Abstract
Alzheimer disease (AD) is highly prevalent in Wadi Ara, despite the low frequency of APOE ε4 in this genetically isolated Arab community in northern Israel. We hypothesized that the reduced genetic variability in combination with increased homozygosity would facilitate identification of genetic variants that contribute to the high rate of AD in this community. AD cases (N=124) and controls (N=142) from Wadi Ara were genotyped for a a genome-wide set of more than 300,000 single nucleotides polymorphisms (SNPs) which were used to calculate measures of population stratification and inbreeding, and to identify regions of autozygosity. Although a high degree of relatedness was evident in both cases and controls, controls were significantly more related and contained more autozygous regions than cases (P = 0.004). Eight autozygous regions on seven different chromosomes were more frequent in controls than cases, and 105 SNPs in these regions, primarily on chromosomes 6 and 9, were nominally associated with AD. Associations with SNPs in NOTCH4 and AGPAT1 (both on chromosome 6) were confirmed in a meta analysis of four genome-wide association study (GWAS) datasets. Analysis of the full Wadi Ara GWAS dataset revealed 99 SNP associations with AD at P ≤ 10−5, however none of these were confirmed in the replication GWAS datasets. The unique population structure of Wadi Ara enhanced efforts to identify genetic variants that might partially explain the high prevalence of AD in the region. Several of these variants show modest evidence for association in other Caucasian populations.
Keywords: Alzheimer Disease, Genome-Wide Association Study, Population Groups, Meta-Analysis
Introduction
Alzheimer's disease (AD) is a common, heritable form of dementia with complex genetic etiology. Several genome-wide association studies have been conducted to identify genes and chromosomal regions that contribute to disease risk [1–9], with varying results. To date, only four genes have been identified with robust, replicable, and large effects on AD risk—mutations in the amyloid precursor protein (APP) and presenilin 1 and 2 (PSEN1, PSEN2) cause rare, early-onset Mendelian forms of the disease, while the ε4 variant of apolipoprotein E (APOE) is the primary genetic susceptibility factor for the more common late onset form of the disease [10]. The primary challenge in identifying genes associated with late onset AD, as demonstrated by recent studies [8, 9, 11] is that 50% or more of the genetic variance of AD risk is not yet accounted for, and likely is due to the effects of multiple loci individually contributing modestly to AD risk. The statistical power to detect disease variants is low under these conditions, highlighting the need for large collaborative genetic association studies.
A classic genetic approach to identify disease variants is to study isolated populations in which the genetic contributions to disease risk may be fewer and more discernable through consanguineous mating patterns. Wadi Ara, an Arab community between Hadera and Afula in northern Israel, is uniquely suited for this purpose due to its genetic isolation and high prevalence of AD. Our initial genetic studies of AD in this community were prompted by the unusually high prevalence of AD (20.5% in those ≥ with age 60 yr, 60.5% in those with age ≥ 85 yr) [12], despite having a lower frequency of the APOE ε4 alleles than other Caucasian populations [13]. Residents of the area belong to one of about 14 hamulas, or tribal groups, and until recently there has been reportedly minimal immigration or emigration from the community since it was founded approximately 300–400 years ago. This geographic isolation, along with the relatively small number of founders has lead to substantial consanguinity among its members.
A consequence of consanguinity is the presence of long stretches of homozygosity throughout the genome, and if the homozygous regions are derived from a common ancestor, the segments are also autozygous. Alternatively, regions of consecutive homozygous markers could arise in the absence of inbreeding through inheritance of two common extended haplotypes [14]. Autozygosity mapping has been successfully used previously to identify risk variants for several complex diseases with appreciable frequency in the population including colorectal cancer [15] and Parkinson’s disease [16].
The Wadi Ara community has already yielded information on the genetics of AD. In a previous low-density genome scan using 375 microsatellite markers, we identified regions on chromosomes 2, 9, 10, and 12 with significant linkage to AD status [17]. The implicated regions on chromosomes 9, 10, and 12 have been reported consistently as linkage peaks in studies of outbred populations [18–23]. We also identified a very strong association of a two-SNP haplotype in the angiotensin-converting enzyme (ACE) gene [24], and a three-SNP haplotype in the neuronal sortilin-related receptor SORL1 gene [11], two of the few genes with robust evidence for association with AD [25, 26]. In this study, we analyzed the Wadi Ara cohort for association of AD with more than 300,000 SNPs covering the entire genome. This approach facilitated a more complete genetic analysis of the population, including tests for differential ancestry (substructure), quantification of inbreeding, and identification of specific genomic regions of autozygosity that might contribute to disease risk.
Materials and Methods
Subject ascertainment and evaluation
Subjects were recruited from Wadi Ara, a geographically defined area in northern Israel comprising three Arab villages [12] during two study periods. The first was in 1995 when the residents ages 60 years and older as of October 1, 1995 were identified. The subjects who agreed to participate were interviewed and examined by an Arabic-speaking physician who had been previously trained in a memory clinic. Each subject underwent a battery of standard cognitive tests modified to fit the cultural and linguistic characteristics of this community. The diagnosis of AD was determined using the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV). Persons with mild cognitive impairment and demented subjects whose medical history and laboratory and cognitive test results suggested the presence of other illnesses such as vascular dementia, Parkinson disease, normal-pressure hydrocephalus or pseudodementia (depression), were excluded. Evidence of ischemic stroke or white matter disease was present in only 2 and 15 AD patients, respectively, who had a brain MRI scan, suggesting that relatively few subjects were misclassified. In the second phase, an independent group of subjects aged 65 years or older was identified. These subjects were examined by an Arabic-speaking neurologist and underwent a battery of cognitive tests similar to the one used in the first phase. Details of the evaluation of these subjects are published elsewhere [27, 28]. Neuropathological confirmation of AD was unavailable owing to the social and religious proscription against autopsy. Peripheral blood samples were obtained for DNA and biochemical analysis from the 650 living phase 1 participants between 1997 and 2000 and from 504 phase 2 participants between 2006 and 2009.
Genotyping and Quality Control
DNA specimens from 219 phase 1 participants (107 AD cases, 112 age-matched cognitively normal controls) and from 88 phase 2 participants (37 AD cases, 51 controls) were genotyped with the Illumina Hap300 (ver.1) or the Human CNV370 (ver.1) high density genotyping arrays according to the manufactures protocol (Illumina). Due to limited amounts of DNA, the phase 1 samples were genome amplified (Genomiphi, GE Healthcare) prior to running the arrays. Genotyping results for 17 individuals were excluded because less than than 95% of their SNPs were successfully called. Also, 22 participants were excluded because they were recruited in both phases and had inconsistent phenotype data. SNPs which had a minor allele frequency (MAF) of less than 1% (n=1159), had a call rate less than 95% (n=18,770, or were not in Hardy-Weinberg equilibrium based on a significance threshold of P < 0.0001 (n=186) were excluded from further analysis.
Statistical Analysis
The inbreeding coefficient (F) was determined for each individual by comparing the observed and expected numbers of homozygous loci [29]. Analysis of variance was used to assess differences in F among hamulas, age groups, and AD status. Substructure within the population was evaluated using several methods. First, we employed PLINK v.1.05 (http://pngu.mgh.harvard.edu/purcell/plink/) [30] to test for clusters within the sample and to determine if the amount of genome wide identity by state (IBS) sharing, or identity by descent (IBD) sharing in related individuals, differed between cases and controls. Second, we applied a model-based clustering approach implemented in the program STRUCTURE [31]. This program assigns a likelihood of population group membership based on marker allele frequencies. We used the smartpca.pl script in the EIGENSOFT package, [32] to determine the principal components of the genotype matrix and detect subtle population substructure.
