Abstract
The process of clonal selection is a central feature of the immune system, but immune specificity is also regulated by receptor selection, in which the fate of a lymphocyte’s antigen receptor is uncoupled from that of the cell itself. Whereas clonal selection controls cell death or survival in response to antigen receptor signaling, receptor selection regulates the process of V(D)J recombination, which can alter or fix antigen receptor specificity. Receptor selection is carried out in both T and B cells and can occur at different stages of lymphocyte differentiation, in which it plays a key role in allelic exclusion, positive selection, receptor editing, and the diversification of the antigen receptor repertoire. Thus, the immune system takes advantage of its control of V(D)J recombination to modify antigen receptors in such a way that self/non-self discrimination is enhanced. New information about receptor editing in T cells and B-1 B cells is also discussed.
Keywords: immunoglobulin genes, T cell receptor, immune tolerance, receptor editing, gene rearrangement
INTRODUCTION
Lymphocytes often express a single antigen receptor despite their genetic potential to simultaneously express many. This fact has focused interest on how expression of multiple receptors is limited. It has recently come to light that cells bearing antigen receptors may actively attempt to express new ones, which can lead to expression of multiple receptors, inactivation of previously expressed receptors, or replacement of old receptors with new ones—a process called receptor editing. Receptor editing occurs through ongoing or renewed antigen receptor gene rearrangement. In this overview, we discuss features of both T cell receptor (TCR) and immunoglobulin (Ig) loci that promote secondary receptor gene rearrangements, and we review the evidence for receptor editing in B and T cells. Receptor editing has recently been the subject of several brief reviews (1–4).
SECONDARY REARRANGEMENTS
V(D)J recombinase rearranges and ligates together dispersed variable (V), diversity (D), and joining (J) minigene elements and brings the newly assembled V(D)J exon in proximity to the constant (C) exons of these genes (see accompanying article by Schatz, “Evolution and Mechanisms of VDJ Recombination,” pp. 495–97). An important feature of some antigen receptor genes is the ability to undergo so-called secondary, or nested, rearrangements, which can salvage the functionality of loci with primary out-of-frame rearrangements and favor receptor editing, i.e., the modification of already functional genes. This process is affected by several factors including the number of gene segments in the locus, their organization along the DNA, and the organization and types of recombination signal sequence (RSSs) adjacent to the gene segments in question. Some typical organizations of receptor genes and their influence on secondary rearrangements are shown schematically in Figures 1 and 2. As we discuss below with respect to the well-studied mouse and human systems, some antigen receptor gene loci appear to be particularly specialized to carry out secondary rearrangements, whereas other loci may disfavor them. The organizations of mouse and human antigen receptor gene loci are shown schematically in Figure 3.
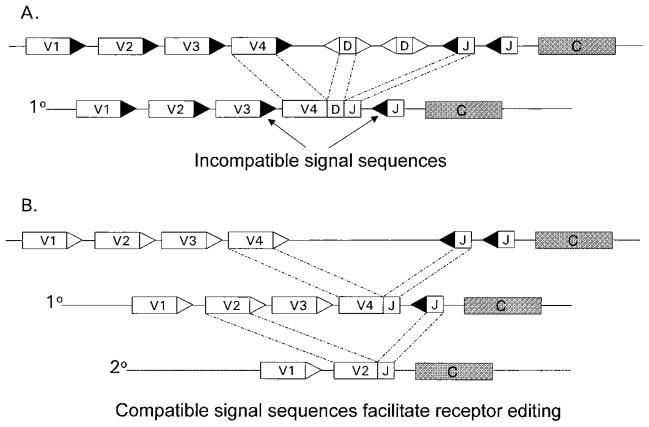
Figure 1.
Role of gene organization in facilitating or inhibiting receptor editing. V, D, and J coding elements are flanked by recombination signal sequences carrying 12-bp spacers (open triangles) or 23-bp spacers (black triangles) that constrain the range of possible rearrangements in cis. Recombinase joins elements with 12-bp spacers to those with 23 bp. Because of their overall organization, loci vary in their abilities to support receptor editing type rearrangements. (A) Cartoon of one type of gene organization similar to mouse and human Ig-H loci (see Figure 3 for details). The presence of D elements along with V genes in the same transcriptional orientation as the J/C cluster forces deletional rearrangements. Primary VDJ assembly cannot be replaced by recombination using conventional signal sequences. (B) In contrast, in loci without D elements, sequential rearrangements are often possible. In this example, a primary (10) V4-to-J join is replaced by a subsequent secondary (20) rearrangement between V2 and the downstream J. Such secondary rearrangement permits the replacement of potentially functional V4-to-J joins, i.e., receptor editing. This type of organization is also seen in mammalian TCRα loci.
Figure 2.
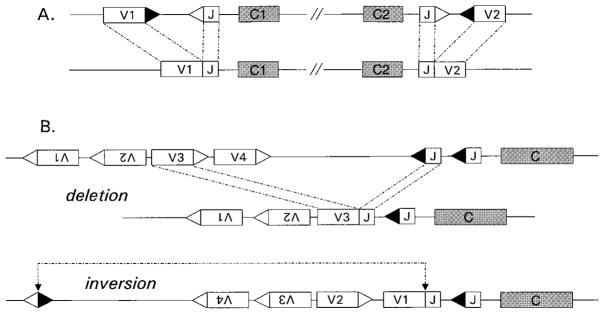
Gene organizations that inhibit or facilitate receptor editing. (A) Cluster type receptor gene organization is used in many lower vertebrates and is retained in certain mammalian receptor gene loci, such as mouse Ig-λ. Rearrangements occur within clusters but not between adjoining clusters, preventing editing and potentially posing problems for isotype exclusion. (B) Inversional rearrangements are dictated by gene orientation. Variable gene segments in inverted transcriptional orientations relative to J/C clusters are indicated by upside-down Vs. Such elements join though inversion rather than excision of intervening DNA. Hypothetical V3 and V4 elements must undergo deletion during primary rearrangement to Js, whereas V1 and V2 elements rearrange by inversion. Note that subsequent secondary rearrangement can again occur through either deletion or inversion, but inversional rearrangements retain more V genes and change the orientations of V elements intervening the break points.
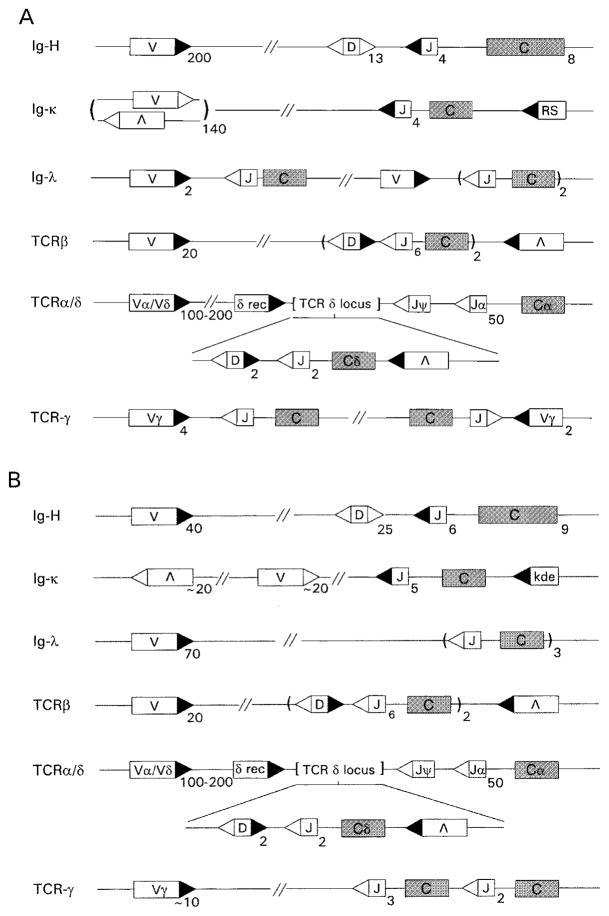
Figure 3.
