Abstract
Psychrophiles thriving permanently at near-zero temperatures synthesize cold-active enzymes to sustain their cell cycle. Genome sequences, proteomic, and transcriptomic studies suggest various adaptive features to maintain adequate translation and proper protein folding under cold conditions. Most psychrophilic enzymes optimize a high activity at low temperature at the expense of substrate affinity, therefore reducing the free energy barrier of the transition state. Furthermore, a weak temperature dependence of activity ensures moderate reduction of the catalytic activity in the cold. In these naturally evolved enzymes, the optimization to low temperature activity is reached via destabilization of the structures bearing the active site or by destabilization of the whole molecule. This involves a reduction in the number and strength of all types of weak interactions or the disappearance of stability factors, resulting in improved dynamics of active site residues in the cold. These enzymes are already used in many biotechnological applications requiring high activity at mild temperatures or fast heat-inactivation rate. Several open questions in the field are also highlighted.
1. Introduction
“Coping with our cold planet” [1], the title of this recent review unambiguously stresses a frequently overlooked aspect: the Earth's biosphere is predominantly cold and permanently exposed to temperatures below 5°C. Such low mean temperature mainly arises from the fact that 71% of the Earth's surface is covered by oceans that have a constant temperature of 2–4°C below 1000 m depth, irrespective of the latitude. The polar regions account for another 15%, to which the glacier and alpine regions must be added as well as the permafrost representing more than 20% of terrestrial soils. Although inhospitable, all these low temperature biotopes have been successfully colonized by cold-adapted organisms (Figure 1). Psychrophiles thrive in permanently cold environments (in thermal equilibrium with the medium) and even at subzero temperatures in supercooled liquid water. Such extremely cold conditions are encountered, for instance, in salty cryopegs at −10°C in the Arctic permafrost [2, 3], in the brine veins between polar sea ice crystals at −20°C [4–6], or in supercooled cloud droplets [7, 8]. Unusual microbiotopes have also been described, such as porous rocks in Antarctic dry valleys hosting microbial communities surviving at −60°C [9, 10]. Cryoconite holes on glacier surfaces represent another permanently cold biotope hosting complex microbial communities [11, 12]. These examples highlight the unsuspected ability of psychrophiles to adapt to low temperatures.
Figure 1.

Harsh conditions and beautiful landscape. Extremely cold environments are not sterile as exemplified by polar fish thriving below the icepack or by active microorganisms found between sea ice crystals (courtesy of Dr. J. C. Marx).
Psychrophiles include a large range of representatives from all three domains: Bacteria, Archaea, and Eukarya. Psychrophiles are mainly represented by microorganisms such as bacteria [13–16], archaea [17], algae [18], or yeast [19] but also by plants and animals [20, 21]. Notably, polar fish thriving beneath the icepack are the biggest psychrophiles [22–24]. Glacier ice worms are also worth mentioning as they complete their life cycle exclusively in glacier ice [25]. All these examples illustrate that psychrophiles are the most abundant extremophiles in terms of biomass, diversity, and distribution.
Psychrophiles do not merely survive or endure such extremely inhospitable conditions but are irreversibly adapted to these environments, as most of them are unable to grow at mild (or mesophilic) temperatures. These extremophiles represent much more than a biological curiosity. Psychrophiles and their biomolecules have already found various applications but their role in biogeochemical cycles is frequently underestimated: the huge biotope volume of oceans represents the largest reservoir of psychrophiles contributing to matter cycling. Various aspects of their ecology, physiology, and molecular adaptations have been reported in previous publications [26–30].
Life in cold environments requires a vast array of adaptive features at nearly all levels of the cell architecture and function. Indeed, cold exerts severe physicochemical constraints on living organisms including increased water viscosity, decreased molecular diffusion rates, reduced biochemical reaction rates, perturbation of weak interactions driving molecular recognition and interaction, strengthening of hydrogen bonds that, for instance, stabilize inhibitory nucleic acid structures, increased solubility of gases, and stability of toxic metabolites as well as reduced fluidity of cellular membranes [1, 31, 32]. Adaptive features to these constraints are frequently detected in the genome sequence of psychrophilic microorganisms [33–36]. Indeed, several genomes from psychrophilic bacteria and archea have been sequenced [37–46] and recently also the first genome of a polar eukaryotic microalga has been reported [47]. In this respect, recent improvements in high throughput sequencing have produced large amounts of data but most of these genomes remain to be analyzed [36, 48–55].
However, a key determinant of adaptation to life in the cold lies in the protein function that drives microbial metabolism and cell cycle. Pioneering studies of psychrophiles at the molecular level were mainly focused on cold-active enzymes because this aspect was regarded as a prerequisite to the environmental adaptation. It has been shown that the high level of specific activity at low temperature of cold-adapted enzymes is a key adaptation to compensate for the exponential decrease in chemical reaction rates as the temperature is reduced. Such high biocatalytic activity arises from the disappearance of various noncovalent stabilizing interactions, resulting in an improved flexibility of the enzyme conformation. It should be noted that this adaptive feature is genetically encoded within the protein sequence and results from a long-term selection. As a general picture, psychrophilic enzymes are all faced to a main constraint, to be active at low temperatures, but the ways to reach this goal are quite diverse. The main functional and structural adaptive properties of cold-active enzymes are presented here as well as the recent advances related to their synthesis, folding and biotechnological applications. This paper is based on updates of previous publications [56–64].
2. Protein Synthesis and Folding
For many years, protein synthesis and protein folding have been regarded as temperature-sensitive cellular processes that severely restrict microbial growth at low temperature in the absence of specific adaptations [65–70]. Despite this well-recognized limitation, the challenge of protein synthesis and folding in psychrophiles has been addressed only recently, mainly via proteomics and, to a lesser extent, transcriptomics [36, 62]. These approaches have produced a huge amount of data with, however, contrasted patterns of cold adaptation [43, 55, 71–95]. Indeed, proteins differentially expressed at low temperature, either cold induced or cold repressed, do not constitute a conserved set of proteins in terms of identification and expression levels in these organisms. It has been suggested that cold adaptation superimposes on preexisting cellular organization and, accordingly, that the adaptive strategies may differ between the various psychrophilic organisms [35, 62]. Nevertheless, and as far as protein synthesis and folding are concerned, some general trends have been noted.
2.1. Translation
In the case of the Antarctic bacterium P. haloplanktis, 30% of the upregulated proteins at 4°C were found to be directly related to protein synthesis [83]. It was concluded that protein synthesis may be a rate-limiting step for growth in the cold, therefore inducing a compensatory cellular response. A similar pattern is indeed observed in several cold-adapted bacteria [76, 78, 80, 84, 85], which overexpress proteic components involved in translation. More specifically, the synthesis of ribosomal proteins and of RNA chaperones [96, 97] appears to be stimulated at low temperature [43, 55, 75, 78–81, 84]. In the archaeon M. burtonii, a notable feature is the upregulation of genes involved in maintaining RNA in a state suitable for translation and for enabling translation initiation [86]. At the genome level, an interesting observation is the relatively high number of rRNA genes and of tRNA genes (up to 106 genes, sometimes organized in long runs of repeated sequences), at least in P. haloplanktis [39], C. psychrerythraea [40], and P. ingrahamii [42]. This could reflect the need for a high capacity for translation in the cold. In addition, RNA helicases have been found to be overexpressed at low temperature in many psychrophilic microorganisms such as M. burtonii [98], E. sibiricum [43], S. alaskensis [84], P. arcticus [80, 81], S. denitrificans [55], and P. haloplanktis [83]. These helicases may help to unwind the RNA secondary structures for efficient translation in the cold [99]. These examples indicate that psychrophiles have indeed evolved adaptive mechanisms to optimize protein synthesis at low temperature.
2.2. Folding Assistance
Some interesting reports have revealed that cold-adapted chaperones such as DnaK [100, 101] and GroEL [102–104] expressed in E. coli provide cold-resistance and improve growth of the mesophilic bacterium at low temperature, therefore highlighting the crucial involvement of chaperones in temperature adaptation of microorganisms.
Amongst the various chaperones involved in protein folding assistance [105], the ribosome-bound trigger factor (TF) deserves a special mention. TF is the first chaperone interacting cotranslationally with virtually all nascent polypeptides synthesized by the ribosome, and most small proteins (~70% of total) may fold rapidly upon synthesis without further assistance. Interestingly, TF is a cold shock protein in E. coli [106], whereas the other chaperones are well-known heat shock proteins (HSPs). This significant distinction appears to be of prime importance for psychrophiles. As a matter of fact, overexpression of TF at low temperature has been observed in several cold-adapted microorganisms [75, 79, 83, 87] while downregulation of HSP chaperones has been noted [43, 73, 88]. Considering such imbalance in the chaperone machinery (Figure 2), it appears that TF rescues the chaperone function and should be regarded as the primary chaperone in these strains [83]. The downregulation of HSP chaperones in the cold suggests that these folding assistants are mainly synthesized at transiently higher environmental temperatures [62].
Figure 2.
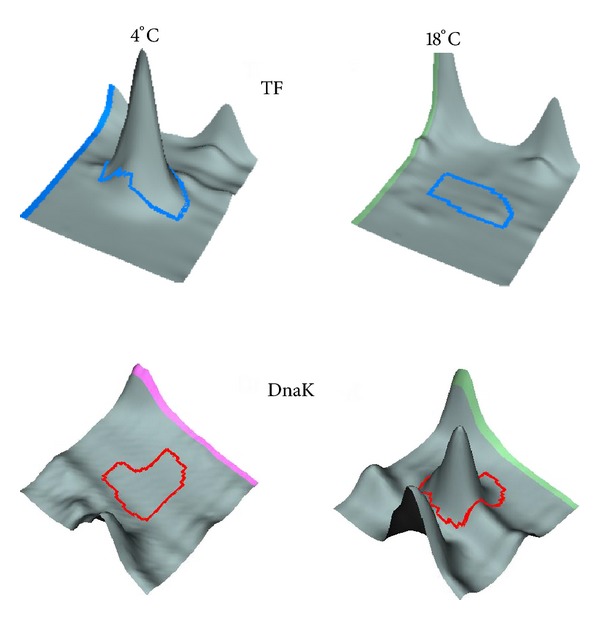
Comparative analysis of electrophoretic spots containing the trigger factor TF and DnaK from P. haloplanktis grown at 4°C (left panels) and 18°C (right panels) illustrating the overexpression of TF (a cold shock protein) and the downregulation of DnaK (a heat shock protein) at low temperature. Adapted from [83].
By contrast, repression of TF synthesis or upregulation of HSP chaperonins has been also reported [80, 81, 84]. Furthermore, S. alaskensis possesses two sets of dnaK-dnaJ-grpE gene clusters (DnaK and its cochaperones). Quantitative proteomics has suggested that one set functions as a low temperature chaperone system, whereas the other set functions at higher growth temperatures [84]. Obviously, distinct strategies have been adopted by psychrophiles to assist proper protein folding.
2.3. Proline Isomerization
The cis-trans-isomerization of the peptidyl-prolyl bond, that is, the peptide bond preceding a proline residue (Figure 3), is an intrinsically slow reaction. As a result, proline isomerization is a rate-limiting step for the folding of most proteins [107]. More specifically for psychrophilic proteins, the spontaneous rate of the proline isomerization reaction should also be slowed down at low temperature. Interestingly, many cold-adapted proteins tend to possess a reduced proline content [57, 60, 108] that should attenuate the negative effect of proline isomerization on folding.
Figure 3.
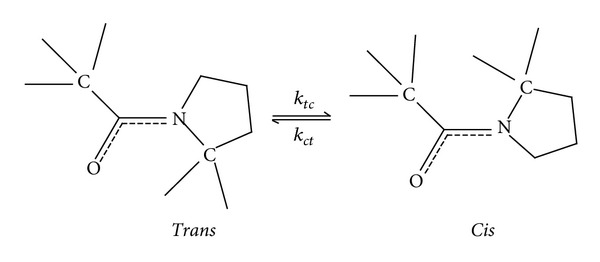
Trans- and cis- isomers of a peptidyl-prolyl peptide bond. Reprinted with permission from [110] © 2009 American Chemical Society.
In addition, living cells are equipped with specialized catalysts, prolyl isomerases (PPiases), which accelerate the isomerization process. Very significantly, PPiases are overexpressed at low temperature in the proteome of most psychrophilic bacteria analyzed so far, and sometimes at very high levels. This involves cytoplasmic PPiases from the cyclophilin family [71, 72, 80, 83], a parvulin-type PPiase [84], and FKBP-type PPiases [71, 109]. Furthermore, the above-mentioned trigger factor contains a PPiase domain and is overexpressed in several psychrophiles. Such recurrent observations strongly suggest that protein folding is a rate-limiting step for psychrophiles, which induces a cellular response aimed at facilitating and accelerating the slowest event in the acquisition of the biologically active conformation of proteins.
2.4. Folding Rate
Following synthesis by the ribosome, the rate of protein folding is intuitively expected to be slowed down at low temperature. In the case of thermophilic proteins, it has been shown that their rate of folding does not differ from the folding rate of mesophilic proteins, whereas thermophilic proteins unfold very slowly therefore revealing the kinetic origin of their stability [111–113]. Such comparative analyses are currently lacking for psychrophilic proteins. Nevertheless, the folding and unfolding rate constants have been recorded for a psychrophilic alpha-amylase and for its stabilized mutants [114]. It was found that despite large differences in stability, the folding rate of these proteins is similar, whereas the unfolding rates correlate with stability. It seems therefore that the folding rate of this cold-adapted enzyme is not altered. There is an obvious need for deeper biophysical studies on protein folding at low temperature.
3. Psychrophilic Enzymes
The previous sections have highlighted the current issues in protein folding at low temperature. The next sections will mainly concentrate on the enzyme function in psychrophiles which occupies a central role in cold adaptation of living organisms.
3.1. 3D Structures of Cold-Active Enzymes
Crystal structures of psychrophilic enzymes were of prime importance to investigate the properties of these cold-active catalysts. Table 1 provides an overview of representative crystal structures. To date, only one report of assignments for NMR structure determination has been published for a thiol-disulfide oxidoreductase from an Antarctic bacterium [115]. Table 1 illustrates that the available structures cover many pathways of the cell metabolism while membrane proteins, for instance, are still lacking. It is worth mentioning that all these structures are closely homologous to mesophilic counterparts: cold-active enzymes only differ by discrete changes that are responsible for their specific properties.
Table 1.
Representative psychrophilic enzymes of known crystal structure.
| Cold-adapted protein | Source | PDB entry | References |
|---|---|---|---|
| Alpha-amylase | Antarctic bacterium Pseudoalteromonas haloplanktis | 1AQH | [116, 117] |
| Xylanase | Antarctic bacterium Pseudoalteromonas haloplanktis | 1H12 | [118] |
| Cellulase | Antarctic bacterium Pseudoalteromonas haloplanktis | 1TVN | [119] |
| Superoxide dismutase | Antarctic bacterium Pseudoalteromonas haloplanktis | 3LJF | [120] |
| S-Formylglutathione hydrolase | Antarctic bacterium Pseudoalteromonas haloplanktis | 3LS2 | [121] |
| Ca2+ Zn2+ protease | Antarctic bacterium Pseudomonas sp. | 1G9K | [122] |
| Beta-lactamase | Antarctic bacterium Pseudomonas fluorescens | 2QZ6 | [123] |
| Citrate synthase | Antarctic bacterium Arthrobacter sp. | 1A59 | [124] |
| Beta-galactosidase | Antarctic bacterium Arthrobacter sp. | 1YQ2 | [125] |
| Subtilisine | Antarctic bacterium Bacillus sp. | 2GKO | [126] |
| Aliphatic amidase | Antarctic bacterium Nesterenkonia sp. | 3HKX | [127] |
| Alkaline phosphatase | Antarctic bacterium | 2IUC | [128] |
| Aminopeptidase | Arctic bacterium Colwellia psychrerythraea | 3CIA | [129] |
| Phenylalanine hydroxylase | Arctic bacterium Colwellia psychrerythraea | 2V27 | [130] |
| Malate dehydrogenase | Arctic bacterium Aquaspirillum arcticum | 1B8P | [131] |
| Serine proteinase | Arctic bacterium Vibrio sp. | 1SH7 | [132] |
| Isocitrate dehydrogenase | Arctic bacterium Desulfotalea psychrophila | 2UXQ | [133] |
| Aspartate carbamoyltransferase | Deep-sea bacterium Moritella profunda | 2BE7 | [134] |
| Adenylate kinase | Bacteria Bacillus globisporus and Marinibacillus marinus | 1S3G, 3FB4 | [135, 136] |
| Triose-phosphate isomerase | Marine bacterium Vibrio marinus | 1AW1 | [137] |
| Ca2+ Zn2+ protease | Marine bacterium Flavobacterium sp. | 1U1R | [138] |
| Tyrosine phosphatase | Bacterium Shewanella sp. | 1V73 | [139] |
| Catalase | Bacterium Vibrio salmonicida | 2ISA | [140] |
| Endonuclease I | Bacterium Vibrio salmonicida | 2PU3 | [141] |
| Lipase | Bacterium Photobacterium lipolytica | 2ORY | [142] |
| Superoxide dismutase | Bacterium Aliivibrio salmonicida | 2W7W | [143] |
| Alkaline phosphatase | Bacteria Vibrio sp. and Shewanella sp. | 3E2D, 3A52 | [144, 145] |
| Alkaline phosphatase | Arctic shrimp Pandalus borealis | 1K7H | [146] |
| Lactate dehydrogenase | Antarctic icefish Champsocephalus gunnari | 2V65 | [147] |
| Trypsin | Atlantic salmon Salmo salar | 2TBS | [148] |
| Elastase | Atlantic salmon Salmo salar | 1ELT | [149] |
| Pepsin | Atlantic cod Gadus morhua | 1AM5 | [150] |
| Uracil-DNA glycosylase | Atlantic cod Gadus morhua | 1OKB | [151] |
3.2. The Low Temperature Challenge
The challenge for a thermophilic enzyme is easily understood: to remain stable and active at high temperatures. By contrast, the challenge for a psychrophilic enzyme is more subtle. Low temperatures strongly reduce the rate of nearly all enzyme-catalyzed reactions and, furthermore, slow down molecular motions associated with protein function. As far as activity is concerned, the catalytic constant k cat corresponds to the maximum number of substrate molecules converted to product per active site per unit of time, and the temperature dependence of the catalytic rate constant is given by the relation
| (1) |
In this equation, κ is the transmission coefficient generally close to 1, k B is the Bolzmann constant (1.38 × 10−23 J K−1), h the Planck constant (6.63 × 10−34 J s), R the universal gas constant (8.31 J K−1 mol−1), and ΔG # the free energy of activation or the variation of the Gibbs energy between the activated enzyme-substrate complex ES* and the ground state ES. Accordingly, the activity constant k cat is exponentially dependent on the temperature. As a rule of thumb, for a biochemical reaction catalyzed by a mesophilic enzyme (from bacteria or warm-blooded animals), a drop in temperature from 37°C to 0°C results in a 20 to 80 times lower activity. This is the main factor preventing the growth of nonadapted organisms at low temperatures.
The effect of temperature on the activity of psychrophilic and mesophilic enzymes is illustrated in Figure 4. Equation (1) is only valid for the exponential rise of activity with temperature on the left limb of the curves. Models have been proposed to simulate the effects of heat-inactivation on activity [152–154] and to take the viscosity of the medium into account [108]. This figure reveals at least two basic features of cold adaptation. (i) In order to compensate for the slow reaction rates at low temperatures, psychrophiles synthesize enzymes having an up to tenfold higher specific activity in this temperature range. This is in fact the main physiological adaptation to cold at the enzyme level; (ii) The temperature for apparent maximal activity for cold-active enzymes is shifted towards low temperatures, reflecting the weak stability of these proteins and their unfolding and inactivation at moderate temperatures. A compilation of 155 characterized psychrophilic enzymes has been reported [155] and can be consulted (http://www2.ulg.ac.be/biochlab/publications_022.htm).
Figure 4.
