Summary
Streptomyces cinnamonensis DSM 1042 produces two types of isoprenoid secondary metabolites: the prenylated naphthalene derivative furanonaphthoquinone I (FNQ I), and isoprenylated phenazines which are termed endophenazines. Previously, a 55 kb gene cluster was identified which contained genes for both FNQ I and endophenazine biosynthesis. However, several genes required for the biosynthesis of these metabolites were not present in this cluster. We now re‐screened the cosmid library for genes of the mevalonate pathway and identified a separate genomic locus which contains the previously missing genes. This locus (15 kb) comprised orthologues of four phenazine biosynthesis genes known from Pseudomonas strains. Furthermore, the locus contained a putative operon of six genes of the mevalonate pathway, as well as the gene epzP which showed sequence similarity to a recently discovered class of prenyltransferases. Inactivation and complementation experiments proved the involvement of epzP in the prenylation reaction in endophenazine biosynthesis. This newly identified genomic locus is more than 40 kb distant from the previously identified cluster. The protein EpzP was expressed in Escherichia coli in form of a his‐tag fusion protein and purified. The enzyme catalysed the prenylation of 5,10‐dihydrophenazine‐1‐carboxylic acid (dihydro‐PCA) using dimethylallyl diphosphate (DMAPP) as isoprenoid substrate. Km values were determined as 108 µM for dihydro‐PCA and 25 µM for DMAPP.
Introduction
Streptomycetes are prolific producers of secondary metabolites, including polyketides, non‐ribosomal peptides and other structural classes. However, isoprenoid secondary metabolites are relatively uncommon in streptomycetes. Isoprenoids identified in Streptomyces comprise, e.g. the sesquiterpene geosmin (Jiang et al., 2007), diterpenes like terpentecin (Dairi et al., 2001) and phenalinolactone (Binz et al., 2008), isoprenylated naphthalene derivatives like naphterpin (Shin‐ya et al., 1990), prenylated phenazines (Gebhardt et al., 2002) and prenylated indole derivatives (Takahashi et al., 2010), and the aminocoumarin antibiotics novobiocin and clorobiocin (Pojer et al., 2003).
By the discovery of the prenyltransferases of clorobiocin and naphterpin biosynthesis (Pojer et al., 2003; Kuzuyama et al., 2005), it has been revealed that the prenylation reactions in the formation of such isoprenylated aromatic compounds are catalysed by a unique, new class of enzymes. Due to their αββα structural motif, these enzymes have been termed ABBA prenyltransferases (Tello et al., 2008; Heide, 2009).
In our search for new members of the ABBA prenyltransferase class, we investigated Streptomyces cinnamonensis DSM 1042 which produces two different types of isoprenylated aromatic secondary metabolites, i.e. furanonaphthoquinone I (FNQ I) and endophenazines (Fig. 1) (Tax et al., 1983; Sedmera et al., 1991). We identified a 55 kb gene cluster which contained genes for both FNQ I and endophenazine biosynthesis (Haagen et al., 2006). Inactivation experiments confirmed the involvement of these genes in the respective pathways. The cluster was found to contain genes for two new members of the ABBA prenyltransferases. One of these, fnq26, was shown to encode the prenyltransferase of FNQ I biosynthesis (Haagen et al., 2007). Unexpectedly, the other gene, fnq28, was proven not to be involved in FNQ I or endophenazine biosynthesis (Haagen et al., 2006). Therefore, the prenyltransferase of endophenazine biosynthesis in S. cinnamonensis remained unknown.
Figure 1.
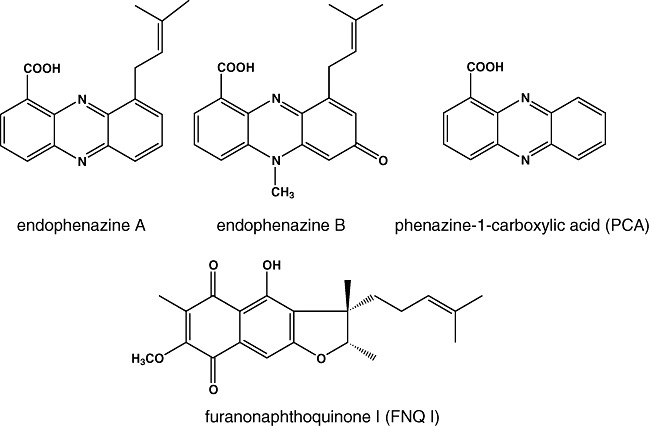
Secondary metabolites of S. cinnamonensis DSM 1042.
In the present study, we identified a genomic locus which contained the gene for the missing prenyltransferase as well as several other genes involved in both endophenazine and FNQ I biosynthesis. The function of the prenyltransferase could be confirmed by inactivation experiments and biochemical investigation. This study provides one of the few examples that the genes of the biosynthesis of a secondary metabolite in a Streptomyces strain are not clustered at a single locus of the genome, but distributed to two different loci.
Results
Identification of the putative ABBA prenyltransferase gene epzP in S. cinnamonensis
The previously identified prenyltransferases with aromatic substrates showed little or no sequence similarity to each other (Tello et al., 2008; Heide, 2009). Therefore, direct screening of a genomic library of S. cinnamonensis for the desired prenyltransferase gene did not appear promising. However, in four Streptomyces strains which form prenylated aromatic secondary metabolites the responsible prenyltransferase gene has been identified in the immediate vicinity of a cluster of genes encoding the enzymes of the mevalonate pathway. In S. cinnamonensis endophenazines and FNQ I are predominantly formed via the mevalonate pathway (Bringmann et al., 2007). Therefore, the presence of a cluster of mevalonate pathway genes was expected in the genome of S. cinnamonensis, and we speculated that the missing prenyltransferase gene may be localized in its vicinity.
Using conserved sequences of mevalonate pathway genes from different streptomycetes, we designed degenerate primers for the hydroxymethylglutaryl‐CoA synthase, hydroxymethylglutaryl‐CoA reductase and mevalonate diphosphate decarboxylase genes, using the codehop (COnsensus‐DEgenerate Hybrid Oligonucleotide Primer) program (Rose et al., 1998; 2003). A cosmid library of S. cinnamonensis had been established previously (Haagen et al., 2006) and was screened with the three primer pairs. This led to the identification of cosmid 8‐4D which gave PCR products for all three mevalonate pathway genes. This cosmid was subjected to full‐length sequencing using a shotgun library of DNA fragments. The cosmid insert comprised 45 167 bp and was deposited in the GenBank database under Accession No. HQ228364. Approximately in the middle of the insert sequence, we found the expected cluster of six mevalonate pathway genes, and directly upstream thereof a gene with obvious sequence similarity to the ABBA prenyltransferase class which was termed epzP (Fig. 2).
