Abstract
Type I Interferons are cytokines of the innate immune system that induce antiviral protein expression in response to viral infection. Various proteins and pathways have been shown to recognize nucleic acids ligands especially from RNA viruses. Here, we will review recent developments including transcription of DNA virus genomes into RNA ligands, and the recognition of viruses by TLR2 for interferon induction. The induced IFNs activate many interferon stimulated genes (ISGs) that have direct anti-viral effects. Recent studies have identified IFITM proteins as the first ISG to inhibit viral entry processes and revealed mechanistic understanding of known anti-viral ISGs such as ISG15 and Viperin.
Introduction
Type I Interferon (IFN) is a key innate immune cytokine produced by cells to combat viral infections. Intricate sensory mechanisms detect invading viruses and rapidly trigger interferon production. Recognition of distinctive viral nucleic acids as a pathogen associated molecular patterns (PAMPs) by cellular pattern recognition receptors (PRRs) will lead to IFN induction. While RNA virus recognition is well understood, new pathways are constantly being elucidated and the receptor for DNA viruses is a subject of intense research. The first part of this review will discuss recent advances in understanding how virus infection leads to IFN production.
Release of interferon after viral recognition signals to cells to induce the expression of a set of Interferon stimulated genes (ISGs) that activate anti-viral processes including amplification of interferon signaling, production of cytokines that activate adaptive immunity, and many factors that directly inhibit viruses. ISGs with direct anti-viral functions remain poorly understood, largely because they are virus-specific and can have multiple mechanisms. The second part of this review will cover well-known and novel ISGs focusing on recent developments in understanding their anti-viral function.
Old and New Paths to IFN Induction
The mechanism involved in how cells exposed to viruses or virion components “know” to release IFN has not been well understood until recently. The discovery of the Toll like receptors (TLRs) as receptors for extracellular or endocytosed viral components was a major advance in understanding viral recognition in the IFN process1. Likewise, the recent discovery of the RIG-I-like RNA Helicases as RNA virus sensors has elucidated how a cell detects an active intracellular virus infection2. Signaling downstream of these receptors has been well studied and although some questions on the biochemical level remain, the signaling pathways generally converge on the activation of TANK-Binding Kinase 1 (TBK1) that phosphorylates and activates the Interferon Regulatory Factors (IRF 3 and/or 7) (Figure 1).
Figure 1. Old and New Players in the Induction of IFN.
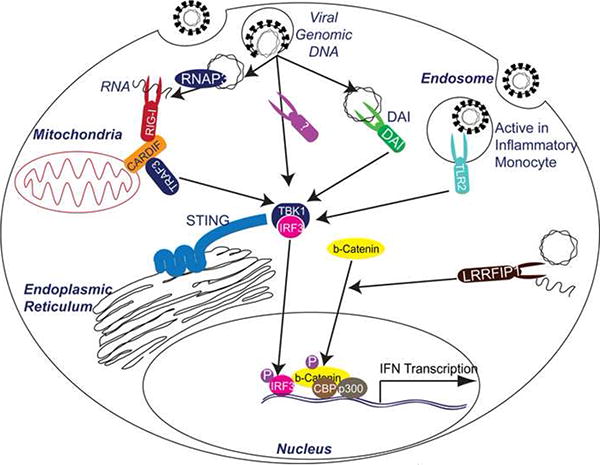
TANK-binding kinase-1 (TBK1) is the primary IRF3 activating kinase. IRF3 is activated by phosphorylation, after which is translocates to the nucleus and induce Interferon gene transcription. DNA induction of this pathway may occur by RNA Polymerase III (RNAP3) transcription of abundant DNA into RNA, which can then function as a RIG-I substrate. RIG-I is well known in RNA induction pathways to signal through CARDIF, a mitochondrial adaptor to TRAF3 and downstream to TBK1. In addition, DAI has been found to recognize DNA and induce signals downstream to TBK1. Further studies may reveal new receptors that are either parallel or a more primary receptor for DNA that either DAI or RNAP3. However, STING has recently been found to be an ER associated multi-membrane protein that is required for signaling by these nucleic acid receptors, potentially by serving as a signaling docking and coordination center. In addition, TLR2 has recently been found to induce IRF3 activation in inflammatory monocytes by signaling from endosomal compartments. Finally, LRRFIP1 has recently been found to respond to cytoplasmic nucleic acids and signal to induce β-catenin phosphorylation. Phosphorylated β-catenin translocate to the nucleus and is recruited to interferon promoters to activate CBP/p300 that then induce acetylation and activation of the interferon promoter in a mechanism whose importance is still being dissected.