Antigen receptor loci of (A) mouse and (B) human. Note that Ig-κ, TCR-α, TCR-δ, and TCR-β have structures that are compatible with secondary, replacement rearrangements in both mouse and human. Conventional V(D)J recombination disfavors receptor editing at the Ig-H locus of mouse or human because of the 12/23 rule and the arrangement of VH elements in the same transcriptional orientation as the JH/CH cluster. In the mouse Ig-λ and TCR-γ loci, functionally rearranged genes cannot efficiently be altered by secondary rearrangements because of their cluster type organization, whereas editing is possible in the human versions of these loci provided that the 3′ most J’s are not initially used. In the TCR-δ locus of both mouse and human, TCR-α rearrangements exclude TCR-δ expression.
Loci That Favor Secondary Rearrangements
Ig-κ
Rearrangements in the mouse involve initial joining of one of ~140 Vκ elements to one of four functional Jκ elements (5) (Figure 3A) (6–12); a similar organization is seen in humans, who have about 66 Vκ elements and 5 Jκs (13) (Figure 3B) (14). The κ locus lacks D gene segments; consequently, upon primary VκJκ joining on one allele, secondary rearrangements between remaining upstream Vκs and downstream Jκs can occur in a single step (shown schematically in Figure 1B). In the mouse, Jκ1 and Jκ2 rearrangements are preferred (15), which retains downstream Js available for secondary rearrangement (16). Furthermore, because many Vκ genes are placed in a transcriptional orientation opposite to the Jκ elements, κ loci often rearrange by inversion, retaining thereby the entire repertoire of Vκs for subsequent rearrangements (shown schematically in Figure 2B, bottom). When the V gene segments are in the same transcriptional orientation as the segments to which they rearrange, deletional rearrangements excise intervening DNA, which is permanently lost from the chromosome (Figure 2B, top). Analysis of the Ig-κ loci in mouse or human B cell lines has shown that a single allele can undergo two or more successive V-J recombinations (16–23). Isolation of the excised circular DNA from sequences intervening in the V/J recombination revealed that they often contained independent V/J joins, indicating that secondary κ rearrangements are common in the mouse splenic B cells analyzed (24, 25). B cells of mice lacking the κ-locus on one chromosome have increased usage of downstream Jκs on the remaining allele, consistent with displacement of out-of-frame VJ joins by nested rearrangement (23). Thus, κ locus structure permits replacement reactions, allowing receptor editing.
TCRα
At the TCRα locus, any one of about 100–200 Vα segments can be joined to one of 50 Jα elements (Figure 3A, B) (26–29). A primary VJ rearrangement may yield an out-of-frame join that can be replaced by subsequent rearrangement of an upstream Vα segment with one of the remaining downstream Jα loci (30). T cells may undergo many nested Vα-Jα recombinations (31, 32). In addition, thymocytes rearranging TCRα loci retain excised DNA circles containing the TCRδ locus, usually in a rearranged form (30, 33). Since these episomal DNAs are lost with cell proliferation (30, 34, 35), TCRα rearrangements functionally exclude TCRδ expression. Furthermore, TCRα rearrangements favor initial use of Jα’s at the 5′ end of the locus (36), possibly suggesting progressive use over time of 5′-to-3′ Jα’s. The TCRα locus organization constrains rearrangement to occur by deletion rather than inversion (27) (shown schematically in Figure 1B) (28, 29).
Loci That Disfavor Secondary Rearrangements
Ig-H
In contrast to the Ig-κ and TCRα loci, some loci are so constructed that they tend to suppress secondary rearrangements, at least through conventional recombination signal sequences. VH genes are arranged in the same transcriptional orientation as the D and J elements (37, 38), forcing VDJ assembly at the Ig-H locus to occur by deletion of the intervening DNA (38) (Figure 3). As a consequence, correction of nonfunctional VDJ rearrangements by secondary rearrangement is prevented because no more D regions are available for recombination to upstream VHs. While unrecombined upstream VHs and downstream JHs are typically retained, they both have recombination signal sequences carrying 23bp spacers, and therefore they cannot be joined together by recombinase (see Figure 1A).
Despite this, secondary rearrangements can sometimes occur on the Ig-H locus. Early in mouse B cell development, secondary D-to-J joining can replace primary DH-to-JH rearrangements by deletion or inversion (39), but this apparently does not occur in humans (40). Furthermore, VH elements can sometimes undergo secondary recombination to preformed VDJ elements through VH replacement by targeting noncanonical, but conserved, RSS-like sequences that are present in the 3′ end of the rearranged VH coding sequence (41). VH replacement is discussed in a later section.
Ig-L λ Loci
The lambda locus in the laboratory mouse is composed of two linked miniclusters spanning about 200 kb (42–47). Vλ2, Vλx/Jλ2 Cλ2 lies upstream of Vλ1/Jλ3-Cλ3, Jλ1–Cλ1, but most rearrangements occur between V and J regions of the same cluster (Figure 3A). All rearrangements involve DNA deletions. The recombined Vλ1-Jλ1-Cλ1 gene is the most common type of λ rearrangement (48). Thus, combinatorial diversity is extremely limited in the mouse λ locus, and receptor editing is precluded (shown schematically in Figure 2A). The human λ-locus has an array of ~70 Vλ genes upstream of seven Jλ-Cλ clusters; each J-C cluster has a single J, and only three of the clusters are functional (Figure 3B) (49). Unlike in the mouse λ-locus, this organization allows nested recombination and receptor editing (50).
Suppressive Rearrangements
RS/kde
An additional mechanism facilitated by receptor gene organization is the ability to preclude function of certain loci by nested rearrangements that can themselves have no functional product. One important example of a rearranging, but noncoding, gene element that illustrates this mechanism is the RS/kde element (“recombining sequence” in mouse, “kappa deleting element” in human) (51). This element inactivates the Ig-κ locus by deletional recombination (52). RS is found ~25 kb downstream of the mouse Cκ exon (53) (Figure 3A). (RS should not be confused with the RSS, recombination signal sequence, carried by all rearranging receptor genes). The kde is in a similar position in the human κ-locus (54) (Figure 3B). Both RS and kde contain an RSS sequence with a 23-bp spacer that can recombine through a V(D)J recombinase–dependent process either to unrearranged Vκs or to noncanonical RSS sites in the Jκ-Cκ intron (25, 51, 55). RS/kde rearrangements occur in ~75% of mouse B cells that express λ L-chain (56) and in an even higher fraction of human λ-producing cells (13, 57). RS rearrangements also occur on the second allele of approximately 12% of mouse κ-expressing cells (58). Because the RS/kde encodes no protein, these joins are nonfunctional (55, 59). It appears that the RS/kde elements have no other purpose than to inactivate κ-genes, many of which were previously functional (57). Since RS rearrangement appears concurrently with Ig-λ locus recombination, they may be predicted to clear the way for λ expression (59, 60). Thus, κ locus structure favors replacement reactions that allow receptor editing.