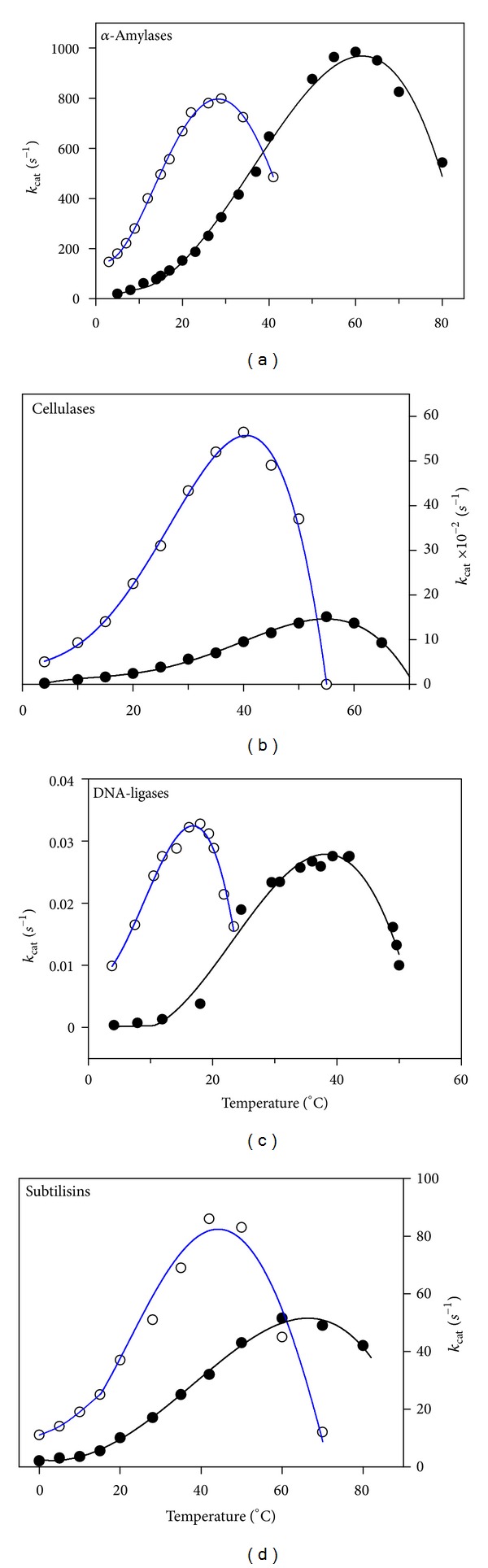
Temperature dependence of activity. The activity of psychrophilic (open symbols, blue lines) and mesophilic (closed symbols) enzymes recorded at various temperatures illustrates the main properties of cold-adapted enzymes: cold activity and heat lability. Adapted from [32, 156–158].
Many observations similar to those shown in Figure 4 have suggested relationships between the activity of the enzyme, the flexibility of the protein, and its stability. Indeed, the high activity at low temperatures seems to arise from an increased flexibility of the protein structure, especially at temperatures that strongly slow down molecular motions, but the consequence of this improved mobility of the protein structure is of course a weak stability. These tradeoffs are developed in the next sections.
4. Cold-Adapted Activity
4.1. Heat-Labile and Unstable Enzymes
Most psychrophilic enzymes share at least one property: a heat-labile activity, irrespective of the protein structural stability. Furthermore, the active site appears to be the most heat-labile structural element of these proteins [158–161]. Figure 5 illustrates this significant difference between the stability of the active site and the stability of the structure. The lower panel shows the stability of the structure as recorded by fluorescence. As expected, the structure of the cold-active enzyme is less stable than that of the mesophilic and thermophilic counterparts. In the upper panel, the activity is recorded under the same experimental conditions and it can be seen that the mesophilic and thermophilic enzymes initiate heat-inactivation when the protein starts to unfold. By contrast, activity of the cold-active enzyme is lost before the protein unfolds. This means that the active site is even more heat-labile than the whole protein structure. All these aspects point to a very unstable and flexible active site and illustrate a central concept in cold adaptation: localized increases in flexibility at the active site are responsible for the high but heat-labile activity [162], whereas other regions of the enzyme might or might not be characterized by low stability when not involved in catalysis [163]. For instance, psychrophilic carbonic anhydrase [164] and isocitrate dehydrogenase [133] are highly stable enzymes with, however, improved flexibility in regions driving catalysis. In multidomain psychrophilic enzymes containing a catalytic and a noncatalytic domain, the catalytic domain is always heat labile, whereas the noncatalytic domain can be as stable as mesophilic proteins [165–167].
Figure 5.
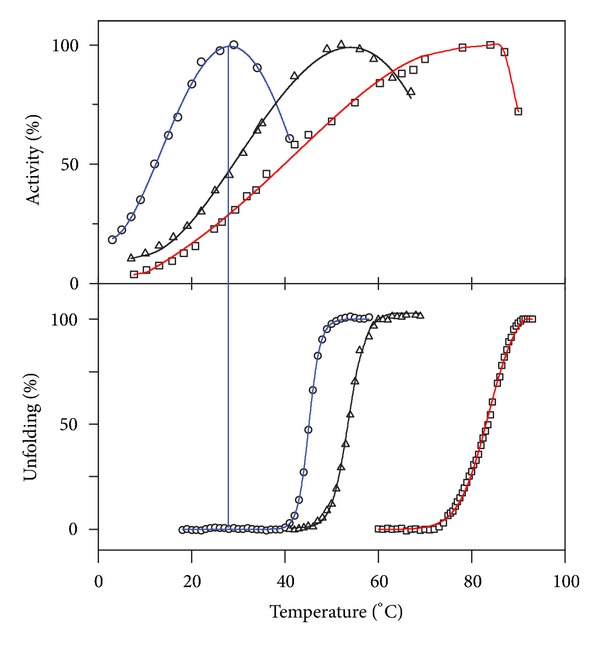
Inactivation and unfolding of psychrophilic enzymes. The activity of most psychrophilic enzymes (upper panel, blue curve) is inactivated by temperature before unfolding of the protein structure (lower panel) illustrating the pronounced heat lability of the active site. By contrast, inactivation of mesophilic (black curves) or thermophilic (red curves) enzymes closely corresponds to the beginning of protein unfolding. Adapted from [160].
4.2. Active Site Architecture
With the availability of crystal structures, the cold-active catalytic centers of psychrophilic enzymes were carefully scrutinized. The first basic observation is that all side chains involved in the catalytic mechanism are strictly conserved. This aspect is not really surprising as the specific reaction mechanism of enzymes is not prone to drastic variation. Furthermore, comparison of the first X-ray structure of a psychrophilic enzyme, a cold-active α-amylase [116, 117], and of its closest mesophilic structural homologue, both in complex with an inhibitor mimicking the transition state intermediate [168, 169], has revealed that all 24 residues forming the catalytic cleft are strictly conserved in the cold-active α-amylase (Figure 6). This outstanding example of active site identity demonstrates that the specific properties of psychrophilic enzymes can be reached without any amino acid substitution in the reaction center. As a consequence, changes occurring elsewhere in the molecule are responsible for optimization of the catalytic parameters and improved dynamic of active site residues.
Figure 6.
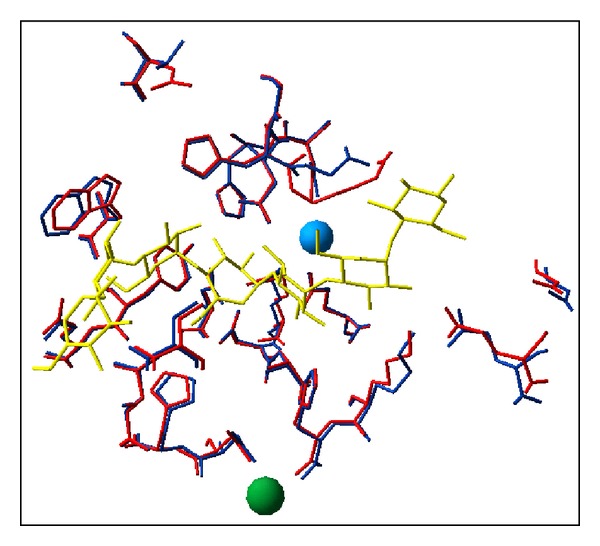
Structure of the active site. Superimposition of the active site residues in psychrophilic (blue) and mesophilic α-amylases (red). The chloride and calcium ions are shown as blue and green spheres, respectively. The 24 residues performing direct or water-mediated interactions with a substrate analog (yellow) are identical and superimpose almost perfectly within the resolution of the structures, demonstrating a structural identity in these psychrophilic and mesophilic enzymes [59].
Nevertheless, significant structural adjustments at the active site of psychrophilic enzymes have been frequently reported. In many cases, a larger opening of the catalytic cleft is observed [142] and is achieved by various ways, including replacement of bulky side chains for smaller groups, distinct conformation of the loops bordering the active site, or small deletions in these loops, as illustrated by a cold-active citrate synthase [124]. In the case of a Ca2+, Zn2+ protease from a psychrophilic Pseudomonas species, an additional bound Ca2+ ion pulls the backbone forming the entrance of the site and markedly increases its accessibility when compared with the mesophilic homologue [122]. As a result of such a better accessibility, cold-active enzymes can accommodate substrates at lower energy cost, as far as the conformational changes are concerned, and therefore reduce the activation energy required for the formation of the enzyme-substrate complex [170]. The larger active site may also facilitate easier release and exit of products and thus may alleviate the effect of a rate-limiting step on the reaction rate [134, 171].
In addition, differences in electrostatic potentials in and around the active site of psychrophilic enzymes appear to be a crucial parameter for activity at low temperatures. Electrostatic surface potentials generated by charged and polar groups are an essential component of the catalytic mechanism at various stages: as the potential extends out into the medium, a substrate can be oriented and attracted before any contact between enzyme and substrate occurs. Interestingly, the cold-active citrate synthase [124], malate dehydrogenase [131], uracil-DNA glycosylase [151, 172, 173], elastase [174], and trypsin [175–177] are characterized by marked differences in electrostatic potentials near the active site region compared to their mesophilic or thermophilic counterparts that may facilitate interaction with ligand. In the case of malate dehydrogenase, for example, the increased positive potential at and around the oxaloacetate binding site and the significantly decreased negative surface potential at the NADH binding region may facilitate the interaction of the oppositely charged ligands with the surface of the enzyme [131]. In all cases, the differences were caused by discrete substitutions in nonconserved charged residues resulting in local electrostatic potential differing in both sign and magnitude. Involvement of electrostatic potentials in cold-activity was also supported by a mutational study [178].
Finally, some unsuspected strategies have been shown to improve the activity in psychrophilic enzymes. With few exceptions, β-galactosidases are homotetrameric enzymes bearing four active sites. However, the crystal structure of a cold-active β-galactosidase revealed that it is a homohexamer, therefore possessing six active sites [125]. This unusual quaternary structure, containing two additional active sites, certainly contributes to improve the activity at low temperatures. Cellulases are microbial enzymes displaying a modular organization made of a globular catalytic domain connected by a linker to a cellulose binding domain. Psychrophilic cellulases were found to possess unusually long linkers of more than 100 amino acid residues, that is, about five times longer than in mesophilic cellulases [119, 179]. It was shown that the long linker adopts a large number of conformations between the fully extended and bended conformations, in caterpillar-like motions [119] and it was calculated that the catalytic domain has a 40-fold higher accessible surface area of substrate when compared with a mesophilic cellulase possessing a much shorter linker. Here also, increasing the surface of the insoluble substrate available to the catalytic domain should improve the activity of this enzyme at low temperatures.
4.3. Active Site Dynamics
The heat-labile activity of psychrophilic enzymes suggests that the dynamics of the functional side chains at the active site is improved in order to contribute to cold activity and the above-mentioned structural adaptations seem to favor a better accessibility to the substrate and release of the product. This view is strongly supported by the enzymological properties of cold-active enzymes. Nonspecific psychrophilic enzymes accept various substrates and have a broader specificity than the mesophilic homologues, because substrates with slightly distinct conformations or sizes can fit and bind to the site. For instance, the observed differences in substrate specificity between Atlantic salmon and mammalian elastases have been interpreted to be based on a somewhat wider and deeper binding pocket for the cold-adapted elastase [176]. The broad specificity of a psychrophilic alcohol dehydrogenase that oxidizes large bulky alcohols was also assigned to a highly flexible active site [180]. This active site dynamics in solution was also well demonstrated by a psychrophilic α-amylase [181]. Being more flexible, the active site can accommodate easily macromolecular polysaccharides and is more active on these substrates than a mesophilic homologue. By contrast, the flexible active site accommodates less efficiently short oligosaccharides and is less active on these substrates. Furthermore, the inhibition pattern of the psychrophilic α-amylase indicates that it can form a ternary complex (enzyme, substrate, and inhibitor), whereas the more rigid mesophilic homologue can only form binary complexes (enzyme and substrate or inhibitor). Several crystal structures of psychrophilic enzymes also point to an increased flexibility at or near the active site, as also supported by molecular dynamic simulations.
4.4. Adaptive Drift and Adaptive Optimization of Substrate Affinity
As a consequence of the improved active site dynamics in cold-active enzymes, substrates bind less firmly in the binding site (if no point mutations have occurred) giving rise to higher K m values. An example is given in Table 2 showing that a psychrophilic α-amylase is more active on its macromolecular substrates, whereas the K m values are up to 30-fold larger; that is, the affinity for the substrates is up to 30-fold lower. Ideally, a functional adaptation to cold would mean optimizing both k cat and K m. However, a survey of the available data on psychrophilic enzymes indicates that optimization of the k cat/K m ratio is far from a general rule but on the contrary that the majority of cold-active enzymes improve the k cat value at the expense of K m, therefore leading to suboptimal values of the k cat/K m ratio, as also shown in Table 2. For instance, high K m values have been reported for psychrophilic aspartate carbamoyltransferase [182, 183], triose-phosphate isomerase [137], subtilisin [184], lactate dehydrogénase [147, 162], DNA ligase [185], elongation factor Tu [186], glutamate dehydrogenase [187, 188], α-amylase [189], dihydrofolate reductase [190, 191], cellulase [179], endonuclease I [192], aspartate aminotransferase [193], isocitrate dehydrogenase [194], xylanase [195], ornithine carbamoyltransferase [196], citrate synthase [197], purine nucleoside phosphorylase [198], DEAD-Box proteins [99], and acetate kinase [199]. There is in fact an evolutionary pressure on K m to increase in order to maximize the overall reaction rate. Such adaptive drift of K m has been well illustrated by the lactate dehydrogenases from Antarctic fish [162] and by the psychrophilic α-amylase [189].
Table 2.
Kinetic parameters for the hydrolysis of polysaccharides at 25°C by psychrophilic and mesophilic α-amylases. Adapted from [181].
| Substrate | Psychrophilic α-amylase | Mesophilic α-amylase | ||||
|---|---|---|---|---|---|---|
| k cat | K m | k cat/K m | k cat | K m | k cat/K m | |
| s−1 | mg L−1 | s−1 mg−1 L | s−1 | mg L−1 | s−1 mg−1 L | |
| Starch | 663 | 155 | 4.3 | 327 | 41 | 8.0 |
| Amylopectin | 636 | 258 | 2.5 | 222 | 53 | 4.2 |
| Amylose | 2148 | 178 | 12.1 | 700 | 36 | 19.4 |
| Dextrin | 716 | 586 | 1.2 | 311 | 61 | 5.1 |
| Glycogen | 491 | 1344 | 0.3 | 193 | 46 | 4.2 |
Several enzymes, especially in some cold-adapted fish, counteract this adaptive drift of K m in order to maintain or to improve the substrate binding affinity by amino acid substitutions within the active site [176, 200]. The first reason for these enzymes to react against the drift is obvious when considering the regulatory function associated with K m, especially for intracellular enzymes. The second reason is related to the temperature dependence of weak interactions. Substrate binding is an especially temperature-sensitive step because both the binding geometry and interactions between binding site and ligand are governed by weak interactions having sometimes opposite temperature dependencies. Hydrophobic interactions form endothermically and are weakened by a decrease in temperature. By contrast, interactions of electrostatic nature (ion pairs, hydrogen bounds, and Van der Waals interactions) form exothermically and are stabilized at low temperatures. Therefore, low temperatures do not only reduce the enzyme activity (k cat), but can also severely alter the substrate binding mode according to the type of interaction involved. A comprehensive example was provided by a cold-active chitobiase: substitutions in the substrate binding site tend to replace hydrophobic residues (found in mesophilic chitobiases) by polar residues that are able to perform stronger interactions at low temperature [165].
4.5. Energetics of Cold Activity
Referring to (1), the high activity of cold-adapted enzymes corresponds to a decrease of the free energy of activation ΔG #. Two strategies have been highlighted to reduce the height of this energy barrier. Figure 7 illustrates the first strategy where an evolutionary pressure increases K m in order to maximize the reaction rate. According to the transition state theory, when the enzyme encounters its substrate, the enzyme-substrate complex ES falls into an energy pit. For the reaction to proceed, an activated state ES# has to be reached, that eventually breaks down into the enzyme and the product. The height of the energy barrier between the ground state ES and the transition state ES# is defined as the free energy of activation ΔG #: the lower this barrier, the higher the activity as reflected in (1). In the case of cold active enzymes displaying a weak affinity for the substrate, the energy pit for the ES complex is less deep (dashed in Figure 7). It follows that the magnitude of the energy barrier is reduced and therefore the activity is increased. This thermodynamic link between affinity and activity is valid for most enzymes (extremophilic or not) under saturating substrate concentrations and this link appears to be involved in the improvement of activity at low temperatures in numerous cold-active enzymes [162, 196, 201].
Figure 7.
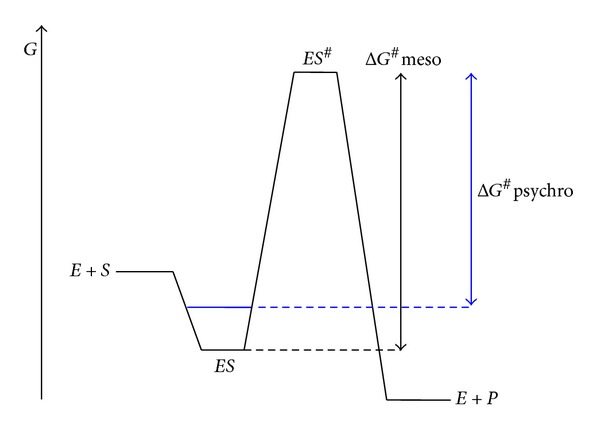
Optimization of activity by decreasing substrate affinity in psychrophilic enzymes. Reaction profile for an enzyme-catalyzed reaction with Gibbs energy changes under saturating substrate concentration. Weak substrate binding (in blue) decreases the activation energy (ΔG # psychro) and thereby increases the reaction rate. Adapted from [202].
The second and more general strategy involves the temperature dependence of the reaction catalyzed by cold-active enzymes. The free energy of activation ΔG # contains both enthalpic and entropic terms according to the classical Gibbs-Helmholtz relation:
| (2) |
Accordingly, (1) can be rewritten as
| (3) |
Equation (3) shows that both ΔH # and ΔS # have the potential to alter k cat. Table 3 reports the enthalpic and entropic contributions to the free energy of activation in extremophilic α-amylases. Similar data have been compiled for other psychrophilic enzymes [147, 158, 159, 163, 179, 192, 203, 204]. The enthalpy of activation ΔH # depicts the temperature dependence of the activity: the lower this value, the lower the variation of activity with temperature. The low value found for almost all psychrophilic enzymes demonstrates that their reaction rate is less reduced than for other enzymes when the temperature is lowered. Accordingly, the decrease of the activation enthalpy in the enzymatic reaction of psychrophilic enzymes can be considered as the main adaptive character to low temperatures [162, 201]. This decrease is structurally achieved by a decrease in the number of enthalpy-driven interactions that have to be broken during the activation steps. These interactions also contribute to the stability of the protein folded conformation and, as a corollary, the structural domain of the enzyme bearing the active site should be more flexible. It is interesting to note that such a macroscopic interpretation of the low activation enthalpy in cold-active enzymes fits with the experimental observation of a markedly heat-labile activity illustrated in Figure 5. Table 3 shows that the entropic contribution TΔS # for the cold-active enzyme is larger and negative. This has been interpreted as a large reduction of the apparent disorder between the ground state with its relatively loose conformation and the well-organized and compact transition state [163]. The heat-labile activity of cold-active enzymes suggests a macroscopic interpretation for this thermodynamic parameter. As a consequence of active site flexibility, the enzyme-substrate complex ES occupies a broader distribution of conformational states translated into increased entropy of this state, compared to that of the mesophilic or thermophilic homologues. This assumption has received strong experimental support [160, 205]. Furthermore, a broader distribution of the ground state ES should be accompanied by a weaker substrate binding strength, as indeed observed for numerous psychrophilic enzymes. Finally, it should be mentioned that the typical activation parameters of psychrophilic enzymes are well reproduced by reaction kinetic simulations [170].
Table 3.
Activation parameters of the hydrolytic reaction of α-amylases at 10°C. Adapted from [160].
| k cat | ΔG # | ΔH # | TΔS # | |
|---|---|---|---|---|
| s−1 | kcal mol−1 | kcal mol−1 | kcal mol−1 | |
| Psychrophile | 294 | 13.3 | 8.3 | −5.0 |
| Mesophile | 97 | 14.0 | 11.1 | −2.9 |
| Thermophile | 14 | 15.0 | 16.8 | 1.8 |
5. Global and Local Dynamics
The previous sections have highlighted the recurrent involvement of dynamics or flexibility in cold-activity of psychrophilic enzymes. Determination of molecular flexibility is complex as it requires the definition of the types and amplitudes of atomic motions as well as a timescale for these motions [206–208]. Nevertheless, various biophysical studies have revealed a less compact conformation of psychrophilic enzymes, undergoing frequent microunfolding events.
