The transcriptional landscape of eukaryotic genomes can be bewildering. A recent estimate holds that processed RNA transcripts cover more than 60% of the human genome [1]; protein-coding exons, in contrast, constitute a mere 1.2% at last report [2]. In addition, genome-wide discoveries of “abnormal” transcripts, such as chimeric [3] and circular RNAs [4] hint at further layers of complexity in our transcriptome. The question that naturally follows, is whether these non-protein coding, sometimes scrambled transcripts have any function, particularly in influencing events of genome evolution or somatic differentiation.
Although studies of non-coding RNAs have clearly demonstrated an expanding catalog of functional RNAs that regulate genes and genomes [5], the ability of RNA to program genome rearrangement remains largely unexplored. Testing such a possibility is important because DNA rearrangements contribute tremendously to genome evolution, and occur frequently on a developmental scale in cancerous cells [6] as well as healthy tissue [7]. The unique biology of nuclear dimorphism in ciliated protozoa provides excellent models for studying genome rearrangement, and previous studies demonstrated a role for non-coding RNA in elaborately regulating the process of genome remodeling [8–11]. Here, we first briefly describe RNA-guided genome rearrangements in ciliates, and then discuss both the possibilities of and evidence for the presence of similar phenomena in other eukaryotes.
Non-coding RNAs guide genome rearrangement in ciliates
Ciliates are single-celled eukaryotes that harbor two structurally and functionally different nuclei. The somatic macronucleus is transcriptionally active and responsible for asexual reproduction, whereas the germline micronucleus is transcriptionally silent during asexual growth, but transmits genetic information to the next sexual generation. The micronuclear genome is disrupted by non-coding sequences, including transposons, satellite repeats, and internally-eliminated sequences, all of which undergo programmed deletion when the macronucleus develops from a copy of micronucleus during conjugation, the sexual phase of the ciliate life cycle. Furthermore, gene pieces that constitute the functional somatic chromosomes can be scrambled in the germline genome of some species, including Oxytricha. Therefore, macronuclear development in these species requires complex genome rearrangements to sort and reorder thousands of DNA segments [12].
In the ciliate species Tetrahymena and Paramecium, where genome rearrangement requires DNA elimination but not unscrambling, a class of non-coding small RNAs called scan RNAs promotes the deletion of homologous genomic regions [8,11]. In contrast, small RNAs in Oxytricha appear to specify DNA sequences for retention [13]. Consistent with these models, injections of synthetic small RNA could lead to either the deletion or retention of corresponding DNA segments [11,13], respectively, in the different model systems. Furthermore, the correct assembly of gene pieces in Oxytricha relies on a maternal supply of RNA templates, essentially a cached copy of the somatic genome [10]. Supporting this RNA template model, injection of artificial RNA templates can alter the pattern of DNA segments to match the synthetic template, demonstrating the ability of RNA to reprogram genome rearrangements [10]. In addition, substitutions close to recombination junctions occasionally transfer from RNA templates to the DNA product, suggesting local RNA-templated DNA repair at the junctions [10].
From genetic and evolutionary perspectives, the ability of RNA to program genome rearrangements and to transfer substitutions to the somatic genome of the next sexual generation offers a particular pathway for the epigenetic inheritance of certain acquired traits. For example, if rearrangements or single-nucleotide substitutions occur either in the somatic genome during vegetative growth, or during RNA transcription of templates, then these mutations have the opportunity to transfer to the somatic genome of the sexual progeny, bypassing inheritance via the germline. Provocatively, such a mechanism could, in principle, allow epigenetic fixation of those somatic mutations that confer a selective advantage to the host. Multiple experiments [10,13,14] documenting transgenerational inheritance of RNA-mediated somatic features in Oxytricha support the conclusion that its somatic nucleus is truly an epigenome.
RNA-mediated genome rearrangement may occur in other eukaryotes
The phenomenon of RNA-mediated genome rearrangement may not be unique to ciliates. Mechanistically, direct RNA-templated DNA synthesis is not only manifest in classic examples of reverse transcription in viruses, retrotransposons, and eukaryotic telomere maintenance, but also in the context of double-stranded DNA break repair in yeast [15] and humans [16]. Furthermore, yeast DNA polymerases α and δ have an ability to copy short RNA tracts in vitro [15], suggesting that RNA-templated DNA synthesis can occur via routes other than classic reverse transcription.
