Abstract
Chromosome duplication normally initiates via the assembly of replication fork complexes at defined origins1,2. DNA synthesis by any one fork is thought to cease when it meets another travelling in the opposite direction, at which stage the replication machinery may simply dissociate before the nascent strands are finally ligated. But what actually happens is not clear. Here we present evidence consistent with the idea that every fork collision has the potential to threaten genomic integrity. In Escherichia coli this threat is kept at bay by RecG DNA translocase3 and by single-strand DNA exonucleases. Without RecG, replication initiates where forks meet via a replisome assembly mechanism normally associated with fork repair, replication restart and recombination4,5, establishing new forks with the potential to sustain cell growth and division without an active origin. This potential is realised when roadblocks to fork progression are reduced or eliminated. It relies on the chromosome being circular, reinforcing the idea that replication initiation is triggered repeatedly by fork collision. The results reported raise the question of whether replication fork collisions have pathogenic potential for organisms that exploit multiple origins to replicate each chromosome.
In Escherichia coli, the number of head-on fork collisions is kept to a single event by replicating the circular chromosome from a single origin (oriC). Chromosome duplication is completed when the two forks established meet in a specialised termination zone (Fig. 1a). This zone is flanked by ter sequences bound by Tus protein, forming polar traps that restrict fork movement1. Highly expressed genes are transcribed co-directionally with replication, minimising the negative impact of conflict between transcription and replication6–8.
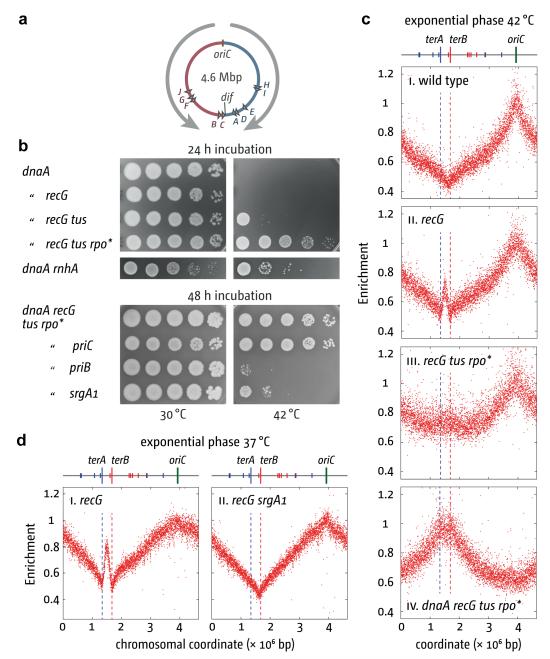
Figure 1. PriA triggers DnaA-independent chromosome replication in the absence of RecG.
(a) Replichore arrangement of the E. coli chromosome. Grey arrows indicate the normal direction of replication and polarity of major transcription. Triangles indicate ter sites.
(b) Genetic analysis of growth without DnaA (dnaA46 at 42°C). The strains were AU1054, AU1091, RCe205, RCe268, AU1066, RCe268, N8201, N8205 and N8206.
(c) and (d) Replication profiling of exponential phase cells. Normalized numbers of reads are plotted against chromosomal coordinate. Sequencing templates for (c) were from MG1655, N8226, RCe261 and RCe268, all cultured at 42°C, and for (e) were from N6576 and JJ1268 cultured at 37°C.