In this respect, the first experimental approach is apparently the pioneering work of Privalov [209] reporting an extensive analysis of collagen structure from organisms adapted to different temperatures (including Antarctic and Arctic fish) and using kinetics of hydrogen/deuterium exchange to quantify flexibility. The author concluded that the correlation between physiological temperature and collagen stability can be explained by a definite level of motility of protein structure required for its efficient functioning under physiological temperatures. This suggests that adjustment of conformational flexibility is a key event in the thermal adaptation of proteins [210–212]. Such differences in flexibility have received further support by the quantification of macromolecular dynamics in the whole protein content of psychrophilic, mesophilic, thermophilic, and hyperthermophilic bacteria by neutron scattering [213]. Neutron spectroscopy provides a unique tool to study thermal atomic motions in macromolecules because neutron wavelengths and energies match motion amplitudes and frequencies, respectively [213, 214]. These experiments have indeed revealed that the resilience (equivalent to macromolecular rigidity in terms of a force constant) increases with physiological temperatures. Furthermore, it was also shown that the atomic fluctuation amplitudes (equivalent to macromolecular flexibility) were similar for each microorganism at its physiological temperature. This is in full agreement with Somero's “corresponding state” concept [215] postulating that protein homologues exhibit comparable flexibilities to perform their biological function at their physiologically relevant temperatures. Fluorescence quenching of extremophilic proteins was also used as this method averages most of the dynamic parameters into a single signal. This technique utilizes increasing concentrations of a small quencher molecule (acrylamide in Figure 8). The decrease of fluorescence arising from diffusive collisions between the quencher and protein fluorophores reflects the ability of the quencher to penetrate the structure and can be viewed as an index of protein permeability. It was found that the structure of a psychrophilic protein has an improved propensity to be penetrated by the small quencher molecule, whereas a thermophilic protein only displays moderate quenching effect. Accordingly, a psychrophilic protein is a flexible molecule displaying numerous opening and closing motions or frequent microunfolding events [158–160, 216].
Figure 8.
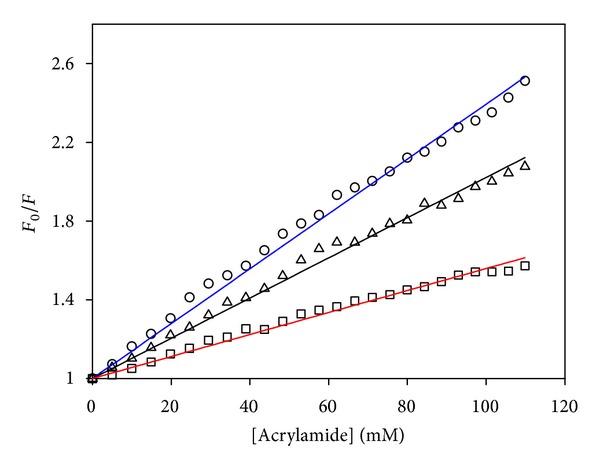
Permeability of the protein structure at room temperature. Fluorescence quenching experiments on psychrophilic (circles, blue) mesophilic (triangles, black), and thermophilic (squares, red) proteins. The steep slope recorded for the psychrophilic protein indicates that its structure is easily penetrated by a small quencher molecule (acrylamide), resulting in a larger attenuation of the intrinsic fluorescence (F 0/F), whereas the thermophilic protein is more rigid and displays fewer internal motions. Adapted from [160].
Local flexibility, as opposed to global flexibility, was predicted to play a pivotal role in enzyme cold activity [162, 163]. This local flexibility was first suggested by the 3D structure of a cold-active trypsin [148] and later observed in stability studies of psychrophilic enzymes showing that, in some cases, the structural domain bearing the active site is less stable than the remaining of the protein [161, 165–167, 217]. This was further evidenced by hydrogen/deuterium exchange studies of a psychrophilic alcohol dehydrogenase [218]. It was shown that functional regions involved in substrate and cofactor binding exhibit greater flexibility compared to a thermophilic homologue and, furthermore, that local flexibility can be uncoupled from global thermal stability. This local flexibility was also correlated with activity parameters [164, 219–222]. Several X-ray structures of psychrophilic enzymes have also pointed to a local flexibility at or around the active site as well as in functional regions [120, 124, 130, 133, 139, 145, 147]. Furthermore, molecular dynamics simulations have frequently highlighted local flexibility as a strategy for cold adaptation [174, 222–232].
The requirement for either global or local flexibility is still under debate. Enzymes displaying global flexibility frequently process large substrates, presumably involving concerted motions of the whole molecules, whereas local flexibility is generally observed for enzymes acting on small substrates [57]. There are, however, too many exceptions to conclude definitively.
6. Conformational Stability of Psychrophilic Enzymes
The numerous insights for close relationships between activity and stability in psychrophilic enzymes have prompted investigations of their conformational stability in comparison with mesophilic and thermophilic counterparts. The low thermal stability of cold-adapted proteins, known for decades, has been highlighted by various techniques such as intrinsic fluorescence spectroscopy (Figure 5). However, the energetics of structure stability was essentially revealed by microcalorimetry [158–160, 189, 203, 216, 233, 234].
6.1. Microcalorimetric Studies
Microcalorimetric records of heat-induced unfolding for psychrophilic, mesophilic and thermophilic proteins are shown in Figure 9. These enzymes clearly show distinct stability patterns that evolve from a simple profile in the unstable psychrophilic proteins to a more complex profile in very stable thermophilic counterparts. The unfolding of the cold-adapted enzymes occurs at lower temperatures as indicated by the temperature of half-denaturation T m, given by the top of the transition. The calorimetric enthalpy ΔH cal (area under the curves in Figure 9) corresponding to the total amount of heat absorbed during unfolding reflects the enthalpy of disruption of bonds involved in maintaining the compact structure and is markedly lower for the psychrophilic enzymes (Table 4). In addition, there is a clear trend for increasing ΔH cal values in the order psychrophile < mesophile < thermophile. The transition for the psychrophilic enzymes is sharp and symmetric, whereas other enzymes are characterized by a flattening of the thermograms. This indicates a pronounced cooperativity during unfolding of the psychrophilic enzymes: the structure is stabilized by fewer weak interactions and disruption of some of these interactions strongly influences the whole molecular edifice and promotes its unfolding. The psychrophilic enzymes unfold according to an all-or-none process, revealing a uniformly low stability of the architecture. By contrast, all other homologous enzymes display two to three transitions (indicated by deconvolution of the heat capacity function in Figure 9). Therefore, the conformation of these mesophilic and thermophilic enzymes contains structural blocks or units of distinct stability that unfold independently. Finally, the unfolding of the psychrophilic proteins is frequently more reversible than that of other homologous enzymes that are irreversibly unfolded after heating. The weak hydrophobicity of the core clusters in cold-adapted enzymes and the low temperature for unfolding, which prevent aggregation, certainly account for this reversible character.
Figure 9.
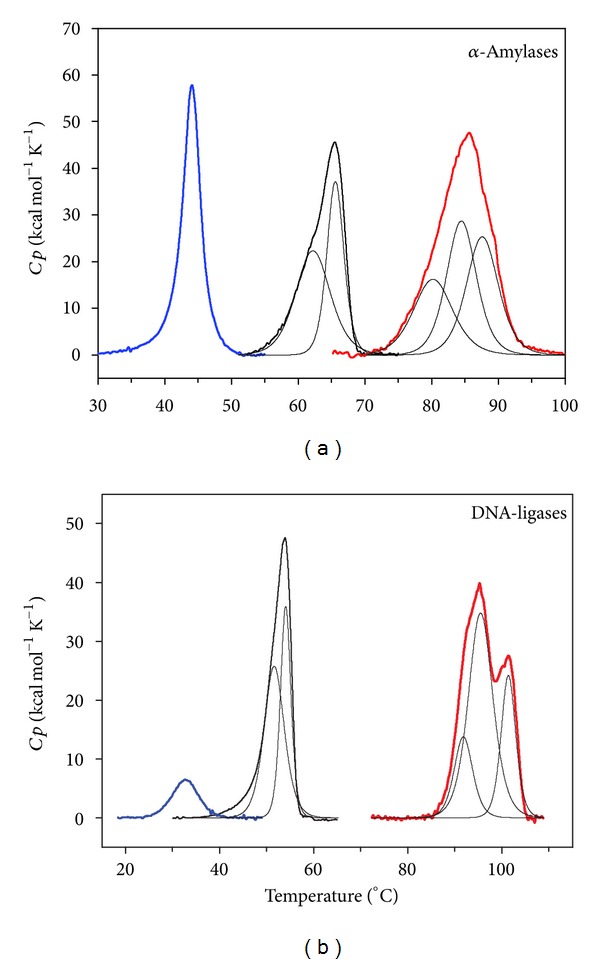
Thermal unfolding of extremophilic proteins. Thermograms of α-amylases and DNA-ligases recorded by differential scanning microcalorimetry showing, from left to right on each panel, psychrophilic (blue), mesophilic (black), and (hyper)thermophilic (red) proteins. The cold-adapted proteins are characterized by a lower T m (top of the transition) and ΔH cal (area under the transition), by a sharp and cooperative transition and by the lack of stability domains (indicated by thin lines in stable proteins). Adapted from [158, 189].
Table 4.
Microcalorimetric parameters of thermal unfolding shown in Figure 9.
| Proteins | T m (°C) | ΔH cal (kcal mol−1) |
|---|---|---|
| α-Amylases | ||
| Psychrophilic | 44 | 214 |
| Mesophilic | 66 | 319 |
| Thermophilic | 86 | 487 |
| DNA-ligases | ||
| Psychrophilic | 33 | 46 |
| Mesophilic | 54 | 253 |
| Thermophilic | 95–101 | 413 |
6.2. Stability Curves
The thermodynamic stability of a protein that unfolds reversibly according to a two-state mechanism is described by the classical Gibbs-Helmholtz relation:
| (4) |
The latter relation can be rewritten for any temperature (T) using the parameters determined experimentally by DSC:
| (5) |
where ΔCp is the difference in heat capacity between the native and the unfolded states. This parameter reflects the hydration of nonpolar groups that are exposed to water upon unfolding. Computing (5) in a temperature range where the native state prevails provides the protein stability curve [235], that is, the free energy of unfolding as a function of temperature (Figure 10). In other words, this is the work required to disrupt the native state at any given temperature [236] and is also referred to as the thermodynamic stability. By definition, this stability is zero at T m (equilibrium constant K = [U]/[N] = 1 and ΔG = − RTlnK). At temperatures below T m, the stability increases, as expected, but perhaps surprisingly for the nonspecialist, the stability reaches a maximum close to room temperature then it decreases at lower temperatures (Figure 10). In fact, this function predicts a temperature of cold unfolding, which is generally not observed because it occurs below 0°C [237]. Increasing the stability of a protein is essentially obtained by lifting the curve towards higher free energy values [238], whereas the low stability of a psychrophilic protein is reached by a global collapse of its curve. As far as extremophiles are concerned, one of the most puzzling observations of the last decade is that most proteins obey this pattern; that is, whatever the microbial source is, the maximal stability of their proteins is clustered around room temperature [238–240], although exceptions have been reported [241]. Accordingly, the environmental temperatures for mesophiles and thermophiles lie on the right limb of the bell-shaped stability curve and obviously, the thermal dissipative force is used to promote molecular motions in these molecules. By contrast, the environmental temperatures for psychrophiles lie on the left limb of the stability curve. It follows that molecular motions in proteins at low temperatures are gained from the factors ultimately leading to cold unfolding [233], that is, the hydration of polar and nonpolar groups [242]. The origin of flexibility in psychrophilic enzymes at low temperatures is therefore drastically different from mesophilic and thermophilic proteins, the latter taking advantage of the conformational entropy rise with temperature to gain in mobility.
Figure 10.
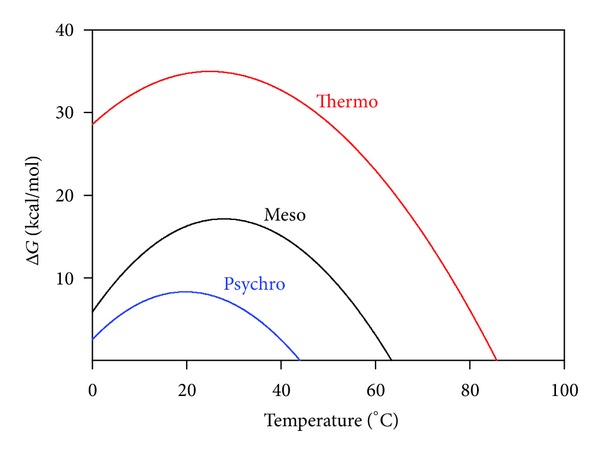
Gibbs free energy of unfolding, or conformational stability, of homologous extremophilic proteins. The work required to disrupt the native state is plotted as a function of temperature. The high stability of the thermophilic protein is reached by lifting the curve towards higher free energy values, whereas the low stability of the psychrophilic proteins corresponds to a global collapse of the bell-shaped stability curve. Adapted from [160].
A surprising consequence of the free energy function for the psychrophilic protein shown in Figure 10 is its weak stability at low temperatures when compared with mesophilic and thermophilic proteins, whereas it was intuitively expected that cold-active proteins should also be cold stable. This protein is in fact both heat and cold labiles. Therefore cold denaturation of some key enzymes in psychrophiles can be an additional, though unsuspected factor fixing the lower limit of life at low temperatures.
6.3. Structural Origin of Low Stability
Most of X-ray crystal structures from psychrophilic enzymes listed in Table 1 have been analyzed in order to decipher the structural origins of both cold activity and weak stability. However, the interpretation of these data is frequently difficult because these structural adaptations are extremely discrete and can easily escape the analysis, as exemplified in Figure 11. For instance, it has been shown that difference in distance interaction between enzyme and substrate as low as 0.1 Å (i.e., well below of the resolution of most crystal structures) can lead to substantial difference in catalytic efficiency [243]. Furthermore, these structural adaptations are very diverse, reflecting the complexity of factors involved in the stability of a macromolecule at the atomic level. As a general picture, it was found that all structural factors currently known to stabilize the protein molecule could be attenuated in strength and number in the structure of cold-active enzymes [176, 244–247]. Two review articles can be consulted for a comprehensive discussion of this topic [108, 176].
Figure 11.
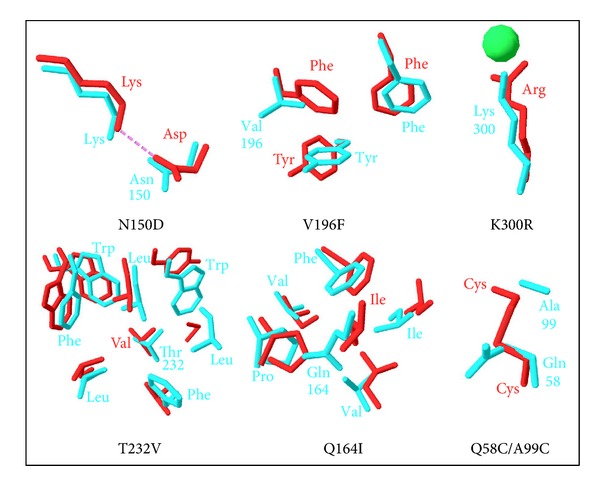
Amino acid substitutions in a psychrophilic alpha-amylase (blue) in comparison with the structure of its mesophilic homologue (red). The substitution N150D disrupts a salt bridge with the corresponding Lys side chain in the cold-adapted protein. The substitution V196F disrupts a triple face-to-edge aromatic interaction. Substitution K300R results in a monodentate coordination of the chloride ion, instead of a bidentate coordination by Arg in the mesophilic protein, as demonstrated by the crystal structure of the single mutant [272]. Substitutions T232V and Q164I decrease the apolarity of hydrophobic core clusters in the psychrophilic enzyme and the double substitution Q58C/A99C eliminates a disulfide bond. Adapted from [114].
The determinants of protein stability include structural factors, hydrophobic effects, and mainly weak interactions between atoms of the protein structure. In psychrophilic proteins, this involves the clustering of glycine residues, providing local mobility [248, 249]; the disappearance of proline residues in loops, enhancing chain flexibility between secondary structures [250]; a reduction in arginine residues which are capable of forming multiple salt bridges and H-bonds [251] as well as a lower number of ion pairs, aromatic interactions, or H-bonds, compared to mesophilic enzymes [148, 252]. The size and relative hydrophobicity of nonpolar residue clusters forming the protein core is frequently smaller, lowering the compactness of the protein interior by weakening the hydrophobic effect on folding [253]. Psychrophilic proteins have larger cavity sizes sufficient to accommodate water molecules: these cavities and embedded water molecules can play a significant role in structural flexibility [254]. The N- and C-caps of α-helices are also altered, weakening the charge-dipole interaction [255] and loose or relaxed protein extremities appear to be preferential sites for unzipping [148, 256]. The binding of stabilizing ions, such as calcium, can be extremely weak, with binding constants differing from mesophiles by several orders of magnitude [157, 257]. Insertions and deletions are sometimes responsible for specific properties such as the acquisition of extrasurface charges via insertions [157] or the weakening of subunit interactions via deletions [255]. Occasionally, disulfide linkages are lacking in cold-adapted proteins [234, 257, 258].
Calculation of the solvent accessible area showed that some psychrophilic enzymes expose a higher proportion of nonpolar residues to the surrounding medium [116, 124, 158]. This is an entropy-driven destabilizing factor caused by the reorganization of water molecules around exposed hydrophobic side chains. Calculations of the electrostatic potential revealed in some instances an excess of negative charges at the surface of the protein and, indeed, the pI of cold-active enzymes is frequently more acidic than that of their mesophilic or thermophilic homologues. This has been related to improved interactions with the solvent, which could be of prime importance in the acquisition of flexibility near zero degrees [233]. Besides the balance of charges, the number of salt bridges covering the protein surface is also reduced. There is a clear correlation between surface ion pairs and temperature adaptation, since these electrostatic interactions significantly increase in number from psychrophiles to mesophiles, to thermophiles, and hyperthermophiles, the latter showing arginine-mediated multiple ion pairs and interconnected salt bridge networks [259, 260]. Such an altered pattern of electrostatic interactions is thought to improve the dynamics or the “breathing” of the external shell of cold-active enzymes.
However, each enzyme adopts its own strategy by using one or a combination of these altered structural factors in order to improve the local or global mobility of the protein edifice. Accordingly, a general theory for structural adaptations cannot be formulated but nevertheless, enzyme families sharing the 3D fold can be compared as reviewed in [231]. Comparative structural analyses of psychrophilic, mesophilic, and thermophilic enzymes indicate that each protein family displays different structural strategies to adapt to temperature [135, 231, 245, 261–266]. However, some common trends are observed: the number of ion pairs, the side-chain contribution to the exposed surface, and the apolar fraction of the buried surface show a consistent decrease with decreasing optimal temperatures [245]. The multitude of structural strategies in cold-adapted proteins has also complicated statistical analyses aimed at delineating general trends in temperature adaptation. Various trends have been reported for psychrophilic proteins using different methodologies and datasets: a preference for smaller-size and less hydrophobic residues [267]; a less hydrophobic core, less charged, and long-chained surface residues [268]; a decrease of solvent accessible surface contribution of charged residues and an increase of hydrophobic surface contribution [37] or various patterns of preferential amino acid substitutions [269–271]. As a result of the diversity of structural factors involved in cold adaptation, the mean amino acid composition of whole genomes does not provide clear trends of preferential residue utilization [37–40].
7. Activity-Flexibility-Stability Relationships: Current Issues
7.1. Experimental Insights
In order to check the validity of the proposed relationships between the activity and the stability in cold-active enzymes, a psychrophilic α-amylase has been used as a model because the identical architecture of its active site, when compared with a close mesophilic homologue (Figure 6), indicates that structural adaptations affecting the active site properties occur outside from the catalytic cavity. On this basis, 17 mutants of this enzyme were constructed, each of them bearing an engineered residue forming a weak interaction found in mesophilic α-amylases but absent in the cold-active α-amylase as well as combinations of up to six stabilizing structural factors [114, 189, 234, 273]. It was shown that these engineered interactions improve all the investigated parameters related to protein stability: the compactness, the kinetically driven stability, the thermodynamic stability, the resistance towards chemical denaturation, and the kinetics of unfolding/refolding.Therefore, these mutants of the psychrophilic α-amylase consistently approximate and reproduce the stability and unfolding patterns of the heat-stable enzymes. These effects were also analyzed by molecular dynamics [274]. Concomitantly to this improved stability, stabilized mutants have lost the kinetic optimization to low temperature activity displayed by the parent psychrophilic enzyme. As shown in Figure 12, stabilizing the cold-active α-amylase tends to decrease the k cat values and concomitantly the K m values of the mutant enzymes, revealing the high correlation between both kinetic parameters. In fact, in addition to an engineered mesophilic-like stability, the multiple-mutant bearing six stabilizing structural factors also displays an engineered mesophilic-like activity in terms of alterations in k cat and K m values and even in thermodynamic parameters of activation [114, 273]. These results provide strong experimental support to the hypothesis assuming that the disappearance of stabilizing interactions in psychrophilic enzymes increases the amplitude of concerted motions required by catalysis and the dynamics of active site residues at low temperature, leading to a higher activity.
Figure 12.
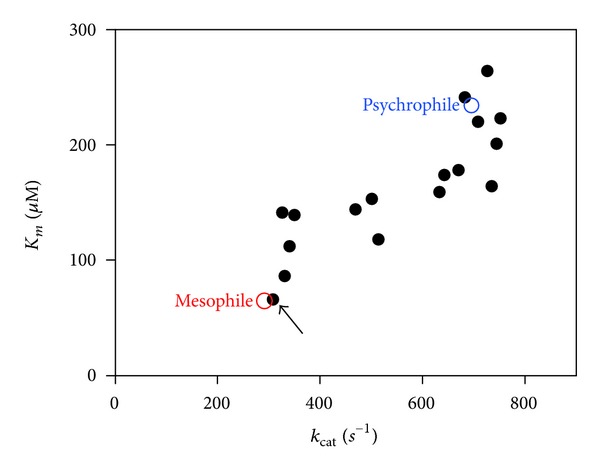
Engineering mesophilic-like activity in mutants of a psychrophilic α-amylase. This plot of the kinetic parameters for the stabilized mutants (filled symbols) shows that the general trend is to decrease the activity and to increase the affinity for the substrate of the wild-type psychrophilic enzyme (open symbol). The most stable mutant bearing six additional interactions (arrow) displays kinetic parameters nearly identical to those of the mesophilic homologue (open symbol). Adapted from [189, 273].