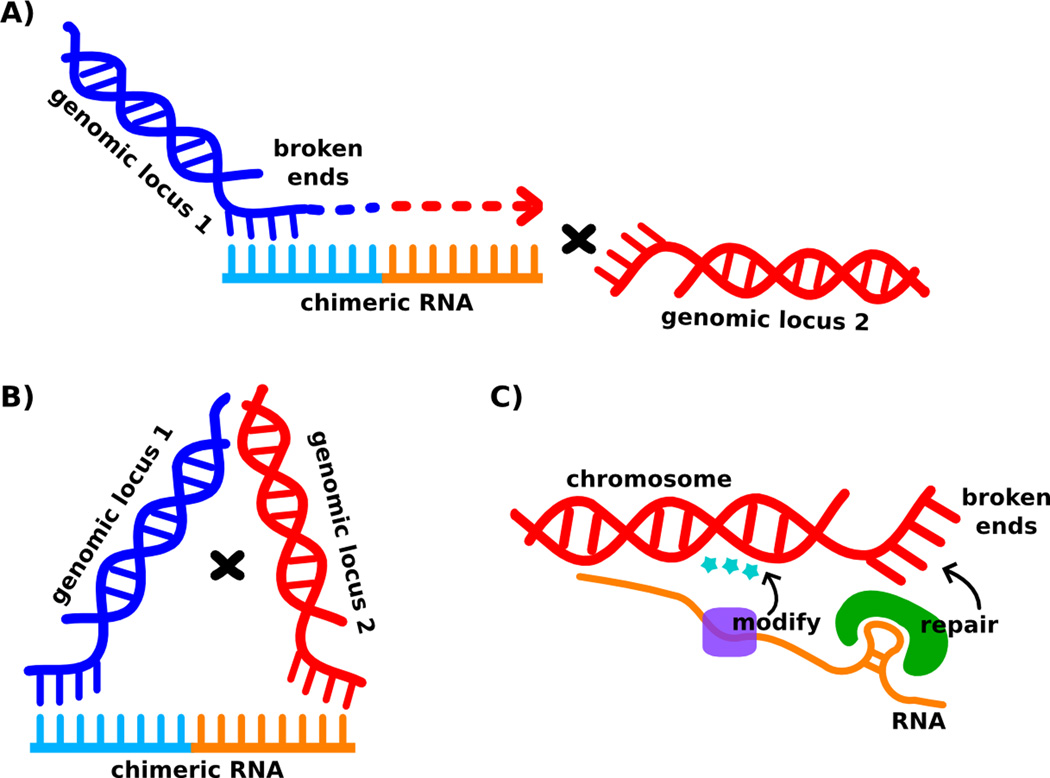
Since DNA breaks usually precede genome rearrangements, RNA-templated DNA repair provides a direct mechanism for RNA-guided genome rearrangement (Figure 1A). In this mechanism, DNA synthesis at broken ends uses a chimeric RNA as a template, thus extending the ends into sequences that share homology with other genomic regions. Subsequent DNA repair via single-stranded annealing or non-homologous end joining pathways may lead to DNA fusion. Intriguingly, multiple recent studies showed that in both plants and animals, double-stranded DNA breaks induce small RNA production at or around the breakage site [17–19]. Furthermore, both double-stranded DNA break repair in Arabidopsis and the DNA damage response in humans depend on small RNA pathway proteins, including Dicer, suggesting a function of the DNA break-induced small RNAs in the repair process [17,18]. It would be informative to test whether any of these small RNAs can serve as templates for DNA repair.
Figure 1.
Putative mechanisms of RNA-mediated DNA rearrangements. A) RNA serves as a repair template for double-stranded DNA break repair. When RNA-templated DNA synthesis occurs across an RNA chimera, then the newly synthesized DNA strand will share sequence similarity with a different genomic locus, and DNA repair through established mechanisms such as single-stranded annealing will result in DNA fusion. B) RNA serves as a scaffold to bring two genomic loci into proximity in space, increasing the chance of recombination between respective loci. C) RNA serves as a recruiting agent for DNA repair complexes, or chromatin modification complexes required for DNA repair. Genomic DNA loci are depicted schematically as double helices. Single stranded DNA and RNA regions that complement each other are drawn as combs, with blue strands annealing to cyan strands, and red strands annealing to orange strands. Dotted line with an arrow in A) indicates DNA synthesis. “×” denotes recombination. Stars in C) indicate chromatin modification. Purple and green modules indicate protein complexes for modification and repair respectively.
