Abstract
The past two decades have seen an increasing number of virulent infectious diseases in natural populations and managed landscapes. In both animals and plants, an unprecedented number of fungal and fungal-like diseases have recently caused some of the most severe die-offs and extinctions ever witnessed in wild species, and are jeopardizing food security. Human activity is intensifying fungal disease dispersal by modifying natural environments and thus creating new opportunities for evolution. We argue that nascent fungal infections will cause increasing attrition of biodiversity, with wider implications for human and ecosystem health, unless steps are taken to tighten biosecurity worldwide.
Emerging infectious diseases (EIDs) caused by fungi are increasingly recognized as presenting a worldwide threat to food security1,2 (Table 1 and Supplementary Table 1). This is not a new problem and fungi have long been known to constitute a widespread threat to plant species. Plant disease epidemics caused by fungi and the fungal-like oomycetes have altered the course of human history. In the nineteenth century, late blight led to starvation, economic ruin and the downfall of the English government during the Irish potato famine and, in the twentieth century, Dutch elm blight and chestnut blight laid bare urban and forest landscapes. The threat of plant disease has not abated, in fact it is heightened by resource-rich farming practices and exaggerated in the landscape by microbial adaptation to new ecosystems, brought about by trade and transportation3, and by climate fluctuations4,5.
Table 1.
Major fungal organisms posing threats to animal and plant species.
| Host | Pathogen (Phylum) | Disease dynamics leading to mass mortality in animal and plant hosts | |
|---|---|---|---|

|
Amphibian species (for example, the common midwife toad, Alytes obstetricans) | Batrachochytrium dendrobatidis (Chytridiomycota) | Worldwide dispersal of a hypervirulent lineage by trade64. Ultra-generalist pathogen manifesting spillover between tolerant/susceptible species. Extent of chytridiomycosis is dependent on biotic and abiotic context15,82. |

|
Rice (Oryza sativa); Magnaporthe grisea species complex on 50 grass and sedge species, including wheat and barley | Magnaporthe oryzae (Ascomycota) | Rice blast disease in 85 countries, causing 10–35% loss of harvest. Global blast population structure determined by deployment of seeds with inbred race-specific disease resistance (RSR). Invasions occur by ‘host hops’ and altered pathogen demographics. |

|
Bat spp. (little brown bats, Myotis lucifugus) | Geomyces destructans (Ascomycota) | New invasion of North American bat roosts occurred in approximately 2006, and disease is spreading rapidly8. Pathogen reservoir may exist in cave soil. Disease is more aggressive compared to similar infections in European bats, possibly owing to differences in roosts and host life histories65. |

|
Wheat (Triticum aestivum); 28 Puccinia graminis f. tritici species, but P. graminis is found on 365 cereal or grass species | Puccinia graminis (Basidiomycota) | Wheat stem rust is present on six continents. Population structure is determined by deployment of RSR cultivars and long-distance spread of aeciospores. Strain Ug99 poses a notable threat to resistant wheat varieties, causing up to 100% crop loss. |

|
Coral species (for example, the sea fan, Gorgonia ventalina) | Aspergillus sydowii (Ascomycota) | Sea-fan aspergillosis caused by a common terrestrial soil fungus21,86. Epizootics are associated with warm-temperature anomalies. Coral immunosuppression is probably a factor causing decline. |

|
Bee species (for example, the hive of the domestic honeybee (Apis mellifera) suffering colony collapse disorder) | Nosema species (Microsporidia) | Microsporidian fungal infections are associated with colony collapse disorder and declining populations. Pathogen prevalence is probably a part of a multifactorial phenomenon that includes environmental stressors and polyparasitism87,88. |

|
Sea turtle species (the loggerhead turtle, Caretta caretta) | Fusarium solani (Ascomycota) | Soil-dwelling saprotroph and phytopathogenic fungus. Infection causes hatch failure in loggerhead turtle nests and suboptimal juveniles44. The disease dynamics fulfil Koch’s postulates. Environmental forcing is suspected but not proven. |
Images in Table 1, with permission: A. obstetricans chytridiomycosis mortalities, M.C.F.; M. oryzae, N. Talbot; WNS-affected little brown bats, A. Hicks; P. graminis, R. Mago; G. ventalina infected with A. sydowii, D. Harvell; A. mellifera hive suffering from colony collapse disorder, J. Evans; sea turtle eggs infected with F. solani, J. Diéguez-Uribeondo and A. Marco.
However, pathogenic fungi (also known as mycoses) have not been widely recognized as posing major threats to animal health. This perception is changing rapidly owing to the recent occurrence of several high-profile declines in wildlife caused by the emergence of previously unknown fungi6,7. For example, during March 2007, a routine census of bats hibernating in New York State revealed mass mortalities8. Within a group of closely clustered caves, four species of bats were marked by a striking fungus growing on their muzzles and wing membranes, and the name ‘white nose syndrome’ (WNS) was coined. After the initial outbreak, the ascomycete fungus Geomyces destructans was shown to fulfil Koch’s postulates and was described as the cause of WNS in American bat species9,10. Mortalities exhibiting WNS have subsequently been found in an increasing number of bat overwintering sites and, by 2010, the infection was confirmed to have emerged in at least 115 roosts across the United States and Canada, spanning over 1,200 km (ref. 11). Bat numbers across affected sites have declined by over 70% and analyses have shown that at least one affected species, the little brown bat Myotis lucifugus, has a greater than 99% chance of becoming locally extinct within the next 16 years (ref. 11). Other species of bats across this region are declining as a consequence of this infection, and the prognosis for their survival and their role in supporting healthy ecosystems, is poor12.
Cases of this sort are no longer perceived to be atypical. The probability of extinction is increasing for some species of North American bats, but another fungal infection has caused the greatest disease-driven loss of biodiversity ever documented. The skin-infecting amphibian fungus Batrachochytrium dendrobatidis was discovered in 1997 (ref. 13) and named in 1999 (ref. 14). B. dendrobatidis has been shown to infect over 500 species of amphibians in 54 countries, on all continents where amphibians are found15,16, and is highly pathogenic across a wide diversity of species. Studies using preserved amphibian specimens showed that the first appearance of B. dendrobatidis in the Americas coincided with a wave of population declines that began in southern Mexico in the 1970s and proceeded through Central America to reach the Panamanian isthmus in 2007 (ref. 17). As a consequence of the infection, some areas of central America have lost over 40% of their amphibian species18, a loss that has resulted in measurable ecosystem-level changes19. This spatiotemporal pattern has been broadly mirrored in other continents15, and ongoing reductions in amphibian diversity owing to chytridiomycosis have contributed to nearly half of all amphibian species being in decline worldwide20.
Fungal infections causing widespread population declines are not limited to crops, bats and frogs; studies show that they are emerging as pathogens across diverse taxa (Table 1), including soft corals (for example, sea-fan aspergillosis caused by Aspergillus sydowii)21, bees (the microsporidian fungus Nosema sp. associated with colony collapse disorder)22, and as human and wildlife pathogens in previously non-endemic regions (for example, the emergent virulent VGII lineage of Cryptococcus gattii in the northwest America23 and Cryptococcus neoformans across southeast Asia24). The oomycetes have life histories similar to those of fungi and are also emerging as aggressive pathogens of animals, causing declines in freshwater brown crayfish (for example, the crayfish plague caused by Aphanomyces astaci)25, Tilapia fish (for example, epizootic ulcerative syndrome caused by A. invadans)26 and many species of plants27,28. Although the direct causal relationship is uncertain in some of these diverse host–pathogen relationships, it seems that pathogenic fungi are having a pronounced effect on the global biota1.