It should be mentioned that other mutational studies of cold-active enzymes have been less conclusive. Various psychrophilic enzymes [178, 184, 248–250, 275–282] have been engineered in order to check the involvement of specific adaptive mutations within or close to the active site. These studies have revealed a complex pattern of effects on kinetic parameters, activation energy, and stability that cannot be interpreted in simple terms.
7.2. Directed Evolution
The activity-stability relationships in cold-adapted proteins have been challenged from an evolutionary point of view. As a matter of fact, directed evolution [283, 284] and protein engineering [285] of enzymes have demonstrated that activity and stability are not physically linked in protein, as reviewed in [286]. Accordingly, it has been proposed that the low stability of cold-active enzymes is the result of a genetic drift related to the lack of selective pressure for stable proteins. Although this seems to be obvious, several lines of evidence indicate that the situation is more subtle. As already mentioned, in multidomain psychrophilic enzymes, the catalytic domain is always heat labile, whereas the noncatalytic domain can be as stable as mesophilic proteins [165–167]. It is therefore unlikely that a genetic drift only affects the catalytic domain without modifying other regions of the protein. Furthermore, several directed evolution experiments have shown that when libraries of randomly mutated enzymes are only screened for improved activity at low temperatures without any other constraints, the best candidates invariably display the canonical properties of psychrophilic enzymes, as discussed in [287]. Examination of the activity versus stability plots for hundreds of mutants shows that random mutations improving both activity and stability are rare [288, 289]. It follows that improvement of activity at low temperatures associated with loss of stability appears to be the most frequent and accessible event. In conclusion, the current view suggests that the strong evolutionary pressure on psychrophilic enzymes to increase their activity at low temperatures can be accommodated for by the lack of selection for stability and represents the simplest adaptive strategy for enzyme catalysis in the cold.
7.3. Noticeable Exceptions
Considering the above-mentioned lessons from laboratory evolution, one should expect that psychrophilic enzymes, which do not share the canonical properties, can be discovered. Indeed, some reports suggest the occurrence of noticeable exceptions to the general rule. For instance, single-point mutations can account for most of the adaptive traits. A single mutation in the calcium-binding site of a psychrophilic subtilisin [184] and in the phosphate-binding helix of triose phosphate isomerase [137] increases drastically the thermostability of the psychrophilic mutants. Conversely, a single amino acid substitution near the active site of a thermostable α-glucosidase provides cold activity [290]. The chaperonin and heat-shock protein GroEL from an Antarctic bacterium is not cold adapted and displays similar stability and activity than that of its E. coli homologue [291]. It has been suggested that this chaperonin remains suited to function during sudden temperature increases of the environment [83]. Similarly, activity of thioredoxin from the same bacterium is much more heat stable than that of E. coli [292, 293]. One cannot exclude that enzymes involved in electron transfer do not require the same type of adaptations because the rate of electron flow is not significantly affected by the low biological temperatures. Isocitrate dehydrogenase from a psychrophilic bacterium is more stable than its mesophilic homologue, while cold activity was attributed to local flexibility at the active site [133].
8. Folding Funnel Model for Psychrophilic Enzymes
The various properties of psychrophilic enzymes have been integrated in a model based on folding funnels [294, 295] to describe the activity-stability relationships in extremophilic enzymes [160]. The energy landscapes of psychrophilic and thermophilic enzymes are shown in Figure 13. The top of the funnel is occupied by the unfolded state having a high free energy (considering the spontaneous folding reaction), whereas the bottom of the funnel is occupied by the stable (low free energy) native state. The height of the funnel, that is, the free energy of folding, also corresponding to the conformational stability, has been fixed here in a 1 to 5 ratio (Figure 10). The upper edge of the funnels is occupied by the unfolded state in random coil conformations but it should be noted that psychrophilic enzymes tend to have a lower proline content than mesophilic and thermophilic enzymes, a lower number of disulfide bonds and a higher occurrence of glycine clusters. Accordingly, the edge of the funnel for the psychrophilic protein is slightly larger (broader distribution of the unfolded state) and is located at a higher energy level. When the polypeptide is allowed to fold, the free energy level decreases as well as the conformational ensemble. However, thermophilic proteins pass through intermediate states corresponding to local minima of energy. These minima are responsible for the ruggedness of the funnel slopes and for the reduced cooperativity of the folding-unfolding reaction, as demonstrated by heat-induced unfolding (Figure 9). By contrast, the structural elements of psychrophilic proteins generally unfold cooperatively without intermediates, as a result of fewer stabilizing interactions and stability domains; therefore, the funnel slopes are steep and smooth. The bottom of the funnel depicts the stability of the native state ensemble. The bottom for a very stable and rigid thermophilic protein can be depicted as a single global minimum or as having only a few minima with high energy barriers between them, whereas the bottom for an unstable and flexible psychrophilic protein is rugged and depicts a large population of conformers with low-energy barriers to flip between them. Rigidity of the native state is therefore a direct function of the energy barrier height [296, 297] and is drawn here according to a global flexibility of cold-adapted proteins. In this context, the activity-stability relationships in these extremophilic enzymes depend on the bottom properties. Indeed, it has been argued that upon substrate binding to the association-competent subpopulation, the equilibrium between all conformers is shifted towards this subpopulation, leading to the active conformational ensemble [296–299]. In the case of the rugged bottom of psychrophilic enzymes, this equilibrium shift only requires a modest free energy change (low energy barriers), a low enthalpy change for interconversion of the conformations but is accompanied by a large entropy change for fluctuations between the wide conformer ensemble. The converse picture holds for thermophilic enzymes, in agreement with the activation parameters shown in Table 3 and with the proposed macroscopic interpretation. Such energy landscapes integrate nearly all biochemical and biophysical data currently available for extremophilic enzymes but they will certainly be refined by future investigations of other series of homologous proteins from psychrophiles, mesophiles, and thermophiles. This model has nevertheless received support from several experimental and computational studies [170, 205, 207, 252, 299–302].
Figure 13.
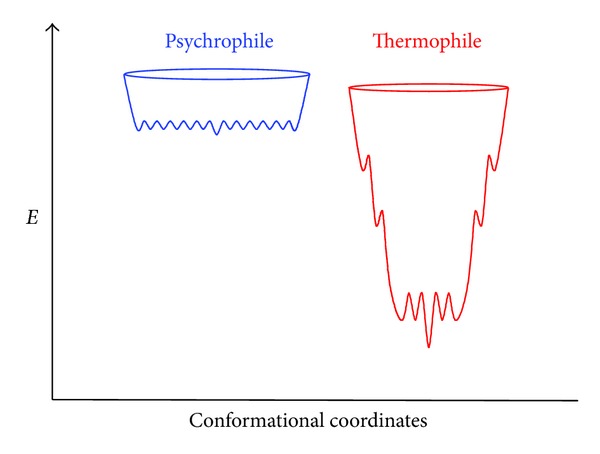
Folding funnel model of enzyme temperature adaptation. In these schematic energy landscapes for extremophilic enzymes, the free energy of folding (E) is depicted as a function of the conformational diversity. The height of the funnels is deduced from the determination of the conformational stabilities. The top of the funnels is occupied by the unfolded states in the numerous random coil conformations, whereas the bottom of the funnels corresponds to native and catalytically active conformations. The ruggedness of the bottom depicts the energy barriers for interconversion, or structural fluctuations of the native state [160].
9. Psychrophilic Enzymes in Biotechnology
As already mentioned, most enzymes from psychrophiles are cold active and heat labile. These specific traits are responsible for the three main advantages of cold active enzymes in biotechnology: (i) as a result of their high activity, a lower concentration of the enzyme catalyst is required to reach a given activity, therefore reducing the amount of costly enzyme preparation in a process; (ii) as a result of their cold activity, they remain efficient at tap water or ambient temperature, therefore avoiding heating during a process, either at domestic (e.g., washing machine) or industrial levels; and (iii) as a result of heat lability, they can be efficiently and sometime selectively inactivated after a process by moderate heat input. Beside these traits, enzymes from organisms endemic to cold environments can be a valuable source of new catalysts possessing useful enzymological characteristics such as novel substrate specificities or product properties. Previous reviews should be consulted for a complete coverage of this topic [16, 28, 303–308]. Bioprospector, an online database (http://www.bioprospector.org/bioprospector/) provides a survey of patents, commercial products, and companies involved in applied research using genetic resources from both the Antarctic and the Arctic. Some specific examples are provided below.
9.1. Heat Lability in Molecular Biology
Alkaline phosphatases are mainly used in molecular biology for the dephosphorylation of DNA vectors prior to cloning to prevent recircularization or for the dephosphorylation of 5′-nucleic acid termini before 5′-end labeling by polynucleotide kinase. However, the phosphatase has to be carefully removed after dephosphorylation to avoid interferences with the subsequent steps. It follows that heat-labile alkaline phosphatases are excellent alternatives as they are inactivated by moderate heat treatment allowing to perform the subsequent steps in the same test tube and minimizing nucleic acid losses [309]. An alkaline phosphatase from an Antarctic bacterium has been fully characterized [128, 310, 311]. This heat-labile alkaline phosphatase, sold as Antarctic phosphatase, is now proposed on the market by New England Biolabs Inc. (Ipswich, MA, USA). In the same context, the heat-labile alkaline phosphatase from the Arctic shrimp Pandalus borealis is also available for instance from Biotec Pharmacon ASA (Tromsø, Norway) or GE Healthcare Life Sciences (Little Chalfont, UK).
Two other psychrophilic enzymes are also marketed for molecular biology applications taking advantage of the heat-labile property. Shrimp nuclease selectively degrades double-stranded DNA: for instance, it is used for the removal of carry-over contaminants in PCR mixtures, and then it is heat inactivated prior addition of the template. This recombinant enzyme is available from Biotec Pharmacon ASA (Tromsø, Norway), USB Corporation (Santa Clara, CA, USA), or Thermo Scientific (Waltham, MA, USA). Heat-labile uracil-DNA N-glycosylase from Atlantic cod (Gadus morhua), that presents typical cold adaptation features [151], is also used to remove DNA contaminants in sequential PCR reactions. Following degradation of contaminants, the enzyme is completely and irreversibly inactivated after heat treatment. Heat-labile uracil-DNA N-glycosylase is available from Biotec Pharmacon ASA (Tromsø, Norway).
9.2. Low Temperature Activity
The market for enzymes used in detergents represents 30%–40% of all enzymes produced worldwide. Amongst these enzymatic cleaning agents, subtilisin (an alkaline serine protease predominantly produced by Bacillus species) largely dominates this market. At the domestic level, the current trend is, however, to use detergents at lower washing temperatures because of the associated reductions in energy consumption and costs as well as to protect texture and colors of the fabrics. Accordingly, cold-active subtilisins are required for optimal washing results at tap water temperatures and the current advertisements for cold-active detergents indicate that this goal has been reached. The first psychrophilic subtilisins isolated from Antarctic Bacillus species have been extensively characterized to comply with such requirements [157, 184]. Subtilisins currently incorporated in cold-active detergents are engineered enzymes that combine storage stability, alkaline stability, and activity and cold activity. Although psychrophilic subtilisins are not components per se of cold-active detergents, they have largely contributed to the advancement of this economically attractive concept.
Beta-galactosidase, or lactase, is a glycoside hydrolase that specifically hydrolyzes the milk sugar lactose into galactose and glucose. It should be stressed that 75% of the world population suffers from lactose intolerance arising from deficient synthesis of intestinal lactase in adults and resulting in digestive disorders. In this context, a cold-active lactase from an Antarctic bacterium has been patented (WO 01/04276A1) for its capacity to hydrolyze lactose during milk storage at low temperatures [204]. This cold-active lactase will be also produced soon in large quantities by Nutrilab NV (Bekkevoort, Belgium) to hydrolyze lactose (as a by-product of the dairy industry) in the process of the high value sweetener D-tagatose, a natural monosaccharide with low caloric value and glycemic index.
Protein chaperones assist the folding of nascent polypeptides, preventing misfolding or even repairing misfolding. Taking advantage of these properties, the ArcticExpress E. coli cells from Stratagene (USA) have been engineered to coexpress cold-active chaperonins from an Antarctic bacterium [102] with the recombinant protein of interest, therefore improving protein processing at low temperatures (that prevent inclusion bodies formation) and increasing the yield of active, soluble recombinant protein.
9.3. New Specificities in Psychrophilic Enzymes
At the industrial level, the yeast Candida antarctica produces two lipases, A and B, the latter being sold for instance as Novozym 435 by Novozymes (Bagsvaerd, Denmark). As a result of its substrate and stereospecificity, lipase B is involved in a very large number of organosynthesis applications related to food/feed processing, pharmaceuticals, or cosmetics [312]. In a survey of patents related to Antarctica [313], it was shown that lipases from C. antarctica by far dominate the number of process- or product-based patents. This is a significant example of the potential for novel catalysts from genetic resources in cold environments.
Xylanases are glycoside hydrolases that degrade the polysaccharide beta-1,4-xylan, thus breaking down hemicellulose, one of the major components of plant cell walls. Xylanases are a key ingredient of industrial dough conditioners used to improve bread quality. It was found that the xylanase from an Antarctic bacterium belongs to a new class of xylanases [118, 159, 171, 195, 314]. Furthermore, baking trials have revealed that the psychrophilic xylanase was very effective in improving the dough properties and final bread quality with, for instance, a positive effect on loaf volume, as shown in Figure 14 [315, 316]. This efficiency appears to be related to the high activity of the psychrophilic xylanase at cool temperatures required for dough resting and to its specific mode of xylan hydrolysis. This xylanase is now sold by Puratos (Grand-Bigard, Belgium). This is apparently the psychrophilic enzyme produced at the highest amounts to date.
Figure 14.

Effects of xylanases on bread volume and cut width (arrows) as compared to a negative control without added xylanase (loaf 1). Adapted from [315].
9.4. Other Psychrophilic Proteins in Biotechnology
In this context, some cold-adapted proteins are also worth mentioning. This includes a recombinant antifreeze protein from an Arctic fish added in several edible ice cream brands from Unilever (The Netherlands, England) for its ice-structuring properties; the bacterial antifreeze Antarticine-NF3 (sometimes under the name Antarctilyne), which is effective for scar treatments and reepithelialization of wounds, or extracts of the Antarctic algae Durvillea antarctica, which are included in cosmetics to improve skin vitality such as in the Extra Firming Day Cream, a top seller of Clarins (France). Improved cold activity by chemical modification of mesophilic enzymes has also been reported [317–319] and was proven successful in some specific cases.
10. Conclusions
Psychrophiles thriving permanently at near-zero temperatures synthesize cold-active enzymes to sustain their cell cycle. Most psychrophilic enzymes optimize a high activity at low temperature at the expense of substrate affinity, therefore reducing the free energy barrier of the transition state. Furthermore, a weak temperature dependence of activity ensures moderate reduction of the catalytic activity in the cold. In these naturally evolved enzymes, the optimization to low temperature activity is reached via destabilization of the structures bearing the active site or by destabilization of the whole molecule. This involves a reduction in the number and strength of all types of weak interactions or the disappearance of stability factors, resulting in improved dynamics of active site residues in the cold. These enzymes are already used in many biotechnological applications requiring high activity at mild temperatures or fast heat-inactivation rate.
11. Future Avenues
Although significant progresses have been recently achieved in the understanding of protein adaptation to extreme environmental temperatures, many questions remain to be addressed with regards to structure-function relationships in psychrophilic proteins. In a previous review on cold-active enzymes [108], various open questions have been also highlighted and this reference should be consulted for the sake of completeness.
Folding at Low Temperature. The folding reaction of a protein should be strongly dependent of the temperature at which any organism synthesizes the polypeptide: how are these polypeptides synthesized in the cold, is the amino acid sequence adapted to cold folding, what is the rate-limiting step of folding, and are cold-adapted chaperones involved?
Kinetic Parameters. Most cold-active enzymes display improved k cat values at low temperature associated with increased (unfavorable) K m values, whereas in some cases the K m is improved. What are the selective pressure and physiological requirements for such adjustments in substrate binding? Which types of psychrophilic enzymes do require such adjustments? Surprisingly, no detailed enzymological analysis of psychrophilic enzymes has been reported, except one [200]. Accordingly, what are the fine adjustments of enzyme activity at low temperature?
Global versus Local Flexibility. Structural adjustments around the active site render the catalytic center more dynamic but in many cases the whole molecule is more dynamic. What are the requirements for local or global flexibility? In parallel with the previous question, some psychrophilic enzymes are uniformly unstable (structure and activity), whereas others only display heat-labile activity: what are the tradeoffs in activity and stability?
Natural Selection. To what extent the weak stability of psychrophilic enzymes is the result of the lack of selection for stable proteins or of the strong selection for high activity? What is the balance between both selective pressures?
Macromolecular Dynamics. The improved dynamics of psychrophilic enzymes is a central theme of previous investigations but what are the most reliable techniques to investigate this aspect and how to discriminate between local and global dynamics? X-ray structures provide a static picture when comparing psychrophilic, mesophilic, or thermophilic proteins. More sophisticated and insightful methods are needed such as NMR, neutron scattering, and H/D exchanges.
Folding Funnels. The global model depicting the activity-flexibility-stability relationships in psychrophilic enzymes in Figure 13 is attractive but it was built on a limited number of case studies: can we generalize this model or are there some variations and refinements to this model?
Extreme Environmental Temperatures. How do psychrophilic proteins compare with thermophilic proteins? Are there specific adaptive mechanisms to either low or high temperatures? Or, is there a continuum in the adaptive strategies from low to moderate and high temperatures?
It is the wish of the present author that such open questions will stimulate young researchers to embark in this field in order to refine our understanding of cold-adapted proteins and to contribute to this fascinating topic which encompass microbiology, enzymology, biochemistry, or biophysics.
Note: The genome sequence of the Antarctic bacterium Oleispira antarctica has been recently deposited to GenBank, as well as the crystal structure of eleven proteins from this bacterium in the PDB. NMR assignments of a new psychrophilic protein have been also reported [320].
Acknowledgments
Research in the authors' laboratory was supported by the European Union, the European Space Agency, the Région Wallonne (Belgium), the Fonds National de la Recherche Scientifique (Belgium), and the University of Liège. The facilities offered by the Institut Polaire Français are also acknowledged.
