles (siYB-1/NP) to ensure similar levels of cell exposures to siYB-1; specifically, 90 µl of antibiotic free media and 10 µl of siYB-1/NP were added to wells (final concentration of siYB-1 added to cells was 50 nm) and incubated for 4 h before replacing with serum media. The next day, 10 µl of micelle formulations containing either: empty NP, siYB-1/NP, RAP formulated with DMSO (RAP/DMSO), RAP/NP, or siYB-1/RAP/NP were added to wells at a final concentration of 0.01–1 nM RAP. The cells were incubated for another 24 or 48 h before measuring cell viability with the resazurin assay [36].
2.2.11. Antitumor activity
Approximately 1 × 106 PC3 cells were inoculated subcutaneously into the right flanks of anesthetized mice. When the tumor size reached about 50 mm3, 200 µl of the various formulations (siYB-1/NP, RAP/NP, siYB-1/RAP/NP, or empty NP) were injected into mice on days 1, 3 and 5 (RAP at 30 mg/kg, siYB-1 at 2 mg/kg). To investigate the effect of sequential therapy, the fifth group was administered siYB-1/ NP on day 0 and treated with RAP/NP on days 1, 3 and 5. The tumor size and bw of animals were monitored daily for 10 days.
3. Results and discussion
3.1. Synthesis of the triblock polymer PEG-b-PAsp(DET)-b-PCL
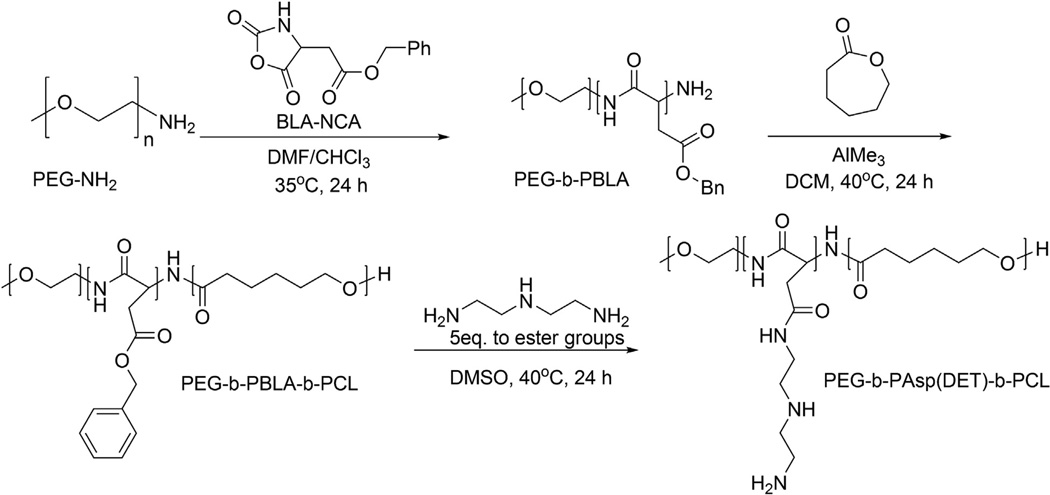
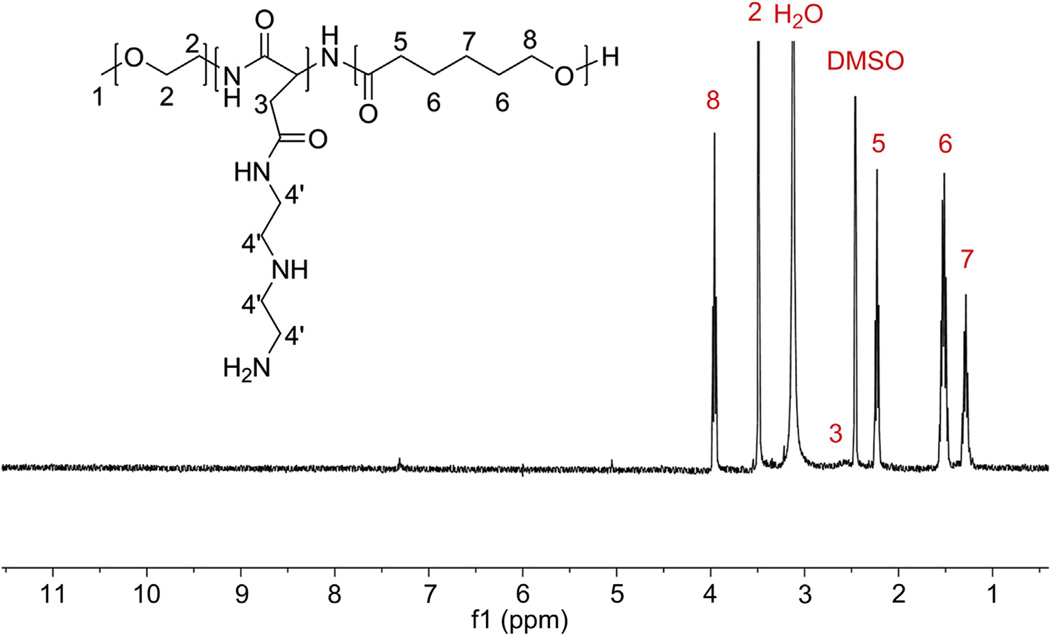
The triblock polymer PEG-b-PAsp(DET)-b-PCL was synthesized via a two-step ring-opening polymerization (Scheme 1). More specifically, PEG5k–NH2 was used as a macro-initiator to polymerize BLA-NCA, to yield amine-terminated PEG-b-PBLA-NH2. The DP of PBLA in the diblock polymer was checked with 1H NMR and found to be 36 (Fig. 2) with a PDI of 1.07. Similarly, for the second ring opening polymerization step, the terminal amino group of PEG-b-PBLA-NH2 was used as an initiator for the polymerization of ε-caprolactone to yield PEG-b-PBLA-b-PCL. 1H NMR (Fig. 3) and GPC revealed a DP of 36 for PBLA, 88 for PCL, and a PDI of 1.24. To obtain the final triblock polymer, the side chains of the PBLA block were substituted with DET groups by an aminolysis reaction in the presence of excess DET. The polymer was then dialyzed against ddH2O to remove excess DET and DMSO. 1H NMR (Fig. 4) of the resulting PEG-b-PAsp(DET)-b-PCL polymer revealed that the side chain substitution level was 100% complete, by noting the disappearance of the aryl protons corresponding to the benzyl groups of PBLA. In addition, 1H NMR and GPC also re-affirmed that the number of repeating units for both PAsp(DET) and PCL were unchanged, 36 and 88 respectively, following the aminolysis reaction. This is important because it was found previously that PAsp(DET) could cleave itself upon prolonged incubation in PBS in a time dependent manner [37]. We did not observe this phenomenon with PEG-b-PAsp(DET)-b-PCL polymers; this is most likely due to the insoluble nature of the polymer in water (due to PCL which preferentially leads to formation of micelles in aqueous media) and thus may have helped prevent self-induced chain cleavage of the PAsp(DET) block. The triblock polymer PEG-b-PAsp(DET)-b-PCL had a final PDI of 1.24 and was estimated to have a total molecular weight of ca. 22 kDa.
Scheme 1.
The triblock polymer PEG-b-PAsp(DET)-b-PCL was synthesized via a two step ring-opening polymerization. More specifically, PEG5k–NH2 was used as a macroinitiator to polymerize BLA-NCA to yield amine-terminated PEG-b-PBLA-NH2. Next, the terminal amino group of PEG-b-PBLA-NH2 was used as an initiator for the polymerization of ε-cap-rolactone to yield PEG-b-PBLA-b-PCL. Finally, to obtain the triblock polymer, the side chains of the PBLA block were substituted with DET groups by an aminolysis reaction.