Under the hypothesis that the high rates of AD in this population may be due to rare risk alleles amplified by inbreeding, we used PLINK to identify stretches of homozygosity across the genome in each individual. This method applies a sliding window approach to identify sequences of at least 100 consecutive homozygous SNPs. We then tested the presence or absence of these homozygous regions, as well as the individual SNPs within these regions for association with AD using logistic regression adjusting for sex and age at ascertainment. In addition to the genotyped SNPs, we imputed genotypes for 2.54 million SNPs using the Markov Chain haplotyping (MaCH) software (http://www.sph.umich.edu/csg/abecasis/MACH/) [33]. Unrelated individuals from the CEPH Utah pedigrees (CEU) in the HapMap consortium were used as a reference haplotype set. We observed a drop in imputation accuracy when we used a mixture of CEU (Caucasians) and Yoruban YRI (Africans) populations as reference haplotypes. The entire SNP set was subsequently analyzed using age- and sex-adjusted logistic regression models. The presence or absence of a homozygous region was modeled as a binary variable, and SNPs were modeled assuming an additive effect of the minor allele dose. The imputed SNPs were also modeled additively, but a non-integral dosage value (between 0 and 2) was used to incorporate the uncertainty in the imputation algorithm. By limiting our initial analysis to the genotyped and imputed SNPs (N = 2070) within homozygous stretches whose frequency differed significantly between cases and controls, we focused our search for population-specific risk loci and reduced the statistical penalty for multiple tests. To assess the significance of the initial associations and correct for the multiple tests, we created 1000 simulated null datasets by permuting individuals’ AD status and covariates as blocks to preserve their correlation structure. We then tested these datasets with randomized AD status for association with the SNPs in disease associated runs of homozygosity. We took the smallest P-value from each replicate and compared the observed P-values to the resulting empirical null distribution. The simulations indicate that a raw P-value ≤ 0.0004 would be required to exceed an experiment wide threshold of P ≤ 0.05. For the remaining association tests that included all the genotyped and imputed SNPs, we used the standard P ≤ 5 × 10−8 threshold for genome wide significance.
To bolster the SNP association findings in the Wadi Ara sample, we conducted meta-analysis on this set of SNPs using four independent Western European datasets described in Table 1. Association results obtained from each of these datasets using logistic regression models were combined using the inverse variance method implemented in the software package METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html).
Table 1.
Datasets contributing to the meta-analysis of AD association
Results
The GWAS sample contained 124 cases of AD with a mean age of 78.4 years and 142 controls with a mean age of 72.1 years. Five cases and two controls carried a single copy of the APOE ε4 allele. Participants represented nine different hamulas, four of which contained 87% of the sample. Although a lack of genealogical records limited our ability to construct extensive pedigrees, 39 pairs of individuals (not all unique) shared at least 25% of their alleles identical by state (IBS). Fourteen of these shared at least 50% of their alleles IBS, which confirmed the known sibpairs in the sample. Age at ascertainment and gender were significantly associated with AD (ORfemale = 3.13, P = 0.0005, ORage = 1.13 per additional year, P < 0.0001). Table 1 shows demographic characteristics of the sample by AD status.
Population Stratification
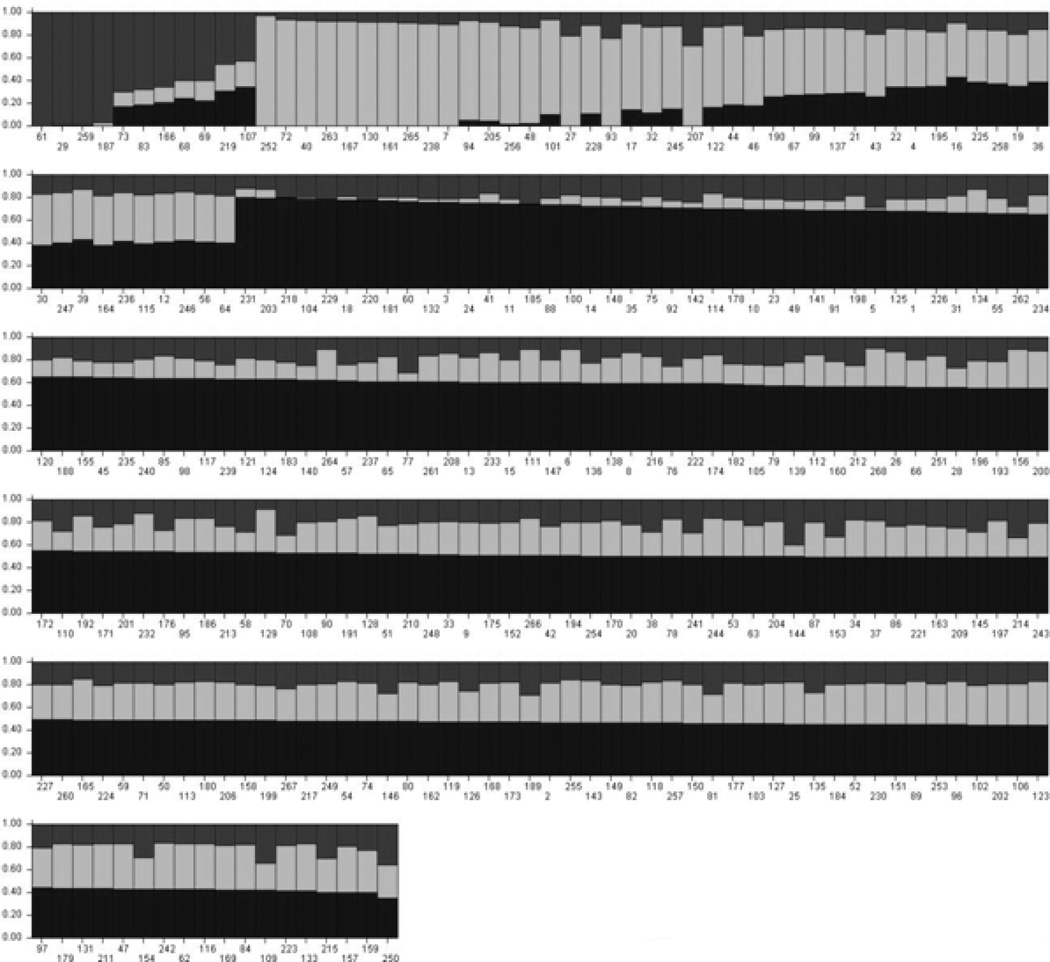
Despite its isolation and purported homogeneity, we found evidence for a relatively high degree of admixture and stratification within the population. STRUCTURE analysis indicated that a three cluster solution fit the data significantly better than two or four-cluster solutions. In the three cluster solution, the average proportions of population membership across individuals were 48%, 31%, and 21% in the first, second, and third cluster, respectively, indicating admixture. On an individual level, several people appeared to share almost no ancestry with the primary population. Seven individuals had at least 90% of their population membership contained within the second cluster and four had over 95% membership in the third cluster, indicating population stratification. Figure 1 shows the proportion of membership in each of the three ancestral populations for each individual.
Figure 1. Individual level proportions of membership in the three ancestral populations.
Individuals are on the X-axis and the proportion of their ancestry in each of the three clusters is on the Y-axis. Estimates were calculated using STRUCTURE software. The black area shows the predominant cluster, from which individuals derive 48% of their ancestry, on average. The gray area shows the second cluster, which contains, on average, 31% of the ancestry in the sample; the third cluster (in dark gray) contains 21%.