Increasing risk of biodiversity loss by Fungi
For infectious disease systems, theory predicts that pathogens will co-evolve with, rather than extirpate, their hosts29,30. Such evolutionary dynamics mirror population-level processes in which density dependence leads to the loss of pathogens before their hosts are driven extinct31. For these reasons, infection has not been widely acknowledged as an extinction mechanism owing to such intrinsic theoretical biotic limitations32. Inspection of species conservation databases would seem to confirm this idea. The International Union for Conservation of Nature (IUCN) red list database details threats to species worldwide, and analysis of the database has shown that of the 833 recorded species extinctions, less than 4% (31 species) were ascribed to infectious disease7. Ecological studies on host–pathogen relationships support this finding by showing that lower parasite richness occurs in threatened host species, suggesting that parasite decline and ‘fade out’ occurs when hosts become rare33. Therefore, given that macroevolutionary and ecological processes should promote diversity and prevent infectious diseases from driving their host species to extinction, we posed the question of whether we are witnessing increasing disease and extinction events driven by fungi on an increasingly large scale, or, alternatively, if there is evidence that a reporting bias has skewed our opinion of the current level of threat.
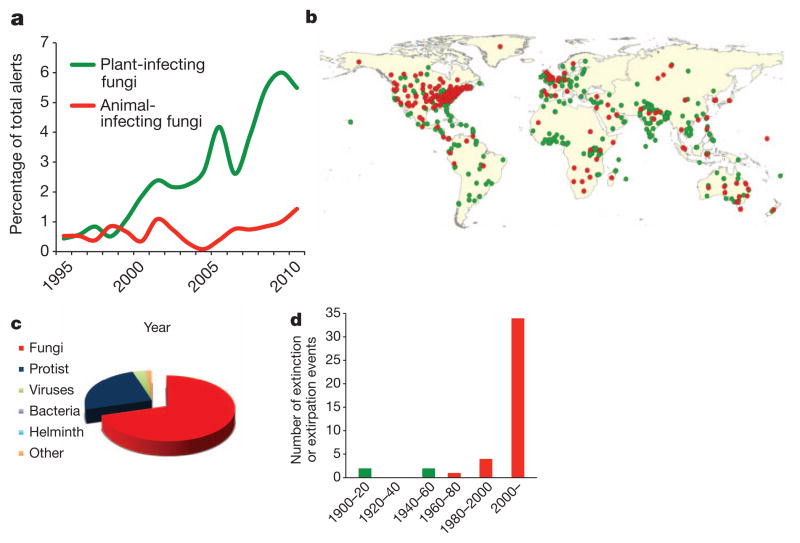
EIDs are those pathogens that are increasing in their incidence, geographic or host range, and virulence34,35. Current attempts to detect EID events centre on capturing changes in the patterns of disease alerts recorded by disease monitoring programmes. ProMED (the Program for Monitoring Emerging Diseases; http://www.promedmail.org) and HealthMap (http://healthmap.org) have two approaches for detecting and monitoring outbreaks worldwide in plant and animal hosts: first, by active reporting of disease outbreaks, and second, by capturing diverse online data sources. To ascertain whether there are changing patterns of fungal disease, we reviewed all disease alerts in ProMED (1994–2010) and HealthMap (2006–10) for combinations of search terms to catalogue fungal alerts. We then classified these according to their relative proportion against the total number of disease alerts, and discriminated between plant- or animal-associated fungal pathogens (Supplementary Table 2). We also searched the primary research literature for reports in which EIDs have caused host extinction events, either at the regional scale (extirpations) or globally (Supplementary Table 3). These analyses show a number of positive trends associated with infectious fungi. Overall, fungal alerts comprise 3.5% of the ~38,000 ProMED records screened. However, over the period from 1995 to 2010, the relative proportion of fungal alerts increased in the ProMED database from 1% to 7% of the database total (Fig. 1a and Supplementary Table 2). This trend is observed for both plant-infecting (0.4% to 5.4%) and animal-infecting (0.5% to 1.4%) fungi. HealthMap shows a recent (2007–11) positive trend in the proportion of records of fungi infecting animals (0.1% to 0.3%) and plants (0.1 to 0.2%), and fungal disease alerts were shown to occur worldwide (Fig. 1b). Web of Science literature searches and compilation of previous meta-analyses of infection-related species extinction and regional extirpation events show that fungi comprise the highest threat for both animal-host (72%) and plant-host (64%) species (Fig. 1c and Supplementary Tables 3 and 4). This effect is more pronounced for animal hosts (39 animal species affected versus 4 plant species); moreover, there is a notable increase in host loss during the second half of the twentieth century, driven mainly by the emergence of B. dendrobatidis (Fig. 1d). This effect is moderated after correcting for mass-species loss in regions of high epizootic loss (such as the mass extirpations of amphibians in Central America). However, fungi remain the major cause (65%) of pathogen-driven host loss after this correction. Our estimates are probably conservative owing to the cryptic nature of most disease-driven species impacts. For example, the lack of disease-related IUCN red list records is due to a lack of baseline data on the incidence of pathogens in natural systems compounded by inadequate disease diagnostics, reporting protocols and a lack of centralized recording mechanisms. Hence, the true numbers of extinctions and extirpations caused by fungi and oomycetes are likely to be greater as we have not been able to categorize the probably high levels of species loss in major plant (such as the Phytophthora dieback in Australia caused by Phytophthora cinnamomi; Supplementary Table 3) or animal outbreaks (for example, the effects of B. dendrobatidis emergence in the American wet tropics). We cannot discount the idea that sampling bias owing to increasing awareness of pathogenic fungi as EIDs may contribute to the patterns that we document. However, because of our observation that increases in the amount of disease caused by fungi are seen across many sources of data, including disease alerts, the peer-reviewed literature and previously noted patterns in human fungal EIDs35, we believe that these trends are real. Therefore, the answer to our question seems to be that the data do indeed support the idea that fungi pose a greater threat to plant and animal biodiversity relative to other taxonomic classes of pathogen and hosts, and that this threat is increasing.
Figure 1. Worldwide reporting trends in fungal EIDs.
a, b, Disease alerts in the ProMED database for pathogenic fungi of animals and plants (a), and the spatial location of the associated reports (b). c, d, Relative proportions of species extinction and/or extirpation events for major classes of infectious disease agents (c) and their temporal trends for fungal pathogens (d). Primary data sources are given in the Supplementary Information.
Fungal-disease dynamics leading to host extinction
Here we illustrate several key biological features of fungi that contribute to the epidemiological dynamics underlying contemporary increases in disease emergence and host extinction (Box 1).
BOX 1. Modelling host extinctions caused by pathogenic fungi.