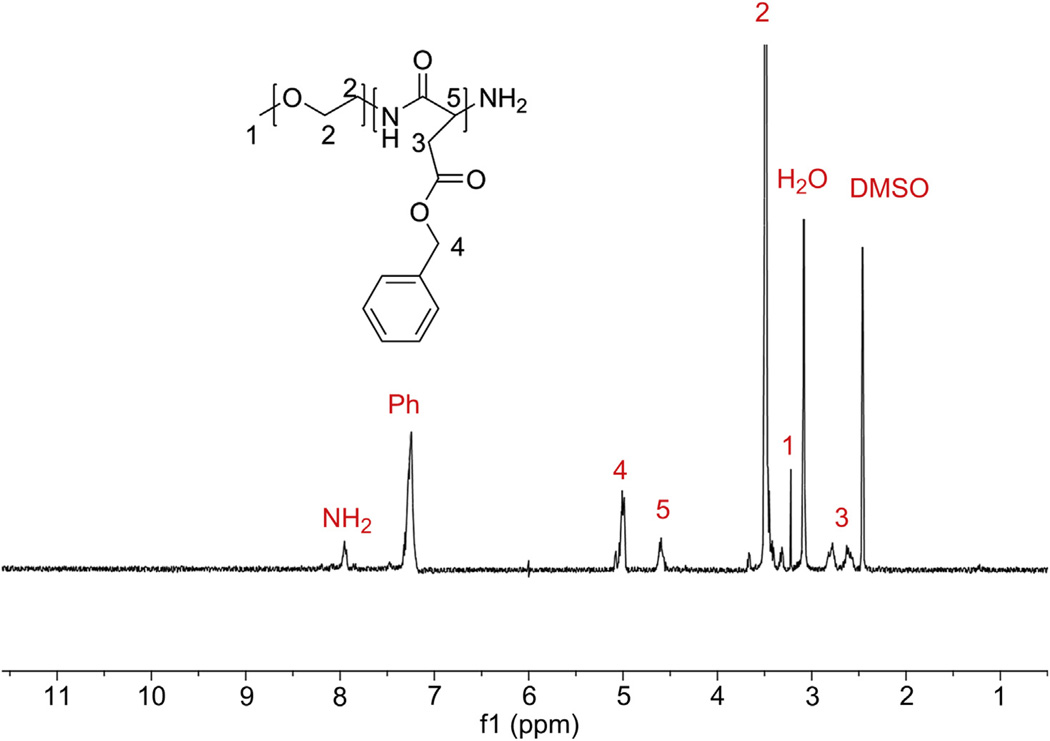
Fig. 2.
1H NMR spectrum of PEG-b-PBLA in DMSO-d6 at 80 °C Degree of polymerization was calculated from integration of the phenyl protons (Ph) with respect to the methyl protons of PEG (peak 2) and found to be 36. When the polymer was run through GPC, there was only one peak with a PDI of 1.07.
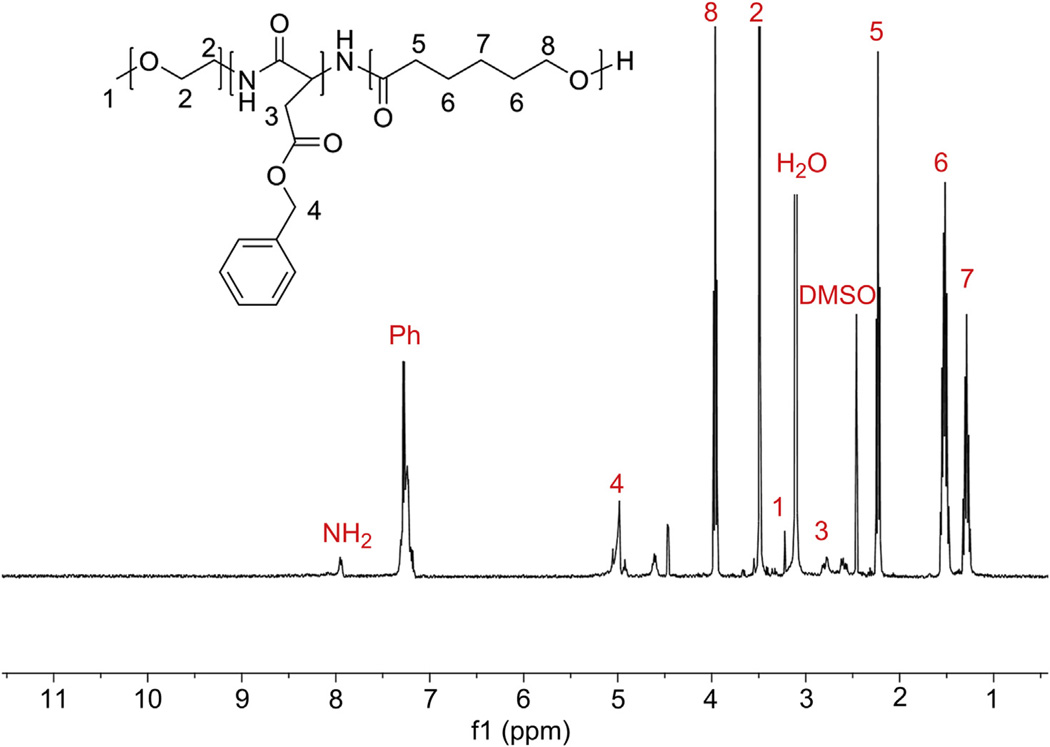
Fig. 3.
1H NMR spectrum of the triblock polymer backbone PEG-b-PBLA-b-PCL in DMSO-d6 at 80 °C DP of PCL was calculated from integration of the corresponding proton peak intensities of PCL (peak 8) with respect to the methyl protons of PEG (peak 2) and found to be 88. When the polymer was run through GPC, there was only one peak with a PDI of 1.24.
Fig. 4.
1H NMR spectrum of PEG-b-PAsp(DET)-b-PCL in DMSO-d6 at 80 °C. Substitution of DET was confirmed by the disappearance of the benzyl groups of PBLA. DP of PCL was recalculated from integration of the corresponding proton peak intensities of PCL and found to be 88, as before. When the triblock polymer was run through GPC, there was only one peak with a PDI of 1.24 which confirmed that there was no chain cleavage of the backbone.
3.2. Preparation and characterization of RAP/siRNA micelles
RAP and/or siRNA were loaded into trilayer micelles based on different partitioning of the two drugs within the micellar core. It has previously been suggested that RAP preferentially partitions into the hydrophobic PCL core or PCL-PEG interface of PEG-b-PCL micelles [17], whereas siRNA is well-known to interact with cationic polymers [38] and therefore should preferentially partition into the cationic PAsp(DET) layer. Both loading of drug and siRNA happened spontaneously upon self-assembly of PEG-b-PAsp(DET)-b-PCL into micelles as acetone was removed.
For the encapsulation of RAP, PCL is an attractive polymer due to the biocompatible nature of the degradation products [39]. In addition, unstable micelles can fall apart rapidly in plasma [40] and lead to excessive drug loss, but it has been shown that micelles with PCL cores (e.g. PEG-b-PCL) can maintain sustained drug release in the plasma due to enhanced circulation [46,47] and low critical micelle concentrations (CMCs), which are indicative of high stability against dilution effects. Other studies have further validated the stability of PCL-based micelles with in vivo assays [48,49]. Of particular interest to us is the encapsulation of RAP into PEG-b-PCL micelles, which was characterized by high loading efficiencies and sustained in vitro drug release [17]. Therefore, we investigated optimal loading of RAP into the PCL core of trilayer micelles by varying the ratio of RAP to triblock polymer (w/w) (Table 1). More specifically, the concentration of the polymer was kept constant at 0.5 mm while RAP was increased (w/w). For each formulation, RAP encapsulation was quantified by RP-HPLC and compared to a standard drug calibration curve. We obtained a max 63% loading (based on initial amount of RAP added) for 25% w/w RAP to polymer, which is higher than the previous report of 50% RAP loading into PEG-b-PCL micelles [17]. This increased loading is likely due to the larger overall size of trilayer micelles at ca. 82 nm compared to PEG-b-PCL micelles at 56 nm (Supporting Info, Fig. S1), leading to RAP encapsulation both within the PCL core and at the PAsp(DET)-PCL interface. This was verified by our attempts to load RAP into PEG-b-PAsp(DET) micelles which was not successful and confirms that PCL is required for efficient encapsulation of RAP.