References
- 1.Rodrigues DF, Tiedje JM. Coping with our cold planet. Applied and Environmental Microbiology. 2008;74(6):1677–1686. doi: 10.1128/AEM.02000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilichinsky D, Rivkina E, Bakermans C, et al. Biodiversity of cryopegs in permafrost. FEMS Microbiology Ecology. 2005;53(1):117–128. doi: 10.1016/j.femsec.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Steven B, Léveillé R, Pollard WH, Whyte LG. Microbial ecology and biodiversity in permafrost. Extremophiles. 2006;10(4):259–267. doi: 10.1007/s00792-006-0506-3. [DOI] [PubMed] [Google Scholar]
- 4.Deming JW. Psychrophiles and polar regions. Current Opinion in Microbiology. 2002;5(3):301–309. doi: 10.1016/s1369-5274(02)00329-6. [DOI] [PubMed] [Google Scholar]
- 5.Junge K, Eicken H, Deming JW. Bacterial activity at -2 to -20°C in arctic wintertime sea ice. Applied and Environmental Microbiology. 2004;70(1):550–557. doi: 10.1128/AEM.70.1.550-557.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakermans C, Skidmore ML. Microbial metabolism in ice and brine at -5°C. Environmental Microbiology. 2011;13:2269–2278. doi: 10.1111/j.1462-2920.2011.02485.x. [DOI] [PubMed] [Google Scholar]
- 7.Sattler B, Puxbaum H, Psenner R. Bacterial growth in supercooled cloud droplets. Geophysical Research Letters. 2001;28(2):239–242. [Google Scholar]
- 8.Vaïtilingom M, Amato P, Sancelme M, Laj P, Leriche M, Delort AM. Contribution of microbial activity to carbon chemistry in clouds. Applied and Environmental Microbiology. 2010;76(1):23–29. doi: 10.1128/AEM.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedmann EI. Endolithic microorganisms in the Antarctic cold desert. Science. 1982;215(4536):1045–1053. doi: 10.1126/science.215.4536.1045. [DOI] [PubMed] [Google Scholar]
- 10.Cary SC, McDonald IR, Barrett JE, Cowan DA. On the rocks: the microbiology of Antarctic Dry Valley soils. Nature Reviews Microbiology. 2010;8(2):129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- 11.Stibal M, Šabacká M, Kaštovská K. Microbial communities on glacier surfaces in Svalbard: impact of physical and chemical properties on abundance and structure of cyanobacteria and algae. Microbial Ecology. 2006;52(4):644–654. doi: 10.1007/s00248-006-9083-3. [DOI] [PubMed] [Google Scholar]
- 12.Cameron KA, Hodson AJ, Osborn AM. Structure and diversity of bacterial, eukaryotic and archaeal communities in glacial cryoconite holes from the Arctic and the Antarctic. FEMS Microbiology Ecology. 2012;82(2):254–267. doi: 10.1111/j.1574-6941.2011.01277.x. [DOI] [PubMed] [Google Scholar]
- 13.Morita RY. Psychrophilic bacteria. Bacteriological reviews. 1975;39(2):144–167. doi: 10.1128/br.39.2.144-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell NJ. Cold adaptation of microorganisms. Philosophical Transactions of the Royal Society. 1990;326:595–611. doi: 10.1098/rstb.1990.0034. [DOI] [PubMed] [Google Scholar]
- 15.Gounot AM. Bacterial life at low temperature: physiological aspects and biotechnological implications. Journal of Applied Bacteriology. 1991;71(5):386–397. doi: 10.1111/j.1365-2672.1991.tb03806.x. [DOI] [PubMed] [Google Scholar]
- 16.Russell NJ. Molecular adaptations in psychrophilic bacteria: potential for biotechnological applications. Advances in Biochemical Engineering/Biotechnology. 1998;61:1–21. doi: 10.1007/BFb0102287. [DOI] [PubMed] [Google Scholar]
- 17.Cavicchioli R. Cold-adapted archaea. Nature Reviews Microbiology. 2006;4(5):331–343. doi: 10.1038/nrmicro1390. [DOI] [PubMed] [Google Scholar]
- 18.Morgan-Kiss RM, Priscu JC, Pocock T, Gudynaite-Savitch L, Huner NPA. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiology and Molecular Biology Reviews. 2006;70(1):222–252. doi: 10.1128/MMBR.70.1.222-252.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buzzini P, Branda E, Goretti M, Turchetti B. Psychrophilic yeasts from worldwide glacial habitats: diversity, adaptation strategies and biotechnological potential. FEMS Microbiology Ecology. 2012;82(2):217–241. doi: 10.1111/j.1574-6941.2012.01348.x. [DOI] [PubMed] [Google Scholar]
- 20.Margesin R, Neuner G, Storey KB. Cold-loving microbes, plants, and animals—fundamental and applied aspects. Naturwissenschaften. 2007;94(2):77–99. doi: 10.1007/s00114-006-0162-6. [DOI] [PubMed] [Google Scholar]
- 21.Doucet D, Walker VK, Qin W. The bugs that came in from the cold: molecular adaptations to low temperatures in insects. Cellular and Molecular Life Sciences. 2009;66(8):1404–1418. doi: 10.1007/s00018-009-8320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eastman JT. Antarctic Fish Biology: Evolution in a Unique Environment. San Diego, Calif, USA: Academic Press; 1993. [Google Scholar]
- 23.di Prisco G, Pisano E, Clarke A. Fishes of AntarcticA: A Biological Overview. Milano, Italy: Springer; 1998. [Google Scholar]
- 24.Giordano D, Russo R, di Prisco G, Verde C. Molecular adaptations in Antarctic fish and marine microorganisms. Marine Genomics. 2012;6:1–6. doi: 10.1016/j.margen.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Dial CR, Dial RJ, Saunders R, et al. Historical biogeography of the North American glacier ice worm, Mesenchytraeus solifugus (Annelida: Oligochaeta: Enchytraeidae) Molecular Phylogenetics and Evolution. 2012;63:577–584. doi: 10.1016/j.ympev.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Margesin R, Schinner F. Cold-Adapted Organisms: Ecology, Physiology, Enzymology and Molecular Biology. Heidelberg, Germany: Springer; 1999. [Google Scholar]
- 27.Gerday C, Glansdorff N. Physiology and Biochemistry of Extremophiles. Washington, DC, USA: ASM Press; 2007. [Google Scholar]
- 28.Margesin R, Schinner F, Marx JC, Gerday C. Psychrophiles, from Biodiversity to Biotechnology. Berlin, Germany: Springer; 2008. [Google Scholar]
- 29.Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO, editors. Extremophiles Handbook. Berlin, Germany: Springer; 2011. [Google Scholar]
- 30.Miller RV, Whyte LG. Polar Microbiology. Life in a Deep Freeze. Washington, DC, USA: ASM Press; 2012. [Google Scholar]
- 31.Margesin R, Feller G, Gerday C, Russell NJ. Cold-adapted microorganisms: adaptation strategies and biotechnological potential. In: Bitton G, editor. EncyclopEdia of Environmental Microbiology. New York, NY, USA: John Wiley & Sons; 2002. pp. 871–885. [Google Scholar]
- 32.D’Amico S, Collins T, Marx JC, Feller G, Gerday C. Psychrophilic microorganisms: challenges for life. EMBO Reports. 2006;7(4):385–389. doi: 10.1038/sj.embor.7400662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Methé B, Nelson KE, Fraser CM. It’s a cold world out there (but the prospects are hot) Trends in Microbiology. 2004;12(12):532–534. doi: 10.1016/j.tim.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 34.Danchin A. An interplay between metabolic and physicochemical constrains: lessons from the psychrophilic prokaryote genomes. In: Gerday C, Glansdorff N, editors. Physiology and Biochemistry of Extremophiles. Washington, DC, USA: ASM Press; 2007. pp. 208–220. [Google Scholar]
- 35.Bowman JB. Genomic analysis of psychrophilic prokaryotes. In: Margesin R, Schinner F, Marx JC, Gerday C, editors. Psychrophiles, from Biodiversity to Biotechnology. Berlin, Germany: Springer; 2008. pp. 265–284. [Google Scholar]
- 36.Casanueva A, Tuffin M, Cary C, Cowan DA. Molecular adaptations to psychrophily: the impact of “omic” technologies. Trends in Microbiology. 2010;18(8):374–381. doi: 10.1016/j.tim.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Saunders NFW, Thomas T, Curmi PMG, et al. Mechanisms of thermal adaptatation revealed from genomes of the anatarctic Archaea Methanogenium frigidum and Methanacoccoides burtonii . Genome Research. 2003;13(7):1580–1588. doi: 10.1101/gr.1180903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabus R, Ruepp A, Frickey T, et al. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environmental Microbiology. 2004;6(9):887–902. doi: 10.1111/j.1462-2920.2004.00665.x. [DOI] [PubMed] [Google Scholar]
- 39.Médigue C, Krin E, Pascal G, et al. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Research. 2005;15(10):1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Methé BA, Nelson KE, Deming JW, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(31):10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duchaud E, Boussaha M, Loux V, et al. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum . Nature Biotechnology. 2007;25(7):763–769. doi: 10.1038/nbt1313. [DOI] [PubMed] [Google Scholar]
- 42.Riley M, Staley JT, Danchin A, et al. Genomics of an extreme psychrophile, Psychromonas ingrahamii . BMC Genomics. 2008;9, article 210 doi: 10.1186/1471-2164-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodrigues DF, Ivanova N, He Z, Huebner M, Zhou J, Tiedje JM. Architecture of thermal adaptation in an Exiguobacterium sibiricum strain isolated from 3 million year old permafrost: a genome and transcriptome approach. BMC Genomics. 2008;9, article 547 doi: 10.1186/1471-2164-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen MA, Lauro FM, Williams TJ, et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. ISME Journal. 2009;3(9):1012–1035. doi: 10.1038/ismej.2009.45. [DOI] [PubMed] [Google Scholar]
- 45.Ayala-Del-Río HL, Chain PS, Grzymski JJ, et al. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Applied and Environmental Microbiology. 2010;76(7):2304–2312. doi: 10.1128/AEM.02101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng C, Demaere MZ, Williams TJ, et al. Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. ISME Journal. 2010;4(8):1002–1019. doi: 10.1038/ismej.2010.28. [DOI] [PubMed] [Google Scholar]
- 47.Blanc G, Agarkova I, Grimwood J, et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biology. 2012;13, article R39 doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jang SH, Kim J, Hong S, Lee C. Genome sequence of cold-adapted Pseudomonas mandelii strain JR-1. Journal of Bacteriology. 2012;194:p. 3263. doi: 10.1128/JB.00517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SJ, Shin SC, Hong SG, et al. Genome sequence of Janthinobacterium sp. strain PAMC, 25724, isolated from alpine glacier cryoconite. Journal of Bacteriology. 2012;194:p. 2096. doi: 10.1128/JB.00096-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margolles A, Gueimonde M, Sanchez B. Genome sequence of the antarctic psychrophile Bacterium Planococcus antarcticus DSM, 14505. Journal of Bacteriology. 2012;194:p. 4465. doi: 10.1128/JB.00888-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park HJ, Shin SC, Kim D. Draft genome sequence of Arctic marine bacterium Pseudoalteromonas issachenkonii PAMC, 22718. Journal of Bacteriology. 2012;194:p. 4140. doi: 10.1128/JB.00744-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin SC, Kim SJ, Ahn-do H, et al. Genome sequence of a Salinibacterium sp. isolated from Antarctic soil. Journal of Bacteriology. 2012;194:p. 2404. doi: 10.1128/JB.00235-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin SC, Kim SJ, Hong SG, et al. Genome sequence of Pseudomonas sp. strain PAMC, 25886, isolated from alpine glacial cryoconite. Journal of Bacteriology. 2012;194:p. 1844. doi: 10.1128/JB.00057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tribelli PM, Iustman LJR, Catone MV, et al. Genome sequence of the polyhydroxybutyrate producer Pseudomonas extremaustralis, a highly stress-resistant Antarctic bacterium. Journal of Bacteriology. 2012;194:2381–2382. doi: 10.1128/JB.00172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe T, Kojima H, Fukui M. Draft genome sequence of a psychrotolerant sulfur-oxidizing bacterium, Sulfuricella denitrificans skB26, and proteomic insights into cold adaptation. Applied and Environmental Microbiology. 2012;78:6545–6549. doi: 10.1128/AEM.01349-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feller G. Molecular adaptations to cold in psychrophilic enzymes. Cellular and Molecular Life Sciences. 2003;60(4):648–662. doi: 10.1007/s00018-003-2155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feller G, Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nature Reviews Microbiology. 2003;1(3):200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 58.Feller G. Life at low temperatures: is disorder the driving force? Extremophiles. 2007;11(2):211–216. doi: 10.1007/s00792-006-0050-1. [DOI] [PubMed] [Google Scholar]
- 59.Feller G. Enzyme function at low temperatures in Psychrophiles. In: Siddiqui KS, Thomas T, editors. Protein Adaptation in Extremophiles. New York, NY, USA: Nova Science Publishers; 2008. pp. 35–69. [Google Scholar]
- 60.Feller G. Protein stability and enzyme activity at extreme biological temperatures. Journal of Physics: Condensed Matter. 2010;22, article 323101 doi: 10.1088/0953-8984/22/32/323101. [DOI] [PubMed] [Google Scholar]
- 61.Margesin R, Feller G. Biotechnological applications of psychrophiles. Environmental Technology. 2010;31(8-9):835–844. doi: 10.1080/09593331003663328. [DOI] [PubMed] [Google Scholar]
- 62.Piette F, Struvay C, Feller G. The protein folding challenge in psychrophiles: facts and current issues. Environmental Microbiology. 2011;13:1924–1933. doi: 10.1111/j.1462-2920.2011.02436.x. [DOI] [PubMed] [Google Scholar]
- 63.Roulling F, Piette F, Cipolla A, Struvay C, Feller G. Psychrophilic enzymes: cool responses to chilly problems. In: Horikoshi K, editor. Extremophiles Handbook. Berlin, Germany: Springer; 2011. pp. 891–916. [Google Scholar]
- 64.Struvay C, Feller G. Optimization to low temperature activity in psychrophilic enzymes. International Journal of Molecular Sciences. 2012;13:11643–11665. doi: 10.3390/ijms130911643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Araki T. Changes in rates of synthesis of individual proteins in a psychrophilic bacterium after a shift in temperature. Canadian Journal of Microbiology. 1991;37(11):840–847. doi: 10.1139/m91-145. [DOI] [PubMed] [Google Scholar]
- 66.Araki T. The effect of temperature shifts on protein synthesis by the psychrophilic bacterium Vibrio sp. strain ANT-300. Journal of General Microbiology. 1991;137(4):817–826. doi: 10.1099/00221287-137-4-817. [DOI] [PubMed] [Google Scholar]
- 67.Araki T. An analysis of the effect of changes in growth temperature on proteolysis in vivo in the psychrophilic bacterium Vibrio sp. strain ANT-300. Journal of General Microbiology. 1992;138(10):2075–2082. doi: 10.1099/00221287-138-10-2075. [DOI] [PubMed] [Google Scholar]
- 68.Hebraud M, Dubois E, Potier P, Labadie J. Effect of growth temperatures on the protein levels in a psychrotrophic bacterium, Pseudomonas fragi . Journal of Bacteriology. 1994;176(13):4017–4024. doi: 10.1128/jb.176.13.4017-4024.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hébraud M, Potier P. Cold shock response and low temperature adaptation in psychrotrophic bacteria. Journal of Molecular Microbiology and Biotechnology. 1999;1(2):211–219. [PubMed] [Google Scholar]
- 70.Hébraud M, Potier P. Cold acclimation and cold shock response in psychrotrophic bacteria. In: Inouye M, Yamanaka K, editors. Cold Shock, Response and Adaptation. Norfolk, UK: Horizon Scientific Press; 2000. pp. 41–60. [Google Scholar]
- 71.Goodchild A, Raftery M, Saunders NFW, Guilhaus M, Cavicchioli R. Biology of the cold adapted archaeon, Methanococcoides burtonii determined by proteomics using liquid chromatography-tandem mass spectrometry. Journal of Proteome Research. 2004;3(6):1164–1176. doi: 10.1021/pr0498988. [DOI] [PubMed] [Google Scholar]
- 72.Goodchild A, Saunders NFW, Ertan H, et al. A proteomic determination of cold adaptation in the Antarctic archaeon, Methanococcoides burtonii . Molecular Microbiology. 2004;53(1):309–321. doi: 10.1111/j.1365-2958.2004.04130.x. [DOI] [PubMed] [Google Scholar]
- 73.Goodchild A, Raftery M, Saunders NFW, Guilliaus M, Cavicchioli R. Cold adaptation of the antarctic archaeon, Methanococcoides burtonii assessed by proteomics using ICAT. Journal of Proteome Research. 2005;4(2):473–480. doi: 10.1021/pr049760p. [DOI] [PubMed] [Google Scholar]
- 74.Saunders NFW, Goodchild A, Raftery M, Guilhaus M, Curmi PMG, Cavicchioli R. Predicted roles for hypothetical proteins in the low-temperature expressed proteome of the antarctic archaeon Methanococcoides burtonii . Journal of Proteome Research. 2005;4(2):464–472. doi: 10.1021/pr049797+. [DOI] [PubMed] [Google Scholar]
- 75.Qiu Y, Kathariou S, Lubman DM. Proteomic analysis of cold adaptation in a Siberian permafrost bacterium—Exiguobacterium sibiricum 255-15 by two-dimensional liquid separation coupled with mass spectrometry. Proteomics. 2006;6(19):5221–5233. doi: 10.1002/pmic.200600071. [DOI] [PubMed] [Google Scholar]
- 76.Reva ON, Weinel C, Weinel M, et al. Functional genomics of stress response in Pseudomonas putida KT2440. Journal of Bacteriology. 2006;188(11):4079–4092. doi: 10.1128/JB.00101-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saunders NFW, Ng C, Raftery M, Guilhaus M, Goodchild A, Cavicchioli R. Proteomic and computational analysis of secreted proteins with type I signal peptides from the antarctic archaeon Methanococcoides burtonii . Journal of Proteome Research. 2006;5(9):2457–2464. doi: 10.1021/pr060220x. [DOI] [PubMed] [Google Scholar]
- 78.Bakermans C, Tollaksen SL, Giometti CS, Wilkerson C, Tiedje JM, Thomashow MF. Proteomic analysis of Psychrobacter cryohalolentis K5 during growth at subzero temperatures. Extremophiles. 2007;11(2):343–354. doi: 10.1007/s00792-006-0042-1. [DOI] [PubMed] [Google Scholar]
- 79.Kawamoto J, Kurihara T, Kitagawa M, Kato I, Esaki N. Proteomic studies of an Antarctic cold-adapted bacterium, Shewanella livingstonensis Ac10, for global identification of cold-inducible proteins. Extremophiles. 2007;11(6):819–826. doi: 10.1007/s00792-007-0098-6. [DOI] [PubMed] [Google Scholar]
- 80.Zheng S, Ponder MA, Shih JY, Tiedje JM, Thomashow MF, Lubman DM. A proteomic analysis of Psychrobacter articus 273-4 adaptation to low temperature and salinity using a 2-D liquid mapping approach. Electrophoresis. 2007;28(3):467–488. doi: 10.1002/elps.200600173. [DOI] [PubMed] [Google Scholar]
- 81.Bergholz PW, Bakermans C, Tiedje JM. Psychrobacter arcticus 273-4 Uses Resource Efficiency and Molecular Motion Adaptations for Subzero Temperature Growth. Journal of Bacteriology. 2009;191(7):2340–2352. doi: 10.1128/JB.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wecker P, Klockow C, Ellrott A, et al. Transcriptional response of the model planctomycete Rhodopirellula baltica SH1T to changing environmental conditions. BMC Genomics. 2009;10, article no. 1471:p. 410. doi: 10.1186/1471-2164-10-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Piette F, D’Amico S, Struvay C, et al. Proteomics of life at low temperatures: trigger factor is the primary chaperone in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Molecular Microbiology. 2010;76(1):120–132. doi: 10.1111/j.1365-2958.2010.07084.x. [DOI] [PubMed] [Google Scholar]
- 84.Ting L, Williams TJ, Cowley MJ, et al. Cold adaptation in the marine bacterium, Sphingopyxis alaskensis, assessed using quantitative proteomics. Environmental Microbiology. 2010;12(10):2658–2676. doi: 10.1111/j.1462-2920.2010.02235.x. [DOI] [PubMed] [Google Scholar]
- 85.Williams TJ, Burg DW, Raftery MJ, et al. Global proteomic analysis of the insoluble, soluble, and supernatant fractions of the psychrophilic archaeon Methanococcoides burtonii part I: the effect of growth temperature. Journal of Proteome Research. 2010;9(2):640–652. doi: 10.1021/pr900509n. [DOI] [PubMed] [Google Scholar]
- 86.Campanaro S, Williams TJ, Burg DW, et al. Temperature-dependent global gene expression in the Antarctic archaeon Methanococcoides burtonii . Environmental Microbiology. 2011;13:2018–2038. doi: 10.1111/j.1462-2920.2010.02367.x. [DOI] [PubMed] [Google Scholar]
- 87.Mykytczuk NCS, Trevors JT, Foote SJ, Leduc LG, Ferroni GD, Twine SM. Proteomic insights into cold adaptation of psychrotrophic and mesophilic Acidithiobacillus ferrooxidans strains. Antonie van Leeuwenhoek. 2011;100(2):259–277. doi: 10.1007/s10482-011-9584-z. [DOI] [PubMed] [Google Scholar]