Figure 2.
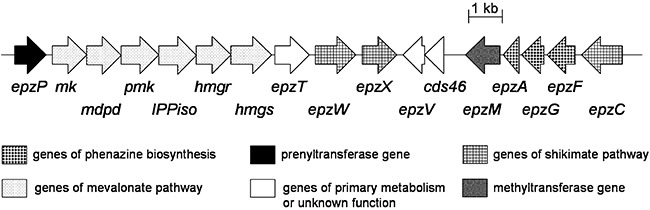
Gene cluster on cosmid 8‐4D of S. cinnamonensis DSM 1042, containing phenazine biosynthesis genes, mevalonate pathway genes and the prenyltransferase gene epzP. Further genes of endophenazine and furanonaphthoquinone I biosynthesis are contained in another genomic locus (Haagen et al., 2006).
The exact distance between the previously identified gene cluster (cosmid 3‐6H) and the newly identified locus (cosmid 8‐4D) in the genome of S. cinnamonensis is not known. We screened the cosmid library for cosmids overlapping with cosmid 3‐6H, and found all overlapping cosmids not to contain genes of the mevalonate pathway or of phenazine biosynthesis. It can therefore be concluded that those two gene clusters must be more than 40 kb apart.
Sequence analysis of the insert of cosmid 8‐4D
The central part of the insert of cosmid 8‐4D contained 17 genes apparently related to the biosynthesis of prenylated phenazines. The results of the comparisons of these genes with database entries are listed in Table 1.
Table 1.
Deduced functions of genes in the insert of cosmid 8‐4D from a genomic library of S. cinnamonensis DSM 1042.
| Gene | AA | Protein homologue, organism | Accession No. | Identity (%) | Proposed function |
|---|---|---|---|---|---|
| epzP | 302 | 5,10‐Dihydrophenazine‐1‐carboxylate‐9‐dimethylallyltransferase, S. anulatus 9663 | CAX48655.1 | 56 | Prenyltransferase of endophenazine biosynthesis |
| mk | 345 | Mevalonate kinase, S. anulatus 9663 | CAX48656.1 | 75 | Mevalonate kinase |
| mdpd | 351 | Diphosphomevalonate decarboxylase, Streptomyces sp. CL190 | BAB07791.1 | 75 | Diphosphomevalonate decarboxylase |
| pmk | 371 | Phosphomevalonate kinase, Streptomyces sp. CL190 | BAB07792.1 | 68 | Phosphomevalonate kinase |
| IPPiso | 363 | Isopentenyl diphosphate isomerase, Streptomyces sp. CL190 | Q9KWG2.1 | 78 | IPP isomerase |
| hmgr | 353 | 3‐Hydroxy‐3‐methylglutaryl CoA reductase, Streptomyces sp. CL190 | BAB70975.1 | 91 | HMG‐CoA reductase |
| hmgs | 391 | 3‐Hydroxy‐3‐methylglutaryl CoA synthase, Streptomyces sp. CL190 | BAB07795.1 | 82 | HMG‐CoA synthase |
| epzT | 333 | 3‐Oxoacyl‐[acyl‐carrier‐protein] synthase, S. anulatus 9663 | CAX48662.1 | 80 | Unknown |
| epzW | 425 | 3‐Phosphoshikimate‐1‐carboxyvinyltransferase, Streptomyces sp. Mg1 | ZP_04998765.1 | 64 | EPSP synthase |
| epzX | 361 | Chorismate synthase, Streptomyces sp. e14 | ZP_06706902.1 | 85 | Chorismate synthase |
| epzV | 203 | PpzV, S. anulatus 9663 | CAX48664.1 | 63 | Unknown |
| cds46 | 176 | Tail sheath protein, Natrialba magadii ATCC 43099 | YP_003478909.1 | 47 | Viral protein |
| epzM | 345 | PpzM, S. anulatus 9663 | CAX48665.1 | 76 | N‐methyltransferase of endophenazine biosynthesis |
| epzA | 169 | EphzA, S. cinnamonensis DSM 1042 | CAL34112.1 | 98 | Oxidoreductase of phenazine biosynthesis |
| epzG | 213 | EphzG, S. cinnamonensis DSM 1042 | CAL34111.1 | 93 | FMN‐dependent oxidase of phenazine biosynthesis |
| epzF | 278 | PpzF, S. anulatus 9663 | CAX48668.1 | 86 | Trans‐2,3‐dihydro‐3‐hydroxyanthranilate isomerase |
| epzC | 391 | EphzC, S. cinnamonensis DSM 1042 | CAL34108.1 | 97 | DAHP synthase |
The sequence of the insert of cosmid 8‐4D has been deposited in the GenBank database under Accession No. HQ228364.
The genes upstream of the putative prenyltransferase gene epzP show very high similarity to genes found in many actinomycetes and probably code for primary metabolic enzymes. The gene epzP is therefore likely to mark the left border of the identified secondary metabolic gene cluster.
Immediately downstream of epzP, the six mevalonate pathway genes are situated. These genes apparently form a single operon, as the first two genes are only separated by 3 bp, while the following genes overlap, suggesting a translational coupling. Highly similar operons have been reported from several other Streptomyces strains forming isoprenoid secondary metabolites (Dairi, 2005). It appears likely that these genes are responsible for the formation of dimethylallyl diphosphate and isopentenyl diphosphate as precursors of the isoprenoid moieties of the endophenazines and of FNQ I in S. cinnamonensis (Bringmann et al., 2007), although no direct experimental proof has been provided.
Approximately 7 kb downstream of the mevalonate pathway genes, and orientated in the opposite direction, is a group of genes with high similarity to genes of phenazine biosynthesis which have previously been examined in Pseudomonas strains (Mavrodi et al., 2008). These genes include epzF, with high similarity to phzF which in Pseudomonas has been shown to code for an essential enzyme of phenazine biosynthesis, catalysing the isomerization of trans‐2,3‐dihydro‐3‐hydroxyanthranilic acid (DHHA, Fig. 3) to a highly reactive aminocyclohexenone derivative. A second gene, epzA, showed similarity to phzA of Pseudomonas, involved in the condensation of two aminocyclohexenone moieties to a tricyclic phenazine precursor (Fig. 3) (Ahuja et al., 2008). This reaction requires a second, similar protein, PhzB, in Pseudomonas. In S. cinnamonensis, a phzB orthologue, as well as an additional phzA orthologue, are found in the previously identified locus for endophenazine and FNQ I biosynthesis genes (Haagen et al., 2006).
