Abstract
We describe genome mapping on nanochannel arrays. In this approach, specific sequence motifs in single DNA molecules are fluorescently labeled, and the DNA molecules are uniformly stretched in thousands of silicon channels on a nanofluidic device. Fluorescence imaging allows the construction of maps of the physical distances between occurrences of the sequence motifs. We demonstrate the analysis, individually and as mixtures, of 95 bacterial artificial chromosome (BAC) clones that cover the 4.7-Mb human major histocompatibility complex region. We obtain accurate, haplotype-resolved, sequence motif maps hundreds of kilobases in length, resulting in a median coverage of 114× for the BACs. The final sequence motif map assembly contains three contigs. With an average distance of 9 kb between labels, we detect 22 haplotype differences. We also use the sequence motif maps to provide scaffolds for de novo assembly of sequencing data. Nanochannel genome mapping should facilitate de novo assembly of sequencing reads from complex regions in diploid organisms, haplotype and structural variation analysis and comparative genomics.
Despite recent advances in base-calling accuracy and read length, de novo genome assembly and structural variant analysis using ‘short read’ shotgun sequencing remain challenging. Most resequencing projects rely on mapping the sequencing data to the reference sequence to identify variants of interest1. When whole-genome assembly is attempted, paired-end sequencing of long DNA fragments provides scaffolds for assembly2. As cloning of large DNA fragments is difficult, small-insert libraries of varying sizes may be prepared for paired-end sequencing, thus limiting the resolution of haplotypes and increasing the complexity, time and cost of the sequencing project. In addition, complex genomic loci, such as the major histocompatibility (MHC) region, which is important for studying infectious and autoimmune diseases3, contain highly repetitive sequences and are particularly difficult to assemble. Robust technologies that can aid de novo sequence assembly are therefore sorely needed as whole-genome sequencing becomes more widely adopted.
Whole-genome scanning techniques have revealed the prevalence and importance of structural variation. Detecting copy number variation often relies on detection of relative signal intensities by array-based or quantitative PCR-based technologies. Array-based methods, such as array-based comparative genomic hybridization, have been used extensively to interrogate copy number variation in the human genome4,5. However, except for deletions, these methods do not provide positional information regarding the locations of copy number variants, and they cannot detect balanced structural variation, such as inversions or translocations6. Paired-end mapping techniques, traditionally using Sanger sequencing and now using next-generation sequencing7, generally have low sensitivity in repetitive regions, where much structural variation lies8. Recent efforts to characterize copy number variants in human genomes at high resolution involved paired-end mapping of clones, but this approach, although useful for exploratory studies on a small sample set, is too labor intensive and time consuming to be applicable to analysis of large numbers of individuals, and the resolution was no better than 8 kb9.
Restriction mapping was instrumental in the Human Genome Project. One approach to address drawbacks of traditional restriction mapping is ‘combing’-based optical mapping10. In this approach, large DNA fragments are stretched and immobilized on glass slides and cut in situ with restriction enzymes. Optical mapping was used to construct ordered restriction maps for whole genomes11–14, and it provided scaffolds for shotgun sequence assembly and validation15,16. However, this method is limited by its low throughput, nonuniform DNA stretching, imprecise DNA length measurement and high error rates.
Another approach developed for mapping of large DNA fragments is based on a hydrodynamic focus technique in microscale fluidics. In this method, DNA molecules labeled with fluorescent peptide nucleic acid are transiently stretched in a microchannel and detected as the fluorophores pass by a sensor17. Because the linearity or quasi-linearity of polymers are maintained through a shear force driven by a pump or motion of the laminar fluid flow, the DNA molecules cannot be held constant for imaging, reducing the resolution of the map and limiting the throughput of the single-channel microfluidic device. Although this method is suitable for generating maps of small genomes for matching against maps of known genomes, its throughput and resolution impose severe limits for large-scale genome mapping.
Here we report an accurate, high-throughput genome mapping technique that has been optimized for general use. The core technology is a nanofluidic chip that contains nanochannels that keep long DNA molecules in a consistent, uniformly elongated state. Fluorescently labeled DNA molecules are drawn into the nanochannels, held still and imaged automatically on the multicolor Irys instrument. After imaging, additional sets of DNA molecules are streamed into the nanochannels for imaging. This process is repeated many times until the DNA is depleted or the nanochannels are rendered unusable as a result of clogging.
The nanofluidic chip contains three sets of nanochannels, each consisting of ~4,000 channels that are 0.4 mm in length and 45 nm in diameter. Using 193-nm lithography in a nanofabrication process on the surface of a silicon substrate, nanochannel array chips are produced with precise diameters. DNA molecules in the 45-mm nanochannels cannot fold back on themselves and are forced by physical confinement to be in an elongated, linearized state18,19.
As long DNA molecules in solution exist as coiled balls, a gradient region consisting of pillars and wider channels is placed in front of the nanochannels to allow the DNA molecules to uncoil as they flow toward the array (Fig. 1)20. In this region, the physical confinement is sufficiently dense that the molecules are forced to interact with the pillars, yet sufficiently sparse that the DNA is free to uncoil. Once uncoiled, the DNA can then be efficiently flowed into the array in a linear manner.
Figure 1.
