Abstract
A prompt and efficient DNA damage response (DDR) eliminates the detrimental effects of DNA lesions in eukaryotic cells. Basic and preclinical studies suggest that the DDR is one of the primary anti-cancer barriers during tumorigenesis. The DDR involves a complex network of processes that detect and repair DNA damage, in which long non-coding RNAs (lncRNAs), a new class of regulatory RNAs, may play an important role. In the current study, we identified a novel lncRNA, lncRNA-JADE, that is induced after DNA damage in an ataxia-telangiectasia mutated (ATM)-dependent manner. LncRNA-JADE transcriptionally activates Jade1, a key component in the HBO1 (human acetylase binding to ORC1) histone acetylation complex. Consequently, lncRNA-JADE induces histone H4 acetylation in the DDR. Markedly higher levels of lncRNA-JADE were observed in human breast tumours in comparison with normal breast tissues. Knockdown of lncRNA-JADE significantly inhibited breast tumour growth in vivo. On the basis of these results, we propose that lncRNA-JADE is a key functional link that connects the DDR to histone H4 acetylation, and that dysregulation of lncRNA-JADE may contribute to breast tumorigenesis.
Keywords: ATM, DNA damage response, histone H4 acetylation, long non-coding RNA, lncRNA-JADE
Introduction
Recent high-throughput transcriptome analyses have identified a substantial portion of transcripts that are non-protein-coding RNAs in mammalian genomes (Bertone et al, 2004; Rinn and Chang, 2012). In addition to functionally important transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), these non-coding RNAs are also composed of small non-coding RNAs such as microRNAs (miRNAs) and piwi-interacting RNAs (piRNAs), and long non-coding RNAs (lncRNAs) which are generally over 200 nucleotides in length. A variety of lncRNAs have been so far found, including antisense, bidirectional, intronic, and intergenic RNA transcripts and those transcripts from pseudogenes and retrotransposons. While only <2% of the mammalian genome encodes proteins, as much as 70–90% of the genome is transcribed to a large transcriptome of lncRNAs (Jia et al, 2010; Cabili et al, 2011; Guttman and Rinn, 2012). Recent studies have demonstrated the importance of lncRNAs in regulating gene expression programme during differentiation, development, and metabolism (Lee, 2012). For example, the X-inactive-specific transcript (Xist) was the lncRNA that controls dosage compensation in female mammals by silencing one X chromosome (Brown et al, 1992; Chaumeil et al, 2006; Zhang et al, 2007). The Xist RNA binds PRC2, an epigenetic complex responsible for trimethylation of histone H3 at lysine 27 (H3K27me3), and directs it to the inactive X chromosome (Zhao et al, 2008). HOTAIR, residing at the HOXC locus, is expressed in cells with distal and posterior positional identities. HOTAIR inhibits gene expression at many loci through its interaction with various repressive chromatin-remodelling complexes such as LSD1, CoREST, REST, and PRC2 (Rinn et al, 2007; Gupta et al, 2010; Tsai et al, 2010). Similar to HOTAIR, another lncRNA Kcnq1ot1 negatively regulates the Kcnq1 imprinting region by recruiting repressive chromatin remodelling complexes G9a, PRC1 and PRC2, and the DNA methyltransferase Dnmt1 (Pandey et al, 2008; Terranova et al, 2008; Redrup et al, 2009; Mohammad et al, 2010). Increasing evidence has suggested that the central dogma should be extended to include lncRNAs. However, underlying mechanisms for the regulation and function of lncRNAs remain poorly understood.
In response to intrinsic and extrinsic genotoxic lesions, cells initiate a number of stress response pathways, collectively termed as DNA damage response (DDR), to safeguard the genome and the survival of eukaryotes (Bartek and Lukas, 2007; Ciccia and Elledge, 2010; Kumar et al, 2012). Ataxia-telangiectasia mutated (ATM) is a master kinase to initiate and mediate a cascade of cellular signals in response to double-stranded DNA breaks (DSBs), the most lethal DNA lesions. Upon DNA damage, ATM is activated by autophosphorylation at damage sites and in turn phosphorylates a large number of downstream substrates, including the tumour suppressor p53, breast cancer type 1 susceptibility protein (Brca1), and checkpoint kinase 2 (Chk2). These effectors transduce the DNA damage signal and activate cell-cycle checkpoints, DNA repair, and apoptosis (Kitagawa and Kastan, 2005; Matsuoka et al, 2007; Ciccia and Elledge, 2010). In addition to those signalling proteins, non-coding RNAs are new players in the regulatory network of the DDR. A novel p53-induced intergenic lncRNA named lincRNA-p21 (long intergenic RNA p21) was recently identified in the DNA damage-induced p53 signalling pathway. LincRNA-p21 is physically associated with hnRNPK and participates in the p53-mediated transcriptional repression (Huarte et al, 2010). A subsequent study by Chang and colleagues identified another lncRNA, PANDA (p21-associated ncRNA DNA damage activated) in the p53 signalling pathway. It modulates the p53-dependent apoptosis by interacting with the transcription factor NF-YA (Hung et al, 2011). Our recent study also showed that a DNA damage-induced lncRNA, ANRIL (antisense non-coding RNA in the INK4 locus), mediates the repression of INK4b-ARF-INK4a locus at the late stage of the DDR (Wan et al, 2013). These studies highlight the crucial function of lncRNAs in the transcriptional regulation during the DDR.
In the current study, we examined genome-wide lncRNA expression profiles in Atm+/+ and Atm−/− mouse embryonic fibroblasts (MEFs) following DNA damage treatment, from which we identified a novel lncRNA, designated as lncRNA-JADE. Upon DNA damage, lncRNA-JADE is induced in an ATM-dependent but p53-independent manner. We found that lncRNA-JADE is crucial for the DNA damage-induced histone H4 acetylation. H4 acetylation occurs at multiple lysine sites, which often results in chromatin remodelling and transcriptional activation (Shahbazian and Grunstein, 2007; Campos and Reinberg, 2009; Suganuma and Workman, 2011). The HBO1 (human acetylase binding to ORC1) complex is responsible for the acetylation of histone H4 at K5, K8, and K12 sites, in which Jade1, a PHD zinc finger protein, modulates the histone acetyltransferase activity (Tzouanacou et al, 2003; Foy et al, 2008; Saksouk et al, 2009). In addition to HBO1, the histone acetylase MOF also mediated the K16 acetylation of histone H4 after DNA damage. Li et al (2010) found that H4 K16 acetylation positively regulated DNA damage repair by recruiting the DNA damage repair protein Mdc1. In this study, we demonstrated that histone H4 acetylation is dramatically increased following DNA damage. The DNA damage-induced lncRNA-JADE upregulates the transcription of Jade1, leading to increased acetylation of histone H4. This study provides an important mechanism that connects the DNA damage signalling to histone acetylation and gene expression programme.
Results
ATM-mediated DNA damage signalling regulates the expression of lncRNAs
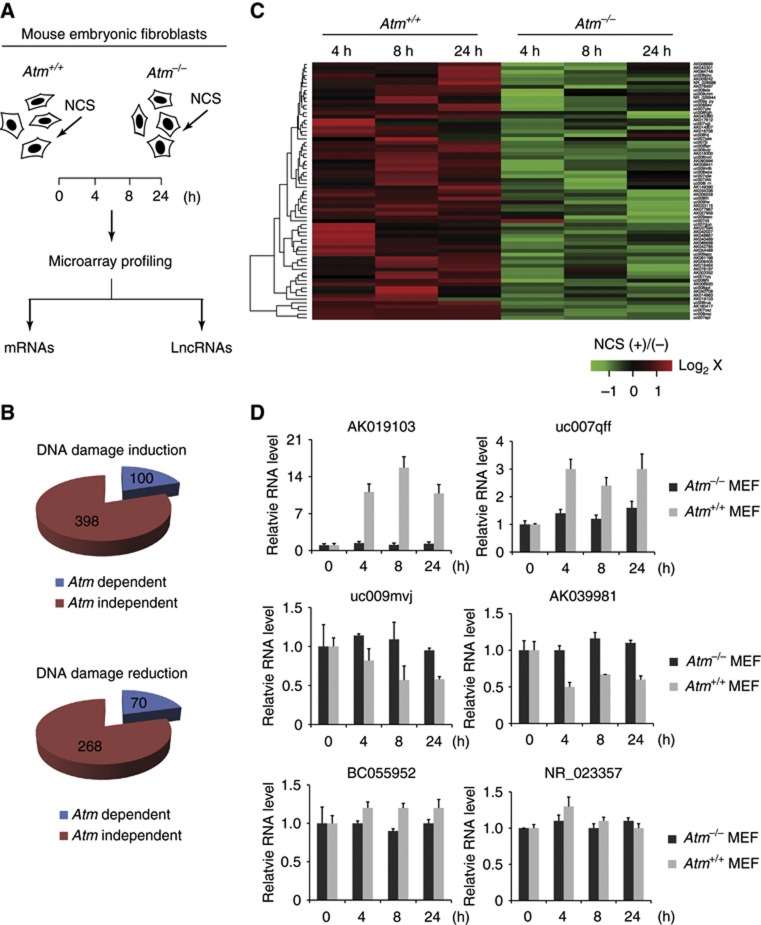
Functional lncRNAs can be studied through bioinformatical analysis and high-throughput assays such as microarray and transcriptome analyses. To examine how lncRNAs are regulated in the DDR, we examined genome-wide lncRNA expression in the Atm+/+ and Atm−/− littermate MEFs. MEFs were treated with a radiomimetic drug, neocarzinostatin (NCS) that generates DSBs. Cells were harvested at varying time points (0–24 h) and the lncRNA expression profile in each sample was determined by mouse lncRNA microarray (probes for 13 800 lncRNAs) analysis (Figure 1A). A total of 498 (100 ATM dependent, 398 ATM independent) were significantly upregulated (cutoff≥1.5-fold for both 4 and 8 h time points), while 338 (70 ATM dependent, 268 ATM independent) lncRNAs were downregulated (cutoff≤0.5-fold for both 4 and 8 h time points) (Figure 1B and C). The results suggest that DSBs caused widespread changes in lncRNA expression. We were particularly interested in those ATM-dependent lncRNAs because ATM is a primary kinase that initiates the DDR in response to DSBs. We next performed quantitative RT–PCR (qRT–PCR) to validate the expression changes for those positive hits in the ATM-dependent group (Figure 1D). A novel lncRNA, annotated as AK019103, was identified as one of the most significantly induced lncRNAs in the MEFs. In the Atm+/+ MEFs, AK019103 was induced 8- to 15-fold after NCS treatment. However, this remarkable induction disappeared in the Atm−/− MEFs. LncRNAs that were repressed or unchanged after DNA damage were also verified in the qRT–PCR assays as shown in Figure 1D. These results suggest that the ATM-mediated DNA damage signalling modulates lncRNA expression.