Figure 3.
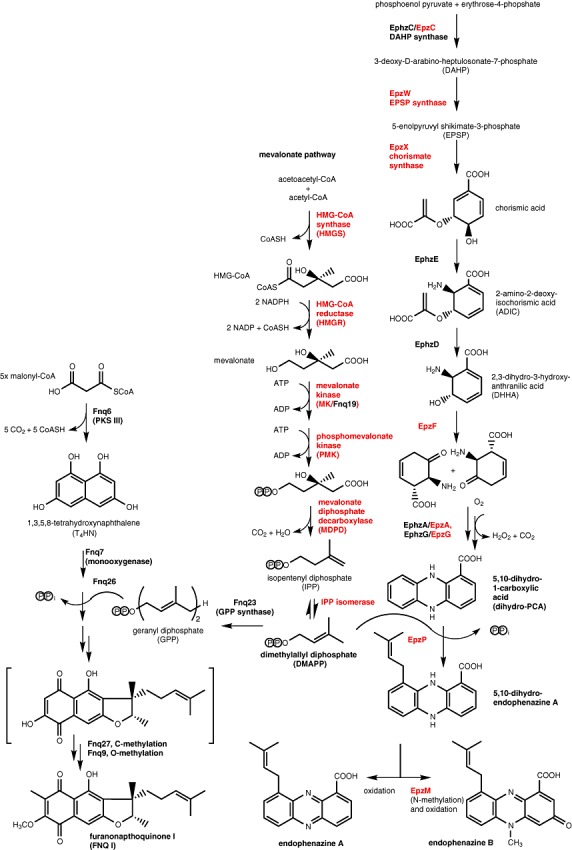
Biosynthetic pathway to endophenazine A, endophenazine B and furanonaphthoquinone I. The enzymes which are encoded on cosmid 8‐4D are marked in red, and those encoded at a previously identified locus (Haagen et al., 2006) are marked in black.
In the biosynthesis of phenazines, the initial tricyclic phenazine precursor is oxidized to 5,10‐dihydroxyphenazine‐1‐carboxylic acid (Fig. 3). epzG (similar to phzG in Pseudomonas) is likely to be involved in this oxidation (Parsons et al., 2004). An additional copy of this gene is contained in the previously identified locus (Haagen et al., 2006).
Most, but not all phenazine gene clusters identified so far contain a gene (phzC) coding for a DAHP synthase (Fig. 3), i.e. the first enzyme of the shikimate pathway (Mavrodi et al., 2010). A similar gene, epzC, was found in the present gene locus. Additionally, and for the first time in any phenazine biosynthetic gene cluster, we found two further genes with obvious sequence similarity to shikimate pathway genes: epzW, coding for a putative 5‐enolpyruvyl shikimate‐3‐phosphate synthase (EPSP), and epzX, with similarity to chorismate synthase (Fig. 3). It appears likely that these three genes contribute to the generation of chorismate for phenazine biosynthesis.
Streptomyces cinnamonensis produces the N‐methylated compound endophenazine B (Figs 1 and 3). Correspondingly, the identified gene cluster contained the gene epzM, with high similarity to a known N‐methyltransferase of phenazine biosynthesis (Parsons et al., 2007). Two further genes, designated as epzT and epzV, have close orthologues in a recently identified gene cluster for prenylated phenazine biosynthesis (Saleh et al., 2009a). It is yet unknown which function, if any, they may have in this pathway.
The genes located upstream of epzC could not be unambiguously assigned to either primary or secondary metabolism, and therefore the position of the right border of the identified gene cluster cannot be decided at present.
Inactivation of the prenyltransferase gene epzP
A principal aim of the present study was the identification of the prenyltransferase of endophenazine biosynthesis in S. cinnamonensis. The predicted gene product of epzP showed sequence similarity to aromatic prenyltransferases of the ABBA family and presented a very likely candidate for the desired gene. In order to confirm the involvement of epzP in endophenazine biosynthesis, we carried out an inactivation experiment (Fig. 4A). Using Red/ET‐mediated recombination, epzP was replaced on cosmid 8‐4D by an apramycin resistance gene (Gust et al., 2004). The modified cosmid was introduced into S. cinnamonensis by conjugation. Integration mutants, resulting from homologous recombination, were selected using their apramycin resistance, and the desired double‐cross‐over event was confirmed by the absence of the kanamycin resistance, encoded in the cosmid backbone. Two independent mutant strains were generated, and the replacement of epzP by the apramycin resistance gene was confirmed by PCR (Fig. 4B).
Figure 4.
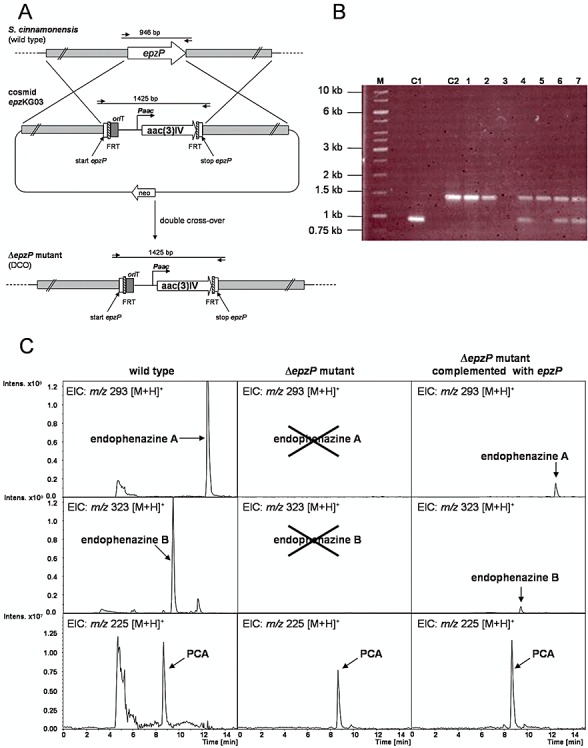
A. Scheme of the gene inactivation of epzP. aac(3)IV, apramycin resistance gene; Paac, promoter of the apramycin resistance gene; FRT, FLP recognition target; oriT, origin of transfer from RK2; neo, kanamycin resistance gene. Out of scale. B. Confirmation of the genotype of single‐cross‐over mutants (SCO) and double‐cross‐over mutants (DCO). Lane M: marker, C1: cosmid 8‐4D (wild type), C2: cosmid epzKG03 (ΔepzP). Lanes 4, 6 and 7: SCO mutants. Lanes 1, 2 and 5: DCO mutants. C. HPLC/MS analysis of the formation of endophenazine A, endophenazine B and phenazine‐1‐carboxylic acid (PCA) in S. cinnamonensis DSM 1042 (wild type), the ΔepzP mutant and the ΔepzP mutant complemented with intact epzP.