Initiation at oriC is controlled by DnaA9. However, replication can initiate independently of both DnaA and oriC. This stable DNA replication (SDR) is robust enough to sustain growth in strains lacking RNase HI, especially in minimal media10 (Supplementary Fig. 1). SDR is also elevated in strains lacking RecG3, but in this case appears unable to sustain growth (Fig. 1b)10. Forks must either be established in too small a fraction of cells or they cannot proceed far enough to complete chromosome duplication. Replication profiling of recG cells by deep sequencing suggested the latter. Logarithmically growing wild type cells showed a clearly defined origin and termination region (Fig. 1c and Supplementary Figs 4 and 8)11. The profile of recG cells on the other hand revealed a high incidence of additional initiation at a sharply focussed site in the terminus area (Fig. 1c–d, 2d, 3c). Initiation at this site is exceptionally frequent in the genetic background of strain AB1157; it approaches that at oriC (Supplementary Fig. 2a). Replication is evident in both directions, but limited to amplification of the terA–terB interval. However, a combination of two mutations, one (Δtus) eliminating Tus/ter traps, and the other (rpoB*35) destabilising transcription complexes12,13, enables replication to extend further, leading to a broader amplification (Fig. 1c). Significantly, these two mutations together enable recG cells to grow without DnaA or oriC (Fig. 1b and Supplementary Figs 1 and 3), demonstrating that SDR is now able to duplicate the entire chromosome. Viability assays on 17 independent log phase cultures revealed that 58% ± 17% of dnaA recG tus rpo* cells detected by plating on LB agar at 30°C establish colonies at 42°C, which is a much higher viability than we see with dnaA rnhA cells (Fig. 1b). The colonies are small, but continue growing (Fig. 1b, Supplementary Fig. 4). The oriC deletion derivative exhibits similar slow growth, with a doubling time of 100 min as opposed to 21 min for the wild type. Marker frequency analysis revealed an inverted profile for dnaA recG tus rpo* cells at 42°C, with a peak at the terminus and no evidence of initiation at oriC (Fig. 1c).
Figure 2. Replisome collision triggers DnaA-independent replication.
(a) Replication profile of exonuclease-depleted cells. Sequencing templates were from exponential phase N6953 cultured at 37°C.
(b) DnaA-independent growth. The strains analysed were RCe268, RCe384, RCe385 and RCe387.
(c) BrdU labelling of a fragment of NotI-digested DNA (b) located near the replication terminus relative to a distant reference fragment (a). The strains were RCe405, RCe409 and RCe418. Cultures of dnaA46 strains of the genotypes indicated were pulse labelled with BrdU at the indicated times after the shift to 42°C.
(d) Replication profiling showing effect of chromosome linearisation on replication. Sequencing templates were from N8226, RCe391 and RCe399 cultured at 37°C.
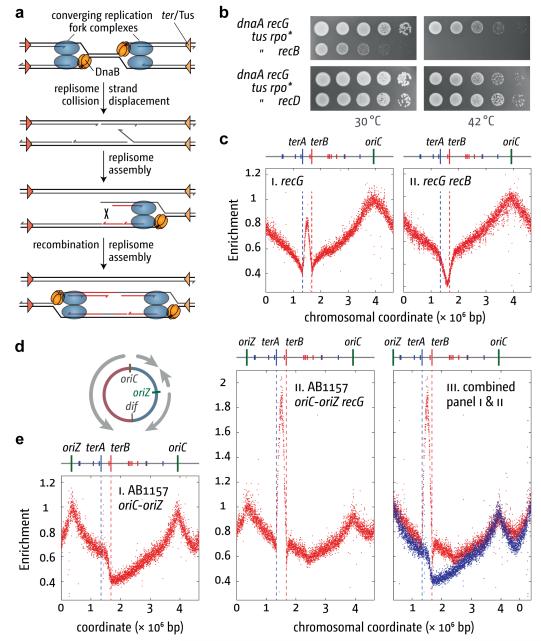
Figure 3. Effect of RecBCD activity and oriC duplication on DnaA-independent replication.
(a) Model illustrating how collision of replication forks within the termination area might lead to the formation of two new divergent forks via PriA-mediated replisome assembly and RecBCD-mediated recombination.
(b) DnaA-independent growth. The strains were RCe268, RCe435 and RCe437.
(c) Effect of recB on the replication profile of recG cells. Sequencing templates were from N8226 and AM1581 cultured at 37°C.
(d) Chromosome map of a double origin strain showing the positions of oriC and oriZ and illustrating where the forks assembled are likely to converge.
(e) Marker frequency analysis of chromosome replication in a double origin strain. Sequencing templates were from WX296 and RCe455 cultured at 37°C. Chromosome coordinates in panel iii are shifted to highlight the elevated marker frequency between oriC and oriZ.