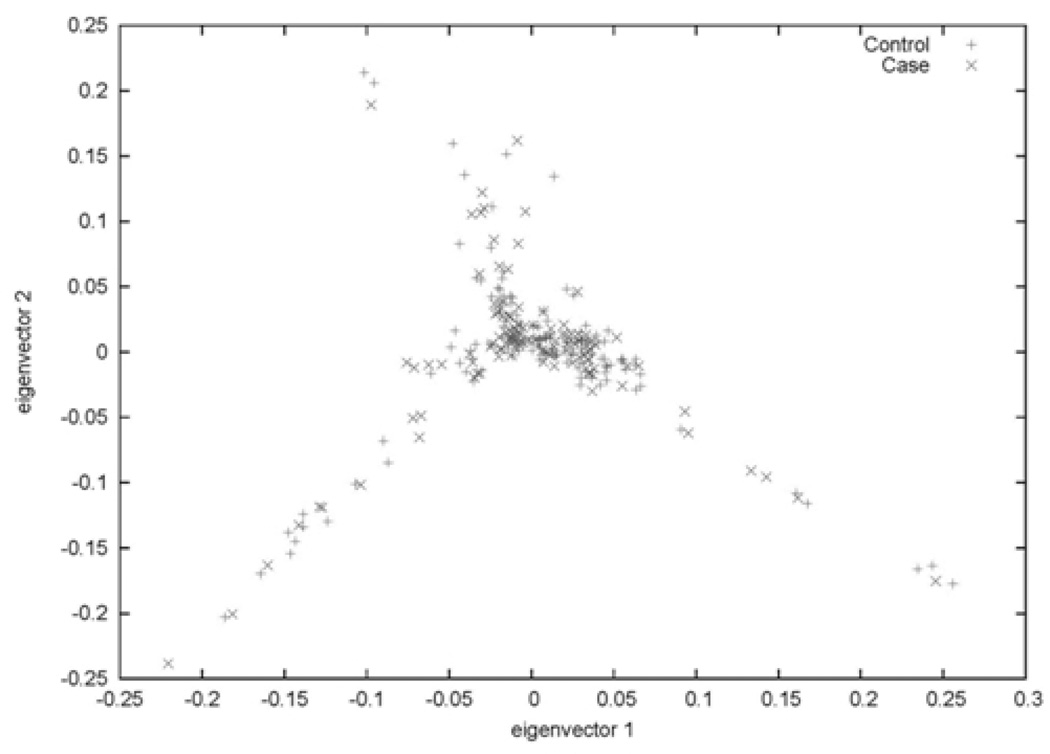
The principal component (PC) analysis also showed evidence of admixture. The plot of each subject’s values for the first two PCs (eigenvectors) by disease status indicates four trends in ancestry—a primary cluster of individuals centered around zero on both PCs, a group with higher values on only the second PC, a group with low values on both components, and another with high values on the first PC and low values on the second (Figure 2). We found no evidence that the values of any of the first three PCs differed by disease status.
Figure 2. Study sample plotted on their values for the first two principal components (eigenvectors) of ancestry.
Individuals’ values on the first two principal components of ancestry by AD status. Neither of these components significantly predicted AD.
Autozygosity Mapping
We found a modest level of excess homozygosity in the sample. The maximum genome-wide F statistic for an individual was 0.15, a value slightly greater than that for a child of an avuncular pair. The maximum mean F value within a hamula was 0.03 which is equivalent to that for a child of a half first-cousin pair. Analysis of variance showed that neither hamula nor age was a significant predictor of inbreeding level. Comparison of F and the first three PCs from EIGENSTRAT analysis revealed that only the third PC was significantly correlated with F (r2=.15, P = 0.01), indicating that individuals with high values on this axis of variation are less consanguineous and may be descendants of more recent migrants to the region. The F values also differed significantly between cases and controls. Fifty-nine percent of cases and 61% of controls had positive F statistics (i.e. higher genome-wide homozygosity than would be expected by chance), and the average degree of inbreeding was significantly higher in controls (0.02 vs. 0.01, P = 0.004; see table 2).
Table 2.
Demographic characteristics of the study sample.
| Cases (N = 124) | Controls (N = 142) | |||
|---|---|---|---|---|
| Variable (units) | Mean or N | SD | Mean or N | SD |
| Age (years) | 78.6 | 7.9 | 72.0 | 6.1 |
| SBP (mmHg) | 158.2 | 14.9 | 154.6 | 9.4 |
| DBP (mmHg) | 81.0 | 9.7 | 78.8 | 3.9 |
| F | 0.01 | 0.03 | 0.02 | 0.04 |
| Gender (M/F) | 59/65 | NA | 76/66 | NA |
| ApoE ε4 (N) | 5 | NA | 2 | NA |
F is measured as the proportion of excess homozygosity over that expected by chance based on allele frequency. ApoE ε4 indicates the number of individuals carrying a single copy of the ε4 allele.
As expected based on the level of genome-wide inbreeding observed, there were several large regions of homozygosity in the study sample. Several of these regions were observed in multiple individuals, indicating inheritance through a common ancestor (autozygosity). The consensus regions (i.e., the region shared by all individuals with a stretch of homozygosity in a given chromosomal segment) ranged in size from a single SNP to 3.4 Mb. The frequency of eight of these stretches differed significantly between cases and controls (Table 2). Consistent with our observation that the level of homozygosity was greater in controls, each of these homozygous regions was more frequent in controls.
Not all of the individuals with runs of homozygosity in a given region were homozygous for the same alleles, indicating that these segments of DNA were not inherited through a single ancestor. To determine if specific allelic configurations, rather than the presence or absence of homozygosity, affected AD risk, we also tested a model in which people with rare (present in less than 5 people) allelic configurations in their homozygous stretches were ignored, comparing the remaining configurations to the most common configuration in a logistic regression model. None of these tests were significant at P < 0.05.
Single SNP Association
The initial set of association tests included only SNPs in the consensus regions of the runs of homozygosity that were significantly associated with AD. Of the 2,194 SNPs in disease-associated homozygous regions, 106 were nominally associated with AD and are shown in Table S1. These SNPs are located primarily in two regions on chromosomes 6 and 9, although one SNP each on chromosomes 8 and 15 was nominally associated with AD. The SNP with the strongest effect was rs1800684 (OR = 4.36, P = 0.002), a synonymous coding SNP in the AGER gene on chromosome 6. Two other genes in this region also have disease associated SNPs and potential biological links to AD, NOTCH4 and AGPAT1. None of the SNPs in homozygous regions exceeded the experiment-wide significance threshold (P ≤ 0.0004) determined by the null simulations.