In addition, to TBK1 in this pathway, IFN induction through a LRRFIP1 mediated pathway was demonstrated in mouse peritoneal macrophages exposed to intracellular DNA or RNA. LRRFIP1 is a leucine rich repeat domain containing protein similar to the TLRs but cytoplasmic in localization3. Intriguingly, induction of IFN-β by LRRFIP1 recognition of free nucleic acids is dependent on β-catenin, a well-known coactivator of transcription4. LRRFIP1 recognition of bacterial DNA or Vesicular Stomatitis Virus (VSV) infection recruits β-catenin to the nucleus in an IRF-3 dependent manner (Figure 1). Nuclear β-catenin can function to activate CBP/p300 that enhances acetylation and activation of the IFN- β promoter. These findings show a novel IFN induction pathway that works with the canonical TBK1/IRF pathways. In fact, most of the recent advancements in understanding IFN induction underscore the need for critical examination of structured paradigms of innate immunity.
New Players in Recognition of DNA Virus Infection
A search for the primary DNA virus receptor has fueled much research over the past years. From the discovery of DAI, a protein that seemed critical for DNA induced IFN to the in vivo finding that DAI may be redundant, many groups have searched for the “key” DNA receptor or sought to understand how DNA recognition occurs5,6. A major development is the discovery that the protein STING is necessary for IFN induction by exposure to B-DNA and the DNA virus HSV-17,8. STING is an ER-localized, multi-transmembrane domain protein that interacts with IRF-3, TBK1, CARDIF and RIG-I, and seems to coordinate the signaling of IFN induction. Though, this protein is not a DNA receptor, STING deficient mice represents one of the first knockouts that are compromised in IFN induction capacity by all exogenous DNA or DNA viruses.
In the past year, the hypothesis that the DNA receptor directly binds to DNA was found to be insufficient. Two reports show that rather than DNA recognition, RNA transcribed from cytoplasmic DNA can function as a ligand for RIG-I induced IFN9. Here abundant cytoplasmic DNA containing AT-rich regions can be transcribed by RNA Polymerase III (RNAP3) and the RNA transcripts are recognized by the RIG-I pathway, in effect turning viral DNA into RNA PAMPs. RNAP3 is important in cells transfected with B-DNA or infected at high MOI with HSV-19. Epstein-Barr Virus (EBV) also transcribes small RNAs via RNAP3 from viral DNA into RIG-I ligands. Whether RNAP3 transcription of viral DNA is physiologically relevant remains an central question.
TLR2 as a Virus Receptor for Interferon Induction
The endosomal Toll-like Receptors (TLR) 3, 7, 8 recognize extracellular viral RNA PAMPs, while TLR9 recognizes CpG DNA and can lead to IFN production upon activation. Other TLRs were thought to primarily be inducers of inflammatory cytokines, none more so than TLR2.
However, in inflammatory monocytes, a small distinct fraction of bone marrow, TLR2 was found to be required for vaccinia virus induced IFN induction10. Ablation of this cell type, which is not present in standard bone marrow derived macrophages or DCs, leads to increased susceptibility to vaccinia virus infection in vivo. TLR2 localizes to endosomal compartments in this cell type where it can induce IFN. This observation suggests that cellular localization of TLR2 can alter its downstream signaling potential. Classical bacterial ligands at the cell surface ligate to TLR2 where a distinct array of signaling adaptors induce inflammatory genes. However, signaling proteins localized to the endosome may be specialized to signal from TLR2 to induce IFN. As most of the nucleic acid sensing TLRs (3/7/8/9) are localized to the endosome, this model is consistent with the paradigm that the endosomal system represents a major hub of virus recognition and signaling.