A second way in which RNA can influence genome rearrangement is by acting as a template catalyst, providing a scaffold to bring two genomic loci into proximity, and thereby promoting rearrangement between them (Figure 1B). While the exact sequence at the resulting DNA recombination junction may not always precisely follow an RNA template, the presence of the hybrid RNA increases the probability of rearrangement between the two loci to which it has partial complementarity. Increasing evidence suggests that RNA can more generally organize nuclear architecture [20], but direct evidence is lacking for a model where genomic loci are brought together by an RNA bridge. Nevertheless, a highly suggestive finding, compatible with an RNA-templated mechanism, is that normal human cells produce a chimeric RNA that mimics a gene fusion in cancerous cells [21]. In ref. [21], Li et al. detected an RNA fusion between JAZF1 located on chromosome 7 and JJAZ1 on chromosome 17 in normal human endometrial cells. Curiously, this same fusion is present in human endometrial stromal sarcomas that typically contain the corresponding translocation between chromosomes 7 and 17. Since this translocation is not detectable in normal endometrial cells, and the RNA fusion occurs at canonical splice sites, the most likely origin for the JAZF1-JJAZ1 mRNA fusion is trans-splicing, an activity thought to be low in human. Indeed, the authors demonstrated trans-splicing between rhesus JAZF1 and human JJAZ1 using in vitro cell extracts [21]. Furthermore, the chimeric RNA appears to be expressed in a physiologically-regulated manner, suggesting that the mRNA trans-splicing is regulated and not merely noise or error [21]. It would be useful to test whether the corresponding JAZF1 and JJAZ1 loci in cells expressing the chimeric RNA are brought together in three-dimensional space, and whether an excess supply of the chimeric RNA in cytogenetically normal cells increases the frequency of chromosome translocation.
The two mechanisms discussed above suggest a direct role for RNA during some types of genome rearrangement: RNA molecules program either the sequence or location of junctions or the general rearrangement patterns. A third, less direct, role that RNA may play is to recruit repair proteins to broken DNA ends (Figure 1C). Under this model, the DNA repair process is dependent on RNA, or an RNA-protein complex, but RNA does not directly influence the recombination outcome. This mechanism may account for observations of small RNA-dependent DNA repair, as Wei et al. suggest [17] and mentioned above. A crucial link would be to identify integral RNA components in DNA repair complexes.
A fourth mechanism for RNA-mediated genome rearrangement, closely related to the third one we suggest, is that RNA may lead to differences in chromatin structure that facilitate DNA repair and recombination (Figure 1C). In Tetrahymena, for example, scan RNAs help introduce heterochromatic modifications, which are sufficient to mark genomic regions for elimination [22]. Both the DNA damage response [23] and V(D)J recombination that generates vertebrate immune cell diversity [24] signal histone modification changes that are important for DNA repair and recombination. Because RNA often acts upstream to signal histone modifications and to establish chromatin domains [20], it would be one step away to test the role of RNA in defining chromatin structures required for DNA repair and recombination.
Future experiments to test RNA-mediated genome rearrangement
With these possibilities in mind (Figure 1), where should one look for evidence of RNA-mediated genome rearrangement? Recurrent chromosomal translocations in cancer cells would be an important test ground. On one hand, normal cells may contain chimeric transcripts that resemble gene fusions in cancer cells. On the other hand, direct tests of RNA-guided DNA recombination in mammalian cells would be key. Indeed, recent experiments do demonstrate RNA-templated DNA repair in human cells, with increased repair of a single chromosomal double-stranded break upon delivery of RNA-containing oligonucleotides to human cells [16]. A possible extension of this study would be to introduce two double-stranded breaks in two different human chromosomes, and to test whether the availability of RNA oligonucleotides may influence chromosomal translocations.
Unless an RNA template is supplied exogenously, via tools such as injection or infection with a viral vector, any spontaneous event of RNA-guided genome rearrangement would require the rearrangement to appear first at the RNA level. Such events, however, might not be as rare as previously thought. Examples of chimeric RNAs derive from both trans-splicing [21,25] and cis-splicing [3]. Furthermore, evidence is accumulating for the production of circular RNAs via non-canonical splicing events [4] or self-ligation [26]. All three types of “abnormal” transcripts (incorrectly spliced in trans or cis or circularized) can influence interchromosomal translocation, intrachromosomal deletion, and the formation of circular extra chromosomes [7], respectively. We anticipate that further analysis of deep sequencing transcriptome data will reveal the presence of more scrambled transcripts (which require validation through other means), and that knowledge of these aberrant RNAs will permit us to test their ability to guide rearrangements at the genomic level. So far, the example of circularly-permuted tRNA genes [26] is consistent with a model in which the observed circular RNA intermediates could have influenced the evolutionary rearrangements.