Figure 1.
ATM-dependent regulation of lncRNA expression in response to DNA damage. (A) Experimental layout to identify ATM-dependent lncRNAs. Atm+/+ and Atm−/− mouse embryonic fibroblasts (MEFs) were treated with NCS (200 ng/ml) and harvested at indicated time points for microarray analyses. (B) The number of ATM-dependent lncRNAs upon DNA damage. (C) A representative group of ATM-dependent and DNA damage-induced lncRNAs. Green or red colour on the heat map indicates a decrease or an increase in the lncRNA level and colour intensities correspond to relative signal levels on a logarithmic scale. (D) Quantitative PCR validation of representative lncRNAs. Data represent the mean of three experimental replicates, with error bars depicting s.d.
LncRNA-JADE is transcriptionally activated after DNA damage
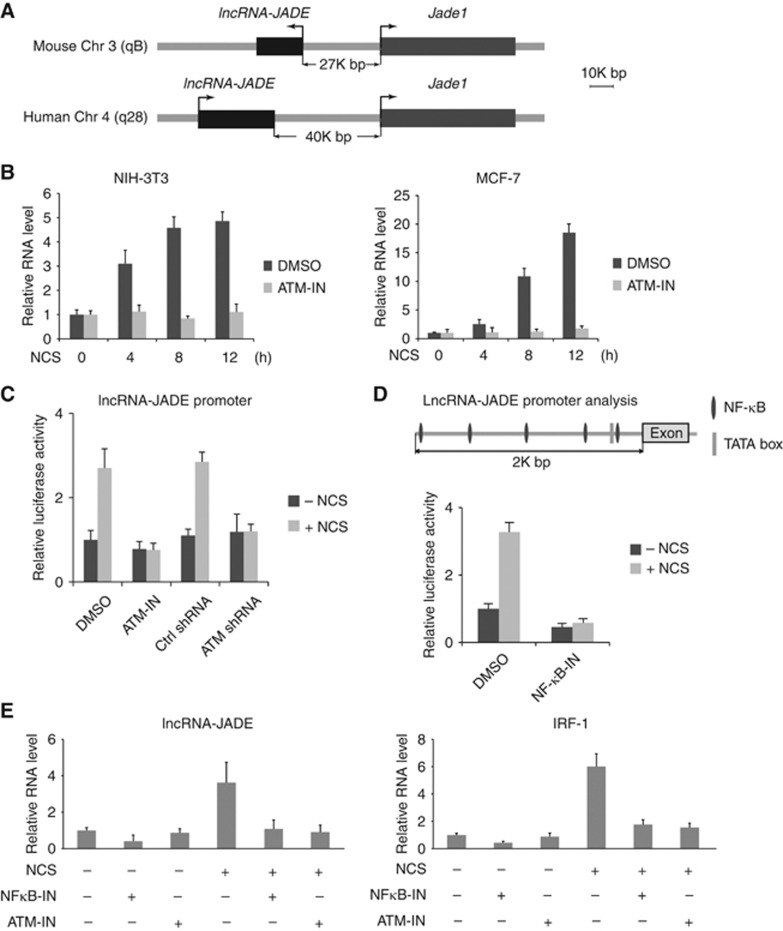
A common feature for the majority of lncRNAs is their poor conservation across species. However, the DNA damage signalling pathway is highly conserved in eukaryotic cells. We assumed that only a small number of conserved lncRNAs might function as major players in the DDR. Therefore, we focussed on those conserved lncRNAs that are differentially expressed after DNA damage, one of which was mouse AK019103 RNA. High level of conservation was found in the predicted AK019103 gene transcripts across mammalian species including rat, rabbit, human, pig, sheep, cow, and dog, suggesting that it may play a fundamental role in the DDR (Supplementary Figure S1A). Because human AK019103 transcript had not previously been identified, we used the RNA sequence of mouse AK019103 to clone the full-length transcript of the human AK019103 by 5′ and 3′ RACE (rapid amplification of cDNA ends)-PCR assays (Schaefer, 1995). The human AK019103 transcript contains only one exon with 1721 nucleotides (nt) in full length (GenBank accession no. KC469579; Supplementary Figure S1B). In both mouse and human genomes, the AK019013 gene is adjacent to Jade1 gene (Figure 2A). Therefore, we designated AK019013 RNA as lncRNA-JADE. LncRNA-JADE is an independent transcript as it contains no overlapping sequence with the transcripts from its neighbouring genes. Mouse lncRNA-JADE is transcribed in the opposite orientation to that of the Jade1 gene, while its human homologue has the same orientation as Jade1 (Figure 2A). Analysis of chromatin structure in mouse indicates that lncRNA-JADE and Jade1 genes have distinct promoters at the chromosomal region 3qB (Mikkelsen et al, 2007).
Figure 2.
LncRNA-JADE is induced after DNA damage. (A) Schematic illustration showing Jade1 and lncRNA-JADE genes in mouse and human. (B) Mouse and human lncRNA-JADE are induced in an ATM-dependent manner after DNA damage. (C) DNA damage positively regulates lncRNA-JADE promoter activity in an ATM-dependent manner. ATM-IN: ATM inhibitor. (D) Schematic illustration showing the NF-κB binding elements in the Jade1 promoter. DNA damage induces the activity of lncRNA-JADE promoter in an NF-κB-dependent manner. NF-κB-IN: NF-κB inhibitor. (E) Expression of LncRNA-JADE is regulated by ATM and NF-κB after DNA damage. IRF-1 (interferon response factor-1) is served as a positive control in the NF-κB signalling. Graphic data in this figure present the mean of three experimental replicates and error bars depict s.d.
We next validated the DNA damage induction of lncRNA-JADE in mouse fibroblast NIH3T3 and human breast cancer MCF7 cells that were pretreated with DMSO or ATM inhibitor KU-55933 (Figure 2B; Supplementary Figure S2A). We observed a significant induction of lncRNA-JADE in both cell lines, but inhibiting ATM almost abolished this induction completely, suggesting that an ATM-dependent mechanism is involved in the lncRNA-JADE expression. Because the tumour suppressor p53 is a central player in the ATM signalling pathway, we tested whether knockdown of p53 in MCF7 cells affected the induction of lncRNA-JADE after DNA damage (Supplementary Figure S2B and C). The results showed that p53 is dispensable for the induction of lncRNA-JADE in the DDR. In addition, the basal expression levels of lncRNA-JADE are not significantly different in the isogenic HCT116p53+/+ and HCT116p53−/− cells (Supplementary Figure S2D). LncRNA-JADE was predominantly localized in the nucleus, and relative intracellular distributions of lncRNA-JADE seemed to be unchanged after DNA damage (Supplementary Figure S2E).
We cloned the putative promoter of lncRNA-JADE and inserted it into a luciferase construct. The promoter activity was increased after DNA damage in the control MCF7 cells, but not in the cells with ATM inhibition (ATM inhibitor or shRNA) (Figure 2C). We analysed the promoter sequence of human lncRNA-JADE and found five putative NF-κB-binding sites (Figure 2D, upper panel). Using the luciferase assay, we found that inhibiting NF-κB activity suppressed the promoter activity of lncRNA-JADE and abolished its induction after DNA damage (Figure 2D). Previous reports have shown the molecular linkage between ATM and NF-κB signalling in response to genotoxic stimuli (Ahmed and Li, 2007; Miyamoto, 2011). ATM-mediated phosphorylation of NEMO (NF-κB essential modulator) promotes its nuclear export, which results in the activation of NF-κB (Wu et al, 2006). To test whether the ATM-NF-κB signalling induces the expression of lncRNA-JADE, we measured the transcriptional activity of lncRNA-JADE in MCF7 cells with or without ATM and NF-κB inhibition (Figure 2E). Inhibiting ATM or NF-κB markedly suppressed the DNA damage induction of lncRNA-JADE and the positive control IRF-1 (an NF-κB target) (Harada et al, 1994; Robinson et al, 2006).These results suggest that transactivation of lncRNA-JADE is regulated by the ATM-NF-κB signalling in the DDR.
