Abstract
MicroRNAs (miRNAs) are non-coding small RNA ∼22 nucleotides in length that can regulate the expression of a wide range of coding genes at the post-transcriptional level. Visceral adipose tissues (VATs) and subcutaneous adipose tissues (SATs), the two main fat compartments in mammals, are anatomically, physiologically, metabolically, and clinically distinct. Various studies of adipose tissues have focused mainly on DNA methylation, and mRNA and protein expression, nonetheless little research sheds directly light on the miRNA transcriptome differences between these two distinct adipose tissue types. Here, we present a comprehensive investigation of miRNA transcriptomes across six variant porcine adipose tissues by small RNA-sequencing. We identified 219 known porcine miRNAs, 97 novel miRNA*s, and 124 miRNAs that are conserved to other mammals. A set of universally abundant miRNAs (i.e., miR-148a-3p, miR-143-3p, miR-27b-3p, miR-let-7a-1-5p, and miR-let-7f-5p) across the distinct adipose tissues was found. This set of miRNAs may play important housekeeping roles that are involved in adipogenesis. Clustering analysis indicated significant variations in miRNA expression between the VATs and SATs, and highlighted the role of the greater omentum in responding to potential metabolic risk because of the observed enrichment in this tissue of the immune- and inflammation-related miRNAs, such as the members of miR-17-92 cluster and miR-181 family. Differential expression of the miRNAs between the VATs and SATs, and miRNA target prediction analysis revealed that the VATs-specific enriched miRNAs were associated mainly with immune and inflammation responses. In summary, the differences of miRNA expression between the VATs and SATs revealed some of their intrinsic differences and indicated that the VATs might be closely associated with increased risk of metabolic disorders.
Introduction
MicroRNAs (miRNAs) are a family of small single-stranded non-coding RNAs, which are known to function in a sequence-specific manner to silence specific protein-coding genes at the post-transcriptional level by targeting the 3′ untranslated region of mRNAs [1]. With the rapid increase in knowledge that has accumulated over the last decade, various miRNAs have been shown to play vital regulatory roles in adipose deposition and adipogenesis. Typically, miR-143 is a potent pro-adipogenic regulator during pre-adipocyte differentiation [2], [3], [4], and the miR-17-92 cluster [5] and miR-103 [6] can accelerate adipocyte differentiation, while miR-27a [7], miR-27b [8] and miR-15a [9] can suppress adipogenic differentiation. In addition, miR-519d [10], miR-335 and miR-377 [11] are strongly associated with lipid metabolism disorders.
Adipose tissues (ATs) are currently recognized as an endocrine organ, and the number of adipokines that have been identified in ATs is expanding rapidly. ATs are deeply involved in the development of metabolic disorders, such as cardiovascular disease and type 2 diabetes mellitus, which are connected to obesity [12], [13], [14]. Nonetheless, the different fat compartments may be associated with differential metabolic risk. Visceral adipose tissues (VATs), which are located within the abdominal and thoracic cavities, have been recognized to be more strongly associated with metabolic risk factors than the subcutaneous adipose tissues (SATs) [15], [16], [17]. It was suggested that the different impacts that VATs and SATs have on metabolic risk may be because of diverse gene expression profiles that lead to differences in lipolysis and in the production and release of adipokines and cytokines [17]. However, miRNA-based gene regulatory mechanisms in the distinct ATs are yet to be investigated. The results will be of interest for the development of diagnostics and therapeutics for metabolic diseases.
Pigs have considerable agricultural significance and are an important model system for human biomedical research, including the study of obesity and energy metabolism [18]. To explore the molecular mechanisms that underlie the metabolic and functional differences between SATs and VATs, we performed a comprehensive comparison of the miRNA transcriptomes from six types of porcine ATs and identified various known and conserved miRNAs. We found that the SATs-specific enriched miRNAs were associated primarily with lipid metabolic homeostasis, whereas the VATs-specific enriched miRNAs were related mainly to the immune and inflammatory responses, which indicated the metabolic risk roles of the VATs. We envision that these findings will contribute to the further understanding of the biological functions of miRNAs in ATs, and the molecular mechanisms behind the distinct metabolic and physiological roles of the SATs and VATs.
Materials and Methods
Ethics statement
All research involving animals were conducted according to the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in June 2004) and approved by the Institutional Animal Care and Use Committee in College of Animal Science and Technology, Sichuan Agricultural University, Sichuan, China under permit No. DKY-B20110807. Animals were allowed free access to food and water under normal conditions, and were humanely sacrificed as necessary, to ameliorate suffering.
Animals and sample collection
Three 210-days old female Landrace pigs with normal weight (111.67±1.15 kilograms) were used. A starter diet provided 3.40 Mcal·kg−1 metabolisable energy (ME), 20.00% crude protein and 1.15% lysine from the thirtieth to sixtieth day after weaning. From the 61st to the 120th day, the diet contained 3.40 Mcal·kg−1 ME, 17.90% crude protein and 0.83% lysine. From the 121st to 210th day, the diet contained 3.40 Mcal·kg−1 ME, 15.00% crude protein and 1.15% lysine.
Four VATs (i.e. greater omentum (GOM), mesenteric adipose (MAD), pericardial adipose (PAD) and retroperitoneal adipose (RAD)), and two SATs (i.e. upper layer of back fat (ULB) and inner layer of back fat (ILB)) were rapidly separated from each carcass, immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
Small RNA libraries construction and deep sequencing
The total RNA of 18 tissue samples were extracted with mirVana™ miRNA isolation kit (Ambion, Austin, USA), and further purified with Rneasy column (QIAGEN, Hilden, Germany). The quantity and purity of total RNA were monitored via analysis by NanoDrop ND-1000 spectrophotometer (Nano Drop, DE, USA) at 260/280 nm (ratio>2.0). The integrity of total RNA was also tested via analysis by Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN number>6.0.
