Abstract
We have developed cross-genotype and genotype-specific quantitative reverse-transcription PCR (qRT-PCR) assays to detect and quantify the number of parasites, transmission stages (gametocytes) and male gametocytes in blood stage Plasmodium chabaudi infections. Our cross-genotype assays are reliable, repeatable and generate counts that correlate strongly (R2s > 90%) with counts expected from blood smears. Our genotype-specific assays can distinguish and quantify different stages of genetically distinct parasite clones (genotypes) in mixed infections and are as sensitive as our cross-genotype assays. Using these assays we show that gametocyte density and gametocyte sex ratios vary during infections for two genetically distinct parasite lines (genotypes) and present the first data to reveal how sex ratio is affected when each genotype experiences competition in mixed-genotype infections. Successful infection of mosquito vectors depends on both gametocyte density and their sex ratio and we discuss the implications of competition in genetically diverse infections for transmission success.
Keywords: Plasmodium chabaudi, Gametocyte, Sex ratio, qRT-PCR
1. Introduction
The application of molecular methods to detect and quantify different life stages of malaria (Plasmodium) parasites has provided novel insights into infection dynamics of relevance to evolutionary biology, epidemiology and medical disciplines [1]. Unlike traditional microscopy, molecular methods, including quantitative PCR (qPCR) and nucleic acid sequence-based amplification (QT-NASBA), are sensitive enough to reveal subtle variations in infection dynamics and detect parasites infections considered sub-patent by microscopy [2–6]. Furthermore, traditional microscopy cannot be used to follow the progress of individual genotypes (clones) in mixed infections that contain multiple distinct genotypes. Genotype-specific quantitative reverse-transcription PCR (qRT-PCR) methods have been developed for the rodent malaria, Plasmodium chabaudi, to quantify the number of parasites belonging to different genotypes within the same infection [7–9]. Application of these methods has revealed that genotypes of P. chabaudi suffer from competition with con-specific genotypes and these effects can be linked to the virulence of co-infecting genotypes [7–11]. Despite genetically diverse infections being the rule in nature, we are yet to examine how competition in genetically diverse infections influences sex ratios produced by individual genotypes [12].
To transmit to vectors, malaria and related parasites produce gametocytes, and these stages are either male or female. The gametocytes differentiate into gametes as soon as they are taken up in a vector’s blood meal. Each male gametocyte can produce up to eight male gametes and each female gametocyte differentiates into a single female gamete. Male gametes locate and fertilise female gametes and the resulting zygotes undergo several development stages in their vectors before being ready to re-infect new vertebrate hosts [13]. Successful transmission to vectors depends on the number of gametocytes produced and the ratio of males to females (sex ratio: proportion of gametocytes that are male) [14–16]. While gametocytes can now be quantified by PCR, their sex is traditionally assigned by microscopy. This method of sexing has been shown to overestimate the proportion of female gametocytes, as maturing male gametocytes resemble females and can be incorrectly sexed [17,18]. In addition, gametocyte densities are often extremely low which results in variable and inaccurate sex ratio estimates [19]. Furthermore, microscopy cannot be used to distinguish between male and female gametocytes produced by different genotypes in mixed-genotype infections.
To overcome these issues we developed cross-genotype quantitative reverse-transcription PCR assays for following gametocyte density, male gametocyte density and asexual parasite density throughout P. chabaudi infections. We used information derived from recent proteome analysis of male and female gametocytes in the closely related rodent malaria parasite Plasmodium berghei, which identified a large cohort of proteins which were pre-synthesised exclusively by male or female gametocytes [20] in preparation for a swift differentiation into gametes. We then extended these assays to be genotype-specific so we could follow the gametocyte density and sex ratio of individual genotypes in mixed-genotype infections. We used our assays to determine how gametocyte production and sex ratio vary throughout infections in two genetically distinct lines (genotypes) and how these traits are influenced by competition in mixed-genotype infections comprising both of these genotypes.
2. Materials and methods
2.1. Parasites and hosts
In all experiments male MF1 mice (in house supplier, University of Edinburgh) were infected with clonal P. chabaudi lines AS, AJ or CR (WHO Registry of Standard Malaria Parasites, The University of Edinburgh). All mice were 6–8 weeks old and received an intra-peritoneal inoculation of 106 parasitized red blood cells (RBC) in a 0.1 μl dose as previously described [21]. Mice were housed at 21 °C with a 12 h light cycle, and maintained on a diet of SDS41B food pellets (Harlan Scientific, UK) and 0.05% PABA supplemented drinking water to enhance parasite growth. Mice were sampled in the morning when the circulating parasites were in ring or early trophozoite stages. Blood samples were taken via a tail snip. Thin blood smears were made and allowed to air dry before being fixed in methanol and stained for 20 min in Giemsa. For DNA extraction, 5 μl of blood was taken from each mouse and added to 200 μl of citrate saline (0.85% NaCl, 1.5% tri-sodium citrate) on ice. Blood samples for DNA were subsequently pelleted by centrifugation and the supernatant removed before being stored at −80 °C until required. For RNA extraction 10 μl of blood was taken from each mouse and added to 90 μl of a 1:2 mix containing 30 μl Ca/Mg free RNase/DNase free phosphate buffered saline (PBS) (Gibco BRL) and 60 μl of 2× Nucleic Acid Purification Lysis Solution (Applied Biosystems) on ice. Blood samples for RNA were immediately mixed by pipetting to allow complete lysis to occur and subsequently stored at −80 °C until required.
2.2. Identification of P. chabaudi target genes
A recent study [20] reported an effective method for the separation and purification of both male and female gametocyte populations. This was based the isolation of subpopulations of P. berghei parasite lines via flow cytometry. Mass spectrometry of these purified populations recorded the male and female gametocyte specific proteomes and established an accurate gene expression profile for P. berghei gametocytes. To design quantitative reverse-transcription PCR assays capable of detecting total gametocyte numbers (the combined numbers of both male and female gametocytes, referred to from now on as “common gametocytes”) and the sex ratio of gametocytes (defined as the proportion of male gametocytes in the total gametocyte population), we examined the P. berghei gametocyte proteomes [20]. From this large cohort of proteins we chose to use the common gametocyte gene PB000198.00.0 and the male gametocyte gene PB000791.00.0 as the target genes for developing our assays.
The DNA sequences of P. chabaudi AS homologues of PB000198.00.0 and PB000791.00.0 were obtained by BLAST searches of the P. chabaudi genome in PlasmoDB (http://www.plasmodb.org/plasmo/home.jsp). The genebank accession number for the P. chabaudi homologue of PB00198.00.0 is PC302249.00.0 (called common gametocyte gene 1/CG1 from now on in the text). Both PB00198.00.0 and PC302249.00.0 encode a conserved hypothetical protein and are closely related to homologues in Plasmodium falicparum 3D7 (PF07 0089), Plasmodium vivax (Pv096260) and Plasmodium yoelii (PY02842). The genebank accession number for the P. chabaudi homologue of PB000791.00.0 is PC000513.00 (called male gametocyte gene 1/MG1 from now on in the text). Both PB000791.00.0 and PC000513.00 encode a putative dynein heavy chain protein likely to be involved in the formation of the flagellum of male gametes during exflagellation, and are closely related to a homologue in P. falciparum (PFI0260c).