Interferon Stimulated Genes
Viral recognition induces the release of IFN that signals to surrounding cells creating the “antiviral state” that was described as far back as the original IFN studies. Expression array studies have shown that hundreds of genes are induced by IFN. While some ISGs such as Protein Kinase R (PKR), 2′5-oligoadenylate synethetase, and Mx GTPases have well described anti-viral functions and mechanisms11-13, functions of most ISGs are poorly characterized with little or no mechanistic understanding. Table 1 summarizes most of the known anti-viral ISGs. Here, we review recent developments in ISG function.
Table 1. Summary of known anti-viral functions of ISGs.
| Interferon Stimulated Gene | Viruses inhibited and Method of Study | Anti-viral mechanism |
|---|---|---|
| Protein Kinase R | Overexpression of wild-type but not mutant PKR inhibits ECMV, vaccinia, HIV-135-37 PKR deficient mice deleted are susceptible to VSV and influenza infections and increased HSV-1 susceptibility in neurons38,39. |
Translation Inhibition Binds to dsRNA and ssRNA and phosphorylate EIF2a, which prevents its guanine nucleotide exchange activity that is required for translational activity12. |
| 2′5-OAS and RNaseL | ssRNA viruses Picornaviridae, Reoviridae, Togaviridae, Paramyxoviridae, Orthomyxoviridae, Flaviviridae and Retroviridae13,40,41 |
RNA Degradation Form short oligoadenylates from ATP, which activates RNaseL to degrade viral RNA12. |
| TRIM5a | Stable expression of TRIM5a from Rhesus monkey in HeLa cells inhibits HIV-1 and SIV42 |
Inhibition of viral cDNA synthesis and nuclear import43
Viral Protein Degradation Target HIV capsids and RT products proteosomal degradation44. |
| APOBEC3G and APOBEC3F | Inhibits HIV-1; APOBEC3G and APOBEC3F deficient cell supports Vif-deficient HIV-145 Expression inhibits parvoviruses and retrotransposons that is deaminase independent45,46 |
Mutation of HIV DNA A cytidine deaminase that converts cytidine to uracil in the viral RNA, which subsequently leads to T/A hypermutation in the viral DNA after reverse transcription47,48. Catalytic activity not required for antiviral function49. Inhibition of HIV-1 provirus formation A3G inhibits minus-strand to plus-strand step in reverse transcription50. A3F inhibits viral 3′ DNA processing51. Inhibition of viral assembly A3G interacts with HIV RNA and Gag and packaged into viral particles52. |
| ISG15 | Influenza, Sinbis, HSV1, MHV6853,54. |
Modifying ubiquitination on many cellular and viral targets ISGylation by ISG15 prevents IRF3 degradation17. Indirectly Prevents Virion release. Inhibits ubiquitination of HIV Gag and Tsg101 and prevents virion release53. |
| ISG20 | Overexpression inhibits VSV, EMCV, influenza, HIV55,56 | A 3-5 exonuclease; mechanism is unclear12. |
| IFITM1,2,3 | Overexpression Inhibits influenza, Dengue, West Niles virus, and VSV. Knockdown of IFITM3 increased susceptibility to influenza, WNV, and Dengue infection in vitro14. |
Inhibition of Viral Entry14 Transmembrane protein. Antiviral mechanism unknown. |
| Mx GTPases | orthomyxoviruses, paramyxoviruses, rhabdoviruses, togaviruses, bunyaviruses including HBV, influenza, coxackie virus11,26 |
Inhibition of vRNP trafficking Human MxA targets viral necleocapsid structures and traps viral components11,26 Inhibition of viral transcription MxA associates with influenza PB2 and prevents transcription of viral genome11,26. |
| Viperin (Cig5) | Overexpression of Viperin inhibits hCMV57 and HCV replication24,58 Induction of Viperin in HeLa cells inhibits influenza budding22. Viperin knockdown reduces TLR3 mediated inhibition of HIV-1 in astrocytes59. |
Inhibition of Budding Disrupts lipid rafts22 |
IFITM3