ψJα/δrec
A similar process of deletional recombination can occur in the TCRδ locus, where the ψJα element joins to δrec, eliminating the entire TCRδ locus that is embedded in the TCRα locus (Figure 3A,B) (61, 62). This type of non-protein coding recombination is reported to occur in ~70% of human T cells that go on to express TCRα (63). Like the ψJα element, RS and kde have homology to J regions (64), but unlike most antigen receptor pseudogenes, they retain consensus RSSs (51, 52, 55), implying evolutionary conservation of rearrangement function.
QUASI-ORDERED REARRANGEMENTS IN DEVELOPMENT
B Cells
Rearrangements generating antigen receptor genes generally occur in a particular temporal sequence regulated by control of gene accessibility. In mouse B cell development, H-chain genes are assembled first. PreB cells bearing in-frame, functional IgM H-chains (μ-chains) are selected for clonal expansion and differentiation, whereas cells lacking μ-chains are eventually targeted for elimination (65). Cells lacking μ-chains often attempt rearrangements on the second allele, which, if functional, can rescue the cells. This selection process is a major developmental checkpoint that involves the testing of μ-chain based on its ability to associate with surrogate L-chain components λ5 (66) and VpreB (67) and to mediate transmembrane signals through associated Ig-α/β signal transducers (68–71). Only after a proliferative burst of approximately six cell divisions (72) do cells exit cell cycle; then recombination activity is redirected to the Ig L-chain loci generating sIgM+ B cells (73).
Gene Assembly and Lineage Commitment in T Cells
In αβT cell development, TCR loci are similarly rearranged in a temporal sequence, beginning with DJ rearrangements at the TCRβ loci, followed by Vβ-to-DJ rearrangements, a proliferative burst of those cells expressing functional β-chains, and subsequent redirection of recombinase to the TCRα loci (74–77). In a further parallel to B cell development, testing of the β-chain in T cell development involves protein association with a surrogate chain, pTα (78), and signaling through a preTCR complex (79). Thymocytes lacking β-chains, pTα, or molecules involved in preTCR signaling fail to develop further (80–82). Enforced expression of β-chain transgenes blocks endogenous β-chain gene assembly (83) and promotes development to TCRα gene rearrangement and expression (84).
Expression of a complete α/βTCR is clearly insufficient for RAG downregulation (31, 32, 85, 86). α/β T cells are subject to the process of positive selection (87), in which cells with a receptor biased to weak MHC self-reactivity are selected to advance in development. This maturation is coincident with the cessation of TCRα rearrangement (88, 89). RAG downregulation can be experimentally induced in thymocytes by antibody cross-linking of the TCR (90). The abundance of V genes and J genes in the TCRα locus allows cells to undergo multiple rearrangement attempts, which may be necessary for efficient positive selection (31, 32, 86).
T cell development in the thymus is further complicated by two factors. First, common precursors can give rise to either γδ or αβ T cells, a situation that is regulated in part by the functionality of rearrangements. And, although the antigen receptors of these cells use different protein chains, the TCRδ locus is embedded in the TCRα locus (Figure 3) and is physically excised in the process of TCRα rearrangements (27, 29, 91). The δ and γ loci rearrange prior to α and β loci (92, 93), and an intact γδ receptor likely promotes development in much the same way as β/pTα heterodimers. Lineage determination is also under control of the Notch pathway (94). Likewise, it has been proposed that γδ- or β/pTα-mediated signals may affect the Notch pathway itself (95).
Regardless of the details of αβ vs. γδ lineage commitment, it is of interest to observe patterns common to the organization and rearrangement of different antigen receptor genes. First, in all lymphocytes, one chain partner gene is preferentially rearranged and expressed before the other. Two waves of RAG gene expression correspond to the points in development in which each locus is recombined (96, 97). After expression of the first chain, cells are selected for proliferative expansion (78, 96). The initially expressed Ig-H, TCR-β, and TCR-δ loci invariably include D elements, whereas loci rearranging later lack D regions (Figure 3). Absence of D regions facilitates receptor editing (Figure 1) but limits potential receptor gene diversity, suggesting that receptor editing may be an important selective force in evolution. The initially rearranging chains are each encoded in a single locus, as is the TCR-α locus, which should theoretically limit dual expression. However, in humans and mice the Ig-L chain is encoded by two or three loci, respectively, each with two alleles that rearrange independently of one another, compounding the problem of allelic exclusion with one of isotypic exclusion.
ALLELIC EXCLUSION
The vast majority of mammalian B cells bear a single antibody heavy and light chain (98–103), despite the ability to produce two, one encoded by each allele. In the mouse, most cells express a single light chain (104, 105), despite the ability to express six (2 alleles × 3 isotype loci: κ, λ2/λx, λ1/λ3). As discussed below, allelic and isotypic exclusion are ensured by a variety of active processes. The evolutionary selective forces that make allelic exclusion desirable are not particularly clear. It has been argued that B cell monospecificity is important for both immune self-tolerance and efficient antibody effector function. In T lymphocytes, several mechanisms ensure that γδ receptors are excluded from αβT cells and vice versa, despite the ability of both types of receptor to rearrange in the same cells (95). Furthermore, in αβT cells the lack of allelic exclusion (allelic inclusion) of TCRβ chains is rare. On the other hand, in these same cells coexpression of two TCRα chains is common (86, 106, 107). Allelic inclusion is also observed in TCRδ (108) and TCRγ (109). Why exclusion should be strict in some situations but not in others is unclear. The presumed risk of autoimmunity that may be caused by relaxed allelic exclusion remains to be proved.
Mechanisms of Allelic Exclusion
Stochastic Factors
Several mechanisms in B cells have been proposed to explain the phenomena of allelic and isotypic exclusion (e.g., expression of only Ig-κ or Ig-λ, but not both). One major mechanism is inherent to the recombination machinery itself. Because recombination introduces small deletions and insertions at the coding join, about two thirds of rearrangements are out of frame. In some cases, the V, D, or J elements themselves may harbor stop codons, or such codons may be created in the process of recombination. In theory, these stochastic mechanisms alone reduce allelic inclusion of a single locus to less than 20%. This frequency drops further if a time limit is imposed on the rearrangement process, and further still if rearrangement is intrinsically preferred in one allele over another. Some evidence suggests both of these factors play a role in L-chain allelic and isotypic exclusion. In the mouse κ-light chain locus, DNA demethylation and concomitant accessibility to recombination is, at least initially, monoallelic (110), though this is clearly not a strict rule since about half of all B cells have rearrangements on both κ alleles (104). Furthermore, most κ-expressing cells lack λ rearrangements, whereas virtually all λ-producing cells bear κ rearrangements, usually on both alleles (13, 56, 111–113). During B cell development in the bone marrow, λ-bearing B cells emerge 24 h later than κ+ B cells (114), and analysis of λ-producing hybridomas revealed that most λ+ cells had rearranged a single λ-gene, suggesting that λ gene recombination is both inefficient and time limited (57). While these stochastic factors reduce allelic and isotypic inclusion, they are complemented by more active regulatory processes.