Table 1.
Drug loading in micelles was investigated by increasing RAP to polymer ratio (w/w) from 5 to 50%, while maintaining the concentration of the polymer at 0.5 mm and siRNA at 3 µm. RP-HPLC was used to quantify RAP encapsulation levels, and to calculate loading efficiency.
| w/w RAP/ polymer |
Initial RAP, mg/ml |
RAP solubilized, mg/ml |
RAP loaded, mg/mg |
RAP: polymer, mol: mol |
Efficiency, % loaded |
|---|---|---|---|---|---|
| 5% | 0.55 | 0.47 ± 0.02 | 0.041 ± 0.002 | 1.01 ± 0.05 | 85 ± 4 |
| 10% | 1.10 | 0.69 ± 0.03 | 0.057 ± 0.002 | 0.43 ± 0.06 | 63 ± 3 |
| 25% | 2.75 | 1.72 ± 0.10 | 0.125 ± 0.008 | 3.13 ± 0.19 | 63 ± 4 |
| 50% | 5.50 | 3.19 ± 0.03 | 0.194 ± 0.002 | 4.84 ± 0.05 | 58 ± 1 |
At the polymer concentration of 0.5 mm, the amount of siRNA entrapped in the trilayer micelle was estimated by measuring absorption at λ = 260 nm using a NanoDrop UV–Vis spectrophotometer and found to be ca. 80% loading (based on initial 3 µm siRNA added). Furthermore, we found that this amount of siRNA entrapped into micelles was similar and was not hindered by increasing RAP concentrations. To further verify that siRNA only interacted with the PAsp(DET) segment of micelles, attempts were made to encapsulate siRNA into PEG-b-PCL micelles but this was not successful as expected. This confirms that the high loading of siRNA into trilayer micelles was most likely due to siRNA entrapped within the PAsp(DET) segment and also at the PEG-PAsp(DET) and PAsp(DET)-PCL interfaces.
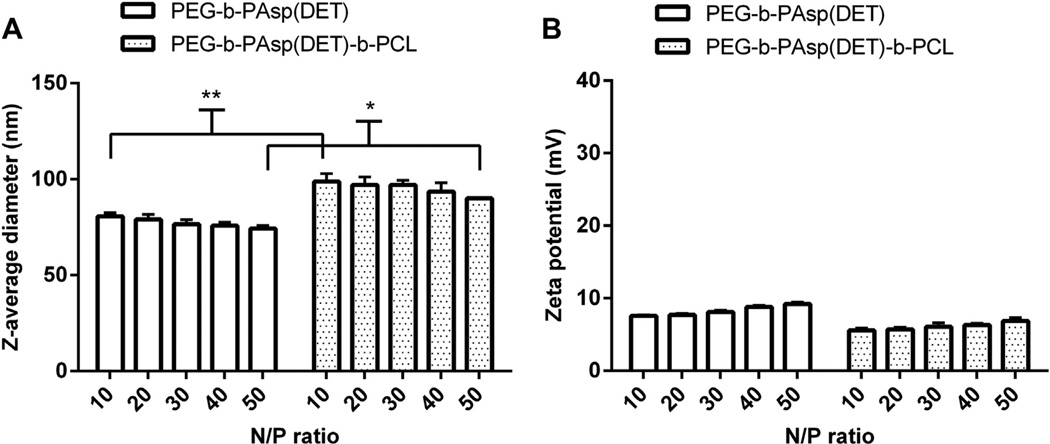
The size and zeta potential of empty NP were characterized at N/ P 10–50. A minimum N/P 10 was chosen due to this value corresponding to 0.5 mm triblock polymer (Table 1). As can be seen in Fig. 5A, the average diameter of control PEG-b-PAsp(DET) micelles averaged ca. 70 nm over N/P 10 to 50, whereas trilayer NP were slightly larger and hovered near ca.100 nm for all N/P. Furthermore, the zeta potential of control micelles (Fig. 5B) ranged from 7 to 10 mV over the range of N/P tested and ranged from 5 to 7 mV for trilayer micelles, being essentially neutral for both formulations due to presence of PEG on the surface of NP.
Fig. 5.
The size and zeta potential of resulting NP were monitored at N/P 10–50. A minimum N/P 10 was chosen due to this value corresponding to 0.5 mm triblock polymer (Table 1 ). As can be seen in (A), the diameter of control PEG-b-PAsp(DET) micelles averaged ca. 70 nm over N/P 10–50, whereas trilayer PEG-b-PAsp(DET)-b-PCL micelles hovered near ca. 100 nm at increasing N/P. On the other hand, the zeta potential (B) of control micelles ranged from 7 to 10 mV and 5–7 mV for trilayer micelles; both formulations are neutral in charge due to presence of PEG on the surface of micelles. *p < 0.01 and **p < 0.001.
Finally, when RAP and/or siRNA were loaded into trilayer micelles at N/P 10, DLS revealed that empty trilayer micelles averaged 70 nm, 81 nm when loaded with RAP (RAP/NP), 113 nm when loaded with siRNA (siRNA/NP), and 108 nm when loaded with both siRNA and RAP (siRNA/RAP/NP). We also investigated the stability of the NP by monitoring their size when incubated in PBS for 7 days. We found that all the NP were stable at room temperature, with no change in their sizes and no formation of aggregates (Supporting Info, Fig. S2). For example, at day 7 of incubation at room temperature, NP averaged 70, 82, 111, and 109 nm for NP, RAP/NP, siRNA/ NP, and siRNA/RAP/NP respectively.
3.3. Complexation studies
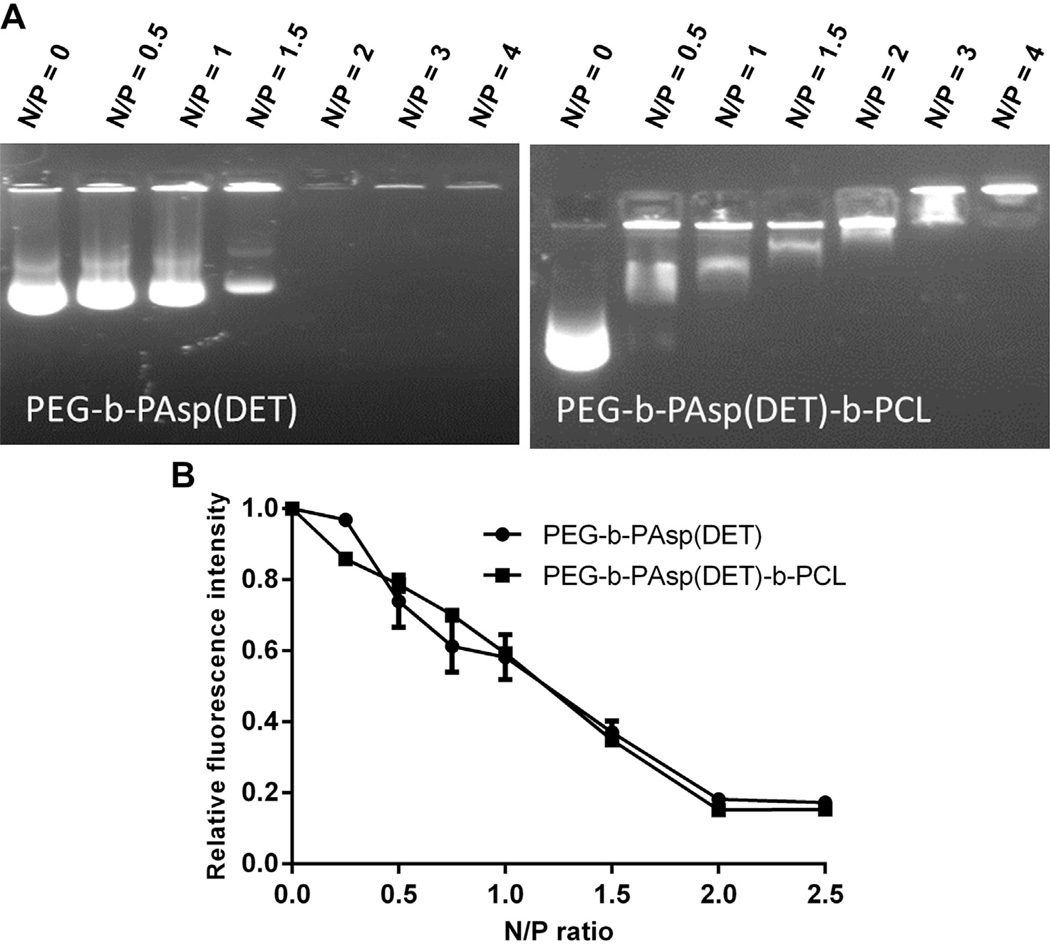
We investigated further the ability of the triblock polymer to interact with nucleic acids by running gel retardation and EtBr fluorescence assays. To limit cost and ensure a good gel picture, we opted to use pDNA. Fig. 6A reveals that the isoelectric point for both the control diblock polymer PEG-b-PAsp(DET) and the triblock PEG-b-PAsp(DET)-b-PCL polymer occurred at N/P 2. The results validate that nucleic acids preferentially interact with the cationic PAsp(DET) segment of the polymers. To further quantify the condensation degree of NP in solution, an EtBr exclusion assay was used to monitor the fluorescence at increasing N/P. Similar to the gel retardation data, Fig. 6B reveals that both polymers were able to interact with pDNA at a minimum N/P 2 and condensed approximately 80% of the pDNA.