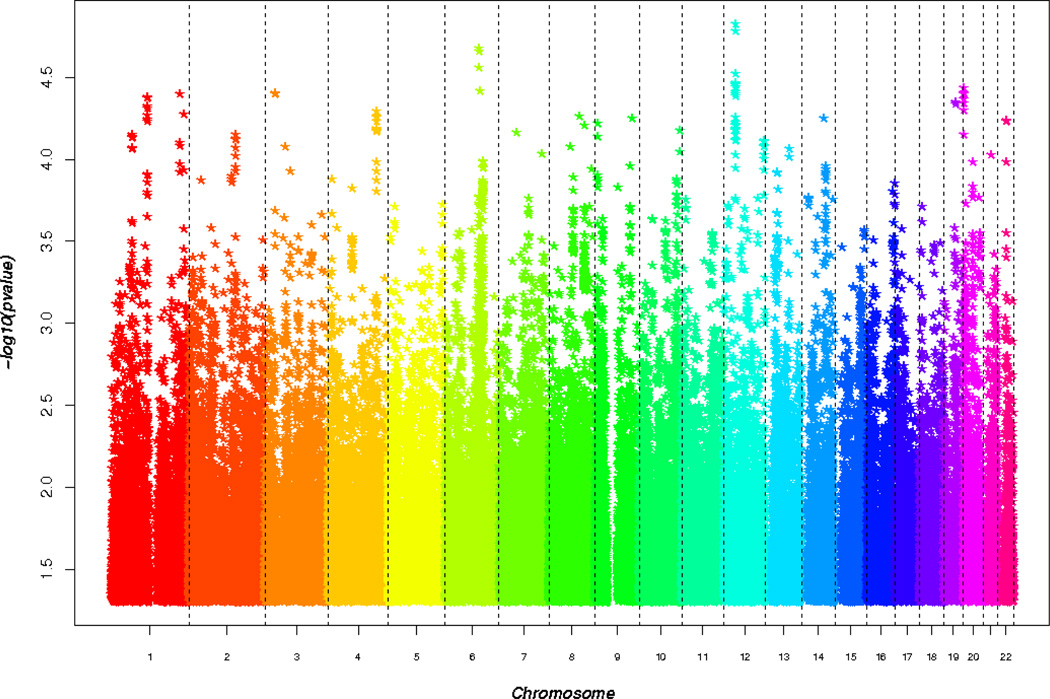
Results for the entire (i.e., genome-wide) set of genotyped and imputed SNPs are shown in Figure 3. While none of the results were genome-wide significant, many of the 30 top-ranked SNPs are in genes of potential interest to AD pathology. These results are shown in Table 4. The strongest association was observed with rs7308580 on chromosome 12 (OR = 0.18, P = 1.5 × 10−5) which is located in an intron of copine 8 (CPN8). Two SNPs (rs2300186, rs4813945) in the proteasome inhibitor subunit 1 (PSMF1) gene on chromosome 20 were associated with AD (OR = 0.41, P = 3.9 × 10−5). A comparable level of significance was obtained with rs2025148 in FIG4 on chromosome 6 (OR = 2.47, P = 3.8 × 10−5) and with rs7028544 in ASTN2 on chromosome 9 (OR = 0.39, P = 5.6 × 10−5).
Figure 3. Age and sex adjusted SNP association results in Wadi Ara.
Manhattan plot summarizing GWAS results in Wadi Ara. SNPs were modeled as the additive effect of minor allele dose. The most significant associations were in the genes CPNE8, PSMF1, FIG4, and CMTM8.
Table 4.
Top 30 SNPs associated with AD overall in Wadi Ara
| Chr | SNP | Position (BP) |
MAF case |
MAF control |
OR | P | Quality | GENE | Type |
|---|---|---|---|---|---|---|---|---|---|
| 12 | rs7308580 | 37571225 | 0.15 | 0.05 | 0.18 | 1.49E-05 | 0.99 | CPNE8 | intron |
| 12 | rs7307936 | 37571373 | 0.15 | 0.05 | 5.60 | 1.51E-05 | 0.99 | CPNE8 | intron |
| 12 | rs1516555 | 37583295 | 0.15 | 0.05 | 5.55 | 1.65E-05 | 0.99 | CPNE8 | intron |
| 6 | rs7761791 | 106683679 | 0.45 | 0.25 | 2.38 | 2.09E-05 | NA | NA | NA |
| 6 | rs4946727 | 106680065 | 0.21 | 0.07 | 0.27 | 2.21E-05 | 0.99 | NA | NA |
| 6 | rs6903903 | 106684200 | 0.45 | 0.26 | 0.41 | 2.76E-05 | 0.97 | NA | NA |
| 12 | rs12423647 | 37593034 | 0.14 | 0.05 | 5.17 | 3.01E-05 | 1.00 | NA | NA |
| 12 | rs12425783 | 37536951 | 0.19 | 0.08 | 4.04 | 3.42E-05 | 0.98 | CPNE8 | intron |
| 12 | rs10876185 | 37544916 | 0.19 | 0.08 | 0.25 | 3.47E-05 | 0.98 | CPNE8 | intron |
| 12 | rs12422883 | 37543217 | 0.19 | 0.08 | 4.03 | 3.49E-05 | 0.98 | CPNE8 | intron |
| 12 | rs9943730 | 37529460 | 0.19 | 0.08 | 0.25 | 3.60E-05 | 0.98 | CPNE8 | intron |
| 12 | rs1878224 | 37595981 | 0.13 | 0.05 | 5.08 | 3.60E-05 | NA | NA | NA |
| 20 | rs2072965 | 1094136 | 0.30 | 0.46 | 0.41 | 3.64E-05 | 0.92 | PSMF1 | UTR-3 |
| 6 | rs2025148 | 110134636 | 0.43 | 0.27 | 2.47 | 3.82E-05 | NA | FIG4 | intron |
| 12 | rs10876062 | 37454453 | 0.18 | 0.07 | 4.28 | 3.87E-05 | 0.98 | CPNE8 | intron |
| 20 | rs2300186 | 1082900 | 0.33 | 0.48 | 0.41 | 3.93E-05 | 0.92 | PSMF1 | intron |
| 3 | rs7644602 | 32295412 | 0.48 | 0.30 | 2.46 | 3.95E-05 | NA | CMTM8 | intron |
| 3 | rs6783478 | 32297979 | 0.49 | 0.32 | 2.60 | 3.97E-05 | 0.91 | CMTM8 | intron |
| 1 | rs2791559 | 217655426 | 0.37 | 0.23 | 2.82 | 3.97E-05 | NA | NA | NA |
| 20 | rs4813044 | 1096783 | 0.34 | 0.51 | 0.44 | 3.97E-05 | NA | PSMF1 | UTR-3 |
| 12 | rs17126713 | 37505285 | 0.19 | 0.08 | 3.94 | 3.99E-05 | 0.99 | CPNE8 | intron |
| 3 | rs11708149 | 32296603 | 0.49 | 0.32 | 2.58 | 4.01E-05 | 0.92 | CMTM8 | intron |
| 12 | rs12424244 | 37501919 | 0.19 | 0.08 | 0.25 | 4.11E-05 | 0.99 | CPNE8 | intron |
| 1 | rs10108 | 114317853 | 0.29 | 0.17 | 0.34 | 4.16E-05 | 0.92 | HIPK1 | UTR-3 |
| 1 | rs11102709 | 114313606 | 0.29 | 0.17 | 2.96 | 4.25E-05 | 0.92 | HIPK1 | intron |
| 20 | rs6074191 | 1084156 | 0.31 | 0.47 | 0.41 | 4.31E-05 | 0.91 | PSMF1 | intron |
| 19 | rs11084710 | 38344196 | 0.48 | 0.30 | 2.43 | 4.50E-05 | 0.98 | WDR88 | intron |
| 20 | rs2300185 | 1083413 | 0.34 | 0.49 | 0.42 | 4.51E-05 | 0.94 | PSMF1 | intron |
| 20 | rs4813945 | 1084656 | 0.34 | 0.49 | 0.42 | 4.51E-05 | 0.95 | PSMF1 | intron |
| 20 | rs6134051 | 1085204 | 0.34 | 0.50 | 0.43 | 4.55E-05 | 0.97 | PSMF1 | intron |
OR = odds ratio
Meta-analysis
Meta-analysis of the 106 nominally significant SNPs in the disease-associated homozygous regions was performed in four independent GWAS datasets (Table 1). Three SNPs in NOTCH4 were nominally associated with AD in both Wadi Ara and the meta-analysis. The A allele of an intronic SNP in NOTCH4, rs3132946, was associated with increased risk of AD in each of the four datasets (ORmeta = 1.14, Pmeta = 0.02). The T allele of rs1044506, a synonymous coding SNP also in NOTCH4, was also associated with greater risk of AD in each of the four datasets (ORmeta = 1.13, Pmeta = 0.04), as was the A allele of rs3131294 (ORmeta = 1.14, Pmeta = 0.03). A fourth SNP in the same region of chromosome 6 as NOTCH4, rs3130286, in an intron of TNXB was also replicated in the meta-analysis (ORmeta= 1.12, Pmeta = 0.03), as was intergenic SNP rs13271711 on chromosome 8 (ORmeta = 1.13, Pmeta = 0.04). Finally, the A allele of rs3130283, which is 50 kb upstream from rs3132946 in an intron of AGPAT1, was also associated with increased AD risk in each dataset, although it narrowly missed the cutoff for nominal significance (ORmeta = 1.13, Pmeta = 0.06). In contrast, none of the most significant associations from the whole genome analysis were replicated in the meta-analysis.