The interferon induced transmembrane proteins (IFITM) 1, 2, and 3 were identified as the first host factors that restrict viral entry14. Brass et al., showed that overexpression of IFITM 2 and 3 significantly inhibited influenza, VSV, West Nile, and Dengue virus14. Conversely, knockdown of IFITM3 or deletion of the Ifitm locus in murine embryonal fibroblast (MEFs) increased susceptibility of the cells to viral infections. IFITM3 inhibited influenza pseudoviruses but not Machupo pseudoviruses, suggesting that IFITM3 inhibits viral entry processes because these pseudoviruses differ only in their envelopes. Ifitm deficient mice are viable, yet their susceptibility to viral infection is not known. Recently, IFITM3 was shown to be modified by S-palmitoylation, a post-translational modification that can regulate localization and function of membrane associated proteins. Interestingly, deletion of the palmitoylation site on IFITM3 abrogates its anti-viral effect on influenza suggesting localization specific function15.
The precise mechanism of the antiviral activity of IFITM3 awaits further studies. Overexpression and knockdown studies suggest that IFITM1, 2, and 3 may have non-redundant functions, but their effects on different viruses need to be further delineated. How IFITM3 affects entry steps, such as binding and fusion, is still unknown. Does it physically interact with influenza virions or does it recruits complexes to affect viral entry? IFITM3 may also have additional anti-viral effects on assembly and budding.
ISG15
ISG15 is a 17kD ubiquitin-like protein that has been shown to inhibit replication of several viruses including influenza, sindbis, herpes, HIV, HPV, and Ebola. ISG15 modification, called ISGylation, occurs on over 100 cellular proteins and is catalyzed by the sequential action of the interferon-inducible E1, E2, and E3 ubiquitin ligases called UBE1L, UbcH8/Ube2L6, and Herc5, respectively12,16. Unlike canonical ubiquitination that targets proteins for degradation, ISGylation can have diverse effects. For example, ISGylation of IRF-3 inhibits its degradation and causes increase in its transcriptional activity17. ISGylation inhibits Ebola by blocking ubiquitin ligase Nedd4, which is required viral budding18.
One of the recent novel discoveries of the anti-viral mechanism of ISG15 is the ISGylation of viral proteins. ISGylation of the influenza protein NS1 nuclear localization domain prevents its association with importin-alpha. Mutation of the ISGylation site conferred increased resistance of influenza virus in the presence of interferon19,20. Other ISGylation sites have been found yet their functional significance is unclear. Interestingly, the amount of ISGylation of NS1 changes across different strains of influenza, which opens the question whether the propensity for ISGylation correlates with virulence21. While many proteins can be modified by ISG15, ISGylation seems to specifically modify newly synthesized host and viral proteins16. This mechanism may help confer specific anti-viral effects without causing global protein modifications in the cell.
Viperin
Viperin is an ER-associated ISG that inhibits HCV, HCMV, influenza, and HIV-1 through several mechanisms. Wang et al. showed that Viperin disrupts cell plasma membrane and lipid raft integrity and inhibits influenza virion budding22. Overexpression of farnesyl diphosphate synthase (FPPS), an enzyme required for isoprenoid synthesis and lipid metabolism, reversed this anti-viral effect, suggesting that Viperin prevents viral budding through inhibition of FPPS22.
Viperin may inhibit HCV replication through a different mechanism. HCV core and nonstructural (NS) proteins associate with lipid droplets, ER-associated organelles important for cellular protein and lipid trafficking that are thought to be a site of HCV replication in the cell23. Both Viperin and NS protein have an N-terminal amphipathic, alpha-helical domain required for localization to lipid droplets. More importantly, the N-terminal domain of Viperin is required for inhibition of HCV24. Although these data suggest that Viperin can inhibit HCV in lipid droplets, it remains unclear whether there is direct association of Viperin with HCV proteins or whether the amphipathic sequence is necessary for its inhibitory activity.