Fig. 6.
(A) Gel retardation assay of pDNA and control diblock polymer PEG-b-PAsp(DET) (left) and triblock polymer PEG-b-PAsp(DET)-b-PCL (right) both show retardation at N/P 2. (B) EtBr exclusion assay revealed that the condensation degree for both polymers was approximately 80% at N/P 2.
To investigate further siRNA loading capacity into trilayer micelles, we found that at 300 µg/ml polymer concentration, loading of siLuc saturated above 30 µg/ml, (Supporting Info, Fig. S3A) with siLuc encapsulation efficiency decreasing below 80% above ca. 35 µg/ml (Supporting Info, Fig. S3B). When siLuc was kept constant at 39 µg/ml (3 µm) and polymer concentration was increased (355– 1772 µg/ml corresponding to N/P 10–50), there were only slight differences in the percentage of siLuc encapsulated (95.6%–89.8%) (Supporting Info, Fig. S3B).
3.4. Drug release kinetics
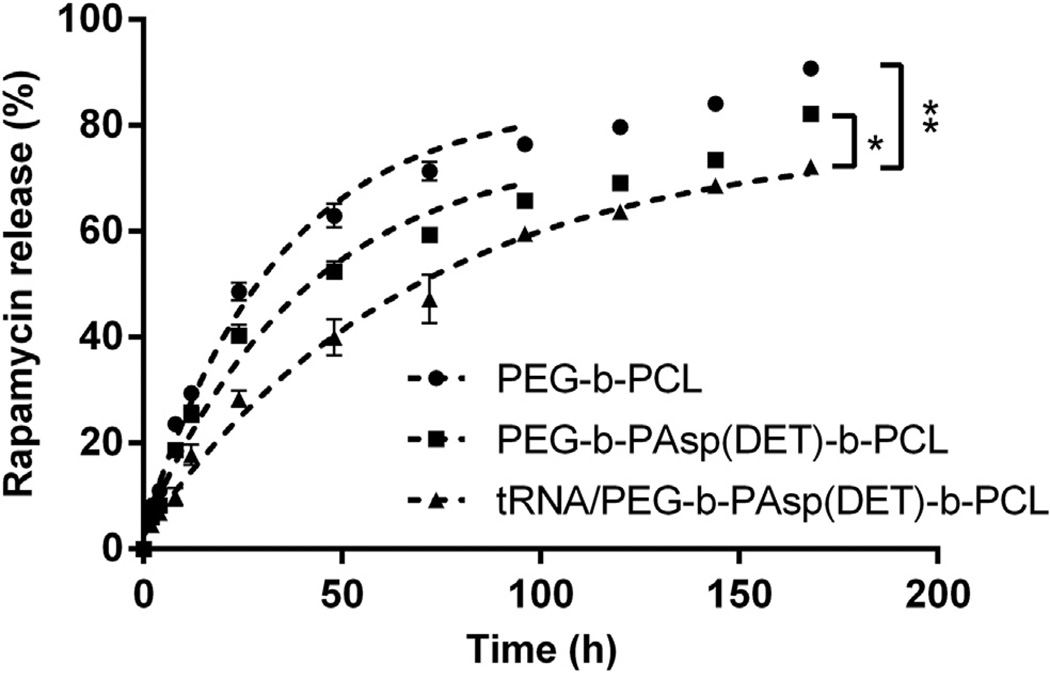
The release kinetics of RAP from PEG-b-PCL, PEG-b-PAsp(DET), and PEG-b-PAsp(DET)-b-PCL micelles was evaluated by incubating the formulations in dialysis cassettes at 37 °C and dialyzing against ddH2O for a week. RAP was formulated into all micelles at the 5% (w/w) RAP to polymer ratio to ensure all of the RAP molecules would be encapsulated in the PCL core. To investigate RAP release in the presence of nucleic acids (and in order to limit cost of siRNA), we encapsulated tRNA and RAP into micelles. tRNA used contained mouse ribosomal RNAs, including fragments at 1.9 kb for 18S and 4.7 kb for 28S ribosomal RNAs. When RAP was formulated into control PEG-b-PCL micelles, we obtained a t1/2 of ca. 36 h for 50% drug release which is similar to a previous report [17]. RAP formulated into trilayer micelles (RAP/NP) revealed a t1/2 of ca. 48 h. When RAP was formulated with tRNA into trilayer micelles (RAP/ tRNA/NP), we obtained an even longer t1/2 of ca. 77 h. Clearly, the presence of tRNA in the middle layer of the micelle provided an extra barrier and slowed down the release of RAP sufficient to increase the t1/2 from 48 to 77 h (Fig. 7). It was not possible to monitor the release of tRNA from micelles because the concentration of nucleic acids in the samples taken was too low for detection by absorbance.
Fig. 7.
For the drug release study, micelles were prepared at the 5% w/w RAP to polymer ratio and dialyzed against ddH2O at 37 °C for a week. RAP in PEG-b-PCL micelles (PEG-b-PCL), RAP in triblock PEG-b-PAsp(DET)-b-PCL micelles (PEG-b-PAsp(DET)- b-PCL), and tRNA/RAP in triblock PEG-b-PAsp(DET)-b-PCL micelles (tRNA/ PEG-b- PAsp(DET)-b-PCL) all exhibited slow and sustained release of RAP over 7 days. Control PEG-b-PCL micelles had a t1/2 of ca. 36 h to 50% drug release; RAP formulated into trilayer micelles revealed a t1/2 of ca. 48 h; when tRNA and RAP were formulated into trilayer micelles, an even longer t1/2of ca. 77 h was obtained. *p < 0.01 and **p < 0.001.
3.5. Gene silencing efficiency
For the delivery of siRNA, the cationic polyaspartamide derivative PAsp(DET) is a promising candidate since it possesses pH-sensitive endosome destabilizing activity at low pH. More specifically, at neutral pH the 1,2-diaminoethane moieties of PAsp(DET) side chains become mono-protonated, but under acidic pH conditions of endosomes they become di-protonated and lead to enhanced buffering properties [41], which may help NP escape into the cytoplasm of cells. PAsp(DET)-based polyplexes formed with pDNA have shown high transfection efficiency with negligible cytotoxicity in various cultured cells, including primary cells [42,43]. However, its application for delivery of siRNA was appreciably limited due to the unstable nature of the polymer-siRNA complex formed. One main reason for the lower stability of the PAsp(DET)/siRNA complex is due to the short and rigid structure of siRNA (21 base pairs) compared to pDNA (>3000 base pairs) [44]. Therefore, an additional association force is needed to enhance the stability of siRNA complexes formed with PAsp(DET). It turns out that the incorporation of a hydrophobic force is a promising candidate for stabilizing siRNA complexes, as has been demonstrated by various studies of siRNA complexes possessing hydrophobic properties [31,45,46]. For example, stearic acids (as a hydrophobic moiety) were previously conjugated to the side chains of PAsp(DET) at various substitution degrees to generate stearoyl PAsp(DET)/siRNA complexes; this modification resulted in efficient endogenous gene kd (in the human pancreatic adenocarcinoma Panc-1 cells) in vitro compared to un-modified PAsp(DET)/siRNA complexes [47], and may explain why our PEG-b-PAsp(DET)-b-PCL micelles are similarly able to kd genes so efficiently in cells.