Nanochannel arrays. (a) In a microfluidic environment, long (>100 kb) DNA fragments (in green in bottom panel) are in the coiled ball form and clog the entrance of the nanochannel array, as it is energetically unfavorable for the molecules to uncoil and enter the nanochannels. (b) A gradient region is placed in front of the nanochannels. Here, the physical confinement is sufficiently dense that the molecules are forced to flow by the pillars, where they uncoil and stream into the nanochannels without clogging. (c) Fabrication of the nanochannel array using interference lithography to produce 120-nm channels in silicon followed by tuning to a smaller diameter with material deposition and capping with a glass cover to allow for fluorescence imaging. (d) A profile scanning electron microscopy image of 45-nm channels. (e) An s.e.m. image of the 45-nm channels patterned on the silicon substrate before bonding to the glass.
Our genome mapping approach combines robust, sequence-specific labeling, consistent linearization of extremely long DNA molecules in nanochannel arrays, automated imaging, high-resolution imaging, size measurements of single molecules and map construction. It provides a simple technique for mapping complex regions or whole genomes, and facilitates sequence assembly with long-range scaffolding information and structural variation analysis.
RESULTS
To demonstrate the utility of our approach, we used genome mapping to construct sequence motif maps of 95 bacterial artificial chromosome (BAC) clones covering the 4.7-Mb MHC region from two individuals (PDF and COX libraries used by the MHC Haplotype Consortium21,22). Subsequently, we performed de novo sequence assembly using next-generation sequencing reads. The sequence-motif maps and sequencing contigs were then compared to the reference sequences reported by the MHC Haplotype Consortium as confirmation and to uncover potential differences.
Generation of sequence motif maps by genome mapping
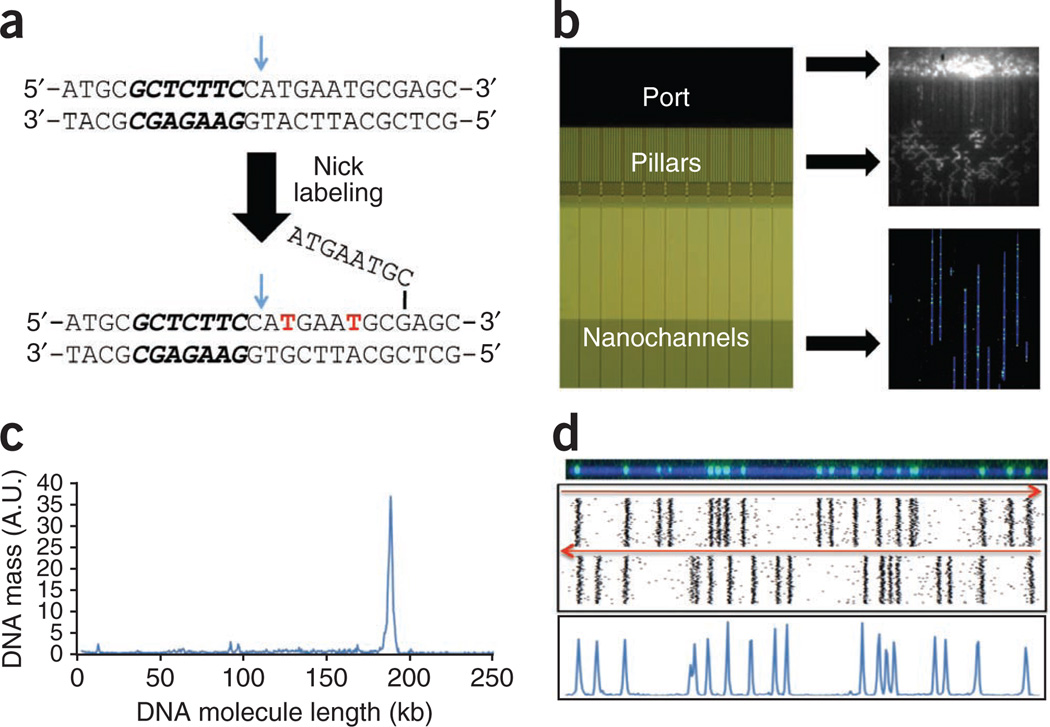
Genome mapping consists of four steps: sequence-specific labeling, linearization of the labeled long DNA molecules, imaging and map construction. The process is illustrated in Figure 2 for a 183-kb BAC clone. We used a nicking endonuclease to introduce single-strand nicks in the double-stranded DNA (dsDNA) at specific sequence motifs. Fluorescent dye conjugated nucleotides (Alexa 546 dUTP) were then incorporated at these sites by Vent (exo−) polymerase (Fig. 2a)23,24. Next, we stained the labeled DNA molecules with the DNA-intercalating dye, YOYO-1, which facilitates visualization of the DNA molecule and measurement of its size. Then, we loaded the DNA onto a nanochannel array chip and applied an electric field, which gradually drives the long, coiled DNA molecules in free suspension through a series of micro- and nanofluidic structures. The molecules stretched and linearized as they moved through entropic-confinement inside the nanochannels20. Once the nanochannels were populated by a set of linearized DNA molecules, we imaged them with automated high-resolution fluorescent microscopy (Fig. 2b).
Figure 2.