For a certain adipose tissue, equal amounts (5 µg) of small-RNA-enriched total RNA isolated from three pigs were mixed and prepared for Illumina sequencing. In general, the processing by Illumina consisted of the following successive steps: the small RNA ranged from 14 to 40 nt were purified by polyacrylamide gel electrophoresis (PAGE) and ligated specific adapters followed by polyacrylamide gel purification. Then the modified small RNA was reverse transcripted and amplificated by RT-PCR. Finally, the enriched cDNA was sequenced on Genome Analyzer Instrument (GAI, Illumina). The small RNA sequence data have been uploaded to NCBI's Gene Expression Omnibus (GEO) [accession number GSE30334].
In silico analysis of small RNA-sequencing data
The raw reads were processed using Illumina's Genome Analyzer Pipeline software and subsequently handled as described by Li et al. [19], [20] with some improvement. After trimming off the sequencing adapters, the resulting reads was successively filtered by read length (only read length ranged from 14 to 27 nt were retained) and sequence component (containing no more than 80% A, C, G or T; containing no more than two N (undetermined bases)). Given the confidence of analysis results, the resulting set of distinct reads was subsequently filtered by copy numbers (the low-abundance reads (copy number<3) was excluded). Then the retained reads were searched against the NCBI [21], Rfam [22] and Repbase database [23] to remove porcine known classes of RNAs (i.e. mRNA, rRNA, tRNA, snRNA, snoRNA and repeats). The sequencing reads survived from above strict filter rules were deemed to ‘high-quality reads’.
The high-quality reads were mapped to the pig genome (∼2.26 Bbp) (Sscrofa9) using NCBI Local BLAST. The mapping process included two steps: (1) map the high-quality reads to the 228 known porcine pre-miRNAs (encoding 257 miRNAs) and then to 6,716 known pre-miRNAs (encoding 7952 miRNAs) from 24 other mammals in miRBase 18.0 [24]; (2) map the mapped high-quality reads to pig genome to obtain their genomic locations and annotations in Ensembl release 59 (Sscrofa9, April 2009).
MiRNA differential expression analysis
Program IDEG6 [25] was employed for detecting the differentially expressed miRNAs in the pairwise comparison between the VATs and SATs. If a unique miRNA simultaneously obtains a P<10−5 under each of the three statistical tests (a Audic-Claverie test, a Fisher exact test and a Chi-squared 2×2 test) with the Bonferroni correction, it should been termed as a differentially expressed miRNA.
Prediction and functional annotation of miRNA target genes
The potential targets of a certain miRNA were predicted using the PicTar [26], TargetScan human 6.2 [27] and, MicroCosm Targets (version 5.0) [28], and the pairwise overlaps of the results from three programs were composed the final predicted targets. The Gene Ontology-biological process (GO-BP) and KEGG pathway terms enriched in the predicted target genes were determined using a DAVID bioinformatics resources [29].
Q-PCR validation
The expression changes of 18 selected miRNAs were validated by an EvaGreen-based High-Specificity miRNA qRT-PCR Detection Kit (Stratagene, La Jolla, USA) on the CFX96™ Real-Time PCR Detection System (Bio-Rad, CA, USA). The q-PCR validation were carried out on three biological replicates. The primer pairs were available in Table S1. Three endogenous control genes (porcine U6 snRNA, 18S rRNA and Met-tRNA) [20] were used in this assay. The ΔΔCt method was used to determine the expression level differences between surveyed samples. Normalized factors (NF) of three endogenous control genes and relative quantities of objective miRNAs were analyzed using the qBase software [30].
Results
Identification of miRNAs in six distinct ATs
Six small RNA libraries, two SATs (i.e. ULB andILB) and four VATs (i.e. GOM, RAD, MAD and PAD), were constructed and sequenced. We obtained an average of 21.10 million (M) (21.10±5.95 M, n = 6) raw reads from each library. More than 70% (77.62±7.15%, n = 6) of the raw reads in each library passed the quality filter (see Methods) and were defined as ‘high-quality reads’ (Figure S1A), which is consistent with the canonical size range of mammalian miRNAs (Figure S1B). The vast majority of high-quality reads were 21−23 nt in length (92.04±0.47%, n = 6); more than half of them were 22 nt in length (66.45±1.44%, n = 6), followed by 21 nt (14.42±1.75%, n = 6) and 23 nt (11.17±1.44%, n = 6). These reads were selected as the reliable miRNA candidates for subsequent analysis.
A total of 440 mature miRNAs corresponding to 271 miRNA precursors (pre-miRNAs) were identified in the six libraries. The identified precursor and mature miRNAs were divided into two groups using the following alignment criteria (Table S2): (1) Known porcine miRNAs: 316 of the miRNAs mapped to 184 known porcine pre-miRNAs. Specifically, 219 miRNAs were documented in miRBase [28], and 97 were newly identified miRNA*s; (2) Conserved miRNAs: 124 reads mapped to 87 mammalian (other than porcine) pre-miRNAs in miRBase. The conserved miRNAs mapped to the pig genome and were labeled with the names of the corresponding miRNAs. Among them were distinct pre-miRNAs that coded identical mature miRNAs, resulting in the 440 mature miRNAs corresponding to 409 unique miRNA sequences (Table S3).
