Abstract
Nfu-type proteins are essential in the biogenesis of iron-sulfur (Fe-S) clusters in numerous organisms. A number of phenotypes including low levels of Fe-S cluster incorporation are associated with deletion of the gene encoding a chloroplast-specific Nfu-type protein, Nfu2 from Arabidopsis thaliana (AtNfu2). Here we report that recombinant AtNfu2 is able to assemble both [2Fe-2S] and [4Fe-4S] clusters. Analytical data and gel filtration studies support cluster/protein stoichiometries of one [2Fe-2S] cluster/homotetramer and one [4Fe-4S] cluster/homodimer. The combination of UV-visible absorption and circular dichroism, resonance Raman and Mössbauer spectroscopies has been employed to investigate the nature, properties and transfer of the clusters assembled on Nfu2. The results are consistent with subunit-bridging [2Fe-2S]2+ and [4Fe-4S]2+ clusters coordinated by the cysteines in the conserved CXXC motif. The results also provided insight into the specificity of Nfu2 for maturation of chloroplastic Fe-S proteins via intact, rapid and quantitative cluster transfer. [2Fe-2S] cluster-bound Nfu2 is shown to be an effective [2Fe-2S]2+ cluster donor for glutaredoxin S16, but not glutaredoxin S14. Moreover, [4Fe-4S] cluster-bound Nfu2 is shown to be a very rapid and efficient [4Fe-4S]2+ cluster donor for adenosine 5′-phosphosulfate reductase (APR1) and yeast two-hybrid studies indicate that APR1 forms a complex with Nfu2, but not with Nfu1 and Nfu3, the two other chloroplastic Nfu proteins. This cluster transfer is likely to be physiologically relevant and is particularly significant for plant metabolism as APR1 catalyzes the second step in reductive sulfur assimilation which ultimately results in the biosynthesis of cysteine, methionine, glutathione, and Fe-S clusters.
Iron-sulfur (Fe-S) centers constitute a ubiquitous class of protein prosthetic groups involving clusters of non-heme iron and bridging sulfides that are attached to the protein primarily by cysteinyl ligands. The resulting Fe-S proteins constitute one of the largest classes of proteins with versatile functions including electron transport, substrate binding and activation, regulation of gene expression and enzyme activity in response to external stimuli, and radical generation (1). In vivo maturation of Fe-S proteins involves delivery of sulfur (via a cysteine desulfurase) and iron to scaffold proteins, cluster assembly on the scaffold proteins, and transfer to the acceptor proteins either directly or via intermediate carrier or storage proteins (2–8).
The Nfu (aka NifU-like) class of proteins constitute a ubiquitous class of intermediate carrier proteins that appear to function in delivering clusters assembly on primary scaffold proteins to specific target proteins (9–11). They were first identified as the C-terminal domain of NifU which constitutes the primary scaffold protein for the maturation of proteins and enzymes associated with nitrogen fixation in nitrogen-fixing bacteria (12–14). As shown in Figure 1, NifU is a modular protein that is composed of three functional domains (13;15;16). In addition to the central ferredoxin (Fdx) domain which constitutively binds a redox-active [2Fe-2S]2+,+ cluster, NifU also comprises an N-terminal IscU domain and a C-terminal Nfu domain, both of which have been shown to be able to assemble [4Fe-4S] clusters and subsequently activate apo nitrogenase Fe-protein by direct cluster transfer (17;18).
Figure 1.
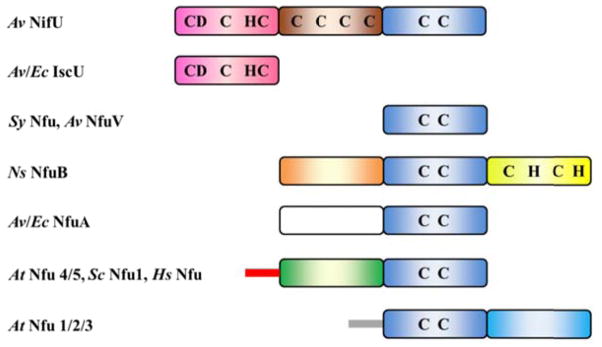
Schematic representations of the domain structures of IscU, NifU and Nfu-type proteins. Known or potential Fe-S cluster binding residues are labeled for each domain when applicable. Pink boxes, IscU domains; blue boxes, Nfu domains; brown box, Fdx domain specific for NifU; orange and yellow boxes, unique N-terminal extension and C-terminal Rieske domain specifically from nitrogen-fixing cyanobacterial NfuB; white box, IscA domain lacking the active site cysteine residues that is specific to bacterial-type NfuA; green box, the N-terminal domain that is unique to mitochondrial, nuclear, or cytosolic types of eukaryotic Nfu; cyan box, Nfu domain lacking the active site CXXC motif specifically from chloroplastic type Nfu; red and grey bars, mitochondrial and chloroplastic targeting sequences. Av, Azotobacter vinelandii; Ec, Escherichia coli; Sy, Synechocystis; Ns, Nodularia spumigena; At, Arabidopsis thaliana; Sc, Saccharomyces cerevisiae; Hs, Homo sapiens.
Orthologs of IscU and Nfu proteins were subsequently identified as separate proteins or domains in a wide range of prokaryotic and eukaryotic organisms (11;15;19;20). IscU was shown to be the primary Fe-S cluster scaffold associated with the ISC machinery for Fe-S cluster assembly that is responsible for cluster assembly in most bacteria and in eukaryotic mitochondria (2;6). Except for NifU, all other IscU-type proteins are single domain proteins. In contrast, Nfu-type proteins are known for having diverse domain structures, which have been proposed to be relevant to their functional specificity (11;21). Figure 1 shows the domain structures of several distinct types of Nfu proteins. Despite the differences in additional domain(s), all Nfu-type proteins share sequence homology in the Nfu-domain, along with the rigorously conserved CXXC motif. Cyanobacteria and many other bacteria encode the simplest form of Nfu-type protein that comprises only the Nfu-domain (22). Another common type of bacterial Nfu protein, namely NfuA, contains an N-terminal domain that is similar in sequence to the A-type class of Fe-S cluster biogenesis protein, but lacks the active site cysteine residues (9;23). Some nitrogen-fixing cyanobacteria such as Nodularia spumigena encode an NfuB protein whose gene is usually clustered with Ni-Fe hydrogenase structural genes. NfuB contains an extra N-terminal domain of unknown function and a C-terminal Rieske domain (24). In humans, alternative splicing of pre-mRNA results in the translation of eukaryotic mitochondrial, cytosolic and nuclear Nfu proteins, all of which comprise a unique N-terminal domain (25). Chloroplastic-type Nfu proteins comprise an extra C-terminal domain, which almost repeats the sequence of the Nfu domain, but lacks the active site cysteine residues (26). Recently a comprehensive phylogenomic classification of Nfu proteins into four subfamilies has been published (11).
A handful of Fe-S cluster-bound forms of Nfu-type proteins have been spectroscopically and analytically characterized. Recombinant AvNfuA and EcNfuA, Synechococcus sp. PCC 7002 Nfu, and human mitochondrial Nfu are purified under aerobic and anaerobic conditions as apo proteins devoid of any significant amount of bound Fe-S clusters. However, each has been shown to assemble up to one [4Fe-4S] cluster/dimer after cysteine desulfurase-mediated reconstitution (9;23–25). Cysteine mutagenesis, oligomeric state studies, and analytical data for NfuA indicate that the [4Fe-4S] cluster is coordinated by the cysteines in the CXXC motif at the subunit interface of the homodimer. In addition, [4Fe-4S] cluster-loaded NfuA was shown to be capable of activating aconitase by cluster transfer at a physiologically relevant rate (9) and [4Fe-4S] cluster-loaded Synechococcus Nfu was shown to rapidly and efficiently transfer clusters to apo PsaC in Photosystem I (PSI) (24). Based on the available evidence, AvNfuA was proposed to be a cluster carrier for clusters assembled on IscU, that is required for the maturation of [4Fe-4S] clusters under oxidative stress conditions (9). This is supported by recent preliminary cluster transfer studies involving EcNfuA, monitored by UV-visible absorption spectroscopy and analytical data, which suggest that NfuA can accept clusters from IscU and SufBCD, the primary scaffolds in the E. coli ISC and SUF systems for Fe-S cluster assembly, respectively, and transfer them to IscA or SufA (11). In vivo studies also indicate a cluster carrier role for Nfu1 in yeast mitochondria for the delivery of Fe-S clusters assembled on Isu1 to specific acceptor proteins such as lipoate synthase, which contains two [4Fe-4S] clusters, and succinate dehydrogenase, which contains [2Fe-2S], [3Fe-4S] and [4Fe-4S] clusters(10).
On the other hand, recombinant cyanobacterial Nfu from Synechocystis PCC 6803 and the chloroplastic Nfu2 (aka CNfu-V) from A. thaliana (AtNfu2) were purified under anaerobic conditions with substoichiometric [2Fe-2S] clusters that can be transferred to the apo form of the chloroplast [2Fe-2S] Fdx (22;27). Moreover, the crystal structure of apo AtNfu2 has revealed a novel homodimeric structure with an extensive dimer interface and the two subunits aligned anti-parallel to each other (28). The CXXC motifs from both subunits point to the center of the molecule and form inter-subunit disulfide bonds. Taken together with Fe-EXAFS data of the [2Fe-2S] cluster-bound form (28), which are best interpreted in terms of four cysteinyl ligands, the results indicate a subunit–bridging [2Fe-2S] cluster with each Fe ligated by CXXC cysteines from different subunits. A. thaliana has five Nfu proteins (26): Nfu1, Nfu2 and Nfu3 are located in the chloroplasts and Nfu4 and Nfu5 are located in mitochondria. Extensive studies on Nfu2 gene knockout strains of Arabidopsis have revealed phenotypes including retarded growth, pale green leaves, poorly developed roots and root hairs, decreased chlorophyll level, and low levels of the chloroplastic [2Fe-2S] ferredoxin and the [4Fe-4S] cluster subunits in photosystem I due to post-transcriptional defects (27;29). However, defects in Rieske [2Fe-2S] proteins and some [3Fe-4S] cluster-containing proteins were not observed. These results leave open the question of how AtNfu2 is involved in the maturation of both [2Fe-2S] and [4Fe-4S] cluster-containing proteins in plant chloroplasts.
In this work, we report in vitro studies designed to address the nature and function of the Fe-S cluster-bound forms of AtNfu2. Spectroscopic and analytical results show that AtNfu2 can accommodate either one [2Fe-2S] cluster per tetramer or one [4Fe-4S] cluster per dimer, with the type of cluster formed via cysteine desulfurase-mediated reconstitution depending on the reaction conditions. Thus far, cluster transfer reactions from cluster-loaded AtNfu2 to appropriate acceptor proteins have been limited to using apo chloroplast [2Fe-2S] Fdx as the acceptor using semi-quantitative methods. The results reported herein provided new insights into the specificity of AtNfu2 for the maturation of chloroplastic acceptor proteins by using CD spectroscopy to monitor the rates of intact cluster transfer to specific [2Fe-2S] and [4Fe-4S] cluster acceptor proteins and yeast-two hybrid studies to identify interprotein interactions.