- 88.Piette F, D’Amico S, Mazzucchelli G, Danchin A, Leprince P, Feller G. Life in the cold: a proteomic study of cold-repressed proteins in the antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Applied and Environmental Microbiology. 2011;77(11):3881–3883. doi: 10.1128/AEM.02757-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Williams TJ, Lauro FM, Ertan H, et al. Defining the response of a microorganism to temperatures that span its complete growth temperature range (-2°C to 28°C) using multiplex quantitative proteomics. Environmental Microbiology. 2011;13:2186–2203. doi: 10.1111/j.1462-2920.2011.02467.x. [DOI] [PubMed] [Google Scholar]
- 90.Jagannadham MV, Chowdhury C. Differential expression of membrane proteins helps Antarctic Pseudomonas syringae to acclimatize upon temperature variations. Journal of Proteomics. 2012;75:2488–2499. doi: 10.1016/j.jprot.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 91.Park J, Kawamoto J, Esaki N, Kurihara T. Identification of cold-inducible inner membrane proteins of the psychrotrophic bacterium, Shewanella livingstonensis Ac10, by proteomic analysis. Extremophiles. 2012;16:227–236. doi: 10.1007/s00792-011-0422-z. [DOI] [PubMed] [Google Scholar]
- 92.Pereira-Medrano AG, Margesin R, Wright PC. Proteome characterization of the unsequenced psychrophile Pedobacter cryoconitis using 15N metabolic labeling, tandem mass spectrometry, and a new bioinformatic workflow. Proteomics. 2012;12:775–789. doi: 10.1002/pmic.201100159. [DOI] [PubMed] [Google Scholar]
- 93.Piette F, Leprince P, Feller G. Is there a cold shock response in the Antarctic psychrophile Pseudoalteromonas haloplanktis? Extremophiles. 2012;16:681–683. doi: 10.1007/s00792-012-0456-x. [DOI] [PubMed] [Google Scholar]
- 94.Piette F, Struvay C, Godin A, Cipolla A, Feller G. Life in the cold: proteomics of the Antarctic bacterium Pseudoalteromonas haloplanktis . In: Heazlewood JL, Petzold CJ, editors. Proteomic Applications in Biology. Rijeka, Croatia: InTech; 2012. pp. 93–114. [Google Scholar]
- 95.Shin SC, Kim SJ, Lee JK, et al. Transcriptomics and comparative analysis of three antarctic notothenioid fishes. PLoS One. 2012;7, article e43762 doi: 10.1371/journal.pone.0043762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jung YH, Yi JY, Jung HJ, et al. Overexpression of Cold shock protein A of Psychromonas arctica KOPRI 22215 confers cold-resistance. Protein Journal. 2010;29(2):136–142. doi: 10.1007/s10930-010-9233-9. [DOI] [PubMed] [Google Scholar]
- 97.Uh JH, Jung YH, Lee YK, Lee HK, Im H. Rescue of a cold-sensitive mutant at low temperatures by cold shock proteins from Polaribacter irgensii KOPRI 22228. Journal of Microbiology. 2010;48(6):798–802. doi: 10.1007/s12275-010-0402-5. [DOI] [PubMed] [Google Scholar]
- 98.Lim J, Thomas T, Cavicchioli R. Low temperature regulated DEAD-box RNA helicase from the Antarctic archaeon, Methanococcoides burtonii . Journal of Molecular Biology. 2000;297(3):553–567. doi: 10.1006/jmbi.2000.3585. [DOI] [PubMed] [Google Scholar]
- 99.Cartier G, Lorieux F, Allemand F, Dreyfus M, Bizebard T. Cold adaptation in DEAD-box proteins. Biochemistry. 2010;49(12):2636–2646. doi: 10.1021/bi902082d. [DOI] [PubMed] [Google Scholar]
- 100.Yoshimune K, Galkin A, Kulakova L, Yoshimura T, Esaki N. Cold-active DnaK of an Antarctic psychrotroph Shewanella sp. Ac10 supporting the growth of dnaK-null mutant of Escherichia coli at cold temperatures. Extremophiles. 2005;9(2):145–150. doi: 10.1007/s00792-004-0429-9. [DOI] [PubMed] [Google Scholar]
- 101.Sung M, Im H, Lee K. Molecular cloning and chaperone activity of DnaK from cold-adapted bacteria, KOPRI22215. Bulletin of the Korean Chemical Society. 2011;32(6):1925–1930. [Google Scholar]
- 102.Ferrer M, Chernikova TN, Yakimov MM, Golyshin PN, Timmis KN. Chaperonins govern growth of Escherichia coli at low temperatures. Nature Biotechnology. 2003;21(11):1266–1267. doi: 10.1038/nbt1103-1266. [DOI] [PubMed] [Google Scholar]
- 103.Ferrer M, Lünsdorf H, Chernikova TN, Yakimov M, Timmis KN, Golyshin PN. Functional consequences of single:double ring transitions in chaperonins: life in the cold. Molecular Microbiology. 2004;53(1):167–182. doi: 10.1111/j.1365-2958.2004.04077.x. [DOI] [PubMed] [Google Scholar]
- 104.Strocchi M, Ferrer M, Timmis KN, Golyshin PN. Low temperature-induced systems failure in Escherichia coli: insights from rescue by cold-adapted chaperones. Proteomics. 2006;6(1):193–206. doi: 10.1002/pmic.200500031. [DOI] [PubMed] [Google Scholar]
- 105.Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nature Structural and Molecular Biology. 2009;16(6):574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 106.Kandror O, Goldberg AL. Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(10):4978–4981. doi: 10.1073/pnas.94.10.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baldwin RL. The search for folding intermediates and the mechanism of protein folding. Annual Review of Biophysics. 2008;37:1–21. doi: 10.1146/annurev.biophys.37.032807.125948. [DOI] [PubMed] [Google Scholar]
- 108.Siddiqui KS, Cavicchioli R. Cold-adapted enzymes. Annual Review of Biochemistry. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 109.Suzuki Y, Haruki M, Takano K, Morikawa M, Kanaya S. Possible involvement of an FKBP family member protein from a psychrotrophic bacterium Shewanella sp. SIB1 in cold-adaptation. European Journal of Biochemistry. 2004;271(7):1372–1381. doi: 10.1111/j.1432-1033.2004.04049.x. [DOI] [PubMed] [Google Scholar]
- 110.Zoldák G, Aumüller T, Lücke C, et al. A library of fluorescent peptides for exploring the substrate specificities of prolyl isomerases. Biochemistry. 2009;48(43):10423–10436. doi: 10.1021/bi9014242. [DOI] [PubMed] [Google Scholar]
- 111.Luke KA, Higgins CL, Wittung-Stafshede P. Thermodynamic stability and folding of proteins from hyperthermophilic organisms. FEBS Journal. 2007;274(16):4023–4033. doi: 10.1111/j.1742-4658.2007.05955.x. [DOI] [PubMed] [Google Scholar]
- 112.Mukaiyama A, Takano K. Slow unfolding of monomeric proteins from hyperthermophiles with reversible unfolding. International Journal of Molecular Sciences. 2009;10(3):1369–1385. doi: 10.3390/ijms10031369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Okada J, Okamoto T, Mukaiyama A, et al. Evolution and thermodynamics of the slow unfolding of hyperstable monomeric proteins. BMC Evolutionary Biology. 2010;10(1, article 207) doi: 10.1186/1471-2148-10-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cipolla A, D'Amico S, Barumandzadeh R, Matagne A, Feller G. Stepwise adaptations to low temperature as revealed by multiple mutants of psychrophilic alpha-amylase from Antarctic Bacterium. The Journal of Biological Chemistry. 2011;286:38348–38355. doi: 10.1074/jbc.M111.274423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Collins T, Matzapetakis M, Santos H. Backbone and side chain 1H, 15N and 13C assignments for a thiol-disulphide oxidoreductase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Biomolecular NMR Assignments. 2010;4(2):151–154. doi: 10.1007/s12104-010-9230-0. [DOI] [PubMed] [Google Scholar]
- 116.Aghajari N, Feller G, Gerday C, Haser R. Structures of the psychrophilic Alteromonas haloplanctisα-amylase give insights into cold adaptation at a molecular level. Structure. 1998;6(12):1503–1516. doi: 10.1016/s0969-2126(98)00149-x. [DOI] [PubMed] [Google Scholar]
- 117.Aghajari N, Feller G, Gerday C, Haser R. Crystal structures of the psychrophilic α-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Science. 1998;7(3):564–572. doi: 10.1002/pro.5560070304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Van Petegem F, Collins T, Meuwis MA, Gerday C, Feller G, Van Beeumen J. The structure of a cold-adapted family 8 xylanase at 1.3 Å resolution. Structural adaptations to cold and investigation of the active site. Journal of Biological Chemistry. 2003;278(9):7531–7539. doi: 10.1074/jbc.M206862200. [DOI] [PubMed] [Google Scholar]
- 119.Violot S, Aghajari N, Czjzek M, et al. Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering. Journal of Molecular Biology. 2005;348(5):1211–1224. doi: 10.1016/j.jmb.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 120.Merlino A, Krauss IR, Castellano I, et al. Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis . Journal of Structural Biology. 2010;172(3):343–352. doi: 10.1016/j.jsb.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 121.Alterio V, Aurilia V, Romanelli A, et al. Crystal structure of an S-formylglutathione hydrolase from Pseudoalteromonas haloplanktis TAC125. Biopolymers. 2010;93(8):669–677. doi: 10.1002/bip.21420. [DOI] [PubMed] [Google Scholar]
- 122.Aghajari N, Van Petegem F, Villeret V, et al. Crystal structures of a psychrophilic metalloprotease reveal new insights into catalysis by cold-adapted proteases. Proteins. 2003;50(4):636–647. doi: 10.1002/prot.10264. [DOI] [PubMed] [Google Scholar]
- 123.Michaux C, Massant J, Kerff F, et al. Crystal structure of a cold-adapted class C β-lactamase. FEBS Journal. 2008;275(8):1687–1697. doi: 10.1111/j.1742-4658.2008.06324.x. [DOI] [PubMed] [Google Scholar]
- 124.Russell RJM, Gerike U, Danson MJ, Hough DW, Taylor GL. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure. 1998;6(3):351–362. doi: 10.1016/s0969-2126(98)00037-9. [DOI] [PubMed] [Google Scholar]
- 125.Skalova T, Dohnalek J, Spiwok V, et al. Cold-active beta-galactosidase from Arthrobacter sp. C2-2 forms compact 660 kDa hexamers: crystal structure at 1. 9 Å resolution. Journal of Molecular Biology. 2005;353:282–294. doi: 10.1016/j.jmb.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 126.Almog O, González A, Godin N, et al. The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state. Proteins. 2009;74(2):489–496. doi: 10.1002/prot.22175. [DOI] [PubMed] [Google Scholar]
- 127.Nel AJM, Tuffin IM, Sewell BT, Cowan DA. Unique aliphatic amidase from a psychrotrophic and haloalkaliphilic Nesterenkonia isolate. Applied and Environmental Microbiology. 2011;77(11):3696–3702. doi: 10.1128/AEM.02726-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang E, Koutsioulis D, Leiros HKS, et al. Crystal Structure of Alkaline Phosphatase from the Antarctic Bacterium TAB5. Journal of Molecular Biology. 2007;366(4):1318–1331. doi: 10.1016/j.jmb.2006.11.079. [DOI] [PubMed] [Google Scholar]
- 129.Bauvois C, Jacquamet L, Huston AL, Borel F, Feller G, Ferrer JL. Crystal structure of the cold-active aminopeptidase from Colwellia psychrerythraea, a close structural homologue of the human bifunctional leukotriene A4 hydrolase. Journal of Biological Chemistry. 2008;283(34):23315–23325. doi: 10.1074/jbc.M802158200. [DOI] [PubMed] [Google Scholar]
- 130.Leiros HKS, Pey AL, Innselset M, et al. Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. Journal of Biological Chemistry. 2007;282(30):21973–21986. doi: 10.1074/jbc.M610174200. [DOI] [PubMed] [Google Scholar]
- 131.Kim SY, Hwang KY, Kim SH, Sung HC, Han YS, Cho Y. Structural basis for cold adaptation: sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum . Journal of Biological Chemistry. 1999;274(17):11761–11767. doi: 10.1074/jbc.274.17.11761. [DOI] [PubMed] [Google Scholar]
- 132.Arnórsdóttir J, Kristjánsson MM, Ficner R. Crystal structure of a subtilisin-like serine proteinase from a psychrotrophic Vibrio species reveals structural aspects of cold adaptation. FEBS Journal. 2005;272(3):832–845. doi: 10.1111/j.1742-4658.2005.04523.x. [DOI] [PubMed] [Google Scholar]
- 133.Fedøy AE, Yang N, Martinez A, Leiros HKS, Steen IH. Structural and Functional Properties of Isocitrate Dehydrogenase from the Psychrophilic Bacterium Desulfotalea psychrophila Reveal a Cold-active Enzyme with an Unusual High Thermal Stability. Journal of Molecular Biology. 2007;372(1):130–149. doi: 10.1016/j.jmb.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 134.De Vos D, Xu Y, Hulpiau P, Vergauwen B, Van Beeumen JJ. Structural investigation of cold activity and regulation of aspartate carbamoyltransferase from the extreme psychrophilic bacterium Moritella profunda . Journal of Molecular Biology. 2007;365(2):379–395. doi: 10.1016/j.jmb.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 135.Bae E, Phillips GN., Jr. Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. Journal of Biological Chemistry. 2004;279(27):28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- 136.Davlieva M, Shamoo Y. Structure and biochemical characterization of an adenylate kinase originating from the psychrophilic organism Marinibacillus marinus . Acta Crystallographica F. 2009;65(8):751–756. doi: 10.1107/S1744309109024348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Alvarez M, Zeelen JP, Mainfroid V, et al. Triose-phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio marinus: kinetic and structural properties. Journal of Biological Chemistry. 1998;273(4):2199–2206. doi: 10.1074/jbc.273.4.2199. [DOI] [PubMed] [Google Scholar]
- 138.Zhang SC, Sun M, Li T, et al. Structure analysis of a new psychrophilic marine protease. PLoS One. 2011;6, article e26939 doi: 10.1371/journal.pone.0026939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tsuruta H, Mikami B, Aizono Y. Crystal structure of cold-active protein-tyrosine phosphatase from a psychrophile, Shewanella sp. Journal of Biochemistry. 2005;137(1):69–77. doi: 10.1093/jb/mvi010. [DOI] [PubMed] [Google Scholar]
- 140.Riise EK, Lorentzen MS, Helland R, Smalås AO, Leiros HKS, Willassen NP. The first structure of a cold-active catalase from Vibrio salmonicida at 1.96 Å reveals structural aspects of cold adaptation. Acta Crystallographica D. 2007;63(2):135–148. doi: 10.1107/S0907444906043812. [DOI] [PubMed] [Google Scholar]
- 141.Altermark B, Helland R, Moe E, Willassen NP, Smalås AO. Structural adaptation of endonuclease I from the cold-adapted and halophilic bacterium Vibrio salmonicida . Acta Crystallographica D. 2008;64(4):368–376. doi: 10.1107/S0907444908000097. [DOI] [PubMed] [Google Scholar]
- 142.Jung SK, Jeong DG, Lee MS, et al. Structural basis for the cold adaptation of psychrophilic M37 lipase from Photobacterium lipolyticum . Proteins. 2008;71(1):476–484. doi: 10.1002/prot.21884. [DOI] [PubMed] [Google Scholar]
- 143.Pedersen HL, Willassen NP, Leiros I. The first structure of a cold-adapted superoxide dismutase (SOD): biochemical and structural characterization of iron SOD from Aliivibrio salmonicida . Acta Crystallographica F. 2009;65(2):84–92. doi: 10.1107/S1744309109001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Helland R, Larsen RL, Ásgeirsson B. The 1.4Å crystal structure of the large and cold-active Vibrio sp. alkaline phosphatase. Biochimica et Biophysica Acta. 2009;1794(2):297–308. doi: 10.1016/j.bbapap.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 145.Tsuruta H, Mikami B, Higashi T, Aizono Y. Crystal structure of cold-active alkaline phosphatase from the psychrophile Shewanella sp. Bioscience, Biotechnology and Biochemistry. 2010;74(1):69–74. doi: 10.1271/bbb.90563. [DOI] [PubMed] [Google Scholar]
- 146.De Backer M, McSweeney S, Rasmussen HB, Riise BW, Lindley P, Hough E. The 1.9 Å crystal structure of heat-labile shrimp alkaline phosphatase. Journal of Molecular Biology. 2002;318(5):1265–1274. doi: 10.1016/s0022-2836(02)00035-9. [DOI] [PubMed] [Google Scholar]
- 147.Coquelle N, Fioravanti E, Weik M, Vellieux F, Madern D. Activity, Stability and Structural Studies of Lactate Dehydrogenases Adapted to Extreme Thermal Environments. Journal of Molecular Biology. 2007;374(2):547–562. doi: 10.1016/j.jmb.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 148.Smalas AO, Heimstad ES, Hordvik A, Willassen NP, Male R. Cold adaption of enzymes: structural comparison between salmon and bovine trypsins. Proteins. 1994;20(2):149–166. doi: 10.1002/prot.340200205. [DOI] [PubMed] [Google Scholar]
- 149.Berglund GI, Willassen NP, Hordvik A, Smalas AO. Structure of native pancreatic elastase from North Atlantic salmon at 1.61 Angstrom resolution. Acta Crystallographica D. 1995;51(6):925–937. doi: 10.1107/S0907444995004835. [DOI] [PubMed] [Google Scholar]
- 150.Karlsen S, Hough E, Olsen RL. Structure and proposed amino-acid sequence of a pepsin from Atlantic cod (Gadus morhua) Acta Crystallographica D. 1998;54(1):32–46. doi: 10.1107/s090744499700810x. [DOI] [PubMed] [Google Scholar]
- 151.Leiros I, Moe E, Lanes O, Smalås AO, Willassen NP. The structure of uracil-DNA glycosylase from Atlantic cod (Gadus morhua) reveals cold-adaptation features. Acta Crystallographica D. 2003;59(8):1357–1365. doi: 10.1107/s0907444903011144. [DOI] [PubMed] [Google Scholar]
- 152.Peterson ME, Eisenthal R, Danson MJ, Spence A, Daniel RM. A new intrinsic thermal parameter for enzymes reveals true temperature optima. Journal of Biological Chemistry. 2004;279:20717–20722. doi: 10.1074/jbc.M309143200. [DOI] [PubMed] [Google Scholar]
- 153.Lee CK, Daniel RM, Shepherd C, et al. Eurythermalism and the temperature dependence of enzyme activity. FASEB Journal. 2007;21(8):1934–1941. doi: 10.1096/fj.06-7265com. [DOI] [PubMed] [Google Scholar]
- 154.Peterson ME, Daniel RM, Danson MJ, Eisenthal R. The dependence of enzyme activity on temperature: determination and validation of parameters. Biochemical Journal. 2007;402(2):331–337. doi: 10.1042/BJ20061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Collins T, Roulling F, Piette F, et al. Fundamentals of cold-adapted enzymes. In: Margesin R, Schinner F, Marx JC, Gerday C, editors. Psychrophiles, from Biodiversity to Biotechnology. Berlin, Germany: Springer; 2008. pp. 211–227. [Google Scholar]
- 156.Feller G, Lonhienne T, Deroanne C, Libioulle C, Van Beeumen J, Gerday C. Purification, characterization, and nucleotide sequence of the thermolabile α-amylase from the antarctic psychrotroph Alteromonas haloplanctis A23. Journal of Biological Chemistry. 1992;267(8):5217–5221. [PubMed] [Google Scholar]
- 157.Davail S, Feller G, Narinx E, Gerday C. Cold adaptation of proteins. Purification, characterization, and sequence of the heat-labile subtilisin from the Antarctic psychrophile Bacillus TA41. Journal of Biological Chemistry. 1994;269(26):17448–17453. [PubMed] [Google Scholar]
- 158.Georlette D, Damien B, Blaise V, et al. Structural and functional adaptations to extreme temperatures in psychrophilic, mesophilic, and thermophilic DNA ligases. Journal of Biological Chemistry. 2003;278(39):37015–37023. doi: 10.1074/jbc.M305142200. [DOI] [PubMed] [Google Scholar]
- 159.Collins T, Meuwis MA, Gerday C, Feller G. Activity, stability and flexibility in glycosidases adapted to extreme thermal environments. Journal of Molecular Biology. 2003;328(2):419–428. doi: 10.1016/s0022-2836(03)00287-0. [DOI] [PubMed] [Google Scholar]
- 160.D’Amico S, Marx JC, Gerday C, Feller G. Activity-stability relationships in extremophilic enzymes. Journal of Biological Chemistry. 2003;278(10):7891–7896. doi: 10.1074/jbc.M212508200. [DOI] [PubMed] [Google Scholar]
- 161.Siddiqui KS, Feller G, D’Amico S, Gerday C, Giaquinto L, Cavicchioli R. The active site is the least stable structure in the unfolding pathway of a multidomain cold-adapted α-amylase. Journal of Bacteriology. 2005;187(17):6197–6205. doi: 10.1128/JB.187.17.6197-6205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fields PA, Somero GN. Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(19):11476–11481. doi: 10.1073/pnas.95.19.11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lonhienne T, Gerday C, Feller G. Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochimica et Biophysica Acta. 2000;1543(1):1–10. doi: 10.1016/s0167-4838(00)00210-7. [DOI] [PubMed] [Google Scholar]
- 164.Chiuri R, Maiorano G, Rizzello A, et al. Exploring local flexibility/rigidity in psychrophilic and mesophilic carbonic anhydrases. Biophysical Journal. 2009;96(4):1586–1596. doi: 10.1016/j.bpj.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lonhienne T, Zoidakis J, Vorgias CE, Feller G, Gerday C, Bouriotis V. Modular structure, local flexibility and cold-activity of a novel chitobiase from a psychrophilic antarctic bacterium. Journal of Molecular Biology. 2001;310(2):291–297. doi: 10.1006/jmbi.2001.4774. [DOI] [PubMed] [Google Scholar]
- 166.Claverie P, Vigano C, Ruysschaert JM, Gerday C, Feller G. The precursor of a psychrophilic α-amylase: structural characterization and insights into cold adaptation. Biochimica et Biophysica Acta. 2003;1649(2):119–122. doi: 10.1016/s1570-9639(03)00184-5. [DOI] [PubMed] [Google Scholar]