Genome mapping. (a) Nick-labeling by Nt.BspQI and DNA polymerase is accomplished by top-strand DNA cleavage (blue arrow), one nucleotide 3′ from the recognition sequence (in bold italics), followed by incorporation of fluorescent nucleotide analogs (in red) with concomitant DNA strand displacement. (b) The DNA molecule is stained with YOYO-1 and loaded into the port of a nanoarray flowcell (left panel). The DNA molecules are introduced into the region with pillars and micrometer-scale relaxation channels by an electric field where they unwind and linearize (top right panel). Finally, the DNA molecules are pushed by a low-voltage electrical pulse, and they enter the 45-nm nanochannels, where they are stretched uniformly to 85% of the length of perfectly linear B-DNA (bottom right panel). The DNA is visualized as blue linear structures in the nanochannels, with green labels marking the Nt.BspQI nick sites. (c) The length of the DNA molecules and the positions of nick labels on each DNA molecule are determined after automated image capture. The fragment size profile of a 183-kb BAC is shown, with the narrow peak width indicating uniform DNA linearization. (d) The DNA molecules are clustered into groups (representing individual BACs) based on nick-labeling pattern similarity. As BAC molecules can enter the nanochannels in either orientation, each BAC is represented by two clusters with opposite orientations (top panel). After combining the two clusters, histogram plots of nick-labeled DNA (bottom panel) are used to define the locations of Nt.BspQI sites. n ≈ 100 molecules.
We determined the size of each DNA molecule by directly measuring its contour length18. The measured length of this clone was 50.5 µm, corresponding to 85% of the theoretical maximal stretching (complete elongation of a 183-kb DNA molecule is expected to be 59.4 µm, assuming 0.34 nm/bp). Based on measurements of 1,251 molecules, the DNA length measurement had a s.d. of 1.3 kb (or 0.36 µm, Fig. 2c).
We then marked the positions of the fluorescent labels along the DNA molecule, which yielded the distinct distribution of the sequence motifs recognized by the nicking endonuclease for each fragment. We constructed consensus sequence motif maps by comparing and clustering DNA molecules with the same sequence motif patterns. Molecules can enter into the nanochannels in either orientation; clusters that are mirror images of each other originating from the same BAC were recovered for each clone (Fig. 2d, top panel). The histogram peaks (Fig. 2d, bottom panel) represent the location of each sequence motif (GCTCTTC) along the molecules. A total of 18 nicking sites >1.5 kb apart were detected, and the pattern is in concordance with the reference in silico map.
Overall, individual molecules were labeled with 79% efficiency at nicking sites (true positives) and with a 4% false-positive rate. To determine the effect of missing labels, we constructed consensus maps using different numbers of single sequence motif maps (depth fold-coverage). We generated consensus maps from 100 data sets of 10, 20, 30, 40, 50, 60, 70, 80, 90 or 100× coverage and compared each one with the reference map. At 20× coverage, false negative nick-label sites occurred at a rate of 0.26% on the consensus map. At higher coverage, there were essentially no missed label sites in the consensus. The s.d. of consensus peak position measurements was 0.9 pixels (1 pixel corresponds to 492 bp).
In a typical experiment, images of 21,000 DNA fragments >50 kb in length and with three or more labels were acquired automatically (covering 2,060 Mb of sequence, or 433× of the 4.75 Mb region being mapped). After removing the lower quality maps of individual fragments in the clustering and mapping process, we found that the median coverage for the BACs was 114×, with minimal 54× and maximal 358× coverage.
We performed genome mapping of ten individual BACs, and the resulting sequence motif maps were highly consistent with the reference maps for the MHC region (data not shown).
Genome mapping of the MHC region with 95 BACs
We generated motif maps of the MHC region for two haploid clone libraries from the MHC Haplotype Consortium collection. We used 49 and 46 BAC clones from the PGF and COX libraries, respectively. We mixed samples of all the clones for each library, extracted DNA, nick-labeled each mixture with Nt.BspQI, and divided it into two aliquots. We linearized one aliquot from each mixture with the NotI restriction enzyme and the other with BsiWI. Thus, in total, we generated four nick-labeled, linearized mixtures. We loaded each mixture in the nanochannel array separately and imaged it, yielding 108 images that covered 27 horizontal field-of-view regions across the 2-mm width of the array with four contiguous fields of view vertically spanning 0.4 mm (Fig. 3a). The contiguous images were stitched together to produce a longer field of view. In total, we collected images of 23,000 molecules corresponding to 3 Gb of DNA sequence. The size of each molecule ranged from 20–220 kb, with a large fraction of the molecules >100 kb (Fig. 3b).
Figure 3.
Genome mapping of mixtures of 95 BACs from the PGF and COX libraries. (a) Image of a single field of view (FOV 73 × 73 µm) containing a mixture of nick-labeled DNA molecules in the nanoarray. This FOV is part of 108 FOVs shown in the bottom part of the panel (outlined in green). Each FOV can accommodate up to 250 kb of a DNA molecule from top to bottom. The images of four FOVs are stitched together so that longer molecules (up to 1 Mb) in a single channel can be analyzed whole. In all, there are 27 sets of four vertical FOVs per array scan. (b) The distribution of the DNA molecules imaged on the nanoarray by length. The majority of the molecules are 100–170 kb in length as expected from the BAC-clone sizes. (c) After clustering of DNA molecules based on nick-labeling patterns, consensus maps with overlapping patterns are assembled into contiguous-sequence motif maps. In this example, three overlapping consensus maps (each ~150 kb long) are assembled into a 300-kb map.