2.3. Amplification and sequencing of CG1 and MG1 genes in AS, AJ and CR P. chabaudi parasite lines
In Experiment 1, we infected mice with P. chabaudi lines in order to harvest parasite DNA that could be used analyse the sequence of the CG1 and MG1 genes across P. chabaudi parasite lines. Two mice per group were infected with P. chabaudi AS, AJ or CR parasites on day 0, and 10 μl of blood collected following the establishment of a patent parasitaemia 5 days post inoculation. We chose AS and AJ as we intended to use these two lines in competition our experiments. We sequenced CR because it normally produces higher densities of gametocytes than AS or AJ, and is more useful for testing the accuracy of our gametocyte specific assays. For general DNA extraction, 10 μl of blood was added to 200 μl of ice cold citrate saline and DNA then extracted as previously described [22]. A 659 base pair (bp) region of the CG1 gene was amplified from 100 ng of AS, AJ and CR template DNA using the PCR primers CG1 SEQFP.1 (5′-CCC TGA AAT GAA AAA TGC ACC ATA TC-3′) and CG1 SEQRP.1 (5′-GCT CGA TAC AAA CAT CGA TAA GTT GTT TAC-3′). A 924 bp region of the MG1 gene was amplified from AS, AJ and CR template DNA using the PCR primers MG1 SEQFP.1 (5′-CAT CAT TAC ATC AAT TTC TTG AGC AAT TTG ATT TAT C-3′) and MG1 SEQRP.1 (5′-GTT TCT ATT TAA TAC TTC ATC TAC AAA TTT GTT TGC ACA C-3′). All oligonucleotide primers were supplied by MWG Biotech UK. PCR amplification was performed as previously described [22] using the following amplification conditions: denaturation at 94 °C/300 s, 35 cycles of amplification at [94 °C/30 s, 55 °C/30 s, 68 °C/180 s] followed by a terminal extension at 72 °C/300 s. After confirmation by gel electrophoresis that a single DNA product of expected size had been amplified in each reaction, PCR products were purified using a QIAquick PCR purification kit (Cat. No. 28104, Quiagen GmbH) and 20 ng of purified template used for sequencing. DNA sequencing was performed by the School of Biological Sciences DNA Sequencing service (Ashworth Laboratories, The University of Edinburgh, UK), using the relevant CG1SEQFP.1, CG1SEQRP.1, MG1SEQFP.1, MG1SEQRP.1 oligonucleotide primers. DNA sequences obtained were aligned online using ClustalW at http://align.genome.jp/. Sequence polymorphisms between parasite lines were identified and used as the basis for designing primer sets which were either clone specific or bound universally across AJ, AS and CR clones.
2.4. Primer and probe sets for used quantitative real-time PCR
Universal and strain-specific PCR primers and common minor groove-binder (MGB) probes targeting the P. chabaudi CG1 and MG1 genes were designed using Primer Express software (Applied Biosystems). The sequence of the CG1 MGB probe used for all CG1 qRT-PCR assays is FAM-CAA AGT AGA CAA TGC ACC AAT-MGB. The amplicon length for the CG1 universal assay is 157 base pairs (bp) and is generated using the CG1 universal forward primer (CG1 UniFP: 5′-GCC CGA ATC ATT ATG TTT TAC TAT AAT GG-3′) and the CG1 universal reverse primer (CG1 UniRP: 5′-AGA TAA AAT TGC ATA TAT TCC TAA AGT ACT ATC TAC TTC-3′). The AJ strain-specific amplicon length is 151 bp and is generated using the CG1 AJ forward primer (CG1 AJFP: 5′-CCG AAT CAT TAT GTT TTA CTA TAA TGG GAA TA-3′) and CG1 AJ reverse primer (CG1 AJRP: 5′-AAA ATT GCA TAT ATT CCT AAA GTA CTA TCT ACT TCA TTA T-3′). The AS strain-specific amplicon length is 149 bp and is generated using the CG1 AS forward primer (CG1 ASFP: 5′-CGA ATC ATT ATG TTT TAC TAT AAT GGG AAT G-3′) and the CG1 AS reverse primer (CG1 ASRP: 5′-AAA TTG CAT ATA TTC CTA AAG TAC TAT CTA CTT CAT TAC-3′).
It should be noted that the strain-specific CG1 primers only differ by two nucleotides. The 3′ terminal nucleotide highlighted with bold text represents the site of a true polymorphism between AS and AJ target sequence. The nucleotide underlined in each primer represents a single mismatched nucleotide introduced into the primers to increase the level of binding disparity between the primer and the two strains. Thus, the AS strain-specific CG1 forward and reverse primers each have a single nucleotide mismatch with the AS CG1 gene, but a two nucleotide mismatch with the AJ CG1 gene, and vice versa for the AJ strain-specific primers. While very small, this is sufficient to allow strain-specific detection of parasites and gametocytes, but prevented cross-strain detection.
The sequence of the MG1 MGB probe used for all MG1 qRT-PCR assays is FAM-CAC TTG AAT CTA AAC GCT CATMGB. The amplicon length for the MG1 universal assay is 150 bp and is generated using the MG1 universal forward primer (MG1 UniFP: 5′-TGA TGT ATT AGA TAT TGA AAA TAG AGA CAT TCC T-3′) and the MG1 universal reverse primer (MG1 UniRP: 5′-TCA TCT ACA AAT TTG TTT GCA CAC AAT A-3′). The AJ strain-specific amplicon length is 131 bp and is generated using the MG1 UniFP and MG1 AJ reverse primer (MG1 AJRP: 5′-CAC ACA ATA TTG TTC TGT CTT CTC TTA AAA AG-3′). The AS strain-specific amplicon length is 131 bp and is generated using MG1 UniFP and the MG1 AS reverse primer (MG1 ASRP: 5′-CAC ACA ATA TCG TTC TAT CTT CTC TCA AAA AT-3′). All MGB probes were supplied by Applied Biosystems, and all oligonucleotide primers supplied by MWG biotech, UK. It should be noted that the strain-specific MG1 assays use the universal forward MG1 primer and strain-specific MG1 reverse primers. These strain-specific MG1 reverse primers differ by five oligonucleotides. Four of these nucleotide differences, highlighted in bold text, represent the sites of true polymorphisms between AS and AJ target sequence. The single underlined nucleotide in each primer represents a single mismatched nucleotide introduced to increase the level of binding disparity between the primer and the two strains, just as was done with the strain-specific CG1 primers. Again the introduction of the single nucleotide mismatch is sufficient to allow strain-specific detection of male gametocytes, but prevented cross-strain detection.