Recent structural and biochemical studies identified Viperin is an S-adenosyl-L-methionine (SAM) enzyme that binds Fe-S clusters and catalyzes SAM to form 5′-deoxyadenosyl radicals25. The significance of this C-terminal catalytic domain is unknown and may be required for other cellular anti-viral processes.
Interferon inducible GTPases
Both type I and type II interferon significantly induce expression of the Mx, p47, and p65 families of GTPases, which hydrolyze GTP and are well-known to confer resistance against a wide range of pathogens. The Mx proteins inhibits replication of orthomyxoviruses, Thogoto virus, bunyaviruses and rhabdoviruses11,26. The family of p47 GTPases, which consists of Iigp, Lrg47, Irg47, Tgtp, Iigp, and Gtpi, predominantly inhibits bacteria and protozoa growth27. Only Tgtp and Igtp overexpression in vitro have been shown to inhibit VSV28 and Coxackie viral replication29, respectively. The family of p65 GTPases, also known as the Guanylate-Binding Proteins (GBPs), is induced by all interferons, with more robust induction by interferon gamma. Overexpression of GBP-1 and GBP-2 inhibited VSV and encephalomyocarditis virus (EMCV) replication30. GBP-1 also reduces HCV replication, but replication competent HCV expresses NS5B that inhibit GBP-1 GTPase activity31. The functions of GBPs are largely unknown. GBP-2 can target to intracellular vesicles32 and GBP-1 can form oligomers like Mx proteins33, which may provide clues to their anti-viral function. There may be more than one anti-viral mechanism as exemplified by the fact that GTP binding activity is required for inhibition of EMCV but not VSV34.
Concluding Remarks
The complex host-virus interactions involved in mounting and executing an effective response to viral infections represents one of the major directions in innate immunity research. While the general scheme for viral detection has been unraveled, much in terms of the actual ligands during an infection as well as the relative contribution of specific receptor signaling remains to be determined. One particularly important point will be defining the definitive detection pathway(s) for DNA viruses and testing whether detection through RNAP3 holds up in in vivo infections. While viral recognition remains an important subject of research, the elusive anti-viral functions of ISGs against specific viruses are getting increased attention. Many studies have shown sufficiency of anti-viral activity of ISGs, such as GBP-1, in vitro, but not necessity. The next level of studies will be to define physiological roles of such ISGs as is the case for IFITM3. Understanding of the interactions between ISGs and particular viral lifecycle processes will be particularly informative but not trivial. Nearly all the ISGs described here can inhibit viruses in more than one way and many of them may have redundant functions. In addition, how viruses have evolved ways to escape anti-viral detection and effectors is equally important and lends another level of complexity to host-pathogen interactions. Elucidation of ISG and viral interaction may allow for identification of susceptibility mutations and provide new approaches for viral therapy.
Figure 2. General Anti-viral Mechanisms of Interferon Stimulated Genes.
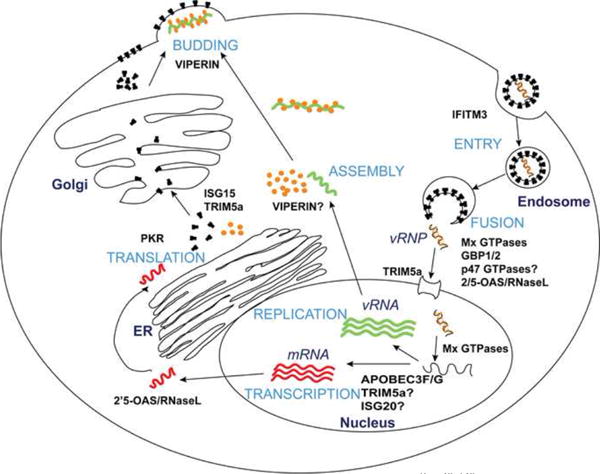
The general lifecycle stages of an enveloped virus are depicted here (light blue letters). The virus binds to specific receptors on the cell surface and often enters the cell through endocytosis. Viral genetic material is released into the cytoplasm through pH-dependent or -independent fusion and may be subsequently transported to the nucleus. Replication of viral genetic material ensues along with mRNA transcription followed by transport to the ER for protein translation. Envelope proteins are transported to the cell surface while core viral proteins assemble with the viral genetic material. New virion particles are enveloped as they bud out of the plasma membrane. ISGs (in capital black bold letters) can inhibit viruses differently and at one or more stages of the viral lifecycle, see Table 1.