Selective Factors
A second mechanism that could enforce allelic exclusion in mature cells is counter selection against double producing cells. As originally conceived, excess H-chain expression was thought to be toxic to cells (115). Evidence suggesting this possibility came from analysis of myeloma cells, in which L-chain loss is toxic if H-chains are produced. While this toxicity explanation is not supported by experiments in which double H-chain expressing B cells were generated by transgenesis in vivo (116), it is nevertheless possible that double producers are counterselected for reasons other than toxicity. If random H/L pairs are frequently self-reactive, as has been predicted on a priori grounds (117), cells bearing two different receptors would be much more likely to be autoreactive than cells with a single H/L pair. Consequently, counterselection of double producers may occur as a consequence of tolerance induction. This might explain the curious finding that in λ5-deficient mice, which manifest poor allelic exclusion of H-chains in the preB compartment (118), allelic exclusion is restored in peripheral B cells (66).
Feedback Suppression of Recombination
Ig-H
While selective forces obviously play a major role in lymphocyte biology in general, a considerable body of evidence argues that instructive mechanisms have a dominant role in establishing allelic exclusion. In preB cells, functional μ-chains actively block further H-chain rearrangements (66, 119–121). This inhibition occurs through assembly of the surrogate L-chain components λ5 (120, 122) and VpreB (67) with the membrane-bound form of μ-chain (121, 123–125), followed by signaling via the associated Ig-α/β complex (125–127). Forced expression of a heavy chain by transgenesis substantially blocks VDJ assembly on the H-chain locus (119, 123, 128, 129). Disruption of the preB cell receptor (preBCR) signaling complex inhibits both B cell development and allelic exclusion at this stage (103, 118). H-chains that fail to associate with surrogate L-chain, such as certain chains that include VH81X sequences, are essentially absent from the peripheral B cell pool (72, 130, 131). In normal B cells, many cells have incomplete, DJ rearrangements on the nonfunctional H-chain gene allele, suggesting that the rate of V-to-DJ recombination is slow compared to the ability of the cell to perceive functional H-chain assembly in a preBCR complex (132, 133). In addition, in the mouse, otherwise nonfunctional Dμ chains generated from partial D-J rearrangements that involve D reading frame 2 can terminate recombination and block further development (65, 134, 135). Overall, there is compelling evidence for active feedback regulation of H-chain gene recombination that contributes to allelic exclusion.
TCRβ
An analogous feedback process in thymocytes regulates allelic exclusion of the TCRβ locus. Pairing of TCRβ chain with the invariant preTCRα chain generates a signaling complex with CD3 that signals cessation of V(D)J recombination and developmental progression (78, 79, 136, 137). Enforced expression of a β-chain transgene (Tg) blocks endogenous rearrangements and promotes developmental progression (83, 138).
Ig-L-Chain Allelic Exclusion
That Ig-L chain loci must similarly be subject to feedback suppression was predicted on a priori grounds, but the evidence for feedback suppression in light chain gene rearrangements is much less compelling or clear than the evidence for H-chain or TCR-β allelic exclusion. B cells from normal animals manifest substantial allelic exclusion (102), but sensitive techniques are able to identify double producing cells above background (139–141). In contrast to the ability of H-chain transgenic constructs to suppress H-chain rearrangements, enforced expression of L-chain transgenes led to mixed results. Initial experiments by Storb and colleagues using the MOPC 21 κ-chain Tg were consistent with a strict feedback regulation model, as hybridomas generated from these mice appeared to lack endogenous κ expression when H-chain was also expressed (142). Curiously, in this study many hybridomas lacked H-chain expression but expressed endogenous κ-chains (142). It was later found that λ-chain-producing hybridomas from these mice not only coexpressed the κ Tg but also often rearranged and expressed endogenous κ-loci (139). Some other transgenic mouse lines apparently failed to exclude endogenous rearrangements because of insufficient protein expression (119), but this quantitative effect could not explain how in the MOPC 21 mice endogenous rearrangements were excluded in some B cells but not in others. Similarly, in MOPC-167 κ-gene transgenic mice, endogenous κ expression was suppressed in only a subset of B cells (123). It is unlikely, but not formally excluded, that the incomplete allelic exclusion observed in these κ-transgenic mouse experiments was solely the result of defective Tg expression. Certain conventional κ transgenic mice manifest excellent allelic exclusion in a substantial proportion of cells (143, 144).
Several lines of evidence suggest that in some cells even normal L-chain gene expression is not sufficient to suppress further L-chain gene rearrangement. Several sIg + B cell tumor lines and long-term IL-7-dependent bone marrow B cell lines express new receptor chains through secondary rearrangements (22, 145–149). In an analysis of episomal DNA present in mouse spleen cells, Harada & Yamagishi found that of 16 clones containing VκJκ joins excised by nested κ rearrangements, 5 were in-frame and theoretically functional (24), suggesting that in these B cells the natural genes were frequently incapable of suppressing further rearrangements. Perhaps more convincingly, in mice in which functional VκJκ-genes were targeted to the natural locus, endogenous κ rearrangements were inhibited in some, but not all, B cells (144, 150). Again, only a subset of cells, which varied in frequency depending upon the variable region gene used, appeared subject to feedback suppression. A criticism of these experiments is that even the targeted κ-genes may be aberrantly expressed owing to the premature juxtaposition of a Vκ promoter near the Cκ locus. This could conceivably target recombination to the κ locus abnormally early in development, perhaps in cells that lack H-chains and that therefore are unable to perceive the presence of light chain by BCR signaling. Conversely, if DNA accessibility in the κ-locus is stochastically controlled (110), the targeted gene may at first be transcriptionally silent in some cells that make the other allele accessible. The targeted allele might then be coexpressed at a later time, resulting in allelic inclusion of a significant fraction of cells, an unexpected but well-documented, property of targeted L-chain mice (144, 150, 151).
In contrast to these results, it appears that when expressed as transgenes, certain H/L pairs are extremely efficient at suppressing endogenous L- and H-chain protein expression and RAG expression in the bone marrow (152), suggesting that BCR specificity plays a critical role in development. One interpretation of these results is that, like T cells, B cells are positively selected by self-antigens that downregulate RAG expression. Some studies with cell lines are consistent with this idea (145, 146). However, if antigen-specific positive selection was associated with cessation of V(D)J recombination in B cells, then allelic inclusion would be predicted to occur frequently among normal B cells in vivo (as is the case for TCRα exclusion in T cells). On the contrary, as we discuss below, there is substantial evidence from the study of autoantibody transgenic mice that negative selection, i.e. tolerance mediated by encounter with self-antigens, blocks developmental progression and prevents recombinase downregulation. These results can be reconciled with the above-mentioned results by taking into account the influence of immune tolerance on feedback suppression of recombination.