HPLC‐UV and HPLC‐ESI‐MS analysis of cultures grown in endophenazine production medium confirmed the formation of both endophenazine A (m/z 293, [M+H+]) and FNQ I (m/z 371, [M+H+]) in the wild‐type strain. HPLC‐ESI‐MS also confirmed the presence of the N‐methylated compound endophenazine B (m/z 323, [M+H+]) (Fig. 4C). In both ΔepzP mutant strains, the production of endophenazine A and B was completely abolished (Fig. 4C), but FNQ I was still produced (wild type 3.4 µmol l−1; ΔepzP 6.6 µmol l−1).
To confirm that the abolishment of endophenazine production was indeed due to the inactivation of epzP, both mutants were complemented with an intact copy of this gene. For this purpose, epzP was amplified by PCR (see Experimental procedures) and cloned into the Escherichia coli–Streptomyces shuttle vector pUWL‐hygR/oriT which contains the strong ermE* promoter. DNA sequencing confirmed the absence of mutations. The resulting construct pKG08 was introduced into the ΔepzP mutants by conjugation, and two independent exconjugants were cultivated in production medium and analysed by HPLC‐UV and HPLC‐ESI‐MS. In both cases, the production of both endophenazine A and endophenazine B was restored (Fig. 4C). The amount of endophenazine A in the complemented mutants, however, reached only 20% of the wild‐type strain, possibly due to an inappropriate regulation of the expression of the introduced gene.
These experiments strongly supported the hypothesis that epzP codes for the prenyltransferase of phenazine biosynthesis in S. cinnamonensis.
Biochemical investigation of EpzP
For the expression and purification of EpzP, its structural gene was again amplified by PCR and cloned into two different expression vectors, pET28a and pHis8. The correct sequence was confirmed for the insert of both constructs and the protein was expressed in E. coli as a fusion protein with an N‐terminal His6 or His8 tag respectively. Both constructs generated equal amounts of protein, and the activity of both fusion proteins were very similar. Ni2+ affinity chromatography resulted in a protein of apparent homogeneity. When this protein was incubated with 5,10‐dihydrophenazine‐1‐carboxylic acid (dihydro‐PCA) and dimethylallyl diphosphate (DMAPP) as substrates, the enzyme‐dependent formation of a single prenylated product was observed (Fig. 5A). The instable prenylated dihydro‐PCA was oxidized to the stable endophenazine A using sodium peroxodisulfate, and the identity of the resulting compound to endophenazine A was confirmed by HPLC‐UV and HPLC‐ESI‐MS in comparison with an authentic reference sample. Both compounds gave identical fragmentation patterns in mass spectrometry.
Figure 5.
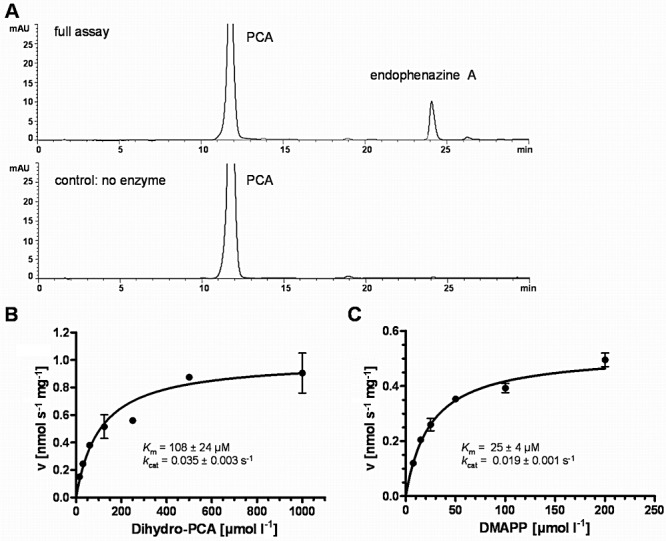
A. HPLC analysis of prenyltransferase assays with purified EpzP, dihydro‐PCA and DMAPP. UV chromatograms were recorded at 365 nm. B and C. Determination of Km values of EpzP for dihydro‐PCA and DMAPP. In (B), DMAPP was kept constant at 0.4 mM. In (C), dihydro‐PCA was kept constant at 0.8 mM. Km and kcat values were determined by non‐linear regression, using GraphPad Prism software (GraphPad Software, La Jolla, CA).
EpzP was specific for dihydro‐PCA as aromatic substrate. No reaction product was obtained with PCA. In contrast to the prenyltransferase Fnq26 from the same organism (Haagen et al., 2007), no product was obtained with flaviolin, using either DMAPP or GPP as isoprenoid substrates; also 4‐hydroxyphenylpyruvate, substrate of the ABBA prenyltransferases CloQ and NovQ (Pojer et al., 2003), was not accepted by EpzP (Fig. S1).
As is typical for most members of the ABBA prenyltransferase family, the enzymatic activity of EpzP did not depend on the presence of Mg2+ or other divalent cations. Addition of MgCl2 up to 10 mM did not increase the activity significantly, and upon addition of EDTA (10 mM) 77% of the activity was retained (Table S1).
Using a constant concentration of DMAPP (0.4 mM) and varying concentrations of dihydro‐PCA, the Km value for the aromatic substrate was determined as 108 ± 24 µM. And correspondingly, using a constant concentration of dihydro‐PCA (0.8 mM) and varying concentrations of DMAPP, the Km value for DMAPP was determined as 25 ± 4 µM (Fig. 5B and C).
Discussion
In a previous study, we identified a cluster of genes for endophenazine and FNQ I biosynthesis in S. cinnamonensis DSM 1042 (Haagen et al., 2006). This cluster will hereafter be referred to as locus A. Locus A did not contain a gene for the prenyltransferase which has to be postulated for endophenazine biosynthesis. In the present study, we could identify this missing prenyltransferase gene in a separate genomic locus, together with mevalonate pathway and phenazine biosynthesis genes. This genomic locus will hereafter be called locus B. The presence of genes for enzymes of the mevalonate pathway was expected in S. cinnamonensis, as feeding studies in this organism had confirmed that the isoprenoid moieties of endophenazines and FNQ I were derived from mevalonate (Bringmann et al., 2007). Likewise, the presence of the putative phenazine‐N‐methyltransferase EpzM was expected, as S. cinnamonensis produces the N‐methylated compound endophenazine B (Figs 1 and 3), but the two methyltransferase genes present in locus A (fnq9 and fnq27) had been experimentally assigned to methyltransferase reactions in FNQ I biosynthesis (Haagen et al., 2006).