2.5. Preparation of DNA and cDNA from blood samples
DNA extraction was performed on 10 μl blood samples previously added to citrate saline and stored at −80 °C. The pelleted blood was thawed on ice and DNA extraction performed using the BloodPrep Kit (Applied Biosystems) on the ABI Prism 6100 Nucleic Acid PrepStation, according to manufacturer’s instructions. DNA was eluted in a total volume of 200 μl and stored at −80 °C until used for PCR quantification. RNA extraction was performed on 5 μl blood samples that had previously been lysed in 1× Nucleic Acid Purification Lysis Solution (Applied Biosystems). After thawing on ice total RNA extraction was performed on the ABI Prism 6100 Nucleic Acid PrepStation, using the RNA blood–DNA method and reagents (Applied Biosystems). RNA was eluted in a total volume of 100 μl and converted into cDNA using the Applied Biosytems high-capacity cDNA archive kit. Briefly, to generate cDNA, a 100 μl reverse-transcription reaction was performed using 50 μl of eluted RNA and 50 μl of reverse-transcription mix (5 μl 10× RT buffer, 2 μl 25× dNTP mixture, 5 μl 10× random primers, 5 U/μl multiscribe reverse transcriptase and 10.5 μl RNase free water). The cDNA reaction was performed at 25 °C for 10 min, 37 °C for 2 h. All cDNA samples were then store at −80 °C until used for PCR quantification.
2.6. Quantitative RT-PCR
To measure the total density of P. chabaudi asexual parasites in blood samples, qRT-PCR was performed on DNA using the CG1 universal or strain-specific primer pairs. To measure total gametocyte densities (combined count of male and female gametocytes) in blood samples, qRT-PCR was performed on cDNA using the CG1 universal or strain-specific primer pairs. To measure the density of male gametocytes in blood samples, qRT-PCR was also performed on cDNA, but this time using the MG1 universal or strain-specific primer pairs.
All quantitative qRT-PCR reactions were performed in a final volume of 25μl. All reactions used 7μl of cDNA or DNA, 250nM of MGB probe, 900nM of forward and reverse primers and a 1× final concentration of Taqman Universal PCR Master Mix (Applied Biosystems). All qRT-PCR reactions were run on an ABI Prism 7000 sequence detection system with an initial denaturation step of 50 °C/2 min and 95 °C/10 min, followed by 45 cycles of denaturation at 95 °C/15 s followed by annealing/extension at 60 °C/1 min. Standard samples in a dilution series were included in all qRT-PCR runs. Standard curves were calibrated for each qRT-PCR run by regressing the threshold cycle values obtained by qRT-PCR against the relative densities of asexual parasites, total gametocytes or male gametocytes obtained from smear counts. Absolute quantification of experimental samples was then achieved by comparing the threshold cycle numbers against this standard curve. Each qRT-PCR run included DNA or cDNA samples isolated from uninfected blood samples as a negative control.
2.7. Determining the minimum sensitivity and strain-specificity of qRT-PCR assays
In Experiment 2, we determined the sensitivity of the CG1 and MG1 qRT-PCR assays. To elevate gametocyte densities obtained during P. chabaudi infection, we pre-treated six mice with phenylhydrazine hydrochloride (PHZ) (Sigma) to induce a hyper-reticulocytosis, which has previously been shown to increase the production of gametocytes by P. chabaudi [23] (100 μl of a 25 mg/ml solution injected intraperitoneally). Two mice per group were then infected with P. chabaudi AJ, AS or CR parasites 48 h post PHZ treatment. The total parasitaemias and gametocyte densities of individual mice were followed daily by taking thin blood smears early in the morning. When individual mice developed a gametocytaemia above 1% of circulating red blood cells, mice were smeared again and a coulter count of RBC density made in triplicate. Following this approximately, 500 μl of blood was obtained from euthanased mice via cardiac puncture and aliquoted into ice cold citrate saline and 1× Nucleic Acid Purification Lysis Solution, for subsequent purification of DNA and RNA respectively. New thin blood smears from these infected mice were examined and the proportion red blood cells infected with asexual parasites, total gametocytes and male gametocytes were determined by microscopy. Serial dilutions of DNA and cDNA from these mice were then prepared over 8 orders of magnitude. These serial dilutions were analysed by qRT-PCR using the universal and strain-specific CG1 and MG1 assays. The highest serial dilution for which a positive threshold cycle was achieved was recorded. This was then used to calculate the minimum sensitivity of the CG1 and MG1 assays. To confirm that the strain-specific qRT-PCR assays only detected DNA or cDNA generated from their target P. chabaudi strain, AS and AJ strain-specific CG1 and MG1 assays were run against DNA and cDNA isolated from all three P. chabaudi strains. The AJ, AS and CR DNA and cDNA standards generated in this experiment were used to calibrate all subsequent qRT-PCR assays in this study.
2.8. Determining correlations between slide and qRT-PCR generated data
Having determined the minimum sensitivity of the qRT-PCR assays, in Experiment 3 we determined how well the parasite densities generated by qRT-PCR correlated with those generated by slide counting. We used genotype CR as it produces high densities of gametocytes throughout the course of an infection to maximise the accuracy of our slide counts. We pre-treated a group of 5 mice with PHZ as described in Section 2.7, and infected these mice with P. chabaudi CR parasites 48 h later. RBC density coulter counts, thin blood smears and blood samples for DNA and RNA were collected from individual mice every morning on days 3–8 post infection. The total densities of asexual parasites, total gametocytes and male gametocytes were determined for each individual, on each day by both microscopy and qRT-PCR.
2.9. Comparing cross- and genotype-specific qRT-PCR assays
In Experiment 4, we investigated whether parasite density data generated using universal primer sets and strain-specific sets were equivalent. To achieve this we infected two groups of mice (n = 5) with either AJ or AS P. chabaudi parasites. Infected mice were sampled on days 5–12 post infection. Parasite densities for AJ infected mice were determined using the universal and AJ strain-specific qRT-PCR assays, calibrated with AJ standards. Parasite densities for AS infected mice were determined using the universal and AS strain-specific qRT-PCR assays calibrated with AS standards.
2.10. Investigating the effects of competition on sex ratio and gametocyte density
In Experiment 5, we examined the dynamics of parasite investment in gametocytes and their sex ratio in single- and mixed-genotype infections of P. chabaudi. To do this we infected three groups of mice (n = 10) with either 106 AJ parasites, 106 AS parasites or a combined dose of 106 AJ and 106 AS parasites. Infected mice were sampled on days 5–16 post infection. Red blood cell densities were calculated by daily coulter counts and reticulocyte densities determined by examination of Giemsa stained thin blood smears. Parasite densities of mice infected with either AJ or AS parasites alone were determined by qRTPCR using universal primer sets and both AJ and AS standards. Mice infected with both AJ and AS parasites, had the AJ-specific parasite densities determined by qRT-PCR using AJ-specific primer sets and AJ standards, while the AS-specific parasite densities were determined by qRT-PCR using AS-specific primer sets and AS standards.