MATERIALS AND METHODS
Materials
All the chemicals used in this work were purchased from Fisher Scientific International Inc. or Sigma-Aldrich Chemical Co. unless otherwise specified. Chromatography instruments and columns were purchased from GE Healthcare. Anaerobic operations were performed in a glovebox (Vacuum Atmospheres) under an argon atmosphere with oxygen levels less than 2 ppm.
Plasmid Construction
The A. thaliana Nfu2 (At5g49940) coding sequence was amplified by PCR from rosette cDNAs using the following primers: AtNfu2for (5′CCCCCCATGGCTGTAGCAACCCCAGACCCC3′) and AtNfu2rev (5′CCCCGGATCCCTATATAAGTTGAACAGC3′). The PCR product was digested with NcoI and BamHI and cloned into the pET3d vector. The amplified sequence encodes a protein deprived of the first 72 amino acids corresponding to the putative plastidial targeting sequence, and thus started with MAVATPDP.
Protein Overexpression and Purification
AtNfu2 was heterologously expressed in E. coli C41 [DE3] (Invitrogen) transformed with the constructed plasmid. One colony was grown overnight at 37°C in 100 mL LB media containing 100 μg/mL ampicillin, and 15 mL of the overnight grown culture was used to inoculate 1 L of the same media. The culture was incubated at 37 °C until reaching exponential growth phase, then isopropyl 1-thio-β-D-galactopyranoside (IPTG) was added to a final concentration of 200 μg/mL in order to induce protein expression. The cells were allowed to grow for an additional 4 hours at 37 °C before harvesting by centrifugation at 6690 × g and storage at −80 °C for later use.
For purification of AtNfu2, 20 g of cell paste was resuspended in 60 mL of 100 mM Tris-HCl, pH 7.5 (buffer A) containing 150 μg/mL PMSF, 2 mU/mL DNase (Roche) and 0.5 μg/mL RNase (Roche), and lysed by intermittent sonication. Cell debris was removed by centrifugation at 39800 × g at 4 °C for 1 hr. The supernatant was subjected to 40% ammonium sulfate cut and the precipitate was removed by centrifugation at 39800 × g for another 20 minutes. The supernatant containing AtNfu2 was loaded onto an 80 mL Phenyl Sepharose hydrophobic interaction column equilibrated with buffer A containing 1.0 M ammonium sulfate, and eluted by a gradient of 1.0 to 0 M of ammonium sulfate. Fractions containing Nfu2, determined by SDS-PAGE, were pooled and concentrated using Amicon ultrafiltration equipped with a 10 kDa membrane. The concentrated fraction was then loaded onto a 25 mL Q Sepharose anion exchange column and eluted with 0 to 1.0 M gradient of sodium chloride. A 300 mL Superdex S75 size exclusion column was subsequently used for further purification if needed. All as-isolated samples used for spectroscopic studies were prepared anaerobically. Apo AtNfu2 was prepared by incubating as isolated samples with 50-fold excess of EDTA and 20-fold excess of potassium ferricyanide for 30 minutes, followed by purification anaerobically with desalting columns to remove residual iron and sulfide. The as-isolated samples were >90% pure for spectroscopic studies or in vitro reconstitution, or >99% pure for amino acid analysis, as estimated by SDS-PAGE.
His-tagged At adenosine 5′-phosphosulfate reductase 1 (AtAPR1) was expressed and purified as a [4Fe-4S] cluster-containing protein using the following procedure. A culture of E. coli strain TL3 cells harboring the plasmid for his-tagged pBAD18-APR1 (30) was grown in LB medium with 35 μg/mL kanamycin, 34 μg/mL chloramphenicol, and 100 μg/mL ampicillin at 30 °C to an optical density at 600 nm of 0.6. To induce the protein expression, 0.2% (w/v) arabinose was added, and growth continued overnight at 30 °C. Cells were then harvested by centrifugation, resuspended in 30 mM Tris-HCl buffer (pH 8.0) containing 200 mM NaCl, passed three times through a French Press at 18,000 psi and then ultracentrifuged. The supernatant was filtered through a 0.45 μm pore-size membrane and applied to a Ni2+ affinity column (HiTrap Chelating HP, obtained from GE Healthcare) incorporated into a BioLogic LP chromatography system (BioRad). The column was washed with 200 mL of 30 mM Tris-HCl buffer (pH 8.0) containing 500 mM NaCl that was supplemented with 20 mM imidazole. The column was then washed with a linear 20 to 250 mM imidazole gradient in buffer B and the His-tagged recombinant protein eluted at an imidazole concentration of approximately 100 mM. All solutions used for the Ni2+ affinity chromatography were degassed and purged with nitrogen gas. The imidazole was removed by buffer exchange against 30 mM Tris-HCl buffer (pH 8.0) using an Amicon YM10 membrane. The purity of the preparations was estimated to be greater than 95% based on a single Coomassie Blue-stained band after SDS-PAGE. Apo AtAPR1 was prepared by incubating the protein with 50-fold excess EDTA and 50-fold excess of potassium ferricyanide, followed by repeated dialysis against with buffer A containing 100 mM NaCl and 10% glycerol, using a Slide-A-Lyzer dialysis cassette with a 10 kDa MW membrane. The apo protein was treated with 2 mM dithiothreitol (DTT) and repurified by anaerobic gel filtration to ensure cleavage of disulfides prior to use in cluster transfer reactions.
AvIscS, AtGrxS16 and AtGrxS14 were heterologously expressed in E. coli and purified to homogeneity according to previously published procedures (19;31). The samples used in this work were all >95% pure based on SDS-PAGE. Apo forms of AtGrxS16 and AtGrxS14 were generated by purification under aerobic conditions and were treated with 2 mM DTT and repurified by anaerobic gel filtration to ensure cleavage of disulfides prior to use in cluster transfer reactions.
IscS-Mediated In Vitro Reconstitution
Reconstitution of clusters on apo AtNfu2 was conducted by incubating 0.1–0.6 mM of apo Nfu2 with 8–12 fold excess of ferrous ammonium sulfate (FAS), 10–16 fold excess of L-cysteine and catalytic amount of IscS under strictly anaerobic environment for approximately 1 hour. The cluster-bound AtNfu2 was purified by loading onto 3 in-line assembled 5 mL HiTrap Q Sepharose HP columns and eluting with 0 to 1.0 M gradient of NaCl. Fractions containing [4Fe-4S] and [2Fe-2S] clusters-bound AtNfu2 were collected at 0.40 to 0.50 M, and 0.55 to 0.65 M of NaCl, respectively. 57Fe-enriched FAS (>95% enrichment) was used in place of the natural abundance FAS for Mössbauer samples.
Analytical and Spectroscopic Methods
Approximate protein concentrations were determined by DC protein assay (Bio-rad), using bovine serum albumin as a standard, as described by Brown (32). The results of the DC protein assay were calibrated by quantitative amino acid analysis carried out by the Protein Chemistry Laboratory, Dept. Biochemistry & Biophysics, Texas A&M University (College Station, TX). A correction factor of 1.33 (concentration determined by amino acid analysis divided by those determined by DC protein assay) was applied to all subsequent DC protein concentration determinations. Iron concentrations were determined colorimetrically with bathophenanthroline under reducing conditions, after digesting proteins with KMnO4/HCl, as described by Fish (33). A series of dilutions of a 1000 ppm atomic absorption iron standard were used to construct a standard curve. All samples used for spectroscopic studies were prepared under strictly anaerobic conditions. UV-visible absorption spectra were recorded with septum sealed quartz cuvettes of 1 mm or 1 cm path length at room temperature, using a Shimadzu UV-3101 PC scanning spectrophotometer. Circular dichroism (CD) spectra were recorded with the same cuvettes using a JASCO J-715 spectropolarimeter (Jasco, Easton, MD). For low temperature resonance Raman (RR) spectra, samples were concentrated to ~2 mM in [2Fe-2S] or [4Fe-4S] clusters and frozen as droplets on an O-ring-sealed gold-plated copper sample holder (34) mounted to the cold finger of a Displex Model CSA-202E closed cycle refrigerator (Air Products, Allentown, PA) at 17 K. The RR spectra were acquired using a Ramanor U1000 spectrometer (Instruments SA, Edison, NJ), coupled with a Sabre argon laser (Coherent, Santa Clara, CA). The spectra were recorded by photon counting at 0.5 cm−1 increments with 7 cm−1 resolution, and each spectrum was a sum of 80 – 120 scans. Mössbauer spectra were recorded on a weak-field Mössbauer spectrometer equipped with a Janis 8DT variable temperature cryostat, operated in a constant acceleration mode in transmission geometry. The data was analyzed with the WMOSS program (Web Research). The zero velocity refers to the centroid of the room temperature spectrum of a metallic iron foil.
Oligomerization State Determination
The oligomerization states of apo and cluster-bound forms of AtNfu2 were determined using an FPLC system equipped with a 25 mL Superdex 75 gel filtration column pre-equilibrated with buffer A containing 100 mM KCl and 1 mM DTT. Elution of protein was monitored by UV absorption at 280 nm. Fractions of distinct oligomerization states were collected manually, and analyzed by absorption and CD spectroscopies to determine the cluster content. Molecular markers employed included blue dextran (Mr 2,000 kDa), alcohol dehydrogenase (Mr 150 kDa), bovine serum albumin (Mr 66 kDa), carbonic anhydrase (Mr 29 kDa), cytochrome C (Mr 12.4 kDa), and aprotinin (Mr 6.5 kDa). The elution profiles and calibration plot derived from the molecular markers are shown in the supporting information (Figure S1)
Cluster Transfer from [2Fe-2S] Cluster-Bound AtNfu2 to Apo AtGrxS16 and AtGrxS14
For cluster transfer from Nfu2 to GrxS16, reconstituted Nfu2 (39 μM in [2Fe-2S] clusters) was incubated with 162 μM apo GrxS16 monomer in buffer A containing 1 mM glutathione (GSH) for 70 min at room temperature. The reaction was initiated by the addition of apo GrxS16 and the time course of cluster transfer was monitored by CD spectroscopy at room temperature using a septum-sealed quartz cuvette with a 1 cm path length. A similar procedure was used for cluster transfer from [2Fe-2S]-Nfu2 to GrxS14 except that 150 μM apo GrxS14 monomer was used in the reaction.
Cluster Transfer from [4Fe-4S] Cluster-Bound AtNfu2 to Apo AtAPR1
10 μM apo APR1 was added to Nfu2 (10 μM in [4Fe-4S] clusters) in buffer A. The reaction was initiated by the addition of apo APR1 and the time course of cluster transfer was monitored by CD spectroscopy at room temperature using a septum-sealed quartz cuvette with a 1 cm path length.