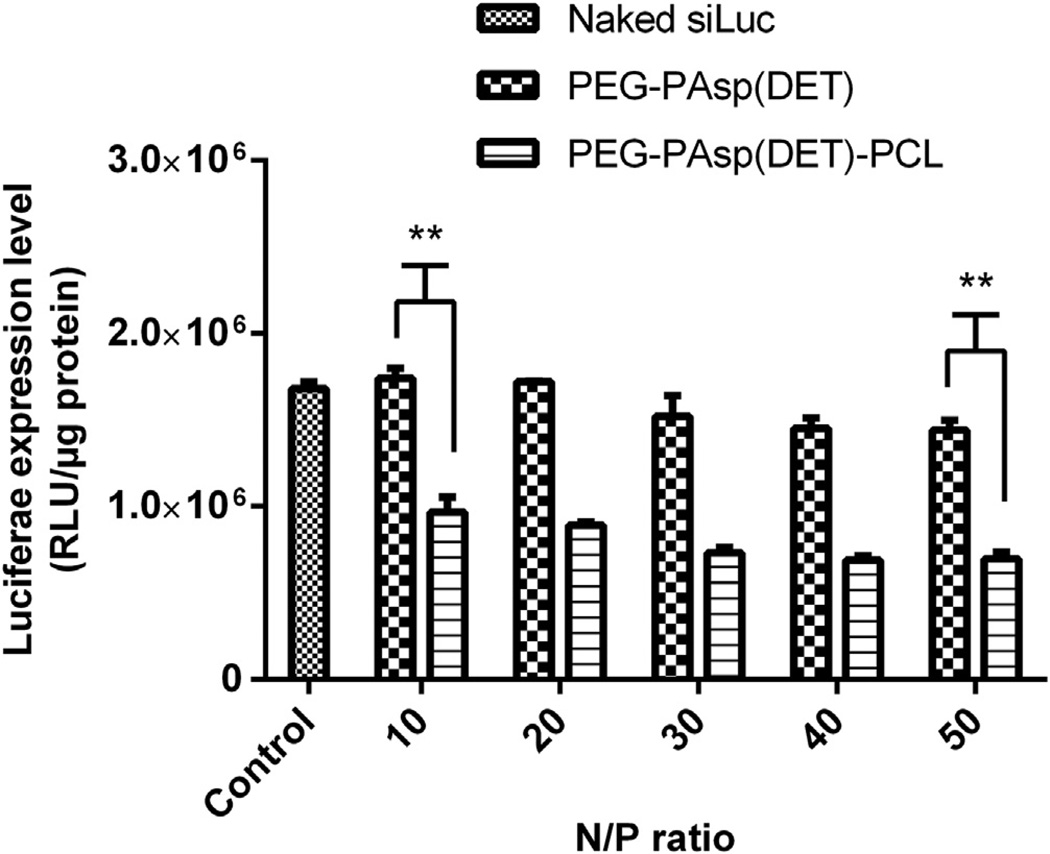
The gene silencing efficiency of control PEG-b-PAsp(DET) vs. PEG-b-PAsp(DET)-b-PCL was investigated by complexing siLuc into resulting micelles and testing on a prostate cancer cell line expressing luciferase, LNCaP-Luc. Micelles were formed at N/P 10– 50 while keeping the concentration of siLuc at 50 nm. As previously reported for PEG-b-PAsp(DET) [47], control PEG-b-PAsp(DET) micelles were not able to efficiently kd luciferase expression compared to trilayer micelles (Fig. 8). The silencing activity of trilayer micelles ranged from 43% to 59% kd efficiency at N/P 10–50, whereas it hovered from 0 to 14% for control micelles. Though both polymers interacted with nucleic acids in a similar manner (Fig. 6), formed NP under ca. 100 nm and had neutral surface charges (Fig. 5), control NP could not mediate efficient siRNA delivery into cells due to particle instability. In contrast, the triblock micelles significantly improved the transfection and kd efficiency of siLuc into LNCaP-Luc cells at N/P 10–50 (p < 0.001), most likely due to presence of the hydrophobic PCL core which helped stabilize the NP (see Supporting Info, Fig. S2).
Fig. 8.
In vitro luciferase kd assay in LNCap-Luc prostate cancer cells after 24 h, comparing control diblock PEG-b-PAsp(DET) and triblock PEG-b-PAsp(DET)-b-PCL micelles complexed with 50 nm siLuc. The kd efficiency for siLuc/triblock micelles ranged from 43 to 59% at N/P 10–50, whereas the kd for siLuc/diblock micelles ranged from 0 to 14% at N/P 10–50. **p < 0.001.
3.6. Combination therapy in vitro
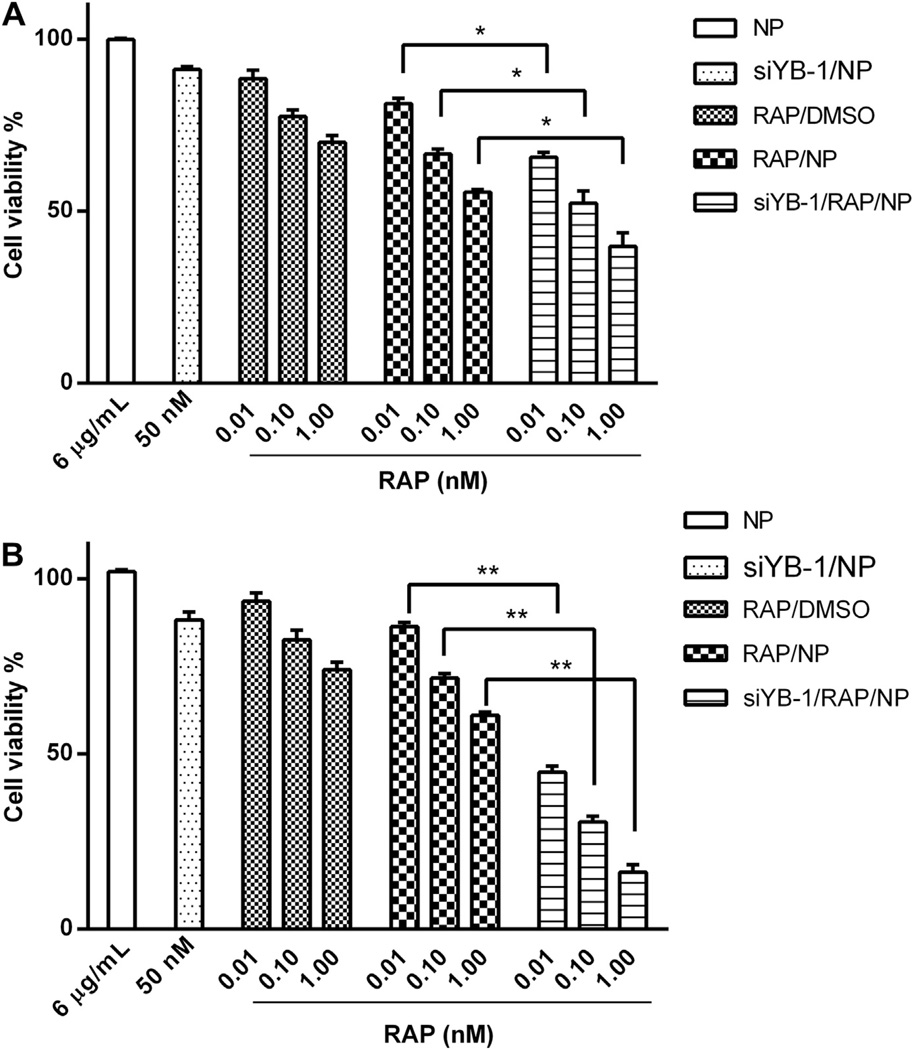
To investigate the in vitro cytotoxicity of combination RAP and siYB-1 in PC3 prostate cancer cells, we looked at sequential and simultaneous dosings of the drugs at both 24 and 48 h time points (Fig. 9). It is important to note that at the polymer concentration investigated (6 µg/ml) there was no cytotoxicity to cells observed up to 48 h (see Supporting Info, Fig. S4), with cell viability similar to untreated cells. When siYB-1/NP at 50 nM was administered to PC3 cells, we found that over >91% of cells were still viable after 48 h thus downregulation of YB-1 alone did not appear to be detrimental to PC3 cells. Because downregulation of genes in vitro by siRNA sometimes takes 24–72 h for full effect, it was necessary to downregulate YB-1 in cells with siYB-1/NP at least 24 h prior to treatment with the RAP formulations. The next day, RAP in DMSO (RAP/DMSO), RAP/NP, or dual loaded siYB-1/RAP/NP at 0.01, 0.1 or 1 nm final RAP concentrations were added to cells. After waiting another 24 and 48 h, the cell viability was measured. We found that cell viability decreased significantly at 48 h for dual loaded siYB-1/ RAP/NP. For example, we obtained similar results for RAP/DMSO and RAP/NP formulations at 24 and 48 h, whereas dual-loaded NP cell viability decreased significantly from 24 to 48 h at all RAP concentrations tested: from 81% to 42% for 0.01 nm RAP (p < 0.001), 65%–45% for 0.1 nm RAP (p < 0.001), and 67%–38% for 1 nm RAP (p < 0.001), respectively.