RECEPTOR EDITING
Receptor Editing Monitored In Vivo in Transgenic Models of Immune Tolerance
The fact that H+L antibody transgenes could be used to generate mice in which most B cells had a defined specificity (119) stimulated studies analyzing immune tolerance in B and T cells (153). Transgenic mouse models using autoantibodies to HEL, MHC class I alloantigens, DNA, erythrocytes, and other antigens have been useful in defining a number of ways that self-reactivity is controlled (154). In transgenic autoantibody models in which developing B cells were confronted with antigen in a multivalent form, autoreactive cells were absent from the peripheral lymphoid system, but a population of cells carrying a low level of receptor was present in the bone marrow (155–157). Two lines of evidence suggested that receptor editing, i.e. autoantigen-induced secondary Ig gene rearrangements, might be a mechanism of immune tolerance in these models.
In mice transgenic for the 3–83 antibody (anti-H-2Kk,b), B cells were numerous and they almost exclusively expressed the Tg-encoded antibody as a consequence of feedback suppression of recombination (158). But when cognate antigen was introduced by breeding the 3–83 transgenes to the appropriate MHC background, spleen and lymph node B cells were reduced in number, and those cells that remained lacked self-reactivity. B cells in the peripheral lymphoid organs retained surface expression of the 3–83 H-chain, but not the 3–83 L-chain, and an extremely high percentage of cells expressed endogenous λ-chain. In the bone marrows of antigen-expressing mice, transgenic sIg + B cells expressed high levels of recombinase mRNA and manifested rearrangements at the endogenous L-chain loci. Based on these findings, it was suggested that autoantigen binding by immature bone marrow B cells could reinduce or prolong L-chain gene rearrangements, allowing a cell to alter specificity and escape death (158). It was further postulated that the ability to undergo receptor editing was limited to early stages of B cell development because 3–83 mice that expressed cognate antigen only in the periphery, under control of a liver-specific Kb Tg, underwent profound B cell deletion without appreciable receptor editing.
Independent evidence for receptor editing was obtained in transgenic mice carrying H + L genes of dsDNA antibody 3H9 (159). Spleen cells of H + L Tg mice and hybridomas generated from them were analyzed for antibody specificity and endogenous rearrangements. While dsDNA-specific B cells were lacking from the hybridoma sample, indicating that specific tolerance was induced, splenic B cell numbers were surprisingly normal, particularly in older mice. Splenic B cells retained transgenic H-chain expression on the cell surface but lacked 3H9 L-chain expression as detected with specific anti-idiotypic antibody. The hybridomas expressed endogenous L-chains that apparently altered B cell specificity and extinguished dsDNA binding. Suppression of dsDNA specificity was reversible in one hybridoma, when the endogenous κ was lost, suggesting that binding of the transgenic H-chain with the endogenous L-chain outcompeted transgenic L-chain. In addition to this evidence for “phenotypic” allelic exclusion, among these hybrids an excessive use of endogenous Vκ12/13 genes associated with downstream Jκs was observed. In control mice transgenic for just the 3H9 L-chain, only 6/25 hybrids excluded endogenous L-chain expression, but the endogenous L-chains that were expressed had the typical distribution of Vκ and J usage.
Radic et al (160) and Prak et al (23) extended this work to test for tolerance-induced receptor editing in 3H9 H-chain-only Tg mice. This test was possible because 3H9 H-chain binds DNA in association with diverse L-chain partners (161), but spleen hybridomas from 3H9 H-chain Tg mice lack reactivity to dsDNA (160, 162). Hybridoma analysis again revealed a strikingly limited usage of Vκ genes in splenic B cells with excess use of Vκ12/13 and skewing to downstream Jκ’s, particularly Jκ5. These studies also documented that in many cells multiple rearrangement attempts were made on a single chromosome. Assuming that additional attempts were required to replace κ-chains that conferred auto-reactivity, these results provide an explanation for skewing of Jκ usage. This would predict that many VJ joins displaced by secondary rearrangements were in-frame and functional, an assumption that was not directly tested. An independent study by Eilat and colleagues of a different set of VH11 anti-DNA H-chain mice yielded similar results (163). Since in these studies no L-chain Tg was present, the later stages of B cell development should have been relatively normal. Furthermore, because simple apoptosis of autoreactive cells from an initially random population would be unlikely to account for the observed skewing in Jκ usage, these results suggested that autoreactive cells were rescued by ongoing Ig-κ recombination.
Analysis of Receptor Editing in Gene Targeted Antibody Gene Mice
VH Replacement
Gene targeted autoantibody Tg mice were generated to more closely mimic the natural genomic context and to test for VH replacement in vivo. In theory, gene targeting to the natural locus should facilitate editing because secondary rearrangements on the targeted allele, should they occur, can silence or modify antibody expression. In conventional Tg mice, the suppression of Tg expression is highly inefficient because transgenes are typically inserted on chromosomes without access to potential rearrangement partners. A complicating feature of mice with targeted H-chain loci is that they retain upstream D regions, which are normally absent after VDJ assembly. These dangling Ds can rearrange to remaining J elements of the constructs or to the heptamers embedded in the targeted VH region. Early studies (164) failed to identify use of the conserved heptamer embedded in the expressed VH, but destructive rearrangements caused by D joining to other heptamer-like sites in the gene were observed. Another study (165) found frequent editing by D-regions, but also found VH-to-VDJ replacement events that rescued H-chain function, a result predicted from prior work with cell lines (41, 166). This and subsequent results from studies of other H-chain targeted mice (116, 163, 167, 168) have verified that such replacement events at the Ig-H locus occur at detectable frequency in these animals, indicating that nested rearrangements alter specificity in vivo. But there is debate about the frequency, stage specificity, and tolerance inducibility of the VH replacement reaction (169). In vitro analyses indicate that the embedded heptamer is extremely inefficient at targeting V(D)J recombination relative to conventional RSSs (170). When VH-to-VDJ replacements have been observed in mouse B cells, they almost always include additional “N” nucleotides derived from TdT activity, indicating that most VH replacements occur at the proB stage, prior to L-chain gene rearrangements (116, 165, 167). This in turn suggests that tolerance signaling through intact surface immunoglobulin (Ig) does not drive the VH-to-VDJ replacement response.