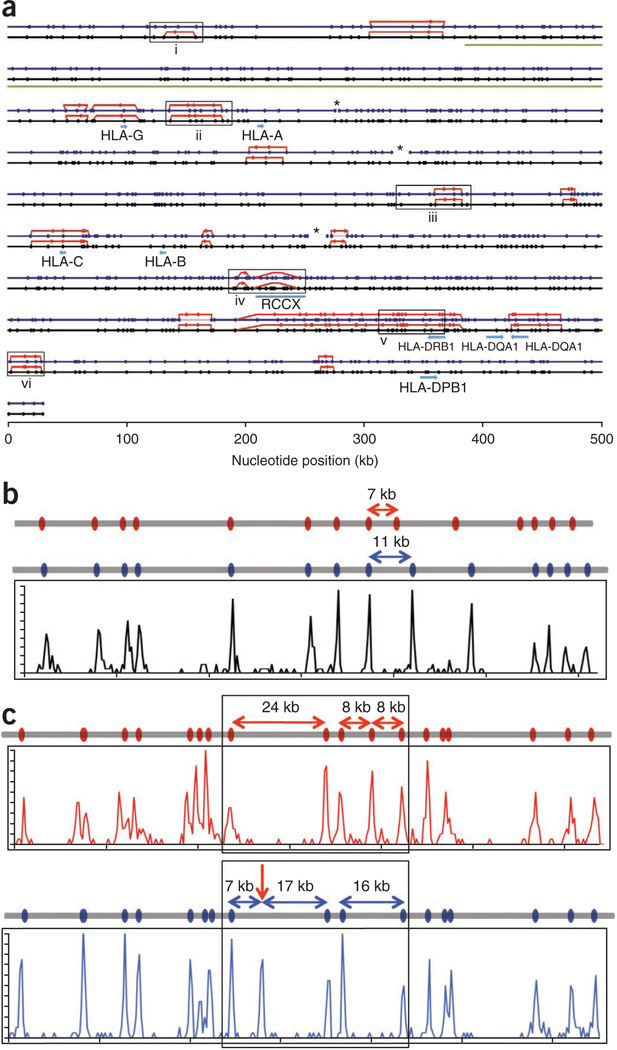
To simulate a data set obtained from a diploid DNA sample, we combined image data from all four mixtures before analysis. We calculated distances between each label for all the molecules and performed unsupervised clustering analysis to produce a total of 140 independent clusters, each with >100× coverage. We then used these clusters to construct consensus sequence motif maps for individual BAC clones, and joined maps of overlapping BACs to produce contig maps (Fig. 3c). In all, we obtained three contigs across the 4.7-Mb MHC region (Fig. 4a). We also identified regions harboring haplotype differences between the two haploid genomes. We confirmed all the differences identified by analyzing the haploid PGF and COX data sets independently. Consistent with previous reports22, differences between the haploid maps were concentrated around the HLA genes.
Figure 4.
Sequence motif map of the MHC region. (a) Alignment of the in silico reference sequence motif map for the PGF library (black line with the Nt.BspQI sites marked with black dots) and the map of the same region produced by genome mapping (blue line with blue dots). Where there are motif variations between COX and PGF, the COX motif is represented with red lines and red dots. Asterisks mark the gaps in the Nt.BspQI map produced by genome mapping. Gene locations and the location of the variable RCCX module are noted. Additional loci of special interest are marked with boxes and are discussed in detail in the text. The green bar from ~400 kb to 1,000 kb represents the region assembled from sequence data displayed in Figure 5. (b,c) Discrepancies between the reference Nt.BspQI map and that produced by genome mapping. (b) The reference Nt.BspQI maps of the region (a,i) indicate that the COX genome (gray line with red dots) has a 4-kb deletion as compared with the PGF genome (gray line with blue dots), with a 7-kb and an 11-kb fragment between two neighboring sites in the COX and PGF genomes, respectively. The map of the same region produced by genome mapping from both libraries (histogram plot in black) shows the same haplotype for both COX and PGF genomes, with an 11-kb fragment between the corresponding two sites. (c) An Nt.BspQI site identified in the region (a,iii) (arrow) is found in the PGF genome (blue histogram plot) by genome mapping, splitting the 24-kb fragment in the reference map (black line) into 7-kb and 17-kb fragments. The COX reference map (red line) and the COX map produced by genome mapping (red histogram plot) are also displayed to show that the COX genome has the 24-kb fragment and a haplotype variation in the adjacent region.
The maps produced by genome mapping generally matched well with the in silico reference maps, although we detected discrepancies at Chr 6: 28.78–28.88 Mb and Chr 6: 30.98–31.11 Mb (Fig. 4a). At Chr 6: 28.78–28.88 Mb, there is a 4-kb insertion in the PGF reference map relative to the COX reference map (Fig. 4b). However, genome mapping produced a single map that is identical to the PGF reference map, suggesting that the COX reference was erroneous. This was confirmed by analyzing the PGF and COX haploid data set separately; they had identical maps for the region. Subsequent sequencing of the clones confirmed the error in the COX reference detected by genome mapping.
At Chr 6: 30.98–31.11 Mb, genome mapping produced two haplotypes from the diploid data set. One haplotype matched perfectly with the COX reference (Fig. 4c, top panel), but the second did not completely match either the COX or PGF reference sequences (Fig. 4c, bottom panel). Further analysis of the haploid data set confirmed that the top map was derived from the COX library and that the bottom map did come from the PGF library. However, the bottom map contained an extra nicking site within the 24-kb fragment found in the reference sequence, splitting it into a 17-kb and a 7-kb fragment. In this case, genome mapping not only uncovered the two different haplotypes, it also identified a nicking site left out in the original reference sequence. Sequencing of the clones revealed that an additional nick site is created by a single base difference (AAAGAGC in the reference and GAAGAGC from our sequencing data).