Locus A contains a contiguous DNA region comprising orthologues of most genes required for phenazine biosynthesis in Pseudomonas (Mavrodi et al., 1998), however with the notable exception of a phzF orthologue. phzF has invariably been found in all functional biosynthetic gene clusters for phenazine biosynthesis identified in Pseudomonas, Burkholderia, Pectobacterium and others (Mavrodi et al., 2010). Locus A of S. cinnamonensis was the first phenazine biosynthetic gene cluster published from any Streptomyces strain, and the absence of phzF raised the question whether the essential biosynthetic step catalysed by PhzF in Pseudomonas may be accomplished in a different way in Streptomyces. However, as we now identified a phzF orthologue in locus B of S. cinnamonensis, and also in an endophenazine gene cluster from Streptomyces anulatus (Saleh et al., 2009a), it appears likely that the phenazine biosynthetic pathway in the Gram‐positive actinobacteria (including Streptomyces) is identical to that in the Gram‐negative gammaproteobacteria (including Pseudomonas spp.) and betaproteobacteria (including Burkholderia spp.).
In S. anulatus, we found all genes for endophenazine biosynthesis to be clustered in a single genomic locus (Saleh et al., 2009a). Likewise, in all previously investigated microbial producer strains of phenazines, all genes required for phenazine biosynthesis were invariably found to be clustered in a single locus (Mavrodi et al., 2010). The present study now provides the first example that the genes for phenazine biosynthesis are distributed to two different loci of the genome of a phenazine‐producing strain.
Nearly all gene clusters for phenazine biosynthesis contain a paralogue of the first gene of the shikimate pathway, i.e. DAHP synthase (Fig. 3). This gene is usually termed PhzC. However, the present study provides the first example that two further genes of the shikimate pathway are contained in a phenazine gene cluster, i.e. epzW and epzX, with obvious similarity to EPSP synthase and chorismate synthase (Fig. 3). It appears likely that these genes have a similar role as generally assumed for the phzC, i.e. to ensure the supply of chorismate for phenazine biosynthesis, independently from the tightly regulated pathway to the aromatic amino acids.
The family of aromatic prenyltransferases to which EpzP, CloQ, NphB, Fnq26 and Fnq28 belong has originally been found to prenylate only phenolic substrates (Heide, 2009; Saleh et al., 2009b). EpzP now provides the second example of an enzyme of this family which prenylates not a phenolic substrate but a dihydrophenazine derivative. The first such example was PpzP (Saleh et al., 2009a).
Experimental procedures
Bacterial strains, plasmids and culture conditions
Streptomyces cinnamonensis DSM 1042 was grown in liquid YMG medium or on solid MS medium (Kieser et al., 2000) at 30°C. The medium described by Sedmera and colleagues (1991) was used for production of secondary metabolites. Escherichia coli XL1 Blue MRF' (Stratagene, Heidelberg, Germany) was used for cloning, and was grown in liquid or on solid (2% agar) Luria–Bertani or SOB medium at 37°C. The REDIRECT technology kit for PCR targeting was obtained from Plant Bioscience Limited (Norwich, UK). The aac(3)IV/oriT (apramycin resistance) cassette from pIJ773 (Gust et al., 2004) was used. For the selection of the recombinant mutants carbenicillin (50 µg ml−1), apramycin (50 µg ml−1), kanamycin (50 µg ml−1), chloramphenicol (25 µg ml−1) and nalidixic acid (25 µg ml−1) were added to DNA (Kieser et al., 2000) and MS medium respectively.
Chemicals and enzymes
Carbenicillin and kanamycin were purchased from GenAxxon BioSciences GmbH, Biberach, Germany; apramycin, nalidixic acid and sodium persulfate (Na2S2O8) from Sigma‐Aldrich, Steinheim, Germany; chloramphenicol and sodium dithionite (Na2S2O4) from Merck, Darmstadt, Germany; and phenazine‐1‐carboxylic acid from InFarmatik, Hungary. Dimethylallyl diphosphate was synthesized as described by Woodside and colleagues (1993) and endophenazines were isolated according to Sedmera and colleagues (1991), with modification described in Production and analysis of secondary metabolites. Restriction enzymes were purchased from New England BioLabs, Ipswich, MA.
Genetic procedures
Standard methods for DNA isolation and manipulation were performed as described by Kieser and colleagues (2000) and Sambrook and Russell (2001). DNA fragments were isolated from agarose gels by using the GFX PCR and gel band purification kit (Amersham Biosciences). Chromosomal DNA was isolated by lysozyme treatment and phenol/chloroform extraction as described by Kieser and colleagues (2000).
Screening of the cosmid library
The preparation of the cosmid library has been described previously (Haagen et al., 2006). Screening was performed by PCR with degenerated primers (codehop) (Rose et al., 1998; 2003) for genes of the mevalonate pathway. The primer for the HMG‐CoA synthase gene were HMGS_for (5′‐GCC AAG TCC GCC GG(A/C/G/T) GT(A/C/G/T) TA(C/T) GT‐3′) and HMGS_rev (5′‐AGC CGG AAG GGG CC(A/C/G/T) GT(A/C/G/T) GT(C/T) TG‐3′); for the HMG‐CoA reductase gene CodeHMG_for (5′‐GGC CAC CTA CGA GAC CCC (A/C/G/T)(C/T)T (A/C/G/T)TG (A/C/G/T)TG GCC‐3′) and CodeHMG_rev (5′‐CGC ATC AGC TCG CCG (G/C)(G/T)(A/G) TT(A/C/G/T) GT(C/T) TG‐3′); for mevalonate diphosphate decarboxylase gene MDPD_for (5′‐GAC CCT GGA CGT CTT CCC (A/C/G/T)AC (A/C/G/T)AC (A/C/G/T)AC‐3′) and MDPD_rev (5′‐GCG TTC CGC TCG GC(A/G/T) AT(C/T) TC(A/C/G/T) CC‐3′).