Editing at the κ-Locus
The possibility that editing on the H- or L-chain loci could be tolerance induced was more directly tested in models of H+L autoantibody gene targeted mice. Chen and colleagues analyzed mice coexpressing heavy and light chain targeted constructs encoding the 3H9H/Vκ4 anti-dsDNA antibody or the 3H9 H-chain with a different targeted L-chain (Vκ8) that conferred reactivity to ssDNA (152). The 3H9/Vκ4 dsDNA reactive combination was subject to extensive κ-chain editing in the absence of editing on the H-chain locus. Ninety-eight percent of cells retained the expression of the 3H9 H-chain, and the vast majority of these failed to express the other H-chain allele, but at the L-chain locus extensive rearrangements occurred on both the targeted and untargeted κ-alleles. Only 4% of hybridomas lacked detectable secondary κ-rearrangements. L-chain editing occurred by inversional rearrangements in 57% of the hybridomas, deletional type rearrangement events in 28% of the cells, and inclusional rearrangements (i.e. on the opposite allele with an unaffected targeted allele) in only 11% of cells. In the 3H9H/Vκ8 mice, B cells bearing this receptor are often seen in the periphery, in an anergic state (162). But suggestive evidence of receptor editing was also observed in the 3H9H/Vκ8 B cells because in both conventional and 3H9H/Vκ8 gene-targeted mice a subset of B cell hybridomas showed exaggerated usage of Vκ12/13 along with skewed Jκ usage on the untargeted allele (152). (In the targeted 3H9H/Vκ8 mice the Vκ8 gene was joined to Jκ5, providing no additional functional rearrangement possibilities on that allele.) These results suggested that tolerance-induced receptor editing was focused on the L-chain loci and could be induced by both strong and weak toleragens, dsDNA and ssDNA, respectively.
In a second study, targeted κ- and H-chain gene mice were generated encoding the 3–83 antibody, specific for self-MHC molecules H-2Kk and H-2Kb, and light chain editing in B cells was monitored (151). Quantitative southern blotting assessed the extent of recombination in the κ-loci and indicated that in the auto-reactive combination at least 30% of targeted alleles and 59% of wild-type alleles were rearranged, while in B cells of κ-only mice, these percentages were 10% and 27%, respectively. As a control, the 3–83 κ mice were bred with H-chain gene mice of an irrelevant, nonautoreactive specificity. (Unfortunately, in this study, it was not possible to compare 3–83 H+L targeted mice with and without antigen because the targeted H-chain gene was linked to a cross-reactive antigen locus contributed by the embryonic stem cell strain–129.) Most control cells appeared to retain expression of the targeted L-chain, whereas at least 85% of the autoreactive B cells lost expression of the autoreactive specificity on the cell surface. RS-type recombinations inactivating the κ-loci were 60-fold more prevalent in the autoreactive context than in the presence of an innocuous receptor. In the bone marrows of mice with an innocuous H+L receptor, small preB cells were absent owing to accelerated developmental progression. However, in mice with autoreactive receptors that manifested receptor editing at the DNA level, this compartment was very large, suggesting that receptor editing specifically caused a retrograde step in development from sIg+ to sIg− stages. This is as one might predict from the high frequency of nonfunctional secondary rearrangements associated with inactivation of the targeted κ-gene. In both this study and that of Chen et al (152), it appeared that receptor editing could allow a rather efficient rescue of previously autoreactive cells because peripheral B cell numbers were relatively normal.
Central B Cell Tolerance Is Associated with Developmental Block
It has been known for a long time that B cell production can be suppressed by anti-IgM treatment (171). Early transgenic mouse studies on B cell tolerance to membrane MHC molecules indicated that self-reactive B cells were absent from the peripheral lymphoid organs, but large numbers of newly formed autoreactive cells bearing a low density of sIg were retained in the bone marrow (155, 156). Because these data were superficially consistent with the clonal selection hypothesis and classical studies on B cell tolerance (172), they were interpreted to mean that autoreactive cells were eliminated at the preB-to-B cell transition. It now appears most likely that autoreactivity initially prevents B cell developmental progression and promotes receptor editing that allows some cells to change their specificities. While cells failing to alter their receptors eventually die, this tempo of cell death is essentially identical to the rate of turnover of preB cells that fail to generate any receptor at all. Thus, a cell death program succeeds a receptor editing phase.
An early indication that this might occur was provided in studies performed in the 1970s, in which bone marrow B cells were challenged with anti-IgM antibodies (173, 174). In these cultures, immature B cells rapidly lost sIg but were not reduced in number, as assessed by quantitation with anti-MHC class II antibodies; after removal of the anti-Ig stimulus, only a small subset reexpressed sIg. In contrast, mature splenic B cells rapidly reexpressed sIg after similar treatment. This “irreversible receptor modulation,” as it was called at the time, was probably the first evidence for editing, which is predicted to frequently lead to nonfunctional secondary rearrangements. More recently, in experiments in which highly purified bone marrow B cells, rather than unpurified bone marrow preparations, were challenged with anti-IgM antibodies, this stimulus was found to result in apoptosis (172, 175, 176). A possible resolution of the discrepancy between the two types of study is that the presence of bone marrow accessory cells may be required for the editing response (177). Experiments studying antigen-induced apoptosis in immature B cells of autoantibody transgenic mice are consistent with a model in which cell death is slow and delayed. Culture of anti-HEL Tg bone marrow B cells with membrane-bound HEL blocked developmental progression at the fraction E (immature B cell) stage but failed to cause rapid cell death (178). The block was reversible upon antigen removal during the first 1–2 days, but at later times B cell death occurred. Enforced expression of Bcl2 in immature auto-reactive B cells was shown to further prolong survival at this developmental stage (178) but was unable to relieve the developmental block.
The receptor editing model provided a rationale for this delayed and reversible death program in immature B cells. In short-term cultures of 3–83 (anti-MHC) B cells, anti-BCR antibodies failed to accelerate B cell death during the initial 48 h of culture but stimulated secondary L-chain gene rearrangements in a considerable proportion of cells, estimated to be from 25% to 50% of all cells (179). This same study reported that anti-BCR treatment could also induce receptor editing in normal, nontransgenic bone marrow B cells. In another model system, immature bone marrow B cells of 3–83 Tg mice were expanded in IL-7 and challenged with antigen. Under these conditions, autoantigen blocked developmental progression and promoted receptor editing but did not appreciably accelerate cell death (180, 181). Consistent with this idea, Bcl-2 overexpression did not allow autoreactive receptor-bearing B cells to escape from the bone marrow but appeared to improve the efficiency of their escape by receptor editing, probably by prolonging the time window of secondary rearrangement (182). In an independent study, Bcl-2 overexpressing immature B cells spontaneously switched from κ to λ L-chain expression in vitro (146). Somewhat analogous results were obtained in Bcl-xl-overexpressing anti-HEL/mHEL double Tg mice, in which central tolerance was apparently disrupted, yielding enhanced peripheralization of autoreactive B cells with an anergic phenotype along with enhanced receptor editing (183). Curiously, however, the same Bcl-xl Tg had minimal effects in the 3–83 model of central tolerance (J Lang, D Nemazee, unpublished data). Overall, these data suggest that receptor editing is associated with a developmental checkpoint.
The notion that through secondary recombination autoreactive cells can overcome a developmental block implies that inhibition of secondary recombination in autoantibody Tg mice should prevent B cells from maturing to populate the periphery. This idea was directly tested in both the anti-H-2Kk,b and anti-DNA models by breeding Ig-H+L transgenes to a RAG-1- or RAG-2-deficient background (184, 185). B cell development did not advance past the late bone marrow stages, and B cells did not populate the peripheral lymphoid organs. The developmentally arrested RAG-deficient autoreactive cells underwent apoptosis in situ (185). In contrast, on an antigen-free (H-2d), RAG-deficient background anti-H-2Kk,b B cells did develop and populated the spleen. These studies are consistent with a model in which developmental arrest is coupled to receptor editing, and cells that fail to appropriately modify their receptors die.