Assessment of Interaction Between AtNfu proteins and AtAPR1 by Yeast Two-Hybrid Studies
The coding sequences for APR1 (At4g04610) and Nfu1, Nfu2, and Nfu3 (At4g01940, At5g49940, At4g25910, respectively) were cloned into pGAD.T7 and pGBK.T7 (Clontech), respectively, in frame with corresponding Gal4 domains, by introducing APR1 and Nfu CDS at NcoI (5′) and BamHI (3′) restriction sites. Constructs were introduced into strain CY306 (35) using the lithium acetate method according to (36). Transformed cells were selected on a minimal YNB medium (0.7% yeast extract w/o amino acids, 2% glucose, 2% agar) with required amino acids (His, Ade, Lys, Ura, Met), and plated as serial dilutions (optical density at 600 nm of 5.10−2 to 5.10−4) on His-containing (control plate) and His-lacking (Y2H interaction plate) YNB media. Interactions were visualized as growing cells on YNB without His, in the presence of 2 mM 3-aminotriazol (3AT).
RESULTS
Nature and Properties of Cluster-bound Forms of AtNfu2
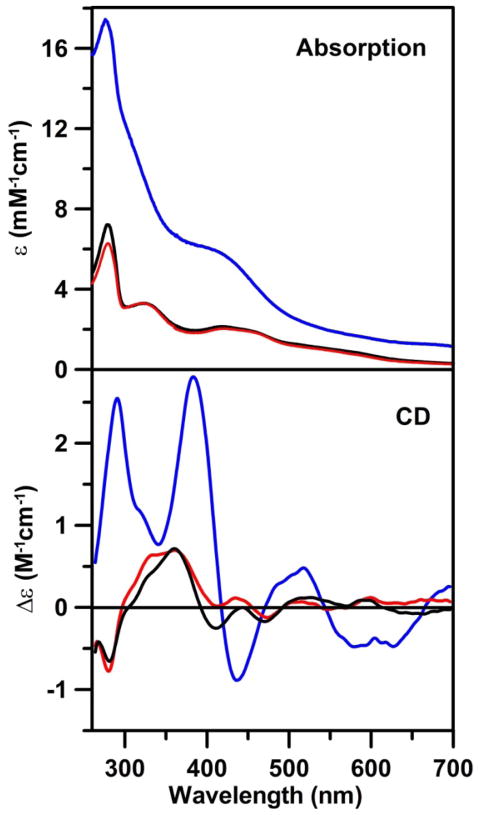
Previous studies have shown that recombinant AtNfu2 can be isolated as a mixture of apo and [2Fe-2S] cluster-loaded forms (27). In agreement with previous reports, the recombinant AtNfu2 purified under strictly anaerobic conditions contained 0.22 ± 0.03 [2Fe-2S]2+ cluster per protein monomer according to protein and iron analysis, whereas aerobic purification resulted in loss of the colored fraction. As-isolated, AtNfu2 exhibited the characteristic S-to-Fe(III) charge transfer bands of a [2Fe-2S]2+ center between 300 and 700 nm, with maxima centered at around 320 and 420 nm, and shoulders centered near 460 and 550 nm, see Figure 2 (black line). Based on the ε280 value (3.5 mM−1cm−1) determined for apo AtNfu2 monomer by quantitative amino acid analysis, which agrees with theoretical predictions based on the amino acid sequence, the ε280, ε320 and ε420 values per Nfu2 monomer arising from the [2Fe-2S] clusters were estimated to be 3.7, 3.3 and 2.1 mM−1cm−1, respectively. Since [2Fe-2S]2+ clusters typically have ε420 values between 7 and 11 mM−1cm−1 (31;37), the absorption data are consistent with the analytical data which indicated 0.22 ± 0.03 [2Fe-2S]2+ clusters per protein monomer. Compared to most other [2Fe-2S] cluster-bound proteins characterized to date, [2Fe-2S] cluster-loaded AtNfu2 exhibits a weak circular dichroism (CD) spectrum and lacked the commonly observed prominent positive band between 450 and 500 nm (31;38). The most prominent CD band for the [2Fe-2S]2+ center of Nfu2, Figure 2 black line, is a positive band centered at 360 nm, with a few minor positive or negative CD bands in the higher wavelength region. The [2Fe-2S]2+ center in Nfu2 was also found to reductively labile as judged by complete loss of the visible absorption and CD on exposure to 1 mM dithionite for 10 min.
Figure 2.
Room temperature UV-visible absorption and CD spectra of [2Fe-2S] cluster-bound as-isolated AtNfu2 (black lines), [2Fe-2S] cluster-bound reconstituted AtNfu2 (red lines) and [4Fe-4S] cluster-bound reconstituted AtNfu2 (blue lines). All ε and Δε values were calculated based on the concentrations of AtNfu2 monomer.
Cysteine desulfurase-mediated in vitro reconstitution of apo AtNfu2 under anaerobic conditions in the absence of DTT monitored by UV-visible absorption spectroscopy was characterized by a gradual increase of a band at ~420 nm which shifts to ~400 nm as the reaction progresses. Purification of the reaction mixture yielded two colored fractions. The second fraction appeared to accommodate the same chromophore as in the as-isolated protein, as evidenced by absorption and CD spectroscopies (Figure 2, red line). Bands arising from [2Fe-2S] cluster charge transfer transitions showed similar extinction coefficients of 2.8, 3.3 and 2.0 mM−1cm−1 for ε280, ε320 and ε420, respectively, which agree with the 0.21 ± 0.04 [2Fe-2S] cluster per protein monomer determined based on iron and protein determinations. The first fraction from the reconstitution exhibits an absorption spectrum comprising a very broad shoulder centered at ~400 nm with a broad tail that extends to the low wavelength region and a band at ~300 nm that is overlapped with the protein absorption band at 280 nm (Figure 2, blue line), and is indicative of a [4Fe-4S]2+ cluster. Protein and iron analyses indicated 0.51 ± 0.03 [4Fe-4S]2+ cluster per monomer, which is in good agreement with the extinction coefficients of 14.0 and 6.5 mM−1cm−1 at ε280 and ε420, respectively, determined for the [4Fe-4S]2+ center based on the Nfu monomer concentration. Compared to other biological [4Fe-4S]2+ centers the 300-nm band of the [4Fe-4S] center in Nfu proteins appears to be abnormally intense compared to the 400-nm band. For example, after subtraction of the protein contribution at 280 nm, the [4Fe-4S]2+ center in bacterial [4Fe-4S]-NfuA has ε280 = 36 mM−1cm−1 and ε400 = 15 mM−1cm−1 (9) compared to ε280 = 28 mM−1cm−1 and ε400 = 13 mM−1cm−1 in AtNfu2, based on cluster concentration. The anomalous electronic properties of the [4Fe-4S]2+ center in AtNfu2 are also reflected in the CD and resonance Raman spectra (see below). The CD spectrum is exceptionally intense compared to other CD spectra reported for [4Fe-4S] centers and is dominated by two positive bands located at 290 and 384 nm, respectively (Figure 2, blue line). Although various reaction times and concentrations of reactants were tried, we were never able to incorporate more than 0.27 [2Fe-2S] cluster per protein monomer, corresponding to one [2Fe-2S]2+ cluster per protein tetramer. On the other hand, extended reaction times or addition of 1–5 mM DTT resulted in the formation of homogeneous [4Fe-4S]2+ cluster-containing Nfu2 containing one [4Fe-4S]2+ cluster per protein dimer.
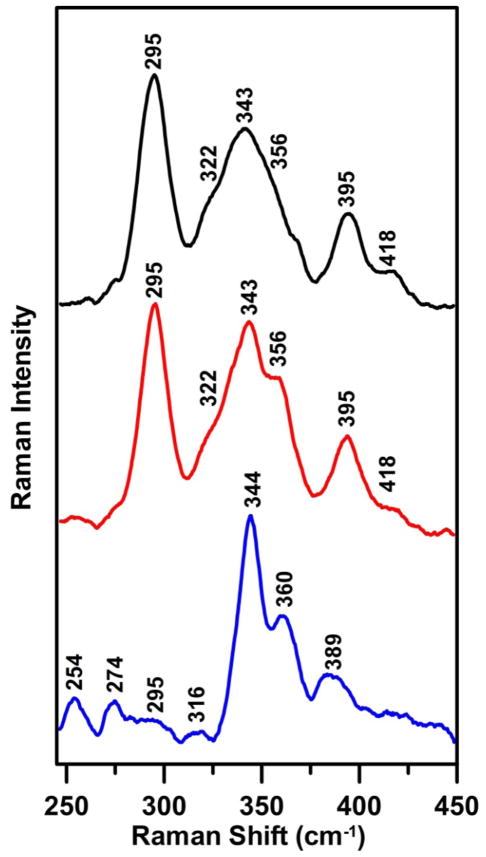
Resonance Raman (RR) spectroscopy in the Fe-S stretching region provides information on cluster type and the nature of the coordination environment. For each cluster type, the frequencies are found to be sensitive to the nature of the ligands (39;40). As shown in Figure 3, the RR spectra of the as-isolated and reconstituted [2Fe-2S] cluster-loaded Nfu2 are very similar and can be readily assigned to the Fe-S stretching modes of [2Fe-2S]2+ clusters (41;42). However, the anomalously high frequencies of the asymmetric (B3ut = 295 cm−1) and symmetric (Agt = 343 cm−1) terminal breathing modes compared to the regular all-cysteinyl coordination [2Fe-2S] cluster such as Fdxs (B3ut = 281–291 cm−1 and Agt = 326–340 cm−1) suggested an unusual coordination environment. There are at least two possible reasons that may account for the anomalously high frequencies. The first could be partial non-cysteinyl ligation with an oxygenic ligand, as has been observed for [2Fe-2S]2+ Fdx variants with one serinate ligand (39;43). The second could be the existence of unique H-bonding interactions or a unique set of cysteinyl Fe-Sγ-Cβ-Cα dihedral angles resulting from ligation by two CXXC motifs at the subunit interface. Anomalous cysteinyl Fe-Sγ-Cβ-Cα dihedral angles were found to be responsible for the anomalous RR spectra of the all-cysteinyl-ligated [2Fe-2S] cluster in human ferrochelatase (44–46).
Figure 3.
Low temperature resonance Raman spectra of [2Fe-2S] cluster-bound as-isolated AtNfu2 (black line), [2Fe-2S] cluster-bound reconstituted AtNfu2 (red line), and [4Fe-4S] cluster-bound reconstituted AtNfu2 (blue line). Spectra were recorded at 17 K using 457.9-nm laser excitation. Each spectrum is the sum of 80–120 scans, with each scan involving counting photons for 1 s every 0.5 cm−1 with 7 cm−1 spectral resolution. Bands resulting from the frozen buffer solution have been subtracted from all spectra.