Acknowledgments
The authors were supported by funding from the National Institutes of Health (R01 AI069120, R01 AI078389 and PN2EY018228). We also thank the members of the Cheng laboratory for helpful discussions on the topic. We apologize for not recognizing authors who have made important contributions to the field of type I interferon induction and anti-viral function due to reference limitation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kawai T, Akira S. Toll-like Receptor and RIG-1-like Receptor Signaling. Annals of the New York Academy of Sciences. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 2.Baum A, García-Sastre A. Induction of type I interferon by RNA viruses: cellular receptors and their substrates. Amino Acids. 2010;38:1283–1299. doi: 10.1007/s00726-009-0374-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang P, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a [beta]-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. In addition to the importance of LRRFIP1 in IFN induction, this paper strengthens the key role of β-catenin in IFN induction. [DOI] [PubMed] [Google Scholar]
- 4.Stadeli R, Hoffmans R, Basler K. Transcription under the Control of Nuclear Arm/[beta]-Catenin. Curr Biol. 2006;16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 5.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proceedings of the National Academy of Sciences. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. 7 and 8. Glen Barber's group defines and studies STING as a key molecule in DNA induced IFN. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu Y, MacMillan JB, Chen ZJ. RNA Polymerase III Detects Cytosolic DNA and Induces Type I Interferons through the RIG-I Pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10:1200–1207. doi: 10.1038/ni.1792. Though TLR2 is canonically inflammatory, here we see that TLR2 can induce IFN and protect from vaccinia virus infection by trafficking to the endosomes of inflammatory monocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haller O, Staeheli P, Kochs G. Interferon-induced Mx proteins in antiviral host defense. Biochimie. 89:812–818. doi: 10.1016/j.biochi.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hovanessian AG. On the discovery of interferon-inducible, double-stranded RNA activated enzymes: The 2′-5′oligoadenylate synthetases and the protein kinase PKR. Cytokine & Growth Factor Reviews. 18:351–361. doi: 10.1016/j.cytogfr.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Brass AL, et al. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. This is the first study that demonstrates IFITM family of proteins inhibit entry processes of several RNA viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yount JS, et al. Palmitoylome profiling reveals S-palmitoylation–dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6:610–614. doi: 10.1038/nchembio.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durfee LA, Lyon N, Seo K, Huibregtse JM. The ISG15 Conjugation System Broadly Targets Newly Synthesized Proteins: Implications for the Antiviral Function of ISG15. Mol Cell. 2010;38:722–732. doi: 10.1016/j.molcel.2010.05.002. This study provides insight on how specificity is conferred by ISGylation. The authors show ISGylation occurs only on newly synthesized host protein and HPV L1 protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi H, et al. Positive Regulation of Interferon Regulatory Factor 3 Activation by Herc5 via ISG15 Modification. Mol Cell Biol. 2010;30:2424–2436. doi: 10.1128/MCB.01466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okumura A, Pitha PM, Harty RN. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proceedings of the National Academy of Sciences. 2008;105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiang T, Zhao C, Krug RM. Interferon-Induced ISG15 Conjugation Inhibits Influenza A Virus Gene Expression and Replication in Human Cells. J Virol. 