Locus Specificity of B Cell Receptor Editing
Several lines of evidence suggest that tolerance-induced receptor editing in immature B cells is focused primarily on the Ig-L chain loci, although some editing at the H-chain locus may also occur. One indication of this is that most autoantibody Tg mice exhibit excellent H-chain exclusion, but poor L-chain allelic exclusion (152, 158, 159). Other evidence has been gleaned from cell culture models of receptor editing. Bone marrow B cells of 3–83 Tg mice cultured in IL-7 maintain excellent H and L-chain exclusion in the absence of cognate antigen, but κ-rearrangements could be induced in over two thirds of cells upon anti-BCR treatment (181). In these same cells, VDJ assembly on the H-chain locus was not detectable by a sensitive PCR assay. However, as discussed above, conventional H-chain transgenic mice do not allow an assessment of VH replacement events, and when targeted H-chain autoantibody mice have been analyzed, VH-to-VDJ replacement occurred infrequently and usually included N-region additions indicative of recombination at the proB stage prior to sIgM expression (152).
Developmental Stage Specificity of Receptor Editing
Immature bone marrow B cells sensitive to tolerance-induced receptor editing rapidly lose this sensitivity as they mature, and that stage is succeeded by an apoptosis-sensitive one. These cells can be distinguished by a series of surface markers (186) and by the ability of the latter cells to migrate to the spleen (187). More detailed analysis has been facilitated by the ability to selectively expand receptor editing–sensitive B cells from bone marrow using IL-7 and to follow their differentiation after IL-7 withdrawal (146, 188). In these cultures, IL-7 withdrawal is required for recombinase expression, possibly because cell cycle events in proliferating cells suppress recombinase expression (189). RAG-2 protein is selectively degraded in cycling cells (189). Upon IL-7 withdrawal, cells express increasing amounts of sIg, and the ability to respond to antigen challenge by editing decreases and apoptosis sensitivity increases concomitantly. Since more mature cells presumably also must retain the option to fix their receptors and to participate in the immune response, the inability of these cells to be induced to receptor editing by antigen alone makes sense.
Is there a role for receptor editing in receptor diversification? Besides providing an elegant self-tolerance mechanism, receptor editing may have an additional biological benefit. V(D)J recombination is often far from random because differences exist within sets of variable and joining segments in their recombination signal sequences, promoter regions, and proximity to other cis-acting elements that affect the efficiency of recombination. As a result, overrepresentation of certain genes is common and perhaps inevitable. Secondary and higher order rearrangements may play an important role in promoting a higher degree of randomness in V and J gene usage than would occur if each locus underwent only a single recombination.
PERIPHERAL EDITING
Receptor Revision
Even mature B cells, it was shown recently, can undergo receptor editing during the immune response in germinal centers (190–193). The first indications of this were that RAG mRNA and protein were inducibly expressed in mature B cells under certain culture conditions or upon a germinal center immune reaction (190, 194). Histological analysis of spleens and lymph nodes of immunized mice indicated that the cell type expressing RAG in vivo was the centrocyte (190, 195). The centrocyte is a nondividing cell that has recently proliferated in the germinal center reaction and is intimately contacted and selected by the follicular dendritic cell network (196). Concerted treatment of splenic B cells with CD40 agonist and IL-4, agents that probably mimic T cell help and induce heavy chain class switching, could rapidly induce upregulation of RAG genes (193, 195, 197). Combined treatment with bacterial lipopolysaccharides and IL-4 had a similar effect (190, 195). Later studies suggested that IL-7 could substitute for IL-4 and was probably the critical cytokine in vivo because antibodies to IL-7R, though they permitted germinal center formation, prevented RAG expression in centrocytes (197).
RAG expression in mature B cells was strongly associated with B cell death both in vivo and in vitro and was often present in cells that were engulfed by macrophages (190, 195). In histological sections of germinal centers, RAG protein expression appeared to be localized to the cytoplasm rather than the nucleus (190, 194). RAG gene products induced in vivo and in vitro were nevertheless shown to cause double-stranded DNA breaks adjacent to recombination signal sequences (191) and new L-chain protein expression (192, 193, 198). Curiously, the cells undergoing these recombination reactions and receptor alterations did not manifest a loss of surface Ig expression (192), which would be predicted to be a common occurrence, suggesting rapid death of sIg-cells in these cultures.
Perhaps most intriguingly, the cell fractions that expressed recombinase also expressed other markers characteristic of B cell precursors, including surrogate L-chain components (199), IL-7R (197), the surface marker GL-7 (194), and, in human germinal center cells, TdT (199). The reexpression of surrogate L-chain components is particularly interesting because of the likelihood that many editing events would initially silence L-chain production, perhaps requiring surrogate L-chain to temporarily pair with H-chain. Expression of surrogate L-chain components is also observed in cycling, centroblast cells (199). These unexpected similarities between bone marrow and germinal center cells, first pointed out by Han, Kelsoe, and colleagues on the basis of RAG expression (194), suggest that many elements of B cell development are recapitulated during the immune response.
However, in the regulation of recombinase expression, there are critical differences as well. In the bone marrow microenvironment, immature B cells require only BCR ligation to undergo receptor editing, whereas different stimuli drive receptor editing in germinal center cells, and BCR stimulation under such conditions actively blocks the recombinase response (198, 199). In one study, in vivo RAG expression appeared to be present in a subset of germinal center centrocytes distinguishable by reduced CD45 levels and nonoptimal receptor gene usage for cognate immunogen (191). It is therefore unlikely that immune tolerance induces V(D)J recombination in germinal center cells. Instead, the data are most consistent with a role for receptor editing in the diversification of the receptors of antigen-reactive cells. However, as we discuss below, this interpretation also presents problems.
Very recently, data has been presented suggesting that receptor revision may occur concurrently with somatic hypermutation. Weigert and colleagues have identified a clone of anti-dsDNA reactive cells captured as hybridomas from MRL/lpr mice harboring targeted L-chain and H-chain genes encoding anti-DNA (F Brard, M Shannon, EL Park, S Litwin, M Weigert, in press). In this clone the L-chain Tg was heavily mutated, and all clone members shared a lethal stop mutation. The clone expressed a second L-chain on the other kappa allele that had acquired fewer mutations. Assuming that the new κ-gene rearrangement appeared some time after the initiation of somatic mutation, these results imply that revision can occur in cells that undergo point hypermutation and that editing may be specifically stimulated by the loss of BCR expression. In a second study, de Wildt et al (200) identified among 365 human primary IgG+ clones a pair of cells expressing an identical hypermutated H chain gene and two different expressed L-chain genes, which were presumably rearranged after these mutations. Since the L-chain genes themselves showed evidence of somatic mutation, hypermutation and receptor editing can apparently occur concurrently in these cells.