Replication initiated in the terminus area in recG cells is abolished by mutations that modulate PriA helicase activity (srgA1, priA300) or which eliminate PriB (Fig. 1d and Supplementary Fig. 4). Both proteins facilitate DnaB loading and replisome assembly at branched DNAs4. The same mutations prevent dnaA recG tus rpo* cells from establishing colonies at 42°C. Eliminating the restart protein PriC does not (Fig. 1b). BrdU-labelling confirmed that priA300 reduces SDR (Supplementary Fig. 4b). Thus the DNA replication initiated at the terminus in cells lacking RecG must be the result of a PriA- PriB-mediated loading of DnaB at a branched DNA structure generated in this region. The ability of srgA1 to prevent initiation (Fig. 1d) indicates that the branched DNA is a 3′ flap. This allele encodes a PriA protein that has specifically lost the ability to unwind such a structure14. So 3′ flaps might normally be unwound by RecG3 and eliminated by ssDNA exonucleases, reducing the likelihood of PriA targeting the structure and triggering replication.
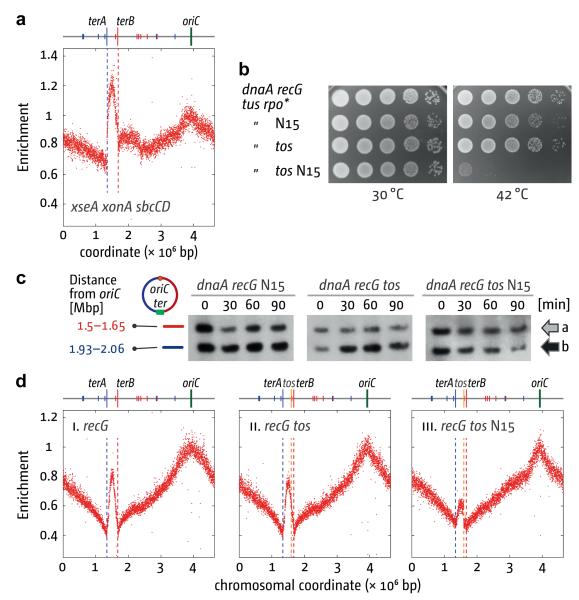
This scenario fits with the fact that 3′ ssDNA exonuclease activity is needed to keep recG cells alive, and that priA300 overcomes this requirement15. We profiled replication in log phase cells lacking ExoI, ExoVII and SbcCD and observed that replication initiates with high frequency in the terminus area despite the presence of RecG (Fig. 2a). This strengthens the notion that replication initiates at 3′ flaps. More importantly, it demonstrates that the re-replication observed without RecG is not a peculiarity of recG cells. Rather, the generation of branched DNA structures with a capacity to trigger replication appears to be a regular feature of the cell cycle triggered each time forks collide to complete replication.
We investigated what happens when forks are prevented from colliding using strains in which the chromosome is linearised by phage N15 telomerase at a tos site inserted near dif16 (Supplementary Fig. 5). Linearisation has no effect on the growth of wild type cells (Supplementary Fig. 5h)16. However, it abolishes the ability of dnaA recG tus rpo* cells to establish colonies at 42°C and reduces amplification of the terminus area (Figs. 2b–d). The residual amplification observed depends on forks established at oriC (Fig. 2c, Supplementary Fig. 6), indicating that it is most likely the result of some replication through the hairpin (Supplementary Fig. 5a and Supplementary Discussion). Amplification is evident on both sides of the tos site (Fig. 2d), making it unlikely that it is due to activation of a dormant origin or of a hotspot for initiation at R-loops10.
But why should fork collisions trigger initiation? We have proposed that when replisome complexes meet, the DnaB helicase of one fork often displaces the nascent leading strand of the opposing fork, generating a 3′ flap. DnaB would most likely collide with and dislodge the leading strand polymerase of the opposing fork to which the 3′ end of the nascent leading strand is engaged (Fig. 3a and Supplementary Discussion)3. This flap would be degraded by 3′ ssDNA exonucleases or converted by RecG to a 5′ flap and subsequently degraded by a 5′ exonuclease. Without RecG, a displaced 3′ flap might persist, providing a substrate for PriA-mediated replisome assembly. Progression of this fork generates a duplex arm with a free end, which invades the homologous duplex behind the fork via RecBCD- and RecA-mediated recombination5. This creates a D-loop that PriA could also target to establish another fork moving in the opposite direction. Divergent replication initiated via recombination is consistent with our finding that growth of dnaA recG tus rpo* cells at 42°C is prevented by inactivating RecA, the RuvABC Holliday junction resolvase or the RecBCD recombinase (Fig. 3b and data not shown). Eliminating just the ExoV activity of RecBCD has no such effect (Fig. 3b, recD derivative5). Marker frequency analysis confirmed that inactivating RecB prevents over-replication of the terminus. Indeed, marker frequency is reduced in this region (Fig. 3c). This is a feature of cells lacking RecBCD as it is also seen in a recB single mutant (Supplementary Fig. 7a). BrdU incorporation confirmed that inactivating RecB reduces SDR in recG cells (Supplementary Fig. 7b). Taken together, these data indicate that without RecG, fork collisions often lead to the generation of dsDNA ends that are then targeted by RecBCD. The viability of recG recB double mutant cells14 excludes the possibility that these ends are the result of fork or chromosome breakage.
