Abstract
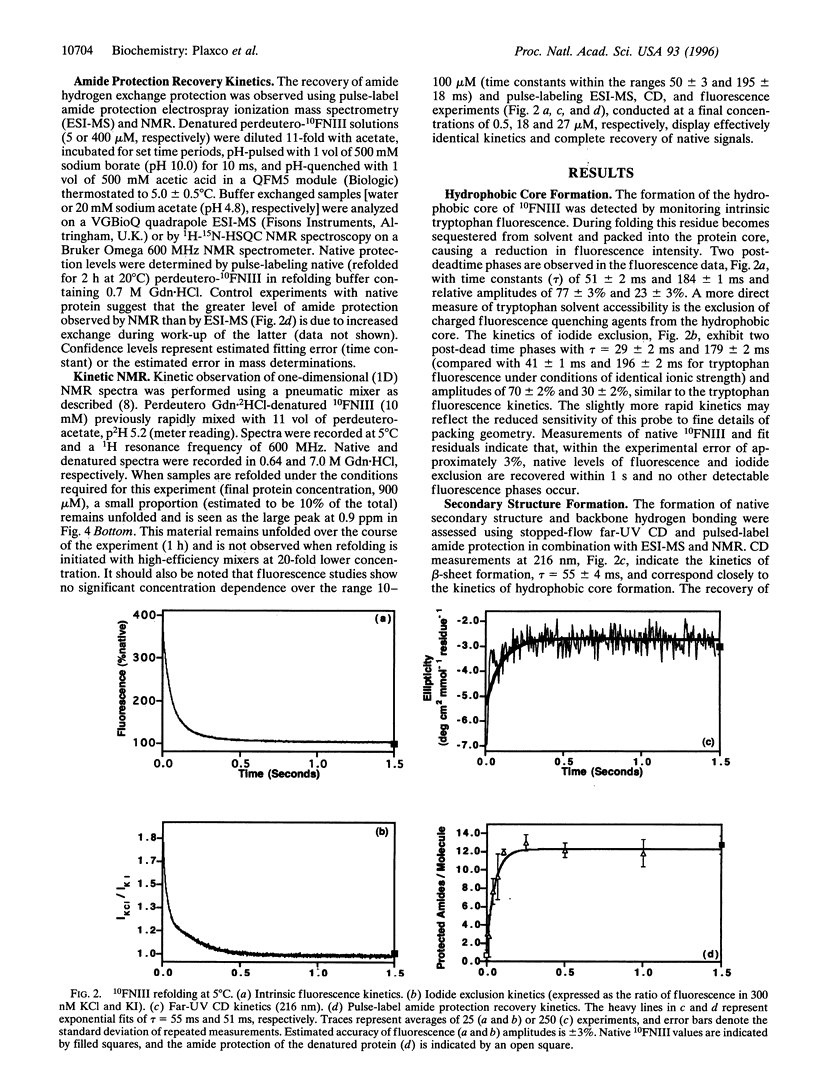
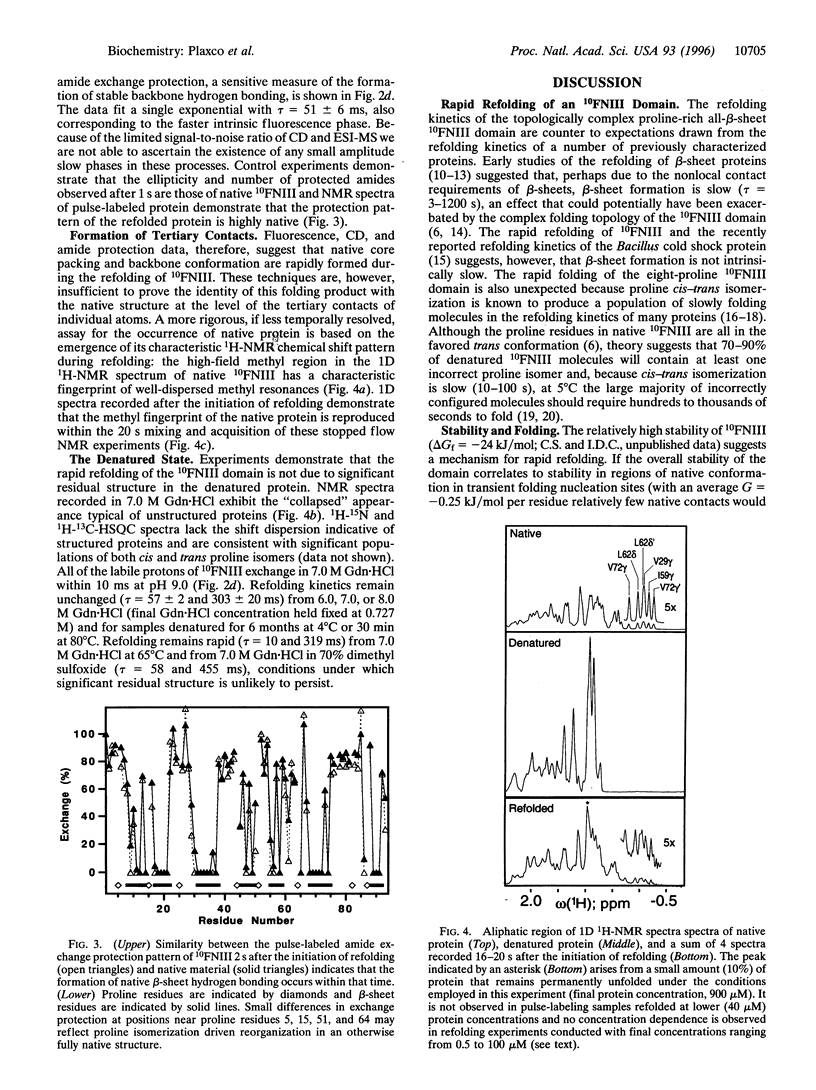
Fibronectin type III modules contain approximately 90 residues and are an extremely common building block of animal proteins. Despite containing a complex all-beta-sheet topology and eight prolines, the refolding of the 10th type III module of human fibronectin has been found to be very rapid, with native core packing, amide hydrogen bonding, and backbone conformation all recovered within 1 s at 5 degrees C. These observations indicate that this domain can overcome many structural characteristics often thought to slow the folding process.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balbach J., Forge V., van Nuland N. A., Winder S. L., Hore P. J., Dobson C. M. Following protein folding in real time using NMR spectroscopy. Nat Struct Biol. 1995 Oct;2(10):865–870. doi: 10.1038/nsb1095-865. [DOI] [PubMed] [Google Scholar]
- Bork P., Doolittle R. F. Proposed acquisition of an animal protein domain by bacteria. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8990–8994. doi: 10.1073/pnas.89.19.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazin W. J., Kördel J., Drakenberg T., Thulin E., Brodin P., Grundström T., Forsén S. Proline isomerism leads to multiple folded conformations of calbindin D9k: direct evidence from two-dimensional 1H NMR spectroscopy. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2195–2198. doi: 10.1073/pnas.86.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook K. H., Schmid F. X., Baldwin R. L. Role of proline isomerization in folding of ribonuclease A at low temperatures. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6157–6161. doi: 10.1073/pnas.76.12.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton E. T. Possible implications of many proline residues for the kinetics of protein unfolding and refolding. J Mol Biol. 1978 Nov 5;125(3):401–406. doi: 10.1016/0022-2836(78)90411-4. [DOI] [PubMed] [Google Scholar]
- Erickson H. P., Carrell N., McDonagh J. Fibronectin molecule visualized in electron microscopy: a long, thin, flexible strand. J Cell Biol. 1981 Dec;91(3 Pt 1):673–678. doi: 10.1083/jcb.91.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H. P. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. A., Kautz R. A., Fox R. O., Dobson C. M. A magnetization-transfer nuclear magnetic resonance study of the folding of staphylococcal nuclease. Biochemistry. 1989 Jan 10;28(1):362–370. doi: 10.1021/bi00427a050. [DOI] [PubMed] [Google Scholar]