2.11. Statistical analysis
We used R version 2.5.0 (The R Foundation for Statistical Computing, 2007, http://www.R-project.org) for all analyses. Statistics are presented from linear regression and linear mixed-effects models. We used linear regression to evaluate and describe the correlations between parasite counts generated by different methods. We used linear mixed-effects models to analyse the patterns of gametocyte production and their sex ratio during single- and mixed-genotype infections. Gametocyte density and sex ratio models included the number of genotypes in infections and day post infection as factors, as well as the interaction between these terms. Covariates known to influence these traits including anaemia, reticulocyte and parasite density were also included in the maximal models. Mixed-effects models account for repeated measures on the same infection by treating each mouse as a ‘random’ effect and overcome problems associated with pseudoreplication in longitudinal analysis of infections [24]. The significance of fixed effects included in the models was evaluated by comparing models using log-likelihood ratio tests following stepwise deletion of the least significant term. Specifically, we compared the change in model deviance, following term deletion, to χ2 distributions with degrees of freedom corresponding to the difference in number of terms in the models. Maximal models were simplified using maximum likelihood techniques, until only significant terms remained in the model (α < 0.05; [25]).
3. Results
3.1. Sensitivity and strain specificity of qRT-PCR assays
The universal total parasite qRT-PCR assay could detect a minimum density of approximately 25 parasites/μl of blood and the AJ and AS-specific assays for measuring total parasites detected approximately two-fold fewer parasites (Table 1). These sensitivities are comparable to a previous study [5], which reported a total parasite detection sensitivity of 740 parasites/μl of blood, using a qRT-PCR assay based on the measurement of MSP-1 gene copies in DNA isolated from P. chabaudi infected blood. When the AS-specific CG1 assay was run against AJ DNA, no parasites were detected in any sample over any dilution series. As expected, when the AJ-specific CG1 assay was run against AS DNA, no parasites were detected in any sample over any dilution series. When the AJ-specific CG1 assay was run against CR DNA, parasites were detected, as CR carries the AJ allele of CG1, but as expected, when the AS-specific CG1 assay was run against CR DNA, no parasites were detected. This confirmed the strain-specificity of these total parasite assays, an observation that was confirmed several times over.
Table 1. Minimum sensitivities of qRT-PCR assays.
| Universal assay mean | AJ-specific assay mean | AS-specific assay mean | |
|---|---|---|---|
| Total parasites | 24.5 (±29.7) | 43.4 (±50.2) | 46.9 (±65.1) |
| Total gametocytes | 0.205 (±0.14) | 1.26 (±1.46) | 1.98 (±2.29) |
| Male gametocytes | 0.054 (±0.04) | 0.031 (±.004) | 0.055 (±0.06) |
The average minimum density of total parasites, total gametocytes or male gametocytes detectable by qRT-PCR assays, per μl of blood, are presented. Standard deviations of the mean values are given in brackets.
The sensitivities of common gametocyte qRT-PCR assays showed a similar pattern. The universal assay was able to detect 0.205(±0.14) gametocytes/μl of blood, while the AJ and AS-specific assays were slightly less sensitive, being able to detect 1.26(±1.46) and 1.98(±2.29) gametocytes/μl of blood respectively (Table 1). These sensitivities are close to those reported for the detection of total gametocytes in P. falciparum infection [4] and P. chabaudi [21] using conventional nested RT-PCR techniques. In contrast the sensitivity of the MG1 assay was approximately the same for both the universal, AJ and AS-strain-specific assays, which could detect a minimum density of approximately 0.046(±0.04) male gametocytes/μl of infected blood (Table 1). When the AS-specific CG1 and MG1 assays were run against AJ cDNA, no gametocytes were detected in any sample over any dilution series. When the AJ-specific CG1 and MG1 assays were run against AS cDNA, no gametocytes were detected in any sample over any dilution series. When the AJ-specific CG1 and MG1 assays were run against CR cDNA, common gametocytes were detected, but not male gametocytes, as CR carries the AJ allele of CG1 and the AS allele of MG1. Conversely, when the AS-specific CG1 and MG1 assays were run against CR cDNA, no common gametocytes were detected, but male gametocytes were. This confirmed the strain-specificity of the common gametocyte and male gametoctye assays. As above, these observations were confirmed several times over.
3.2. Correlations between PCR and slide generated parasite counts
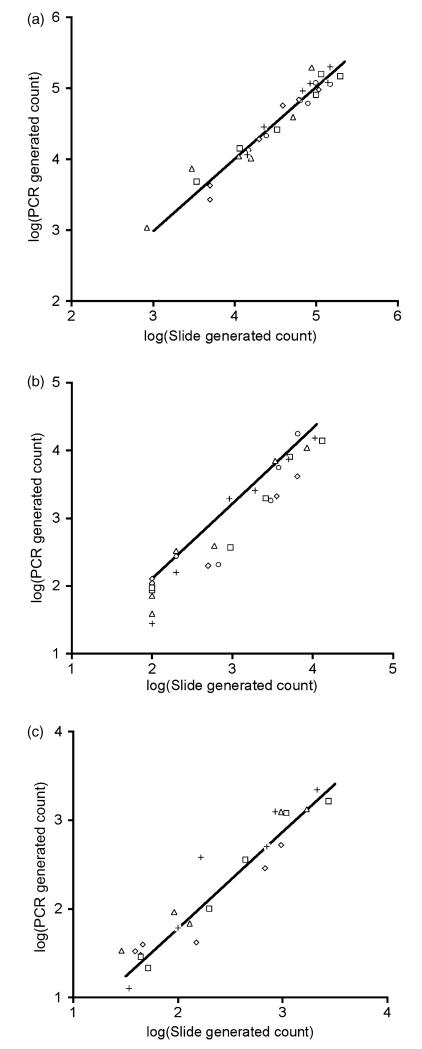
There were significant correlations between slide generated counts using thin blood smears and qRT-PCR counts for total parasites (F1,28 = 442; P < 0.0001; Fig. 1a), total gametocytes (F1,28 = 355; P < 0.0001; Fig. 1b) and male gametocytes (F1,28 = 228; P < 0.0001; Fig. 1c). For all assays the mean R2 within mice was >88% and the mean slope of the relationship between qRT-PCR and slide counts was not significantly different from 1 (Table 2). For total parasites and total gametocytes the mean intercept of the relationship between qRT-PCR and slide counts was not significantly different from 0 but for male gametocytes the intercept was <0, indicating that slightly more male gametocytes were counted from thin smears than qRT-PCRs (Table 2). For all assays, the relationships between slide and PCR counts did not differ significantly across mice (all P > 0.22).
Fig. 1. Correlations between slide and qPCR generated parasite counts.
(a) Total parasites; (b) total gametocytes; (c) male gametocytes. Different symbols represent the five different mice that we measured over 6 days. Fitted lines are the average relationships between slide and qPCR counts for all mice; see Table 2 for details.
Table 2. Within infection correlations between slide and qRT-PCR data.
|
R
2
|
Slope |
Intercept |
||||
|---|---|---|---|---|---|---|
| Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | |
| Total parasites | 0.94 | (0.91, 0.97) | 1.02 | (0.92, 1.11) | −0.06 | (−0.51, 0.39) |
| Total gametocytes | 0.93 | (0.89, 0.97) | 1.11 | (0.96, 1.26) | −0.11 | (−0.81, 0.59) |
| Male gametocytes | 0.88 | (0.81, 0.95) | 1.09 | (0.95, 1.22) | −0.39 | (−0.67, −0.12) |
Within infection correlations between slide and qRT-PCR data generated parasite counts for all three assays (total parasites, total gametocytes and male gametocytes). The mean and 95% confidence intervals for R2 values, slopes and intercepts are presented.