Inactivation of the gene epzP
The inactivation was carried out with the REDIRECT technology kit for PCR targeting. The cosmid 8‐4D was transformed into E. coli BW25113 (pIJ790) by electroporation and the bacteria were grown at 30°C. We used the following primers for the amplification of the apramycin resistance cassette (aac(3)IV) from pIJ773: orf16_PT_for_1 (5′‐TTC GCC AAA TTC GAT CAT TCG ATC AGT GGA GGA ACC ATG ACT AGT ATT CCG GGG ATC CGT CGA CC‐3′) and orf16_PT_rev (5′‐CCT TTT GAA TGC CCG CCC CGG CGG GCC GGA GCG TGG TCA TCT AGA TGT AGG CTG GAG CTG CTT C‐3′). The resulting PCR product had the size 1436 bp and contained restriction sites (underlined) for XbaI and SpeI. The PCR product was used to replace the gene epzP on cosmid 8‐4D by Red/ET mediated recombination, resulting in cosmid epzKG03. The resulting cosmid was transformed into the non‐methylating E. coli ET12567 (pUZ8002), and subsequently the non‐methylated DNA was introduced by conjugation into S. cinnamonensis DSM1042 (wild type). Double‐cross‐over (DCO) mutants were selected by replica plating (DCO mutants are kanamycin‐sensitive and apramycin‐resistant) on solid DNA medium with kanamycin or apramycin respectively. DNA was isolated of resulting mutant strains and analysed by PCR with the primers epzKG03_for (5′‐CAT TCG ATC AGT GGA GGA ACC ATG‐3′) and epzKG03_rev (5′‐GGC GGG CCG GAG CGT GGT CA‐3′). Gene replacement mutants showed a single PCR band with the size of 1400 bp, whereas the wild‐type gene resulted in a band at 950 bp.
Production and analysis of secondary metabolites
Mutants and wild‐type S. cinnamonensis strains were pre‐cultured for 48 h in liquid YMG medium (50 ml) at 30°C and 180 r.p.m. Fifty millilitres of production medium (Sedmera et al., 1991) was inoculated with 3 ml of the pre‐culture in a 300 ml Erlenmeyer flask with spring and baffle and cultivated for 120 h. In case of the ΔepzP mutants apramycin (50 µg ml−1) was added to the medium. For isolation of endophenazines and FNQ I, 50 ml of culture was centrifuged at 3500 g for 10 min. The supernatant was discarded and the cells were extracted with methanol (10 ml) by vortexing and treatment in an ultrasonic bath for 5 min. The extract was mixed with sodium acetate buffer (10 ml; 1 M, pH 4.0) and extracted with dichloromethane (5 ml). After separation of the organic phase, the solvent was evaporated and the residue was dissolved in methanol (50 µl). Extracts were analysed with HPLC (Agilent 1100 series; Waldbronn, Germany) by using an Eclipse XDB‐C18 column (4.6 × 150 mm, 5 µm; Agilent) at a flow rate of 1 ml min−1 with a linear gradient from 10% to 100% of solvent B in 30 min (solvent A: 1% formic acid in water; solvent B: 1% formic acid in acetonitrile). Detection was carried out at 252 and 365 nm. Additionally, a UV spectrum from 200 to 400 nm was logged by a photodiode array detector (DAD). The absorbance at 365 nm was used for quantitative analysis, employing authentic samples of PCA and endophenazine as external standards.
Complementation of ΔepzP mutants
The ΔepzP mutants were complemented with plasmid pKG08, carrying an intact copy of epzP in the shuttle vector pUWL‐hygR/oriT (Zhao et al., 2010). The gene epzP was amplified from cosmid 8‐4D by PCR with the primers K_orf16_for (5′‐AAG CTT ATG TCG GAA AGC GCC GAC‐3′) with a HindIII restriction site (underlined) and K_orf16_rev (5′‐ACT AGT TCA GCC GTC GGA ACG CAG‐3′) with a SpeI restriction site. The PCR product was first cloned into pGEM®‐T (Promega Corporation, Madison, WI) to give plasmid pKG03. After restriction with HindIII and SpeI, isolation of the 915 bp fragment, it was ligated into pUWL‐hygR/oriT (linearized with HindIII and SpeI), resulting in plasmid pKG08. The plasmid was introduced into the non‐methylating E. coli ET12567 (pUZ8002) and then transferred into ΔepzP mutants via conjugation. For cultivation of the complemented ΔepzP mutants apramycin (50 µg ml−1) and hygromycin (40 µg ml−1) were added to the production medium. The extraction procedure for the endophenazines and FNQ I has been described above.
LC‐ESI‐MS and ‐MS/MS analysis
The extracts were examined with LC‐ESI‐MS using a Nucleosil 100‐C18 column (3 µm, 100 × 2 mm) coupled to an ESI mass spectrometer (LC/MSD Ultra Trap System XCT 6330; Agilent Technology). Analysis was performed at a flow of 0.4 ml min−1 with a linear gradient from 10% to 100% of solvent B in 15 min (solvent A: 0.1% formic acid in water; solvent B: 0.06% formic acid in acetonitrile). Detection was carried out at 230, 260, 280, 360 and 435 nm (± 10 nm). Electron spray ionization (positive and negative ionization) in Ultra Scan mode with capillary voltage of 3.5 kV and heated temperature of 350°C was used. LC‐MS/MS analysis was carried out in positive ionization mode with the same capillary voltage and temperature. For endophenazine A, the mass 293 ± 0.5 Da was selected for fragmentation.
Expression and purification of EpzP
For the construction of the expression plasmids pBB09 (epzP in pET28a) and pKG14 (epzP in pHis8), epzP was amplified with Phusion® DNA Polymerase (Finnzymes, Woburn, MA) using the cosmid 8‐4D as template. The following primers were used: for construction of pBB09, orf16_NdeI_F (5′‐GGG AAT TCC ATA TGT CGG AAA GCG CCG ACC‐3′) and orf16_XhoI_Stop_R (5′‐GCC CTC GAG TCA GCC GTC GGA ACG CAG‐3′), for construction of pKG14, epz16_EcoRI_F (5′‐GTG CCG CGC GAA TTC CAT ATG TCG‐3′) and the same reverse primer as above, i.e. orf16_XhoI_Stop_R. The resulting PCR products were cloned into pGEM®‐T (Promega). After sequencing (Eurofins MWG Operon, Martinsried, Germany) epzP was cloned into pET28a to give pBB09, and into pHis8 to give pKG14. The plasmid pKG14 was transformed into E. coli Rosetta 2 (DE3) pLysS (Stratagene) and a pre‐culture of 100 ml of liquid LB medium was cultured overnight at 37°C and 200 r.p.m. Thirty‐five millilitres of the pre‐culture were added to liquid TB medium (1 l) containing kanamycin (50 µg ml−1) and chloramphenicol (25 µg ml−1) and grown at 37°C to an A600 of 0.6. The temperature was lowered to 20°C and isopropyl 1‐thio‐β‐d‐galactopyranoside (IPTG) was added to a final concentration of 0.5 mM. After 20 h of cultivation at 20°C, the cells were harvested by centrifugation for 10 min at 2700 g at 4°C. The cells (45 g from 2 l of culture) were resuspended in 110 ml of lysis buffer (50 mM Tris‐HCl pH 8.0, 1 M NaCl, 10% glycerol, 10 mM β‐mercaptoethanol, 20 mM imidazole, 0.5 mg ml−1 lysozyme, 0.5 mM phenylmethylsulfonyl fluoride, PMSF). After stirring at 4°C for 30 min, cells were ruptured with a sonifier (Branson W‐250 D, Branson, Danbury, CT) and centrifuged for 45 min at 55 000 g at 4°C. The supernatant was purified by nickel affinity chromatography (5 ml HisTrapTM HP column, GE Healthcare). For elution of the protein imidazole buffer [50 mM Tris‐HCl pH 8.0, 500 mM NaCl, 10% (v/v) glycerol, 10 mM β‐mercaptoethanol, 250 mM imidazole] was used. The buffer was changed using PD‐10 desalting columns according to the protocol (GE Healthcare Life Sciences), equilibrated with 100 mM Tris pH 7.5, 15% (v/v) glycerol, 2 mM DTT. Ninety‐three milligrams of His8‐EpzP could be purified.