LncRNA-JADE positively regulates H4 acetylation
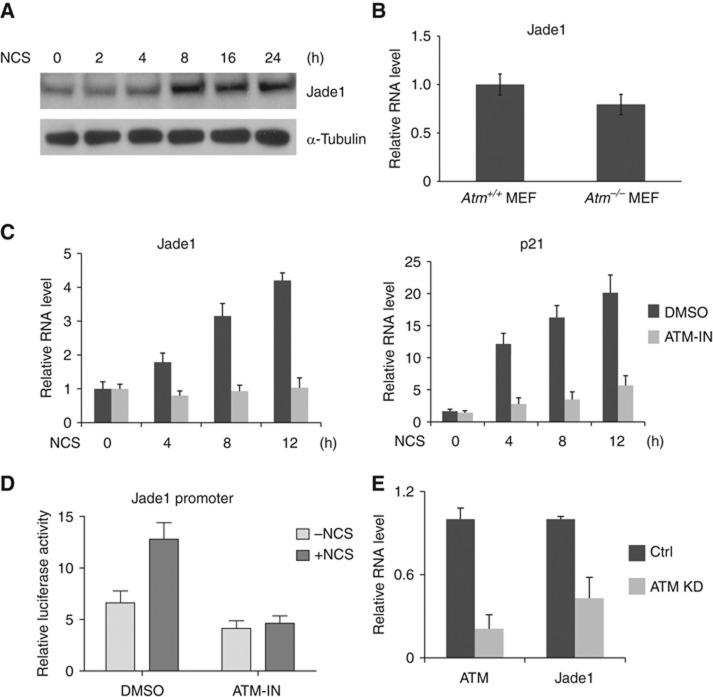
Recent studies revealed that lncRNA transcripts in some cases function as enhancer RNAs that promote messenger RNA synthesis at nearby genes (De et al, 2010; Kim et al, 2010; Wang et al, 2011a; Melo et al, 2013). These findings suggest that lncRNA genes have an active ‘promoter-like’ role in regulating adjacent gene expression. We attempted to investigate the functional interaction between lncRNA-JADE and the neighbouring Jade1 gene. Jade1 is an essential component of the HBO1 complex that is responsible for histone H4 acetylation. Similar to the induction of lncRNA-JADE, the Jade1 protein was also induced after DNA damage (Figure 3A). The basal expression level of Jade1 was only slightly reduced in Atm−/− MEFs in the absence of DNA damage (Figure 3B). To determine whether the induction of Jade1 protein is due to transcriptional regulation or post-transcriptional regulation, we checked Jade1 mRNA levels in MCF7 cells with or without pretreatment of ATM inhibitor (Figure 3C). Jade1was transcriptionally induced 1.8- to 4.2-fold following DNA damage. As a positive control for the functionality of the DDR, p21 levels were significantly increased after DNA damage and this increase was dramatically abrogated in the presence of the ATM inhibitor. We cloned the Jade1 promoter into a luciferase vector and measured its activity in the cells treated with or without NCS (Figure 3D). While the basal activity of the Jade promoter was not much affected by ATM inhibition, the promoter activity was profoundly upregulated after DNA damage in an ATM-dependent manner. Consistent with it, knockdown of ATM in the NCS-treated MCF7 cells had severely inhibited Jade 1 expression (Figure 3E). These results suggest that transcription of Jade1 is activated after DNA damage in an ATM-dependent manner.
Figure 3.
Jade1 is transcriptionally induced after DNA damage. (A) Jade1protein is induced after DNA damage. (B) Jade1 expression is slightly reduced in Atm−/− MEFs. (C) The level of Jade1mRNA is induced in an ATM-dependent manner after DNA damage. P21 mRNA is served as an indicator for the functionality of the DNA damage response. (D) Activity of the Jade1 promoter is upregulated in ATM-dependent manner after DNA damage. MCF-7 cells were transfected with the Jade1 promoter-driven firefly luciferase expression vector and Renilla luciferase expression vector. Firefly luciferase activity was measured and normalized to the activity of Renilla luciferase. (E) Knockdown of ATM in MCF7 cells inhibits Jade1 expression. Graphic data in this figure present the mean of three experimental replicates and error bars depict s.d.
Source data for this figure is available on the online supplementary information page.
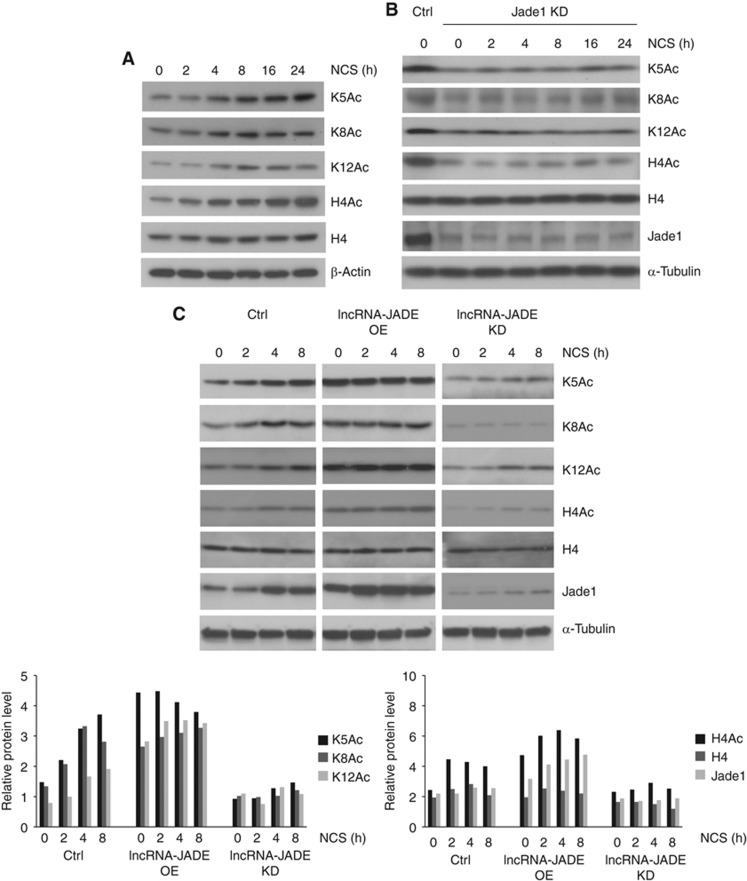
Jade1 is essential for the HBO1complex-mediated histone H4 acetylation (Foy et al, 2008). We next checked whether the DNA damage-induced Jade1 had an impact on the histone H4 acetylation. We measured the levels of total H4 protein, total acetylated H4 protein, and H4 proteins specifically acetylated at sites K5, K8, and K12 in the control or NCS-treated MCF7 cells. A rapid increase in the acetylation of H4 (global and K5/K8/K12) was observed after DNA damage, and the peak level of acetylation occurred around 8 h post damage (Figure 4A), consistent with the temporal pattern for Jade1 induction (Figure 3A). Knockdown of Jade1 (knockdown efficiency ∼80%) resulted in dramatic decreases in total and site-specific H4 acetylation and abolished the DNA damage-induced H4 acetylation (Figure 4B; Supplementary Figure S3A). To determine the functional connection between lncRNA-JADE and H4 acetylation, we examined whether altering lncRNA-JADE levels affects Jade1 expression and histone H4 acetylation. We found that overexpression of lncRNA-JADE boosted up Jade1 mRNA levels whereas knockdown of lncRNA-JADE significantly inhibited Jade1 transcription (Supplementary Figure S3B and C). These transcriptional changes were translated to protein level changes of Jade1 (Figure 4C, upper and left bottom panels). As a consequence, overexpression of lncRNA-JADE increased the H4 acetylation to much higher levels in the control and DNA damage-treated cells. In contrast, silencing lncRNA-JADE led to profound reductions in total and site-specific H4 acetylation (Figure 4C, upper and right bottom panels). It was also noted that the lncRNA-JADE had no effect on the total histone H4 protein levels. Collectively, these results defined lncRNA-JADE as an important link that connects the DNA damage signalling to the Jade1-mediated H4 acetylation.
Figure 4.
LncRNA-JADE positively regulates histone H4 acetylation through Jade1. (A) H4 acetylation is induced after DNA damage. MCF7 cells were treated with NCS (500 ng/ml). H4Ac: total histone H4 acetylation; K5, K8, K12: Histone H4 acetylation at lysine 5, 8, or 12. (B) Jade1 knockdown abolishes the induction of H4 acetylation after DNA damage. (C) LncRNA-JADE positively regulates H4 acetylation and Jade1. Overexpression of lncRNA-JADE enhanced the induction of H4 acetylation and Jade1, and knockdown of lncRNA-JADE abolished the induction of H4 acetylation and Jade1. Semi-quantification of proteins is shown at the bottom.
Source data for this figure is available on the online supplementary information page.
