Abstract
Imprinted genes are often grouped in clusters at defined chromosomal locations. Long-range regulatory effects are implicated in the control of imprinting and these could be co-opted in the emergence of novel imprinted genes during evolution. We present a detailed analysis of a novel imprinted GFP mouse line. Tel7KI is a new insertion allele near the Ins2 locus within a cluster of imprinted genes on distal mouse Chr 7. The GFP reporter becomes regulated by the host domain in two notable fashions. First, transcription of GFP is imprinted and active exclusively from the maternally inherited allele in the embryo. Second, the expressed maternal allele is subject to position effects reflecting a distinct pattern of expression. The GFP reporter acquires silencing DNA methylation marks on the paternal allele after fertilization. This imprinting is not acquired in the placenta, where GFP is active from both parental alleles, demonstrating key epigenetic differences between embryonic and extra-embryonic lineages. Our analysis shows that imprinted clusters can provide environments conducive to the acquisition of imprinting upon novel inserted transcriptional units. The Tel7KI line offers new powerful avenues to explore both genetic and environmental factors implicated in the acquisition and maintenance of imprinted transcription in mammals.
Keywords: genomic imprinting, transgene, GFP, DNA methylation, Ins2, placenta, mouse Chr 7
Introduction
Genomic imprinting represents a unique epigenetic regulatory mechanism operating in placental mammals and originating from differential epigenetic marks inherited from the paternal and maternal genomes at fertilization (Reik and Walter, 2001a). The most widely studied manifestation of these allelic differences is the parent-of-origin specific monoallelic expression of so-called imprinted genes. As a group, these genes share a number of important characteristics which provide some insight into the molecular mechanisms involved in their regulation (Wood and Oakey, 2006). Imprinted genes tend to be grouped together in large chromosomal domains, suggesting that some aspect of the imprinting mechanism is mediated via cis interactions between loci. At the epigenetic level, DNA methylation at promoter regions has been associated with silencing at imprinted genes, transcripts on the inactive X chromosome, as well as during programmed or pathological silencing of gene expression in mammals (Weber and Schübeler, 2007).
The imprinted domain located close to the telomeric end of mouse chromosome 7 (Chr 7) has provided an important model for the study of imprinting (Reik et al., 2004). This region, covering ~1 Mb, contains two differentially methylated regions (DMRs) which inherit their DNA methylation imprints directly from one of the parental germlines. At the proximal end, the imprinting centre 1 (IC1, or H19 DMR) located 2.3 kb upstream of the maternally expressed H19 gene acquires a DNA methylation imprint during spermatogenesis but remains unmethylated in the maternal germline (Bartolomei et al., 1993; Ferguson-Smith et al., 1993; Tremblay et al., 1995). The DNA methylation germline imprint at IC1 spreads to the H19 promoter and is responsible for silencing of the paternal allele of H19 (Srivastava et al., 2000). Furthermore, IC1 is implicated in the long-range regulation of Igf2 and Ins2 via the formation of a DNA-methylation sensitive insulator requiring the binding of CTCF to the unmethylated maternal IC1 (Leighton et al., 1995; Bell and Felsenfeld, 2000; Hark et al., 2000; Szabó et al., 2000). The mechanisms by which the effects of this epigenetically controlled insulator are restricted along Chr 7 are unknown, thus it is not known whether it can bias allelic usage at sites distal of Ins2.
Regulating the more distal imprinted domain, IC2 (KvDMR1) acts at least in part as the CpG-rich promoter for the long non-coding RNA Kcnq1ot1 (Fitzpatrick et al., 2002). Since IC2 is specifically methylated during oogenesis (Smilinich et al., 1999), only the paternally inherited allele of Kcnq1ot1 is expressed, leading to paternal allele-specific recruitment of Polycomb group proteins and repressive histone marks which are implicated in the silencing of at least ten neighboring protein-coding genes (Lewis et al., 2004; Umlauf et al., 2004; Terranova et al., 2008). The exact function of Kcnq1ot1 is still unknown, though both the presence of IC2 and proper paternal expression of the transcript are required for silencing in cis of two categories of imprinted genes found in this cluster: the ubiquitously imprinted genes, monoallelically expressed in both embryo and placenta, and the placentally-imprinted genes, which show monoallelic expression only the placenta (Lewis et al., 2006; Shin et al., 2008). Proximally, silencing from IC2 spreads ~330 kb to the Ascl2 locus, which is exclusively expressed from the maternal allele in the placenta (Guillemot et al., 1995). The recent identification of Th and Dhcr7 as preferentially expressed from the maternal allele in the placenta (Schulz et al., 2006) has led to the proposition of a broader domain of IC2-regulated genes, mediated by a Polycomb group protein-dependent compaction of the paternal chromosome expressing Kcnq1ot1 (Terranova et al., 2008). As in the case of IC1, the extent of the spreading of this ncRNA-mediated silencing is currently unknown.
We present here the initial characterization of a novel transgenic mouse line carrying an insertion on Chr 7 between the IC1- and IC2-regulated clusters and expressing the fluorescent reporter EGFP. This line, called Tel7KI (official name Ins2tm1Lef), was obtained in the course of experiments aimed at truncating Chr 7 using a linear telomere seed vector (Oh et al., 2008) by a Cre-mediated trans reaction involving a targeted loxP-containing promoter-less neo cassette, the I2loxP allele (Ins2tm1.1Nagy, MGI: 3807183) located 2.6 kb upstream of the Ins2 gene (Fig. 1A) and a vector containing the CAG-EGFP reporter and a Pgk-1 promoter (Pgk) followed by a loxP site (Fig. 1B). Using this approach, we have previously described the generation of two modifications of distal Chr7, the chromosomal truncation DelTel7 (discussed in detail in (Oh et al., 2008)) and the conditional insertion Tel7KI, which is the subject of the current study. Previously the CAG-EGFP construct, driving widespread embryonic and postnatal GFP expression in the mouse (Okabe et al., 1997), has been shown to be capable of undergoing epigenetic silencing on the inactive X chromosome (Hadjantonakis et al., 2001). Other than this case, several transgenic lines which have been derived with the same promoter-enhancer combination showed no evidence of epigenetic regulation. This includes EGFP, EYFP, ECFP, dsRed variants as well as CAG-based conditional constructs. These results imply the the CAG enhancer-promoter combination does not contain the necessary signals to direct its own imprinting at ectopic sites in the genome (Okabe et al., 1997; Novak et al., 2000; Hadjantonakis et al., 2001; Nakanishi et al., 2002; Hadjantonakis and Papaioannou, 2004).
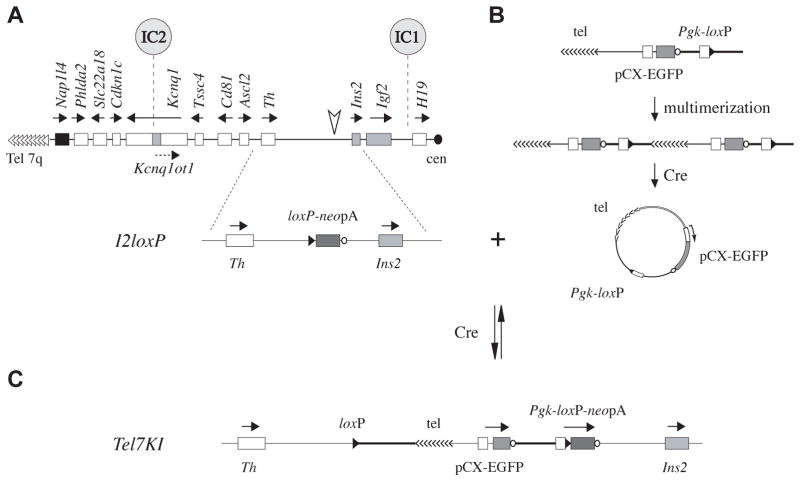
Fig. 1. Cre-mediated insertion at the Ins2 locus and structure of the Tel7KI allele.