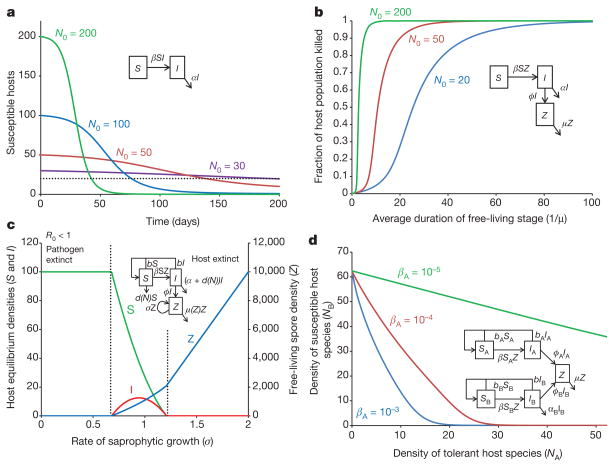
A simple susceptible–infected model shows that the presence of a threshold host population size for disease persistence does not prevent host extinction during a disease outbreak, especially in cases in which a lethal pathogen invades a large host population. In a large host population transmission is rapid and all hosts can become infected before the hostpopulation is suppressed below the threshold. The model follows the dynamics of susceptible (S) and infected (I) hosts during the short time duration of an epidemic (deaths that are not due to disease, and births, are ignored): dS/dt = − βSI; dI/dt = βSI − αI, where β is the pathogen transmission rate, and α is the disease-induced death rate. For the parameters shown in Fig. 2a, the threshold population size below which the pathogen has a negative growth rate is NT = 20 individuals. Figure 2a shows that large host populations are rapidly driven extinct, but only a fraction of individuals are killed in small host populations.
Pathogens with a long-lived infectious stage have an increased potential to cause host extinction. In this model, the disease is transmitted through contact between susceptible hosts and free-living infectious spores (Z), resulting in infected hosts, I: dS/dt = − βSZ; dI/dt = βSZ − αI; dZ/dt = ϕI − μZ − βNZ, where β is the transmission rate, α is the pathogen-induced death rate and ϕ is the rate of release of spores from infected hosts. Figure 2b shows that fraction of hosts killed in a disease outbreak increases with the duration of the free-living infectious spore stage (1/μ, where μ is the spore mortality rate).
Saprophytic growth by a pathogen can lead to extinction of the host, and even allow the pathogen to persist in the absence of its host. In this model, free-living infectious spores are released from infected hosts (with rate ϕ), and can increase in abundance through saprophytic growth, with rate σ. To illustrate the effects of saprophytic pathogen growth on host and pathogen equilibria (Fig. 2c), density-independent host reproduction (with rate b), density-dependent host mortality (with rate d0 + d1N, where N = S + I), and density-dependent spore mortalities (at rate μ0 + μ1Z) were included: dS/dt = bN − (d0 + d1N) S − βSZ; dI/dt = βSZ − αI − (d0 + d1N)I; dZ/dt = ϕI + σZ − (μ0 + μ1Z) Z − βNZ.
The presence of a tolerant host species can lead to the extinction of a susceptible host species. In this model, species A is the tolerant host species, which can become infected and shed infectious spores but does not die as a result of the disease, whereas the susceptible host species (species B) has a disease-induced per-capita mortality rate of αB. Figure 2d shows that species B is driven extinct at high densities of species A. dSA/dt = bANA − (dA0 + dA1NA) SA − βASAZ; dIA/dt = βASAZ − (dA0 + dA1NA) IA; dSB/dt = bBNA − (dB0+ dB1NB) SB − βBSBZ; dIB/dt = βBSBZ − (dB0 + dB1NB) IB − αBIB; dZ/dt = ϕAIA + ϕBIB − μZ − βANAZ − βBNBZ, where all parameters are as previously defined, but with the subscripts A or B referring to host species A or B, respectively.
High virulence
Fungi, like some bacterial and viral infections, can be highly lethal to naive hosts with rates of mortality approaching 100% (for example, B. dendrobatidis in amphibians, G. destructans in bats and Ophiostoma ulmi in elm trees). Virulence is a measure of the relative capacity of a microbe to cause damage to a host36, and high virulence is associated with rapid intra-host growth rates, ultimately leading to rapid inter-host transmission37,38. Fungi have a high reproductive potential and in a large host population this effect can result in all individuals becoming infected before the population is driven to the low densities at which the pathogen can no longer spread (Fig. 2a). Thus, host extirpation can occur before density dependence limits the rate of transmission, a feature that has contributed to the mass extirpations seen in frog populations across the US Sierra Nevada mountains39. Similarly, even if the pathogen does not drive the host to complete extinction, it may severely reduce the population size to the point at which the species is vulnerable owing to catastrophic collapses as a result of stochastic32 or Allee effects40.
Figure 2. Fungal disease dynamics leading to host extinction.
a, The presence of a threshold host population size for disease persistence does not prevent host extinction during a disease outbreak, especially in cases in which a lethal pathogen invades a large host population. In a large host population transmission is rapid and all hosts can become infected before the host population is suppressed below the threshold (pathogen transmission rate, β = 0.001 per individual per day; disease induced-death rate, α = 0.02 per day; simulations start with one infected individual and N0 susceptible individuals). b, Long-lived infectious stages can increase the potential for host extinction. The fraction of hosts killed in a disease outbreak is shown as a function of the duration of the free-living infectious spore stage (pathogen transmission rate, β = 5 × 10−6; disease-induced death rate, α = 0.02; rate of release of spores from infected hosts, ϕ = 10; outbreaks initiated with one infected host in a population of N0 susceptible individuals). c, Saprophytic growth: equilibrium densities of susceptible and infected hosts and free-living spores as a function of the rate of saprophytic growth, σ. With no (or low levels of) saprophytic growth, the basic reproductive rate of the pathogen (R0) is less than 1, the pathogen cannot invade the system and the host persists at its disease-free equilibrium density. Intermediate levels of saprophytic growth allow the pathogen to invade and persist with the host. High levels of saprophytic growth lead to extinction of the host, and the pathogen persists in the absence of the host (host intrinsic rate of increase, r = b − d0 = 0.01; density-independent host death rate, d0 = 1 × 10−3; strength of density dependence in host death rate, d1 = 1 × 10−4; pathogen transmission rate, β = 1 × 10−5; disease-induced death rate, α = 0.02; rate of release of spores from infected hosts, ϕ = 10, density-independent spore mortality rate, μ0 = 1; strength of density-dependence in spore mortality rate, μ1 = 1 × 10−4). d, The presence of a tolerant host species (host species A), which can become infected and shed infectious spores can lead to the extinction of a susceptible host species (host species B). Species A does not die because of the disease, but species B has a disease-induced per-capita mortality rate of αB. Species B is driven extinct at high densities of species A. For all parameters, subscripts A or B indicate the host species. Host intrinsic rates of increase, rA = rB = 0.01; density-independent host death rates, dA0 = dB0 = 1 × 10−3; host birth rates, bA = bB = rA + dA0; density-independent death rate for species, B dB1 = 1 × 10−4; rate of release of spores from infected hosts, ϕA = ϕB = 10; αB = 0.05; spore mortality rate, μ = 1. The density of tolerant species NA was varied by varying dA1 (the strength of density-dependence in host species A), such that NA = rA/dA1.