Universally abundant miRNAs across distinct ATs
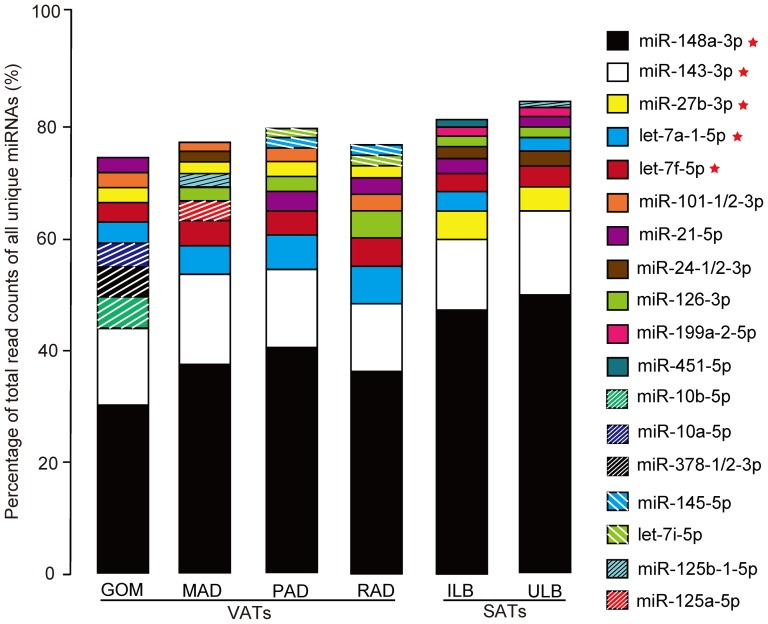
In this study, we observed that the majority of abundant miRNAs were from a few miRNA species. As shown in Figure 1, the top 10 unique miRNAs with the highest expression levels account for more than 74.42%, by total read counts, of the 409 unique miRNAs. The unified set of top 10 unique miRNAs over the six ATs correspond to 18 unique miRNAs, five of which (miR-148a-3p, miR-143-3p, miR-27b-3p, let-7a-1-5p and let-7f-5p) have the highest abundance in all six libraries. These five miRNAs may play potential housekeeping roles in the adipocytes, and hence may be important regulators involved in adipogenesis. For example, miR-148a-3p [31] was reported to be up-regulated during the differentiation of 3T3-L1 pre-adipocyte, whereas miR-27b-3p was suggested to be a negative regulator during adipogenesis [8]. MiR-143 was elevated in adipogenesis and the loss of function of miR-143 resulted in the prevention of adipocyte differentiation [32]. In addition, let-7a and let-7f (members of the let-7 family) are well-known regulators in development, and in cellular basal metabolism, and are present in abundance in various species including mammals, flies, worms, and plants [33]. Furthermore, the expression levels of the 18 most abundant miRNAs show good correlation (Person's r = 0.875±0.121, n = 18) between the q-PCR and small RNA-seq sequencing results, which highlights the high confidence of the results obtained using the deep-sequencing approach (Figure S2).
Figure 1. Top 10 unique miRNAs with the highest expression level in six adipose tissues.
Five miRNAs that have the highest abundance in all six libraries were marked with the symbol of red stars.
Extraordinary miRNA expression implied a role for GOM in metabolic risk
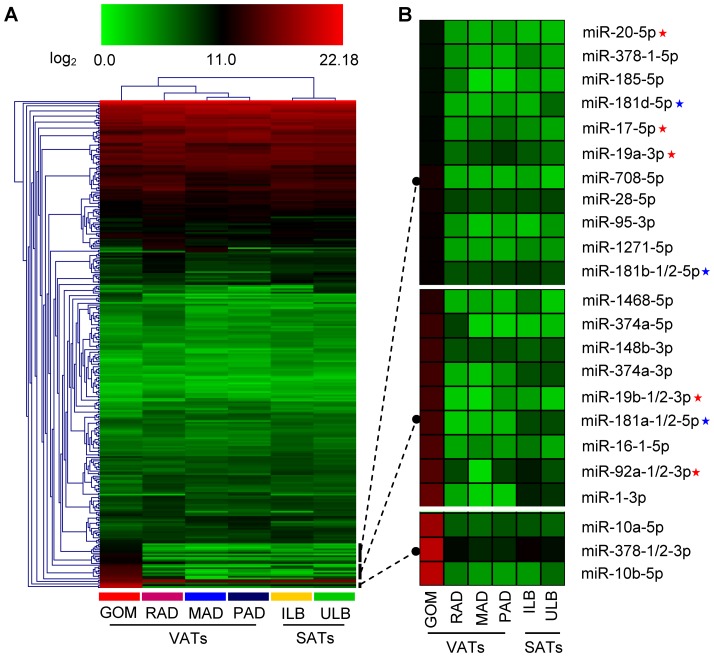
The 409 unique miRNAs were divided into six categories according to their tissue-related expression (Table 1 and Table S4). Category 1 consisted of 284 (69.44%) unique miRNAs that were co-expressed in all six ATs. Category 6 consisted of 53 (12.96%) unique miRNAs which were expressed specifically in only one of the six ATs. The total number of miRNAs expressed in more than one and less than six ATs (category 2, 3, 4 and 5) was 72 (17.60%). Notably, the vast majority (29/53, 54.71%) of the tissue-specific miRNAs were specifically expressed in GOM, and included three members of the miR-17-92 cluster (miR-18a-3p, miR-20-3p and miR-19b-1-5p) and two members of the miR-181 family (miR-181a-2-3p and miR-181b-2-3p), both of which are known to be important for the development and production of the pro-inflammatory B-cells and T-cells [34], [35], [36].
Table 1. miRNAs classified into six categories according to the status of tissue-related expression.