(A) Diagram of the imprinted domain on distal Chr7 showing the location of the insertion upstream of Ins2 (arrowhead), relative to the two imprinting centers (IC). Maternally and paternally expressed genes are in white and grey, non-imprinted genes in black; arrows show transcriptional orientation. Structure of the targeted I2loxP allele, with its promoterless loxP-neo-polyA (loxP-neopA) cassette inserted 2.6 kb telomeric to Ins2 (below). (B) Proposed mechanism to generate a circular intermediate in I2loxP/+ ES cells. The linear targeting vector is postulated to have multimerized, then a circular intermediate was resolved by Cre from this array. This provided a substrate for Cre-mediated insertion at I2loxP. (C) Resulting structure of the loxP-flanked Tel7KI allele showing the telomeric repeats (tel), the ubiquitous EGFP reporter (pCX-EGFP), and active Pgk-loxP-neopA marker.
We show here that the Tel7KI allele is regulated by genomic imprinting in the embryo and is exclusively expressed from the maternal allele. It provides a sensitive and non-invasive assay to study the epigenetic regulation of imprinted transcription throughout mammalian development. The paternal allele acquires repressive DNA methylation marks post-fertilization, which are present in the embryo but not in the extra-embryonic tissues. Accordingly, Tel7KI is not imprinted in the placenta. Our findings show that long-range signals can impart a complex tissue-specific imprinted regulation to an inserted transcriptional unit. This line provides a powerful model for genetic studies of genomic imprinting in vivo and raises important issues on the tissue-specific spreading of epigenetic signals on distal Chr 7.
Materials and methods
Tel7KI line: ES cells and mice
Derivation of Tel7KI by a recombinase-mediated insertion at the Ins2 locus was previously described (Oh et al., 2008). The culture and electroporation of embryonic stem (ES) cells followed standard protocols (Nagy et al., 2003). 129S1/SvImJ mice are from The Jackson Laboratory (stock number: 002448), CD-1 mice from the UBC Animal Care Centre. The congenic mouse line with distal Chr 7 M. m. castaneus SNPs on the 129S1 background was derived in our laboratory (129S1cCAST7/Lef). All animal experimentation followed the guidelines from the Canadian Council on Animal Care (CCAC) under UBC animal care license numbers A03-0289 and A03-0292.
Embryos and genotyping
Heterozygous Tel7KI male and/or female mice were mated to 129S1, CD-1 or 129S1cCAST7 animals. For timed mating, the day of the vaginal plug is E0.5. Pre-implantation embryos were collected at E3.5 as described (Nagy et al., 2003). Females were sacrificed at the desired day from E7.5 to E18.5, embryos were scored for GFP phenotype and yolk sac samples were taken for PCR genotyping (Nagy et al., 2003). Photographs at low magnification were taken on a Leica MS5 dissecting microscope equipped with a Q-imaging Micropublisher 3.3 RTC colour camera and the fluorescent light source MAA-03 (BLS Ltd.). Genotyping of the Tel7KI allele was performed with the Δ5′ PCR reaction (Oh et al., 2008). Homozygous mutant embryos were detected by a positive Δ5′ PCR reaction and absence of a wild-type band (I2wt). Primer sequences for genotyping are given in Table S1.
DNA bisulfite modification and sequencing
DNA was extracted from the remaining Trizol fraction or from tissues digested with proteinase K (Lefebvre et al., 1997). 1–2 μg of genomic DNA was treated with sodium bisulfite (Davis et al., 1998; Umlauf et al., 2004; Oh et al., 2008) and 1 μl of the treated DNA was used in the first round of nested PCR as follows: 40 cycles of 95°C for 30s, 52°C for 30s and 72°C for 30 s. 1 μl of the first round product was used in a 50-μl reaction for a second PCR with an annealing temperature of 55°C for 35 cycles. Bisulfite PCR for the β-actin promoter (36 CpGs) was a semi-nested reaction, using primers BABF6 and BABR5d in round 1 (299 bp), followed by round 2 with BABF6 and BABR4c (253 bp). Bisulfite sequencing primers for the GFP open-reading-frame were a gift from M. Lorincz and analyze 37 CpGs in a 430-bp fragment. For each sample, four independent PCR reactions were performed and four cloned strands were sequenced per PCR. All primer sequences are in Table S1.
Flow cytometry
E9.5 embryos were washed in PBS and passed through a p200 pipette tip in the presence of trypsin (0.25%, Gibco). Suspensions were pipetted up and down to disaggregate cells after 2 and 5 minutes at 37°C, the trypsin was inactivated by the addition of FACS buffer (PBS with 2mM EDTA, 20% FCS (Gibco), and 2mg/ml Propidium Iodide (Gibco)), and cells passed through a 26-gauge needle. For sorting cells from E10.5 embryos, the procedure was identical, though collagenase/dispase (Roche, 1mg/ml) was used in place of trypsin. Analyses were performed on a BD LSRII and sorting on a BDFacsARIA. Data analysis was performed with FlowJo 8.0.
RNA extraction and RT-PCR
RNA was extracted from snap-frozen tissues by a single-step isolation using Trizol (Invitrogen Life Technologies, CA) according to manufacturer’s directions. Approximately 1μg of RNA was reverse transcribed as per Invitrogen SSII protocol. Q-PCR on 1μl of cDNA was performed on an Bio-Rad Opticon 2: 95°C 2 minutes followed by 35 cycles of 95°C 30 sec, 58°C 30 sec, 72°C 30 sec, 85°C 1sec with plate read. Ct values of triplicate samples were averaged and used to calculate relative amounts of transcript, normalized to Gapdh. EGFP transcript from Tel7KI was detected with the BAE1F and BAE1R primers (Table S1).
Immunohistochemistry
Embryos and placentae were fixed in 4% paraformaldehyde (PFA, Sigma) and embedded as described (Cowan et al., 2001). Frozen blocks were sectioned on a Leica CM3050 cryostat and sections placed on Superfrost slides (Fisher). Detection of epitopes was performed as described (Cowan et al., 2001) with chicken anti-GFP (1:500, AbCam) or rat anti-CD34 (1:75, BD) primary antibodies and A594 anti-rat (1:200, Molecular Probes) and A488 anti-chicken (1:200, Molecular Probes) secondary antibodies. Sections were coverslipped with vectashield (Vector Labs) and imaged on a Leica DMI6000B inverted fluorescent microscope. Images were captured with a Q-imaging Retiga 4000R monochrome camera and processed with Openlab (Improvision). Whole embryo and placenta photos were stitched together from 6–8 overlapping photos in Photoshop. Overlay analysis on placental sections was performed with “Colocalization Highlighter” plugin for ImageJ (created by Pierre Bourdoncle, Institut Jacques Monod, Service Imagerie, Paris, bourdoncle@ijm.jussieu.fr) with default values.
Whole EPCs cultured on coverslips were treated similarly with a few modifications. Coverslips were blocked in 3% BSA with 0.2% Triton-X for 30 minutes, then incubated with chicken anti-GFP (1:500, AbCam) and rabbit anti-placental lactogen I (Pl-I, or PRL3D1,1:250, Chemicon) for 2 hours at room temperature. After washing with PBS-T and re-blocking for 10 minutes, they were incubated with secondary antibodies, washed, counterstained, mounted and imaged as above.
Ectoplacental cone collection and culture
Ectoplacental cones (EPCs) were isolated from E8.5 conceptuses heterozygous for Chr 7 SNPs as described (Nagy et al., 2003). EPCs were cultured in a 4-well dish as described (El-Hashash and Kimber, 2004). For immediate RNA and DNA preparation, 5–6 EPCs were pooled and frozen on dry ice. EPCs were cultured on fibronectin-coated 12 mm glass coverslips (VWR) in DMEM supplemented with 20% FCS, penicillin/streptomycin (Gibco), and L-glutamine (Gibco) for 5 days before either being collected with trypsin, pooled, and frozen for RNA and DNA extraction, or fixed for 30 minutes at room temperature in 4% PFA for IHC.