Long-lived environmental stages
Fungi have remarkably resilient dispersal stages (a feature that they share with some spore-forming bacteria, such as Bacillus anthracis). The ability to survive independently outside of their host, as a free-living saprophyte or as durable spores in the environment, is probably the most important feature in driving the emergence of pathogenic fungi, owing to an increased risk of transporting the inocula to naive hosts (Fig. 2b)41. Furthermore, pathogenic fungi with a saprophytic stage (called sapronoses; Fig. 2c) can lead to host extirpation because their growth rate is decoupled from host densities and many fungal diseases threatening natural populations are caused by opportunistic fungi with long-lived environmental stages. Many fungi in the phylum Ascomycota are common soil organisms and are tolerant of salinity with the consequence that, when they enter the marine system through freshwater drainage, they are able to infect susceptible hosts such as corals (A. sydowii42), sea otters (Coccidioides immitis43) and the nests of loggerhead turtles (Fusarium solani44). In terrestrial environments, potentially lethal fungi are ubiquitous, such as the causative agent of aspergillosis, Aspergillus fumigatus, and soil surveys have shown that Geomyces spp. are common soil organisms. Viable G. destructans has been recovered from the soil of infected bat caves45, showing that the pathogen is able to survive and persist in infected roosts when the bats are absent. Likewise, long-term persistence of fungal inoculum in the agricultural landscape is achieved by quiescent survival on plant debris, such as the spores of wheat stem rust (Puccinia graminis), which overwinter on straw stubble before infecting a secondary host.
Generalist pathogens and opportunistic pathogens
Although many fungi demonstrate extreme host specialization, exemplified by the gene-for-gene interactions between biotrophic fungi and their plant hosts, broad host ranges twinned with high virulence can be a lethal combination. Fungi exhibit the broadest spectrum of host ranges for any group of pathogens, and B. dendrobatidis and the oomycete Phytophthora ramorum (the cause of sudden oak death and ramorum blight) are known to infect 508 (ref. 16) and 109 (ref. 3) host species, respectively. Different host species vary in their susceptibility to infection and these differences create the potential for parasite-mediated competition when the pathogens concerned are generalists46. Host species that can tolerate high infection loads while serving as a source of infectious stages (known as pathogen spill-over) act as community ‘super spreaders’ by maintaining persistent infectious stages in the system (Fig. 2d). Invasive North American signal crayfish, which tolerate infection by the oomycete A. astaci, force the infection into more susceptible European species that then decline25, and similarly, although P. ramorum is deadly to Notholithocarpus densiflorus (tanoak) and several Quercus species47, many of its other hosts survive infection but generate inoculum themselves for new infections. Furthermore, disease-tolerant life-history stages of otherwise susceptible species can maintain high pathogen levels leading to extinction dynamics. In chytridiomycosis, the long-lived multi-year tadpole stages of amphibians such as the mountain yellow-legged frog Rana muscosa and the midwife toad Alytes obstetricans are not killed by chytrid infections, but they can build up high loads of B. dendrobatidis that can infect and overwhelm juvenile metamorphs of the same species, leading to rapid population loss39. Ultimately, when host-generalist pathogens manifest long-lived environmental stages, conditions may occur that lead to long-distance dispersal and infection of naive hosts and environments5.
Trade and transport promotes globalization of fungi
Fungi comprise most of the viable biomass in the air, with an average human breath containing between one and ten fungal spores48. This ability of fungi to disperse results in some species with cosmopolitan distributions5,49,50. However, these species are in the minority and it is noticeable that few fungi exhibit truly globally distributions; instead they exhibit spatially restricted endemic ranges51. In many cases, local adaptation and host specificity are thought to underlie fungal endemicity51,52. Nevertheless, when local climatic and vegetative constraints are projected globally it becomes clear that potential ranges of pathogenic fungi may be much larger than their realized range53. If fungi are contained spatially by the combination of physical limits on dispersal, abiotic conditions, host distributions and genetic limits on adaptation, then how are pathogenic fungi able to overcome these barriers? Although fungi have shown the ability to undergo range expansions in response to environmental shifts54, human-mediated intercontinental dispersal of unrecognized fungal pathogens is the major component in initiating new chains of transmission.
Pathogenic fungi have dispersed alongside early human migrations, and several thousand years ago two of these fungi, Coccidioides immitis and C. neoformans lineage VNI, seem to have invaded South America and southeast Asia, respectively, vectored by humans and their domesticated animals24,55. Similar ancient patterns of human-associated disease spread are detected by studies of the genome diversity of many plant fungal pathogens56. However, more recent increases in fungal disease are attributable to the many-fold increase in fungal-infected trade products and food57. The consequences of recent introductions of pathogens in association with trade are well known; examples include the Irish Famine4 (a consequence of Phytophthora infestans late blight introduction from South America), the destruction of the North American chestnuts58 (caused by the importation of Cryphonectria parasitica-infected Asian chestnut trees to the east coast of the United States in the early twentieth century) and the Second World War introduction of Heterobasidion annosum into Italy from the USA (vectored by untreated wooden transport crates)59. Human-mediated intercontinental trade has also been linked clearly to the spread of animal-pathogenic fungi through the transportation of infected vector species. B. dendrobatidis has been introduced repeatedly to naive populations worldwide as a consequence of the trade in the infected, yet disease-tolerant species such as North American bullfrogs (Rana catesbeiana)60–62 and African clawed frogs (Xenopus laevis)63,64. Whether the emergence of bat WNS constitutes an introduction of G. destructans into North America from Europe or elsewhere remains to be shown. However, the widespread but apparently non-pathogenic nature of the infection in European bats tentatively suggests that the disease may have been vectored from this region in contaminated soil65.
Accelerated evolution of virulence in pathogenic fungi
Human activities are not only associated with the dispersal of pathogenic fungi, they also interact with key fungal characteristics, such as habitat flexibility, environmental persistence and multiple reproductive modes, to cause the emergence of disease. Importantly, many fungi are flexible in their ability to undergo genetic recombination, hybridization or horizontal gene transfer66, causing the clonal emergence of pathogenic lineages but also allowing the formation of novel genetic diversity leading to the genesis of new pathogens56, 67,. Reproductive barriers in fungi are known to evolve more rapidly between sympatric lineages that are in the nascent stages of divergence than between geographically separated allopatric lineages, in a process known as reinforcement68,69. As a consequence, anthropogenic mixing of previously allopatric fungal lineages that still retain the potential for genetic exchange can drive rapid macroevolutionary change. Although many hybrids are inviable owing to genome incompatibilities, large phenotypic leaps can be achieved by the resulting ‘hopeful monsters’, leading to host jumps and increased virulence70. Such mechanisms are thought to drive the formation of new pathotypes in plant pathogens52, and oomycetes as well as fungi exhibit the genesis of new interspecific hybrids as lineages come into contact71,72. Evidence of the effect of multiple fungal co-dispersal events and recombination can also be seen in the recent C. gattii outbreaks in northwestern North America. In this case, strains that do not normally recombine have increased their virulence by undergoing recombination and adaptation to overcome mammalian immune responses23,67. Recent studies based on the resequencing of B. dendrobatidis genomes have shown that, although several lineages exist, only a single lineage (known as the B. dendrobatidis global panzootic lineage) has emerged in at least five continents during the twentieth century to cause epizootic amphibian declines64. Notably, the genome of the B. dendrobatidis global panzootic lineage shows the hallmarks of a single hybrid origin and, when compared against other newly discovered lineages of B. dendrobatidis, is more pathogenic, suggesting that transmission and onward spread of the lineage has been facilitated by an increase in its virulence. Given that the rate of intra- and inter-lineage recombination among fungi will be proportional to the contact rates between previously geographically separate populations and species, these data from across plant and animal fungal pathosystems suggest that the further evolution of new races is inevitable given current rates of homogenization of previously allopatric, geographically separated, fungal lineages.