Profiling cells carrying two copies of oriC (Fig. 3d) revealed that fork collisions trigger initiation of replication wherever forks meet. As expected17, both origins fire with equal efficiency on a population basis (Fig. 3e), leading to the termination of replication in two distinct zones, one in the area flanked by ter sites and a second in the shorter of the two intervals between the origins. The latter is shallower and less focussed, which is to be expected given the absence of Tus/ter traps, while the former is deeper and more sharply defined. There is also a clear step between terA and terB, consistent with the clockwise fork from oriZ being blocked by Tus bound at terB, having reached this point before meeting the anticlockwise fork from oriC.
In the recG derivative, we observed a strong increase in amplification of the terA–terB interval (Fig. 3e and Supplementary Discussion). More significantly, the termination region in the shorter interval is also noticeably shallower. This is precisely what one might expect if fork collisions were prone to trigger initiation. Without Tus/ter complexes to focus events, such initiation would lead to a wide region of amplification rather than to a distinct peak, as is seen when Tus is eliminated from a recG strain (cf Figs. 3c and 4a). Incomplete synchronisation of origin firing17 would also reduce collisions in this region. These data support the notion that replication fork collisions have the potential to trigger new replication wherever forks meet.
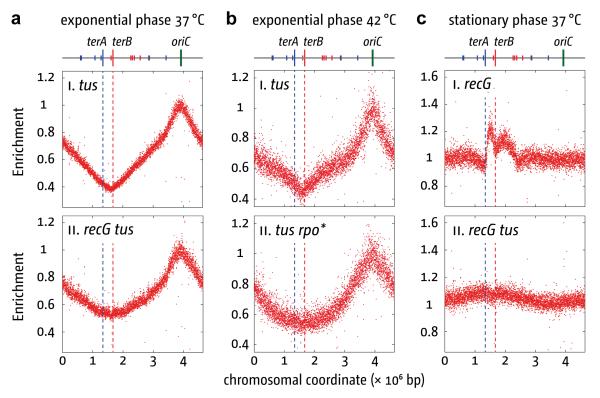
Figure 4. Effect of recG, tus and growth phase on chromosome marker frequencies.
(a) and (b) Effect of tus and rpo* on marker frequency in exponential phase cells cultured at 37°C (a) or 42°C (b). Sequencing templates were from N8227, N7957, JJ1378 and RCe260.
(c) Effect of recG and tus on marker frequency in stationary phase cells at 37°C. Sequencing templates were from cultures incubated with vigorous aeration until well after no further increase in cell density was detectable. The strains were, N6576 and N7957.
But how is replication maintained in dnaA recG tus rpo* cells without oriC firing? Shifting the cells from 30°C to 42°C would not affect existing forks. When these collide at the terminus and trigger initiation, the new forks would proceed towards oriC. Any subsequent fork collisions would create an opportunity to repeat the cycle, potentially ad infinitum. With no Tus/ter complexes, collisions might become increasingly random. However, a broad peak of increased terminal markers is evident (Fig. 1c), suggesting perhaps that initiation is not random. Therefore, it is significant that the replication profile of tus cells in exponential phase is identical to wild type (Figs 4a, b). Even in recG tus cells the termination area is still evident, though more broadly delineated (Fig. 4a). Thus, the sharp focussing of termination in wild type cells is likely due to some factor in addition to Tus/ter. We suggest that the polarity of transcription impedes forks moving beyond the terminus towards oriC. Broadening of the terminus area in tus rpo* cells supports this idea (Fig. 4b). However, rpo* is unlikely to eliminate this problem entirely. Therefore, dnaA recG tus rpo* cells with forks moving towards oriC, having duplicated the terminus, might be overrepresented, giving the marker frequency observed (Fig. 1c). Also, if forks moving towards oriC failed to complete duplication of the chromosome because of the conflicts with transcription, DNA from the affected (dead) cells would further bias the profile in favour of terminal markers.