Assay for prenyltransferase activity
One hundred microlitres of the reaction mixture contained 100 mM Na‐TAPS pH 7.5 (Sigma‐Aldrich, Steinheim, Germany), 0.4 mM freshly prepared dihydro‐PCA, 1 mM DMAPP and 0.5 µg of EpzP. Dihydro‐PCA was prepared by using 90 µl of freshly dissolved 50 mM sodium dithionite and 10 µl of 100 mM PCA (in 1 M Tris‐HCl pH 8). After incubation of the assay for 4 min at 30°C, 15 µl of 100 mM sodium persulfate were added to oxidize dihydro‐PCA to PCA and dihydro‐endophenazine A to endophenazine A. The mixture was extracted with 200 µl of ethylacetate : formic acid (40:1) and after centrifugation 175 µl of the organic phase was evaporated. The residue was dissolved in 100 µl of methanol. Ninety microlitres were investigated by HPLC analysis (Eclipse XDB‐C18 column, 4.6 × 150 mm, 5 µm, Agilent 1200 series, Waldbronn, Germany) with the same method and liquid phase as described for the analysis of the secondary metabolites. The analysis by LC‐MS was the same as for extracts.
Acknowledgments
We thank SPP1152 EVOMET (DFG) for funding, Björn Boll for plasmid pBB09 and assistance with protein purification, the students of the course Pharmaceutical Biology III in summer 2008 for preparing pKG08, Ute Metzger for the synthesis of DMAPP, Inge Unsöld for the synthesis of GPP, Philipp Zeyhle for preparation of flaviolin, Joseph P. Noel (Salk Institute, La Jolla, CA) for plasmid pHis8, and Yvonne Haagen for assistance with screening of the cosmid library and annotation of the genes.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Activity of EpzP with different substrates. EpzP prenylated 9,10‐dihydrophenazine‐1‐carboxylic acid (H2PCA), but not phenazine‐1‐carboxylic acid (PCA), ‐hydroxyphenylpyruvate (4‐HPP) or flaviolin. The prenylation of flaviolin with GPP by Fnq26 results in two structurally different prenylated products (Haagen et al., 2007). 4‐HPP and its prenylated product show two peaks due to keto‐enol tautomerism. Detection in HPLC was carried out at 365 nm for PCA and endophenazine A, 308 nm for 4‐HPP and its products, and 306 nm for flaviolin and its products. All assays contained 0.4 mM aromatic substrate and 0.4 mM isoprenoid substrate and were incubated for 30 min at 30°C. EpzP assays were performed with 500 mM NaCl and 100 mM TAPS pH 7.5, the assay with CloQ with 2 mM MgCl2 and 75 mM Tris‐HCl pH 7.5 and the assay with Fnq26 with 2 mM MgCl2 and 100 mM TAPS pH 8.5 respectively.
Table S1. Influence of MgCl2 and EDTA on the activity of EpzP. Endophenazine A formation was assayed with 0.4 mM H2PCA, 1 mM DMAPP, 10 μg of EpzP in 100 mM TAPS pH 7.5 (100 μl, 30 min, 30°C).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ahuja E.G., Janning P., Mentel M., Graebsch A., Breinbauer R., Hiller W. PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis. J Am Chem Soc. 2008;130:17053–17061. doi: 10.1021/ja806325k. et al. [DOI] [PubMed] [Google Scholar]
- Binz T.M., Wenzel S.C., Schnell H.J., Bechthold A., Müller R. Heterologous expression and genetic engineering of the phenalinolactone biosynthetic gene cluster by using red/ET recombineering. Chembiochem. 2008;9:447–454. doi: 10.1002/cbic.200700549. [DOI] [PubMed] [Google Scholar]
- Bringmann G., Haagen Y., Gulder T.A., Gulder T., Heide L. Biosynthesis of the isoprenoid moieties of furanonaphthoquinone I and endophenazine A in Streptomyces cinnamonensis DSM 1042. J Org Chem. 2007;72:4198–4204. doi: 10.1021/jo0703404. [DOI] [PubMed] [Google Scholar]
- Dairi T. Studies on biosynthetic genes and enzymes of isoprenoids produced by actinomycetes. J Antibiot. 2005;58:227–243. doi: 10.1038/ja.2005.27. [DOI] [PubMed] [Google Scholar]
- Dairi T., Hamano Y., Kuzuyama T., Itoh N., Furihata K., Seto H. Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J Bacteriol. 2001;183:6085–6094. doi: 10.1128/JB.183.20.6085-6094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt K., Schimana J., Krastel P., Dettner K., Rheinheimer J., Zeeck A., Fiedler H.P. Endophenazines A–D, new phenazine antibiotics from the arthropod associated endosymbiont Streptomyces anulatus. I. Taxonomy, fermentation, isolation and biological activities. J Antibiot. 2002;55:794–800. doi: 10.7164/antibiotics.55.794. [DOI] [PubMed] [Google Scholar]
- Gust B., Chandra G., Jakimowicz D., Yuqing T., Bruton C.J., Chater K.F. Lambda red‐mediated genetic manipulation of antibiotic‐producing Streptomyces. Adv Appl Microbiol. 2004;54:107–128. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- Haagen Y., Glück K., Fay K., Kammerer B., Gust B., Heide L. A gene cluster for prenylated naphthoquinone and prenylated phenazine biosynthesis in Streptomyces cinnamonensis DSM 1042. Chembiochem. 2006;7:2016–2027. doi: 10.1002/cbic.200600338. [DOI] [PubMed] [Google Scholar]
- Haagen Y., Unsöld I., Westrich L., Gust B., Richard S.B., Noel J.P., Heide L. A soluble, magnesium‐independent prenyltransferase catalyzes reverse and regular C‐prenylations and O‐prenylations of aromatic substrates. FEBS Lett. 2007;581:2889–2893. doi: 10.1016/j.febslet.2007.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide L. Prenyl transfer to aromatic substrates: genetics and enzymology. Curr Opin Chem Biol. 2009;13:171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Jiang J., He X., Cane D.E. Biosynthesis of the earthy odorant geosmin by a bifunctional Streptomyces coelicolor enzyme. Nat Chem Biol. 2007;3:711–715. doi: 10.1038/nchembio.2007.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A. John Innes Foundation; 2000. [Google Scholar]