Genome mapping for de novo sequencing assembly
To demonstrate that genome mapping can provide useful scaffolds for de novo sequence assembly of short reads, we pooled the BAC clone DNA from each library and prepared two sequencing libraries for next-generation sequencing. We performed paired-end, 100-bp sequencing of the BACs, and obtained 131 million and 142 million reads for the PGF and COX clones, respectively. For quality control, we first aligned the reads to the reference genome, hg19. Of the uniquely aligned reads, 93% of the PGF reads and 95% of the COX reads aligned to the MHC region, confirming that most BACs originated from the MHC region. The sequence motif maps suggested that a small subset of the BACs might be mislabeled by our supplier and did not map to the MHC region. Indeed, there was an excess of sequencing reads mapping to chromosomes 3, 7, 9 and 18, corresponding to the five BAC clones that did not belong to the MHC region.
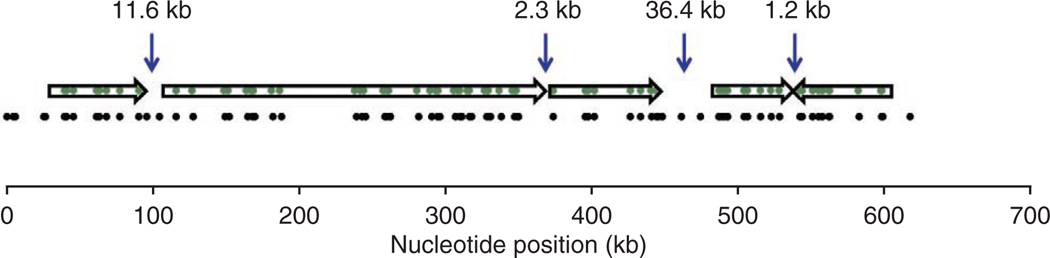
We used SOAPdenovo2 to separately assemble the reads for each of the two libraries (Supplementary Table 1). Figure 5 shows the results of sequence assembly of a 575-kb region with the use of long-range sequence motif maps. Four sequencing contigs were oriented and placed on the scaffold generated by genome mapping, with a good estimate of the gap sizes between contigs (from left: 11.6 kb, 2.3 kb, 36.4 kb and 1.2 kb, respectively). Three of the four gaps may be closed by designing PCR assays that bridge them. A number of sequencing contigs could not be mapped on the sequence motif map, indicating that they were assembly errors. BLAST analysis confirmed that these erroneous contigs do not properly align to the human reference sequence.
Figure 5.
De novo sequence assembly of the MHC region. DNA of 95 BACs from the PGF and COX libraries was sequenced and the sequence reads were assembled into contigs (arrows). The contigs were aligned to the Nt.BspQI map produced by genome mapping, providing information on the relationship and orientation of contigs together with the location and size of each gap between contigs. Shown are in silico sequence motif maps of the contigs (green dots in arrows) and of the reference sequence (black dots) of a 575-kb region marked in green in Figure 4a.
Genome mapping for haplotype and structural variation analysis
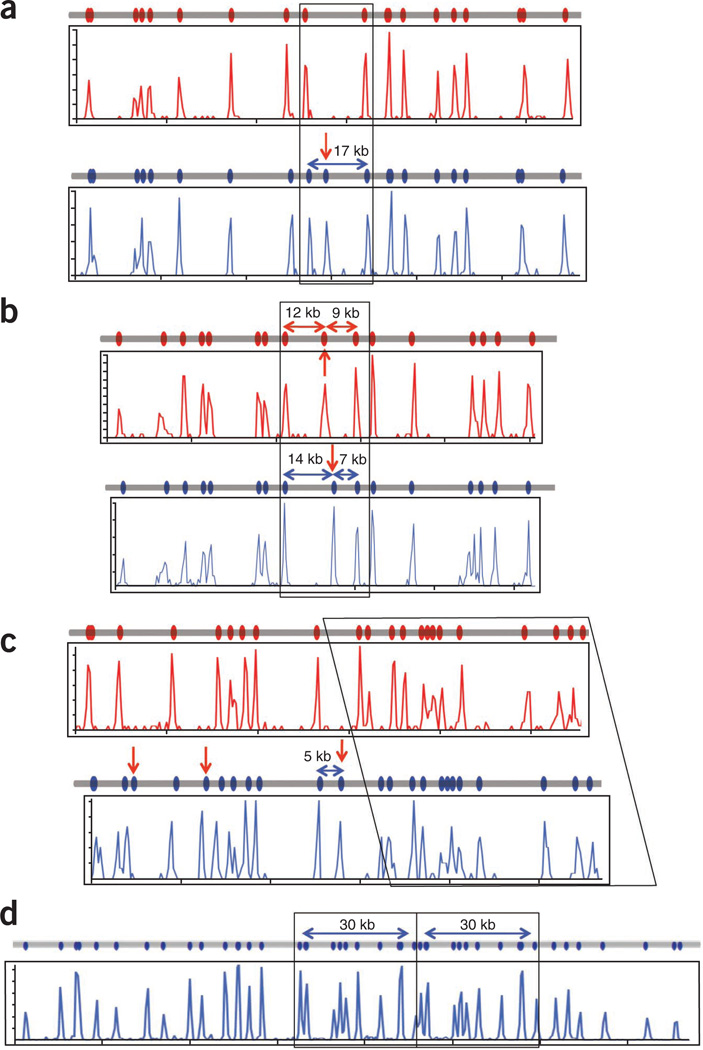
As genome mapping produced data on molecules hundreds of thousands of base pairs in length, it is useful for long-range haplotype and structural variation analysis. Analyzing the mixed PGF and COX data, we resolved the two haplotypes across the MHC region. The most common haplotype difference detected was the presence or absence of nicking sites. An example of the presence of a nicking site in one but not the other haplotype was found at Chr 6: 29.77–29.92 Mb, where the PGF sequence had an Nt.BspQI site that was not found in the COX sequence (Fig. 6a).
