Abstract
Blueberry is an economically and nutritionally important small fruit crop, native to North America. As with many crops, extreme low temperature can affect blueberry crop yield negatively and cause major losses to growers. For this reason, blueberry breeding programs have focused on developing improved cultivars with broader climatic adaptation. To help achieve this goal, the blueberry genomic database (BBGD454) was developed to provide the research community with valuable resources to identify genes that play an important role in flower bud and fruit development, cold acclimation and chilling accumulation in blueberry. The database was developed using SQLServer2008 to house 454 transcript sequences, annotations and gene expression profiles of blueberry genes. BBGD454 can be accessed publically from a web-based interface; this website provides search and browse functionalities to allow scientists to access and search the data in order to correlate gene expression with gene function in different stages of blueberry fruit ripening, at different stages of cold acclimation of flower buds, and in leaves.
Availability
It can be accessed from http://bioinformatics.towson.edu/BBGD454/
Keywords: Blueberry, database, 454, transcriptomics
Background
Blueberry (Vaccinium corymbosum) is one of the major berry crops grown in the United States [1]. North America, in fact, is the world's leading blueberry producer, accounting for nearly 90% of world production at the present time. Total area devoted to growing commercial blueberries in North America is approximately 74,000 hectares. Blueberry is a high value crop, often times grown in acidic and imperfectly drained soils that would otherwise be considered unfit for agricultural production [1]. Blueberry is also an important fruit crop because of its nutritional value. Of all fresh fruits and vegetables, blueberries are one of the richest sources of antioxidants [2, 3, 4]. Blueberry is a model organism for the heath family Ericaceae, which also includes the economically important, closely related cranberry as well as the economically important, more distantly related ornamentals, rhododendron, azalea, and mountain laurel. For all these related species, genomic studies, including EST generation and microarray analyses, are lacking or completely absent. Functional genomic studies on berry crops are lacking, especially studies dealing with the molecular impacts of low temperature on berry crop yield and chilling accumulation. Low temperature extremes reduce blueberry yields and impact the profitability and competitiveness of U.S. producers. Enhanced cold tolerance during the winter and early spring of elite varieties would be of great value to the blueberry industry. New low-chilling varieties are also desirable for the southern U.S. BBGD454 is a public database that houses 454 transcriptome sequences from numerous blueberry cDNA library samples. The ultimate goal of these experiments is to better understand the genetic control of cold hardiness, chilling requirement, and various fruit quality traits and apply the information to develop new cultivars for the blueberry industry.
Methodology
BBGD454 database was designed, implemented and hosted using Microsoft SQL Server 2008 Enterprise Edition. Microsoft Visual Studio 2008 was used to design and implement the web pages, which were programmed using ASP.NET framework 2.35 with C# programming language. Real time charts were built using a charting library called Highcharts written in pure JavaScript. Both the database and the website are on the same server at Towson University in Baltimore, MD, USA. This server is running Microsoft Windows Server 2003 and Internet Information Services (IIS V6.0). BBGD454 stores unique putative transcripts (contigs and singletons) that resulted from assembling the normalized, corrected and filtered 454 sequences using the GS DeNovo Assembler (454 Life Sciences, Roche). In addition to the sequences, BBGD454 also houses information on taxonomy, gene function, tissue specificity, gene ontology, and blast results of those contigs with high number of reads in one library against all the other libraries. All blast results were obtained using Blast2Go [5].
Utility to the biological community
The database contains 454 sequences from nine cDNA libraries that were constructed from mRNA from various organs collected from plants of the highbush blueberry (V. corymbosum) variety Bluecrop. Organs included young, fully expanded leaves, flower buds collected at various stages of cold acclimation (0, 397, 789, and 1333 chill units), and fruit collected at various stages of ripening (green, white, pink, and blue) [6]. The nine libraries (leaves, buds at four different stages of cold acclimation, and fruit at four different stages of ripening) were multiplexed and sequenced on two plate runs of the 454-GS FLX Titanium platform. A summary of the sequencing and assembly results is shown in Table 1 (see supplementary material). Overall, 1,348,819 reads were generated, with an average read length of 287 nucleotides (nt). This yielded a total of ~390 megabases of cDNA sequence. The BBGD454 web-accessible interface (Figure 1) provides an easy way to search, browse and download the sequences and expression data stored in the database. The following are the main functions the website provides:
Figure 1.