| Category | Number of miRNAs | Defination |
| 1 | 284 | miRNAs co-expressed in six libraries |
| 2 | 23 | miRNAs absent in one library |
| 3 | 13 | miRNAs absent in two libraries |
| 4 | 17 | miRNAs absent in three libraries |
| 5 | 19 | miRNAs absent in four libraries |
| 6 | 53 | miRNAs specifically expressed in only one library |
Subsequently, we performed hierarchical clustering based on the expression profiles of the 284 miRNAs that were co-expressed in all six ATs, and observed a deep split between the VATs and SATs. The two SATs (ILB and ULB) were tightly clustered into a subgroup, which were clearly distinct from the four VATs (GOM, MAD, RAD and PAD). This significant variation in miRNA expression may be responsible, at least in part, for the phenotypic differences between the SATs and VATs; for example, their anatomical, functional, and metabolic differences (Figure 2A). Of the four VATs, the GOM was clearly separated from the others with several highly expressed miRNAs (Figure 2B). Remarkably, seven members of the miR-17-92 cluster (miR-17-5p, miR-19a-3p, miR-19b-1/2-3p, miR-20-5p and miR-92a-1/2-3p) and five members of the miR-181 family (miR-181a-1/2-5p, miR-181b-1/2-5p and miR-181d-5p) were highly enriched in the GOM; this cluster and family are both important for the development and production of B- and T-cells [34], [35], [36]. These results are consistent with the fact that milky spots, mainly comprising macrophages and B- and T-lymphocytes, are widely distributed in the GOM [37], [38]. Other GOM-enriched miRNAs were also related to various pathological responses. For example, miR-10a-5p as a vital modulator, could suppress the growth of chronic myeloid leukemia CD34+ cells [39], and miR-10b-5p targets the potent inflammatory mediator KLF4 (Kruppel-like factor 4) [40] and has important roles in tumor invasion and the initiation of metastasis in breast cancer [41]. These results are consistent with the critical role of abdominal GOM in immune and inflammatory responses [19], [42], [43].
Figure 2. The expression profiles corresponding to 284 co-expressed miRNAs.
(A) Hierarchical clustering analysis for six adipose tissues. (B) Three miRNA subgroups that specially enriched in GOM. Red stars : the members of the miR-17-92 cluster. Blue stars: the members of the miR-181 family.
Inflammation-related miRNAs were specifically enriched in VATs
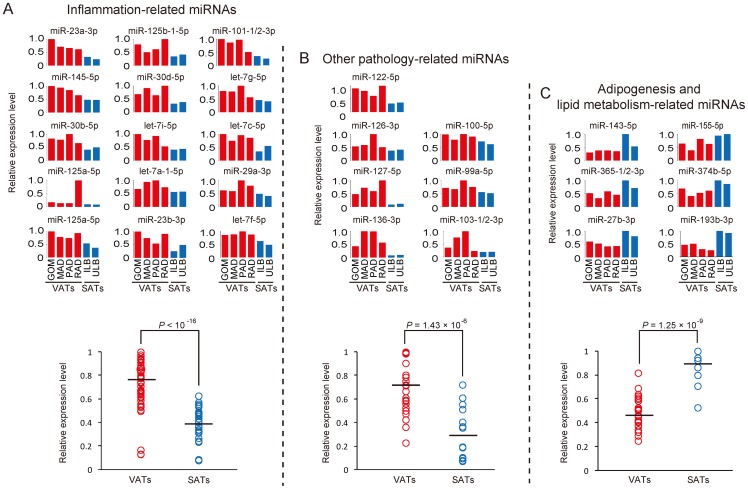
We performed a pairwise comparison analysis between each of the VATs and SATs for the 284 co-expressed miRNAs, and identified 22 and 21 differentially expressed miRNAs that were specifically enriched in the VATs and SATs, respectively (Table 2 and Table S5). Notably, vast majority of the VATs-specific enriched miRNAs (15/22, 68.18%) were associated with inflammation based on the annotation from the Pathway Central database (SABiosciences, MD, USA) (Figure 3A). MiR-let-7a-1-5p, miR-let-7c-5p, miR-let-7f-5p, miR-let-7g-5p and miR-let-7i-5p (members of the miR-let-7 family) are involved in allergic airway inflammation by suppressing the expression of interleukin-13 [44], and also inhibit cell transformation by directly targeting interleukin-6 [45]. MiR-145, a regulator of inflammation, was reported to be down-regulated in longstanding ulcerative colitis [46]. MiR-30b-5p and miR-30d-5p, two members of the miR-30-family, can inhibit mitochondrial fission by suppressing the expression of tumor protein 53 (p53), and can also promote cellular invasion and immunosuppression by targeting GalNAc transferase 7 [47]. MiR-125a-5p is a critical inhibitor of the pro-inflammatory response by targeting oxysterol binding protein-like 9, which resulted in decreased secretion of inflammatory cytokines [48]. MiR-23b-5p suppresses nuclear factor κB (NF-κB) activation and inflammatory cytokine expression by targeting TAB2 (TGF-β-activated kinase 1/MAP3K7 binding protein, TAB3 (TGF-β-activated kinase 1/MAP3K7 binding protein 3), and IKK-α (inhibitor of NF-κB kinase subunit α) [49]. MiR-29a is an important threshold modulator of thymic epithelial cell response to peripheral PAMP (pathogen-associated molecular patterns) signals by targeting INFAR1 (Interferon-α receptor 1) [50]. By targeting MKP-1 (MAPK phosphatase-1) miR-101-1-5p is involved in the regulation of the innate immune responses of macrophages to lipopolysaccharide (LPS) [51].
Table 2. Identification of VATs- and SATs-specific enriched miRNAs.
| Comparison | Up-regulated | Down-regulated |
| GOM vs. ILB | 76 | 120 |
| MAD vs. ILB | 46 | 155 |
| PAD vs. ILB | 52 | 134 |
| RAD vs. ILB | 124 | 56 |
| GOM vs. ULB | 78 | 109 |
| MAD vs. ULB | 57 | 138 |
| PAD vs. ULB | 60 | 114 |
| RAD vs. ULB | 125 | 41 |
| Specifically enriched miRNAs* | 22 (VATs-specific) | 21 (SATs-specific) |
:if a differentially expressed miRNA was identified from all eight comparisons with the same regulation pattern, it was termed as a specifically enriched miRNA for VATs or SATs.
Figure 3. The expression changes of specifically enriched miRNAs across six adipose tissues.
(A) The expression pattern of inflammation-related miRNAs across six ATs. (B) The expression pattern of pathology-related miRNAs (other than inflammation-related miRNAs) across six ATs. (C) The expression pattern of adipogenesis and lipid metabolism-related miRNA across six ATs. The red and blue bars/circles represent the relative expression levels of miRNAs for VATs and SATs, respectively. The black lines across the circles present the average expression level for a certain type of miRNAs, and Student's t test was used for the testing significance of difference between VATs and SATs.