- Finkelstein A. V. Rate of beta-structure formation in polypeptides. Proteins. 1991;9(1):23–27. doi: 10.1002/prot.340090104. [DOI] [PubMed] [Google Scholar]
- Goto Y., Hamaguchi K. Unfolding and refolding of the reduced constant fragment of the immunoglobulin light chain. Kinetic role of the intrachain disulfide bond. J Mol Biol. 1982 Apr 25;156(4):911–926. doi: 10.1016/0022-2836(82)90147-4. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Kohler H. H., Schmid F. X. Kinetic coupling between protein folding and prolyl isomerization. I. Theoretical models. J Mol Biol. 1992 Mar 5;224(1):217–229. doi: 10.1016/0022-2836(92)90585-8. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Quaas R., Hahn U., Schmid F. X. Folding of ribonuclease T1. 2. Kinetic models for the folding and unfolding reactions. Biochemistry. 1990 Mar 27;29(12):3061–3070. doi: 10.1021/bi00464a024. [DOI] [PubMed] [Google Scholar]
- Levitt M. Effect of proline residues on protein folding. J Mol Biol. 1981 Jan 5;145(1):251–263. doi: 10.1016/0022-2836(81)90342-9. [DOI] [PubMed] [Google Scholar]
- Litvinovich S. V., Ingham K. C. Interactions between type III domains in the 110 kDa cell-binding fragment of fibronectin. J Mol Biol. 1995 May 5;248(3):611–626. doi: 10.1006/jmbi.1995.0246. [DOI] [PubMed] [Google Scholar]
- Main A. L., Harvey T. S., Baron M., Boyd J., Campbell I. D. The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell. 1992 Nov 13;71(4):671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Grant K. E. The role of the ninth and tenth type III domains of human fibronectin in cell adhesion. FEBS Lett. 1994 Mar 7;340(3):197–201. doi: 10.1016/0014-5793(94)80137-1. [DOI] [PubMed] [Google Scholar]
- Murray A. J., Lewis S. J., Barclay A. N., Brady R. L. One sequence, two folds: a metastable structure of CD2. Proc Natl Acad Sci U S A. 1995 Aug 1;92(16):7337–7341. doi: 10.1073/pnas.92.16.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo C. A., Jones D. T., Thornton J. M. Protein superfamilies and domain superfolds. Nature. 1994 Dec 15;372(6507):631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- Politou A. S., Gautel M., Pfuhl M., Labeit S., Pastore A. Immunoglobulin-type domains of titin: same fold, different stability? Biochemistry. 1994 Apr 19;33(15):4730–4737. doi: 10.1021/bi00181a604. [DOI] [PubMed] [Google Scholar]