Without means to control initiation, these cells lack the ability to co-ordinate chromosome replication with cell growth. New rounds of replication initiate only following replication fork collision, preventing the increased rate of growth and division made possible in wild type cells by firing oriC before the previous round of replication has been completed3. So, what happens when growth ceases? Marker frequency analysis of recG cells grown to saturation revealed an overrepresentation of the terminus (Fig. 4c), as if they had ceased growing with forks stalled at Tus/ter. An even broader elevation of terminal markers is observed in saturated recG tus cells. Coupled with its absence in tus cells (Supplementary Fig. 8), this persistence of forks suggests that once stationary phase dnaA recG tus rpo* cells are diluted in fresh growth medium replication resumes at the stalled forks.
Taken together, the studies reported here demonstrate that RecG and 3′ ssDNA exonucleases play critical roles in limiting re-replication of the already replicated DNA. It is significant that re-replication is blocked by mutations (priA300, srgA1 and ΔpriB) that suppress the recG mutant phenotype18. This suggests that such replication has pathological consequences that destabilise the genome.
The single fork collision typical of the average E. coli cell cycle is a stark contrast to the multiple events occurring in eukaryotes. Recent studies have revealed that the final stages of replication in eukaryotic cells have pathogenic potential19,20. RecG is absent, but several studies have reflected on the ability of human and yeast helicases to remodel branched DNA structures in a manner reminiscent of RecG21–24. Several ssDNA exonucleases have also been linked with replication, both nuclear and mitochondrial25. The human mitochondrial nuclease MGME1, loss of which is associated with multi-systemic mitochondrial disease, has been shown to process displaced ssDNA flaps25. So, as with E. coli, eukaryotic helicases and ssDNA exonucleases may well turn out to have important roles in making sure that replication is completed without triggering pathological events that destabilise the genome. Indeed, limiting the incidence of pathological events at termination may prove to be as crucial to genome stability as the prevention of re-initiation at replication origins2.
MATERIAL AND METHODS
Bacterial strains and general methods
The Escherichia coli strains used in this study are derivatives of E. coli K-12 MG1655 unless stated otherwise (Table S1). The dnaA46 allele encodes a thermosensitive DnaA protein that is inactive at 42°C. The oriC deletion allele tagged with a sequence encoding resistance to kanamycin (oriC::kan) was generated by the single-step gene replacement method27. LB broth and 56/2 salts media have been cited elsewhere12,28. Strain constructions, genetic manipulations and assays for bacterial and phage growth employed standard microbiological materials and protocols, as described or cited18,29. For assessing growth without DnaA, cultures of dnaA46 constructs grown at 30°C to an A650 of 0.4 (~ 2 × 108 cells ml−1) were diluted in 10-fold steps from 10−1 to 10−5 before spotting 10 μl samples of each dilution on LB agar. Duplicate plates were incubated at 30°C and 42°C. Pictures were taken after 24 h unless stated otherwise.
Replication profiling by marker frequency analysis
Samples from cultures of a strain grown to an A650 of ~1.2 in LB broth were diluted 400-fold in fresh broth and incubated with vigorous aeration until the A650 reached 0.4 at the temperature indicated. Samples from these exponential phase cultures were frozen in liquid nitrogen at this point for subsequent DNA extraction. Incubation of the remaining culture was continued until several hours after the culture had saturated and showed no further increase in the A650. A further sample (stationary phase) was frozen at this point. DNA was then extracted using the GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich). Marker frequency analysis was performed using AB SOLiD sequencing to measure sequence copy number. Enrichment of uniquely mapping sequence tags, in 1 kb windows, was calculated for an exponentially growing (replicating) sample relative to a non-replicating stationary phase wild type sample to correct for differences in read depth across the genome and to allow presentation of the data as a marker frequency, as described previously26.