LncRNA-JADE interacts with Brca1 to promote Jade1 transcription
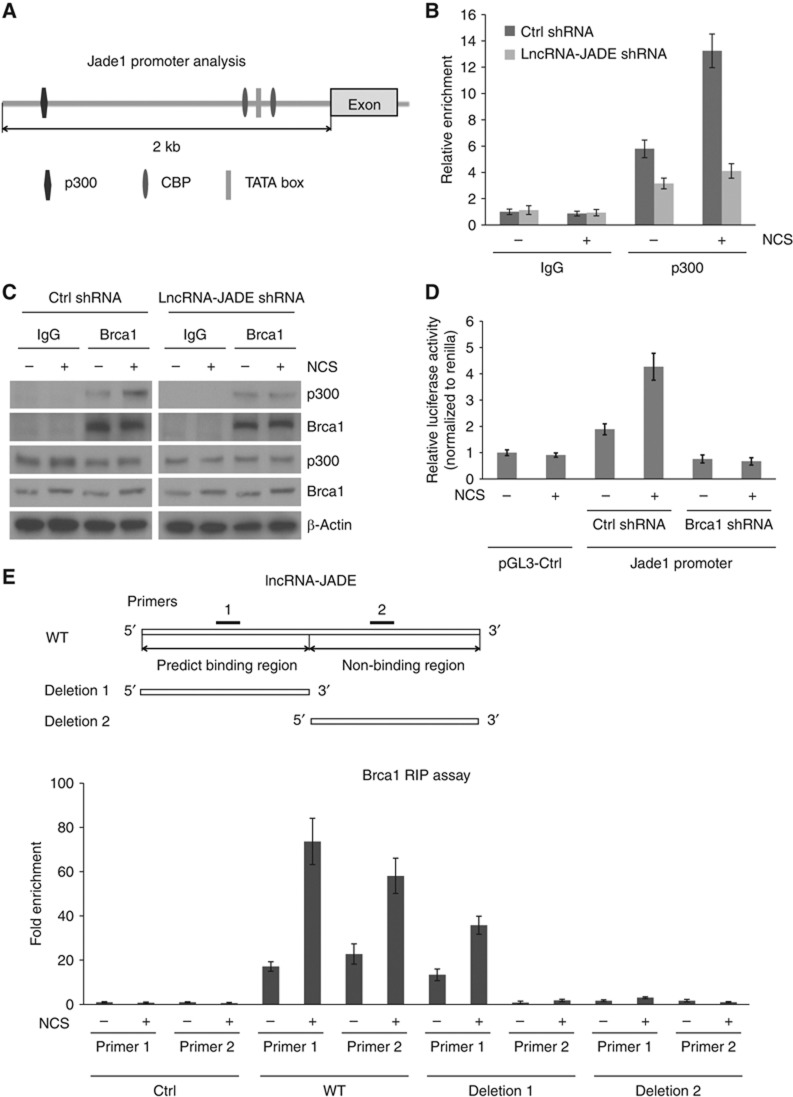
To investigate how lncRNA-JADE interacts with the transcription machinery, we first used a web tool TESS (Transcription Element Search System) to predict transcription factor binding sites in the Jade1promoter sequence. It was predicted that transcriptional cofactors p300 and CBP may interact with CREB and bind to the Jade1 promoter (Figure 5A). Using DNA chromatin immunoprecipitation (ChIP) assays, we found that p300 was indeed recruited to the Jade1 promoter and this recruitment was significantly increased after DNA damage (Figure 5B). Knockdown of lncRNA-JADE inhibited the basal level of p300’s binding with Jade1 promoter DNA and also abolished the induced p300 recruitment following DNA damage. While no direct interaction between p300 and lncRNA-JADE was detected, we asked what protein mediates the effect of lncRNA-JADE in the p300-containing transcription machinery. Previous studies showed that Brca1 interacts with p300/CBP coactivators to transcriptionally activate target gene transcription (Pao et al, 2000; Crowe and Lee, 2006). We confirmed their interaction in the cells treated with or without NCS (Figure 5C). While p300 interacts with Brca1 in the untreated cells, DNA damaging treatment notably increased this interaction as more p300 was detected in the Brca1 immunoprecipitate. To determine whether Brca1 is essential for the Jade1 transactivation in the DDR, we measured the promoter activity of Jade1 in the control and Brca1 knockdown MCF7 cells. The promoter activity of Jade1 was markedly increased after DNA damage, but knockdown of Brca1 depleted this increase (Figure 5D; Supplementary Figure S4A). It is well known that activation of Brca1 was depended on the ATM-mediated phosphorylation in the initiation of the DDR (Cortez et al, 1999). Consistent with this study, we observed that the phosphorylation level of Brca1 protein was increased after DNA damage (indicated by a band shift in western blots) and the increase was dependent on the ATM activity (Supplementary Figure S4B).
Figure 5.
Brca1 binds lncRNA-JADE and mediates Jade1 induction in the DNA damage response via p300-containing transcription complex. (A) Schematic illustration showing the p300/CBP binding elements in the Jade1 promoter. (B) p300 physically interacts with the promoter region of Jade1 gene. Control or lncRNA-JADE knockdown MCF7 cells were treated with or without NCS (200 ng/ml) and cell lysates were immunoprecipitated with control IgG or p300 antibodies. The p300-binding activity of Jade1 promoter DNA was quantified by qPCR. (C) Brca1 interacts with p300 and this interaction is increased after DNA damage. (D) Jade1 promoter activity is induced in a Brca1-dependent manner after DNA damage. MCF-7 cells were infected with lentiviruses expressing control or Brca1 shRNA. The cells were transfected with pGL3-control vector (SV40 promoter) or Jade1 promoter-driven firefly luciferase expression vector and Renilla luciferase expression vector 2 days post infection. They were treated with NCS (500 ng/ml) 24 h after transfection and then harvested 16 h after treatment. Firefly luciferase activity was measured and normalized to the activity of Renilla luciferase. (E) LncRNA-JADE physically interacts with Brca1. 5′- and 3′-deletion mutants of lncRNA-JADE were generated as indicated. Two pairs of primers were used to detect the Brca1-binding sequences of lncRNA-JADE in RIP assays. Graphic data in this figure present the mean of three experimental replicates and error bars depict s.d.
Source data for this figure is available on the online supplementary information page.
Next, we asked whether lncRNA-JADE-dependent Jade1 induction was mediated by Brca1. We analysed the interaction between Brca1 and lncRNA-JADE using the catRAPID software to predict RNA–protein interaction (Bellucci et al, 2011). Brca1 was predicted to bind with 5′-portion of lncRNA-JADE and the interaction was specifically identified as NPInter (ncRNA–protein interactions) according to the discriminative power (Supplementary Figure S4C). To verify the binding prediction, we generated 3′- and 5′-deletion mutants of lncRNA-JADE (upper panels in Figure 5E). PCR primers were designed to specifically detect 5′- or 3′-portion of lncRNA-JADE. Ribonucleoprotein immunoprecipitation (RIP) assays were performed to measure the interaction between Brca1 and lncRNA-JADE with or without NCS treatment. Full-length lncRNA-JADE physically interacted with Brca1 and this interaction was induced after DNA damage in an ATM-dependent manner (Supplementary Figure S4D). The 5′-portion of lncRNA-JADE showed high affinity to Brca1, and the binding was significantly increased after NCS treatment (Figure 5E). In contrast, the 3′-portion of lncRNA-JADE exhibited no binding activity with Brca1 in the absence or presence of DNA damage. Through its interaction with lncRNA-JADE, Brca1 was better recruited to the p300/CBP complex as shown in Figure 5C. This increased binding is dependent on lncRNA-JADE, which was abrogated by knockdown of lncRNA-JADE. These results support a mechanism that lncRNA-JADE interacts with Brca1 to mediate the induction of Jade1 transcription after DNA damage.
Biological functions of lncRNA-JADE
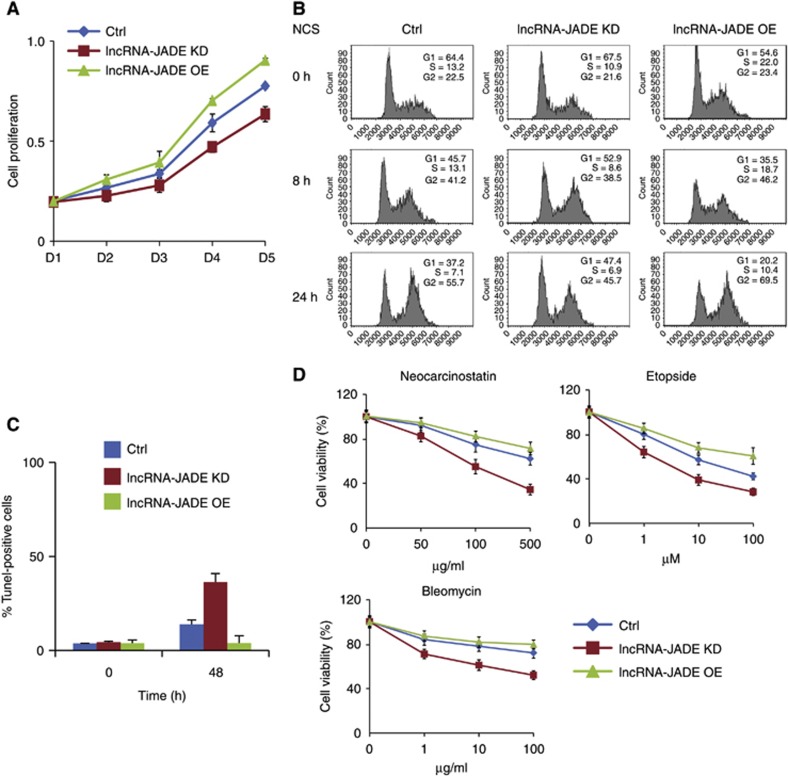
Since lncRNA-JADE is a previously unexplored lncRNA, its biological functions are completely unknown. We first checked whether lncRNA-JADE affected cell proliferation (Figure 6A). Cell growth was moderately slowed down when lncRNA-JADE was stably knocked down in MCF7 cells. In contrast, ectopic expression of lncRNA-JADE resulted in a mild increase in cell proliferation. Cell-cycle analyses demonstrated a weaker G1/S checkpoint when lncRNA-JADE was overexpressed (Figure 6B). Twenty-four hours post NCS treatment, only 20.2% of the lncRNA-JADE-overexpressing cells were arrested in G1 phase in comparison with 37.2% of the control cells in G1 phase, whereas knockdown of lncRNA-JADE significantly enhanced this checkpoint with 47.4% of cells arrested in G1. We also performed TUNEL assay in the control, lncRNA-JADE knockdown or overexpressing MCF7 cells after NCS treatment (Figure 6C; Supplementary Figure S5A). LncRNA-JADE-silenced cells had a much higher percentage of apoptotic cells than the control cells (37.8 versus 15.0%), whereas overexpression of lncRNA-JADE appeared to desensitize the cells with only 3% of apoptotic cells under the same treatment. Overexpression of lncRNA-JADE by itself did not cause detectable DNA damage in MCF7 cells (Supplementary Figure S5B). We tested the sensitivity of the MCF7 cells expressing altered levels of lncRNA-JADE to three DNA damaging agents (Figure 6D). An inverse correlation was observed between lncRNA-JADE levels and cell sensitivity. Knockdown of lncRNA-JADE significantly sensitized MCF7 cells to the DNA damaging treatments. These results suggest that inhibition of lncRNA-JADE may enhance the DNA damage-induced apoptosis and sensitize cells to DNA damaging agents.