Environmental change as a driver of fungal EIDs
Climate fluctuation can be a potent cofactor in forcing changing patterns of fungal phenology73 and are known to govern plant fungal EIDs. Models of climate change for the coming decades predict increases in global temperature, atmospheric CO2, ozone and changes in humidity, rainfall and severe weather74. For this reason, many interactions must be taken into consideration when attempting to predict the future effects of climate change on plant diseases75. First, the physiological and spatial changes that plants may undergo in response to the various different components of climate change and the resulting effects on the pathogen76, and second, the effects on the pathogen’s physiology and dispersal external to their plant hosts75. Frequently, however, experimental models have only taken into account one element of climate change, a common example being the free-air CO2 enrichment (FACE) studies that model the effects of elevated atmospheric CO2 (ref. 77). A notable result here has been rice blast severity being higher at higher CO2 levels78. However, although there has been a general trend for increased disease severity under simulated climate-change conditions79, and although some species are thought to be changing their distribution in response to these changes (for example, P. graminis80), other elements of climate change, such as increased ozone, have been shown to have the opposite effect (for example, in Puccinia recondita77).
Evidence for the idea that climate change has an impact on the dynamics and distribution of animal-infecting fungi is less clear-cut than that in relation to plant-infecting fungi and, although arguments have been made that warming trends may have contributed to the emergence of B. dendrobatidis in Central America and Europe81,82, there is active debate about these conclusions83,84. Regardless, it is clear that the disease state, chytridiomycosis, is linked to environmental factors; regional climate warming can increase the local range of the pathogen54 and disease risk is inversely related to rates of deforestation85. Correlations between ecosystem change and a rise in infection by opportunistic pathogens has been proposed to account for the occurrence of coral reef declines worldwide. For example, disease caused by a variety of microbes threatens hard corals to the extent that two of the most abundant Caribbean reef-builders (staghorn and elkhorn corals) are now listed under the US Endangered Species Act. Across varied reef systems, the occurrence of warm-temperature anomalies leading to bleaching events is associated with increases in disease caused by opportunistic pathogens such as A. sydowii86. In an allied colonial system, colony collapse disorder has resulted in steep declines of the European honeybee Apis mellifera in Europe and North America87. These losses seem to be influenced by a mixture of aetiological agents that are fungal (for example, microsporidian (Nosema ceranae)), viral (for example, Kashmir bee virus and Israeli acute paralysis virus) and ectoparasitic (for example, Varroa destructor) in origin. So far, no single environmental cause has been identified that can account for the apparent reduction in the ability of honeybee colonies to resist these infections, and agricultural chemicals, malnutrition and modern beekeeping practices have all been suggested as potential cofactors for colony-collapse disorder88. The increasing use of azole-based agricultural chemicals has been implicated as a factor underpinning the increase in the frequency of multiple-triazole-resistant (MTR) isolates of A. fumigatus infecting humans89. The widespread agricultural use of azoles as a means of combating crop pathogens is speculated to have led to selection for MTR alleles, an idea that is supported by the recent discovery that resistance clusters onto a single lineage in Dutch populations of the fungus90. Efforts must now be turned to integrating epidemiological studies with those on environmental change so that the many possible interactions and outcomes can be assessed, as making blanket predictions for fungal diseases is currently impossible91. The highly coordinated response to the recent outbreak of wheat stem rust (P. graminis, strain Ug99) is a positive step towards this goal77,92.
Fungal EIDs impact food security and ecosystem services
Impacts of fungal diseases are clearly manifested in crops and there are direct measurable economic consequences associated with die-off in forest and urban environments. Losses that are due to persistent and epidemic outbreaks of fungal and oomycete infection in rice (rice blast caused by Magnaporthe oryzae), wheat (rust caused by P. graminis), maize (smut caused by Ustilago maydis), potatoes (late blight caused by P. infestans) and soybean (rust caused by Phakospora pachyrizi) vary regionally but pose a current and growing threat to food security2. Our estimates of loss of food are based on the 2009–10 world harvest statistics of five of our most important crops and make certain basic assumptions of calorific value and worldwide average production (Supplementary Table 1). Our calculations show that even low-level persistent disease leads to losses that, if mitigated, would be sufficient to feed 8.5% of the 7 billion humans alive in 2011. If severe epidemics in all five crops were to occur simultaneously, this would leave food sufficient for only 39% of the world’s population, but the probability of such an event occurring is very low indeed.
Invasive tree diseases have caused the loss of approximately 100 million elm trees in the United Kingdom and the United States52,93, and 3.5 billion chestnut trees have succumbed to chestnut blight in the United States (Supplementary Table 5). Losses of western Canadian pine trees to the mountain pine beetle–blue-stain fungus association will result in the release of 270 megatonnes of CO2 over the period from 2000 to 2020, with a clearly ascribed economic cost both for the wood itself and the carbon released94. These, and other diseases such as ‘sudden oak death’ in California and ‘foliar and twig blight’ and ‘dieback’ on ornamental trees, woody shrubs and forestry plants in the European Union, affect ecological diversity, are costly to manage and account for huge losses of fixed CO2. Indeed, we calculate regional losses of absorbed CO2 to total 230–580 megatonnes for just a handful of diseases (Supplementary Table 5) with the higher figure equating to 0.069% of the global atmospheric CO2. We have included both emerging (Jarrah dieback, sudden oak death and pine beetle–blue-stain fungus) and emergent diseases (Dutch elm blight and chestnut blight), as these represent the few examples for which informed estimates are possible. We are unable to quantify any of the many other recent emerging diseases, such as red band needle blight of pines, Phytophthora alni on alders or pitch pine canker on Monterey pines, owing to a lack of data and economic interest, both of which are trends that must be reversed. Assessing the economic burden of fungal mycoses in animals is a challenging task. Although the impact of fungal EIDs is manifested in domestic animal settings, particularly the amphibian trade95 and in regions where virulent lineages have established96, reporting mechanisms for outbreaks do not widely exist. In natural settings, valuations have recently estimated the losses to US agriculture that are the result of declines in bat populations at more than US$3.7 billion per year (ref. 12). However, although broad ecosystem-level impacts of other fungal EIDs of wildlife are suspected, economic valuations of the ecosystem services that these species support are wholly lacking.
Mitigating fungal EIDs in animals and plants
The high socioeconomic value of crops means that detection and control of fungal diseases in agriculture far outpaces that in natural habitats. Epidemiological models have been developed to predict the risk of seasonally specific crop pathogens, allowing targeted control, and specific threats are assessed through consortia of research, governmental and global non-governmental organizations, led by the United Nations Food and Agricultural Organization (FAO), and related organizations. Scientifically led development of disease-resistant crop varieties has been mainly successful, although monocultures have in some instances vastly increased the susceptibility of harvests to highly virulent pathogens, a pertinent example being P. graminis Ug99. Conversely, although there have been some attempts to mitigate the fungal disease burden in wildlife in situ—most notably efforts to eliminate B. dendrobatidis in infected populations with the antifungal itraconazole97 and the use of probiotic bacteria98—communicable wildlife EIDs are essentially unstoppable once they have emerged. International biosecurity against the spread of plant fungal pathogens, although not perfect, is more advanced than protocols to protect against the introduction of animal-associated fungi. Fundamentally, this is the result of a financial dynamic: wildlife is not correctly valued economically, whereas crops are.