- Ropson I. J., Gordon J. I., Frieden C. Folding of a predominantly beta-structure protein: rat intestinal fatty acid binding protein. Biochemistry. 1990 Oct 16;29(41):9591–9599. doi: 10.1021/bi00493a013. [DOI] [PubMed] [Google Scholar]
- Rudolph R., Siebendritt R., Nesslaŭer G., Sharma A. K., Jaenicke R. Folding of an all-beta protein: independent domain folding in gamma II-crystallin from calf eye lens. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4625–4629. doi: 10.1073/pnas.87.12.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler T., Herrler M., Marahiel M. A., Schmid F. X. Extremely rapid protein folding in the absence of intermediates. Nat Struct Biol. 1995 Aug;2(8):663–673. doi: 10.1038/nsb0895-663. [DOI] [PubMed] [Google Scholar]
- Schmid F. X. A native-like intermediate on the ribonuclease A folding pathway. 1. Detection by tyrosine fluorescence changes. Eur J Biochem. 1981;114(1):105–109. doi: 10.1111/j.1432-1033.1981.tb06179.x. [DOI] [PubMed] [Google Scholar]
- Schmid F. X., Baldwin R. L. Acid catalysis of the formation of the slow-folding species of RNase A: evidence that the reaction is proline isomerization. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4764–4768. doi: 10.1073/pnas.75.10.4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F. X., Blaschek H. A native-like intermediate on the ribonuclease A folding pathway. 2. Comparison of its properties to native ribonuclease A. Eur J Biochem. 1981;114(1):111–117. doi: 10.1111/j.1432-1033.1981.tb06180.x. [DOI] [PubMed] [Google Scholar]
- Schmid F. X., Mayr L. M., Mücke M., Schönbrunner E. R. Prolyl isomerases: role in protein folding. Adv Protein Chem. 1993;44:25–66. doi: 10.1016/s0065-3233(08)60563-x. [DOI] [PubMed] [Google Scholar]
- Schreiber G., Fersht A. R. The refolding of cis- and trans-peptidylprolyl isomers of barstar. Biochemistry. 1993 Oct 19;32(41):11195–11203. doi: 10.1021/bi00092a032. [DOI] [PubMed] [Google Scholar]
- Soteriou A., Clarke A., Martin S., Trinick J. Titin folding energy and elasticity. Proc Biol Sci. 1993 Nov 22;254(1340):83–86. doi: 10.1098/rspb.1993.0130. [DOI] [PubMed] [Google Scholar]
- Varley P., Gronenborn A. M., Christensen H., Wingfield P. T., Pain R. H., Clore G. M. Kinetics of folding of the all-beta sheet protein interleukin-1 beta. Science. 1993 May 21;260(5111):1110–1113. doi: 10.1126/science.8493553. [DOI] [PubMed] [Google Scholar]