Results
Generation of a GFP insertion on distal chromosome 7
The Tel7KI allele was recovered as a Cre-mediated insertion at the Ins2 allele I2loxP using a linear telomere seed vector (Fig. 1A) (Oh et al., 2008). We hypothesize that a circular intermediate (Fig. 1B) provided a substrate for a Cre-mediated insertion of the vector at the I2loxP site, resulting in G418-resistant ES cell colonies following the reconstitution of a functional Pgk-loxP-neopA marker (Fig. 1C) (Sauer, 1993; Hardouin and Nagy, 2000).
The structure of Tel7KI was confirmed by genomic PCR, Southern blot, and DNA FISH (Oh et al., 2008). The insertion is conditional and can be excised by transient Cre production in ES cells (Supplementary material, Fig. S1). A mouse line carrying this allele was previously derived (Oh et al., 2008). Tel7KI animals have now been maintained on the 129S1/SvImJ background for seven generations with no abnormal phenotype observed. The line was also outcrossed onto the CD-1 outbred background with no noticeable differences in expression phenotypes. The results presented here therefore combine observations made on both strain backgrounds. In this study, by expression of Tel7KI we refer to the transcription of the EGFP reporter from the CAG promoter, detection of GFP fluorescence, or immunological detection of the EGFP protein itself.
Imprinted GFP expression in post-implantation embryos
We hypothesized that the CAG-EGFP reporter might be regulated by imprinting signals in the context of its insertion site within the IC1- and IC2-regulated domains in the Tel7KI line. To address this possibility, we examined pre- and post-implantation embryos carrying either a maternally or paternally derived copy of Tel7KI. In E3.5 blastocysts, GFP fluorescence is observed in inner cell mass and trophectoderm cells upon both maternal and paternal inheritance (Fig. 2A). Starting at E7.5, the GFP reporter is expressed in a parent-of-origin-specific manner and GFP fluorescence is observed only in the embryos inheriting Tel7KI from the maternal germline (KI/+, data not shown and Fig. 2B, 3A). The widespread GFP activity of the maternal allele has been consistently observed at all stages examined, from E7.5 to E18.5 (Fig. 2B, 3A and data not shown), but little GFP expression is observed in transgenic KI/+ neonates or in adult tissues (data not shown).
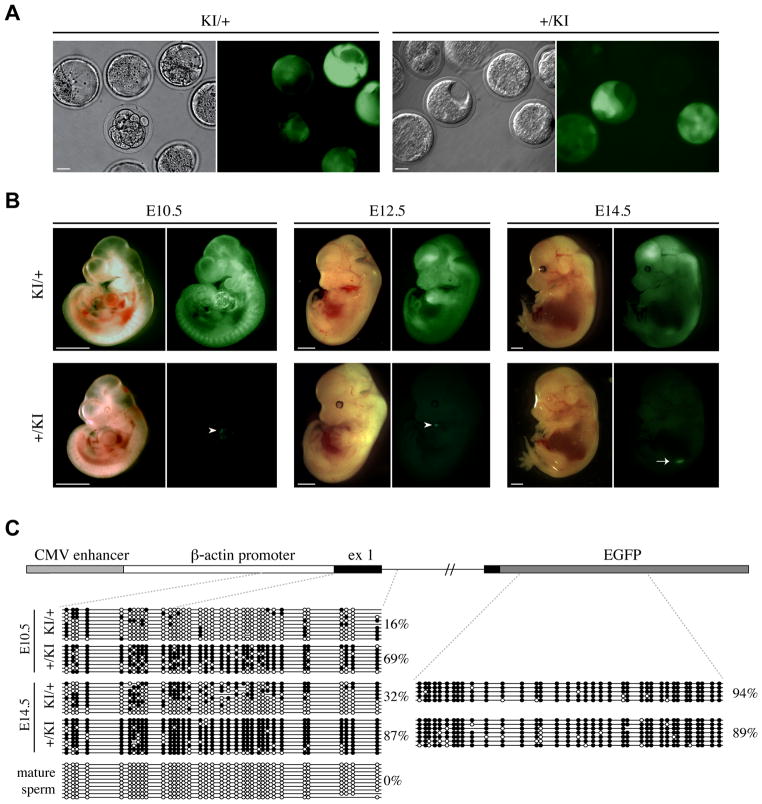
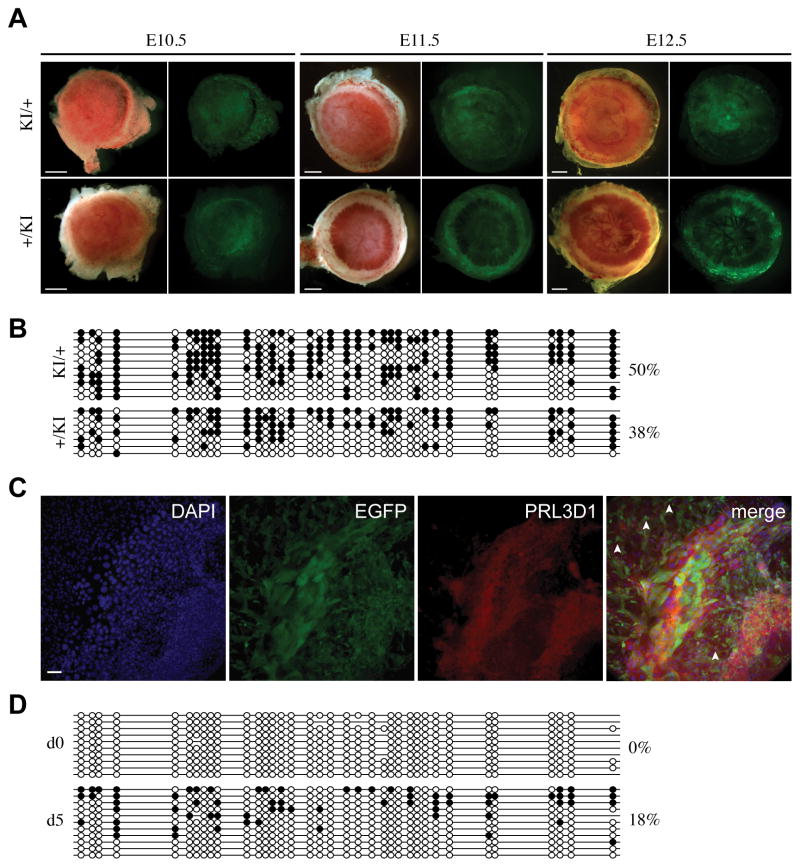
Fig. 2. Relationship between imprinted GFP expression and DNA methylation at the CAG promoter in post-implantation embryos.
(A) E3.5 maternal (KI/+) and paternal (+/KI) heterozygous embryos were collected from crosses between wild-type C57BL/6J and Tel7KI (KI for short) heterozygous mice. Pre-implantation embryos were observed by microscopy under bright field (left) or GFP fluorescence (right). Scale bar: 20 μm. (B) Imprinted expression of Tel7KI in post-implantation embryo. E10.5, E12.5, and E14.5 embryos carrying a maternal (KI/+) or a paternal (+/KI) Tel7KI were visualized under bright field (left) or GFP fluorescence (right). Occasional GFP fluorescence is observed in hearts of +/KI embryos (white arrowheads), and GFP expression from the gonads of +/KI embryos can be seen through the body wall after E13.5 (white arrow). Scale bar: 1 mm. (C) Structure of the CAG-EGFP reporter of Tel7KI (above) showing the CMV enhancer, the chicken β-actin promoter, and a 5′ intron from the chicken β-globin gene, fused to EGFP. Sequencing of sodium bisulfite-modified genomic DNA (below) purified from paternal (+/KI) and maternal (KI/+) transmission embryos and mature sperm from a +/KI adult male. Circles represent methylated (filled) or unmethylated (open) CpGs; absent circles indicate ambiguous sites.