2009;83:5971–5977. doi: 10.1128/JVI.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao C, Hsiang T, Kuo R, Krug RM. ISG15 conjugation system targets the viral NS1 protein in influenza A virus–infected cells. Proceedings of the National Academy of Sciences. 2010;107:2253–2258. doi: 10.1073/pnas.0909144107. This study demonstrated ISGylation on NS1 of influenza in lysine 41 is partially responsible for its direct inhibitory effect on the virus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y, et al. Herc5 Attenuates Influenza A Virus by Catalyzing ISGylation of Viral NS1 Protein. J Immunol. 2010;184:5777–5790. doi: 10.4049/jimmunol.0903588. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Hinson ER, Cresswell P. The Interferon-Inducible Protein Viperin Inhibits Influenza Virus Release by Perturbing Lipid Rafts. Cell Host Microbe. 2007;2:96–105. doi: 10.1016/j.chom.2007.06.009. This study to show mechanism of Viperin inhibition on influenza through changes in membrane integrity. [DOI] [PubMed] [Google Scholar]
- 23.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 24.Jiang D, et al. Identification of Three Interferon-Inducible Cellular Enzymes That Inhibit the Replication of Hepatitis C Virus. J Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaveta G, Shi J, Chow VT, Song J. Structural characterization reveals that viperin is a radical S-adenosyl-l-methionine (SAM) enzyme. Biochemical and Biophysical Research Communications. 2010;391:1390–1395. doi: 10.1016/j.bbrc.2009.12.070. [DOI] [PubMed] [Google Scholar]
- 26.Haller O, Stertz S, Kochs G. The Mx GTPase family of interferon-induced antiviral proteins. Microbes and Infection. 9:1636–1643. doi: 10.1016/j.micinf.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Taylor GA, Feng CG, Sher A. p47 GTPases: regulators of immunity to intracellular pathogens. Nat Rev Immunol. 2004;4:100–109. doi: 10.1038/nri1270. [DOI] [PubMed] [Google Scholar]
- 28.Carlow DA, Teh S, Teh H. Specific Antiviral Activity Demonstrated by TGTP, A Member of a New Family of Interferon-Induced GTPases. J Immunol. 1998;161:2348–2355. [PubMed] [Google Scholar]
- 29.Zhang HM, et al. Overexpression of Interferon-γ-inducible GTPase Inhibits Coxsackievirus B3-induced Apoptosis through the Activation of the Phosphatidylinositol 3-Kinase/Akt Pathway and Inhibition of Viral Replication. Journal of Biological Chemistry. 2003;278:33011–33019. doi: 10.1074/jbc.M305352200. [DOI] [PubMed] [Google Scholar]
- 30.Anderson SL, Carton JM, Lou J, Xing L, Rubin BY. Interferon-Induced Guanylate Binding Protein-1 (GBP-1) Mediates an Antiviral Effect against Vesicular Stomatitis Virus and Encephalomyocarditis Virus. Virology. 1999;256:8–14. doi: 10.1006/viro.1999.9614. [DOI] [PubMed] [Google Scholar]
- 31.Itsui Y, et al. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology. 2009;50:1727–1737. doi: 10.1002/hep.23195. [DOI] [PubMed] [Google Scholar]
- 32.Gorbacheva VY, Lindner D, Sen GC, Vestal DJ. The Interferon (IFN)-induced GTPase, mGBP-2. Journal of Biological Chemistry. 2002;277:6080–6087. doi: 10.1074/jbc.M110542200. [DOI] [PubMed] [Google Scholar]
- 33.Prakash B, Praefcke GJK, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 34.Carter CC, Gorbacheva VY, Vestal DJ. Inhibition of VSV and EMCV replication by the interferon-induced GTPase, mGBP-2: differential requirement for wild-type GTP binding domain. Archives of Virology. 2005;150:1213–1220. doi: 10.1007/s00705-004-0489-2. [DOI] [PubMed] [Google Scholar]
- 35.Meurs EF, et al. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J Virol. 1992;66:5805–5814. doi: 10.1128/jvi.66.10.5805-5814.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SB, Esteban M. The Interferon-Induced Double-Stranded RNA-Activated Human p68 Protein Kinase Inhibits the Replication of Vaccinia Virus. Virology. 1993;193:1037–1041. doi: 10.1006/viro.1993.1223. [DOI] [PubMed] [Google Scholar]