Figure 6.
Haplotype resolution and structural variation detected by genome mapping. (a) Single-site variation resulting from the creation or destruction of an Nt.BspQI site can be identified by genome mapping. The region in Figure 4a, ii shows that the PGF genome (blue line) contains an extra Nt.BspQI site not found in the COX genome (red line) with the maps generated by genome mapping (blue and red histogram plots) showing the expected pattern. (b) Shifting of a site relative to others in two haplotypes may be due to a double mutation or an inversion event. In Figure 4a, vi, the 21-kb region is split into 12- and 9-kb fragments in the COX genome (red line and red histogram plots) but 14- and 7-kb fragments in the PGF genome (blue line and blue histogram plot). (c) Insertions can be identified and localized by genome mapping for haplotyping resolution. In Figure 4a, v, the PGF genome has a 5-kb insertion that also includes an Nt.BspQI site (blue line, blue histogram plot) when compared to the COX genome (red line, red histogram plot). (d) A 30-kb duplication at the RCCX locus (Fig. 4a, iv) is identified and localized in both the reference map (gray line) and that produced by genome mapping (blue histogram plot).
Another form of variation found on these maps is that of a ‘shifted’ nicking site—that is, a nicking site is found at one position in one haplotype but at another position in the other haplotype, whereas the neighboring nicking sites match up perfectly (one such example is at Chr 6: 32.65–32.77 Mb (Fig. 6b)). Shifting of nicking sites could be due to nearby single-nucleotide variants that destroy or create nicking sites, or to single or multiple insertion or deletion events, depending on which allele is defined as the reference allele.
A third type of haplotype difference identified by genome mapping is due to insertions or deletions. A 5-kb insertion at Chr 6: 32.41–32.53 Mb in the PGF clone was not found in the COX clone (Fig. 6c). In this example, there were also extra nicking sites found in the PGF clone.
Structural variants can also be identified by genome mapping, regardless of whether there is a haplotypic difference. We observed a 30-kb tandem duplication at Chr 6: 31.92–31.95 Mb where the two haplotypes were identical (Fig. 6d).
DISCUSSION
We describe genome mapping on nanochannel arrays, which combines the specific labeling of sequence motifs (nicking-endonuclease recognition sites) and automated imaging of long, uniformly stretched DNA molecules. The image data were analyzed to produce sequence motif maps. These maps were used to resolve haplotypes and to identify the presence and location of structural variation. The maps also provided scaffolds for de novo assembly of short reads, to improve assembly contiguity while retaining haplotype and structural information. The importance of phasing haplotypes is well-recognized25. Read-backed phasing is available in the Genome Analysis Tool Kit26; however, the resulting phased segments are short, owing to the limited information provided by the short reads27. Misassembling contigs, misplacing and misorienting contigs, failure to join contigs, inability to accurately measure gaps in the assembled sequence and difficulty in closing the gaps are some of the challenges in de novo sequence assembly of large genomes28. The use of sequence motif maps, especially those spanning hundreds of thousands of bases, can reduce these difficulties. In addition to informing contig order and orientation, sequence motif maps can be used to identify incorrect contigs, which may be disassembled and reassembled in an iterative manner to improve contig fidelity.
The fundamental advance that enables genome mapping is the high-throughput, uniform linearization of long DNA molecules compared with traditional DNA combing methods29,30. The uniformity directly contributes to accurate measurement of the length of a DNA molecule and precise measurements of the distances between labels. Consequently, the accuracy of the sequence motif map of each individual molecule greatly facilitates the deconvolution of mixed clones through unsupervised clustering and formation of unique and accurate consensus maps of each individual clone. Furthermore, this accuracy allows us to detect single-nucleotide variants within nicking sites, duplications and relatively small insertions and deletions. For example, the MHC haplotype maps produced here differentiated the two HLA-DRB1 variants (DRB*150101 and 030101) within the coding region. Differentiation of HLA-DRB1 is difficult for next-generation sequencing because it is a relatively long gene with large introns in a highly repetitive region. Current approaches rely on specially designed PCR reactions31 or target capture followed by long-read sequencing32. However, with genome mapping, the multiple nick sites in this gene, in conjunction with adjacent sequence, were sufficient to differentiate many of the HLA-DRB1 variants.