The RR spectrum of the [4Fe-4S]2+ cluster-bound AtNfu2 in the Fe-S stretching region is characteristic of that observed and assigned for [4Fe-4S]2+ centers in other Fe-S proteins (47), see Figure 3. However, the intense band at 344 cm−1 that is assigned to the totally symmetric breathing mode of the Fe4S4 core (A1b) is out of the normal range for all-cysteinyl ligated [4Fe-4S]2+ centers (333–339 cm−1), but closer to the range established for clusters with oxygenic ligand (OH−, serine or aspartate) at a unique Fe site (340–343 cm−1) (40). The possibility of partial oxygenic ligation in AtNfu2 cannot be ruled out, because the crystal structure of apo Nfu2 shows two conserved serine residues located in close proximity to the cluster coordination pocket (28). However, an alternative interpretation is that the increased frequency reflects cluster distortion associated with a subunit-bridging [4Fe-4S]2+ cluster ligated symmetrically by the cysteines of the conserved CXXC sequence in each subunit. Indeed, the most intense band in RR spectrum of the [4Fe-4S]2+ center in AvNfuA occurs at an even higher frequency (353 cm−1), even though this protein lacks the conserved serines present in AtNfu2 (9). Moreover, in this case, the observed spectrum is very similar to that of the subunit-bridging, all-cysteinyl-ligated [4Fe-4S]2+ center in the MgATP-bound nitrogenase Fe-protein oxidized in the presence of glycerol (most intense RR band at 350 cm−1), which has been attributed to increased distortion of the Fe4S4 core resulting from subunit bridging (48). This conclusion is based on crystallographic studies of the nitrogenase Fe protein trapped in an ATP-bound conformation in the presence of glycerol, which showed that the [4Fe-4S] cluster is cleaved into two [2Fe-2S] fragments separated by 5 Å, each with partial glycerol ligation (48). Hence it is difficult to make definitive conclusions concerning the ligation of the [2Fe-2S] and [4Fe-4S] cluster ligation in AtNfu2 based on the RR data alone.
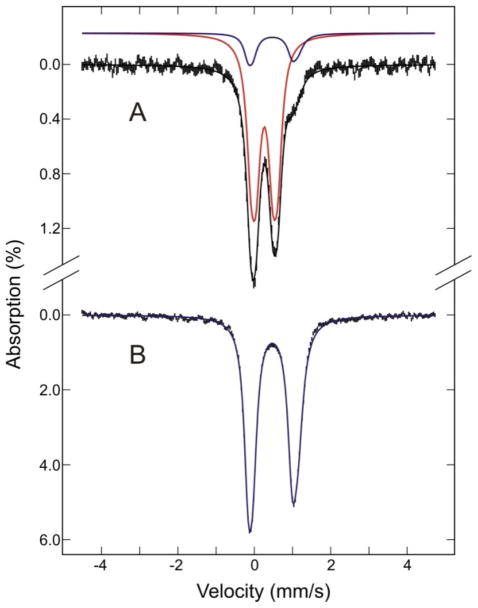
Mössbauer spectroscopy provides more definitive evidence for the ligands to the [2Fe-2S] cluster in Nfu2. As shown in Figure 4A, the major component of the spectrum of the reconstituted [2Fe-2S]2+ cluster-bound AtNfu2, which accounts for 85% of the total absorption, can be well simulated by two quadruple doublets of equal intensity with parameters indicative of [2Fe-2S]2+ clusters with complete cysteinyl ligation, see Table 1. The simulation parameters are very similar to those of [2Fe-2S]2+ centers in structurally characterized Fdxs (49) and monothiol glutaredoxins (Grxs) (31;50) and therefore are interpreted as two high spin Fe(III) centers tetrahedrally coordinated by two cysteines and two bridging sulfides. The minor component that accounts for 15% of the total absorption is identical to the spectrum of the homogeneous reconstituted [4Fe-4S]2+ cluster-containing Nfu2 (Figure 4B). This spectrum is readily simulated with two quadruple doublets of equal intensity arising from two valence delocalized [2Fe-2S]+ pairs. Moreover, the isomer shifts and quadrupole splittings of both doublets, see Table 1, are typical of those observed for structurally characterized [4Fe-4S]2+ clusters with one cysteinyl ligand completing approximately tetrahedral coordination on each Fe (49;51). Hence the Mössbauer data strongly support the assembly of all-cysteinyl-ligated [2Fe-2S]2+ or [4Fe-4S]2+ clusters in AtNfu2.
Figure 4.
Low temperature Mössbauer spectra of [2Fe-2S] cluster-bound reconstituted AtNfu2 (A) and [4Fe57-4S] cluster-bound as-reconstituted AtNfu2 (B). Spectra were recorded at 4.2 K with a weak magnetic field of 50 mT applied parallel to the direction of γ-radiation. The black solid lines overlaid on the experimental spectra (vertical bars) are simulations based on mixtures of [2Fe-2S]2+ and [4Fe-4S]2+ clusters (red and blues lines, respectively), with each comprised of two equal intensity doublets using the parameters listed in Table 1. The [2Fe-2S]2+ and [4Fe-4S]2+ cluster components in (A) account for 85% and 15% of the total Mössbauer absorption, respectively. [4Fe-4S]2+ clusters account for 100% of the Mössbauer absorption in (B).
Table 1.
Mössbauer parameters of [2Fe-2S]2+ and [4Fe-4S]2+ cluster-bound forms of AtNfu2
| Cluster type | Fe Site | δ(mm/s) | ΔEQ(mm/s) |
|---|---|---|---|
| [2Fe-2S]2+ | 1 | 0.27 | 0.70 |
| 2 | 0.27 | 0.43 | |
| [4Fe-4S]2+ | pair 1a | 0.49 | 1.30 |
| pair 2a | 0.48 | 1.03 |
The [4Fe-4S]2+ cluster is comprised of two valence-delocalized [2Fe-2S]+ pairs.
EDTA-induced [4Fe-4S] to [2Fe-2S] Cluster Conversion
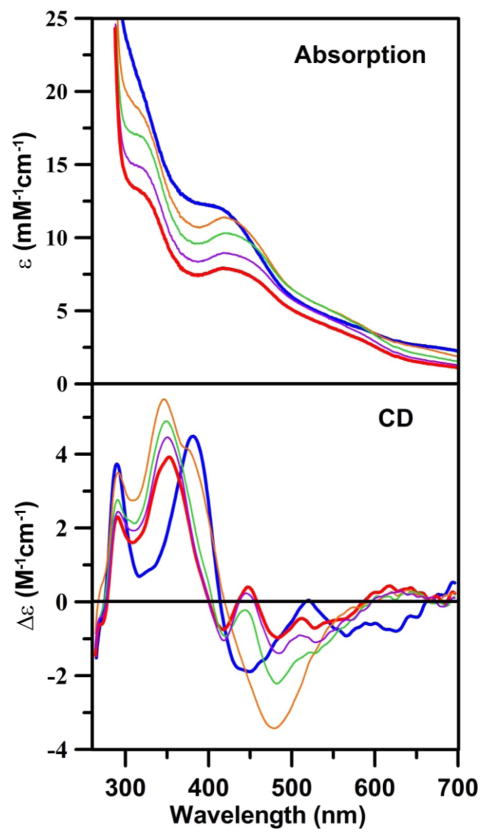
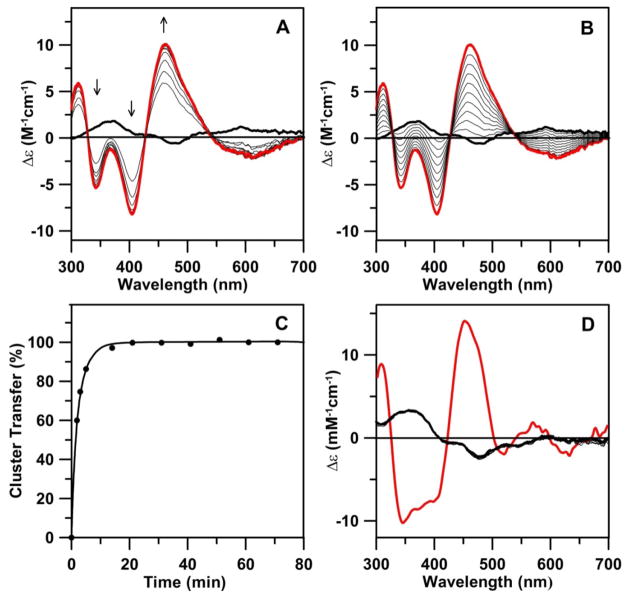
Some [4Fe-4S] cluster-containing proteins undergo cluster degradation to a semi-stable [2Fe-2S] cluster upon treatment with Fe chelators or exposure to air. Two well characterized examples are the selective removal of 2 irons from oxidized MgATP-bound nitrogenase Fe-protein in the absence of glycerol on addition of an iron chelator (52), and oxygen degradation of the [4Fe-4S] cluster in AvIscU (53). To investigate the possibility that [2Fe-2S] cluster-bound Nfu2 is a degradation product of the [4Fe-4S] cluster-bound form, we examined the stability of the [4Fe-4S] cluster assembled on Nfu2 to EDTA and oxygen. The UV-visible absorption and CD spectra shown in Figure 5, indicate that the addition of a 20-fold excess of EDTA to [4Fe-4S]-Nfu2 results in the gradual depletion of the features characteristic of the [4Fe-4S]2+ center and the emergence of features characteristic of [2Fe-2S]-Nfu2, over a period of 90 min. Moreover, the CD spectra, in particular, demonstrate that this transformation occurs via at least one intermediate, with a CD spectrum distinct from that of [4Fe-4S]-Nfu2 and [2Fe-2S]-Nfu2. The nature of this intermediate is unclear at present. Based on analogous UV-visible absorption and CD studies, [4Fe-4S]-Nfu2 was found to be stable in air for approximately 1 hr before starting to undergo gradual degradation without any clear indication of a [2Fe-2S]-Nfu2 intermediate. This relative insensitivity to oxygen makes [4Fe-4S]-Nfu2 a viable candidate for the maturation of [4Fe-4S] cluster-containing proteins in plant chloroplasts. Notable the [4Fe-4S] cluster-bound forms of human Nfu (25) and bacterial NfuA (9) display similar levels of oxygen sensitivity to that observed for AtNfu2, suggesting a similar role in the maturation of [4Fe-4S] cluster- containing proteins under aerobic or oxidative stress conditions.
Figure 5.