The World Organisation for Animal Health (also known as the OIE) and the FAO may be the best-placed authorities to coordinate tighter biosecurity controls for trade-associated fungal pathogens of animals. The OIE has listed B. dendrobatidis and the crayfish pathogen A. astaci in the Aquatic Animal Health Code as internationally notifiable infections, and the FAO compiles outbreak data on transboundary animal diseases using the emergency prevention information system (EMPRES-i). Similarly, the IUCN Wildlife Health Specialist Group determines policy that is specific to combating emerging wildlife disease internationally. On national scales there are a number of initiatives being deployed and in the United States the National Wildlife Health Centre has developed the national federal plan99 to mitigate WNS in bats. Intensive monitoring and surveillance will be increasingly important in the coming years because predictive modelling and small-scale experiments can never fully predict future disease spread and severity. An increased political and public profile for the effects of fungal diseases in natural habitats is needed to highlight the importance of fungal disease control outside of the managed agricultural environment to policy makers. If this occurs, then there will be more sympathy for attempts to improve the regulatory frameworks that are associated with biosecurity in international trade, as this is the most important tool to tackle both plant and animal fungal EIDs now and in the future. The monitoring of fungal inocula in wild populations should be the utmost priority and tighter control of international trade in biological material must be imposed, and with considerable haste. Inadequate biosecurity will mean that new fungal EIDs and virulent races will emerge at an increasingly destructive rate. In addition to better global monitoring and control, attention must also be turned to increasing our understanding of the interactions between hosts, pathogens and the environment, across regional and global scales. Integrated approaches encompassing theoretical and practical epidemiology, climate forecasting, genomic surveillance and monitoring molecular evolution are needed. These should be facilitated by scientists from currently disparate research fields entering into regular global discussions to develop clear and urgent strategies for working towards the elusive magic bullet for emerging fungal diseases: effective prevention and timely control.
Supplementary Material
Acknowledgments
M.C.F. was supported by grants from the Wellcome Trust, Natural Environment Research Council (NERC), and the European Research Area (ERA)-net project BiodivERsA. D.A.H. was supported by a grant from the Leverhulme Trust, C.J.B. was supported by the US National Science Foundation (NSF) Ecology of Infectious Disease grant EF-0723563. S.J.G. was supported by grants from the UK Biotechnology and Biological Sciences Research Council (BBSRC) and the John Fell Fund of the University of Oxford, and S.L.M. was supported by a graduate scholarship from Magdalen College, University of Oxford. J.S.B. was supported by Google.org and the National Institutes of Health grant 5R01LM010812-02. N. Knowlton and J. Heitman provided impetus to develop this review.
Footnotes
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.C. F., D.A.H., C.J.B., S.L.M. and S.J.G designed, analysed and wrote the paper. Data were collected and analysed by J.S.B. and L.C.M.
References
- 1.The Institute of Medicine. Fungal Diseases: an Emerging Threat to Human Animal and Wildlife Health. National Academy of Sciences; 2011. The output of a key workshop assessing the risk of novel fungal diseases. [Google Scholar]
- 2.Pennisi E. Armed and dangerous. Science. 2010;327:804–805. doi: 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- 3.Grünwald NJ, Goss EM, Press CM. Phytophthora ramorum: a pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol Plant Pathol. 2008;9:729–740. doi: 10.1111/j.1364-3703.2008.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson PK, et al. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol. 2004;19:535–544. doi: 10.1016/j.tree.2004.07.021. The first meta-analysis of emerging plant diseases. Reasons for this emergence are proposed and the cost to human welfare and biodiversity is estimated. [DOI] [PubMed] [Google Scholar]
- 5.Brown JKM, Hovmoller MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- 6.Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 7.Smith KF, Sax DF, Lafferty KD. Evidence for the role of infectious disease in species extinction and endangerment. Conserv Biol. 2006;20:1349–1357. doi: 10.1111/j.1523-1739.2006.00524.x. [DOI] [PubMed] [Google Scholar]
- 8.Blehert DS, et al. Bat white-nose syndrome: an emerging fungal pathogen? Science. 2009;323:227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- 9.Gargas A, Trest MT, Christensen M, Volk TJ, Blehert DS. Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon. 2009;108:147–154. [Google Scholar]
- 10.Lorch JM, et al. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature. 2011;480:376–378. doi: 10.1038/nature10590. [DOI] [PubMed] [Google Scholar]
- 11.Frick WF, et al. An emerging disease causes regional population collapse of a common North American bat species. Science. 2010;329:679–682. doi: 10.1126/science.1188594. Population viability analysis showing the high risk of extinction of little brown bats caused by the emergence of a pathogenic fungus. [DOI] [PubMed] [Google Scholar]
- 12.Boyles JG, Cryan PM, McCracken GF, Kunz TH. Economic importance of bats in agriculture. Science. 2011;332:41–42. doi: 10.1126/science.1201366. [DOI] [PubMed] [Google Scholar]
- 13.Berger L, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci USA. 1998;95:9031–9036. doi: 10.1073/pnas.95.15.9031. The first study describing the discovery of amphibian chytridiomycosis in the tropics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longcore JE, Pessier AP, Nichols DK. Batrachochytrium dendrobatidis gen. et sp. Nov., a chytrid pathogenic to amphibians. Mycologia. 1999;91:219–227. [Google Scholar]
- 15.Fisher MC, Garner TWJ, Walker SF. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu Rev Microbiol. 2009;63:291–310. doi: 10.1146/annurev.micro.091208.073435. [DOI] [PubMed] [Google Scholar]
- 16.Bd-Maps. [accessed, February 2012]; 〈 http://www.bd-maps.net/〉.