5-Bromo-2′-deoxyuridine (BrdU) labelling
BrdU labelling and detection via immunostaining was essentially as described30. Cells were grown in 56/2 salts supplemented with 0.2% casamino acids and 0.32% glucose to an A650 of 0.2. The culture was split into various 2 ml aliquots, 5-Bromo-2′-deoxyuridine (BrdU, Sigma) added to the first aliquot to 20 μg/ml and the cultures shifted to 42°C. At the times indicated BrdU was added to one of the remaining aliquots. The aliquots were labelled with BrdU for 8 min, pelleted and resuspended in 85 μl TEE buffer (10 mM Tris • HCl, 10 mM EGTA, 100 mM EDTA, pH 8.0), containing 0.05% lauroylsarcosine and 0.5% SDS. 85 μl of liquid 1.4% low melting point agarose was added and the mixture solidified in a disposable plug former (Bio-Rad) at 4°C. Plugs were treated with 10 mg/ml lysozyme in 3 ml TEE buffer containing 0.05% lauroylsarcosine and 0.5% SDS for 2 h at 37°C and then at 52°C overnight with 5 mg/ml proteinase K in 3 ml TEE containing 1% SDS. Plugs were washed in TEE for 30 min at 37°C, treated with 1 mM phenylmethane sulphonyl fluoride (freshly prepared as 100 mM stock solution in methanol) in fresh TEE for 1 h at 37°C, washed in fresh TEE for 30 min at 37°C and finally in 0.1 × TEE for 30 min at 37°C. The plugs were subsequently transferred into 300 μl restriction enzyme buffer and incubated for 30 min at room temperature, the buffer changed and 25 u of NotI (NEB) added. Chromosomal DNA was digested overnight and the fragments separated on a 0.8% agarose gel (Bio-Rad pulse field certified agarose) in 0.5 × TBE using a CHEF Mapper PFGE system (Bio-Rad), running with a gradient voltage of 6 V/cm, an included angle of 120°, and initial and final switch times of 1.65 and 32.45 sec, respectively, with a run time of 20 h at 14°C. DNA was transferred to a Hybond-N+ Membrane (GE Healthcare) by alkaline vacuum transfer and UV crosslinked (120 mJ/cm2). Blocking was achieved with TBS Tween (50 mM Tris • HCl, 150 mM NaCl, pH 8.0, 0.5% Tween 20) containing milk powder (2%). After blocking the membrane was incubated for 2 h in the presence of mouse anti-BrdU antibody (Santa Cruz), diluted 1:5000 in TBS Tween. Horse radish peroxidase conjugated secondary antibody (goat anti-mouse, Bio-Rad) was used at a dilution in TBS Tween of 1:10,000 for 1.5 h. The membrane was incubated with ECL Plus Western Blotting Detection Reagents (GE Healthcare) and the signal visualised either by exposure to X-Omat UV Plus film (Kodak) or by the ChemiDoc chemiluminescence detection system (Bio-Rad).
Chromosome linearisation
Linearisation of the E. coli chromosome was achieved using the bacteriophage N15 telomere generating system (Supplementary Fig. 5). N15 is a temperate E. coli phage, which does not integrate into the chromosome during lysogenisation31. Instead, the N15 telomerase, TelN, cleaves and processes a specific phage DNA sequence called tos, generating a linear chromosome with hairpin ends (Supplementary Fig. 5). When replication forks reach these structures, the nascent leading and lagging strands are sealed, forming a chromosomal dimer. This restores two tos sites, which are immediately re-processed by TelN16,31, thus allowing segregation of two linear chromosomes. For our studies, we exploited E. coli derivatives that carry a tos site inserted near the dif locus, close to terC16. These were lysogenised with phage N15 to linearise the chromosome (Supplementary Fig. 5). Preparation of phage N15 stocks and E. coli lysogens followed methods and protocols described for phage λ29.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Takashi Horiuchi and David Sherratt for E. coli strains, Akeel Mahdi for help with DNA extractions, Sunir Malla and Martin Blythe for deep sequencing, Shemsi Demolli and Darja Ivanova for control experiments, and Carol Buckman and Lynda Harris for assistance. This work was supported by grants from the MRC (R.G.L., G0800970), The Leverhulme Trust (C.J.R.) and the BBSRC (C.A.N., BB/E023754/1).