In our nick-labeling scheme, the specificity was determined by both the enzymatic nicking reaction and the fluorescent nucleotide incorporation reaction. Nonenzymatic nicking was not extended by DNA polymerase owing to the lack of a functional 3′-hydroxyl group. Furthermore, the fluorescent nucleotides are covalently bound to the dsDNA, and thus, the binding is not subjected to the variation in binding constants for some noncovalent dsDNA labeling33–35. The Nt.BspQI nicking endonuclease has 537 resolvable sites (having at least 1.5 kb between neighboring sites) across the 4.7-Mb MHC region in the PGF genome, yielding roughly 9-kb average spacing. At this resolution, we detected 22 haploid differences between PGF and COX in this highly variable region. However, owing to the size limitation of the clones, the full phase information between the haploid map differences cannot be derived from the ‘diploid’ data set, even though the long-range phasing information represents a substantial advance over what one can derive from next-generation sequencing alone. With additional overlap, longer genomic DNA molecules and finer resolution, a single haplotype map would be achievable across the MHC region. To construct whole-genome haplotype maps, denser nick labeling (e.g., by combining two nicking enzymes with two different-color labels) can be employed. Alternatively, a labeling scheme such as nick-flap labeling23 can provide additional targeted and high-resolution sequence information besides the nicking sequence motifs for constructing a true de novo haplotype map.
The studies reported here demonstrate that genome mapping can be used for a variety of genome analyses. Current throughput of >300 Mb per scan is sufficient for large-scale genome analysis. Hence, coverage of a human genome (3.2 Gb) at 20× depth could be generated in ~13 h at nominal cost. Genome mapping can be paired with next-generation sequencing to produce a fully assembled genome (after gap closing) with all classes of genetic variation characterized. It is also useful for analyses of genomes of other species. It can rapidly and accurately catalog large-insert clone libraries, useful for repositories or individual laboratories. Genome mapping will have special relevance in studies of new pathogens, complex metagenomics and cancer genomes, where copy number variation and structural variation are abundant.
ONLINE METHODS
Sample preparation and data collection
We obtained BAC clones in LB slabs from the BACPAC Resource Center at the Children’s Hospital Oakland Research Institute (http://bacpac.chori.org/) from the BAC libraries CHORI-501 and CHORI-502. All DNA samples used in the study were prepared using Qiagen’s Large-Construct Kit. To prepare BAC mixtures, we grew 8 mL cultures of each BAC in LB containing 20 µg/mL chloramphenicol overnight and combined the separate cultures before proceeding with DNA extraction of the BACs as a pool. The DNA samples were quantified using Nanodrop 1000 (Thermal Fisher Scientific) and their quality assessed using pulsed-field gel electrophoresis. One milligram BAC DNA was linearized with 2 U of NotI or BsiWIand nicked with 0.5 U nicking endonuclease Nt.BspQI (New England BioLabs, NEB) at 37 °C for 2 h in NEB Buffer 3. (Note: NotI and BsiWI both cut in the vector but also cut several times within the clone insert. Linearizing with the two enzymes in separate reactions produces overlapping DNA fragments and minimizes the number of gaps in the final map.) The resultant DNA fragments were labeled with 25 nM Alexa546-dUTP (Invitrogen) and Vent (exo−) (NEB) for 1 h at 72 °C. The backbone of above fluorescently tagged DNA (5 ng/µL) was stained with YOYO-1 (3 nM; Invitrogen).
DNA was loaded in BioNano Genomics nanochannel arrays by electrophoresis of DNA. First, 12 volts were applied to concentrate the DNA at the entrance of the channels. Thirty volts were applied to move DNA into the nanochannels, and 10 V was applied to distribute the DNA in the nanochannels. Linearized DNA molecules were imaged using blue and green lasers for YOYO-1 and Alexa546 on the BioNano Genomics Irys automated imaging system.
Sequence motif map generation
The DNA molecule (YOYO-1) and locations of fluorescent labels (Alexa546) along the length of each molecule were detected using the software package, NanoStudio. A set of label locations of each DNA molecule comprises the individual DNA molecular map.
To take into account sizing errors and differences in the length of molecules, we transformed the data using a sliding window calculation. Each sequence motif map was thus made to have the same dimensions. We performed complete pairwise comparison of all single-molecule sequence motif maps and built a Euclidean distance matrix of the sequence motif maps. We then used the matrix as input for unsupervised hierarchical clustering of individual sequence motif maps based on Ward’s method using the R package fastcluster. After unsupervised clustering, individual sequence motif maps were grouped and analyzed together. Peaks corresponding to signal coming from true nick sites were fit and called based on a Gaussian model. Clusters corresponding to different orientations of otherwise the same BAC are combined. False signal were filtered out based on a set threshold. The consensus clusters were then joined based on overlap to form the sequence motif map for the MHC region36.
While we did not assign quality scores for the clusters, we consolidated the clusters for individual BACs as follows: for each cluster, a density plot was generated and a first-pass preliminary consensus map was constructed. Then, the map of each DNA fragment in the cluster was compared to this first-pass consensus. DNA fragments with maps that differed by more than 3 pixels on average from the consensus were removed from the cluster; clusters with more than 50% of such fragments were discarded.
Next-generation sequencing of BAC clones
We pooled and performed paired-end 100-bp sequencing of the two sets of BACs in one HiSeq 2000 lane each. We obtained 131 million and 142 million reads for PGF and COX, respectively. We first aligned the reads to the current reference genome build hg19 using Bowtie37 allowing only unique, properly paired alignments and for up to 2 mismatches in the alignment. Duplicate reads with identical end-coordinates are removed using Picard (http://picard.sourceforge.net/).
We then performed de novo assembly of the reads using SOAPdenovo2. Multiple k-mer lengths were tested to maximize assembly contiguity, and we chose ones that maximized the assembly N50. The assembly was refined by using GapCloser to further bridge gaps in the contigs. We also ran Velvet with different k-mer lengths for de novo assembly for the same data set. “LONGSEQUENCES” was enabled to optimize assembly of long contigs38. Discussion of assembly throughout the manuscript is based on results from SOAPdenovo since the contigs from Velvet were much shorter.