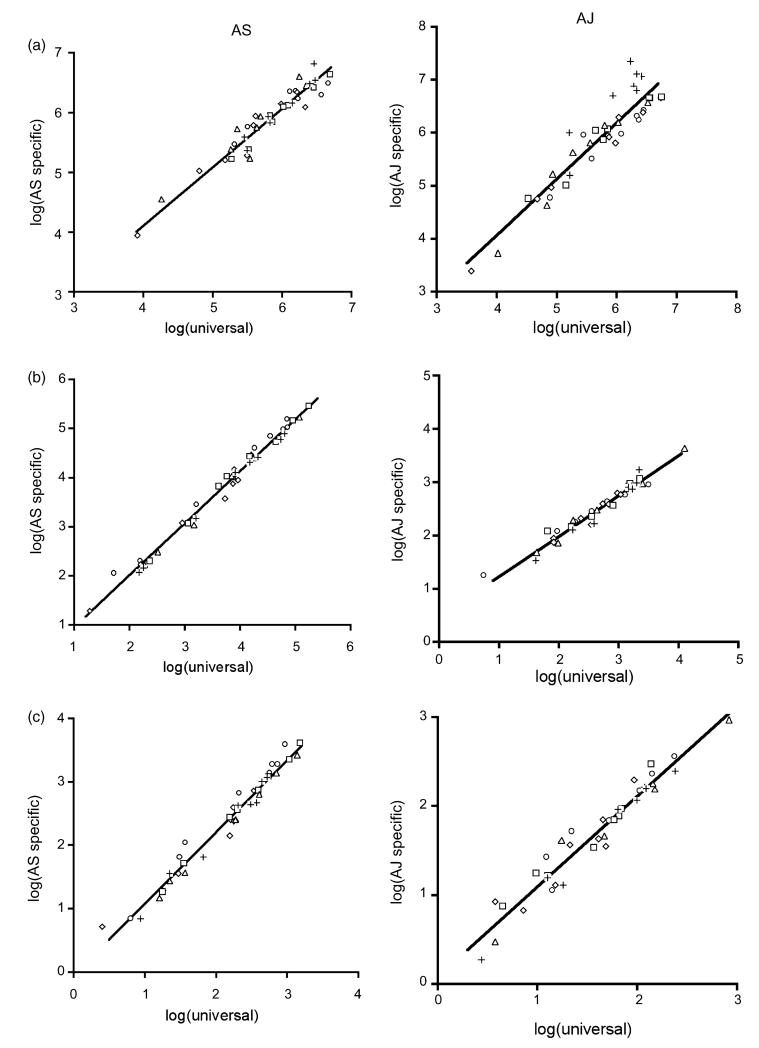
3.3. Correlations between universal and strain-specific assays
There were significant correlations between parasite counts from the universal and genotype-specific qRT-PCR assays (Fig. 2 and Table 3) for both AS and AJ. The mean R2 of the correlations within mice was >91% for both genotypes and all assays. The mean slope of the relationships between universal and genotype-specific assays was not significantly different from 1 for total parasites and AJ male gametocytes. For AS total and male gametocytes the genotype-specific assay detected marginally more parasites than the universal assay and the AJ total gametocyte assay detected marginally fewer parasites than the universal assay.
Fig. 2. Correlations between parasite counts from the universal and strain-specific qRT-PCR assays.
(a) Total parasites; (b) total gametocytes; (c) male gametocytes, with AS as the left hand panel and AJ as the right hand panel. Different symbols represent the five different mice that we measured over 8 days. Fitted lines are the average relationships between universal and strain-specific counts for all mice; see Table 2 for details.
Table 3a. Correlations between universal and AS genotype-specific qRT-PCR assays.
|
R
2
|
Slope |
Intercept |
||||
|---|---|---|---|---|---|---|
| Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | |
| Total parasites | 0.91 | (0.88, 0.95) | 0.98 | (0.87, 1.10) | 0.17 | (−0.50, 0.84) |
| Total gametocytes | 0.99 | (0.99, 1.00) | 1.06 | (1.03, 1.09) | −0.10 | (−0.28, 0.08) |
| Male gametocytes | 0.98 | (0.96, 1.00) | 1.14 | (1.07, 1.20) | −0.07 | (−0.22, −0.08) |
Within infection correlations between universal and AS genotype-specific qRT-PCR assays for total parasites, total gametocytes and male gametocytes. The mean and 95% confidence intervals for R2 values, slopes and intercepts are presented.
Table 3b. Correlations between universal and AJ genotype-specific qRT-PCR assays.
|
R
2
|
Slope |
Intercept |
||||
|---|---|---|---|---|---|---|
| Mean | 95% CI | Mean | 95% CI | Mean | 95% CI | |
| Total parasites | 0.94 | (0.88, 0.99) | 1.06 | (0.93, 1.19) | −0.17 | (−0.78, 0.43) |
| Total gametocytes | 0.96 | (0.94, 0.99) | 0.76 | (0.66, 0.85) | 0.47 | (0.20, 0.75) |
| Male gametocytes | 0.92 | (0.87, 0.98) | 1.02 | (0.96, 1.07) | 0.08 | (−0.06, 0.21) |
Within infection correlations between universal and AJ genotype-specific qRT-PCR assays for total parasites, total gametocytes and male gametocytes. The mean and 95% confidence intervals for R2 values, slopes and intercepts are presented.
3.4. Transmission investment and sex ratio adjustment in mixed-genotype infections
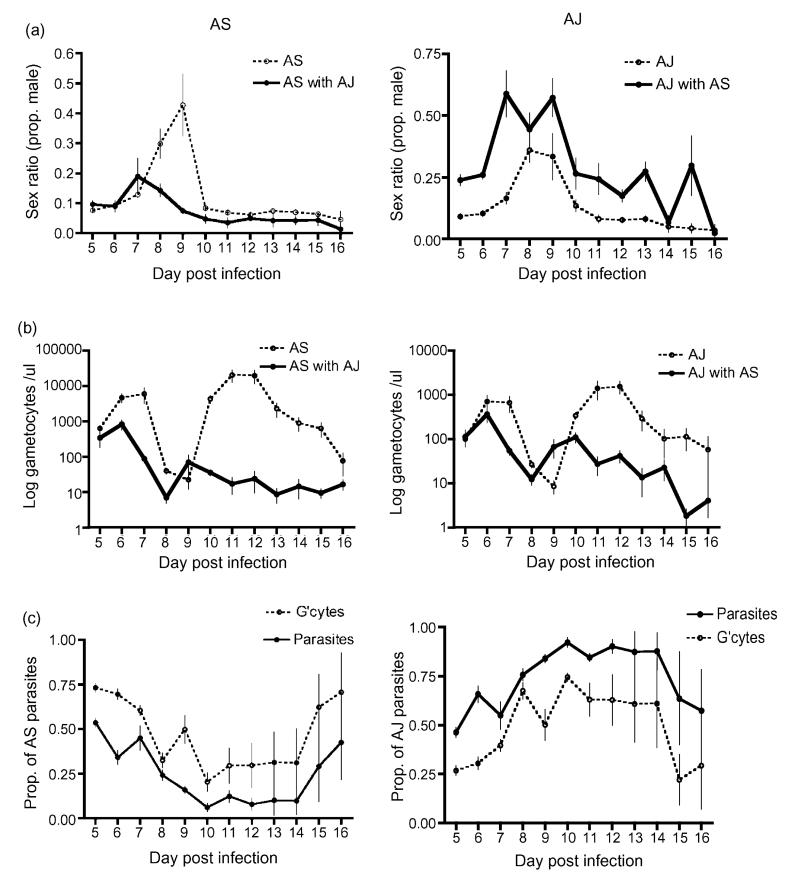
Gametocyte density of AS (Fig. 3 ) varied throughout infections and this variation was influenced by infection genetic diversity ( = 88.14; P < 0.0001). On average, AS gametocyte densities were significantly lower when co-infecting with another genotype ( = 19.47; P < 0.0001). In addition, AS gametocyte densities correlated positively with the densities of AS parasites, reticulocytes and total parasites (Table 4a). Within the mixed-genotype infections the proportion of parasites that AS contributed to infections did not influence gametocyte density ( = 2.19; P = 0.139).