The very low frequency of the APOE ε4 allele and its lack of association with AD in Wadi Ara [34, 35] afforded a unique opportunity to evaluate association of AD with TOMM40 and PVRL2, genes near APOE which have been purported to harbor AD susceptibility alleles whose effects are independent from that of APOE [35, 36]. Alternatively, it has been argued that the observed association with these loci is due to the high correlation of TOMM40 SNPs with APOE ε4, or an extended cis regulatory region affecting APOE expression [36]. In our meta-analysis of other Caucasian data sets, variants in TOMM40 (rs2075650, ORmeta = 0.43, Pmeta = 7.4 × 10−52) and PVRL2 (rs6857, ORmeta = 3.49, Pmeta = 2.6 × 10−44) yielded the two smallest P-values genome wide. Given these effect sizes and the allele frequencies of these SNPs in Wadi Ara (MAF ~ 5%), we have greater than 80% power to detect association with TOMM40 and PVRL2 at P < 0.05. However, we did not observe association with these or any other TOMM40 or PVRL2 SNPs in the Wadi Ara dataset. In fact, the pattern of association with rs2075650 was opposite that in the other datasets.
Discussion
This work represents, to our knowledge, the first GWAS of a complex trait that capitalized on the structure of a consanguineous human population to enhance gene discovery. Our analysis of an elderly cohort in Wadi Ara using genome-wide SNP coverage allowed us to examine formally the population genetic architecture and individual genetic variants that might explain the high rates of AD in this community. We confirmed that there is significant consanguinity among study participants, and found evidence of multiple ancestries likely due to a combination of immigration and heterogeneity in the founder population. The degree of heterogeneity does not appear to be associated with AD risk.
A very surprising observation was that the level of homozygosity across the genome is higher in controls than in AD cases. Comparisons of specific stretches of homozygosity showed the same pattern—every homozygous region whose frequency differed significantly between cases and controls was more common in controls. Single SNP association tests confined to the disease associated homozygous regions showed two nominally significant results near AD related genes that were replicated at P < 0.05 in other GWAS datasets. The fact that these associations are observed in multiple populations, albeit with very modest effect sizes, indicates that they are neither unique to Wadi Ara nor sufficient to explain the high rate of AD in this community. The seven SNPs which are among the most significant in Wadi Ara but only nominally significant in the outbred Caucasian populations had much stronger effects on AD risk in Wadi Ara than in the other datasets. Arguably, these SNPs with larger effect sizes are better candidates than the SNPs in the autozygous regions to explain the AD prevalence in Wadi Ara. However, statistical support is greater for the SNPs emerging from the autozygosity mapping approach than the GWAS after taking into account correction for multiple testing.
We hypothesized that loci associated with AD in this study might act recessively since the SNPs analyzed initially were selected based on their location in disease-associated homozygous regions. However, recessive models for these SNPs were not significant. One explanation for this finding and the increased level of autozygosity in controls is that there are recessively acting protective variants. However, this idea is inconsistent with the higher frequency of AD in Wadi Ara than in other Caucasian populations. Alternatively, the frequency of the risk alleles may have been augmented in this population by inbreeding but the risk model is additive comparable to other AD risk loci [8, 9, 11, 37]. Comparing the allele frequencies of the associated SNPs in homozygous regions in Wadi Ara across datasets revealed the opposite pattern—the frequencies of the minor alleles that increased AD risk were invariably lower in Wadi Ara, and especially so in controls, which led to the AD associations. This observation is consistent with the idea that AD susceptibility alleles were introduced to Wadi Ara through intermarriage. Although the overall frequency of these variants is lower in Wadi Ara compared to other populations, the pattern of association is the same in large outbred Caucasian populations and Wadi Ara. Moreover, since the frequency of these risk alleles are similar among AD cases in Wadi Ara and the other Caucasian populations, the relative risk ascribed to these variants is higher in Wadi Ara which made the detection of associations with these loci easier in this cohort. By comparison, the AD-associated SNPs in Wadi Ara that were not in homozygous regions showed no discernable pattern when their allele frequencies were compared to the other datasets. Table 5 shows the MAF across datasets for the most strongly AD-associated SNP in AGER, RNF5, AGPAT1, NOTCH4, CPN8, FIG4, PSMF1, and ASTN2.
Table 5.
Comparison of minor allele frequencies of AD associated SNPs across datasets.
| TGEN | CAP MIAMI | ADNI | MIRAGE | Wadi Ara | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Case | Control | Case | Control | Case | Control | Case | Control | Case | Control |
| rs1800684* | AGER | 0.15 | 0.14 | 0.16 | 0.14 | 0.11 | 0.10 | NA | NA | 0.09 | 0.03 |
| rs3130283* | AGPAT1 | 0.16 | 0.14 | 0.16 | 0.15 | 0.15 | 0.13 | 0.14 | 0.12 | 0.11 | 0.06 |
| rs3132946* | NOTCH4 | 0.15 | 0.15 | 0.16 | 0.14 | 0.14 | 0.12 | 0.12 | 0.11 | 0.06 | 0.02 |
| rs3134943* | RNF5 | 0.16 | 0.15 | 0.16 | 0.15 | 0.14 | 0.13 | 0.14 | 0.13 | 0.10 | 0.04 |
| rs2025148± | FIG4 | 0.39 | 0.37 | 0.39 | 0.38 | 0.39 | 0.41 | 0.41 | 0.41 | 0.43 | 0.27 |
| rs2072965± | PSMF1 | 0.37 | 0.35 | 0.38 | 0.36 | 0.34 | 0.32 | 0.34 | 0.37 | 0.30 | 0.46 |
| rs7028544± | ASTN2 | 0.39 | 0.37 | 0.38 | 0.38 | 0.39 | 0.38 | 0.40 | 0.38 | 0.26 | 0.44 |
| rs7308580± | CPN8 | 0.06 | 0.05 | 0.06 | 0.05 | 0.06 | 0.05 | NA | NA | 0.15 | 0.05 |
| rs2300186± | PSMF1 | 0.36 | 0.35 | 0.38 | 0.36 | 0.35 | 0.32 | NA | NA | 0.33 | 0.48 |
SNPS were identified through autozygosity mapping
SNPs were among the top 30 in Wadi Ara not associated with AD in meta-analysis
SNPs in bold showed evidence for association with AD in Wadi Ara and meta-analysis
Two regions identified by autozygosity mapping (Table 2) contain at least one strong biological candidate gene for AD, and the chromosome 6 region contains at least three. APBA1 is located in the AD-associated homozygosity stretch on chromosome 9. APBA1 encodes X11α, a member of the X11 family of adaptor proteins which inhibit Aβ production [38]. One study found nominal evidence for association with APBA1 SNP rs1411318 [39]. Unfortunately, genotype data were not available in the Wadi Ara GWAS dataset for any APBA1 SNPs. At present, it is unclear whether the association findings with SNPs in MAMDC2 and TRMP3 are due to LD of these genes with APBA1, functional variants within one of those genes, or chance.