Figure 6.
Biological functions of lncRNA-JADE in human MCF7 cells. (A) LncRNA-JADE positively regulates MCF7 cell proliferation. (B) Altering lncRNA-JADE expression affects DNA damage-induced cell-cycle arrest. Cell-cycle profiles were analysed by flow cytometry using propidium iodide-stained cells. (C) Knockdown of lncRNA-JADE increases cell apoptosis in the control and NCS-treated cells. The percentage of TUNEL-positive cells was summarized in the graph. (D) Knockdown of lncRNA-JADE increases the cell sensitivity to DNA damaging drugs NCS, Etopside, and Bleomycin. MCF7 cells were treated with DNA damaging agents as indicated and cultured for 48 h and cell viability was measured. Graphic data in this figure present the mean of three biological replicates and error bars depict s.d.
LncRNA-JADE is highly expressed in human breast tumours
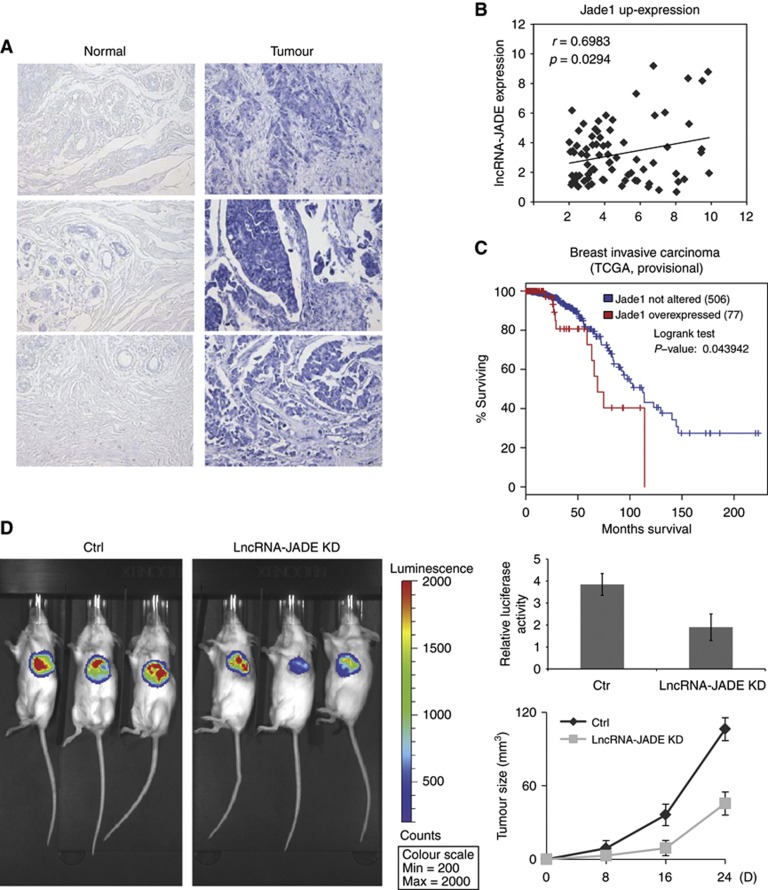
As a functional target of lncRNA-JADE, Jade1 is required for epithelial cell proliferation (Havasi et al, 2013). Downregulation of Jade1 by small-interfering RNAs led to reduced levels of DNA synthesis, HBO1 activity, and chromatin recruitment of replication factor Mcm7. In addition, overexpressed HBO1 proteins were detected in human breast cancer by the immunohistochemistry (Wang et al, 2010). Similarly, we showed that lncRNA-JADE promotes cell proliferation, inhibits DNA damage checkpoints, and reduces the cellular sensitivity to DNA damaging drugs, suggesting that lncRNA-JADE possibly has the proto-oncogene properties. To explore the oncogenic role of lncRNA-JADE, we first analysed the expression of lncRNA-JADE in 100 human breast tumour tissues and 10 control breast tissues in a human breast tumour tissue microarray. In situ hybridization displayed very low or undetectable levels of lncRNA-JADE in all of normal breast tissues. However, 71 out of 100 breast tumours exhibited overexpressed lncRNA-JADE, in which 25 tumour samples had high or very high levels of lncRNA-JADE (Figure 7A; Supplementary Figure S6). Using breast tumour cDNA array, we detected the levels of lncRNA-JADE and Jade1 in normal and breast tumour tissues. A significant correlation between lncRNA-JADE and Jade1 levels was observed in breast tumours (coefficient=0.698, P=0.0294) (Figure 7B; Supplementary Figure S7A and B). We reasoned that lncRNA-JADE is involved in mammary tumorigenesis. While the levels of lncRNA-JADE are not available from current databases, our analysis of Jade1 expression data in The Cancer Genome Atlas (TCGA) revealed that Jade1 levels are negatively correlated with overall survival of patients with breast invasive carcinoma (P-value=0.0439) (Figure 7C). In monolayer and soft agar colony-formation assays, knockdown of lncRNA-JADE significantly inhibited the transforming ability of oncogenic H-RasV12 in human mammary epithelial MCF10A cells (Supplementary Figure S8A and B). We also performed 4T1 orthotopic xenograft experiments in BALB/c SCID mice (Figure 7D). The 4T1 cells were engineered to stably express luciferase for in vivo imaging. Control and lncRNA-JADE knockdown 4T1 cells were orthotopically injected into mouse mammary gland fat pads. Xenograft tumour growth was monitored and mice were euthanized at day 24 after implantation. At 8 days after injection, all mice bearing lncRNA-JADE knockdown cells barely had detected tumours whereas mice bearing control cells showed visible tumour growth. Knockdown of lncRNA-JADE resulted in a significant reduction in tumour growth throughout the whole period. In vivo imaging also showed reduced luciferase signals in the mice bearing lncRNA-JADE knockdown cells.
Figure 7.
Knockdown of lncRNA-JADE inhibits mammary tumour growth in vivo. (A) Higher levels of lncRNA-JADE in human breast cancer tissues in comparison with normal breast tissues. In situ hybridization of lncRNA-JADE was performed on tissue microarray comprised of human normal breast and breast cancer tissues. (B) Correlation between lncRNA-JADE and Jade1 up-expression in breast cancer tissue cDNA array. The level of lncRNA-JADE and Jade1 was measured by RT–PCR. The lncRNA-JADE expression demonstrated a significant correlation with the Jade1 expression according to the Spearman correlation coefficient (r=0.6983 and P=0.0294). (C) Comparison of survival curves between patients with Jade1 overexpression and patients with normal Jade1 expression using TCGA data in breast invasive carcinomas. (D) Knockdown of lncRNA-JADE inhibits xenografted 4T1 tumour growth in vivo. One million luciferase expressing 4T1 cells stably expressing control or lncRNA-JADE shRNA were injected into the mammary fat pad of each Balb/cSCID mouse. Two weeks after injection, luciferase activity was measured and quantified by an IVIS device (left panel and upper right panel). Breast tumour size was measured in the mice for 24 days (bottom right panel). Graphic data present the mean of five mice and error bars depict s.d.