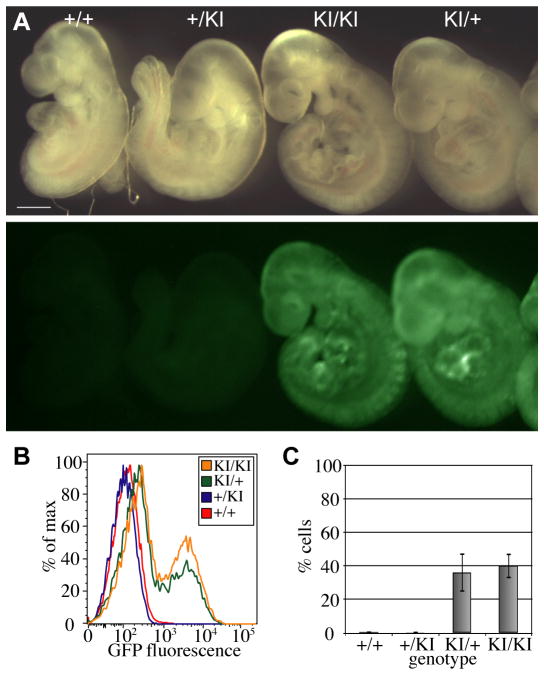
Fig. 3. Regulation of GFP expression from Tel7KI in post-implantation embryos.
(A) E9.5 wild-type (+/+), paternal transmission (+/KI), maternal transmission (KI/+), and homozygous (KI/KI) embryos show imprinted expression of GFP viewed in bright field (above) and under GFP fluorescence (below). Scale bar: 1 mm. (B) Flow cytometry data of single disaggregated E9.5 mouse embryos. GFP expression profiles are from single +/+ (red), +/KI (blue), KI/+ (green), and KI/KI (orange) embryos. (C) Summary of flow cytometry analysis over multiple litters showing the percentage of cells expressing GFP in E9.5 embryos. Bars indicate mean and standard deviation of results obtained from multiple individual embryos: +/+ n=3, +/KI n=9, KI/+ n=14, and KI/KI, n=4.
Upon paternal transmission, the GFP reporter is silenced in most embryonic tissues (+/KI, Fig. 2B, 3A). The exception is the developing gonad, which showed strong GFP expression in all the E11.5 and later stage embryos examined (E14.5 shown in Fig. 2B, white arrow). Furthermore, in some embryos, particularly at later stages, localized foci of GFP-expressing cells are observed in the heart (Fig. 2B, arrowheads), and less frequently and in a more variable pattern, in the brain. Importantly, this parent-of-origin specific expression of GFP from Tel7KI is reversible. Female mice inheriting a silent allele from their fathers give embryos which show high levels of GFP expression and male mice with an active maternal allele give rise to GFP-negative progeny. Our results indicate that the epigenetic parent-of-origin specific marking of Tel7KI is appropriately reset at each generation as observed at endogenous imprinted loci.
Promoter DNA methylation marks are acquired on the silent paternal Tel7KI allele after fertilization
Since the CAG-EGFP reporter is CpG rich we hypothesized that DNA methylation might be implicated in the regulation of its expression in Tel7KI embryos. We devised a sodium bisulfite sequencing assay to examine 36 CpG dinucleotides from the 5′ portion of the reporter, including part of the chicken β-actin promoter, the transcription start site, exon 1 and part of intron 1 (Fig. 2C). First, we analyzed two different developmental stages (E10.5 and E14.5), both of which show high levels of GFP expression from the maternal allele (KI/+) and no GFP in +/KI embryos (Fig. 2A). At E10.5 there is a striking difference in the level of DNA methylation at the CAG promoter region between maternal (16%) and paternal (69%) transmission of Tel7KI (Fig. 2C). This methylation difference is maintained at E14.5, where the paternal allele is methylated at more than 85%. During this period we also observed an increase in the methylation level of the expressed maternal allele which is not fully unmethylated despite the high expression levels.
In order to determine whether the DNA methylation at the promoter driving GFP expression from Tel7KI constitutes a germline imprint, mature sperm collected from a 1-year old transgenic +/KI male was analyzed. No methylation was detected in the epididymal sperm sample (Fig. 2C, bottom left panel). Finally, we examined CpG methylation at the EGFP open reading frame (ORF) at E14.5 to see if the observed differential methylation extended past the promoter region. The EGFP ORF is also CpG-rich (51 CpGs in 732bp), but no differences in DNA methylation levels were observed between maternal and paternal transmission of Tel7KI as both alleles were highly methylated (89% for +/KI and 94% for KI/+; Fig. 2C). Our results are consistent we the observation that mammalian genes are often methylated in the body of the gene (Hellman and Chess, 2007; Ball et al., 2009), and suggest that at Tel7KI only the CAG promoter methylation respond to imprinting signals.
Expression of the maternally inherited Tel7KI is tissue-specific
To analyze the profile of GFP expression during development, we collected E9.5 embryos of all four possible genotypes: wild-type (+/+), paternal hemizygous (+/KI), maternal hemizygous (KI/+), or homozygous mutant (KI/KI). Analysis of GFP fluorescence in whole mount showed a clear pattern of imprinted expression, with fluorescence only detected from the maternal allele in KI/+ and KI/KI embryos (Fig. 3A). These embryos were disaggregated and single-cell suspensions were analyzed by flow cytometry. While no cells expressing GFP above wild-type background were detected in +/KI embryos, approximately 40% of cells analyzed from KI/+ and KI/KI embryos express GFP at high levels (Fig. 3B, C). GFP expression was also analyzed by quantitative RT-PCR (Q-RT-PCR) for two individual embryos of each genotype at E10.5. The results confirmed the absence of GFP expression in wild-type or paternal transmission embryos and detected a variable level of expression from the maternal allele in KI/+ and KI/KI embryos (Supplementary Material, Fig. S2).
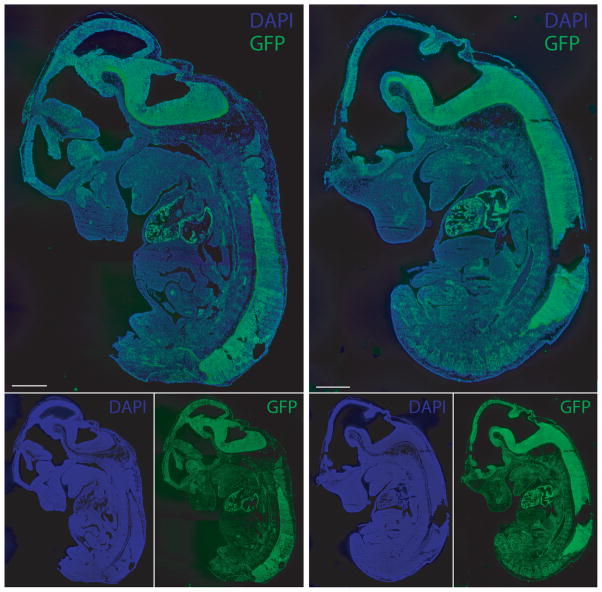
We then asked whether the expression detected upon maternal transmission reflects a reproducible tissue-specific or a stochastic pattern of expression. Fixed E12.5 KI/+ embryos were analyzed by immunohistochemistry on frozen sections using an antibody against GFP. Two independent embryos from two different E12.5 litters were analyzed (only two littermates are shown in Fig.4). The production of GFP was observed most strongly in the heart and central nervous system (CNS), both exhibiting consistently high levels of expression in all embryos examined. In other tissues, GFP expression was lower but still detectable above background (Fig. 4). Most tissues contained at least some cells expressing GFP, but no tissues outside the heart and CNS showed consistent high-level GFP expression. The results show a clear tissue-specific expression acting on the maternally inherited, non-imprinted allele of Tel7KI.
Figure 4. Tissue-specific expression of GFP in maternal transmission Tel7KI embryos.
GFP expression in 14-μm frozen sections of E12.5 maternal transmission embryos. Two embryos were stained with a polyclonal anti-GFP antibody and counterstained with DAPI to examine extent and reproducibility of GFP expression from the maternal allele of Tel7KI. Scale bars: 1 mm.