- Kuzuyama T., Noel J.P., Richard S.B. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi D.V., Ksenzenko V.N., Bonsall R.F., Cook R.J., Boronin A.M., Thomashow L.S. A seven‐gene locus for synthesis of phenazine‐1‐carboxylic acid by Pseudomonas fluorescens 2‐79. J Bacteriol. 1998;180:2541–2548. doi: 10.1128/jb.180.9.2541-2548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrodi D.V., Thomashow L.S., Blankenfeldt W. Biosynthesis and regulation of phenazine compounds in Pseudomonas spp. In: Rehm B.H.A., editor. Wiley‐VCH; 2008. pp. 331–351. [DOI] [PubMed] [Google Scholar]
- Mavrodi D.V., Peever T.L., Mavrodi O.V., Parejko J.A., Raaijmakers J.M., Lemanceau P. Diversity and evolution of the phenazine biosynthesis pathway. Appl Environ Microbiol. 2010;76:866–879. doi: 10.1128/AEM.02009-09. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.F., Calabrese K., Eisenstein E., Ladner J.E. Structure of the phenazine biosynthesis enzyme PhzG. Acta Crystallogr D Biol Crystallogr. 2004;60:2110–2113. doi: 10.1107/S0907444904022474. [DOI] [PubMed] [Google Scholar]
- Parsons J.F., Greenhagen B.T., Shi K., Calabrese K., Robinson H., Ladner J.E. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry. 2007;46:1821–1828. doi: 10.1021/bi6024403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pojer F., Wemakor E., Kammerer B., Chen H., Walsh C.T., Li S.M., Heide L. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci USA. 2003;100:2316–2321. doi: 10.1073/pnas.0337708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose T.M., Schultz E.R., Henikoff J.G., Pietrokovski S., McCallum C.M., Henikoff S. Consensus‐degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose T., Henikoff J., Henikoff S. codehop (COnsensus‐DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh O., Gust B., Boll B., Fiedler H.P., Heide L. Aromatic prenylation in phenazine biosynthesis: dihydrophenazine‐1‐carboxylate dimethylallyltransferase from Streptomyces anulatus. J Biol Chem. 2009a;284:14439–14447. doi: 10.1074/jbc.M901312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh O., Haagen Y., Seeger K., Heide L. Prenyl transfer to aromatic substrates in the biosynthesis of aminocoumarins, meroterpenoids and phenazines: the ABBA prenyltransferase family. Phytochemistry. 2009b;70:1728–1738. doi: 10.1016/j.phytochem.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sedmera P., Pospíšil S., Novák J. New furanonaphthoquinone from Streptomyces cinnamonensis. J Nat Prod. 1991;54:870–872. [Google Scholar]
- Shin‐ya K., Furihata K., Hayakawa Y., Seto H. Biosynthetic studies of naphterpin, a terpenoid metabolite of Streptomyces. Tetrahedron Lett. 1990;31:6025–6026. [Google Scholar]
- Takahashi S., Takagi H., Toyoda A., Uramoto M., Nogawa T., Ueki M. Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN‐593. J Bacteriol. 2010;192:2839–2851. doi: 10.1128/JB.01557-09. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax J., Sedmera P., Vokoun J., Urban J., Karnetová J., Stajner K. Phenazines from Streptomyces cinnamonensis. Collect Czech Chem Commun. 1983;48:527–532. et al. [Google Scholar]
- Tello M., Kuzuyama T., Heide L., Noel J.P., Richard S.B. The ABBA family of aromatic prenyltransferases: broadening natural product diversity. Cell Mol Life Sci. 2008;65:1459–1463. doi: 10.1007/s00018-008-7579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodside A.B., Huang Z., Poulter C.D. Trisammonium geranyl diphosphate [diphosphoric acid, mono(3,7‐dimethyl‐2,6‐octadienyl) ester (E)‐, trisammonium salt] Org Synth. 1993;66:211–211. [Google Scholar]
- Zhao X.Q., Gust B., Heide L. S‐adenosylmethionine (SAM) and antibiotic biosynthesis: effect of external addition of SAM and of overexpression of SAM biosynthesis genes on novobiocin production in Streptomyces. Arch Microbiol. 2010;192:289–297. doi: 10.1007/s00203-010-0548-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Activity of EpzP with different substrates. EpzP prenylated 9,10‐dihydrophenazine‐1‐carboxylic acid (H2PCA), but not phenazine‐1‐carboxylic acid (PCA), ‐hydroxyphenylpyruvate (4‐HPP) or flaviolin. The prenylation of flaviolin with GPP by Fnq26 results in two structurally different prenylated products (Haagen et al., 2007). 4‐HPP and its prenylated product show two peaks due to keto‐enol tautomerism. Detection in HPLC was carried out at 365 nm for PCA and endophenazine A, 308 nm for 4‐HPP and its products, and 306 nm for flaviolin and its products. All assays contained 0.4 mM aromatic substrate and 0.4 mM isoprenoid substrate and were incubated for 30 min at 30°C. EpzP assays were performed with 500 mM NaCl and 100 mM TAPS pH 7.5, the assay with CloQ with 2 mM MgCl2 and 75 mM Tris‐HCl pH 7.5 and the assay with Fnq26 with 2 mM MgCl2 and 100 mM TAPS pH 8.5 respectively.
Table S1. Influence of MgCl2 and EDTA on the activity of EpzP. Endophenazine A formation was assayed with 0.4 mM H2PCA, 1 mM DMAPP, 10 μg of EpzP in 100 mM TAPS pH 7.5 (100 μl, 30 min, 30°C).