Discussion
The DDR is a major protective mechanism against genomic instability in eukaryotic cells. In the past couple of decades, a large number of protein components have been identified in the DDR pathways, including DNA damage sensors, signalling PIKKs (phosphatidylinositol 3-kinase-related kinases) and effectors (Ciccia and Elledge, 2010). The ATM kinase is a primary PIKK that responds to DSBs and activates a variety of cell activities (Matsuoka et al, 2007). The ATM-p53 signalling plays a central role in cell-cycle checkpoints and cell death pathways following DNA damage and oncogenic stresses (Toledo and Wahl, 2006). Regulators in the ATM-p53 signalling modulate not only the activity and stability of signalling proteins but also their mutual recognition, interaction, and signalling complex formation. In additional to canonical protein components, recent studies have revealed essential functions of non-coding RNAs in the DDR (Liu and Lu, 2012). MiRNAs, a type of small non-coding RNAs, join the DDR by regulating the expression of their target mRNA transcripts. Defects in miRNA biogenesis lead to genomic instability and are often associated with human tumorigenesis (Pecot et al, 2011; Wan et al, 2011). Recently, a large transcriptome of lncRNAs was found in eukaryotic cells with total number varying from 10 000 to 200 000 (Rinn and Chang, 2012). These lncRNA transcripts are often poorly conserved and present at low abundance. Emerging evidence has illustrated how this class of RNA establishes a new paradigm for gene expression programme. However, the functional role of lncRNAs in the DDR is largely unknown.
The first identified p53-dependent lncRNA was lincRNA-p21. The Rinn group deciphered a molecular mechanism by which lincRNA-p21 participates in the p53-mediated transrepression machinery through its physical association with hnRNPK (Huarte et al, 2010). In response to DNA damage, p53 also transcriptionally induces PANDA and TUG1 that are involved in apoptosis and cell-cycle checkpoint, respectively (Hung et al, 2011; Yang et al, 2011). In the current study, we identified an ATM-dependent lncRNA expression profile in the DDR. While several hundred of mouse lncRNAs are either induced or repressed after NCS treatment, only a small number of these lncRNAs are conserved in humans, one of which is lncRNA-JADE. LncRNA-JADE is unique because its induction is dependent on ATM, but not on p53. Consistent with it, no p53-responsive elements were found on the putative promoter region of lncRNA-JADE. Whereas the ATM kinase is not directly recruited to the promoter, it was believed that one or more ATM targets are associated with the transcription of the lncRNA-JADE gene and mediate the transactivating effect. We found that the transcription factor NF-κB appears to be an important player that translates the ATM signalling to the expression of lncRNA-JADE. In response to DNA damage, NEMO, the regulatory subunit of IκB kinase (IKK), is phosphorylated by ATM (Wu et al, 2006; Miyamoto, 2011). This phosphorylation promotes the translocalization of NEMO from the nucleus to the cytoplasm and its subsequent interaction with the catalytic IKK subunit in the cytoplasm, resulting in activation of NF-κB signalling and increased lncRNA-JADE expression. In addition to NF-κB binding sites, responsive element (RE) sites of other transcription factors or cofactors are also found in the putative promoter of the lncRNA-JADE gene, implying that other ATM-associated mechanisms may exist in regulating its transcription.
A large body of literature has established a role for chromatin in the DDR pathways. Chromatin structure is subject to histone modifications such as phosphorylation, ubiquitination, acetylation, and methylation. One of the most intensively studied histone modifications in the DDR is the serine 139 phosphorylation of histone variant H2AX, which provides a host of binding sites for the key checkpoint factor Mdc1 and stabilizes its interaction with DSB chromatin (Stewart et al, 2003). In addition to H2AX phosphorylation, higher order organization of H4K16ac has also been suggested to regulate the association of Mdc1 with the DSB domains (Li et al, 2010). Ablation of MOF, a histone acetyltransferase specific for H4K16, leads to peri-implantation lethality in mouse embryos (Li et al, 2010). Conditional knockout of MOF results in global reduction in H4 K16 acetylation, severe G2/M cell-cycle arrest, massive chromosome aberration, and defects in ionizing radiation-induced DNA damage repair. Another histone acetyltransferase complex NuA4 also contributes to chromatin remodelling in the DDR (Allard et al, 1999, 2004). Trrap and Tip60 in the complex bind to the DSB chromatin and induce acetylation of histone H2A and H4. In this study, we showed that DNA damage induces histone H4 acetylation at K5, K8, and K12. This set of acetylation events are primarily catalysed by the HBO1 complex in which Jade1 is a major cofactor. Jade1 levels are remarkably increased following DNA damage. Knockdown of Jade1 abolished the DNA damage-induced histone H4 acetylation. Although little is known about potential post-translational modifications of Jade1 protein, we observed that protein stability of Jade1 was unchanged after DNA damage (data not shown), indicating a molecular mechanism involving transcriptional activation.
We showed that silence of lncRNA-JADE notably inhibited Jade1 induction and histone H4 acetylation, suggesting that this lncRNA is likely an important regulator for Jade1 expression. Since lncRNA-JADE gene is adjacent to the Jade1 gene, it is reasonable to think that newly synthesized lncRNA-JADE is in spatial proximity with the transcription machinery for Jade1. A similar example was previously found in the regulation of lncRNA HOTTIP (Wang et al, 2011b). HOTTIP is encoded on the distal 5′ end of the HOXA gene cluster, but chromosomal looping brings HOTTIP into close proximity to multiple HOXA genes. HOTTIP RNA directly interacts with the adaptor protein WDR5 and targets WDR5/MLL complexes across HOXA, driving histone H3 lysine 4 trimethylation and gene transcription. We propose a molecular mechanism for lncRNA-JADE to transcriptionally activate Jade1 gene. This model is based on the finding that lncRNA-JADE is associated with the p300/CBP coactivator complex via Brca1and promotes Jade1 transcription. The direct interaction of lncRNA-JADE with Brca1 is assumed to recruit lncRNA-JADE to the p300/CBP coactivator complex. Supporting this model is the following evidence: (1) Brca1 was previously shown to interact with p300/CBP complex (Pao et al, 2000), (2) Brca1 is physically associated with lncRNA-JADE, (3) the level of Brca1-assoicated lncRNA-JADE is significantly increased after DNA damage, and (4) overexpression of lncRNA-JADE enhances the Brca1–p300 interaction, and knockdown of lncRNA-JADE abolished the interaction. The 5′ half of the lncRNA-JADE sequence was validated to bind with Brca1. While the ATM signalling is required for Brca1 to be recruited to p300/CBP, it is not known which of ATM-mediated modifications of Brca1 are essential for its interaction with lncRNA-JADE or p300/CBP. In-depth structural analysis of lncRNA-JADE will be required to provide an answer into the question.
Given the ubiquitous nature and abundance of lncRNAs in the cell, it is not surprising that they are involved in many biological processes such as cell proliferation and differentiation, embryo development, tissue regeneration, metabolism, and tumour suppression. However, the role of lncRNAs in tumorigenesis is much less characterized than well-defined small non-coding RNAs, miRNAs. Several lncRNAs have been reported in human cancer. As examples, ANRIL, a lncRNA that negatively regulate the INK4B–ARF–INK4A locus, was found to be underexpressed in neuroblastoma, acute lymphocytic leukaemia, and melanoma (Pasmant et al, 2007; Cunnington et al, 2010; Aguilo et al, 2011; Iacobucci et al, 2011). Aberrant expression of HOTAIR, a lncRNA associated with the HOXD gene cluster, is associated with colorectal cancer and breast cancer (Gupta et al, 2010; Kogo et al, 2011; Lu et al, 2012). In this study, Jade1 is identified as a functional target of lncRNA-JADE in the DNA damage-induced H4 acetylation. In cell studies, the Jade1-HBO1 histone acetylase is required for cell-cycle progression and the majority of histone H4 acetylation (Foy et al, 2008; Havasi et al, 2013). Previous studies have shown that Jade1 plays a key role in the pathogenesis of renal cancer and von Hippel-Lindau (VHL) disease (Zhou et al, 2004). Jade1 is highly expressed in kidney and renal proximal tubule cells and may be involved in renal tubular epithelial cell differentiation, growth suppression, and apoptosis. Truncations and mutations of the tumour suppressor pVHL altered its interaction with Jade1, suggesting a correlation between Jade1 and renal cancer. A recent study demonstrated that Jade1 binds and promotes the ubiquitination and destruction of β-catenin and inhibits canonical Wnt signalling (Chitalia et al, 2008). Because inhibition of β-catenin activity reduces renal cancer cell growth, there may be a pVHL/Jade1/β-catenin axis in suppression of renal tumour. Due to the lack of genomic engineered mouse models, it remains elusive whether Jade1 is a tumour suppressor or an oncogene in different contexts. We showed here that lncRNA-JADE is upregulated in a high percentage of breast tumours, consistent with the previous studies showing hyperactive DDR activity in human breast cancer progression (Bartkova et al, 2006; Halazonetis et al, 2008). Knockdown of lncRNA-JADE inhibited H-RasV12-induced oncogenic transformation in vitro and suppressed xenografted 4T1 mammary tumour growth in vivo. As a direct target of lncRNA-JADE, Jade1 overexpression is correlated with poor survival of patients with breast invasive carcinoma. These results suggest a potential role of lncRNA-JADE in breast carcinogenesis. Sporadic non-hereditary breast cancer is recognized as the most common form of this malignancy. The presence of germ-line mutations in the BRCA1 gene of these tumours is an infrequent event. This work provides a potential mechanism by which an oncogenic lncRNA contributes to breast carcinogenesis by interacting with Brca1 and histone acetylation machinery.
Our current study establishes a functional link that connects DNA damage-induced lncRNA-JADE to histone H4 acetylation. However, questions remain to decipher the biological functions of lncRNA-JADE. To address them, it will be of great interest to further examine Jade1-independent targets and functions of lncRNA-JADE. Based on that, physiological roles of lncRNA-JADE in normal and tumour tissues can be further studied to establish a correlation between aberrant expression lncRNA-JADE and human cancer prognosis.