Fig. 9.
PC3 cells were pre-treated with siYB-1/NP for 24 h, and then treated with: empty NP (at N/P 10), siYB-1/NP, RAP/DMSO, RAP/NP or siYB-1/RAP/NP for another 24 h (A) or 48 h (B) at RAP concentrations of 0.01, 0.1, and 1 nm. Enhanced cytotoxicity was observed after 48 h in cells treated with combination siYB-1/RAP/NP for all 3 RAP concentrations. At the polymer concentration investigated (6 µg/ml) there was no cytotoxicity to cells observed after 24 and 48 h, with cell viability similar to untreated cells. When siYB-1/NP at 50 nm was administered to PC3 cells, we found that over >91% of cells were still viable after 48 h *p < 0.01 and **p < 0.001.
The exact mechanism by which combination siYB-1 and RAP demonstrated enhanced cytotoxicity is unknown, though it is likely that they target the JAK/STAT3 and PI3k/Akt/mTOR signaling pathways that are often up-regulated in cancer. It has previously been found that YB-1 downregulation successfully sensitized breast cancer cells to apoptosis by interfering with the STAT3 pathway [30].The STAT3 pathway is known to promote oncogenesis by being constitutively active and is suggestive of poor cancer prognosis [48,49]. Similarly, the PI3K/Akt/mTOR pathway is an intracellular signaling pathway that is important in cell proliferation and its up-regulation often confers resistance to many types of cancers [50,51]. The mammalian target of rapamycin (mTOR) is a key molecule for activation/regulation of various downstream targets responsible for proliferation, transcription, translation, and apoptosis of cells, and is involved in regulating the translation of specific mRNAs such as those of YB-1 [21,52,53] Interestingly, it has been found that YB-1 synthesis and translation was down-regulated by the mTOR inhibitor PP242 but not RAP [21]. The enhanced cytotoxicity of silencing STAT3 and inhibiting mTOR via RAP was demonstrated on hepatocarcinoma cells and characterized by increased cellular apoptosis [54].As such, the downregulation of critical signaling pathways such as STAT3 and mTOR by siYB-1/RAP/NP may help explain the enhanced cytotoxicity observed in PC3 cancer cells, and validates further investigations into animal tumor models.
3.7. Combination therapy in a xenograft mouse model of PC3 human prostate cancer
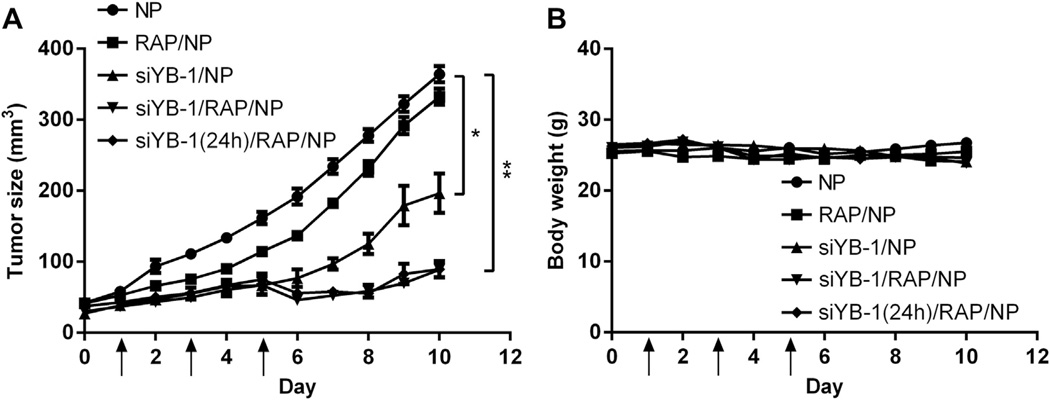
Since not all drug combinations demand simultaneous delivery (timing may play a critical role) [55,56], we decided to investigate the antitumor activity of siYB-1/RAP NP administered both simultaneously and sequentially (1 day apart) against a nude mouse xenograft tumor model of PC3 prostate cancer. All NP were formulated at 50% (w/w) RAP and N/P 50 to maximize RAP and siYB-1 loading. When the tumor size reached ca. 50 mm3, 200 µl of either empty NP, siYB-1/NP, RAP/NP, or dual-loaded siYB-1/RAP/NP were administered via retro-orbital injections into mice on days 1, 3 and 5 (RAP at 30 mg/kg, siYB-1 at 2 mg/kg). For the sequential administration strategy, mice were injected with siYB-1/NP on day 0 and then treated with RAP/NP on days 1, 3 and 5 as before. The tumor size and bw of animals were monitored daily for 10 days to assess the therapeutic efficacy and acute toxicity of the treatments in animals. The empty NP treatments did not inhibit tumor growth, whereas RAP/NP was only slightly effective against this aggressive xenograft tumor model. As previously noted in a breast cancer tumor model [30], we saw significant improved prostate cancer tumor regression with siYB-1/NP. More significantly, the results for combination siYB-1 and RAP NP administered either simultaneously or sequentially revealed dramatic tumor regression, with up to 75% tumor volume reduction by day 10 compared to the NP vehicle (Fig. 10A). In addition, the formulations did not generate acute toxicity in animals based on consistent animal weights throughout the treatment period (Fig. 10B); a bw loss of ±20% would have indicated acute toxicities to animals. It is also important to keep in mind that the bw did not change drastically because the tumors were not allowed to grow to extreme volumes (e.g. 1000–2000 mm3 [57,58],) to avoid tumor necrosis, often characteristic of large tumors. None of the animals died during the course of the experiments and they were all promptly euthanized by day 10.
Fig. 10.
When the PC3 tumor reached 50 mm3, nude mice (5 per group) received three retro-orbital injections on days 1,3 and 5 of: NP vehicle, siYB-1/NP, RAP/NP, orsiYB-1/RAP/NP (RAP at 30 mg/kg, siYB-1 at 2 mg/kg). For the sequential administration strategy, mice were treated with siYB-1/NP on day 0 and then treated with RAP/NP on days 1, 3 and 5. The tumor volume (A) and bw of animals (B) were monitored daily for 10 days. The results shows that combination and simultaneous administration of siYB-1/RAP both resulted in significant tumor regression, with up to 75% tumor volume reduction compared to NP vehicle by day 10, and none of the animals exhibited signs of acute toxicity (based on ±20% bw loss) during the course of the experiment *p < 0.01, **p < 0.001.