UV-visible absorption and CD monitored [4Fe-4S] cluster to [2Fe-2S] cluster conversion on AtNfu2 in the presence of EDTA. The zero time point of the reaction corresponds to the as-reconstituted [4Fe-4S] cluster-bound AtNfu2 (thick blue lines). Spectra were recorded 10 min (thin orange line), 30 min (thin green line), 60 min (thin purple line), and 90 min (thick red line) after addition of a 20-fold excess of EDTA (with respect to initial [4Fe-4S]2+ cluster concentration). ε and Δε values were calculated based on the initial concentration of [4Fe-4S] clusters in the sample.
Oligomeric States of Apo and Cluster-bound Forms of AtNfu2
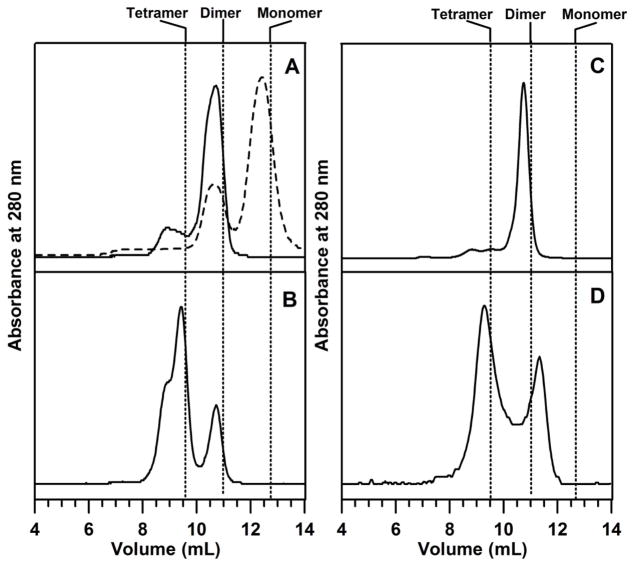
The oligomeric states of apo and cluster-bound forms of AtNfu2 were assessed by gel filtration chromatography using a Superdex 75 column. Previous gel filtration studies of recombinant AtNfu2 indicated a monomeric apo protein and a dimeric [2Fe-2S] cluster-bound form (27). However, apo AtNfu2 was found to be a dimer in the crystalline state (28). Dimeric [4Fe-4S] cluster-bound forms of cyanobacterial Nfu (24) and apo AvNfuA (9) have also been reported.
In this work, as purified apo AtNfu2 was eluted almost exclusively at ~42 kDa, indicating a dimer based on the monomer MW of the recombinant Nfu2 (17.8 kDa), see Figure 6A (solid line). However, a mixture of dimer and monomer apo AtNfu2 was observed in samples of [2Fe-2S]2+-Nfu2 treated overnight with 5 mM DTT, 10 mM EDTA and 1 mM dithionite and repurified by Amicon ultrafiltration, see Figure 6A (broken line). Both the dimer and monomer fractions were devoid of [2Fe-2S] clusters as judged by UV-visible absorption spectra. [4Fe-4S]2+ cluster-bound AtNfu2 eluted at ~42 kDa, see Figure 6C, which is consistent with a dimer and with protein and iron analytical data which indicate one [4Fe-4S] cluster per Nfu2 homodimer. Moreover, the UV-visible absorption and CD spectrum of this fraction were analogous to those of [4Fe-4S]2+-Nfu2 shown as the blue spectrum in Figure 2. In contrast, [2Fe-2S] cluster-bound Nfu2, eluted as two peaks at ~79 and ~41 kDa, corresponding to tetrameric and dimeric forms, respectively, see Figure 6B. The ratio of the two species was estimated to be 3:1 according to the peak area. Both fractions contained [2Fe-2S]2+ clusters exclusively based on comparison with the UV-visible absorption and CD spectra show in Figure 2. The first fraction contained slightly higher cluster content compared to the second fraction (0.27 vs 0.17 [2Fe-2S] cluster/monomer) as estimated by absorption spectroscopy. Analysis of [2Fe-2S] cluster-bound Nfu2 formed by EDTA-induced [4Fe-4S] cluster degradation also resulted in two fractions, see Figure 6D, one of which is clearly a tetramer. This also argues that the tetrameric form is at least one of the intrinsic oligomeric states of the [2Fe-2S] cluster-bound AtNfu2, a result that is also consistent with the analytical data which also indicated stoichiometry of one [2Fe-2S] cluster per tetramer.
Figure 6.
Gel filtration chromatograms of apo and cluster-loaded forms of AtNfu2. The elution of protein was monitored by absorption at 280 nm. Positions of theoretical oligomerization states calculated from the calibration plot shown in the supporting information (Figure S1) are marked by dotted lines. (A) apo as-isolated AtNfu2 (solid line) and apo AtNfu2 generated by degradation of [2Fe-2S]-Nfu2 using DTT, EDTA and dithionite (broken line); (B) [2Fe-2S] cluster-bound reconstituted AtNfu2; (C) [4Fe-4S] cluster-bound reconstituted AtNfu2; (D) [4Fe-4S] cluster-bound AtNfu2 after incubating with EDTA for 30 min.
Incorporation of Clusters on AtGrxS16 and AtAPR1 with Cluster-bound Forms of AtNfu2
In vivo studies have indicated that AtNfu2 is required for the maturation of both [2Fe-2S] and [4Fe-4S] clusters-containing proteins in chloroplasts (27;29), although previous in vitro evidence of cluster transfer from AtNfu2 is limited to [2Fe-2S] cluster transfer to the apo chloroplast ferredoxin (Fdx) (27). Moreover, Fdx is considered a multifunctional electron carrier (54), and may accept clusters from multiple sources. For example, we have previously reported that two plant chloroplastic monothiol Grxs, GrxS14 and GrxS16, can both serve as [2Fe-2S] cluster donors to Fdx (31;55). We therefore sought to identify more specific potential physiological partner proteins from chloroplasts that are likely to accept clusters from Nfu2 in vivo.
We first selected AtGrxS16, which has been proposed to be a cluster carrier protein (31;55), as a potential [2Fe-2S] cluster acceptor of Nfu2. Major differences in the CD spectra between the cluster-loaded forms of the donor and acceptor enabled facile monitoring of cluster transfer by CD spectroscopy as a function of time. Apo GrxS16 was pre-incubated with excess GSH which is required for cluster accommodation on monothiol Grxs (56). The cluster transfer was initiated by addition of [2Fe-2S] cluster-bound Nfu2 to apo GrxS16. Immediately after mixing, the characteristic positive band at 360 nm and the minor negative band at 470 nm arising from Nfu2 disappeared, as the intense 460 nm positive band from GrxS16 appeared and became dominant, see Figure 7A. Comparison to simulated CD changes for intact cluster transfer in 10% increments from 0–100%, see Figure 7B, indicated that more than 70% cluster was transferred in the first 6 min, and that the reaction approached completion after ~20 min. By monitoring the reaction at 461 nm, kinetic plots were generated that are well simulated based on the initial concentrations of [2Fe-2S] clusters on Nfu2 and the concentration of apo GrxS16 using second order kinetics with rate constant of 7,000 M−1min−1, see Figure 7C. This rate constant is significantly higher than the [2Fe-2S] cluster transfer from AvIscU to AvFdx even in the presence of the molecular co-chaperones and ATP (800 M−1min−1) (38), but not as high as the cluster transfer from AtGrxS14 to Synechocystis Fdx (20,000 M−1min−1) (31). This cluster transfer was not significantly perturbed even in the presence of 10 mM EDTA, and the reverse transfer using [2Fe-2S] cluster-bound GrxS16 and apo Nfu2 was not observed over a 60 min time period in the absence of GSH or the presence of DTT. This indicates a unidirectional and intact cluster transfer. No reaction was observed when [4Fe-4S]-Nfu2 was used in place of [2Fe-2S]-Nfu2 indicating that the cluster transfer reaction from Nfu2 to GrxS16 is cluster specific. However, parallel [2Fe-2S] cluster transfer experiments from Nfu2 to GrxS14, using analogous initial concentrations of donor and acceptor, did not show any significant changes in the CD spectrum of the [2Fe-2S]-Nfu2 donor after 51 min, see Figure 7D. Moreover, [2Fe-2S]-GrxS14 was not found to be capable of functioning as a cluster donor to apo-Nfu2 in the absence of GSH or the presence of DTT. Hence only GrxS16 is a competent [2Fe-2S] cluster acceptor for [2Fe-2S] clusters assembled on Nfu2 in these in vitro studies. Taken together, the rapid, quantitative, and specific [2Fe-2S] cluster transfer from Nfu2 to GrxS16 is likely to be a physiological relevant process.
Figure 7.
[2Fe-2S] cluster transfer from AtNfu2 to apo At GrxS16 and GrxS14 monitored by CD spectroscopy at room temperature as a function of time. All Δε values are based on the concentrations of [2Fe-2S] clusters on Nfu2. (A) CD spectra of cluster transfer from Nfu2 (39 μM in [2Fe-2S] clusters) to GrxS16 (162 μM in monomer) were recorded at 2.0, 3.5, 5, 14, 21, 31, 41, 51, 61 and 71 min (thin black lines). The thick black line corresponds to zero reaction time, i.e. [2Fe-2S] cluster-bound Nfu2, and the thick red line corresponds to complete [2Fe-2S] cluster transfer to GrxS16. The arrows indicated the direction of change in CD intensity with time at selected wavelengths. (B) Simulated quantitative changes in CD spectra based on complete cluster transfer from [2Fe-2S]-Nfu2 (thick black line) to GrxS16 (thick red line). The thin black lines correspond to 10 to 90% completion of cluster transfer in 10% increments. (C) Kinetic simulation (black line) of cluster transfer from [2Fe-2S]-Nfu2 to GrxS16, based on the initial concentrations of [2Fe-2S] clusters on Nfu and of apo GrxS16 dimer, using a second order rate constant of 7,000 M−1 min−1. Percentage of cluster transfer (●) was assessed by monitoring CD intensity at 461 nm. (D) CD spectra for attempted cluster transfer from Nfu2 (39 μM in [2Fe-2S] clusters) to GrxS14 (150 μM in monomer) recorded at 3, 11, 21, 31, 41 and 51 min (thin lines). The thick black line corresponds to zero reaction time, i.e., [2Fe-2S] cluster-bound Nfu2, and the thick red line corresponds to complete cluster transfer [2Fe-2S] to GrxS14.