Fig. 3. Infection dynamics of each focal genotype when alone and in competition with a second genotype in mixed-genotype infections.
Left hand panel shows AS and right hand panel shows AJ infection dynamics (a) Sex ratio (proportion of male gametocytes); (b) number of gametocytes and (c) the relative contribution of focal parasites and gametocytes to mixed-genotype infections with both AS and AJ. Means and S.E. are presented.
Table 4a. Influence of infection parameters on AS sex ratios and gametocyte densities.
| χ2 (d.f. = 1) | P | |
|---|---|---|
| Sex ratio | ||
| AS parasites | 0.001 | 0.969 |
| AS gametocytes | 0.028 | 0.866 |
| Total parasites | 2.837 | 0.092 |
| Total gametocytes | 0.022 | 0.838 |
| Reticulocyte density | 0.125 | 0.723 |
| Red blood cell density | 1.979 | 0.160 |
| Gametocyte density | ||
| AS parasites | 5.703 | 0.017 |
| Total parasites | 6.958 | 0.008 |
| Reticulocyte density | 4.160 | 0.041 |
| Red blood cell density | 0.448 | 0.503 |
Influence of infection parameters on sex ratios and gametocyte dynamics of AS alone and when co-infecting with another genotype. AS sex ratios did not correlate significantly with any of these parameters. AS gametocyte densities correlated positively with reticulocyte density and total parasites but negatively with AS parasites. In addition, as described in the text, AS sex ratios and gametocyte dynamics varied significantly throughout infections and were significantly different in single- and mixed-genotype infections.
Gametocyte density of AJ (Fig. 3) also varied throughout infections ( = 45.98; P < 0.0001) but was not significantly influenced by infection genetic diversity ( = 8.67; P = 0.653). Variation in AJ gametocyte densities could not be explained by AJ dynamics, dynamics of whole infections or anaemia (Table 4b). Within the mixed-genotype infections gametocyte density correlated positively with the proportion of parasites that AJ contributed to infections ( = 29.17; P < 0.0001).
Table 4b. Influence of infection parameters on AJ sex ratios and gametocyte densities.
| χ2 (d.f. = 1) | P | |
|---|---|---|
| Sex ratio | ||
| AJ parasites | 0.0445 | 0.834 |
| AJ gametocytes | 0.086 | 0.769 |
| Total parasites | 0.159 | 0.690 |
| Total gametocytes | 0.118 | 0.732 |
| Reticulocyte density | 0.024 | 0.876 |
| Red blood cell density | <0.0001 | 0.999 |
| Gametocyte density | ||
| AJ parasites | 0.168 | 0.682 |
| Total parasites | 0.002 | 0.961 |
| Reticulocyte density | 0.464 | 0.496 |
| Red blood cell density | 1.082 | 0.298 |
Influence of infection parameters on sex ratios and gametocyte dynamics of AJ alone and when co-infecting with another genotype. AJ sex ratios and gametocyte densities did not correlate significantly with any of these parameters. In addition, as described in the text, AJ sex ratios and gametocyte dynamics varied significantly throughout infections and were significantly different in single- and mixed-genotype infections.
Genotype AS produced sex ratios were largely female biased (Fig. 3) but varied during the course of infections according to genetic diversity ( = 49.32; P < 0.0001). On average, AS sex ratios were significantly more female biased in mixed-genotype infections ( = 10.50; P = 0.001). This sex ratio adjustment could not be explained by variation in focal parasite dynamics, dynamics of whole infections or anaemia (Table 4a). Within the mixed-genotype infections the proportion of gametocytes that AS contributed to infections did not influence sex ratio ( = 0.40; P = 0.527).
Genotype AJ produced sex ratios were largely female biased (Fig. 3) but varied during the course of infections according to genetic diversity ( = 25.76; P = 0.007). On average, AJ sex ratios were significantly less female biased in mixed-genotype infections ( = 45.64; P < 0.0001). This sex ratio adjustment could not be explained by focal parasite dynamics, infection dynamics or anaemia parameters (Table 4b). Within the mixed-genotype infections the proportion of gametocytes that AJ contributed to infections did not influence sex ratio ( = 3.24; P = 0.072).
4. Discussion
We have developed qRT-PCR assays to detect and quantify total parasite, total gametocyte and male gametocyte densities throughout the course of P. chabaudi infections. The qRT-PCR assays we developed are based on the detection of mRNA transcripts of P. chabaudi homologues of the P. berghei genes PB00198.00.0 (CG1) and PB000791.00.0 (MG1). These genes were chosen because their proteins were not detected in the proteome of asexual P. berghei parasites and were strongly predicted by the proteomic data to be expressed exclusively in both male and female gametocytes or only male gametocytes respectively. Furthermore, the expression profiles of these two genes were confirmed following examination of the GFP expression profiles of transgenic P. berghei parasites that had been transfected with plasmids containing GFP under the control of either the PB00198.00.0 or PB000791.00.0 promoter regions [20].
When calculating transmission stage parasite densities using the CG1 and MG1 assays, the stage-specific expression of these genes in purified RNA is measured. While the expression profiles of the P. chabaudi homologues have not been determined, parasite densities generated by the qRT-PCR repeatedly showed a highly significant correlations with densities determined from slides. This strongly suggests that the CG1 and MG1 are transcribed exclusively by P. chabaudi transmission stages, and male gametocytes respectively. While differential expression of target genes could present a potential problem to the accuracy of these qRT-PCRs, the high level of correlation between slide and qRT-PCR calculated parasite densities, confirmed that any differential expression is not great enough to effect the accuracy our qRT-PCR assays. Furthermore, because our data show that PCR detected gametocytes correlate with mature gametocytes identified by morphology, our assays are appropriate to detect mature, transmission-ready gametocytes.