Materials and methods
Cell lines, antibodies and western blot analysis
293T, NIH3T3 and MCF7 cell lines were obtained from the American Type Culture Collection. 4T1-luciferase cell line was kindly provided by Dr Fidler at the University of Texas MD Anderson Cancer Center. Cells were cultured and stored according to the supplier’s instructions. Once resuscitated, cell lines are routinely authenticated through cell morphology monitoring, growth curve analysis, species verification by isoenzymology and karyotyping, identity verification using short tandem repeat profiling analysis, and contamination checks. Primary Atm+/+ and Atm−/− MEFs were harvested and cultured as previously described (Zhang et al, 2011b). Western blot analyses were performed by standard methods described previously (Zhang et al, 2011a). Anti-Jade1 antibody (#NBP1-47280) was purchased from Novus Biologicals. Anti-histone H4 (#ab7311), anti-α-tubulin (#ab7291) and anti-histone H4K8Ac (#ab15823) antibodies were purchased from Abcam. Anti-Brca1 (#9010) antibody was purchased from Cell Signaling. Anti-acetylated histone H4 (#06-866), anti-histone H4K5Ac (#39170) and H4K12Ac (#39165) antibodies were purchased from Active Motif. Anti-P53 DO-1 (#sc-126), anti-β-actin (#1616), HRP-anti-goat IgG (#2020), HRP-anti-rabbit IgG (#2054) and HRP-anti-mouse IgG (#2055) antibodies were purchased from Santa Cruz.
RNA isolation, qRT–PCR, and lncRNA microarray
Total RNA was isolated using the Trizol reagent according to the manufacturer’s instructions (Invitrogen) and was then reverse transcribed with an iScript cDNA Synthesis Kit (Bio-Rad). The resulting cDNA was used for qPCR using the Cybergreen (Bio-Rad) with gene-specific primers and data were normalized to endogeneous control, β-actin or GAPDH. Breast Cancer and Normal Tissue cDNA Arrays were purchased from Origene. Real-time PCR and data collection were performed on a realplex2 machine (Eppendorf). The RT–PCR primers are shown below:
Mouse lncRNA-JADE: 5′-TATGTGCAAGATGAGAGCCAGCCA-3′ and 5′-TTCACTGTGGACCCGGTGATTAAC-3′; Mouse Jade1: 5′-AGCTTGTCAACGACATGGTCC-3′ and 5′-AGGCTTTCGATCTTCAGGTCT-3′; Human lncRNA-JADE: 5′-AACCTGAAGAGAGCGT-3′ and 5′-AGGAAGCGAATGCTCA-3′; Human Jade1: 5′-AGTTGGGCTATGTGGACATCC-3′ and 5′-CGTAGCATCGCTGCTCAAATTC-3′; Human p21: 5′-GCAGACCAGCATGACAGATTT-3′ and 5′-GGATTAGGGCTTCCTCTTGGA-3′; Human IRF-1: 5′-TCTTAGCATCTCGGCTGGACTTC-3′ and 5′-CGATACAAAGCAGGGGAAAAGG-3′.
For lncRNA microarray analyses, RNA was extracted from a pair of primary Atm+/+ and Atm−/− MEFs treated with NCS (500 ng/ml) for 4, 8, and 24 h. RNA samples were subjected to mouse genome-wide lncRNA microarray analysis at ArrayStar, Rockville, MD. Differentially expressed lncRNAs with statistical significance were identified. The thresholds we used to screen upregulated or downregulated lncRNAs are fold change (≥1.5 or ≤0.5) and P-value<0.05. The microarray data from this publication have been submitted to the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) and assigned the accession number GSE43425.
Human lncRNA-JADE cloning using RACE
Total RNA extracted from human U2OS and MCF7 cells was used to generate RACE-ready cDNA using the SMARTer RACE cDNA Amplification Kit (Clontech) following the manufacturer’s protocol. cDNA ends were amplified with universal primer mix and gene-specific primers. In case the primary PCR failed to generate distinct DNA fragments, we performed a ‘nested’ PCR with the nested universal primer and the nested gene-specific primers. Primer sequences were as follows (5′ to 3′):
GSP1 (3′ RACE): AGCGAATGCTCAGGGGAAGCTTTAA
GSP2 (5′ RACE): AACTAACTCTCTGAAGTTGGCTGGCTTC
NGSP1 (3′ RACE): CTACTTACAGAAGTTGAATTGCAAGAGC
NGSP2 (5′ RACE): TCACACTAGACCTGGTGATTAATGGTAA the resulting DNA fragments were cloned and sequenced (Lonestar Labs). Sequence data of the full-length human lncRNA-JADE can be found in the GenBank/EMBL database (accession # KC469579).
DNA damaging agents, ATM inhibitor, and NF-κB inhibitor
Cells were treated with neocarzinostatin (NCS, #N9162, Sigma-Aldrich), etoposide (#1383, Sigma-Aldrich) or bleomycin (#2434, Sigma-Aldrich) at indicated concentrations and harvested at indicated time points after treatment for RNA and protein analyses. To examine the role of ATM and NF-κB, cells were pretreated with 10 μM ATM kinase inhibitor CGK733 (no. 118501, Calbiochem) or 5 μM NF-κB inhibitor BAY11-7085 (no. B5681, Sigma-Aldrich) prior to DNA damage treatment.
Lentiviral vectors expressing Jade1 shRNAs, lncRNA-JADE or its shRNAs
Jade1 shRNA-pGIPZ lentiviral plasmids were obtained from the MD Anderson shRNA and ORFeome Core Facility (originally from Open Biosystem). Human lncRNA-JADE targeting shRNA1: 5′-CTGAAGAGAGCGTGAGATA-3′, shRNA2: 5′-GTAAGTAGAAAGAAATTTA-3′, mouse lncRNA-JADE targeting shRNA1: 5′-CAGCCAACTTCAGAGAGTG-3′, shRNA2: 5′-CTGAAGCATTCAAATAGGA-3′, and negative control shRNA 5′-TTCTCCGAACGTG TCACGT-3′ were designed and cloned into a lentiviral vector pLKO.1 or pSIH-H1-Puro shRNA. Human lncRNA-JADE gene was cloned into a pCDN-CMV-MCS-EF1-Puro or pCDN-CMV-MCS-EF1-copGFP vector.
Luciferase reporter assay
Human U2OS cells were transfected with 200 ng of firefly luciferase reporter plasmid DNA (Control, lncRNA-JADE or Jade1 promoter-driven luciferase construct) and 0.6 ng of Renilla luciferase reporter plasmid pRL-TK (Promega) per well in 24-well dish. Cells were lysed 24 h after transfection and luminescence was quantified in a GloMax 20/20n Luminometer (Promega). Experiments were performed in triplicates. The firefly luciferase activity values were normalized to the Renilla luciferase activity values that reflect transfection efficiency. Data are presented as mean values (±s.d.).
RNA RIP assay
RIP assay was performed as described previously (Zhang et al, 2011b). Briefly, cells were crosslinked for 20 min with 1% formaldehyde, and cell pellets were resuspended in buffer B (1% SDS, 10 mM EDTA, 50 mM Tris–HCl (pH 8.1), 1 × protease inhibitor, 50 U/ml RNase inhibitor). Incubated 10 min in ice, the pellets were disrupted by sonication, and the lysates were cleared and subjected to immunoprecipitation with control IgG or anti-Brca1 antibody, followed by stringent washing, elution, and reversal of crosslinking. The RNA was resuspended in 20 μl of TE buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 50 U/ml RNase inhibitor) and incubated with DNase I for 30 min at 37°C to remove any remaining DNA. After extraction with phenol:chloroform:isoamyl alcohol (25:24:1), RNA was precipitated with ethanol and dissolved in 20 μl of DEPC-treated water. RNA (5 μl) was used for the cDNA synthesis reaction. Quantitative PCRs were then performed on real-time PCR machine (Realplex2, Eppendorf). BRCA1 Primer 1F: 5′-AGGCCCCTTTAAACGCAGAA-3′; 1R: 5′-TCAGGCTGCACTTACCCACTT-3′ Primer 2F: 5′-TGGCAGCCAGAGAGAGATTT-3′; 2R: 5′-CCTAACCTCAGGCAATCCAC-3′.
Cell proliferation and apoptosis assays
In cell proliferation assay, cells were seeded at a concentration of 1000 cells/well in 96-well plates. Cell numbers were quantified using reagent WST-1 (no. 05015944001, Roche) at the indicated time according to the manufacturer’s instructions. In apoptosis assay, cells were seeded in 8- or 16-well chamber slides and treated with NCS. Apoptosis was measured using the DeadEnd Fluorometric Tunel Kit (Promega) according to the manufacturer’s instructions.
Monolayer colony-formation assay and soft agar colony-formation assay
For monolayer colony-formation assay, 1000 MCF10A cells stably expressing oncogenes were plated in 35 mm2 plates in triplicate and allowed to grow in appropriate culture medium for 2–3 weeks. Fresh media were supplied every 3 days. Colonies were stained with crystal violet dye after formaldehyde fixation. The soft agar colony-formation assay was performed as previously described (Zhang et al, 2011a). Cells were transduced with lentiviral oncogenes, and plated in 0.3% top agarose in 35 mm2 plates and cultured for 3 weeks. Colonies were counted under a light microscope.