- 167.Suzuki Y, Takano K, Kanaya S. Stabilities and activities of the N- and C-domains of FKBP22 from a psychrotrophic bacterium overproduced in Escherichia coli . FEBS Journal. 2005;272(3):632–642. doi: 10.1111/j.1742-4658.2004.04468.x. [DOI] [PubMed] [Google Scholar]
- 168.Qian M, Haser R, Buisson G, Duee E, Payan F. The active center of a mammalian α-amylase. Structure of the complex of a pancreatic α-amylase with a carbohydrate inhibitor refined to 2.2-Å resolution. Biochemistry. 1994;33(20):6284–6294. doi: 10.1021/bi00186a031. [DOI] [PubMed] [Google Scholar]
- 169.Aghajari N, Roth M, Haser R. Crystallographic evidence of a transglycosylation reaction: ternary complexes of a psychrophilic α-amylase. Biochemistry. 2002;41(13):4273–4280. doi: 10.1021/bi0160516. [DOI] [PubMed] [Google Scholar]
- 170.Bjelic S, Brandsdal BO, Åqvist J. Cold adaptation of enzyme reaction rates. Biochemistry. 2008;47(38):10049–10057. doi: 10.1021/bi801177k. [DOI] [PubMed] [Google Scholar]
- 171.De Vos D, Collins T, Nerinckx W, et al. Oligosaccharide binding in family 8 glycosidases: crystal structures of active-site mutants of the β-1,4-xylanase pXyl from Pseudoaltermonas haloplanktis TAH3a in complex with substrate and product. Biochemistry. 2006;45(15):4797–4807. doi: 10.1021/bi052193e. [DOI] [PubMed] [Google Scholar]
- 172.Olufsen M, Smalås AO, Brandsdal BO. Electrostatic interactions play an essential role in DNA repair and cold-adaptation of Uracil DNA glycosylase. Journal of Molecular Modeling. 2008;14(3):201–213. doi: 10.1007/s00894-007-0261-0. [DOI] [PubMed] [Google Scholar]
- 173.Raeder ILU, Moe E, Willassen NP, Smalås AO, Leiros I. Structure of uracil-DNA N-glycosylase (UNG) from Vibrio cholerae: mapping temperature adaptation through structural and mutational analysis. Acta Crystallographica F. 2010;66(2):130–136. doi: 10.1107/S1744309109052063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Papaleo E, Olufsen M, De Gioia L, Brandsdal BO. Optimization of electrostatics as a strategy for cold-adaptation: a case study of cold- and warm-active elastases. Journal of Molecular Graphics and Modelling. 2007;26(1):93–103. doi: 10.1016/j.jmgm.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 175.Gorfe AA, Brandsdal BO, Leiros HKS, Helland R, Smalås AO. Electrostatics of mesophilic and psychrophilic trypsin isoenzymes: qualitative evaluation of electrostatic differences at the substrate binding site. Proteins. 2000;40(2):207–217. [PubMed] [Google Scholar]
- 176.Smalås AO, Leiros HK, Os V, Willassen NP. Cold adapted enzymes. Biotechnology Annual Review. 2000;6:1–57. doi: 10.1016/s1387-2656(00)06018-x. [DOI] [PubMed] [Google Scholar]
- 177.Brandsdal BO, Smalås AO, Åqvist J. Electrostatic effects play a central role in cold adaptation of trypsin. FEBS Letters. 2001;499(1-2):171–175. doi: 10.1016/s0014-5793(01)02552-2. [DOI] [PubMed] [Google Scholar]
- 178.Moe E, Leiros I, Riise EK, et al. Optimisation of the surface electrostatics as a strategy for cold adaptation of uracil-DNA N-glycosylase (UNG) from Atlantic cod (Gadus morhua) Journal of Molecular Biology. 2004;343(5):1221–1230. doi: 10.1016/j.jmb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 179.Garsoux G, Lamotte J, Gerday C, Feller G. Kinetic and structural optimization to catalysis at low temperatures in a psychrophilic cellulase from the Antarctic bacterium Pseudoalteromonas haloplanktis . Biochemical Journal. 2004;384(2):247–253. doi: 10.1042/BJ20040325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tsigos I, Velonia K, Smonou I, Bouriotis V. Purification and characterization of an alcohol dehydrogenase from the Antarctic psychrophile Moraxella sp. TAE123. European Journal of Biochemistry. 1998;254(2):356–362. doi: 10.1046/j.1432-1327.1998.2540356.x. [DOI] [PubMed] [Google Scholar]
- 181.D’Amico S, Sohier JS, Feller G. Kinetics and energetics of ligand binding determined by microcalorimetry: insights into active site mobility in a psychrophilic α-amylase. Journal of Molecular Biology. 2006;358(5):1296–1304. doi: 10.1016/j.jmb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 182.Sun K, Camardella L, Di Prisco G, Hervé G. Properties of aspartate transcarbamylase from TAD1, a psychrophilic bacterial strain isolated from Antarctica. FEMS Microbiology Letters. 1998;164(2):375–382. doi: 10.1111/j.1574-6968.1998.tb13112.x. [DOI] [PubMed] [Google Scholar]
- 183.Xu Y, Zhang Y, Liang Z, Van De Casteele M, Legrain C, Glansdorff N. Aspartate carbamoyltransferase from a psychrophilic deep-sea bacterium, Vibrio strain 2693: properties of the enzyme, genetic organization and synthesis in Escherichia coli . Microbiology. 1998;144(5):1435–1441. doi: 10.1099/00221287-144-5-1435. [DOI] [PubMed] [Google Scholar]
- 184.Narinx E, Baise E, Gerday C. Subtilisin from psychrophilic antarctic bacteria: characterization and site-directed mutagenesis of residues possibly involved in the adaptation to cold. Protein Engineering. 1997;10(11):1271–1279. doi: 10.1093/protein/10.11.1271. [DOI] [PubMed] [Google Scholar]
- 185.Georlette D, Jónsson ZO, Van Petegem F, et al. A DNA ligase from the psychrophile Pseudoalteromonas haloplanktis gives insights into the adaptation of proteins to low temperatures. European Journal of Biochemistry. 2000;267(12):3502–3512. doi: 10.1046/j.1432-1327.2000.01377.x. [DOI] [PubMed] [Google Scholar]
- 186.Masullo M, Arcari P, De Paola B, Parmeggiani A, Bocchini V. Psychrophilic elongation factor Tu from the antarctic Moraxella sp. Tac II 25: biochemical characterization and cloning of the encoding gene. Biochemistry. 2000;39(50):15531–15539. doi: 10.1021/bi0018133. [DOI] [PubMed] [Google Scholar]
- 187.Ciardiello MA, Camardella L, Carratore V, Di Prisco G. L-Glutamate dehydrogenase from the Antarctic fish Chaenocephalus aceratus: primary structure, function and thermodynamic characterisation: relationship with cold adaptation. Biochimica et Biophysica Acta. 2000;1543(1):11–23. doi: 10.1016/s0167-4838(00)00186-2. [DOI] [PubMed] [Google Scholar]
- 188.Di Fraia R, Wilquet V, Ciardiello MA, et al. NADP+-dependent glutamate dehydrogenase in the Antarctic psychrotolerant bacterium Psychrobacter sp. TAD1: characterization, protein and DNA sequence, and relationship to other glutamate dehydrogenases. European Journal of Biochemistry. 2000;267(1):121–131. doi: 10.1046/j.1432-1327.2000.00972.x. [DOI] [PubMed] [Google Scholar]
- 189.D’Amico S, Gerday C, Feller G. Structural Determinants of Cold Adaptation and Stability in a Large Protein. Journal of Biological Chemistry. 2001;276(28):25791–25796. doi: 10.1074/jbc.M102741200. [DOI] [PubMed] [Google Scholar]
- 190.Xu Y, Feller G, Gerday C, Glansdorff N. Moritella cold-active dihydrofolate reductase: are there natural limits to optimization of catalytic efficiency at low temperature? Journal of Bacteriology. 2003;185(18):5519–5526. doi: 10.1128/JB.185.18.5519-5526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Evans RM, Behiry EM, Tey LH, Guo J, Loveridge EJ, Allemann RK. Catalysis by dihydrofolate reductase from the psychropiezophile Moritella profunda . ChemBioChem. 2010;11(14):2010–2017. doi: 10.1002/cbic.201000341. [DOI] [PubMed] [Google Scholar]
- 192.Altermark B, Niiranen L, Willassen NP, Smalås AO, Moe E. Comparative studies of endonuclease I from cold-adapted Vibrio salmonicida and mesophilic Vibrio cholerae . FEBS Journal. 2007;274(1):252–263. doi: 10.1111/j.1742-4658.2006.05580.x. [DOI] [PubMed] [Google Scholar]
- 193.Birolo L, Tutino LM, Fontanella B, et al. Aspartate aminotransferase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. Cloning, expression, properties, and molecular modelling. European Journal of Biochemistry. 2000;267(9):2790–2802. doi: 10.1046/j.1432-1327.2000.01299.x. [DOI] [PubMed] [Google Scholar]
- 194.Watanabe S, Yasutake Y, Tanaka I, Takada Y. Elucidation of stability determinants of cold-adapted monomeric isocitrate dehydrogenase from a psychrophilic bacterium, Colwellia maris, by construction of chimeric enzymes. Microbiology. 2005;151(4):1083–1094. doi: 10.1099/mic.0.27667-0. [DOI] [PubMed] [Google Scholar]
- 195.Collins T, Meuwis MA, Stals I, Claeyssens M, Feller G, Gerday C. A novel family 8 xylanase, functional and physicochemical characterization. Journal of Biological Chemistry. 2002;277(38):35133–35139. doi: 10.1074/jbc.M204517200. [DOI] [PubMed] [Google Scholar]
- 196.Xu Y, Feller G, Gerday C, Glansdorff N. Metabolic enzymes from psychrophilic bacteria: challenge of adaptation to low temperatures in ornithine carbamoyltransferase from Moritella abyssi . Journal of Bacteriology. 2003;185(7):2161–2168. doi: 10.1128/JB.185.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Gerike U, Danson MJ, Russell NJ, Hough DW. Sequencing and expression of the gene encoding a cold-active citrate synthase from an Antarctic bacterium, strain DS2-3R. European Journal of Biochemistry. 1997;248(1):49–57. doi: 10.1111/j.1432-1033.1997.00049.x. [DOI] [PubMed] [Google Scholar]
- 198.Li X, Jiang X, Li H, Ren D. Purine nucleoside phosphorylase from Pseudoalteromonas sp. Bsi590: molecular cloning, gene expression and characterization of the recombinant protein. Extremophiles. 2008;12(3):325–333. doi: 10.1007/s00792-007-0131-9. [DOI] [PubMed] [Google Scholar]
- 199.Tang MA, Motoshima H, Watanabe K. Fluorescence studies on the stability, flexibility and substrate-induced conformational changes of acetate kinases from psychrophilic and mesophilic bacteria. The Protein Journal. 2012;31:337–344. doi: 10.1007/s10930-012-9408-7. [DOI] [PubMed] [Google Scholar]
- 200.Ásgeirsson B, Cekan P. Microscopic rate-constants for substrate binding and acylation in cold-adaptation of trypsin I from Atlantic cod. FEBS Letters. 2006;580(19):4639–4644. doi: 10.1016/j.febslet.2006.07.043. [DOI] [PubMed] [Google Scholar]
- 201.Low PS, Bada JL, Somero GN. Temperature adaptation of enzymes: roles of the free energy, the enthalpy, and the entropy of activation. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(2):430–432. doi: 10.1073/pnas.70.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Feller G, Gerday C. Psychrophilic enzymes: molecular basis of cold adaptation. Cellular and Molecular Life Sciences. 1997;53(10):830–841. doi: 10.1007/s000180050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Thomas T, Cavicchioli R. Effect of temperature on stability and activity of elongation factor 2 proteins from Antarctic and thermophilic methanogens. Journal of Bacteriology. 2000;182(5):1328–1332. doi: 10.1128/jb.182.5.1328-1332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hoyoux A, Jennes I, Dubois P, et al. Cold-Adapted β-Galactosidase from the Antarctic Psychrophile Pseudoalteromonas haloplanktis . Applied and Environmental Microbiology. 2001;67(4):1529–1535. doi: 10.1128/AEM.67.4.1529-1535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lam SY, Yeung RCY, Yu TH, Sze KH, Wong KB. A rigidifying salt-bridge favors the activity of thermophilic enzyme at high temperatures at the expense of low-temperature activity. PLoS Biology. 2011;9(3) doi: 10.1371/journal.pbio.1001027.e1001027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gurd FR, Rothgeb TM. Motions in proteins. Advances in Protein Chemistry. 1979;33:73–165. doi: 10.1016/s0065-3233(08)60459-3. [DOI] [PubMed] [Google Scholar]
- 207.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450(7171):913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 208.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nature Chemical Biology. 2009;5(11):808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 209.Privalov PL. Stability of proteins. Proteins which do not present a single cooperative system. Advances in Protein Chemistry. 1982;35:1–104. [PubMed] [Google Scholar]
- 210.Závodszky P, Kardos J, Svingor A, Petsko GA. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(13):7406–7411. doi: 10.1073/pnas.95.13.7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Fields PA. Protein function at thermal extremes: balancing stability and flexibility. Comparative Biochemistry and Physiology A. 2001;129:417–431. doi: 10.1016/s1095-6433(00)00359-7. [DOI] [PubMed] [Google Scholar]
- 212.Shlyk-Kerner O, Samish I, Kaftan D, et al. Protein flexibility acclimatizes photosynthetic energy conversion to the ambient temperature. Nature. 2006;442(7104):827–830. doi: 10.1038/nature04947. [DOI] [PubMed] [Google Scholar]
- 213.Tehei M, Franzetti B, Madern D, et al. Adaptation to extreme environments: macromolecular dynamics in bacteria compared in vivo by neutron scattering. EMBO Reports. 2004;5(1):66–70. doi: 10.1038/sj.embor.7400049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tehei M, Zaccai G. Adaptation to extreme environments: macromolecular dynamics in complex systems. Biochimica et Biophysica Acta. 2005;1724(3):404–410. doi: 10.1016/j.bbagen.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 215.Somero GN. Proteins and temperature. Annual Review of Physiology. 1995;57:43–68. doi: 10.1146/annurev.ph.57.030195.000355. [DOI] [PubMed] [Google Scholar]
- 216.Huston AL, Haeggström JZ, Feller G. Cold adaptation of enzymes: structural, kinetic and microcalorimetric characterizations of an aminopeptidase from the Arctic psychrophile Colwellia psychrerythraea and of human leukotriene A4 hydrolase. Biochimica et Biophysica Acta. 2008;1784(11):1865–1872. doi: 10.1016/j.bbapap.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 217.Bentahir M, Feller G, Aittaleb M, et al. Structural, kinetic, and calorimetric characterization of the cold- active phosphoglycerate kinase from the antarctic Pseudomonas sp. TACII18. Journal of Biological Chemistry. 2000;275(15):11147–11153. doi: 10.1074/jbc.275.15.11147. [DOI] [PubMed] [Google Scholar]
- 218.Liang ZX, Tsigos I, Lee T, et al. Evidence for increased local flexibility in psychrophilic alcohol dehydrogenase relative to its thermophilic homologue. Biochemistry. 2004;43(46):14676–14683. doi: 10.1021/bi049004x. [DOI] [PubMed] [Google Scholar]
- 219.Fields PA, Wahlstrand BD, Somero GN. Intrinsic versus extrinsic stabilization of enzymes: the interaction of solutes and temperature on A4-lactate dehydrogenase orthologs from warm-adapted and cold-adapted marine fishes. European Journal of Biochemistry. 2001;268(16):4497–4505. doi: 10.1046/j.1432-1327.2001.02374.x. [DOI] [PubMed] [Google Scholar]
- 220.Svingor A, Kardos J, Hajdú I, Németh A, Závodszky P. A better enzyme to cope with cold: comparative flexibility studies on psychrotrophic, mesophilic, and thermophilic IPMDHs. Journal of Biological Chemistry. 2001;276(30):28121–28125. doi: 10.1074/jbc.M104432200. [DOI] [PubMed] [Google Scholar]
- 221.Liang ZX, Tsigos I, Bouriotis V, Klinman JP. Impact of protein flexibility on hydride-transfer parameters in thermophilic and psychrophilic alcohol dehydrogenases. Journal of the American Chemical Society. 2004;126:9500–9501. doi: 10.1021/ja047087z. [DOI] [PubMed] [Google Scholar]
- 222.Olufsen M, Smalås AO, Moe E, Brandsdal BO. Increased flexibility as a strategy for cold adaptation: a comparative molecular dynamics study of cold- and warm-active uracil DNA glycosylase. Journal of Biological Chemistry. 2005;280(18):18042–18048. doi: 10.1074/jbc.M500948200. [DOI] [PubMed] [Google Scholar]
- 223.Heimstad ES, Hansen LK, Smalas AO. Comparative molecular dynamics simulation studies of salmon and bovine trypsins in aqueous solution. Protein Engineering. 1995;8(4):379–388. doi: 10.1093/protein/8.4.379. [DOI] [PubMed] [Google Scholar]
- 224.Brandsdal BO, Heimstad ES, Sylte I, Smalås AO. Comparative molecular dynamics of mesophilic and psychrophilic protein homologues studied by 1.2 ns simulations. Journal of Biomolecular Structure and Dynamics. 1999;17(3):493–506. doi: 10.1080/07391102.1999.10508380. [DOI] [PubMed] [Google Scholar]
- 225.Adekoya OA, Helland R, Willassen NP, Sylte I. Comparative sequence and structure analysis reveal features of cold adaptation of an enzyme in the thermolysin family. Proteins. 2006;62(2):435–449. doi: 10.1002/prot.20773. [DOI] [PubMed] [Google Scholar]
- 226.Papaleo E, Riccardi L, Villa C, Fantucci P, De Gioia L. Flexibility and enzymatic cold-adaptation: a comparative molecular dynamics investigation of the elastase family. Biochimica et Biophysica Acta. 2006;1764(8):1397–1406. doi: 10.1016/j.bbapap.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 227.Spiwok V, Lipovova P, Skalova T, et al. Cold-active enzymes studied by comparative molecular dynamics simulation. Journal of Molecular Modeling. 2007;13:485–497. doi: 10.1007/s00894-006-0164-5. [DOI] [PubMed] [Google Scholar]
- 228.Papaleo E, Pasi M, Riccardi L, Sambi I, Fantucci P, Gioia LD. Protein flexibility in psychrophilic and mesophilic trypsins. Evidence of evolutionary conservation of protein dynamics in trypsin-like serine-proteases. FEBS Letters. 2008;582(6):1008–1018. doi: 10.1016/j.febslet.2008.02.048. [DOI] [PubMed] [Google Scholar]
- 229.Aurilia V, Rioux-Dubé JF, Marabotti A, Pézolet M, D’Auria S. Structure and dynamics of cold-adapted enzymes as investigated by FT-IR spectroscopy and MD. the case of an esterase from Pseudoalteromonas haloplanktis . Journal of Physical Chemistry B. 2009;113(22):7753–7761. doi: 10.1021/jp901921r. [DOI] [PubMed] [Google Scholar]
- 230.Pasi M, Riccardi L, Fantucci P, De Gioia L, Papaleo E. Dynamic properties of a psychrophilic α-amylase in comparison with a mesophilic homologue. Journal of Physical Chemistry B. 2009;113(41):13585–13595. doi: 10.1021/jp900790n. [DOI] [PubMed] [Google Scholar]
- 231.Papaleo E, Tiberti M, Invernizzi G, Pasi M, Ranzani V. Molecular determinants of enzyme cold adaptation: comparative structural and computational studies of cold- and warm-adapted enzymes. Current Protein and Peptide Science. 2011;12:657–683. doi: 10.2174/1389203711109070657. [DOI] [PubMed] [Google Scholar]
- 232.Tiberti M, Papaleo E. Dynamic properties of extremophilic subtilisin-like serine-proteases. Journal of Structural Biology. 2011;174(1):69–83. doi: 10.1016/j.jsb.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 233.Feller G, D’Amico D, Gerday C. Thermodynamic stability of a cold-active α-amylase from the antarctic bacterium Alteromonas haloplanctis . Biochemistry. 1999;38(14):4613–4619. doi: 10.1021/bi982650+. [DOI] [PubMed] [Google Scholar]
- 234.D’Amico S, Gerday C, Feller G. Dual effects of an extra disulfide bond on the activity and stability of a cold-adapted α-amylase. Journal of Biological Chemistry. 2002;277(48):46110–46115. doi: 10.1074/jbc.M207253200. [DOI] [PubMed] [Google Scholar]
- 235.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26(11):1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 236.Privalov PL. Stability of proteins small globular proteins. Advances in Protein Chemistry. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 237.Privalov PL. Cold denaturation of proteins. Critical Reviews in Biochemistry and Molecular Biology. 1990;25:281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- 238.Kumar S, Nussinov R. Experiment-guided thermodynamic simulations on reversible two-state proteins: implications for protein thermostability. Biophysical Chemistry. 2004;111(3):235–246. doi: 10.1016/j.bpc.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 239.Rees DC, Robertson AD. Some thermodynamic implications for the thermostability of proteins. Protein Science. 2001;10(6):1187–1194. doi: 10.1110/ps.180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Kumar S, Tsai CJ, Nussinov R. Maximal stabilities of reversible two-state proteins. Biochemistry. 2002;41(17):5359–5374. doi: 10.1021/bi012154c. [DOI] [PubMed] [Google Scholar]
- 241.García-Arribas O, Mateo R, Tomczak MM, Davies PL, Mateu MG. Thermodynamic stability of a cold-adapted protein, type III antifreeze protein, and energetic contribution of salt bridges. Protein Science. 2007;16(2):227–238. doi: 10.1110/ps.062448907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Makhatadze GI, Privalov PL. Energetics of protein structure. Advances in Protein Chemistry. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 243.Sigala PA, Kraut DA, Caaveiro JMM, et al. Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole. Journal of the American Chemical Society. 2008;130(41):13696–13708. doi: 10.1021/ja803928m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Russell NJ. Toward a molecular understanding of cold activity of enzymes from psychrophiles. Extremophiles. 2000;4(2):83–90. doi: 10.1007/s007920050141. [DOI] [PubMed] [Google Scholar]