- 17.Cheng TL, Rovito SM, Wake DB, Vredenburg VT. Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen. Batrachochytrium dendrobatidis. Proc Natl Acad Sci USA. 2011;108:9502–9507. doi: 10.1073/pnas.1105538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crawford AJ, Lips KR, Bermingham E. Epidemic disease decimates amphibian abundance, species diversity, and evolutionary history in the highlands of central Panama. Proc Natl Acad Sci USA. 2010;107:13777–13782. doi: 10.1073/pnas.0914115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colón-Gaud C, et al. Assessing ecological responses to catastrophic amphibian declines: patterns of macroinvertebrate production and food web structure in upland Panamanian streams. Limnol Oceanogr. 2009;54:331–343. [Google Scholar]
- 20.Stuart SN, et al. Status and trends of amphibian declines and extinctions worldwide. Science. 2004;306:1783–1786. doi: 10.1126/science.1103538. Analysis describing the high levels of amphibian extinctions caused by many environmental factors and disease. [DOI] [PubMed] [Google Scholar]
- 21.Kim K, Harvell CD. The rise and fall of a six-year coral-fungal epizootic. Am Nat. 2004;164:S52–S63. doi: 10.1086/424609. [DOI] [PubMed] [Google Scholar]
- 22.Cameron SA, et al. Patterns of widespread decline in North American bumble bees. Proc Natl Acad Sci USA. 2011;108:662–667. doi: 10.1073/pnas.1014743108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrnes EJ, III, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6:e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simwami SP, et al. Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathog. 2011;7:e1001343. doi: 10.1371/journal.ppat.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holdich DM, Reynolds JD, Souty-Grosset C, Sibley PJ. A review of the ever increasing threat to European crayfish from non-indigenous crayfish species. Knowl Managt Aquat Ecosyst. 2009;394–395:11. [Google Scholar]
- 26.Andrew TG, Huchzermeyer KDA, Mbeha BC, Nengu SM. Epizootic ulcerative syndrome affecting fish in the Zambezi river system in southern Africa. Vet Rec. 2008;163:629–631. doi: 10.1136/vr.163.21.629. [DOI] [PubMed] [Google Scholar]
- 27.Rizzo DM, Garbelotto M. Sudden oak death: endangering California and Oregon forest ecosystems. Front Ecol Environ. 2003;1:197–204. [Google Scholar]
- 28.Wills RT. The ecological impact of Phytophthora cinnamomi in the Stirling Range National Park, Western Australia. Aust J Ecol. 1993;18:145–159. [Google Scholar]
- 29.Jaenike J. An hypothesis to account for the maintenance of sex within populations. Evol Theor. 1978;3:191–194. [Google Scholar]
- 30.Paterson S, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464:275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCallum H, Dobson A. Detecting disease and parasite threats to endangered species and ecosystems. Trends Ecol Evol. 1995;10:190–194. doi: 10.1016/s0169-5347(00)89050-3. [DOI] [PubMed] [Google Scholar]
- 32.De Castro F, Bolker B. Mechanisms of disease-induced extinction. Ecol Lett. 2005;8:117–126. Theoretical study outlining the conditions under which disease can cause extinction of its host species. [Google Scholar]
- 33.Altizer S, Nunn CL, Lindenfors P. Do threatened hosts have fewer parasites? A comparative study in primates. J Anim Ecol. 2007;76:304–314. doi: 10.1111/j.1365-2656.2007.01214.x. [DOI] [PubMed] [Google Scholar]
- 34.Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 35.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. Macroecological analysis of recent patterns of EIDs worldwide in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casadevall A, Pirofski LA. The damage response framework of microbial pathogenesis. Nature Rev Microbiol. 2003;1:17–24. doi: 10.1038/nrmicro732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Roode JC, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci USA. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowak MA, May RM. Superinfection and the evolution of parasite virulence. Proc R Soc Lond B. 1994;255:81–89. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- 39.Briggs CJ, Knapp RA, Vredenburg VT. Enzootic and epizootic dynamics of the chytrid fungal pathogen of amphibians. Proc Natl Acad Sci USA. 2010;107:9695–9700. doi: 10.1073/pnas.0912886107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens PA, Sutherland WJ, Freckleton RP. What is the Allee effect? Oikos. 1999;87:185–190. [Google Scholar]
- 41.Mitchell KM, Churcher TS, Garner TWG, Fisher MC. Persistence of the emerging pathogen Batrachochytrium dendrobatidis outside the amphibian host greatly increases the probability of host extinction. Proc R Soc B. 2008;275:329–334. doi: 10.1098/rspb.2007.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rypien KL, Andras JP, Harvell CD. Globally panmictic population structure in the opportunistic fungal pathogen. Aspergillus sydowii Mol Ecol. 2008;17:4068–4078. doi: 10.1111/j.1365-294X.2008.03894.x. [DOI] [PubMed] [Google Scholar]
- 43.Jessup DA, et al. Southern sea otter as a sentinel of marine ecosystem health. Eco Health. 2004;1:239–245. [Google Scholar]
- 44.Sarmiento-Ramírez JM, et al. Fusarium solani is responsible for mass mortalities in nests of loggerhead sea turtle, Caretta caretta, in Boavista, Cape Verde. FEMS Microbiol Lett. 2010;312:192–200. doi: 10.1111/j.1574-6968.2010.02116.x. [DOI] [PubMed] [Google Scholar]
- 45.Lindner DL, et al. DNA-based detection of the fungal pathogen Geomyces destructans in soils from bat hibernacula. Mycologia. 2011;103:241–246. doi: 10.3852/10-262. [DOI] [PubMed] [Google Scholar]
- 46.Holt RD, Dobson AP, Begon M, Bowers RG, Schauber EM. Parasite establishment in host communities. Ecol Lett. 2003;6:837–842. [Google Scholar]
- 47.Hansen EM, Parke JL, Sutton W. Susceptibility of Oregon forest trees and shrubs to Phytophthora ramorum: a comparison of artificial inoculation and natural infection. Plant Dis. 2005;89:63–70. doi: 10.1094/PD-89-0063. [DOI] [PubMed] [Google Scholar]
- 48.Fröhlich-Nowoisky J, Pickersgill DA, Despres VR, Poschl U. High diversity of fungi in air particulate matter. Proc Natl Acad Sci USA. 2009;106:12814–12819. doi: 10.1073/pnas.0811003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henk DA, et al. Speciation despite globally overlapping distributions in Penicillium chrysogenum: the population genetics of Alexander Fleming’s lucky fungus. Mol Ecol. 2011;20:4288–4301. doi: 10.1111/j.1365-294X.2011.05244.x. [DOI] [PubMed] [Google Scholar]
- 50.Pringle A, Baker DM, Platt JL, Latge JP, Taylor JW. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus. Aspergillus fumigatus Evolution. 2005;59:1886–1899. [PubMed] [Google Scholar]
- 51.Ellison CE, et al. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci USA. 2011;108:2831–2836. doi: 10.1073/pnas.1014971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giraud T, Gladieux P, Gavrilets S. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol Evol. 2010;25:387–395. doi: 10.1016/j.tree.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Springer DJ, Chaturvedi V. Projecting global occurrence of. Cryptococcus gattii Emerg Infect Dis. 2010;16:14–20. doi: 10.3201/eid1601.090369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seimon TA, et al. Upward range extension of Andean anurans and chytridiomycosis to extreme elevations in response to tropical deglaciation. Glob Change Biol. 2007;13:288–299. [Google Scholar]
- 55.Fisher MC, et al. Biogeographic range expansion into South America by Coccidioides immitis mirrors New World patterns of human migration. Proc Natl Acad Sci USA. 2001;98:4558–4562. doi: 10.1073/pnas.071406098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stukenbrock EH, McDonald BA. The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol. 2008;46:75–100. doi: 10.1146/annurev.phyto.010708.154114. [DOI] [PubMed] [Google Scholar]
- 57.Brasier CM. The biosecurity threat to the UK and global environment from international trade in plants. Plant Pathol. 2008;57:792–808. An analysis of the lack of biosecurity and of the risk of disease introduction associated with the international plant trade. [Google Scholar]
- 58.Milgroom MG, Wang KR, Zhou Y, Lipari SE, Kaneko S. Intercontinental population structure of the chestnut blight fungus. Cryphonectria parasitica Mycologia. 1996;88:179–190. [Google Scholar]