Tel7KI is not imprinted in the placenta
Tel7KI placentae were found to deviate from the pattern of imprinted GFP expression seen in the embryo. Both paternal and maternal transmission placentae show punctate GFP expression throughout the placenta (Fig. 5A). The molecular basis for this deviation from the embryonic patterns was examined by DNA methylation analysis at the GFP promoter of the Tel7KI allele in E14.5 placentae. At E14.5, while the paternal allele is methylated at a much higher level than the maternal allele in the embryo (Fig. 2C), these epigenetic differences are not observed in whole placentae (Fig. 5B). Both maternal and paternal transmission placentae are moderately methylated (38% paternal, 50% maternal), and there is no significant difference between their levels of DNA methylation.
Fig. 5. Lack of imprinted GFP expression and DNA methylation at Tel7KI in the placenta and in cultured trophoblast giant cells.
(A) Placentae carrying maternal (KI/+) or paternal (+/KI) alleles of Tel7KI visualized under bright field (left) and GFP fluorescence (right) at E10.5, E11.5, and E12.5. Scale bars: 1 mm. (B) Promoter DNA methylation analysis by bisulfite sequencing at Tel7KI in whole E14.5 placentae carrying a maternal (KI/+) or paternal (+/KI) Tel7KI. Filled circles represent methylated CpGs, open circles represent unmethylated CpGs and absent circles are CpGs for which data is unavailable. (C) Immunohistochemical detection of GFP in giant cells. Trophoblast giant cells (TGCs) grown from EPCs in culture for 5 days were stained with antibodies against GFP and the TGC marker placental lactogen 1 (PRL3D1). Scale bar: 100 μm. (D) DNA methylation analysis of paternal transmission Tel7KI E8.5 EPCs both uncultured (d0) and cultured in vitro for 5 days (d5). Sodium bisulfite-modified genomic DNA was analyzed for DNA methylation patterns at the CAG promoter of the paternal Tel7KI allele.
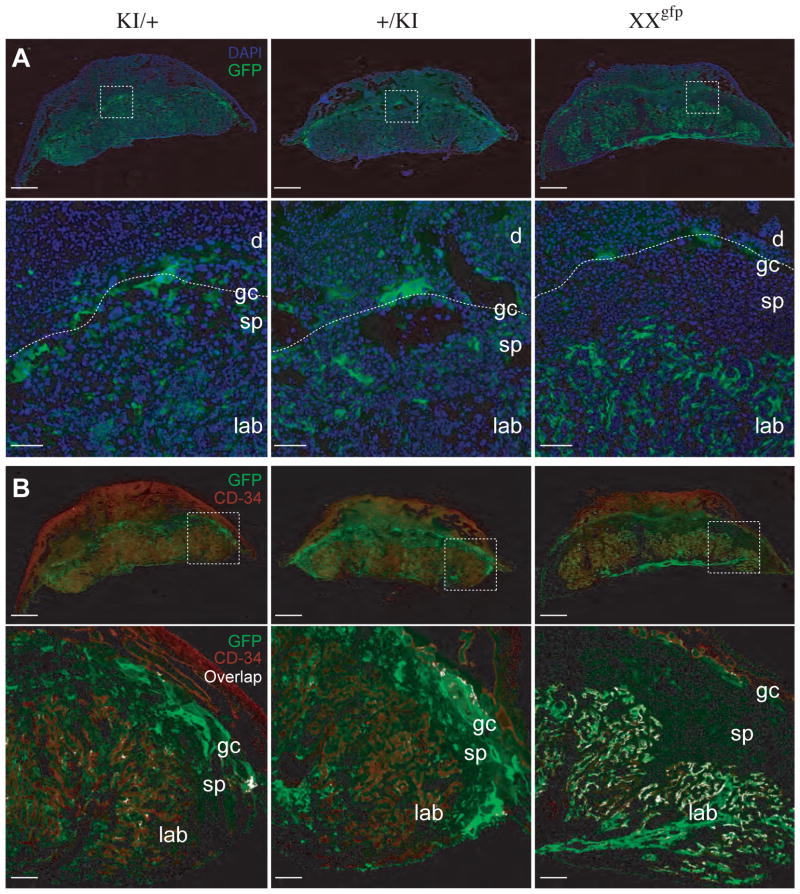
Our results therefore suggest that in the placenta the Tel7KI allele does not acquire the dense DNA methylation mark which characterizes the paternal allele in the embryo. Alternatively, imprinted expression could be lineage-specific in the placenta and restricted, for instance, to the extra-embryonic mesoderm (ExM), with the trophoblast lineage showing a relaxation of imprinting. We addressed this possibility by analyzing sections of E12.5 placentae by immunohistochemistry to determine which placental cell types were producing GFP from Tel7KI. Expression patterns of GFP upon maternal or paternal transmission were similar: a punctate pattern of expression throughout the labyrinth, spongiotrophoblast, and giant cell layer was observed, with the highest level of expression seen in the giant cell layer (Fig. 6A). No major differences were observed between +/KI and KI/+ placentae. This is in sharp contrast with the pattern of GFP expression observed from the X-linked D4 transgene. In the mouse, X chromosome inactivation is imprinted in the trophoblast lineage, with preferential inactivation of the paternally inherited X chromosome. We compared the placental expression of Tel7KI with that of the X-linked EGFP inherited paternally in a female placenta (Fig. 6A, XXgfp). As observed previously, imprinted silencing of the paternally inherited transgene is maintained in most trophoblast cell types, with the exception of giant cells which show abnormal relaxation of silencing and activation of the GFP transgene (Hadjantonakis et al., 2001). Unlike the Tel7KI allele, D4 is broadly expressed in the labyrinth, as would be expected in these epiblast derivatives undergoing random X-inactivation in the ExM.
Fig. 6. Placental expression of Tel7KI.
14-μm frozen sections of E12.5 placentae were analyzed for GFP expression with an anti-GFP antibody (green). Samples are from conceptuses with maternal (KI/+) or paternal (+/KI) Tel7KI and from a female conceptus carrying the pCX-EGFP D4 transgene on its paternal X chromosome (XXgfp). (A) Sections counter-stained with DAPI (blue). Scale bars: 1 mm. Lower panels show higher magnification views of the areas boxed above, highlighting the GFP-positive giant cell layer at the fetal-maternal interface (dotted lines) and the trophoblast glycogen cells which have migrated into the decidua in both Tel7KI placentae. Scale bars: 200 μm. (B) Expression of GFP in the extra-embryonic mesoderm assessed by co-immunohistochemical staining with antibody against CD34 (red). Scale bars: 1 mm. Lower panels: magnification of the areas boxed above, showing co-localization of GFP and CD34 staining with overlap highlighted in white. Scale bars: 200 μm. The structures labeled include the maternal decidua (d), the giant cell (gc), spongiotrophoblast (sp), and labyrinthine (lab) layers.
Using immunohistochemistry for CD34 the pattern of expression of GFP in the ExM of the placenta was analyzed (Sánchez et al., 1996). If Tel7KI is imprinted in all epiblast derivatives we predicted that, as in the embryo, GFP expression should be visible in ExM only upon maternal transmission. However we observed little co-localization between CD34 and GFP, indicating that Tel7KI is not highly expressed in extraembryonic mesoderm in either paternal or maternal hemizygotes (Fig. 6B). In contrast, a placenta from a female embryo carrying the paternally-derived X-linked GFP shows high co-localisation between these two markers.