The ability of Nfu2 to act as a [4Fe-4S] cluster donor was examined by investigating cluster transfer from [4Fe-4S] cluster-bound Nfu2 to apo AtAPR1, a crucial chloroplastic enzyme involved in the sulfur assimilation pathway, that requires a redox-inactive, cysteinyl-ligated [4Fe-4S]2+ cluster for optimal catalytic activity (57–62). APR1 is another example of a [4Fe-4S] cluster-containing protein that exhibits an intense CD signal and the CD spectra of [4Fe-4S]2+-Nfu2 and holo APR1 are partially overlapped, see Figure 8A. The former has a sharp intense positive band at 384 nm and a broad positive feature centered around 512 nm, whereas the latter has a broader intense positive band centered at 374 nm and a sharp minor positive band at 483 nm. The cluster transfer was initiated by addition of [4Fe-4S] cluster-bound Nfu2 to DTT-pretreated apo APR1. The reaction approached completion within 3 min of adding Nfu2, see Figures 8A and 8B. Kinetic simulations of the percent cluster transfer, as monitored by CD changes at 384 nm and based on the initial concentrations of [4Fe-4S] clusters on Nfu2 and the apo APR1 monomer, indicate a second order rate constant ≥ 150,000 M−1min−1, see Figure 8C. This rate constant is significantly greater than that observed for activation of apo aconitase with [4Fe-4S] cluster-loaded AvNfuA (60,000 M−1min−1) (9), and is among the most rapid and efficient [4Fe-4S] cluster transfer reactions reported thus far. No reaction was observed when excess [2Fe-2S]-Nfu2 was mixed with apo-APR1 indicating that the reaction is cluster specific.
Figure 8.
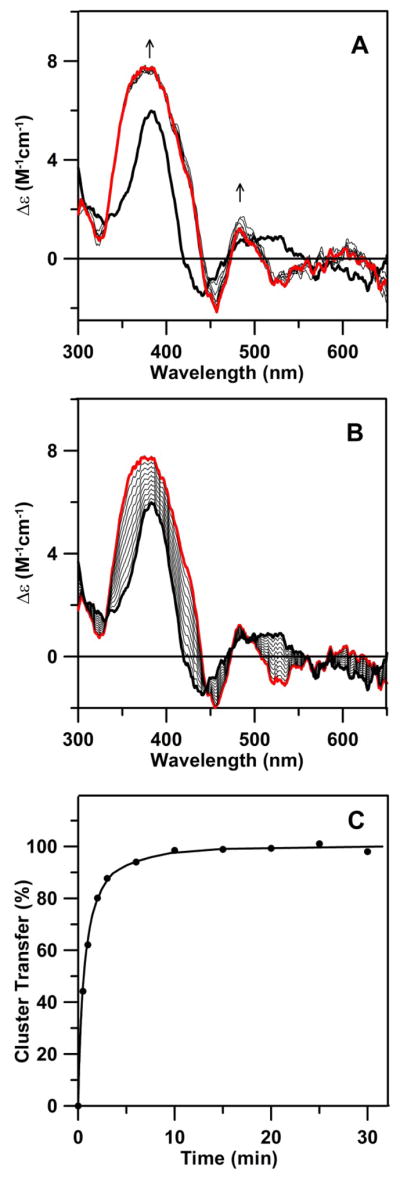
[4Fe-4S] cluster transfer from AtNfu2 (10 μM in [4Fe-4S] clusters) to 10 μM apo APR1 was monitored by CD spectroscopy at room temperature as a function of time. All Δε values are based on the concentration of [4Fe-4S] clusters on Nfu2. (A) CD spectra were recorded at 3, 7, 12, 22, 32, 42, 52 and 62 min (thin black lines). The thick black line corresponds to zero reaction time, i.e, [4Fe-4S] cluster-bound AtNfu2, and the thick red line corresponds to complete [4Fe-4S] cluster transfer to APR1. The arrows indicated the direction of change in CD intensity as a function of increasing time at selected wavelengths. (B) Predicted changes in CD spectra for quantitative cluster transfer using the zero reaction time (thick black line) and holo At APR1 (thick red line). Thin black lines correspond to 10 to 90% completion of cluster transfer in 10% increments. (C) Kinetic simulation (solid line) of cluster transfer from Nfu2 to APR1, based on the initial concentrations of [4Fe-4S] clusters on Nfu and of apo APR1 monomer, based on second order kinetics with a rate constant of 150,000 M−1 min−1. Percentage of cluster transfer (●) was assessed by monitoring CD intensity at 384 nm.
Yeast Two-Hybrid Evidence for Complex Formation Between AtNfu2 and AtAPR1
In order to investigate whether the [4Fe-4S] cluster transfer from AtNfu2 to AtAPR1 involves a direct interaction between the two proteins, the capacity of Nfu2 to physically interact with APR1 was assessed using a yeast two-hybrid binary assay. As shown in Figure 9, the co-expression of Nfu2 and APR1 in the HIS3 reporter gene-based yeast CY306 allowed a strong yeast growth in the absence of histidine, confirming that APR1 and Nfu2 proteins also interact in vivo. Interestingly, among the three chloroplastic Nfu proteins, the Nfu/APR1 interaction was found to be specific to Nfu2, as no yeast growth was observed when Nfu2 was replaced by Nfu1 or Nfu3. This strongly suggests that Nfu2 is the specific [4Fe-4S] cluster donor for maturation of APR1 in plant chloroplasts (Figure 9).
Figure 9.

At APR1 interacts with AtNfu2 in a yeast-two hybrid assay. CY306 strain (35) was co-transformed with pGAD.APR1 and individual pGBK.Nfu constructs and plated as serial dilutions on a control plate containing histidine (+His, left panel) and on a test plate containing 2 mM 3-amino triazol (3AT) without histidine (−His, right panel). Yeast cells were allowed to grow at 30°C for 4 days. Neither pGAD.APR1/empty pGBK nor empty pGAD/pGBK.Nfu2 co-transformations allowed yeast growth (not shown). Results shown here are the means of 4 independent experiments.
DISCUSSION
Based on previous gene knockout studies, AtNfu2 is essential for the effective maturation of various chloroplastic proteins, including [2Fe-2S] cluster-containing chloroplastic Fdx, and [4Fe-4S] cluster-containing PSI complex and sulfite reductase (27;29). However, prior to this work there was no in vivo or in vitro evidence that Nfu2 was able to accommodate [4Fe-4S] clusters. Although various other types of Nfu proteins, such as bacterial NfuA and human mitochondrial Nfu, are capable of assembling [4Fe-4S] clusters via in vitro reconstitution (9;18;25), their domain structures are distinct from those of the plant chloroplast Nfu proteins, see Figure 1. However, the single domain cyanobacterial Synechococcus sp. PCC 7002 Nfu was shown to assemble a mixture of [2Fe-2S] clusters (minor component) and [4Fe-4S] clusters (major component) and to facilitate maturation of [4Fe-4S] clusters in PSI by direct cluster transfer (24). This result is particularly relevant to chloroplastic Nfu proteins, since cyanobacteria are considered to be the evolutionary precursor of plant chloroplasts. The work described in this manuscript provides the first direct evidence that plant chloroplast Nfu2 is capable of accommodating either [2Fe-2S] or [4Fe-4S] clusters and that the [4Fe-4S] clusters can be subsequently transferred rapidly and intact to a physiologically relevant partner protein, APR1. As discussed below, it also contributes significantly to understanding the role and specificity of Nfu2 in the maturation of chloroplastic [2Fe-2S] and [4Fe-4S] cluster-containing proteins.
The spectroscopic and analytical characterization of the clusters assembled on Nfu2 reported in this work reveals the nature and properties of the clusters, and provide insight into the origins of the anomalous spectroscopic features associated with these clusters. The [4Fe-4S] center exhibits anomalously high intensity for the band at ~300 nm. Both the [2Fe-2S] and [4Fe-4S] centers exhibited unexpected CD signals, anomalously low intensity for the former and anomalously high intensity for the latter, and both show notable frequency shifts in characteristic RR bands that could be interpreted in terms of partial non-cysteinyl ligation. However, the site-directed mutagenesis studies on the cysteine residues of AtNfuA, EcNfuA and AtNfu2 have indicated that both of the cysteine residues were required for cluster ligation (9;23;28). Moreover, the Fe EXAFS studies of [2Fe-2S]-Nfu2 (28) and the Mössbauer results shown in this work for [2Fe-2S]-Nfu2 and [4Fe-4S]-Nfu2, taken together with the crystal structure of dimeric apo Nfu2 (28), clearly support complete cysteinyl ligation for [2Fe-2S] and [4Fe-4S] clusters at subunit interfaces involving the CXXC motifs. Hence the anomalous absorption, CD and resonance Raman properties of the [2Fe-2S] and [4Fe-4S] centers in Nfu2 are attributed to cluster distortions associated with the subunit bridging environment.
Our analytical and gel filtration results indicate that Nfu2 can accommodate approximately one [2Fe-2S] cluster per homotetramer both as purified under anaerobic conditions and following in vitro cysteine desulfurase-mediated cluster reconstitution. All attempts to increase the stoichiometry of the [2Fe-2S] clusters were unsuccessful. Although it has been previously proposed that Nfu2 contained one [2Fe-2S] cluster per homodimer (27), Nfu2 and other types of [2Fe-2S] cluster-containing Nfu proteins have only been isolated in forms containing variable amounts of apo protein thus making cluster stoichiometry difficult to assess (22;27). Surprisingly, our gel filtration studies revealed that the [2Fe-2S] cluster-bound Nfu2 can exist as both dimeric and tetrameric forms with the latter being the major and more cluster-replete component. The tetrameric nature of [2Fe-2S]-Nfu2 is further supported by the observation that EDTA-induced [4Fe-4S]-to-[2Fe-2S] cluster conversion is accompanied by a dimer to tetramer conversion. Given that absorption spectra recorded during reconstitution in the absence of DTT indicate that [2Fe-2S] cluster assembly precedes [4Fe-4S] cluster assembly on Nfu2, whereas the reconstitution in the presence of DTT directly yields a homogeneous dimeric [4Fe-4S] cluster form, it is tempting to speculate that the tetrameric [2Fe-2S]-Nfu2 is an oxygen tolerant precursor of [4Fe-4S]-Nfu2 during cysteine desulfurase-mediated reconstitution conditions. However, the significance of this hypothesis is unclear in light of the available in vitro and in vivo evidence that bacterial NfuA and mitochondrial Nfu1 proteins appear to be carrier proteins that accept clusters from primary scaffold proteins (U-type proteins in the ISC system and SufB in the SUF system) (8;18;21), rather than scaffold proteins that assemble de novo clusters via interaction with cysteine desulfurases and/or sulfur transferases.
Higher plants often encode several paralogs of proteins from the same family. For example, A. thaliana encodes 5 Nfus (26) and 33 different Grxs (63), of which 3 Nfus and at least 4 Grxs (including the two monothiol glutaredoxins) are localized in the chloroplasts. While some degree of redundancy is possible, functional specificity is also expected, as evidenced by previous gene knockout and complementation studies (26;27;29). The specificity could include but is not limited to differences in expression regulation pathways, cellular conditions, specific sub-organelle localizations, partner proteins, etc. Here we have explored the specificity of Nfu2 with respect to three potentially significant relevant chloroplastic acceptor proteins, GrxS14, GrxS16, and APR1. It is known that Nfu2 deficiency is responsible for the reduced activity or abundance of chloroplastic Fdx, PSI complex, and sulfite reductase. In this work, we provide in vitro evidence of direct cluster transfer from Nfu2 to a potential carrier protein, GrxS16, and a potential acceptor protein, APR1, at physiologically relevant rates.