- 37.Muto NF, Martinand-Mari C, Adelson ME, Suhadolnik RJ. Inhibition of Replication of Reactivated Human Immunodeficiency Virus Type 1 (HIV-1) in Latently Infected U1 Cells Transduced with an HIV-1 Long Terminal Repeat-Driven PKR cDNA Construct. J Virol. 1999;73:9021–9028. doi: 10.1128/jvi.73.11.9021-9028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balachandran S, et al. Essential Role for the dsRNA-Dependent Protein Kinase PKR in Innate Immunity to Viral Infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- 39.Leib DA, Machalek MA, Williams BRG, Silverman RH, Virgin HW. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin R, et al. Distinct Antiviral Roles for Human 2′,5′-Oligoadenylate Synthetase Family Members against Dengue Virus Infection. J Immunol. 2009;183:8035–8043. doi: 10.4049/jimmunol.0902728. [DOI] [PubMed] [Google Scholar]
- 41.Silverman RH. Viral Encounters with 2′,5′-Oligoadenylate Synthetase and RNase L during the Interferon Antiviral Response. J Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stremlau M, et al. The cytoplasmic body component TRIM5[alpha] restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 43.Berthoux L, Sebastian S, Sokolskaja E, Luban J. Lv1 Inhibition of Human Immunodeficiency Virus Type 1 Is Counteracted by Factors That Stimulate Synthesis or Nuclear Translocation of Viral cDNA. J Virol. 2004;78:11739–11750. doi: 10.1128/JVI.78.21.11739-11750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proceedings of the National Academy of Sciences. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pion M, et al. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. The Journal of Experimental Medicine. 2006;203:2887–2893. doi: 10.1084/jem.20061519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Research. 34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris RS, et al. DNA Deamination Mediates Innate Immunity to Retroviral Infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 48.Strebel K, Luban J, Jeang K. Human cellular restriction factors that target HIV-1 replication. BMC Medicine. 2009;7:48. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bishop KN, Holmes RK, Malim MH. Antiviral Potency of APOBEC Proteins Does Not Correlate with Cytidine Deamination. J Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Guo F, Zhang L, Kleiman L, Cen S. APOBEC3G Inhibits DNA Strand Transfer during HIV-1 Reverse Transcription. Journal of Biological Chemistry. 2007;282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- 51.Mbisa JL, Bu W, Pathak VK. APOBEC3F and APOBEC3G Inhibit HIV-1 DNA Integration by Different Mechanisms. J Virol. 2010;84:5250–5259. doi: 10.1128/JVI.02358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strebel K, Khan M. APOBEC3G encapsidation into HIV-1 virions: which RNA is it? Retrovirology. 2008;5:55. doi: 10.1186/1742-4690-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lenschow DJ, et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proceedings of the National Academy of Sciences. 2007;104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espert L, et al. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J Gen Virol. 2005;86:2221–2229. doi: 10.1099/vir.0.81074-0. [DOI] [PubMed] [Google Scholar]
- 56.Espert L, et al. ISG20, a New Interferon-induced RNase Specific for Single-stranded RNA, Defines an Alternative Antiviral Pathway against RNA Genomic Viruses. Journal of Biological Chemistry. 2003;278:16151–16158. doi: 10.1074/jbc.M209628200. [DOI] [PubMed] [Google Scholar]
- 57.Chin K, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helbig KJ, Lau DT, Semendric L, Harley HAJ, Beard MR. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology. 2005;42:702–710. doi: 10.1002/hep.20844. [DOI] [PubMed] [Google Scholar]
- 59.Rivieccio MA, et al. TLR3 Ligation Activates an Antiviral Response in Human Fetal Astrocytes: A Role for Viperin/cig5. J Immunol. 2006;177:4735–4741. doi: 10.4049/jimmunol.177.7.4735. [DOI] [PubMed] [Google Scholar]