4T1 xenografted tumour growth
Six- to eight-week-old female Balb/c SCID mice were purchased from Jackson Laboratories and housed in pathogen-free conditions. All studies were approved and supervised by the Institutional Animal Care and Use Committee at MD Anderson Cancer Center. After SCID mice were anaesthetized and the skin was incised, control and lncRNA-JADE-silenced 4T1-luciferase cells (1 × 106) in 50 μl Hanks solution were orthotopically injected into mammary fat pads using a 1-ml Hamilton microlitre syringe, and then the incision was closed using wound clips. Five mice were used in each group. Tumour size was measured once a week up to 24 days Mice tumours were also monitored by the IVIS system after luciferin injection for 15 min. Mice were euthanized when they met the institutional euthanasia criteria for tumour size and overall health condition.
LncRNA in situ hybridization
Breast cancer tissue microarray (no. BC081115) was purchased from Biomax, including 100 breast tumour samples and 10 control breast tissue samples. The tissue microarray was dewaxed in xylenes, and rehydrated through an ethanol dilution series. Tissue sections were digested with 15 μg/ml proteinase K for 20 min at RT, were then loaded onto Ventana Discovery Ultra for in situ hybridization analysis. The tissue slides were incubated with double-DIG labelled mercury lncRNA probe (Exiqon) for 2 h at 55°C, and signals were detected with a polyclonal anti-DIG antibody and alkaline phosphatase-conjugated second antibody (Ventana) using NBT-BCIP as the substrate. The double-DIG labelled control U6 snRNA probe was also purchased from Exiqon.
Supplementary Material
Acknowledgments
We thank Li Huang and Dr Bar-Eli for providing us with lentiviral vectors and helping produce viruses. We also thank Fanhao Zhang for his technical assistance on mouse mammary gland xenografts. This research was supported by grants to XL from the National Institutes of Health (R01CA136549), the American Cancer Society (119135-RSG-10-185-10-TBE), and the University of Texas Star Plus award. CH was supported in part by the Odyssey award and The Cockrell Foundation Award for Scientific Achievement at The University of Texas MD Anderson Cancer Center.
Author contributions: XH, XZ, and XL initiated the project. GW, XH, and XL designed the experiments. XH, GW, YL, and XZ performed the experiments. GAC, AKS, CH, and XL analysed the data. XL wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aguilo F, Zhou MM, Walsh MJ (2011) Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res 71: 5365–5369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed KM, Li JJ (2007) ATM-NF-kappaB connection as a target for tumor radiosensitization. Curr Cancer Drug Targets 7: 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard S, Masson JY, Cote J (2004) Chromatin remodeling and the maintenance of genome integrity. Biochim Biophys Acta 1677: 158–164 [DOI] [PubMed] [Google Scholar]
- Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J (1999) NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas J (2007) DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 19: 238–245 [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J et al. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444: 633–637 [DOI] [PubMed] [Google Scholar]
- Bellucci M, Agostini F, Masin M, Tartaglia GG (2011) Predicting protein associations with long noncoding RNAs. Nat Methods 8: 444–445 [DOI] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43: 559–599 [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le BP, Wutz A, Heard E (2006) A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20: 2223–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitalia VC, Foy RL, Bachschmid MM, Zeng L, Panchenko MV, Zhou MI, Bharti A, Seldin DC, Lecker SH, Dominguez I, Cohen HT (2008) Jade-1 inhibits Wnt signalling by ubiquitylating beta-catenin and mediates Wnt pathway inhibition by pVHL. Nat Cell Biol 10: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Wang Y, Qin J, Elledge SJ (1999) Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286: 1162–1166 [DOI] [PubMed] [Google Scholar]
- Crowe DL, Lee MK (2006) New role for nuclear hormone receptors and coactivators in regulation of BRCA1-mediated DNA repair in breast cancer cell lines. Breast Cancer Res 8: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnington MS, Santibanez KM, Mayosi BM, Burn J, Keavney B (2010) Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet 6: e1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De SF, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G (2010) A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foy RL, Song IY, Chitalia VC, Cohen HT, Saksouk N, Cayrou C, Vaziri C, Cote J, Panchenko MV (2008) Role of Jade-1 in the histone acetyltransferase (HAT) HBO1 complex. J Biol Chem 283: 28817–28826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Harada H, Kondo T, Ogawa S, Tamura T, Kitagawa M, Tanaka N, Lamphier MS, Hirai H, Taniguchi T (1994) Accelerated exon skipping of IRF-1 mRNA in human myelodysplasia/leukemia; a possible mechanism of tumor suppressor inactivation. Oncogene 9: 3313–3320 [PubMed] [Google Scholar]
- Havasi A, Haegele JA, Gall JM, Blackmon S, Ichimura T, Bonegio RG, Panchenko MV (2013) Histone acetyl transferase (HAT) HBO1 and JADE1 in epithelial cell regeneration. Am J Pathol 182: 152–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van d Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y et al. (2011) Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43: 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci I, Sazzini M, Garagnani P, Ferrari A, Boattini A, Lonetti A, Papayannidis C, Mantovani V, Marasco E, Ottaviani E, Soverini S, Girelli D, Luiselli D, Vignetti M, Baccarani M, Martinelli G (2011) A polymorphism in the chromosome 9p21 ANRIL locus is associated to Philadelphia positive acute lymphoblastic leukemia. Leuk Res 35: 1052–1059 [DOI] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L (2010) Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16: 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa R, Kastan MB (2005) The ATM-dependent DNA damage signaling pathway. Cold Spring Harb Symp Quant Biol 70: 99–109 [DOI] [PubMed] [Google Scholar]
- Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M (2011) Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 71: 6320–6326 [DOI] [PubMed] [Google Scholar]
- Kumar R, Horikoshi N, Singh M, Gupta A, Misra HS, Albuquerque K, Hunt CR, Pandita TK (2012) Chromatin modifications and the DNA damage response to ionizing radiation. Front Oncol 2: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT (2012) Epigenetic regulation by long noncoding RNAs. Science 338: 1435–1439 [DOI] [PubMed] [Google Scholar]
- Li X, Corsa CA, Pan PW, Wu L, Ferguson D, Yu X, Min J, Dou Y (2010) MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol 30: 5335–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lu X (2012) Non-coding RNAs in DNA damage response. Am J Cancer Res 2: 658–675 [PMC free article] [PubMed] [Google Scholar]
- Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, Mayne ST, Risch HA, Mu L, Canuto EM, Gregori G, Benedetto C, Yu H (2012) Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat 136: 875–883 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Melo CA, Drost J, Wijchers PJ, van de Werken H, de WE, Oude Vrielink JA, Elkon R, Melo SA, Leveille N, Kalluri R, de LW, Agami R (2013) eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49: 524–535 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S (2011) Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res 21: 116–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C (2010) Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137: 2493–2499 [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C (2008) Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Pao GM, Janknecht R, Ruffner H, Hunter T, Verma IM (2000) CBP/p300 interact with and function as transcriptional coactivators of BRCA1. Proc Natl Acad Sci USA 97: 1020–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I (2007) Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 67: 3963–3969 [DOI] [PubMed] [Google Scholar]
- Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK (2011) RNA interference in the clinic: challenges and future directions. Nat Rev Cancer 11: 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W (2009) The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 136: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CM, Hale PT, Carlin JM (2006) NF-kappa B activation contributes to indoleamine dioxygenase transcriptional synergy induced by IFN-gamma and tumor necrosis factor-alpha. Cytokine 35: 53–61 [DOI] [PubMed] [Google Scholar]
- Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Cote V, Yang XJ, Gozani O, Kutateladze TG, Cote J (2009) HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell 33: 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer BC (1995) Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem 227: 255–273 [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M (2007) Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Suganuma T, Workman JL (2011) Signals and combinatorial functions of histone modifications. Annu Rev Biochem 80: 473–499 [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van LM, Orkin SH, Peters AH (2008) Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell 15: 668–679 [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 6: 909–923 [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouanacou E, Tweedie S, Wilson V (2003) Identification of Jade1, a gene encoding a PHD zinc finger protein, in a gene trap mutagenesis screen for genes involved in anteroposterior axis development. Mol Cell Biol 23: 8553–8562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Mathur R, Hu X, Liu Y, Zhang X, Peng G, Lu X (2013) Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal 25: 1086–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan G, Mathur R, Hu X, Zhang X, Lu X (2011) miRNA response to DNA damage. Trends Biochem Sci 36: 478–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD (2011a) Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY (2011b) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WZ, Liu HO, Wu YH, Hong Y, Yang JW, Liu YH, Wu WB, Zhou L, Sun LL, Xu JJ, Yun XJ, Gu JX (2010) Estrogen receptor alpha (ERalpha) mediates 17beta-estradiol (E2)-activated expression of HBO1. J Exp Clin Cancer Res 29: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S (2006) Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 311: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG (2011) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT (2007) Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129: 693–706 [DOI] [PubMed] [Google Scholar]
- Zhang X, Berger FG, Yang J, Lu X (2011a) USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J 30: 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wan G, Berger FG, He X, Lu X (2011b) The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell 41: 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou MI, Wang H, Foy RL, Ross JJ, Cohen HT (2004) Tumor suppressor von Hippel-Lindau (VHL) stabilization of Jade-1 protein occurs through plant homeodomains and is VHL mutation dependent. Cancer Res 64: 1278–1286 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.