ACKNOWLEDGMENTS
The authors thank A. Pool and P. Ling-Fung Tang for their assistance in sequencing. O. Hampton and H. VanSteenhouse provided critical comments. This research is supported in part by US National Institutes of Health (NIH) award to P.-Y.K. and M.X. (R01 HG005946). E.T.L. was supported in part by NIH Training Grant T32 GM007175.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
E.T.L., M.X. and P.-Y.K. conceived the study. M.X. and P.-Y.K. supervised the project. E.T.L., C.L. and D.E. maintained BAC cultures and extracted DNA. A.H., S.K.D., M.D.A., P.D., D.E., N.N., E.L.T., H.C., M.X. and P.-Y.K. participated in data generation and analysis. E.T.L., A.H., M.X. and P.-Y.K. wrote the manuscript. All authors were involved in discussion of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernando MMA, et al. Defining the role of the MHC in autoimmunity: a review and pooled analysis. PLoS Genet. 2008;4:e1000024. doi: 10.1371/journal.pgen.1000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 5.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 6.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat. Genet. 2007;39(suppl):S16–S21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 8.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 9.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jing J, et al. Automated high resolution optical mapping using arrayed, fluid-fixed DNA molecules. Proc. Natl. Acad. Sci. USA. 1998;95:8046–8051. doi: 10.1073/pnas.95.14.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou S, et al. Validation of rice genome sequence by optical mapping. BMC Genomics. 2007;8:278. doi: 10.1186/1471-2164-8-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou S, et al. A single molecule scaffold for the maize genome. PLoS Genet. 2009;5:e1000711. doi: 10.1371/journal.pgen.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Church DM, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teague B, et al. High-resolution human genome structure by single-molecule analysis. Proc. Natl. Acad. Sci. USA. 2010;107:10848–10853. doi: 10.1073/pnas.0914638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C-w, Schramm T, Zhou S, Schwartz D, Talaat A. Optical mapping of the Mycobacterium avium subspecies paratuberculosis genome. BMC Genomics. 2009;10:25. doi: 10.1186/1471-2164-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latreille P, et al. Optical mapping as a routine tool for bacterial genome sequence finishing. BMC Genomics. 2007;8:321. doi: 10.1186/1471-2164-8-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan EY, et al. DNA mapping using microfluidic stretching and single-molecule detection of fluorescent site-specific tags. Genome Res. 2004;14:1137–1146. doi: 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tegenfeldt JO, et al. The dynamics of genomic-length DNA molecules in 100-nm channels. Proc. Natl. Acad. Sci. USA. 2004;101:10979–10983. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reisner W, et al. Statics and dynamics of single DNA molecules confined in nanochannels. Phys. Rev. Lett. 2005;94:196101. doi: 10.1103/PhysRevLett.94.196101. [DOI] [PubMed] [Google Scholar]
- 20.Cao H, Tegenfeldt JO, Austin RH, Chou SY. Gradient nanostructures for interfacing microfluidics and nanofluidics. Appl. Phys. Lett. 2002;81:3058–3060. [Google Scholar]
- 21.Horton R, et al. Variation analysis and gene annotation of eight MHC haplotypes: The MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart CA, et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–1187. doi: 10.1101/gr.2188104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das SK, et al. Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res. 2012;38:e177. doi: 10.1093/nar/gkq673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao M, et al. Rapid DNA mapping by fluorescent single molecule detection. Nucleic Acids Res. 2007;35:e16. doi: 10.1093/nar/gkl1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tewhey R, Bansal V, Torkamani A, Topol EJ, Schork NJ. The importance of phase information for human genomics. Nat. Rev. Genet. 2011;12:215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suk E-K, et al. A comprehensively molecular haplotype-resolved genome of a European individual. Genome Res. 2011;21:1672–1685. doi: 10.1101/gr.125047.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat. Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samad A, Huff EF, Cai W, Schwartz DC. Optical mapping: a novel, single-molecule approach to genomic analysis. Genome Res. 1995;5:1–4. doi: 10.1101/gr.5.1.1. [DOI] [PubMed] [Google Scholar]
- 30.Michalet X, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 31.Erlich R, et al. Next-generation sequencing for HLA typing of class I loci. BMC Genomics. 2011;12:42. doi: 10.1186/1471-2164-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pröll J, et al. Sequence capture and next generation resequencing of the MHC region highlights potential transplantation determinants in HLA identical haematopoietic stem cell transplantation. DNA Res. 2011;18:201–210. doi: 10.1093/dnares/dsr008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dervan PB, Bürli RW. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999;3:688–693. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 34.Felsenfeld G, Rich A. Studies on the formation of two- and three-stranded polyribonucleotides. Biochim. Biophys. Acta. 1957;26:457–468. doi: 10.1016/0006-3002(57)90091-4. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen PE, Egholm M. An introduction to peptide nucleic acid. Curr. Issues Mol. Biol. 1999;1:89–104. [PubMed] [Google Scholar]
- 36.Nagarajan N, Read TD, Pop M. Scaffolding and validation of bacterial genome assemblies using optical restriction maps. Bioinformatics. 2008;24:1229–1235. doi: 10.1093/bioinformatics/btn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]