We next asked whether the GFP expression and lack of methylation upon paternal transmission of Tel7KI in the placenta was due to loss of methylation during placental growth. It is possible that the hypomethylation of the placenta (Rossant, 1986) causes a reduction in methylation at Tel7KI and a concomitant increase in expression of GFP. We isolated ectoplacental cones (EPCs) from E8.5 transgenic embryos and examined DNA methylation at Tel7KI both before and after culturing the EPCs in vitro for 5 days. During this differentiation period, cultured diploid trophoblast cells give rise to polyploid secondary giant cells which show strong GFP fluorescence in +/KI cultures. By immunohistochemistry, the high levels of GFP co-localize with cells expressing placental lactogen 1 (product of the Prl3b1 gene), a giant cell marker (Fig. 5C), although several PRL3B1-negative cells of lower ploidy were also found to be expressing GFP (Fig. 5C, arrowheads). We found no DNA methylation at Tel7KI in the uncultured +/KI E8.5 EPCs (Fig. 5D, d0 panel). However, upon culturing, some de novo DNA methylation was observed at Tel7KI (Fig. 5D, d5 panel). This suggests that the moderate amount of DNA methylation seen in mature paternal transmission placentae is not due to loss of methylation, but rather that the density of methyl groups present in the embryo is in fact never acquired in the placenta on the paternal allele. Our data also show that trophoblast derivatives are capable of methylating Tel7KI and that DNA methylation is not restricted to the epiblast-derived ExM lineage.
Our analysis has also revealed that in two different imprinted GFP transgenic lines, Tel7KI on Chr 7 and D4 on the X chromosome, the trophoblast giant cell lineage demonstrates high levels of GFP expression (Fig. 6A). This reactivation in the D4 line has been hypothesized to reflect loss of imprinted X-inactivation in TGCs (Hadjantonakis et al., 2001). To determine whether this cell lineage shows a general defect in the maintenance of epigenetic silencing we analyzed the status of endogenous imprinted genes in TGCs differentiated from EPCs in vitro (Gonçalves et al., 2003; El-Hashash and Kimber, 2004). The distal Chr 7 imprinted genes H19, Igf2, and Cdkn1c exhibited normal imprinted expression in TGCs, and the H19 DMR (IC1) and KvDMR1 (IC2) maintained their normal allele-specific pattern of DNA methylation (Supplementary material, Fig. S3). Our results show that the Tel7KI line is not imprinted in trophoblast lineages and that relaxation of imprinting is not seen globally at endogenous imprinted loci in trophoblast giant cells. We thus predict that the high level of GFP observed in TGCs in both Tel7KI and D4 is transgene-specific and does not reflect changes in epigenetic instability in this cell type.
Discussion
We have characterized a new GFP transgenic reporter for the epigenetic regulation of gene expression by genomic imprinting in the mouse. Tel7KI is an imprinted allele, allowing easy monitoring of the developmental cycle of imprinting and gene silencing, and offering new opportunities for the study of these phenomena in vivo in the context of the developing embryo. Our analysis of Tel7KI has established the following characteristics for its developmental regulation: (i) Tel7KI behaves as an imprinted allele and its GFP reporter is maternally expressed in the embryo. (ii) DNA methylation at Tel7KI is not inherited from the germline, but is acquired preferentially on the paternal allele post-fertilization. (iii) The maternal allele is broadly expressed in the embryo, following a fixed tissue-specific pattern. (iv) The imprinting of Tel7KI is not maintained in the placenta, where Tel7KI is expressed from both alleles in trophoblast lineages. Our analysis of trophoblast giant cells has also revealed that the expression of autosomal or X-linked imprinted reporters in these cells do not reflect a fundamental instability of imprints in this polyploid lineage. Together these observations shed light on the mechanism of acquisition of imprinted expression by novel transcripts in the mammalian genome and raise important questions on the mechanisms of regulation of imprinting in the context of distal mouse Chr 7.
In the embryo, the inserted GFP reporter behaves as a maternally expressed gene. Several randomly inserted transgenic constructs have previously been shown to respond to parent-of-origin effects (Reik et al., 1987; Sapienza et al., 1987; Swain et al., 1987; Ueda et al., 1992). In contrast to what we observed at Tel7KI, these imprinted transgenes are paternally expressed and acquire a gametic DNA methylation imprint specifically during oogenesis. This silencing pathway is widely used in the regulation of endogenous paternally expressed imprinted genes, characterized by germline DNA methylation imprints of maternal origin (Reik and Walter, 2001b). A related mechanism which has been proposed for the generation of new imprinted transcripts during evolution involves the insertion of processed retrogenes in the genome (Nabetani et al., 1997; Wood et al., 2007). These retrogenes also acquire DNA methylation imprints in the maternal germline, providing the epigenetic mark responsible for their imprinted expression. Examples include U2af1-rs1 (Nabetani et al., 1997), Inpp5f_v2 (Choi et al., 2005), Mcts2 (Wood et al., 2007), and Peg10 (Ono et al., 2003).
Tel7KI provides a paradigm for a different process, namely the acquisition of imprinting upon an inserted transcriptional unit. In evolution this could manifest itself in rearrangements or translocations involving an already imprinted host locus. The imprinting of exogenous sequences inserted within known imprinted regions by gene targeting has previously been documented in the IC1 regulated region close to Tel7KI insertion (Ainscough et al., 1997; Ripoche et al., 1997). The behavior of these transgenes have established that non-imprinted elements, when inserted within an imprinted locus, can acquire functionally relevant epigenetic imprints. In these examples, the inserted element essentially acquires the imprinted pattern of the targeted locus. This is not observed at the Tel7KI allele, inserted less than 3 kb upstream of the Ins2 gene (Oh et al., 2008). Ins2 is biallelically expressed in all tissues other than the developing yolk sac, where it shows preferential expression of the paternal allele (Deltour et al., 1993; Giddings et al., 1994; Deltour et al., 1995). The imprinting of Tel7KI and its exclusive expression from the maternal allele suggest that transcription of the GFP reporter has fallen under the regulation of long-range imprinting signals. Our results show that interactions between Tel7KI and these signals can generate a new imprinted locus with a complex tissue-specific imprinted pattern of expression. This provides a model for the acquisition of imprinted expression by novel genes during evolution and a new framework to dissect the epigenetic differences between embryonic and extraembryonic lineages in maintaining and interpreting the underlying epigenetic signals.
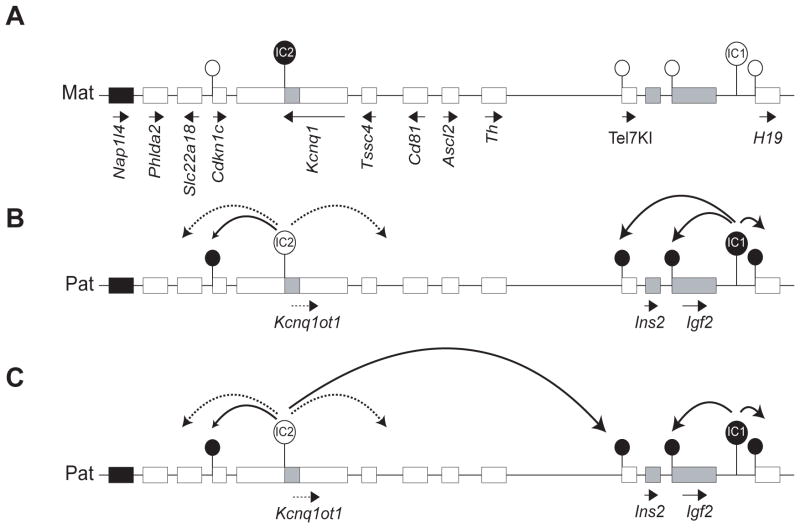
In the context of our current understanding of imprinting on distal Chr 7, the regulation of Tel7KI suggests that the effects of existing imprinting centres can reach loci located further than previous appreciated. Based on the ontogeny of allele-specific methylation described here at the Tel7KI allele, we envisage that either the H19 DMR (IC1) or KvDMR1 (IC2) could be responsible for the imprinted expression observed at Tel7KI and propose two models for its imprinted behavior (Fig. 7).
Fig. 7. Models for long-range regulation of Tel7KI.
(A) The maternal chromosome is marked by a DNA methylation imprint at IC2. The Tel7KI promoter is unmethylated and active. (B) In the first model, the germline imprint on the paternal IC1 spreads not only to Igf2 but also to Tel7KI. Whereas this inhibits the function of Igf2 silencers, the promoter methylation leads to silencing of the paternal Tel7KI allele. (C) Another model implicates long-range silencing on the paternal chromosome by formation of the Kcnq1ot1 non-coding RNA, which bidirectionally spreads repressive histone marks in the IC2 cluster (dotted lines) and silencing DNA methylation at Cdkn1c and Tel7KI (solid lines).