In addition to these well-annotated miRNAs, seven other VATs-specific enriched miRNAs (miR-122-5p, miR-126-3p, miR-127-5p, miR-136-3p, miR-100-5p, miR-99a-5p and miR-103-1-3p) were found to be related to various pathological responses (Figure 3B). By modulating cyclin G1, miR-122-5p influences p53 stability and transcriptional activity and reduces the invasion capability of hepatocellular carcinoma-derived cell lines [52]. MiR-126-3p can inhibit the growth of lung cancer cell lines in vitro and in vivo by down-regulating VEGF (vascular endothelial growth factor) [53]. MiR-127-5p and miR-136-3p (members of the miR-433-127 cluster) are involved in hepatocarcinogenesis [54]. MiR-100-5p targets Plk1 (polo-like kinase 1), a critical regulator of many stages of mitosis, resulting in the inhibition of cancer progression in nasopharyngeal cancer cell lines [55]. MiR-99a-5p, as a potential tumor suppressor, was down-regulated in advanced prostate cancer cell lines relative to the parental cell lines and its overexpression was reported to inhibit the growth of prostate cancer cells and decrease the expression of prostate-specific antigen [56]. MiR-103 decreased early in Alzheimer's disease and accelerated disease progression through the regulation of BACE1 (β-site amyloid precursor protein-cleaving enzyme 1) [57].
Adipogenesis and lipid metabolism-related miRNAs were enriched in SATs
In contrast, the SATs-specific enriched miRNAs (Figure 3C) were related mainly to adipogenesis and lipid metabolism. For example, miR-155-5p has been shown to inhibit adipogenesis by targeting the transcriptional factor C/EBP-β [58]. MiR-143-5p was elevated in adipogenesis and its inhibition with antisense oligonucleotides prevented adipocyte differentiation [4]. MiR-193b-3p and miR-365 (members of the miR-193-365 cluster) were revealed as central regulators of brown fat differentiation and adipogenesis [59]. MiR-27b-3p is a negative regulator of adipocyte differentiation by suppressing PPARγ (peroxisome proliferator-activated receptor γ) expression [8]. In pigs, miR-374b-5p was reported to be involved in the effect of maternal dietary protein on lipid metabolism in the offspring by targeting C/EBP-β [60].
Functional enrichment analysis of the mRNA targets of the differentially expressed miRNAs
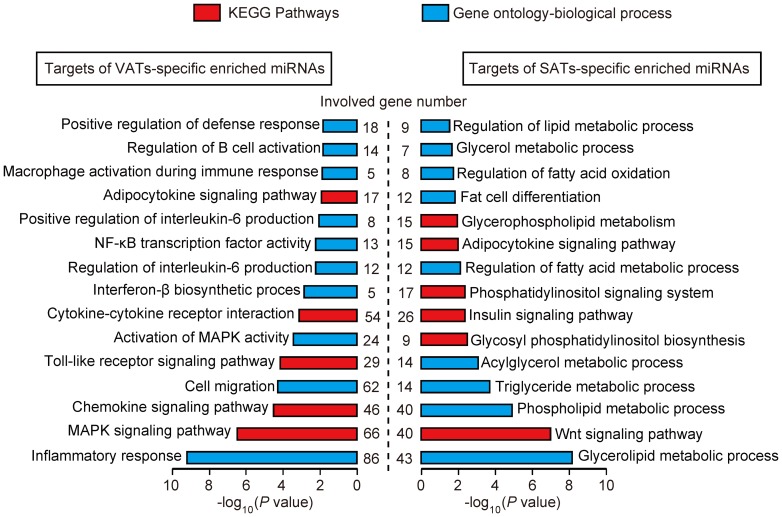
To further understand the distinct functional features between the VATs and SATs, the target protein coding genes of the miRNAs, which were specifically enriched in the VATs (2,572 mRNA genes) and SATs (1,961 mRNA genes), were predicted using PicTar [26], TargetScan human 6.2 [27], and MicroCosm Targets (version 5.0) [28] (Table S6). The predicted target genes were analyzed using the DAVID software [29] to determine whether they were enriched for specific functional categories and pathways. As expected, the target genes of the VATs-specific enriched miRNAs were associated primarily with immune and inflammatory processes, such as ‘inflammatory response’ (86 genes, P = 6.86×10−10), ‘chemokine signaling pathway’ (46 genes, P = .25×10−5), ‘Toll-like receptor signaling pathway’ (29 genes, P = 7.84×10−5), ‘regulation of interleukin-6 production’ (12 genes, P = 6.35×10−3), ‘NF-kB transcription factor activity’ (13 genes, P = 6.51×10−3), and ‘macrophage activation during immune response’ (5 genes, P = 1.48×10−2). In contrast, the target genes of the SATs-specific enriched miRNAs were associated mainly with lipid and energy metabolism, such as ‘glycerolipid metabolic process’ (43 genes, P = 7.66×10−9), ‘lipid biosynthetic process’ (38 genes, P = 7.36×10−6), ‘Wnt signaling pathway’ (40 genes, P = 1.14×10−7), ‘phospholipid metabolic process’ (40 genes, P = 1.45×10−5), ‘triglyceride metabolic process’ (14 genes, P = 2.32×10−4), ‘insulin signaling pathway’ (26 genes, P = 4.76×10−3), and ‘regulation of fatty acid metabolic process’ (12 genes, P = 8.94×10−3) (Figure 4). These results further suggested that SATs are involved mainly in lipid and energy metabolic homeostasis, whereas the VATs are susceptible to inflammation and should be regarded as a potential metabolic risk factor of obesity.
Figure 4. KEGG pathways and Gene Ontology-Biological Processes (GO-BP) enriched for target genes of VATs- and SATs-specific enriched miRNAs.