Among the top results from the autozygosity mapping analyses, the gene with the most obvious biological link to AD is AGER which encodes a receptor for the Aβ peptide whose expression is greatly increased in AD patients, especially in neurons proximal to deposits of Aβ peptide and neurofibrillary tangles [40]. AGER may contribute to AD pathology by transporting Aβ40 and Aβ42 across the blood-brain barrier, and increases the expression of proinflammatory cytokines and endothelin-1, which promotes vasoconstriction [41]. Furthermore, inhibiting the binding between AGER and Aβ suppresses Aβ accumulation in the mouse brain [41]. The only SNPs identified in this manner that replicated across datasets are in AGPAT1 and NOTCH4. AGPAT1 is an endoplasmic reticulum transmembrane protein which catalyzes the conversion of lysophosphatidic acid (LPA) to phosphatidic acid, both of which are involved in cellular signal transduction and in lipid biosynthesis [42]. LPA increases Tau phosphorylation (a commonly observed characteristic in AD brains) during LPA-induced neurite retraction [43]. NOTCH4 is a member of a gene family which is involved in a variety of developmental processes by controlling cell fate decisions. Intramembrane proteolysis of NOTCH4 is regulated by γ-secretase, whose activity is responsible for the final cleavage of the APP releasing Aβ peptide [44]. Several studies have evaluated association of AD with NOTCH4, but the results were equivocal because the sample sizes were small and only a few SNPs in this 33 kb gene were tested [45, 46]. SNPs in TNXB have shown evidence for association with schizophrenia in Japanese and UK populations [47, 48], although the association was not observed in a Chinese cohort [49].
Although none of the associations were replicated in the meta-analysis, several functional candidates were identified through the GWAS approach. The relationship between impaired proteasome function and AD [50–53] makes PSMF1 an attractive candidate gene by impacting disease risk through the intracellular protein degradation pathway. Localized at the nuclear envelope/endoplasmic reticulum membrane, PSMF1 does not inhibit cellular proteasome activity, but instead interferes with the maturation of immunoproteasome precursor complexes [54]. Although the function of CPN8, the gene with the strongest SNP associations in Wadi Ara, has not been well characterized, CPN1 promotes calcium-dependent aggregation of lipid vesicles [55]. The FIG4 gene product forms a regulatory complex with VAC14, controlling the synthesis of phosphatidylinositol 3,5-bisphosphate. Loss of VAC14 function caused neurodegeneration in mice [56], and certain mutations in FIG4 cause neurodegeneration in humans [57]. ASTN2 was identified by its similarity to ASTN1, a neuronal adhesion molecule that mediates the migration of young postmitotic neuroblasts in cortical regions of developing brain [58]. The associations with SNPs in these genes are mostly speculative given the lack of association in the meta-analysis datasets.
Finally, the lack of significant association results for the small number of SNPs typed in the genes surrounding APOE (TOMM40 and PVRL2) are consistent with those regions having a regulatory function on APOE. If these variants in these genes alter the expression levels of non-risk alleles of APOE, the overall disease risk would be unlikely to change. Alternately, the low SNP density or weak linkage disequilibrium (LD) in the region might have prevented us from observing effects of TOMM40 and/or PVRL2. Lastly, APOE might be the true causal variant and the observed effects of SNPs in TOMM40 and PVRL2 might be due the LD with APOE.
Several caveats limit the interpretation of the results in this study. First, the Wadi Ara sample is relatively small compared to other published AD GWAS, a fact reflected in the modest P-values. The size of this sample prohibits detection of genome wide significant associations with variants with small effects, i.e., odds ratios less than 1.2 that were observed in two GWAS each containing more than 10,000 subjects [8, 9]. By comparison, we had 80% power to detect an odds ratio above 3.5 at P ≤ 5 × 10−8 for a SNP with a MAF of 10%. However, our strategy of replicating the top-ranked SNPs from the autozygosity mapping and genome wide approaches in several larger GWAS datasets identified several genes and regions which can be further interrogated in even larger datasets assembled by consortia in the US, Europe and Asia. Second, the lack of data on potential environmental risk factors for AD limited our ability to determine conclusively the cause(s) of the high disease rate in Wadi Ara. Despite these limitations, this work exemplifies the advantages of isolated populations in genetic association studies and shows homozygosity mapping to be a useful analytic tool in studies of these populations.
Supplementary Material
Table 3.
Stretches of homozygosity associated with AD
| Chr | Start (BP) | End (BP) | Size (BP) | Ncase | Ncontrol | OR | P |
|---|---|---|---|---|---|---|---|
| 2 | 62669234 | 62682983 | 13749 | 3 | 12 | 0.27 | 0.05 |
| 3 | 153080000 | 153142464 | 62464 | 1 | 9 | 0.12 | 0.05 |
| 6 | 31747736 | 32300538 | 552802 | 3 | 12 | 0.27 | 0.05 |
| 8 | 49021486 | 49671802 | 650316 | 1 | 10 | 0.11 | 0.03 |
| 9 | 71678237 | 72589570 | 911333 | 1 | 9 | 0.12 | 0.05 |
| 13 | 82166221 | 82729863 | 563642 | 3 | 13 | 0.25 | 0.03 |
| 15 | 86657188 | 86675400 | 18212 | 1 | 11 | 0.10 | 0.03 |
| 15 | 88345490 | 88418543 | 73053 | 0 | 11 | NA | NA |
OR = odds ratio
Acknowledgments
This work was supported in part by NIH grants R01-AG17173, R01-AG09029, and R01-AG025259 and by a grant from an anonymous foundation. We are grateful to Efthymia Melista for expert technical assistance and John Farrell for database management support
References
- 1.Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O'Donovan MC, Williams J, Owen MJ, Kirov G. A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med. Genomics. 2008;1:44. doi: 10.1186/1755-8794-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, Pericak-Vance MA. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am. J. Hum. Genet. 2009;84:35–43. doi: 10.1016/j.ajhg.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, Jiang H, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am. J. Hum. Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD, Ertekin-Taner N, Crook JE, Dickson DW, Petersen RC, Graff-Radford NR, Younkin SG. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat. Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Walker DG, Ravid R, Heward CB, Rogers J, Papassotiropoulos A, Reiman EM, Hardy J, Stephan DA. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J. Clin. Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- 6.Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, Karnoub M, Nowotny P, Tacey K, Catanese J, Sninsky J, Brayne C, Rubinsztein D, Gill M, Lawlor B, Lovestone S, Holmans P, O'Donovan M, Morris JC, Thal L, Goate A, Owen MJ, Williams J. Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Hum. Mol. Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung GY, Johnson J, Kelly DD, Keren R, Kertesz A, King KS, Lovestone S, Loy-English I, Matthews PM, Owen MJ, Plumpton M, Pryse-Phillips W, Prinjha RK, Richardson JC, Saunders A, Slater AJ, St George-Hyslop PH, Stinnett SW, Swartz JE, Taylor RL, Wherrett J, Williams J, Yarnall DP, Gibson RA, Irizarry MC, Middleton LT, Roses AD. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch. Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 8.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O'Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat. Genet. 2009 doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P European Alzheimer's Disease Initiative Investigators. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 10.Avramopoulos D. Genetics of Alzheimer's disease: recent advances. Genome Med. 2009;1:34. doi: 10.1186/gm34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowirrat A, Treves TA, Friedland RP, Korczyn AD. Prevalence of Alzheimer's type dementia in an elderly Arab population. Eur. J. Neurol. 2001;8:119–123. doi: 10.1046/j.1468-1331.2001.00183.x. [DOI] [PubMed] [Google Scholar]
- 13.Bowirrat A, Friedland RP, Chapman J, Korczyn AD. The very high prevalence of AD in an Arab population is not explained by APOE epsilon4 allele frequency. Neurology. 2000;55:731. doi: 10.1212/wnl.55.5.731. [DOI] [PubMed] [Google Scholar]
- 14.Curtis D, Vine AE, Knight J. Study of regions of extended homozygosity provides a powerful method to explore haplotype structure of human populations. Ann. Hum. Genet. 2008;72:261–278. doi: 10.1111/j.1469-1809.2007.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bacolod MD, Schemmann GS, Wang S, Shattock R, Giardina SF, Zeng Z, Shia J, Stengel RF, Gerry N, Hoh J, Kirchhoff T, Gold B, Christman MF, Offit K, Gerald WL, Notterman DA, Ott J, Paty PB, Barany F. The signatures of autozygosity among patients with colorectal cancer. Cancer Res. 2008;68:2610–2621. doi: 10.1158/0008-5472.CAN-07-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Haynes C, Barany F, Ott J. Genome-wide autozygosity mapping in human populations. Genet. Epidemiol. 2009;33:172–180. doi: 10.1002/gepi.20344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum. Mol. Genet. 2003;12:415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- 18.Zuchner S, Gilbert JR, Martin ER, Leon-Guerrero CR, Xu PT, Browning C, Bronson PG, Whitehead P, Schmechel DE, Haines JL, Pericak-Vance MA. Linkage and association study of late-onset Alzheimer disease families linked to 9p21.3. Ann. Hum. Genet. 2008;72:725–731. doi: 10.1111/j.1469-1809.2008.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry RT, Wiener H, Harrell LE, Blacker D, Tanzi RE, Bertram L, Bassett SS, Go RC. Follow-up mapping supports the evidence for linkage in the candidate region at 9q22 in the NIMH Alzheimer's disease Genetics Initiative cohort. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:220–227. doi: 10.1002/ajmg.b.30433. [DOI] [PubMed] [Google Scholar]
- 20.Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 1997;278:1237–1241. [PubMed] [Google Scholar]
- 21.Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze FW, Crook R, Hamshere M, Abraham R, Tunstall N, Rice F, Carty S, Lillystone S, Kehoe P, Rudrasingham V, Jones L, Lovestone S, Perez-Tur J, Williams J, Owen MJ, Hardy J, Goate AM. Susceptibility locus for Alzheimer's disease on chromosome 10. Science. 2000;290:2304–2305. doi: 10.1126/science.290.5500.2304. [DOI] [PubMed] [Google Scholar]
- 22.Rogaeva E, Premkumar S, Song Y, Sorbi S, Brindle N, Paterson A, Duara R, Levesque G, Yu G, Nishimura M, Ikeda M, O'Toole C, Kawarai T, Jorge R, Vilarino D, Bruni AC, Farrer LA, St George-Hyslop PH. Evidence for an Alzheimer disease susceptibility locus on chromosome 12 and for further locus heterogeneity. JAMA. 1998;280:614–618. doi: 10.1001/jama.280.7.614. [DOI] [PubMed] [Google Scholar]
- 23.Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL. Identification of novel genes in late-onset Alzheimer's disease. Exp. Gerontol. 2000;35:1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 24.Meng Y, Baldwin CT, Bowirrat A, Waraska K, Inzelberg R, Friedland RP, Farrer LA. Association of polymorphisms in the Angiotensin-converting enzyme gene with Alzheimer disease in an Israeli Arab community. Am. J. Hum. Genet. 2006;78:871–877. doi: 10.1086/503687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 26.Reitz C, Rogaeva E, Lee J, Tokuhiro S, Bettens K, Sleegers K, Tan E, Kimura R, Shibata N, Kamboh I, Prince J, Maier W, Riemenschneider M, Williams J, van Broeckhoven C, Farrer L, St.George-Hyslop P, Mayeux R. Association of SORL1 gene variants with Alzheimer's disease: A meta-analysis. Arch. Neurol. 2010 (in press) [Google Scholar]
- 27.Inzelberg R, Schechtman E, Abuful A, Masarwa M, Mazarib A, Strugatsky R, Farrer LA, Green RC, Friedland RP. Education effects on cognitive function in a healthy aged Arab population. Int. Psychogeriatr. 2007;19:593–603. doi: 10.1017/S1041610206004327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Israeli-Korn SD, Masarwa M, Schechtman E, Abuful A, Strugatsky R, Avni S, Farrer LA, Friedland RP, Inzelberg R. Hypertension Increases the Probability of Alzheimer's Disease and of Mild Cognitive Impairment in an Arab Community in Northern Israel. Neuroepidemiology. 2010;34:99–105. doi: 10.1159/000264828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavalli-Sforza LL, Bodmer WF. The Genetics of Human Populations. Hoboken: Wiley; 1971. [Google Scholar]
- 30.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Abecasis G. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am. J. Hum. Genet. 2006;S79:2290. [Google Scholar]
- 34.Roses AD, Lutz MW, Amrine-Madsen H, Saunders AM, Crenshaw DG, Sundseth SS, Huentelman MJ, Welsh-Bohmer KA, Reiman EM. A TOMM40 variable-length polymorphism predicts the age of late-onset Alzheimer's disease. Pharmacogenomics J. 2009 doi: 10.1038/tpj.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takei N, Miyashita A, Tsukie T, Arai H, Asada T, Imagawa M, Shoji M, Higuchi S, Urakami K, Kimura H, Kakita A, Takahashi H, Tsuji S, Kanazawa I, Ihara Y, Odani S, Kuwano R Japanese Genetic Study Consortium for Alzheimer Disease. Genetic association study on in and around the APOE in late-onset Alzheimer disease in Japanese. Genomics. 2009;93:441–448. doi: 10.1016/j.ygeno.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Yu CE, Seltman H, Peskind ER, Galloway N, Zhou PX, Rosenthal E, Wijsman EM, Tsuang DW, Devlin B, Schellenberg GD. Comprehensive analysis of APOE and selected proximate markers for late-onset Alzheimer's disease: patterns of linkage disequilibrium and disease/marker association. Genomics. 2007;89:655–665. doi: 10.1016/j.ygeno.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 38.Miller CC, McLoughlin DM, Lau KF, Tennant ME, Rogelj B. The X11 proteins, Abeta production and Alzheimer's disease. Trends Neurosci. 2006;29:280–285. doi: 10.1016/j.tins.