In the IC1 domain, the DNA methylation patterns at Igf2 are reminiscent of what we have observed at Tel7KI, though the parental expression of these two genes is opposite. Both Tel7KI and Igf2 are paternally methylated but Igf2 is also paternally expressed, the hypothesis being that the paternal methylation on this gene represses a silencer element (Constância et al., 2000). The methylation acquired on the paternal allele of Tel7KI could mimic the situation at Igf2, but in the case of Tel7KI the promoter DNA methylation would result in silencing of GFP transcription (Fig. 7B). Not only is the timing of acquisition of DNA methylation similar for Igf2 and Tel7KI (Lopes et al., 2003), but we also note a parallel with regard to imprinted transcription, the Igf2 gene being biallelically expressed in blastocysts, as we observed for Tel7KI (Szabó and Mann, 1995). For Igf2, this is likely to reflect a basal rather than activated biallelic transcription. Whether or not a similar basal transcription is responsible for the observed biallelic expression of Tel7KI in blastocyst remains to be determined.
The current model of how the maternal DMRs of Igf2 remain unmethylated involves chromatin looping, CTCF binding, and epigenetically-mediated contact between distant sites (Lopes et al., 2003; Murrell et al., 2004). The Tel7KI allele is found more than 20 kb away from the Igf2 CpG-rich region involved in this looping. Furthermore, the gene found in between Igf2 and Tel7KI, Ins2, is imprinted only in embryonic yolk sac endoderm, and has not been implicated in this looping model. However, it is interesting to note that circular chromosome conformation capture experiments designed to identify genomic regions physically associated with the CTCF complex at IC1 have uncovered several interacting regions on distal Chr 7, including 3 sites immediately distal of the Tel7KI insertion site and two other sites proximal of Th, located ~300 kb telomeric of Ins2 (Zhao et al., 2006). Our model therefore raises the possibility that the allele-specific regulation mediated by IC1 extends distally beyond Ins2, perhaps as far as the Th locus, which is consistent with the recent finding that Th is preferentially expressed from the maternal allele in the placenta (Schulz et al., 2006). A prediction from this model would be that absence of CTCF binding from the maternal IC1 should lead to acquisition of DNA methylation at the maternal Tel7KI and silencing of the GFP.
The post-fertilization acquisition of DNA methylation on the silent paternal Tel7KI allele is also reminiscent of that observed at the IC2-regulated maternally expressed Cdkn1c, the only imprinted gene regulated by IC2 which contains its own CpG island (Lewis et al., 2004). The pattern of Cdkn1c methylation is similar to that observed at Tel7KI, with paternal methylation acquired between E6.5 and E8.5, though the GFP from Tel7KI becomes monoallelically expressed between E4.5 and E7.5, while Cdkn1c is already preferentially maternally expressed at E4.5 and is imprinted in both embryo and placenta (Hatada and Mukai, 1995; Mager et al., 2003; Bhogal et al., 2004; Lewis et al., 2004; Umlauf et al., 2004). Interestingly, other genes regulated by IC2 are biallelically expressed in blastocysts and acquire their monoallelic expression during post-implantation development (Lewis et al., 2006). These genes, Tssc4 (Paulsen et al., 2000) and Cd81 (Umlauf et al., 2004), are imprinted only in the placenta, which is opposite to what we observed at Tel7KI. Like in the case of Ascl2, these IC2-regulated genes are not known to acquire repressive DNA methylation marks on the paternal allele and their inactive state might rely solely on ncRNA-induced histone modifications. It is possible that the combination of being located at a distance from IC2 and containing a CpG island has resulted in a unique combination of mechanisms regulating Tel7KI.
Unlike the situation at IC1 where long-range effects involve an epigenetically regulated insulator, imprinting in the IC2 sub-domain is dependent on the cis-spreading of repressive chromatin via the action of a large non-coding RNA, Kcnq1ot1 (Lee et al., 1999; Fitzpatrick et al., 2002; Umlauf et al., 2004). In our second model, we propose that Tel7KI is regulated by IC2 through the action of Kcnq1ot1 which would spread a further 300 kb towards the proximal IC1 sub-domain (Fig. 7C). We hypothesize that in the blastocyst, the imprinting signal from IC2 has not yet reached Tel7KI, as is observed by biallelic expression of distal or “placentally-imprinted” IC2-regulated genes (Lewis et al., 2004; Terranova et al., 2008). However, a main difference between Tel7KI and endogenous genes of the IC2 cluster is that Tel7KI contains a CpG island (Lewis et al., 2004). Thus, it is possible that IC2 can act on Tel7KI only in the embryo and only through the presence of sites capable of acquiring DNA methylation. According to this second model, we predict that deletion of IC2 in cis of Tel7KI or truncation of the Kcnq1ot1 ncRNA should prevent silencing of the paternally inherited allele, as indeed observed for endogenous IC2-regulated genes (Fitzpatrick et al., 2002; Mancini-Dinardo et al., 2006; Shin et al., 2008). We have now established ES cell lines hemizygous for the Tel7KI mutation and we are in the process of testing the respective roles of IC1 and IC2 in the regulation of Tel7KI silencing.
Supplementary Material
Acknowledgments
The authors would like to thank the support of Prof. Andras Nagy in whose laboratory this mouse line was originally developed with the help of Marina Gertsenstein. We also thank Dr. Nagy for providing the D4/XEGFP transgenics, M. Lorincz for the GFP ORF bisulfite sequencing oligos, K. McNagny for the CD-34 antibody, J. Roskams for helpful discussion and advice on immunohistochemistry, M. Mann for the sequence of the Cdkn1c allele-specific RT-PCR oligos, Andy Johnston of the UBC Flow Cytometry & Cell Sorting service, as well as Aaron Bogutz for comments. This work was supported in part by operating grants from the Canadian Institutes for Health Research (CIHR) to L.L (MOP-64193 and MOP-82863). L.L. holds a Canada Research Chair and was also supported by a Michael Smith Foundation for Health Research (MSFHR) Scholar Award. M.J was supported by scholarships from the National Science and Engineering Research Council of Canada, CIHR and MSFHR.