4. Conclusion
A three layer polymeric micelle system based on the self-association of the cationic triblock polymer PEG-b-PAsp(DET)-b-PCL has been developed and characterized for combination delivery of siYB-1 and RAP. RAP could be loaded up to 63% (at 25% w/w RAP to polymer) with 80% siRNA (from an initial 3 µm concentration). siYB-1/RAP micelles averaged ca. 100 nm in diameter and were stable up to 7 days in PBS. RAP release from tRNA/RAP micelles at 37 ° C was characterized by a t1/2 of 77 h. The incorporation of PCL into the polymer stabilized resulting NP and generated significant kd of luciferase in LNCap-Luc cells (<59% kd) after 24 h, compared to the diblock polymer PEG-b-PAsp(DET) (<14% kd). Combination siYB-1/RAP micelles exerted enhanced cytotoxicity on PC3 prostate cancer cells cultured in vitro, both at the 24 and 48 h time points for all concentrations of RAP tested (0.01, 0.1, and 1 nm). Simultaneous (siYB-1/RAP/NP on days 1, 3, 5) and sequential retro-orbital injections (siYB-1/NP on day 0, followed by RAP/NP on days 1, 3, 5) generated significant tumor regression in a PC3 xenograft nude mouse model of human prostate cancer.
Supplementary Material
Acknowledgments
This research was supported by NIH grant R00 CA136970 and startup funds from the University of Wisconsin-Madison, School of Pharmacy.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.biomaterials.2013.05.010.
References
- 1.Vézina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 2.Buck E, Eyzaguirre A, Brown E, Petti F, McCormack S, Haley JD, et al. Rapamycin synergizes with the epidermal growth factor receptor inhibitor erlotinib in non-small-cell lung, pancreatic, colon, and breast tumors. Mol Cancer Ther. 2006;5:2676–2684. doi: 10.1158/1535-7163.MCT-06-0166. [DOI] [PubMed] [Google Scholar]
- 3.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 4.Ji J, Zheng PS. Activation of mTOR signaling pathway contributes to survival of cervical cancer cells. Gynecol Oncol. 2010;117:103–108. doi: 10.1016/j.ygyno.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 5.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 6.Gulhati P, Cai Q, Li J, Liu J, Rychahou PG, Qiu S, et al. Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin Cancer Res. 2009;15:7207–7216. doi: 10.1158/1078-0432.CCR-09-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Poel HG, Hanrahan C, Zhong H, Simons JW. Rapamycin induces smad activity in prostate cancer cell lines. Urol Res. 2003;30:380–386. doi: 10.1007/s00240-002-0282-1. [DOI] [PubMed] [Google Scholar]
- 8.Noh WC, Mondesire WH, Peng JY, Jian WG, Zhang HX, Dong JJ, et al. Determinants of rapamycin sensitivity in breast cancer cells. Clin Cancer Res. 2004;10:1013–1023. doi: 10.1158/1078-0432.ccr-03-0043. [DOI] [PubMed] [Google Scholar]
- 9.Guba M, Koehl GE, Neppl E, Doenecke A, Steinbauer M, Schlitt HJ, et al. Dosing of rapamycin is critical to achieve an optimal antiangiogenic effect against cancer. Transpl Int. 2005;18:89–94. doi: 10.1111/j.1432-2277.2004.00026.x. [DOI] [PubMed] [Google Scholar]
- 10.Seeliger H, Guba M, Kleespies A, Jauch K-W, Bruns CJ. Role of mTOR in solid tumor systems: a therapeutical target against primary tumor growth, metastases, and angiogenesis. Cancer Metastasis Rev. 2007;26:611–621. doi: 10.1007/s10555-007-9077-8. [DOI] [PubMed] [Google Scholar]
- 11.Raught B, Gingras AC, Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci U S A. 2001;98:7037–7044. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wanner K, Hipp S, Oelsner M, Ringshausen I, Bogner C, Peschel C, et al. Mammalian target of rapamycin inhibition induces cell cycle arrest in diffuse large B cell lymphoma (DLBCL) cells and sensitises DLBCL cells to rituximab. Br J Haematol. 2006;134:475–484. doi: 10.1111/j.1365-2141.2006.06210.x. [DOI] [PubMed] [Google Scholar]
- 13.Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–897. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simamora P, Alvarez JM, Yalkowsky SH. Solubilization of rapamycin. Int J Pharm. 2001;213:25–29. doi: 10.1016/s0378-5173(00)00617-7. [DOI] [PubMed] [Google Scholar]
- 16.Napoli KL, Wang ME, Stepkowski SM, Kahan BD. Distribution of sirolimus in rat tissue. Clin Biochem. 1997;30:135–142. doi: 10.1016/s0009-9120(96)00157-9. [DOI] [PubMed] [Google Scholar]
- 17.Forrest ML, Won CY, Malick AW, Kwon GS. In vitro release of the mTOR inhibitor rapamycin from poly(ethylene glycol)-b-poly(epsilon-caprolactone) micelles. J Control Release. 2006;110:370–377. doi: 10.1016/j.jconrel.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 18.Law JH, Li Y, To K, Wang M, Astanehe A, Lambie K, et al. Molecular decoy to the Y-box binding protein-1 suppresses the growth of breast and prostate cancer cells whilst sparing normal cell viability. PLoS One. 2010;5:e12661. doi: 10.1371/journal.pone.0012661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Moscow) 2011;76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 20.Skabkin MA, Evdokimova V, Thomas AAM, Ovchinnikov LP. The major messenger ribonucleoprotein particle protein p50 (YB-1) promotes nucleic acid strand annealing. J Biol Chem. 2001;276:44841–44847. doi: 10.1074/jbc.M107581200. [DOI] [PubMed] [Google Scholar]
- 21.Lyabin DN, Eliseeva IA, Ovchinnikov LP. YB-1 synthesis is regulated by mTOR signaling pathway. PLoS One. 7:e52527. doi: 10.1371/journal.pone.0052527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, et al. Trans-lational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Lovett DH, Cheng S, Cape L, Pollock AS, Mertens PR. YB-1 alters MT1-MMP trafficking and stimulates MCF-7 breast tumor invasion and metastasis. Bio-chem Biophys Res Commun. 2010;398:482–488. doi: 10.1016/j.bbrc.2010.06.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Yamada S, Izumi H, Li Z, Shimajiri S, Wang KY, et al. Strong YB-1 expression is associated with liver metastasis progression and predicts shorter disease-free survival in advanced gastric cancer. J Surg Oncol. 2012;105:724–730. doi: 10.1002/jso.23030. [DOI] [PubMed] [Google Scholar]
- 25.Knosel T, Emde A, Schluns K, Chen Y, Jurchott K, Krause M, et al. Immuno-profiles of 11 biomarkers using tissue microarrays identify prognostic subgroups in colorectal cancer. Neoplasia. 2005;7:741–747. doi: 10.1593/neo.05178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bargou RC, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3:447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 27.Ohga T, Koike K, Ono M, Makino Y, Itagaki Y, Tanimoto M, et al. Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 1996;56:4224–4228. [PubMed] [Google Scholar]
- 28.Kuwano M, Oda Y, Izumi H, Yang SJ, Uchiumi T, Iwamoto Y, et al. The role of nuclear Y-box binding protein 1 as a global marker in drug resistance. Mol Cancer Ther. 2004;3:1485–1492. [PubMed] [Google Scholar]
- 29.Yang JY, Ha S-A, Yang Y-S, Kim JW. p-Glycoprotein ABCB5 and YB-1 expression plays a role in increased heterogeneity of breast cancer cells: correlations with cell fusion and doxorubicin resistance. BMC Cancer. 2010;10:388. doi: 10.1186/1471-2407-10-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee C, Dhillon J, Wang MY, Gao Y, Hu K, Park E, et al. Targeting YB-1 in HER-2 overexpressing breast cancer cells induces apoptosis via the mTOR/STAT3 pathway and suppresses tumor growth in mice. Cancer Res. 2008;68:8661–8666. doi: 10.1158/0008-5472.CAN-08-1082. [DOI] [PubMed] [Google Scholar]