GrxS16 is a monothiol Grx that belongs to the Grx superfamily. The Grx superfamily is a group of GSH-dependent thiol oxidoreductases that are previously considered to be involved solely in disulfide bond cleavage as part of the cellular redox regulation (64;65). Subsequently it was discovered that one class of Grxs, namely, monothiol Grxs that are characterized by a conserved CGFS active site motif, are able to accommodate [2Fe-2S] clusters and play essential roles in cellular Fe and Fe-S cluster homeostasis (31;56;66;67). Moreover, in bacteria and mitochondria, recent in vivo and in vitro studies have demonstrated that monothiol Grxs play a specific role in accepting [2Fe-2S] clusters assembled on U-type proteins in the ISC system in a ATP-dependent process mediated by dedicated co-chaperones (HscA/HscB in bacteria and Ssq1/Jac1 in yeast mitochondria) (68;69). In A. thaliana, there are two monothiol Grxs localized in chloroplasts, GrxS14 and GrxS16. In addition to the targeting sequence and the cluster coordination domain which comprises the CGFS motif, GrxS16 has a unique N-terminal domain which differs from GrxS14. Both of the Grxs have been shown to accommodate [2Fe-2S] clusters and were proposed to be cluster carriers (31;55). Our results show that Nfu2 selectively transfers [2Fe-2S] cluster to GrxS16 but not to GrxS14, leading to the hypothesis that Nfu2 functions as a specialized cluster carrier protein, which specifically delivers [2Fe-2S] clusters to various enzymes via the GrxS16 pathway. In contrast, Nfu2 is clearly not a cluster donor for GrxS14 and experiments are in progress to assess if Nfu1 or Nfu3 fulfills this role for GrxS14. The mechanism of the selective [2Fe-2S] cluster transfer from Nfu2 to GrxS16 is not known, but it may involve interaction involving the N-terminal domain of GrxS16.
The most compelling and significant cluster transfer result presented herein is the ability of [4Fe-4S]-Nfu2 to function as a very effective [4Fe-4S] cluster donor for maturation of AtAPR1. Yeast two- hybrid studies provide strong evidence for complex formation between AtNfu2 and AtAPR1, and in vitro studies demonstrate extremely rapid (second order rate constant ≥ 150,000 M−1min−1), quantitative and intact cluster transfer from [4Fe-4S]-Nfu2 to apo APR1. APR1 catalyzes the second step of the sulfur assimilation process of the plants, i.e. the reduction of adenosine 5′-phosphosulfate (APS) to sulfite (70), and contains a [4Fe-4S] cluster ligated by four cysteines in a unique CC…..CXXC motif (71). The sulfite is subsequently reduced to sulfide by sulfite reductase, another [4Fe-4S] cluster-containing protein, before it is incorporated into other sulfur-containing bio-molecules, such as cysteine, methionine, GSH, and Fe-S clusters. Since two of the sulfur assimilation enzymes, APR1 and sulfite reductase, appear to depend on Nfu2 for their Fe-S cluster assembly, the sulfur assimilation and cysteine biosynthesis pathways are likely to be impaired due to a deficiency in Nfu2. Hence, lower levels of cluster insertion into chloroplastic Fe-S proteins in general is presumably responsible for the phenotypes associated with Nfu2 deletion, which include retarded growth, pale green leaves, poor developed roots and root hairs, decreased chlorophyll level, and low levels of the chloroplastic [2Fe-2S] Fdx and the [4Fe-4S] clusters in photosystem I and sulfite reductase due to post-transcriptional defects (27;29). However, since Nfu2 deletion is not lethal, some functional redundancy may exist with Nfu1 and/or Nfu3. In vivo and in vitro experiments are in progress to address the functional redundancy among AtNfu proteins in plant chloroplasts and the putative role of the C-terminal domain in targeting specific acceptor proteins
In summary, we have demonstrated that AtNfu2 is capable of assembling [2Fe-2S] and [4Fe-4S] clusters, and provided in vitro evidence that [2Fe-2S]-Nfu2 is a potential cluster donor to AtGrxS16 and that [4Fe-4S]-Nfu2 is likely to be the physiological cluster donor for the maturation of AtAPR1 in plant chloroplasts. Future work will focus on assessing the ability of the AtSufBCD complex to serve as a cluster donor for AtNfu1/2/3 and addressing the specificity of cluster-loaded forms of AtNfu1/2/3 in the maturation of a wide range of chloroplastic acceptor proteins using yeast two-hybrid studies as a guide to assess complex formation.
Supplementary Material
ABBREVIATIONS
- At
Arabidopsis thaliana
- Av
Azotobacter vinelandii
- Ec
Escherichia coli
- Grx
glutaredoxin
- Fdx
ferredoxin
- APR
adenosine 5′-phosphosulfate reductase
- CD
circular dichroism
- RR
resonance Raman
- DTT
dithiothreitol
- IPTG
isopropyl 1-thio-β-D-galactopyranoside
- FAS
ferrous ammonium sulfate
- PMSF
Phenylmethanesulfonyl fluoride
- GSH
reduced glutathione
- EDTA
ethylenediaminetetraacetic acid disodium salt
- SDS-PAGE
dodecyl sulfate polyacrylamide gel electrophoresis
Footnotes
This work was supported by the National Institutes of Health (GM62524 to M.K.J. and GM47295 to B.H.H.), Agence Nationale de la Recherche (2010BLAN1616 to N.R., J.C., H.C.W. and F.V.), the U.S. Department of Energy (DE-FG03-99ER20346 to D.B.K.), the U.S. Department of Energy (DE-FG03-99ER20346 to D.B.K.), the USDA (2002-35318-12503 to and T.L. and D.B.K.), and a Hatch Grant (NJ 12136 to T.L.)
Calibration data for the analytical gel filtration studies is provided as supporting information. Supporting information may be accessed free of charge online at http://pubs.acs.org.
References
- 1.Johnson MK, Smith AD. In: Encyclopedia of Inorganic Chemistry. 2. King RB, editor. John Wiley & Sons; Chichester: 2005. pp. 2589–2619. [Google Scholar]
- 2.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 3.Fontecave M, Ollagnier-de-Choudens S. Iron-sulfur cluster biosynthesis in bacteria: Mechanisms of cluster assembly and transfer. Arch Biochem Biophys. 2008;474:226–237. doi: 10.1016/j.abb.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Ayala-Castro C, Saini A, Outten FW. Fe-S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev. 2008;72:110–125. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balk J, Pilon M. Ancient and essential: The assembly of iron-sulfur clusters in plants. Trends Plant Sci. 2011;16:218–226. doi: 10.1016/j.tplants.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 7.Py B, Barras F. Building Fe-S proteins: bacterial strategies. Nat Rev Microbiol. 2010;8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- 8.Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Mühlenhoff U. The role of mitochondria in cellular iron-sulfur protein biosynthesis and iron metabolism. Biochim Biophys Acta. 2012;1823:1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Bandyopadhyay S, Naik S, O’Carroll IP, Huynh BH, Dean DR, Johnson MK, Dos Santos PC. A proposed role for the Azotobacter vienlandii NfuA protein as an intermediate iron-sulfur cluster carrier. J Biol Chem. 2008;283:14092–14099. doi: 10.1074/jbc.M709161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro-Sastre A, Tort F, Stehling O, Uzarska MA, Arranz JA, del Torro M, Labayru MT, Landa J, Font A, Garcia-Villoria J, Mennero B, Ugarte M, Guterrrez-Solano LG, Canpistol J, Garcia-Caroria A, Vaquerizo J, Riudor E, Briones P, Elpeleg O, Ribes A, Lill R. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am J Hum Genet. 2011;89:656–667. doi: 10.1016/j.ajhg.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Py B, Gerez C, Angelini S, Planel R, Vinella D, Loiseau L, Talla E, Brochier-Armanet C, Garcia Serres R, Latour JM, Ollagnier-de Choudens S, Fontecave M, Barras F. Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol Microbiol. 2012;86:155–171. doi: 10.1111/j.1365-2958.2012.08181.x. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson MR, Cash VL, Weiss MC, Laird NF, Newton WE, Dean DR. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol Gen Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 13.Fu W, Jack RF, Morgan TV, Dean DR, Johnson MK. nifU gene product from Azotobacter vinelandii is a homodimer that contains two identical [2Fe-2S] clusters. Biochemistry. 1994;33:13455–13463. doi: 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- 14.Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc Natl Acad Sci USA. 2000;97:599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang DM, Dempsey A, Tan KT, Liew CC. A modular domain of NifU, a nitrogen fixation cluster protein, is highly conserved in evolution. J Mol Evol. 1996;43:536–540. doi: 10.1007/BF02337525. [DOI] [PubMed] [Google Scholar]
- 16.Agar JN, Yuvaniyama P, Jack RF, Cash VL, Smith AD, Dean DR, Johnson MK. Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J Biol Inorg Chem. 2000;5:167–177. doi: 10.1007/s007750050361. [DOI] [PubMed] [Google Scholar]
- 17.Dos Santos PC, Smith AD, Frazzon J, Cash VL, Johnson MK, Dean DR. Iron-sulfur cluster assembly: NifU-directed activation of the nitrogenase Fe-protein. J Biol Chem. 2004;279:19705–19711. doi: 10.1074/jbc.M400278200. [DOI] [PubMed] [Google Scholar]
- 18.Smith AD, Jameson GNL, Dos Santos PC, Agar JN, Naik S, Krebs C, Frazzon J, Dean DR, Huynh BH, Johnson MK. NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry. 2005;44:12955–12969. doi: 10.1021/bi051257i. [DOI] [PubMed] [Google Scholar]
- 19.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 20.Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saio T, Kumeta H, Ogura K, Yokochi M, Asayama M, Katoh S, Katoh E, Teshima K, Inagaki F. The cooperative role of OsCnfU-1A domain I and domain II in the iron-sulphur cluster transfer process as revealed by NMR. J Biochem. 2007;142:113–121. doi: 10.1093/jb/mvm120. [DOI] [PubMed] [Google Scholar]
- 22.Nishio K, Nakai M. Transfer of iron-sulfur cluster from NifU to apoferredoxin. J Biol Chem. 2000;275:22615–22618. doi: 10.1074/jbc.C000279200. [DOI] [PubMed] [Google Scholar]