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, Ramasamy K, Mullin K, Menon R, Sampson AJ, Hsiao MY, Elliott KJ, Velicelebi G, Moscarillo T, Hyman BT, Wagner SL, Becker KD, Blacker D, Tanzi RE. Family-based association between Alzheimer's disease and variants in UBQLN1. N. Engl. J. Med. 2005;352:884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 40.Yan SD, Chen X, Fu J, Chen M, Zhu H, Roher A, Slattery T, Zhao L, Nagashima M, Morser J, Migheli A, Nawroth P, Stern D, Schmidt AM. RAGE and amyloid-beta peptide neurotoxicity in Alzheimer's disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- 41.Deane R, Du Yan S, Submamaryan RK, LaRue B, Jovanovic S, Hogg E, Welch D, Manness L, Lin C, Yu J, Zhu H, Ghiso J, Frangione B, Stern A, Schmidt AM, Armstrong DL, Arnold B, Liliensiek B, Nawroth P, Hofman F, Kindy M, Stern D, Zlokovic B. RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nat. Med. 2003;9:907–913. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- 42.Aguado B, Campbell RD. Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J. Biol. Chem. 1998;273:4096–4105. doi: 10.1074/jbc.273.7.4096. [DOI] [PubMed] [Google Scholar]
- 43.Sayas CL, Moreno-Flores MT, Avila J, Wandosell F. The neurite retraction induced by lysophosphatidic acid increases Alzheimer's disease-like Tau phosphorylation. J. Biol. Chem. 1999;274:37046–37052. doi: 10.1074/jbc.274.52.37046. [DOI] [PubMed] [Google Scholar]
- 44.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 45.Lambert JC, Mann D, Harris J, Araria-Goumidi L, Chartier-Harlin MC, Cottel D, Iwatsubo T, Amouyel P, Lendon C. Association study of Notch 4 polymorphisms with Alzheimer's disease. J. Neurol. Neurosurg. Psychiatry. 2004;75:377–381. doi: 10.1136/jnnp.2003.017368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibata N, Ohnuma T, Higashi S, Higashi M, Usui C, Ohkubo T, Watanabe T, Kawashima R, Kitajima A, Ueki A, Nagao M, Arai H. Genetic association between Notch4 polymorphisms and Alzheimer's disease in the Japanese population. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:350–351. doi: 10.1093/gerona/62.4.350. [DOI] [PubMed] [Google Scholar]
- 47.Tochigi M, Zhang X, Ohashi J, Hibino H, Otowa T, Rogers M, Kato T, Okazaki Y, Kato N, Tokunaga K, Sasaki T. Association study between the TNXB locus and schizophrenia in a Japanese population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2007;144B:305–309. doi: 10.1002/ajmg.b.30441. [DOI] [PubMed] [Google Scholar]
- 48.Wei J, Hemmings GP. TNXB locus may be a candidate gene predisposing to schizophrenia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2004;125B:43–49. doi: 10.1002/ajmg.b.20093. [DOI] [PubMed] [Google Scholar]
- 49.Liu LL, Wei J, Zhang X, Li XY, Shen Y, Liu SZ, Ju GZ, Shi JP, Yu YQ, Xu Q, Hemmings GP. Lack of a genetic association between the TNXB locus and schizophrenia in a Chinese population. Neurosci. Lett. 2004;355:149–151. doi: 10.1016/j.neulet.2003.10.059. [DOI] [PubMed] [Google Scholar]
- 50.Thomas SN, Cripps D, Yang AJ. Proteomic analysis of protein phosphorylation and ubiquitination in Alzheimer's disease. Methods Mol. Biol. 2009;566:109–121. doi: 10.1007/978-1-59745-562-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghanevati M, Miller CA. Phospho-beta-catenin accumulation in Alzheimer's disease and in aggresomes attributable to proteasome dysfunction. J. Mol. Neurosci. 2005;25:79–94. doi: 10.1385/JMN:25:1:079. [DOI] [PubMed] [Google Scholar]
- 52.Layfield R, Cavey JR, Lowe J. Role of ubiquitin-mediated proteolysis in the pathogenesis of neurodegenerative disorders. Ageing Res. Rev. 2003;2:343–356. doi: 10.1016/s1568-1637(03)00025-4. [DOI] [PubMed] [Google Scholar]
- 53.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer's disease. J. Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 54.Zaiss DM, Standera S, Kloetzel PM, Sijts AJ. PI31 is a modulator of proteasome formation and antigen processing. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14344–14349. doi: 10.1073/pnas.212257299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maitra R, Grigoryev DN, Bera TK, Pastan IH, Lee B. Cloning, molecular characterization, and expression analysis of Copine 8. Biochem. Biophys. Res. Commun. 2003;303:842–847. doi: 10.1016/s0006-291x(03)00445-5. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, Meisler MH, Weisman LS. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fink JM, Hirsch B, Zheng C, Deitz G, Hatten ME, Ross ME. The CNS neuronal migration gene, astrotactin, is mapped to human chromosome band 1q25 by fluorescence in situ hybridization (FISH) Am. J. Hum. Genet. 1995;5757:A133. [Google Scholar]
- 59.Demissie S, Green RC, Mucci L, Tziavas S, Martelli K, Bang K, Coons L, Bourque S, Buchillon D, Johnson K, Smith T, Sharrow N, Lautenschlager N, Friedland R, Cupples LA, Farrer LA. Reliability of information collected by proxy in family studies of Alzheimer's disease. Neuroepidemiology. 2001;20:105–111. doi: 10.1159/000054768. [DOI] [PubMed] [Google Scholar]
- 60.Farrer LA, Cupples LA, Blackburn S, Kiely DK, Auerbach S, Growdon JH, Connor-Lacke L, Karlinsky H, Thibert A, Burke JR. Interrater agreement for diagnosis of Alzheimer's disease: the MIRAGE study. Neurology. 1994;44:652–656. doi: 10.1212/wnl.44.4.652. [DOI] [PubMed] [Google Scholar]
- 61.Lautenschlager NT, Cupples LA, Rao VS, Auerbach SA, Becker R, Burke J, Chui H, Duara R, Foley EJ, Glatt SL, Green RC, Jones R, Karlinsky H, Kukull WA, Kurz A, Larson EB, Martelli K, Sadovnick AD, Volicer L, Waring SC, Growdon JH, Farrer LA. Risk of dementia among relatives of Alzheimer's disease patients in the MIRAGE study: What is in store for the oldest old? Neurology. 1996;46:641–650. doi: 10.1212/wnl.46.3.641. [DOI] [PubMed] [Google Scholar]
- 62.Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Kukull W, Papassotiropoulos A, Stephan DA. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jack CR, Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, L Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haroutunian V, Perl DP, Purohit DP, Marin D, Khan K, Lantz M, Davis KL, Mohs RC. Regional distribution of neuritic plaques in the nondemented elderly and subjects with very mild Alzheimer disease. Arch. Neurol. 1998;55:1185–1191. doi: 10.1001/archneur.55.9.1185. [DOI] [PubMed] [Google Scholar]
- 65.Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, Hiner BC, Jankovic J, Ondo WG, Allen FH, Jr, Goetz CG, Small GW, Masterman D, Mastaglia F, Laing NG, Stajich JM, Slotterbeck B, Booze MW, Ribble RC, Rampersaud E, West SG, Gibson RA, Middleton LT, Roses AD, Haines JL, Scott BL, Vance JM, Pericak-Vance MA. Complete genomic screen in Parkinson disease: evidence for multiple genes. JAMA. 2001;286:2239–2244. doi: 10.1001/jama.286.18.2239. [DOI] [PubMed] [Google Scholar]
- 66.Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, Wang L, Zuchner S, Konidari I, Wang G, Singer C, Nahab F, Scott B, Stajich JM, Pericak-Vance M, Haines J, Vance JM, Martin ER. Genome-Wide Association Study Confirms SNPs in SNCA and the MAPT Region as Common Risk Factors for Parkinson Disease. Ann. Hum. Genet. 2010 doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.