- 59.Gonthier P, Warner R, Nicolotti G, Mazzaglia A, Garbelotto MM. Pathogen introduction, as a collateral effect of military activity. Mycol Res. 2004;108:468–470. doi: 10.1017/s0953756204240369. [DOI] [PubMed] [Google Scholar]
- 60.Goka K, et al. Amphibian chytridiomycosis in Japan: distribution, haplotypes and possible route of entry into Japan. Mol Ecol. 2009;18:4757–4774. doi: 10.1111/j.1365-294X.2009.04384.x. [DOI] [PubMed] [Google Scholar]
- 61.Garner TWJ, et al. The emerging amphibian pathogen Batrachochytrium dendrobatidis globally infects introduced populations of the North American bullfrog, Rana catesbeiana. Biol Lett. 2006;2:455–459. doi: 10.1098/rsbl.2006.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cunningham AA, et al. Emergence of amphibian chytridiomycosis in Britain. Vet Rec. 2005;157:386–387. doi: 10.1136/vr.157.13.386. [DOI] [PubMed] [Google Scholar]
- 63.Walker SF, et al. Invasive pathogens threaten species recovery programs. Curr Biol. 2008;18:R853–R854. doi: 10.1016/j.cub.2008.07.033. [DOI] [PubMed] [Google Scholar]
- 64.Farrer RA, et al. Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalised hypervirulent recombinant lineage. Proc Natl Acad Sci USA. 2011;108:18732–18736. doi: 10.1073/pnas.1111915108. Population genomics analysis of the generation, and spread, of a hypervirulent fungal lineage in amphibians worldwide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wibbelt G, et al. White-nose syndrome Fungus (Geomyces destructans) in Bats, Europe. Emerg Infect Dis. 2010;16:1237–1243. doi: 10.3201/eid1608.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richards TA, et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc Natl Acad Sci USA. 2011;108:15258–15263. doi: 10.1073/pnas.1105100108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fraser JA, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. Analysis of the evolution of a hypervirulent lineage of human-infecting fungus that invaded British Columbia. [DOI] [PubMed] [Google Scholar]
- 68.Turner E, Jacobson DJ, Taylor JW. Genetic architecture of a reinforced, postmating, reproductive isolation barrier between Neurospora species indicates evolution via natural selection. PLoS Genet. 2011;7:e1002204. doi: 10.1371/journal.pgen.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coyne JA, Orr HA. Speciation. Sinauer Associates; 2004. [Google Scholar]
- 70.Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- 71.Brasier CM, Rose J, Gibbs JN. An unusual phytophthora associated with widespread alder mortality in Britain. Plant Pathol. 1995;44:999–1007. [Google Scholar]
- 72.Inderbitzin P, Davis RM, Bostock RM, Subbarao KV. The ascomycete Verticillium longisporum is a hybrid and a plant pathogen with an expanded host range. PLoS One. 2011;6:e18260. doi: 10.1371/journal.pone.0018260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gange AC, Gange EG, Sparks TH, Boddy L. Rapid and recent changes in fungal fruiting patterns. Science. 2007;316:71. doi: 10.1126/science.1137489. [DOI] [PubMed] [Google Scholar]
- 74.Pachauri RK, Resinger A, editors. Climate change 2007: Synthesis report. (Intergovernmental Panel on Climate Change) 2007. [Google Scholar]
- 75.Newton AC, Johnson SN, Gregory PJ. Implications of climate change for diseases, crop yields and food security. Euphytica. 2011;179:3–18. This paper highlights the importance of understanding the impact of climate change on crops and disease. [Google Scholar]
- 76.Lake JA, Wade RN. Plant–pathogen interactions and elevated CO2: morphological changes in favour of pathogens. J Exp Bot. 2009;60:3123–3131. doi: 10.1093/jxb/erp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chakraborty S, et al. Impacts of global change on diseases of agricultural crops and forest trees. CAB Rev. 2008;3:1–5. [Google Scholar]
- 78.Kobayashi T, et al. Effects of elevated atmospheric CO2 concentration on the infection of rice blast and sheath blight. Phytopathology. 2006;96:425–431. doi: 10.1094/PHYTO-96-0425. [DOI] [PubMed] [Google Scholar]
- 79.Madgwick JW, et al. Impacts of climate change on wheat anthesis and fusarium ear blight in the UK. Eur J Plant Pathol. 2011;130:117–131. [Google Scholar]
- 80.Gregory PJ, Johnson SN, Newton AC, Ingram JSI. Integrating pests and pathogens into the climate change/food security debate. J Exp Bot. 2009;60:2827–2838. doi: 10.1093/jxb/erp080. [DOI] [PubMed] [Google Scholar]
- 81.Pounds JA, et al. Widespread amphibian extinctions from epidemic disease driven by global warming. Nature. 2006;439:161–167. doi: 10.1038/nature04246. [DOI] [PubMed] [Google Scholar]
- 82.Bosch J, Carrascal LM, Duran L, Walker S, Fisher MC. Climate change and outbreaks of amphibian chytridiomycosis in a montane area of Central Spain; is there a link? Proc R Soc B. 2007;274:253–260. doi: 10.1098/rspb.2006.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rohr JR, Raffel TR, Romansic JM, McCallum H, Hudson PJ. Evaluating the links between climate, disease spread, and amphibian declines. Proc Natl Acad Sci USA. 2008;105:17436–17441. doi: 10.1073/pnas.0806368105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garner TWJ, Rowcliffe JM, Fisher MC. Climate change, chytridiomycosis or condition: an experimental test of amphibian survival. Glob Change Biol. 2011;17:667–675. [Google Scholar]
- 85.Becker CG, Zamudio KR. Tropical amphibian populations experience higher disease risk in natural habitats. Proc Natl Acad Sci USA. 2011;108:9893–9898. doi: 10.1073/pnas.1014497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harvell CD, et al. Review: Emerging marine diseases - Climate links and anthropogenic factors. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 87.vanEngelsdorp D, et al. Colony collapse disorder: a descriptive study. PLoS One. 2009;4:e6481. doi: 10.1371/journal.pone.0006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ratnieks FLW, Carreck NL. Clarity on honey bee collapse? Science. 2010;327:152–153. doi: 10.1126/science.1185563. [DOI] [PubMed] [Google Scholar]
- 89.Verweij PE, Mellado E, Melchers WJG. Multiple-triazole-resistant aspergillosis. N Engl J Med. 2007;356:1481–1483. doi: 10.1056/NEJMc061720. [DOI] [PubMed] [Google Scholar]
- 90.Klaassen CHW, Gibbons JG, Fedorova ND, Meis JF, Rokas A. Evidence for genetic differentiation and variable recombination rates among Dutch populations of the opportunistic human pathogen. Aspergillus fumigatus Mol Ecol. 2012;21:57–70. doi: 10.1111/j.1365-294X.2011.05364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miraglia M, et al. Climate change and food safety: an emerging issue with special focus on Europe. Food Chem Toxicol. 2009;47:1009–1021. doi: 10.1016/j.fct.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 92.Stokstad E. The famine fighter’s last battle. Science. 2009;324:710–712. doi: 10.1126/science.324_710a. [DOI] [PubMed] [Google Scholar]
- 93.Loo JA. Ecological impacts of non-indigenous invasive fungi as forest pathogens. Biol Invasions. 2009;11:81–96. [Google Scholar]
- 94.Kurz WA, et al. Mountain pine beetle and forest carbon feedback to climate change. Nature. 2008;452:987–990. doi: 10.1038/nature06777. Study describing pest- and pathogen-induced loss of forest carbon sinks. [DOI] [PubMed] [Google Scholar]
- 95.Mazzoni R, et al. Emerging pathogen of wild amphibians in frogs (Rana catesbeiana) farmed for international trade. Emerg Infect Dis. 2003;9:995–998. doi: 10.3201/eid0908.030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Byrnes EJ, III, Bildfell RJ, Dearing PL, Valentine BA, Heitman J. Cryptococcus gattii with bimorphic colony types in a dog in western Oregon: additional evidence for expansion of the Vancouver Island outbreak. J Vet Diagn Invest. 2009;21:133–136. doi: 10.1177/104063870902100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lubick N. Emergency medicine for frogs. Nature. 2010;465:680–681. doi: 10.1038/465680a. [DOI] [PubMed] [Google Scholar]
- 98.Harris RN, et al. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 2009;3:818–824. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]
- 99.U.S. Fish and Wildlife Service. A national plan for assisting states, federal agencies, and tribes in managing white-nose syndrome in bats. 2011 〈 http://www.fws.gov/WhiteNoseSyndrome/pdf/WNSnationalplanMay2011.pdf〉.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.