We used our assays to follow the gametocyte production and gametocyte sex ratio (proportion of male gametocytes) of two genetically distinct genotypes of the rodent malaria, P. chabaudi (AS and AJ) throughout single- and mixed-genotype infections (Fig. 3). Gametocyte production by both genotypes followed patterns previously described for the acute phase, with an early peak followed by a second and higher peak [21,22]. Genotype AS produced significantly fewer gametocytes than AJ when in competition, which follows the pattern expected because AJ is more virulent than AS [8–11,26]. Both genotypes produced female-biased sex ratios throughout infections when in single-genotype infections. In this case, female-biased sex ratios are predicted because each male gametocyte can produce enough gametes to fertilise several females [32–37]. Each male gametocyte can theoretically produce eight gametes and in this case we would expect sex ratio to be approximately 10% males [28,36]. For the majority of their infections, both genotypes produced sex ratios that are consistent with this prediction. In all cases, sex ratios varied throughout infections and followed the same pattern where sex ratios become less female-biased for several days during the middle of the acute phase. We are yet to understand why sex ratios vary throughout infections but current theory predicts that parasites should produce more males when environmental conditions make transmission more difficult by reducing the ability of males to make viable gametes [15,16,27–31]. Our data indicate that producing extra males is important during the middle of the acute phase and this is the period when asexual densities peak and hosts are at their most anaemic.
Parasites are expected to increase their production of males in mixed-genotype infections according to Local Mate Competition theory [32]. In mixed-genotype infections, each genotype should produce more males, to mate with the females produced by other genotypes, and thus maximise the representation of their genes in the next generation [33,34]. This theory has been supported by data from field studies showing that female-biased sex ratios are associated with higher rates of inbreeding [15,35–38]. However, these data are correlative and using LMC theory to understand parasite sex ratios has proved controversial [39–42]. In our experiment, both genotypes altered their sex ratios when competing in mixed-genotype infections, genotype AS produced fewer males where as AJ produced more males. Therefore, genotype AJ appears to adjust sex ratio as predicted by LMC theory but AS does not. In both cases sex ratio adjustment was not confounded by variation in anaemia, gametocyte dynamics or the contributions of each genotype to their infections. Clearly, experiments following the sex ratios of more genotypes in infections that vary more widely in genetic diversity are now required to resolve whether parasites are able to adjust their sex ratio in accordance with LMC theory.
We can now accurately follow sex ratio and investment in gametocytes and ask questions about how parasites alter these traits in response to changes in their environment, genetic diversity of infections and competition. Malaria parasites are expected to alter their gametocyte production and/or sex ratio in times of stress and such changes have been observed in response to host anaemia [30,16,27]. These results come from experiments where samples contained plenty of gametocytes to sex, but we can now extend these experiments to include more realistic scenarios where gametocyte densities are low, for example, in chronic infections. The development of rapid, efficient and accurate genotype-specific assays allows new questions to be asked about the relationships between transmission and virulence in genetically diverse infections. A better understanding of the ecology and evolution of the complex transmission biology of malaria parasites will help inform medical interventions designed to target this crucial life-stage [43].
Acknowledgements
We thank the NERC, Wellcome Trust and BBSRC for funding; S. Khan, J. Thompson, A. Wargo, A. Bell and D. Nussey for help and discussion. We also thank A. Read for advice and discussion and for allowing us the use of laboratory space and equipment.
Abbreviations
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
- bp
base pairs
- CG1
common gametocyte gene 1
- MG1
male gametocyte gene 1
- PHZ
phenylhydrazine hydrochloride
- RBC
red blood cells
- GFP
green fluorescent protein
References
- [1].Alano P. Molecular approaches to monitor parasite genetic complexity in the transmission of Plasmodium falciparum malaria. Parasitologia. 2005;47(2):199–203. [PubMed] [Google Scholar]
- [2].Abdel-Wahab A, Abdel-Muhsin AM, Ali E, et al. Dynamics of gametocytes among Plasmodium falciparum clones in natural infection in an area of highly seasonal transmission. J Infect Dis. 2002;185(12):1838–42. doi: 10.1086/340638. [DOI] [PubMed] [Google Scholar]
- [3].Ranford-Cartwright LC, Taylor J, Umasunthar T, et al. Molecular analysis of recrudescent parasites in a Plasmodium falciparum drug efficacy trial in Gabon. Trans R Soc Trop Med Hyg. 1997;91(6):719–24. doi: 10.1016/s0035-9203(97)90539-3. [DOI] [PubMed] [Google Scholar]
- [4].Babiker HA, Abdel-Wahab A, Ahmed S, et al. Detection of low level Plasmodium falciparum gametocytes using reverse transcriptase polymerase chain reaction. Mol Biochem Parasitol. 1999;99:143–8. doi: 10.1016/s0166-6851(98)00175-3. [DOI] [PubMed] [Google Scholar]
- [5].Cheeseman SJ, de Roode JC, Read AF, Carter R. Real-time quantitative PCR for analysis of genetically mixed infections of malaria parasites: technique validation and applications. Mol Biochem Parasitol. 2003;131(2):83–91. doi: 10.1016/s0166-6851(03)00195-6. [DOI] [PubMed] [Google Scholar]
- [6].Schneider P, Wolters L, Schoone G, et al. Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. J Clin Microbiol. 2005;43(1):402–5. doi: 10.1128/JCM.43.1.402-405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bell AS, de Roode JC, Sim D, Read AF. Within-host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution. 2006;60(7):1358–71. [PubMed] [Google Scholar]
- [8].Wargo AR, de Roode JC, Huijben S, Drew DR, Read AF. Transmission stage investment of malaria parasites in response to in-host competition. Proceedings of the Royal Society of London. Series B. 2007;274(1625):2629–2638. doi: 10.1098/rspb.2007.0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].De Roode JC, Read AF, Chan BH, Mackinnon MJ. Rodent malaria parasites suffer from the presence of conspecific clones in three-clone Plasmodium chabaudi infections. Parasitology. 2003;127:411–8. doi: 10.1017/s0031182003004001. [DOI] [PubMed] [Google Scholar]