References
- Ainscough JF, Koide T, Tada M, Barton S, Surani MA. Imprinting of Igf2 and H19 from a 130 kb YAC transgene. Development. 1997;124:3621–3632. doi: 10.1242/dev.124.18.3621. [DOI] [PubMed] [Google Scholar]
- Ball M, Li J, Gao Y, Lee J, Leproust E, Park I, Xie B, Daley G, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M, Webber A, Brunkow M, Tilghman S. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 1993;7:1663–1673. doi: 10.1101/gad.7.9.1663. [DOI] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bhogal B, Arnaudo A, Dymkowski A, Best A, Davis T. Methylation at mouse Cdkn1c is acquired during postimplantation development and functions to maintain imprinted expression. Genomics. 2004;84:961–970. doi: 10.1016/j.ygeno.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Choi JD, Underkoffler LA, Wood A, Collins JN, Williams PT, Golden JA, Schuster EF, Loomes KM, Oakey R. A novel variant of Inpp5f is imprinted in brain, and its expression is correlated with differential methylation of an internal CpG island. Mol Cell Biol. 2005;25:5514–5522. doi: 10.1128/MCB.25.13.5514-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constância M, Dean W, Lopes S, Moore T, Kelsey G, Reik W. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- Cowan CM, Thai J, Krajewski S, Reed JC, Nicholson DW, Kaufmann SH, Roskams AJ. Caspases 3 and 9 send a pro-apoptotic signal from synapse to cell body in olfactory receptor neurons. J Neurosci. 2001;21:7099–7109. doi: 10.1523/JNEUROSCI.21-18-07099.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Tremblay KD, Bartolomei M. Imprinted expression and methylation of the mouse H19 gene are conserved in extraembryonic lineages. Dev Genet. 1998;23:111–118. doi: 10.1002/(SICI)1520-6408(1998)23:2<111::AID-DVG3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Deltour L, Leduque P, Blume N, Madsen O, Dubois P, Jami J, Bucchini D. Differential expression of the two nonallelic proinsulin genes in the developing mouse embryo. Proc Natl Acad Sci USA. 1993;90:527–531. doi: 10.1073/pnas.90.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltour L, Montagutelli X, Guenet JL, Jami J, Páldi A. Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev Biol. 1995;168:686–688. doi: 10.1006/dbio.1995.1114. [DOI] [PubMed] [Google Scholar]
- El-Hashash A, Kimber SJ. Trophoblast differentiation in vitro: establishment and characterisation of a serum-free culture model for murine secondary trophoblast giant cells. Reprod. 2004;128:53–71. doi: 10.1530/rep.1.00149. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature. 1993;362:751–755. doi: 10.1038/362751a0. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick G, Soloway P, Higgins M. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- Giddings SJ, King CD, Harman KW, Flood JF, Carnaghi LR. Allele specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat Genet. 1994;6:310–313. doi: 10.1038/ng0394-310. [DOI] [PubMed] [Google Scholar]
- Gonçalves CR, Antonini S, Vianna-Morgante AM, Machado-Santelli GM, Bevilacqua E. Developmental changes in the ploidy of mouse implanting trophoblast cells in vitro. Histochem Cell Biol. 2003;119:189–198. doi: 10.1007/s00418-003-0500-0. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Caspary T, Tilghman S, Copeland NG, Gilbert DJ, Jenkins NA, Anderson DJ, Joyner AL, Rossant J, Nagy A. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat Genet. 1995;9:235–242. doi: 10.1038/ng0395-235. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis A, Papaioannou V. Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol. 2004;4:33. doi: 10.1186/1472-6750-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Cox LL, Tam PP, Nagy A. An X-linked GFP transgene reveals unexpected paternal X-chromosome activity in trophoblastic giant cells of the mouse placenta. genesis. 2001;29:133–140. doi: 10.1002/gene.1016. [DOI] [PubMed] [Google Scholar]
- Hardouin N, Nagy A. Gene-trap-based target site for cre-mediated transgenic insertion. genesis. 2000;26:245–252. doi: 10.1002/(sici)1526-968x(200004)26:4<245::aid-gene50>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman S. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Hatada I, Mukai T. Genomic imprinting of p57KIP2, a cyclin-dependent kinase inhibitor, in mouse. Nat Genet. 1995;11:204–206. doi: 10.1038/ng1095-204. [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, Oshimura M, Feinberg A. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA. 1999;96:5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre L, Viville S, Barton SC, Ishino F, Surani MA. Genomic structure and parent-of-origin-specific methylation of Peg1. Hum Mol Genet. 1997;6:1907–1915. doi: 10.1093/hmg/6.11.1907. [DOI] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman S. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- Lewis A, Green K, Dawson C, Redrup L, Huynh K, Lee J, Hemberger M, Reik W. Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development. 2006;133:4203–4210. doi: 10.1242/dev.02612. [DOI] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forné T, Murrell A, Constância M, Bartolomei M, Walter J, Reik W. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum Mol Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- Mager J, Montgomery N, De Villena F, Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJ, Levorse J, Ingram RS, Tilghman S. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Hatada I, Morisaki H, Oshimura M, Mukai T. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol Cell Biol. 1997;17:789–798. doi: 10.1128/mcb.17.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the mouse embryo: a laboratory manual. 3 2003. [Google Scholar]
- Nakanishi T, Kuroiwa A, Yamada S, Isotani A, Yamashita A, Tairaka A, Hayashi T, Takagi T, Ikawa M, Matsuda Y, Okabe M. FISH analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics. 2002;80:564–574. doi: 10.1006/geno.2002.7008. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Oh R, Ho R, Mar L, Gertsenstein M, Paderova J, Hsien J, Squire JA, Higgins M, Nagy A, Lefebvre L. Epigenetic and phenotypic consequences of a truncation disrupting the imprinted domain on distal mouse chromosome 7. Mol Cell Biol. 2008;28:1092–1103. doi: 10.1128/MCB.01019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Letters. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- Ono R, Shiura H, Aburatani H, Kohda T, Kaneko-Ishino T, Ishino F. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 2003;13:1696–1705. doi: 10.1101/gr.906803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen M, El-Maarri O, Engemann S, Strödicke M, Franck O, Davies K, Reinhardt R, Reik W, Walter J. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum Mol Genet. 2000;9:1829–1841. doi: 10.1093/hmg/9.12.1829. [DOI] [PubMed] [Google Scholar]
- Reik W, Collick A, Norris ML, Barton SC, Surani MA. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328:248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- Reik W, Murrell A, Lewis A, Mitsuya K, Umlauf D, Dean W, Higgins M, Feil R. Chromosome loops, insulators, and histone methylation: new insights into regulation of imprinting in clusters. Cold Spring Harb Symp Quant Biol. 2004;69:29–37. doi: 10.1101/sqb.2004.69.29. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001a;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Evolution of imprinting mechanisms: the battle of the sexes begins in the zygote. Nat Genet. 2001b;27:255–256. doi: 10.1038/85804. [DOI] [PubMed] [Google Scholar]
- Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- Rossant J. Undermethylation of structural gene sequences in extraembryonic lineages of the mouse*1. Dev Biol. 1986;117:567–573. doi: 10.1016/0012-1606(86)90325-8. [DOI] [PubMed] [Google Scholar]
- Sánchez MJ, Holmes A, Miles C, Dzierzak E. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of the mouse embryo. Immunity. 1996;5:513–525. doi: 10.1016/s1074-7613(00)80267-8. [DOI] [PubMed] [Google Scholar]
- Sapienza C, Peterson AC, Rossant J, Balling R. Degree of methylation of transgenes is dependent on gamete of origin. Nature. 1987;328:251–254. doi: 10.1038/328251a0. [DOI] [PubMed] [Google Scholar]
- Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- Schulz R, Menheniott TR, Woodfine K, Wood A, Choi J, Oakey R. Chromosome-wide identification of novel imprinted genes using microarrays and uniparental disomies. Nucleic Acids Res. 2006;34:e88. doi: 10.1093/nar/gkl461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Fitzpatrick G, Higgins M. Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 2008;27:168–178. doi: 10.1038/sj.emboj.7601960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilinich NJ, Day CD, Fitzpatrick GV, Caldwell GM, Lossie AC, Cooper PR, Smallwood AC, Joyce JA, Schofield PN, Reik W, Nicholls RD, Weksberg R, Driscoll DJ, Maher ER, Shows TB, Higgins MJ. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci USA. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Hsieh S, Grinberg A, Williams-Simons L, Huang SP, Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 2000;14:1186–1195. [PMC free article] [PubMed] [Google Scholar]
- Swain JL, Stewart TA, Leder P. Parental legacy determines methylation and expression of an autosomal transgene: a molecular mechanism for parental imprinting. Cell. 1987;50:719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- Szabó P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- Szabó PE, Mann JR. Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev. 1995;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler M, Otte A, Van Lohuizen M, Orkin S, Peters A. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Saam JR, Ingram RS, Tilghman S, Bartolomei M. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet. 1995;9:407–413. doi: 10.1038/ng0495-407. [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamazaki K, Suzuki R, Fujimoto H, Sasaki H, Sakaki Y, Higashinakagawa T. Parental methylation patterns of a transgenic locus in adult somatic tissues are imprinted during gametogenesis. Development. 1992;116:831–839. doi: 10.1242/dev.116.4.831. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Weber M, Schübeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Current Opinion in Cell Biology. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Wood A, Oakey R. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2006;2:e147. doi: 10.1371/journal.pgen.0020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Roberts R, Monk D, Moore G, Schulz R, Oakey R. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 2007;3:e20. doi: 10.1371/journal.pgen.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.