The P values was calculated using Benjamini-corrected modified Fisher's exact test.
Discussion
It have been well documented that the type of adipocytes, their endocrine function, lipolytic activity, response to insulin and other hormones are different between SATs and VATs. The content of non-adipocyte cells from ATs, such as monocytes/macrophages, were also shown to be significant different between SATs and VATs [61], [62]. It was shown that individual tissues maintain a unique miRNA profile, suggesting that miRNAs contribute to specific tissue function by regulating different gene targets [63]. As expected, we found that distinct ATs from different anatomical locations expressed a core number (284/409, 69.44%) of miRNAs that may fundamental for the regulation of genes involved in adipose metabolism (Table 1), but significant differences in miRNA expression were demonstrated. This result suggests that VATs and SATs are two locations of a developmentally homogeneous adipose organ [64].
Currently, it is thought that the increase in VATs rather than SATs best correlates with measures of insulin resistance [65] and cardiovascular diseases [66], and contributed to the chronic low-grade inflammation accompanied by these metabolic disorders in ATs [67], [68], [69]. Growing evidence indicates that deregulation of miRNAs is closely associated with obesity-related metabolic disorders including type 2 diabetes and cardiovascular diseases [70], [71]. Specific miRNAs have been implicated in adipogenesis and mature adipocyte function [32]. Nonetheless, the involvement of miRNAs in adipose tissue inflammation has been scarcely investigated. As yet, only a few miRNAs have been identified as relevant in this field [72], [73]. In this study, we presented a set of inflammation- and pathology-related miRNAs, which were specifically enriched in porcine VATs and could contribute to the special role of VATs in metabolic risk. Further studies are encouraged to validate the role of these miRNAs in metabolic disorders and obesity-associated adipose tissue inflammation in human individuals under physiological and pathological conditions.
In summary, we have presented a comprehensive comparison of miRNA expression among six ATs and focused on the variations in miRNA expression between the VATs and SATs, which reflected the intrinsic differences in their physiological and metabolic roles. Compared with the SATs that were related mainly to adipogenesis and lipid metabolism, the VATs (in particularly, the GOM) were mainly associated with the immune and inflammation responses, and should be deemed as a potential metabolic risk factor of obesity. Our results will benefit future studies into the metabolic role of the distinct AT compartments in obesity-related metabolic dysfunction, and in the further development of the pig model for human metabolic research.
Supporting Information
Characterization of the small RNA-seq data. (A) Filter process of sequencing data. (B) Size distribution of high-quality reads for six small RNA libraries.
(TIF)
Q-PCR validation for 18 miRNAs with the highest expression level across six adipose tissues. The Y-axis on the left represents the percentage of a certain miRNA accounted for in total high-quality reads resulting from small RNA-seq. The Y-axis on the right represents the relative expression levels of a certain miRNA derived from q-PCR. Pearson correlation was used to determine the relation of miRNAs expression changes between the q-PCR and the small RNA-sequencing approaches. Values are means ± SD.
(TIF)
Primer sequences used in q-PCR validation.
(XLS)
Porcine Known and conserved miRNAs identified in this study.
(XLS)
Porcine unique miRNAs identified in this study.
(XLS)
MiRNA categories defined by tissue-related expression pattern.
(XLS)
Expression patterns of VATs- and SATs-specific enriched miRNAs across distinct adipose tissues.
(XLS)
Predicted mRNA targets for the VATs- and SATs-specific enriched miRNAs.
(XLS)
Acknowledgments
We thank Hongmei Wang, Lu Bai and Yihui Liu for help with experiments.
Funding Statement
This work was supported by grants from the National High Technology Research and Development Program of China (863 Program) (2013AA102502) to M.L., the Specialized Research Fund of Ministry of Agriculture of China (NYCYTX-009), the Project of Provincial Twelfth Five Years' Animal Breeding of Sichuan Province (2011YZGG15), and the National Special Foundation for Transgenic Species of China (2011ZX08006-003) to X.L. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z (2003) The microRNA world: small is mighty. Trends in Biochemical Sciences 28: 534–540. [DOI] [PubMed] [Google Scholar]
- 2. Xie H, Lim B, Lodish HF (2009) MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 58: 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ (2009) MicroRNA let-7 regulates 3T3-L1 adipogenesis. Molecular Endocrinology 23: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, et al. (2004) MicroRNA-143 regulates adipocyte differentiation. Journal of Biological Chemistry 279: 52361–52365. [DOI] [PubMed] [Google Scholar]
- 5. Wang Q, Li YC, Wang J, Kong J, Qi Y, et al. (2008) miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proceedings of the National Academy of Sciences of the United States of American 105: 2889–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ortega FJ, Moreno-Navarrete JM, Pardo G, Sabater M, Hummel M, et al. (2010) MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PloS One 5: e9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin Q, Gao Z, Alarcon RM, Ye J, Yun Z (2009) A role of miR-27 in the regulation of adipogenesis. The FEBS Journal 276: 2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, et al. (2009) microRNA miR-27b impairs human adipocyte differentiation and targets PPARγ. Biochemical and Biophysical Research Communications 390: 247–251. [DOI] [PubMed] [Google Scholar]
- 9. Andersen DC, Jensen CH, Schneider M, Nossent AY, Eskildsen T, et al. (2010) MicroRNA-15a fine-tunes the level of Delta-like 1 homolog (DLK1) in proliferating 3T3-L1 preadipocytes. Experimental Cell Research 316: 1681–1691. [DOI] [PubMed] [Google Scholar]
- 10. Martinelli R, Nardelli C, Pilone V, Buonomo T, Liguori R, et al. (2012) miR-519d overexpression is associated with human obesity. Obesity 18: 2170–2176. [DOI] [PubMed] [Google Scholar]