- [10].De Roode JC, Helinski ME, Anwar MA, Read AF. Dynamics of multiple infection and within-host competition in genetically diverse malaria infections. Am Nat. 2005;166(5):531–42. doi: 10.1086/491659. [DOI] [PubMed] [Google Scholar]
- [11].De Roode JC, Pansini R, Cheesman SJ, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci USA. 2005;102(21):7624–8. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Read AF, Taylor LH. The ecology of genetically diverse infections. Science. 2001;292(5519):1099–102. doi: 10.1126/science.1059410. [DOI] [PubMed] [Google Scholar]
- [13].Sinden RE. Sexual development of malarial parasites in their mosquito vectors. Trans Roy Soc Trop Med Hyg. 1981;75:171–2. doi: 10.1016/0035-9203(81)90058-4. [DOI] [PubMed] [Google Scholar]
- [14].Schall JJ. Transmission success of the malaria parasite Plasmodium mexicanum into its vector: role of gametocyte density and sex ratio. Parasitology. 2000;121:575–80. doi: 10.1017/s0031182000006818. [DOI] [PubMed] [Google Scholar]
- [15].West SA, Reece SE, Read AF. Evolution of gametocyte sex ratios in malaria and related apicomplexan (protozoan) parasites. Trends Parasitol. 2001;17:525–31. doi: 10.1016/s1471-4922(01)02058-x. [DOI] [PubMed] [Google Scholar]
- [16].Paul REL, Coulson TN, Raibaud A, Brey PT. Sex determination in malaria parasites. Science. 2000;287:128–31. doi: 10.1126/science.287.5450.128. [DOI] [PubMed] [Google Scholar]
- [17].Reece SE, Duncan AB, West SA, Read AF. Sex ratios in the rodent malaria parasite Plasmodium chabaudi. Parasitology. 2003;127:419–25. doi: 10.1017/s0031182003004013. [DOI] [PubMed] [Google Scholar]
- [18].Schall JJ. The sex ratio of Plasmodium gametocytes. Parasitology. 1989;98:343–50. doi: 10.1017/s0031182000061412. [DOI] [PubMed] [Google Scholar]
- [19].Taylor LH, Read AF. Why so few transmission stages? Reproductive restraint by malaria parasites. Parasitol Today. 1997;13(4):135–40. doi: 10.1016/s0169-4758(97)89810-9. [DOI] [PubMed] [Google Scholar]
- [20].Khan SM, Franke-Fayard B, Mair GR, et al. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell. 2005;121:675–87. doi: 10.1016/j.cell.2005.03.027. [DOI] [PubMed] [Google Scholar]
- [21].Mackinnon MJ, Read AF. Selection for high and low virulence in the malaria parasite Plasmodium chabaudi. Proc R Soc Lond B. 1999;266:741–8. doi: 10.1098/rspb.1999.0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wargo AR, Randle N, Chan BHK, Thompson J, Read AF, Babiker HA. Plasmodium chabaudi: reverse transcription PCR for the detection and quantification of transmission stage malaria parasites. Exp Parasitol. 2006;112:13–20. doi: 10.1016/j.exppara.2005.08.013. [DOI] [PubMed] [Google Scholar]
- [23].Gautret P, Miltgen F, Gantier JC, Chabaud AG, Landau I. Enhanced gametocyte formation by Plasmodium chabaudi in immature erythrocytes: pattern of production, sequestration and infectivity to mosquitoes. J Parasitol. 1996;82:900–6. [PubMed] [Google Scholar]
- [24].Paterson S, Lello J. Mixed models: getting the best use of parasitological data. Trends Parasitol. 2003;19(8):370–5. doi: 10.1016/s1471-4922(03)00149-1. [DOI] [PubMed] [Google Scholar]
- [25].Pinheiro JC, Bates DM. Mixed-effects models in S and S-Plus. Springer-Verlag; New York: 2000. [Google Scholar]
- [26].Taylor LH, Walliker D, Read AF. Mixed-genotype infections of the rodent malaria Plasmodium chabaudi are more infectious to mosquitoes than single genotype infections. Parasitology. 1997;115:121–32. doi: 10.1017/s0031182097001145. [DOI] [PubMed] [Google Scholar]
- [27].Paul REL, Ariey F, Robert V. The evolutionary ecology of Plasmodium. Ecol Lett. 2003;6:866–80. [Google Scholar]
- [28].West SA, Smith TG, Nee S, Read AF. Fertility insurance and the sex ratios of malaria and related hemospororin blood parasites. J Parasitol. 2002;88:258–63. doi: 10.1645/0022-3395(2002)088[0258:FIATSR]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- [29].Gardner A, Reece SE, West SA. Even more extreme fertility insurance and the sex ratios of protozoan blood parasites. J Theor Biol. 2003;223:515–21. doi: 10.1016/s0022-5193(03)00142-5. [DOI] [PubMed] [Google Scholar]
- [30].Reece SE, Duncan AB, West SA, Read AF. Host cell preference and variable transmission strategies in malaria parasites. Proc R Soc B. 2005;272:511–7. doi: 10.1098/rspb.2004.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Reece SE, Read AF. Malaria sex ratios. Trends Ecol Evol. 2000;15(7):259–60. doi: 10.1016/s0169-5347(00)01893-0. [DOI] [PubMed] [Google Scholar]
- [32].Hamilton WD. Extraordinary sex ratios. Science. 1967;156:477–88. doi: 10.1126/science.156.3774.477. [DOI] [PubMed] [Google Scholar]
- [33].Dye C, Godfray HCF. On sex ratio and inbreeding in malaria parasite populations. J Theor Biol. 1993;161:131–4. doi: 10.1006/jtbi.1993.1045. [DOI] [PubMed] [Google Scholar]
- [34].Nee S, West SA, Read AF. Inbreeding and parasite sex ratios. Proceedings of the Royal Society of London. Series B. 2002;269(1492):755–760. doi: 10.1098/rspb.2001.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Paul REL, Packer MJ, Walmsley M, et al. Mating patterns in malaria parasite populations of Papua-New-Guinea. Science. 1995;269:1709–11. doi: 10.1126/science.7569897. [DOI] [PubMed] [Google Scholar]
- [36].Read AF, Narara A, Nee S, Keymer AE, Day KP. Gametocyte sex ratios as indirect measures of outcrossing rates in malaria. Parasitology. 1992;104:387–95. doi: 10.1017/s0031182000063630. [DOI] [PubMed] [Google Scholar]
- [37].Read AF, Anwar M, Shutler D, Nee S. Sex allocation and population structure in malaria and related parasitic protozoa. Proc R Soc B. 1995;260:359–63. doi: 10.1098/rspb.1995.0105. [DOI] [PubMed] [Google Scholar]
- [38].West SA, Smith TG, Read AF. Sex allocation and population structure in Apicomplexan (protozoa) parasites. Proceedings of the Royal Society of London. Series B. 2000;267(1440):257–263. doi: 10.1098/rspb.2000.0995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ferguson DJP. More on Toxoplasma gondii, sex and premature rejection. Trends Parasitol. 2003;19:157–8. doi: 10.1016/s1471-4922(03)00033-3. [DOI] [PubMed] [Google Scholar]
- [40].Ferguson DJP. Toxoplasma gondii and sex: essential or optional extra. Trends Parasitol. 2002;18:355–9. [PubMed] [Google Scholar]
- [41].West SA, Reece SE, Read AF. Toxoplasma gondii, sex and premature rejection. Trends Parasitol. 2003;19:155–7. doi: 10.1016/s1471-4922(03)00033-3. [DOI] [PubMed] [Google Scholar]
- [42].Shutler D, Bennett GF, Mullie A. Sex proportions of Haemoproteus blood parasites and local mate competition. Proc Natl Acad Sci USA. 1995;92:6748–52. doi: 10.1073/pnas.92.15.6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Foster KR. Biomedicine-Hamiltonian medicine: why the social lives of pathogens matter. Science. 2005;308:1269–70. doi: 10.1126/science.1108158. [DOI] [PubMed] [Google Scholar]