Footnotes
SUPPLEMENTAL INFORMATION Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
AUTHOR STATEMENTS Deep sequencing data are deposited with NCBI Gene Expression Omnibus under accession number GSE41975. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
REFERENCES
- 1.Reyes-Lamothe R, Wang X, Sherratt D. Escherichia coli and its chromosome. Trends Microbiol. 2008;16:238–245. doi: 10.1016/j.tim.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Diffley JF. Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc Lond B Biol Sci. 2011;366:3545–3553. doi: 10.1098/rstb.2011.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudolph CJ, Upton AL, Briggs GS, Lloyd RG. Is RecG a general guardian of the bacterial genome? DNA Repair (Amst) 2010;9:210–223. doi: 10.1016/j.dnarep.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Gabbai CB, Marians KJ. Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival. DNA Repair (Amst) 2010;9:202–209. doi: 10.1016/j.dnarep.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 6.Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet. 2012;13:204–214. doi: 10.1038/nrg3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. Accelerated gene evolution through replication-transcription conflicts. Nature. 2013;495:512–515. doi: 10.1038/nature11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudolph CJ, Dhillon P, Moore T, Lloyd RG. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair (Amst) 2007;6:981–993. doi: 10.1016/j.dnarep.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Mott ML, Berger JM. DNA replication initiation: mechanisms and regulation in bacteria. Nat Rev Microbiol. 2007;5:343–354. doi: 10.1038/nrmicro1640. [DOI] [PubMed] [Google Scholar]
- 10.Kogoma T. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol. Molec. Biol. Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skovgaard O, Bak M, Lobner-Olesen A, Tommerup N. Genome-wide detection of chromosomal rearrangements, indels, and mutations in circular chromosomes by short read sequencing. Genome Res. 2011;21:1388–1393. doi: 10.1101/gr.117416.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–543. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregg AV, McGlynn P, Jaktaji RP, Lloyd RG. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell. 2002;9:241–251. doi: 10.1016/s1097-2765(02)00455-0. [DOI] [PubMed] [Google Scholar]
- 15.Rudolph CJ, Mahdi AA, Upton AL, Lloyd RG. RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase activity in Escherichia coli. Genetics. 2010;186:473–792. doi: 10.1534/genetics.110.120691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui T, et al. Escherichia coli with a linear genome. EMBO Rep. 2007;8:181–187. doi: 10.1038/sj.embor.7400880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Lesterlin C, Reyes-Lamothe R, Ball G, Sherratt DJ. Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc Natl Acad Sci U S A. 2011;108:E243–250. doi: 10.1073/pnas.1100874108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahdi AA, Briggs GS, Lloyd RG. Modulation of DNA damage tolerance in Escherichia coli recG and ruv strains by mutations affecting PriB, the ribosome and RNA polymerase. Mol Microbiol. 2012;86:675–691. doi: 10.1111/mmi.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fachinetti D, et al. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinacher R, Osman F, Dalgaard JZ, Lorenz A, Whitby MC. The DNA helicase Pfh1 promotes fork merging at replication termination sites to ensure genome stability. Genes Dev. 2012;26:594–602. doi: 10.1101/gad.184663.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ralf C, Hickson ID, Wu L. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 22.Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Betous R, et al. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Killen MW, Stults DM, Wilson WA, Pierce AJ. Escherichia coli RecG functionally suppresses human Bloom syndrome phenotypes. BMC Mol Biol. 2012;13:33. doi: 10.1186/1471-2199-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornblum C, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller CA, Nieduszynski CA. Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res. 2012;22:1953–1962. doi: 10.1101/gr.139477.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 29.Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 30.Rudolph CJ, Upton AL, Lloyd RG. Replication fork collisions cause pathological chromosomal amplification in cells lacking RecG DNA translocase. Mol Microbiol. 2009;74:940–955. doi: 10.1111/j.1365-2958.2009.06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rybchin VN, Svarchevsky AN. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol Microbiol. 1999;33:895–903. doi: 10.1046/j.1365-2958.1999.01533.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.