- 11. Wang Q, Wang Y, Minto AW, Wang J, Shi Q, et al. (2008) MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. The FASEB Journal 22: 4126–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arner P (2005) Insulin resistance in type 2 diabetes-role of the adipokines. Current Molecular Medicine 5: 333–339. [DOI] [PubMed] [Google Scholar]
- 13. Dogru T, Sonmez A, Tasci I, Bozoglu E, Yilmaz MI, et al. (2007) Plasma visfatin levels in patients with newly diagnosed and untreated type 2 diabetes mellitus and impaired glucose tolerance. Diabetes Research and Clinical Practice 76: 24–29. [DOI] [PubMed] [Google Scholar]
- 14. Ikeoka D, Mader JK, Pieber TR (2010) Adipose tissue, inflammation and cardiovascular disease. Revista da Associacao Medica Brasileira 56: 116–121. [DOI] [PubMed] [Google Scholar]
- 15. Sam S, Haffner S, Davidson MH, D′Agostino RB Sr, Feinstein S, et al. (2008) Relationship of abdominal visceral and subcutaneous adipose tissue with lipoprotein particle number and size in type 2 diabetes. Diabetes 57: 2022–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fox CS, Massaro JM, Hoffmann U, Pou KM, Maurovich-Horvat P, et al. (2007) Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation 116: 39–48. [DOI] [PubMed] [Google Scholar]
- 17.Bjørndal B, Burri L, Staalesen V, Skorve J, Berge RK (2011) Different adipose depots: their role in the development of metabolic syndrome and mitochondrial response to hypolipidemic agents. Journal of Obesity 2011. [DOI] [PMC free article] [PubMed]
- 18. Spurlock ME, Gabler NK (2008) The development of porcine models of obesity and the metabolic syndrome. The Journal of Nutrition 138: 397–402. [DOI] [PubMed] [Google Scholar]
- 19. Li M, Wu H, Luo Z, Xia Y, Guan J, et al. (2012) An atlas of DNA methylomes in porcine adipose and muscle tissues. Nature Communications 3: 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li M, Xia Y, Gu Y, Zhang K, Lang Q, et al. (2010) MicroRNAome of porcine pre-and postnatal development. PloS One 5: e11541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pruitt KD, Tatusova T, Klimke W, Maglott DR (2009) NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Research 37: D32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, et al. (2009) Rfam: updates to the RNA families database. Nucleic Acids Research 37: D136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kohany O, Gentles AJ, Hankus L, Jurka J (2006) Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research 39: D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Romualdi C, Bortoluzzi S, d′Alessi F, Danieli GA (2003) IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiological Genomics 12: 159–162. [DOI] [PubMed] [Google Scholar]
- 26. Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, et al. (2005) Combinatorial microRNA target predictions. Nature Genetics 37: 495–500. [DOI] [PubMed] [Google Scholar]
- 27. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. [DOI] [PubMed] [Google Scholar]
- 28. Griffiths-Jones S, Saini HK, Van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Research 36: D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Da Wei Huang BTS, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 30. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qin L, Chen Y, Niu Y, Chen W, Wang Q, et al. (2010) A deep investigation into the adipogenesis mechanism: Profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/β-catenin signaling pathway. BMC Genomics 11: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alexander R, Lodish H, Sun L (2011) MicroRNAs in adipogenesis and as therapeutic targets for obesity. Expert Opinion on Therapeutic Targets 15: 623–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roush S, Slack FJ (2008) The let-7 family of microRNAs. Trends in Cell Biology 18: 505. [DOI] [PubMed] [Google Scholar]
- 34. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, et al. (2008) Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nature Immunology 9: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, et al. (2008) Targeted deletion reveals essential and overlapping functions of the miR-17-92 family of miRNA clusters. Cell 132: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 37. Yildirim A, Aktaş A, Nergiz Y, Akkuş M (2010) Analysis of human omentum-associated lymphoid tissue components with S-100: an immunohistochemical study. Romanian Journal of Morphology and Embryology 51: 759–764. [PubMed] [Google Scholar]
- 38. Carlow DA, Gold MR, Ziltener HJ (2009) Lymphocytes in the peritoneum home to the omentum and are activated by resident dendritic cells. The Journal of Immunology 183: 1155–1165. [DOI] [PubMed] [Google Scholar]
- 39. Agirre X, Jiménez-Velasco A, San José-Enériz E, Garate L, Bandrés E, et al. (2008) Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Molecular Cancer Research 6: 1830–1840. [DOI] [PubMed] [Google Scholar]
- 40. Tian Y, Luo A, Cai Y, Su Q, Ding F, et al. (2010) MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. Journal of Biological Chemistry 285: 7986–7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449: 682–688. [DOI] [PubMed] [Google Scholar]
- 42. Wang T, Jiang A, Guo Y, Tan Y, Tang G, et al. (2013) Deep sequencing of the transcriptome reveals inflammatory features of porcine visceral adipose tissue. International Journal of Biological Sciences 9: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou C, Zhang J, Ma J, Jiang A, Tang G, et al. (2013) Gene expression profiling reveals distinct features of various porcine adipose tissues. Lipids in Health and Disease 12: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, et al. (2011) Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. Journal of Allergy and Clinical Immunology 128 1077–1085: e1010. [DOI] [PubMed] [Google Scholar]
- 45. Iliopoulos D, Hirsch HA, Struhl K (2009) An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139: 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pekow JR, Dougherty U, Mustafi R, Zhu H, Kocherginsky M, et al. (2011) miR-143 and miR-145 are downregulated in ulcerative colitis: Putative regulators of inflammation and protooncogenes. Inflammatory Bowel Diseases 18: 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, et al. (2011) miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 20: 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen T, Huang Z, Wang L, Wang Y, Wu F, et al. (2009) MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovascular Research 83: 131–139. [DOI] [PubMed] [Google Scholar]