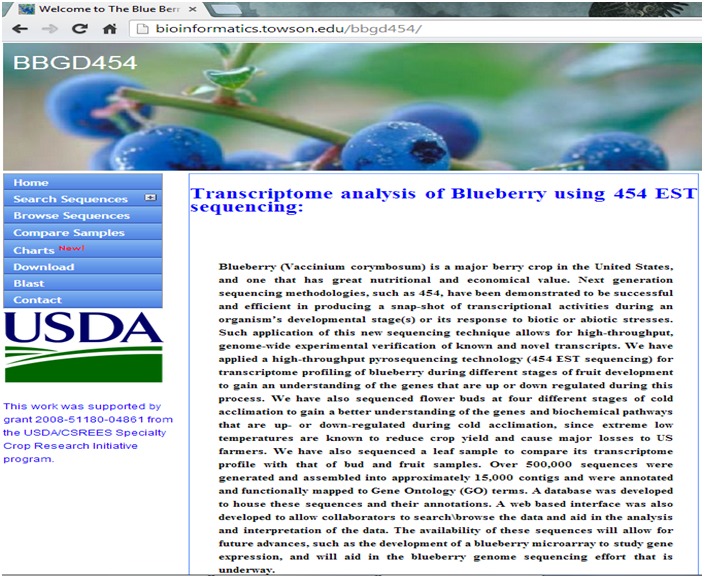
A snapshot of BBGD454 main web page, shows a quick summary about the project and the functions this website provides.
(i) Search:
Users can search by contig number, or by sequence description (Figure 2). Partial numbers and/or characters can be used if one is not sure of the contig number or full gene name. Both the search by contig and search by description return their results in a nice tabular format that allows the user to select any record of the returned search results to see details about that specific sequence. The information includes sequence name, sequence description, sequence length, number of hits, gene ontolgy, RPKM [7], number of reads, and more.
Figure 2.
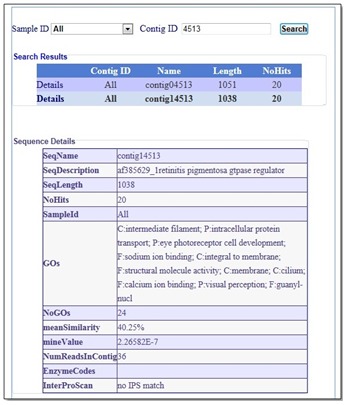
A snapshot of the Search by Contig ID page. Users can use the Search pages (By Contig Id, By Sequence Description) to search for the sequences using any part of their IDs or description.
(ii) Browse:
The webpage displays a table of all the blueberry cDNA samples. The user can click on any of the samples to see all sequences of that specific sample in a table that can be sorted based on many sequence attributes (id, length, description, etc). Sequence details can also be seen from this page by clicking on details next to any sequence id (contig number).
(iii) Compare samples:
Users can compare the expression of genes between two different libraries from the web site. This should help identify genes that are potentially differentially expressed during cold acclimation and chilling accumulation and during fruit development.
(iv) Blast:
We have a Blast application on the site that provides the functionality of blasting a sequence of interest against the sequences stored in BBGD454, the Sanger ESTs stored in BBGD (http://bioinformatics.towson.edu/bbgd/) and/or a collection of sequence data comprising EST and genomic sequences from all plant species, kingdom Viridiplantae.
Caveats
The sequences were de-novo assembled, which may have resulted in a number of false positives. Once the genome of blueberry is completely sequenced, one can redo the analysis, using these sequences, and aligning them to the genome. This would yield more accurate results.
Future Development
A web browser aligning the reads to the genome of blueberry would be extremely beneficial. This is something we might consider implementing once the genome is published and made available.
Authors' contributions
Omar Darwish, a doctoral student in IT, designed and developed the database and user interface, under the guidance of Nadim Alkharouf at Towson University. L. Jeannine Rowland was the PI of the Blueberry transcriptomic project at the USDA and was in charge of library preparation as well as the overall design of the experiments. All authors contributed to the writing of the manuscript.
Supplementary material
Footnotes
Citation:Darwish et al, Bioinformation 9(17): 883-886 (2013)
References
- 1.Yarborough D, et al. Factors contributing to the increase in productivity in the wild blueberry industry. 2004;3:33. [Google Scholar]
- 2.Nile SH, et al. Nutrition. 2013;9007:00220. [Google Scholar]
- 3.Cho E, et al. Arch Ophthalmol. 2004;122:883. doi: 10.1001/archopht.122.6.883. [DOI] [PubMed] [Google Scholar]
- 4.Kalt W, et al. Journal-American Pomological Society. 2007;61:151. [Google Scholar]
- 5.Conesa A, et al. Bioinformatics. 2005;21:3674. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 6.Rowland L, et al. BMC plant biology. 2012;12:46. doi: 10.1186/1471-2229-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortazavi A, et al. Nature methods. 2008;5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