- 23.Angelini S, Gerez C, Ollagnier-de-Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;289:14084–14091. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- 24.Jin Z, Heinnickel M, Krebs C, Shen G, Golbeck JH, Bryant DA. Biogenesis of iron-sulfur clusters in photosystem I. J Biol Chem. 2008;283:28426–28435. doi: 10.1074/jbc.M803395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong WH, Jameson GNL, Huynh BH, Rouault TA. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci USA. 2003;100:9762–9767. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Léon S, Touraine B, Ribot C, Briat JF, Lobréaux S. Iron-sulfur cluster assembly in plants: Distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem J. 2003;371:823–830. doi: 10.1042/BJ20021946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yabe T, Morimoto K, Kikuchi S, Nishio K, Terashima I, Nakai M. The arabidopsis chloroplast NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for the biogenesis of ferredoxin and photosystem I. Plant Cell. 2004;16:993–1007. doi: 10.1105/tpc.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yabe T, Yamashita E, Kikuchi A, Morimoto K, Nakagawa A, Tsukihara T, Nakai M. Structural analysis of Arabidopsis CnfU protein: an iron-sulfur cluster biosynthetic scaffold in chloroplasts. J Mol Biol. 2008;381:160–173. doi: 10.1016/j.jmb.2008.05.072. [DOI] [PubMed] [Google Scholar]
- 29.Touraine B, Boutin JP, Marion-Poll A, Briat JF, Peltier G, Lobréaux S. Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulfur cluster assembly in Arabidopsis chloroplasts. Plant J. 2004;40:101–111. doi: 10.1111/j.1365-313X.2004.02189.x. [DOI] [PubMed] [Google Scholar]
- 30.Bick J-A, Setterdahl AT, Knaff DB, Chen Y, Pitcher LH, Zilinskas BA, Leustek T. Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry. 2001;40:9040–9048. doi: 10.1021/bi010518v. [DOI] [PubMed] [Google Scholar]
- 31.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot J-P, Johnson MK, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown RE, Jarvis KL, Hyland KJ. Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal Biochem. 1989;180:136–139. doi: 10.1016/0003-2697(89)90101-2. [DOI] [PubMed] [Google Scholar]
- 33.Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Meth Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 34.Drozdzewski PM, Johnson MK. A simple anaerobic cell for low temperature Raman spectroscopy. Appl Spectrosc. 1988;42:1575–1577. [Google Scholar]
- 35.Vignols F, Brehelin C, Surdin-Kerjan Y, Thomas D, Meyer Y. A yeast two-hybrid knockout strain to explore thioredoxin-interacting proteins in vivo. Proc Natl Acad Sci USA. 2005;102:16729–16734. doi: 10.1073/pnas.0506880102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425–1426. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agar JN, Krebs B, Frazzon J, Huynh BH, Dean DR, Johnson MK. IscU as a scaffold for iron-sulfur cluster biosynthesis: Sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry. 2000;39:7856–7862. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 38.Chandramouli K, Johnson MK. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry. 2006;45:11087–11095. doi: 10.1021/bi061237w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moulis JM, Davasse V, Golinelli MP, Meyer J, Quinkal I. The coordination sphere of iron-sulfur clusters: Lessons from site-directed mutagenesis. J Biol Inorg Chem. 1996;1:2–14. [Google Scholar]
- 40.Brereton PS, Duderstadt RE, Staples CR, Johnson MK, Adams MWW. Effect of serinate ligation at each of the iron sites of the [Fe4S4] cluster of Pyrococcus furiosus ferredoxin on the redox, spectroscopic and biological properties. Biochemistry. 1999;38:10594–10605. doi: 10.1021/bi990671d. [DOI] [PubMed] [Google Scholar]
- 41.Han S, Czernuszewicz RS, Kimura T, Adams MWW, Spiro TG. Fe2S2 protein resonance Raman revisited: Structural variations among adrenodoxin, ferredoxin, and red paramagnetic protein. J Am Chem Soc. 1989;111:3505–3511. [Google Scholar]
- 42.Fu W, Drozdzewski PM, Davies MD, Sligar SG, Johnson MK. Resonance Raman and magnetic circular dichroism studies of reduced [2Fe-2S] proteins. J Biol Chem. 1992;267:15502–15510. [PubMed] [Google Scholar]
- 43.Meyer J, Fujinaga J, Gaillard J, Lutz M. Mutated forms of the [2Fe-2S] ferredoxin fromClostridium pasteurianum with noncysteinyl ligands to the iron-sulfur cluster. Biochemistry. 1994;33:13642–13650. doi: 10.1021/bi00250a014. [DOI] [PubMed] [Google Scholar]
- 44.Crouse BR, Sellers VM, Finnegan MG, Dailey HA, Johnson MK. Site-directed mutagenesis and spectroscopic characterization of human ferrochelatase: Identification of residues coordinating the [2Fe-2S] cluster. Biochemistry. 1996;35:16222–16229. doi: 10.1021/bi9620114. [DOI] [PubMed] [Google Scholar]
- 45.Wu CK, Dailey HA, Rose JP, Burden A, Sellers VM, Wang BC. The 2.0 Å structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nature Struct Biol. 2001;8:156–160. doi: 10.1038/84152. [DOI] [PubMed] [Google Scholar]
- 46.Albetel A-N. PhD Dissertation. University of Georgia; 2012. Role and spectroscopic charaterization of [2Fe-2S] centers in ferrochelatases. [Google Scholar]
- 47.Czernuszewicz RS, Macor KA, Johnson MK, Gewirth A, Spiro TG. Vibrational mode structure and symmetry in proteins and analogues containing Fe4S4 clusters: Resonance Raman evidence for different degrees of distortion in HiPIP and ferredoxin. J Am Chem Soc. 1987;109:7178–7187. [Google Scholar]
- 48.Sen S, Igarashi R, Smith AD, Johnson MK, Seefeldt LC, Peters JW. A conformational mimic of the MgATP-bound “on state” of the nitrogenase iron protein. Biochemistry. 2004;43:1787–1797. doi: 10.1021/bi0358465. [DOI] [PubMed] [Google Scholar]
- 49.Trautwein AX, Bill E, Bominaar EL, Winkler H. Iron-containing proteins and related analogs. Complementary Mõssbauer, EPR and magnetic susceptibility studies. Struct Bonding. 1991;78:1–95. [Google Scholar]
- 50.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Middleton P, Dickson DPE, Johnson CE, Rush JD. Interpretation of the Mössbauer spectra of the four-iron ferredoxin from Bacillus stearothermophilus. Eur J Biochem. 1978;88:135–141. doi: 10.1111/j.1432-1033.1978.tb12430.x. [DOI] [PubMed] [Google Scholar]
- 52.Anderson GL, Howard JB. Reactions of the oxidized iron protein of Azotobacter vinelandii nitrogenase: Formation of a 2Fe center. Biochemistry. 1984;23:2118–2122. doi: 10.1021/bi00305a002. [DOI] [PubMed] [Google Scholar]
- 53.Chandramouli K, Unciuleac M-C, Naik S, Dean DR, Huynh BH, Johnson MK. Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry. 2007;46:6804–6811. doi: 10.1021/bi6026659. [DOI] [PubMed] [Google Scholar]
- 54.Sticht H, Rösch P. The structure of iron-sulfur proteins. Prog Biophys Mol Biol. 1998;70:95–136. doi: 10.1016/s0079-6107(98)00027-3. [DOI] [PubMed] [Google Scholar]
- 55.Subramanian S. PhD Dissertation. University of Georgia; 2010. Characterization of the properties and roles of Fe-S centers in ferredoxins, glutaredoxins and radical S-adenosyl methionine enzymes. [Google Scholar]
- 56.Iwema T, Picciocchi A, Traore DA, Ferrer JL, Chauvat F, Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48:6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 57.Kim SK, Rahman A, Bick JA, Conover RC, Johnson MK, Mason JT, Hirasawa M, Leustek T, Knaff DB. Properties of the cysteine residues and iron-sulfur cluster of the assimilatory 5′-adenylyl sulfate reductase from Pseudomonas aeruginosa. Biochemistry. 2004;43:13478–13486. doi: 10.1021/bi048811t. [DOI] [PubMed] [Google Scholar]
- 58.Kim SK, Rahman A, Conover RC, Johnson MK, Mason JT, Gomes V, Hirasawa M, Moore ML, Leustek T, Knaff DB. Properties of the cysteine residues and the iron-sulfur cluster of the assimilatory 5′-adenylyl sulfate reductase from Enteromorpha intestinalis. Biochemistry. 2006;45:5010–5018. doi: 10.1021/bi0519250. [DOI] [PubMed] [Google Scholar]
- 59.Kopriva S, Buchert T, Fritz G, Suter F, Weber M, Benda R, Schaller J, Feller U, Schurmann P, Schunemann V, Trautwein AX, Kroneck PMH, Brunold C. Plant adenosine 5′-phosphosulfate reductase is a novel iron-sulfur protein. J Biol Chem. 2001;276:42881–42886. doi: 10.1074/jbc.M107424200. [DOI] [PubMed] [Google Scholar]
- 60.Kopriva S, Buchert T, Fritz G, Suter M, Benda R, Schunemann V, Koprivova A, Schurmann P, Trautwein AX, Kroneck PMH, Brunold C. The presence of an iron-sulfur cluster in adenosine 5′-phosphosulfate reductase separates organisms utilizing adenosine 5′-phosphosulfate and phosphoadenosine 5′-phosphosulfate for sulfate assimilation. J Biol Chem. 2002;277:21786–21791. doi: 10.1074/jbc.M202152200. [DOI] [PubMed] [Google Scholar]
- 61.Chartron J, Carroll KS, Shiau C, Gao H, Leary JA, Bertozzi CR, Stout CD. Substrate recognition, protein dynamics, and iron-sulfur cluster in Pseudomonas aeruginosa adenosine 5′-phosphosulfate reductase. J Mol Biol. 2006;364:152–169. doi: 10.1016/j.jmb.2006.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhave DP, Hong JA, Keller RL, Krebs C, Carroll KS. Iron-sulfur cluster engineering provides insight into the evolution of substrate specificity among sulfonucleoitide reductases. ACS Chem Biol. 2012;7:306–315. doi: 10.1021/cb200261n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Couturier J, Jacquot JP, Rouhier N. Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell Mol Life Sci. 2009;66:2539–2557. doi: 10.1007/s00018-009-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 65.Rouhier N, Gelhaye E, Jacquot JP. Plant glutaredoxins: still mysterious reducing systems. Cell Mol Life Sci. 2004;61:1266–1277. doi: 10.1007/s00018-004-3410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 67.Rouhier N, Couturier J, Johnson MK, Jacquot JP. Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci. 2010;35:43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shakamuri P, Zhang B, Johnson MK. Monothiol glutaredoxins function in storing and transporting [Fe2S2] clusters assembled on IscU scaffold proteins. J Am Chem Soc. 2012;134:15213–15216. doi: 10.1021/ja306061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, Mühlenhoff U. The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol Biol Cell. 2013;24:1830–1841. doi: 10.1091/mbc.E12-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kopriva S, Koprivova A. Plant adenosine 5′-phosphosulphate reductase: the past, the present, and the future. J Exp Bot. 2004;55:1775–1783. doi: 10.1093/jxb/erh185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.