- 245.Gianese G, Bossa F, Pascarella S. Comparative structural analysis of psychrophilic and meso- and thermophilic enzymes. Proteins. 2002;47(2):236–249. doi: 10.1002/prot.10084. [DOI] [PubMed] [Google Scholar]
- 246.Goldstein RA. Amino-acid interactions in psychrophiles, mesophiles, thermophiles, and hyperthermophiles: insights from the quasi-chemical approximation. Protein Science. 2007;16(9):1887–1895. doi: 10.1110/ps.072947007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Alimenti C, Vallesi A, Pedrini B, Wüthrich K, Luporini P. Molecular cold-adaptation: comparative analysis of two homologous families of psychrophilic and mesophilic signal proteins of the protozoan ciliate, Euplotes. IUBMB Life. 2009;61(8):838–845. doi: 10.1002/iub.228. [DOI] [PubMed] [Google Scholar]
- 248.Mavromatis K, Tsigos I, Tzanodaskalaki M, Kokkinidis M, Bouriotis V. Exploring the role of a glycine cluster in cold adaptation of an alkaline phosphatase. European Journal of Biochemistry. 2002;269(9):2330–2335. doi: 10.1046/j.1432-1033.2002.02895.x. [DOI] [PubMed] [Google Scholar]
- 249.Kulakova L, Galkin A, Nakayama T, Nishino T, Esaki N. Cold-active esterase from Psychrobacter sp. Ant300: gene cloning, characterization, and the effects of Gly→Pro substitution near the active site on its catalytic activity and stability. Biochimica et Biophysica Acta. 2004;1696(1):59–65. doi: 10.1016/j.bbapap.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 250.Sakaguchi M, Matsuzaki M, Niimiya K, Seino J, Sugahara Y, Kawakita M. Role of proline residues in conferring thermostability on aqualysin I. Journal of Biochemistry. 2007;141(2):213–220. doi: 10.1093/jb/mvm025. [DOI] [PubMed] [Google Scholar]
- 251.Siddiqui KS, Poljak A, Guilhaus M, et al. Role of lysine versus arginine in enzyme cold-adaptation: modifying lysine to homo-arginine stabilizes the cold-adapted α-amylase from Pseudoalteromonas haloplanktis . Proteins. 2006;64(2):486–501. doi: 10.1002/prot.20989. [DOI] [PubMed] [Google Scholar]
- 252.Xie BB, Bian F, Chen XL, et al. Cold adaptation of zinc metalloproteases in the thermolysin family from deep sea and arctic sea ice bacteria revealed by catalytic and structural properties and molecular dynamics: new insights into relationship between conformational flexibility and hydrogen bonding. Journal of Biological Chemistry. 2009;284(14):9257–9269. doi: 10.1074/jbc.M808421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Sælensminde G, Halskau JO, Jr., Jonassen I. Amino acid contacts in proteins adapted to different temperatures: hydrophobic interactions and surface charges play a key role. Extremophiles. 2009;13(1):11–20. doi: 10.1007/s00792-008-0192-4. [DOI] [PubMed] [Google Scholar]
- 254.Paredes DI, Watters K, Pitman DJ, Bystroff C, Dordick JS. Comparative void-volume analysis of psychrophilic and mesophilic enzymes: structural bioinformatics of psychrophilic enzymes reveals sources of core flexibility. BMC Structural Biology. 2011;11, article 42 doi: 10.1186/1472-6807-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Rentier-Delrue F, Mande SC, Moyens S, et al. Cloning and overexpression of the triosephosphate isomerase genes from psychrophilic and thermophilic bacteria. Structural comparison of the predicted protein sequences. Journal of Molecular Biology. 1993;229(1):85–93. doi: 10.1006/jmbi.1993.1010. [DOI] [PubMed] [Google Scholar]
- 256.Feller G, Zekhnini Z, Lamotte-Brasseur J, Gerday C. Enzymes from cold-adapted microorganisms. The class C β-lactamase from the antarctic psychrophile Psychrobacter immobilis A5. European Journal of Biochemistry. 1997;244(1):186–191. doi: 10.1111/j.1432-1033.1997.00186.x. [DOI] [PubMed] [Google Scholar]
- 257.Feller G, Payan F, Theys F, Qian M, Haser R, Gerday C. Stability and structural analysis of α-amylase from the antarctic psychrophile Alteromonas haloplanctis A23. European Journal of Biochemistry. 1994;222(2):441–447. doi: 10.1111/j.1432-1033.1994.tb18883.x. [DOI] [PubMed] [Google Scholar]
- 258.Siddiqui KS, Poljak A, Guilhaus M, et al. Role of disulfide bridges in the activity and stability of a cold-active α-amylase. Journal of Bacteriology. 2005;187(17):6206–6212. doi: 10.1128/JB.187.17.6206-6212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yip KSP, Stillman TJ, Britton KL, et al. The structure of Pyrococcus furiosus glutamate dehydrogenase reveals a key role for ion-pair networks in maintaining enzyme stability at extreme temperatures. Structure. 1995;3(11):1147–1158. doi: 10.1016/s0969-2126(01)00251-9. [DOI] [PubMed] [Google Scholar]
- 260.Vetriani C, Maeder DL, Tolliday N, et al. Protein thermostability above 100°C: a key role for ionic interactions. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(21):12300–12305. doi: 10.1073/pnas.95.21.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bell GS, Russell RJM, Connaris H, Hough DW, Danson MJ, Taylor GL. Stepwise adaptations of citrate synthase to survival at life’s extremes: from psychrophile to hyperthermophile. European Journal of Biochemistry. 2002;269(24):6250–6260. doi: 10.1046/j.1432-1033.2002.03344.x. [DOI] [PubMed] [Google Scholar]
- 262.Mandrich L, Pezzullo M, Del Vecchio P, Barone G, Rossi M, Manco G. Analysis of thermal adaptation in the HSL enzyme family. Journal of Molecular Biology. 2004;335(1):357–369. doi: 10.1016/j.jmb.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 263.Altermark B, Thorvaldsen S, Moe E, Smalås AO, Willassen NP. Sequence comparison and environmental adaptation of a bacterial endonuclease. Computational Biology and Chemistry. 2007;31(3):163–172. doi: 10.1016/j.compbiolchem.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 264.Thorvaldsen S, Hjerde E, Fenton C, Willassen NP. Molecular characterization of cold adaptation based on ortholog protein sequences from Vibrionaceae species. Extremophiles. 2007;11(5):719–732. doi: 10.1007/s00792-007-0093-y. [DOI] [PubMed] [Google Scholar]
- 265.Tronelli D, Maugini E, Bossa F, Pascarella S. Structural adaptation to low temperatures—analysis of the subunit interface of oligomeric psychrophilic enzymes. FEBS Journal. 2007;274(17):4595–4608. doi: 10.1111/j.1742-4658.2007.05988.x. [DOI] [PubMed] [Google Scholar]
- 266.Zheng B, Yang W, Zhao X, et al. Crystal structure of hyperthermophilic endo-beta-1, 4-glucanase: implications for catalytic mechanism and thermostability. The Journal of Biological Chemistry. 2012;287:8336–8346. doi: 10.1074/jbc.M111.266346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.De Vendittis E, Castellano I, Cotugno R, Ruocco MR, Raimo G, Masullo M. Adaptation of model proteins from cold to hot environments involves continuous and small adjustments of average parameters related to amino acid composition. Journal of Theoretical Biology. 2008;250(1):156–171. doi: 10.1016/j.jtbi.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 268.Sælensminde G, Halskau JO, Jr., Helland R, Willassen NP, Jonassen I. Structure-dependent relationships between growth temperature of prokaryotes and the amino acid frequency in their proteins. Extremophiles. 2007;11(4):585–596. doi: 10.1007/s00792-007-0072-3. [DOI] [PubMed] [Google Scholar]
- 269.Jahandideh M, Barkooie SMH, Jahandideh S, et al. Elucidating the protein cold-adaptation: investigation of the parameters enhancing protein psychrophilicity. Journal of Theoretical Biology. 2008;255(1):113–118. doi: 10.1016/j.jtbi.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 270.Khan MTH, Sylte I. Determinants for psychrophilic and thermophilic features of metallopeptidases of the M4 family. In Silico Biology. 2009;9(3):105–124. [PubMed] [Google Scholar]
- 271.Metpally RPR, Reddy BVB. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: insights into the molecular basis of cold adaptation of proteins. BMC Genomics. 2009;10, article 11 doi: 10.1186/1471-2164-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Aghajari N, Feller G, Gerday C, Haser R. Structural basis of α-amylase activation by chloride. Protein Science. 2002;11(6):1435–1441. doi: 10.1110/ps.0202602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.D’Amico S, Gerday C, Feller G. Temperature adaptation of proteins: engineering mesophilic-like activity and stability in a cold-adapted α-amylase. Journal of Molecular Biology. 2003;332(5):981–988. doi: 10.1016/j.jmb.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 274.Papaleo E, Pasi M, Tiberti M, De Gioia L. Molecular dynamics of mesophilic-like mutants of a cold-adapted enzyme: insights into distal effects induced by the mutations. PLoS One. 2011;6, article e24214 doi: 10.1371/journal.pone.0024214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Laurell H, Contreras JA, Castan I, Langin D, Holm C. Analysis of the psychrotolerant property of hormone-sensitive lipase through site-directed mutagenesis. Protein Engineering. 2000;13(10):711–717. doi: 10.1093/protein/13.10.711. [DOI] [PubMed] [Google Scholar]
- 276.Gerike U, Danson MJ, Hough DW. Cold-active citrate synthase: mutagenesis of active-site residues. Protein Engineering. 2001;14(9):655–661. doi: 10.1093/protein/14.9.655. [DOI] [PubMed] [Google Scholar]
- 277.Ohtani N, Haruki M, Morikawa M, Kanaya S. Heat labile ribonuclease HI from a psychrotrophic bacterium: gene cloning, characterization and site-directed mutagenesis. Protein Engineering. 2001;14(12):975–982. doi: 10.1093/protein/14.12.975. [DOI] [PubMed] [Google Scholar]
- 278.Tsigos I, Mavromatis K, Tzanodaskalaki M, Pozidis C, Kokkinidis M, Bouriotis V. Engineering the properties of a cold active enzyme through rational redesign of the active site. European Journal of Biochemistry. 2001;268(19):5074–5080. doi: 10.1046/j.0014-2956.2001.02432.x. [DOI] [PubMed] [Google Scholar]
- 279.Mavromatis K, Feller G, Kokkinidis M, Bouriotis V. Cold adaptation of a psychrophilic chitinase: a mutagenesis study. Protein Engineering. 2003;16(7):497–503. doi: 10.1093/protein/gzg069. [DOI] [PubMed] [Google Scholar]
- 280.Arnórsdóttir J, Helgadóttir S, Thorbjarnardóttir SH, Eggertsson G, Kristjánsson MM. Effect of selected Ser/Ala and Xaa/Pro mutations on the stability and catalytic properties of a cold adapted subtilisin-like serine proteinase. Biochimica et Biophysica Acta. 2007;1774(6):749–755. doi: 10.1016/j.bbapap.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 281.Yang YR, Zhu H, Fang N, et al. Cold-adapted maturation of thermophilic WF146 protease by mimicking the propeptide binding interactions of psychrophilic subtilisin S41. FEBS Letters. 2008;582(17):2620–2626. doi: 10.1016/j.febslet.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 282.Matsuo S, Shirai H, Takada Y. Isocitrate dehydrogenase isozymes from a psychrotrophic bacterium, Pseudomonas psychrophila . Archives of Microbiology. 2010;192(8):639–650. doi: 10.1007/s00203-010-0595-3. [DOI] [PubMed] [Google Scholar]
- 283.Miyazaki K, Wintrode PL, Grayling RA, Rubingh DN, Arnold FH. Directed evolution study of temperature adaptation in a psychrophilic enzyme. Journal of Molecular Biology. 2000;297(4):1015–1026. doi: 10.1006/jmbi.2000.3612. [DOI] [PubMed] [Google Scholar]
- 284.Wintrode PL, Miyazaki K, Arnold FH. Cold adaptation of a mesophilic subtilisin-like protease by laboratory evolution. Journal of Biological Chemistry. 2000;275(41):31635–31640. doi: 10.1074/jbc.M004503200. [DOI] [PubMed] [Google Scholar]
- 285.Bae E, Phillips GN., Jr. Roles of static and dynamic domains in stability and catalysis of adenylate kinase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2132–2137. doi: 10.1073/pnas.0507527103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Wintrode PL, Arnold FH. Temperature adaptation of enzymes: lessons from laboratory evolution. Advances in Protein Chemistry. 2000;55:161–225. doi: 10.1016/s0065-3233(01)55004-4. [DOI] [PubMed] [Google Scholar]
- 287.D’Amico S, Claverie P, Collins T, et al. Molecular basis of cold adaptation. Philosophical Transactions of the Royal Society B. 2002;357(1423):917–925. doi: 10.1098/rstb.2002.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Giver L, Gershenson A, Freskgard PO, Arnold FH. Directed evolution of a thermostable esterase. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(22):12809–12813. doi: 10.1073/pnas.95.22.12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Cherry JR, Lamsa MH, Schneider P, et al. Directed evolution of a fungal peroxidase. Nature Biotechnology. 1999;17(4):379–384. doi: 10.1038/7939. [DOI] [PubMed] [Google Scholar]
- 290.Noguchi A, Nishino T, Nakayama T. Kinetic and thermodynamic characterization of the cold activity acquired upon single amino-acid substitution near the active site of a thermostable α-glucosidase. Journal of Molecular Catalysis B. 2009;56(4):300–306. [Google Scholar]
- 291.Tosco A, Birolo L, Madonna S, Lolli G, Sannia G, Marino G. GroEL from the psychrophilic bacterium Pseudoalteromonas haloplanktis TAC 125: molecular characterization and gene cloning. Extremophiles. 2003;7(1):17–28. doi: 10.1007/s00792-002-0291-6. [DOI] [PubMed] [Google Scholar]
- 292.Cotugno R, Ruocco M, Marco S, et al. Differential cold-adaptation among protein components of the thioredoxin system in the psychrophilic eubacterium Pseudoalteromonas haloplanktis TAC 125. Molecular BioSystems. 2009;5(5):519–528. doi: 10.1039/b818467d. [DOI] [PubMed] [Google Scholar]
- 293.Falasca P, Evangelista G, Cotugno R, et al. Properties of the endogenous components of the thioredoxin system in the psychrophilic eubacterium Pseudoalteromonas haloplanktis TAC 125. Extremophiles. 2012;16:539–552. doi: 10.1007/s00792-012-0453-0. [DOI] [PubMed] [Google Scholar]
- 294.Dinner AR, Šalib A, Smitha LJ, Dobsona CM, Karplus M. Understanding protein folding via free-energy surfaces from theory and experiment. Trends in Biochemical Sciences. 2000;25(7):331–339. doi: 10.1016/s0968-0004(00)01610-8. [DOI] [PubMed] [Google Scholar]
- 295.Schultz CP. Illuminating folding intermediates. Nature Structural & Molecular Biology. 2000;7:7–10. doi: 10.1038/71197. [DOI] [PubMed] [Google Scholar]
- 296.Tsai CJ, Ma B, Nussinov R. Folding and binding cascades: shifts in energy landscapes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(18):9970–9972. doi: 10.1073/pnas.96.18.9970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Science. 2000;9(1):10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Ma B, Kumar S, Tsai CJ, Hu Z, Nussinov R. Transition-state ensemble in enzyme catalysis: possibility, reality, or necessity? Journal of Theoretical Biology. 2000;203(4):383–397. doi: 10.1006/jtbi.2000.1097. [DOI] [PubMed] [Google Scholar]
- 299.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Free-energy landscape of enzyme catalysis. Biochemistry. 2008;47(11):3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 300.Bandi S, Bowler BE. Probing the bottom of a folding funnel using conformationally gated electron transfer reactions. Journal of the American Chemical Society. 2008;130(24):7540–7541. doi: 10.1021/ja801941r. [DOI] [PubMed] [Google Scholar]
- 301.Schrank TP, Bolen DW, Hilser VJ. Rational modulation of conformational fluctuations in adenylate kinase reveals a local unfolding mechanism for allostery and functional adaptation in proteins. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(40):16984–16989. doi: 10.1073/pnas.0906510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Mereghetti P, Riccardi L, Brandsdal BO, Fantucci P, De Gioia L, Papaleo E. Near native-state conformational landscape of psychrophilic and mesophilic enzymes: probing the folding funnel model. Journal of Physical Chemistry B. 2010;114(22):7609–7619. doi: 10.1021/jp911523h. [DOI] [PubMed] [Google Scholar]
- 303.Margesin R, Schinner F. Biotechnological Applications of Cold-Adapted Organisms. Heidelberg, Germany: Springer; 1999. [Google Scholar]
- 304.Gerday C, Aittaleb M, Bentahir M, et al. Cold-adapted enzymes: from fundamentals to biotechnology. Trends in Biotechnology. 2000;18(3):103–107. doi: 10.1016/s0167-7799(99)01413-4. [DOI] [PubMed] [Google Scholar]
- 305.Allen D, Huston AL, Weels LE, Deming JW. Biotechnological use of psychrophiles. In: Bitton G, editor. Encyclopedia of Environmental Microbiology. New York, NY, USA: John Wiley & Sons; 2002. pp. 1–17. [Google Scholar]
- 306.Cavicchioli R, Siddiqui KS, Andrews D, Sowers KR. Low-temperature extremophiles and their applications. Current Opinion in Biotechnology. 2002;13(3):253–261. doi: 10.1016/s0958-1669(02)00317-8. [DOI] [PubMed] [Google Scholar]
- 307.Marx JC, Collins T, D’Amico S, Feller G, Gerday C. Cold-adapted enzymes from marine Antarctic microorganisms. Marine Biotechnology. 2007;9(3):293–304. doi: 10.1007/s10126-006-6103-8. [DOI] [PubMed] [Google Scholar]
- 308.Cavicchioli R, Charlton T, Ertan H, Omar SM, Siddiqui KS, Williams TJ. Biotechnological uses of enzymes from psychrophiles. Microbial Biotechnology. 2011;4(4):449–460. doi: 10.1111/j.1751-7915.2011.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kobori H, Sullivan CW, Shizuya H. Heat-labile alkaline phosphatase from Antarctic bacteria: rapid 5’ end-labeling of nucleic acids. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(21):6691–6695. doi: 10.1073/pnas.81.21.6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Rina M, Pozidis C, Mavromatis K, Tzanodaskalaki M, Kokkinidis M, Bouriotis V. Alkaline phosphatase from the Antarctic strain TAB5 properties and psychrophilic adaptations. European Journal of Biochemistry. 2000;267(4):1230–1238. doi: 10.1046/j.1432-1327.2000.01127.x. [DOI] [PubMed] [Google Scholar]
- 311.Koutsioulis D, Wang E, Tzanodaskalaki M, et al. Directed evolution on the cold adapted properties of TAB5 alkaline phosphatase. Protein Engineering, Design and Selection. 2008;21(5):319–327. doi: 10.1093/protein/gzn009. [DOI] [PubMed] [Google Scholar]
- 312.Babu J, Ramteke PW, Thomas G. Cold active microbial lipases: some hot issues and recent developments. Biotechnology Advances. 2008;26(5):457–470. doi: 10.1016/j.biotechadv.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 313.Lohan D, Johnston S. UNU-IAS report: bioprospecting in Antarctica. http://www.ias.unu.edu/binaries2/antarctic_bioprospecting.pdf.
- 314.Collins T, De Vos D, Hoyoux A, et al. Study of the active site residues of a glycoside hydrolase family 8 xylanase. Journal of Molecular Biology. 2005;354(2):425–435. doi: 10.1016/j.jmb.2005.09.064. [DOI] [PubMed] [Google Scholar]
- 315.Collins T, Hoyoux A, Dutron A, et al. Use of glycoside hydrolase family 8 xylanases in baking. Journal of Cereal Science. 2006;43(1):79–84. [Google Scholar]
- 316.Dornez E, Verjans P, Arnaut F, Delcour JA, Courtin CM. Use of psychrophilic xylanases provides insight into the xylanase functionality in bread making. Journal of Agricultural and Food Chemistry. 2011;59:9553–9562. doi: 10.1021/jf201752g. [DOI] [PubMed] [Google Scholar]
- 317.Siddiqui KS, Poljak A, Cavicchioli R. Improved activity and stability of alkaline phosphatases from psychrophilic and mesophilic organisms by chemically modifying aliphatic or amino groups using tetracarboxy-benzophenone derivatives. Cellular and Molecular Biology. 2004;50(5):657–667. [PubMed] [Google Scholar]
- 318.Siddiqui KS, Cavicchioli R. Improved thermal stability and activity in the cold-adapted lipase B from Candida antarctica following chemical modification with oxidized polysaccharides. Extremophiles. 2005;9(6):471–476. doi: 10.1007/s00792-005-0464-1. [DOI] [PubMed] [Google Scholar]
- 319.Siddiqui KS, Parkin DM, Curmi PMG, et al. A novel approach for enhancing the catalytic efficiency of a protease at low temperature: reduction in substrate inhibition by chemical modification. Biotechnology and Bioengineering. 2009;103(4):676–686. doi: 10.1002/bit.22300. [DOI] [PubMed] [Google Scholar]
- 320.Smal C, Aran M, Lanzarotti E, et al. 1H, 15N and 13C chemical shift assignments of the BA42 protein of the psychrophilic bacteria Bizionia argentinensis sp. nov. Biomolecular NMR Assignments. 2012;6(2):181–183. doi: 10.1007/s12104-011-9351-0. [DOI] [PubMed] [Google Scholar]