Programmed genome diminution or chromosome elimination also occurs in the nematodes Parascaris and Ascaris [27], the arthropod Cyclops [28], and the vertebrates hagfish [29] and lamprey [30]. These would be other natural places to test for the possibility of RNA-guided genome rearrangement. As in studies of ciliates as lab models for RNA-guided DNA rearrangements, the detection of developmentally-regulated non-coding RNA during genome rearrangement may be a first step, followed by functional studies that either knock down candidate RNAs, or supply artificial long or small noncoding RNAs to test whether they can reprogram the outcome of DNA rearrangement [9–11,13].
It is perhaps not surprising to find that the complexity of the transcriptome is far beyond what the Jacob and Monod gene model predicted. After all, RNA, presumably a more ancient biological molecule, is chemically more reactive than DNA. If there exists a pathway for information to flow from RNA to DNA in the context of genome rearrangement, then this complexity and versatility of RNA leads to a rich source of epigenetic states that can be transgenerationally inherited, as has been so well demonstrated in ciliated protozoa. As we delve deeper into the transcription landscape and functional space of non-coding RNA, it would be crucial to investigate where, when, and how RNA may contribute in a more general context to genome rearrangement and evolution.
Acknowledgements
This study was supported by NIH grant GM59708 and NSF grants 0923810 and 0900544 (to L.F.L.) and a DOD pre-doctoral fellowship W81XWH-10-1-0122 (to W.F.).
Footnotes
Conflict of interest
The authors declare that they have no conflict of interests.
Contributor Information
Wenwen Fang, Email: wenwenf@princeton.edu.
Laura F. Landweber, Email: lfl@princeton.edu.
References
- 1.Djebali S, Davis CA, Merkel A, Dobin A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein BE, Birney E, Dunham I, Green ED, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Djebali S, Lagarde J, Kapranov P, Lacroix V, et al. Evidence for transcript networks composed of chimeric RNAs in human cells. PLoS One. 2012;7:e28213. doi: 10.1371/journal.pone.0028213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salzman J, Gawad C, Wang PL, Lacayo N, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Shibata Y, Kumar P, Layer R, Willcox S, et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–86. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- 9.Yao MC, Fuller P, Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- 10.Nowacki M, Vijayan V, Zhou Y, Schotanus K, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lepere G, Betermier M, Meyer E, Duharcourt S. Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang W, Wang X, Bracht JR, Nowacki M, et al. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowacki M, Haye JE, Fang W, Vijayan V, et al. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci U S A. 2010;107:22140–22144. doi: 10.1073/pnas.1012236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storici F, Bebenek K, Kunkel TA, Gordenin DA, et al. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Nandi P, Taylor MB, Stuckey S, et al. RNA-driven genetic changes in bacteria and in human cells. Mutat Res. 2011;717:91–98. doi: 10.1016/j.mrfmmm.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Wei W, Ba Z, Gao M, Wu Y, et al. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Francia S, Michelini F, Saxena A, Tang D, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michalik KMBR, Förstemann K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012;40:9596–9603. doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caudron-Herger M, Rippe K. Nuclear architecture by RNA. Curr Opin Genet Dev. 2012;22:179–187. doi: 10.1016/j.gde.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Wang J, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Taverna SD, Muratore TL, Shabanowitz J, et al. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn Y, Bera TK, Gehlhaus K, Kirsch IR, et al. Finding fusion genes resulting from chromosome rearrangement by analyzing the expressed sequence databases. Proc Natl Acad Sci U S A. 2004;101:13257–13261. doi: 10.1073/pnas.0405490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soma A, Onodera A, Sugahara J, Kanai A, et al. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007;318:450–453. doi: 10.1126/science.1145718. [DOI] [PubMed] [Google Scholar]
- 27.Tobler H, Etter A, Muller F. Chromatin diminution in nematode development. Trends Genet. 1992;8:427–432. doi: 10.1016/0168-9525(92)90326-y. [DOI] [PubMed] [Google Scholar]
- 28.Beermann S. The diminution of Heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda) Chromosoma. 1977;60:297–344. doi: 10.1007/BF00292858. [DOI] [PubMed] [Google Scholar]
- 29.Kohno S, Nakai Y, Satoh S, Yoshida M, et al. Chromosome elimination in the Japanese hagfish, Eptatretus burgeri (Agnatha, Cyclostomata) Cytogenet Cell Genet. 1986;41:209–214. doi: 10.1159/000132231. [DOI] [PubMed] [Google Scholar]
- 30.Smith JJ, Baker C, Eichler EE, Amemiya CT. Genetic consequences of programmed genome rearrangement. Current biology : CB. 2012;22:1524–1529. doi: 10.1016/j.cub.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]