- 49. Zhu S, Pan W, Song X, Liu Y, Shao X, et al. (2012) The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-α . Nature Medicine 18: 1077–1086. [DOI] [PubMed] [Google Scholar]
- 50. Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, et al. (2011) The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nature Immunology 13: 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu Q-Y, Liu Q, Chen J-X, Lan K, Ge B-X (2010) MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation of MAPKs in macrophages. The Journal of Immunology 185: 7435–7442. [DOI] [PubMed] [Google Scholar]
- 52. Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, et al. (2009) MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Research 69: 5761–5767. [DOI] [PubMed] [Google Scholar]
- 53. Liu B, Peng X-C, Zheng X-L, Wang J, Qin Y-W (2009) MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer 66: 169–175. [DOI] [PubMed] [Google Scholar]
- 54. Tryndyak VP, Ross SA, Beland FA, Pogribny IP (2009) Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Molecular Carcinogenesis 48: 479–487. [DOI] [PubMed] [Google Scholar]
- 55. Shi W, Alajez NM, Bastianutto C, Hui AB, Mocanu JD, et al. (2010) Significance of Plk1 regulation by miR-100 in human nasopharyngeal cancer. International Journal of Cancer 126: 2036–2048. [DOI] [PubMed] [Google Scholar]
- 56. Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, et al. (2011) MiR-99 family of microRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Research 71: 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang W-X, Rajeev BW, Stromberg AJ, Ren N, Tang G, et al. (2008) The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. The Journal of Neuroscience 28: 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu S, Yang Y, Wu J (2011) TNFα-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors. Biochemical and Biophysical Research Communications 3: 618–624. [DOI] [PubMed] [Google Scholar]
- 59. Sun L, Xie H, Mori MA, Alexander R, Yuan B, et al. (2011) Mir193b–365 is essential for brown fat differentiation. Nature Cell Biology 13: 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pan S, Zheng Y, Zhao R, Yang X (2012) MicroRNA-130b and microRNA-374b mediate the effect of maternal dietary protein on offspring lipid metabolism in Meishan pigs. British Journal of Nutrition 1: 1–8. [DOI] [PubMed] [Google Scholar]
- 61. Bruun JM, Lihn AS, Pedersen SB, Richelsen B (2005) Monocyte chemoattractant protein-1 release is higher in visceral than subcutaneous human adipose tissue (AT): implication of macrophages resident in the AT. Journal of Clinical Endocrinology & Metabolism 90: 2282–2289. [DOI] [PubMed] [Google Scholar]
- 62. Curat C, Wegner V, Sengenes C, Miranville A, Tonus C, et al. (2006) Macrophages in human visceral adipose tissue: increased accumulation in obesity and a source of resistin and visfatin. Diabetologia 49: 744–747. [DOI] [PubMed] [Google Scholar]
- 63. Liang Y, Ridzon D, Wong L, Chen C (2007) Characterization of microRNA expression profiles in normal human tissues. BMC genomics 8: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lafontan M (2013) Differences Between Subcutaneous and Visceral Adipose Tissues. Physiology and Physiopathology of Adipose Tissue: Springer. 329–349.
- 65. Piché M-È, Lapointe A, Weisnagel SJ, Corneau L, Nadeau A, et al. (2008) Regional body fat distribution and metabolic profile in postmenopausal women. Metabolism 57: 1101–1107. [DOI] [PubMed] [Google Scholar]
- 66. Maury E, Brichard S (2010) Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Molecular and Cellular Endocrinology 314: 1–16. [DOI] [PubMed] [Google Scholar]
- 67. Ferrante A (2007) Obesity-induced inflammation: a metabolic dialogue in the language of inflammation. Journal of Internal Medicine 262: 408–414. [DOI] [PubMed] [Google Scholar]
- 68. Pou KM, Massaro JM, Hoffmann U, Vasan RS, Maurovich-Horvat P, et al. (2007) Visceral and subcutaneous adipose tissue volumes are cross-sectionally related to markers of inflammation and oxidative stress the Framingham heart study. Circulation 116: 1234–1241. [DOI] [PubMed] [Google Scholar]
- 69. Clément K, Langin D (2007) Regulation of inflammation-related genes in human adipose tissue. Journal of Internal Medicine 262: 422–430. [DOI] [PubMed] [Google Scholar]
- 70. Heneghan H, Miller N, Kerin M (2010) Role of microRNAs in obesity and the metabolic syndrome. Obesity Reviews 11: 354–361. [DOI] [PubMed] [Google Scholar]
- 71. Hulsmans M, De Keyzer D, Holvoet P (2011) MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. The FASEB Journal 25: 2515–2527. [DOI] [PubMed] [Google Scholar]
- 72. Strum JC, Johnson JH, Ward J, Xie H, Feild J, et al. (2009) MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Molecular Endocrinology 23: 1876–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, et al. (2012) A novel regulator of macrophage activation clinical perspective miR-223 in obesity-associated adipose tissue inflammation. Circulation 125: 2892–2903. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of the small RNA-seq data. (A) Filter process of sequencing data. (B) Size distribution of high-quality reads for six small RNA libraries.
(TIF)
Q-PCR validation for 18 miRNAs with the highest expression level across six adipose tissues. The Y-axis on the left represents the percentage of a certain miRNA accounted for in total high-quality reads resulting from small RNA-seq. The Y-axis on the right represents the relative expression levels of a certain miRNA derived from q-PCR. Pearson correlation was used to determine the relation of miRNAs expression changes between the q-PCR and the small RNA-sequencing approaches. Values are means ± SD.
(TIF)
Primer sequences used in q-PCR validation.
(XLS)
Porcine Known and conserved miRNAs identified in this study.
(XLS)
Porcine unique miRNAs identified in this study.
(XLS)
MiRNA categories defined by tissue-related expression pattern.
(XLS)
Expression patterns of VATs- and SATs-specific enriched miRNAs across distinct adipose tissues.
(XLS)
Predicted mRNA targets for the VATs- and SATs-specific enriched miRNAs.
(XLS)