- 31.Sengupta S, Eavarone D, Capila I, Zhao G, Watson N, Kiziltepe T, et al. Temporal targeting of tumour cells and neovasculature with a nanoscale delivery system. Nature. 2005;436:568–572. doi: 10.1038/nature03794. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Radovic-Moreno AF, Alexis F, Gu FX, Basto PA, Bagalkot V, et al. Co-delivery of hydrophobic and hydrophilic drugs from nanoparticle-aptamer bioconjugates. ChemMedChem. 2007;2:1268–1271. doi: 10.1002/cmdc.200700121. [DOI] [PubMed] [Google Scholar]
- 33.Bae Y, Fukushima S, Harada A, Kataoka K. Design of environment-sensitive supramolecular assemblies for intracellular drug delivery: polymeric micelles that are responsive to intracellular pH change. Angew Chem Int Ed Engl. 2003;42:4640–4643. doi: 10.1002/anie.200250653. [DOI] [PubMed] [Google Scholar]
- 34.Kakizawa Y, Furukawa S, Kataoka K. Block copolymer-coated calcium phosphate nanoparticles sensing intracellular environment for oligodeox-ynucleotide and siRNA delivery. J Control Release. 2004;97:345–356. doi: 10.1016/j.jconrel.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 35.Xiong MP, Forrest ML, Ton G, Zhao A, Davies NM, Kwon GS. Poly(aspartate-g-PEI800), a polyethylenimine analogue of low toxicity and high transfection efficiency for gene delivery. Biomaterials. 2007;28:4889–4900. doi: 10.1016/j.biomaterials.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y, Chau M, Boyle AJ, Liu P, Niehoff A, Weinrich D, et al. Effect of pendant group structure on the hydrolytic stability of polyaspartamide polymers under physiological conditions. Biomacromolecules. 2012;13:1296–1306. doi: 10.1021/bm2018239. [DOI] [PubMed] [Google Scholar]
- 37.Gary DJ, Puri N, Won Y-Y. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Woodward SC, Brewer PS, Moatamed F, Schindler A, Pitt CG. The intracellular degradation of poly(epsilon-caprolactone) J Biomed Mater Res. 1985;19:437–444. doi: 10.1002/jbm.820190408. [DOI] [PubMed] [Google Scholar]
- 39.Raffetseder U, Frye B, Rauen T, Jurchott K, Royer HD, Jansen PL, et al. Splicing factor SRp30c interaction with Y-box protein-1 confers nuclear YB-1 shuttling and alternative splice site selection. J Biol Chem. 2003;278:18241–18248. doi: 10.1074/jbc.M212518200. [DOI] [PubMed] [Google Scholar]
- 40.Miyata K, Oba M, Nakanishi M, Fukushima S, Yamasaki Y, Koyama H, et al. Polyplexes from poly(aspartamide) bearing 1,2-diaminoethane side chains induce pH-selective, endosomal membrane destabilization with amplified transfection and negligible cytotoxicity. J Am Chem Soc. 2008;130:16287–16294. doi: 10.1021/ja804561g. [DOI] [PubMed] [Google Scholar]
- 41.Kanayama N, Fukushima S, Nishiyama N, Itaka K, Jang W-D, Miyata K, et al. A PEG-based biocompatible block catiomer with high buffering capacity for the construction of polyplex micelles showing efficient gene transfer toward primary cells. ChemMedChem. 2006;1:439–444. doi: 10.1002/cmdc.200600008. [DOI] [PubMed] [Google Scholar]
- 42.Masago K, Itaka K, Nishiyama N, Chung U-i, Kataoka K. Gene delivery with biocompatible cationic polymer: pharmacogenomic analysis on cell bioactivity. Biomaterials. 2007;28:5169–5175. doi: 10.1016/j.biomaterials.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 43.De Smedt SC, Demeester J, Hennink WE. Cationic polymer based gene delivery systems. Pharm Res. 2000;17:113–126. doi: 10.1023/a:1007548826495. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Gao S, Ye W-H, Yoon HS, Yang Y-Y. Co-delivery of drugs and DNA from cationic core-shell nanoparticles self-assembled from a biodegradable copolymer. Nat Mater. 2006;5:791–796. doi: 10.1038/nmat1737. [DOI] [PubMed] [Google Scholar]
- 45.Alshamsan A, Haddadi A, Incani V, Samuel J, Lavasanifar A, Uludag H. Formulation and delivery of siRNA by oleic acid and stearic acid modified polyethylenimine. Mol Pharm. 2009;6:121–133. doi: 10.1021/mp8000815. [DOI] [PubMed] [Google Scholar]
- 46.Kim WJ, Christensen LV, Jo S, Yockman JW, Jeong JH, Kim Y-H, et al. Cholesteryl oligoarginine delivering vascular endothelial growth factor siRNA effectively inhibits tumor growth in colon adenocarcinoma. Mol Ther. 2006;14:343–350. doi: 10.1016/j.ymthe.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 47.Kim HJ, Ishii A, Miyata K, Lee Y, Wu S, Oba M, et al. Introduction of stearoyl moieties into a biocompatible cationic polyaspartamide derivative, PAsp(-DET), with endosomal escaping function for enhanced siRNA-mediated gene knockdown. J Control Release. 2010;145:141–148. doi: 10.1016/j.jconrel.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 48.Klampfer L. Signal transducers and activators of transcription (STATs): novel targets of chemopreventive and chemotherapeutic drugs. Curr Cancer Drug Targets. 2006;6:1071–21. doi: 10.2174/156800906776056491. [DOI] [PubMed] [Google Scholar]
- 49.Alvarez JV, Greulich H, Sellers WR, Meyerson M, Frank DA. Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor. Cancer Res. 2006;66:3162–3168. doi: 10.1158/0008-5472.CAN-05-3757. [DOI] [PubMed] [Google Scholar]
- 50.Morgensztern D, McLeod HL. PI3K/Akt/mTOR pathway as a target for cancer therapy. Anticancer Drugs. 2005;16:797–803. doi: 10.1097/01.cad.0000173476.67239.3b. [DOI] [PubMed] [Google Scholar]
- 51.LoPiccolo J, Blumenthal GM, Bernstein WB, Dennis PA. Targeting the PI3K/Akt/ mTOR pathway: effective combinations and clinical considerations. Drug Resist Updat. 2008;11:32–50. doi: 10.1016/j.drup.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The trans-lational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Zhang JW, Lv GY, Xie SL, Wang GY. Effects of STAT3 gene silencing and rapamycin on apoptosis in hepatocarcinoma cells. Int J Med Sci. 2012;9:216–224. doi: 10.7150/ijms.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bevins RL, Zimmer SG. It’s about time: scheduling alters effect of histone deacetylase inhibitors on camptothecin-treated cells. Cancer Res. 2005;65:6957–6966. doi: 10.1158/0008-5472.CAN-05-0836. [DOI] [PubMed] [Google Scholar]
- 56.Sato T, Suzuki M, Sato Y, Echigo S, Rikiishi H. Sequence-dependent interaction between cisplatin and histone deacetylase inhibitors in human oral squamous cell carcinoma cells. Int J Oncol. 2006;28:1233–1241. [PubMed] [Google Scholar]
- 57.Dalla Pozza E, Lerda C, Costanzo C, Donadelli M, Dando I, Zoratti E, et al. Targeting gemcitabine containing liposomes to CD44 expressing pancreatic adenocarcinoma cells causes an increase in the antitumoral activity. Biochim Biophys Acta. 2013;1828:1396–1404. doi: 10.1016/j.bbamem.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 58.Caso J, Masko EM, Ii JAT, Poulton SH, Dewhirst M, Pizzo SV, et al. The effect of carbohydrate restriction on prostate cancer tumor growth in a castrate mouse xenograft model. Prostate. 2013;73:449–454. doi: 10.1002/pros.22586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.