A second recent study argues that receptor revision can occur in mouse B-1 cells (201), which are not thought to take part in germinal center reactions. IgM+B220low peritoneal cells, particularly those from the autoimmune-prone NZB strain, express RAG mRNA and possess double-strand breaks at Jκ RSSs. In analysis of mice carrying functional replacements of IgH and IgL variable genes, B-1 cells lost idiotypic determinants, indicative of receptor editing at the protein level. The function of such modification is unclear and, as in the case of germinal center B cells, has important implications for the generation of autoantibodies.
Locus Specificity of Receptor Revision
Several studies documented renewed recombination of L-chain genes in mature B cells (191–193, 197, 199, 201), and a single study provided possible evidence of rearrangements at the IgH locus (192). This latter result was obtained in a VDJ replacement mouse in which both nonphysiological D-to-VDJ joins and potentially physiological V-to-VDJ replacement reactions are possible. Thus, either this experimental system may represent the ideal model to reveal physiologically relevant receptor revision on the IgH-locus, or it may allow recombination events on the IgH-locus that rarely occur in normal cells. It will be important to determine if receptor revision can occur at the H-loci through V-to-VDJ replacements.
RECEPTOR EDITING IN T CELLS
As discussed earlier, TCRα allelic exclusion essentially fails to occur because of ongoing rearrangements in cells prior to positive selection. This raises the question of whether an editing mechanism may play a role in thymocyte negative selection. Wang et al (202) established a targeted replacement mouse using a rearranged TCRα VJ gene from a pigeon cytochrome c (PCC)–specific T cell hybridoma inserted in the 5′ region of the germ-line J locus. This gene was expressed in most double positive cells but was lost, presumably through nested rearrangements, in all but ~3% of peripheral T cells. Retention of gene expression was greatly improved when a TCRβ Tg encoding the original hybridoma was coexpressed on a positively selecting background but was not improved on a nonselecting background. Most importantly, in the presence of PCC, negative selection was not associated with loss of thymic cellularity, in contrast to what was seen with conventional anti-PCC TCR transgenic mice, and nonautoreactive cells populated the periphery. While these results failed to prove that autoreactivity promoted further rearrangements, the possibility is not excluded, and this model should allow this question to be further tested. It is equally possible that the autoreactive cells in this system were positively selected and subsequently lost by apoptosis, but the compartment was filled with competing cells that had lost expression of the targeted TCRα gene prior to antigen encounter.
Perhaps more remarkable is the recent suggestion that receptor editing can occur during peripheral T cell tolerance (203). In this experiment, a TCRβ-chain (Vβ5) Tg mouse was constructed and tolerance to the endogenous mouse mammary tumor virus superantigen MTV-8 was assessed. In antigen-expressing mice most Vβ5+ cells were deleted, and the remainder were functionally inactivated. However, over time, a Vβ5− population emerged. It was concluded that in a subset of the tolerant cells, extra-thymic V(D)J recombination occurred, altering and replacing their receptors because RAG expression and doublestranded breaks adjacent to TCR Dβ2 and Jα50 were detected in a subset of peripheral T cells. Furthermore, the appearance of Vβ5− cells, but not tolerance, was dependent on the presence of B cells, suggesting that a B:T interaction, such as a germinal center reaction, might be required for this process. A potential inconsistency between the data and this model was that the RAG-expression was detected in Vβ5− rather than Vβ5+ cells. These points will no doubt be addressed in the near future as experimental systems are designed with these questions in mind (202).
Finally, it should be noted that the structure of the TCRβ loci is consistent with receptor editing, as active genes using the 5′-most D/J/Cβ cluster can in theory be subsequently eliminated and replaced by V-to-DJ rearrangements to the 3′ D/J/Cβ cluster on the same chromosome (Figure 3). In addition, the spacers of their RSS elements are theoretically compatible with direct Vβ-to-Jβ rearrangement, but this has never been observed.
CONCLUSION
The ability of immune cells to turn V(D)J recombinase on and off, or to redirect its activity to new gene loci, appears to regulate lymphocyte specificity in a novel way distinct from, and complementary to, the control of cellular growth and survival. This regulation of recombinase is largely controlled by signaling through the antigen receptor itself and constitutes a feedback mechanism that allows cells to “repair” receptor genes whose products are dysfunctional, or of inappropriate specificity, including those with too high or too low affinity for self-antigens. Receptor editing facilitates immune self-tolerance during B cell development and also permits developing thymocytes to screen multiple TCRs for potential positive selection. Mature B lymphocytes (and possibly also T cells) may re-express recombinase upon foreign antigenic stimulation, possibly allowing more rapid clonal evolution to high-affinity reactivity. The organization of antigen receptor genes appears to facilitate these processes, often at the cost of limiting potential receptor gene diversity and lymphocyte monospecificity. The full extent to which these receptor selection processes normally occur, or benefit the organism, remains to be elucidated.
Acknowledgments
This work was supported by grants from the National Institutes of Health and the Arthritis Foundation. The author thanks Anne Feeney, Norman Klinman, Martin Weigert, and members of the laboratory for their comments on the manuscript, and Kathy Offerding for secretarial assistance.
Footnotes
Since the submission of the manuscript, the cloning by Zachau’s group of the murine κ-locus has made extraordinary progress. For information on this fast-moving area and associated recent publications, the reader is referred to their web site: http://www.med.uni-muenchen.de/biochemie/zachau/kappa,htm.
LITERATURE CITED
- 1.Radic MZ, Zouali M. Receptor editing, immune diversification, and self-tolerance. Immunity. 1996;5:505–11. doi: 10.1016/s1074-7613(00)80266-6. [DOI] [PubMed] [Google Scholar]
- 2.Hertz M, Nemazee D. Receptor editing and commitment in B lymphocytes. Curr Opin Immunol. 1998;10:208–13. doi: 10.1016/s0952-7915(98)80250-1. [DOI] [PubMed] [Google Scholar]
- 3.Nussenzweig MC. Immune receptor editing: revise and select. Cell. 1998;95:875–78. doi: 10.1016/s0092-8674(00)81711-0. [DOI] [PubMed] [Google Scholar]
- 4.Ohmori H, Hikida M. Expression and function of recombination activating genes in mature B cells. Crit Rev Immunol. 1998;18:221–35. doi: 10.1615/critrevimmunol.v18.i3.30. [DOI] [PubMed] [Google Scholar]
- 5.Kofler R, Geley S, Kofler H, Helmberg A. Mouse variable-region gene families: complexity, polymorphism and use in non-autoimmune responses. Immunol Rev. 1992;128:5–21. doi: 10.1111/j.1600-065x.1992.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 6.Tonegawa S, Maxam AM, Tizard R, Bernard O, Gilbert W. Sequence of a mouse germ-line gene for a variable region of an immunoglobulin light chain. Proc Natl Acad Sci USA. 1978;75:1485–89. doi: 10.1073/pnas.75.3.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cory S, Tyler BM, Adams JM. Sets of immunoglobulin V kappa genes homologous to ten cloned V kappa sequences: implications for the number of germline V kappa genes. J Mol Appl Genet. 1981;1:103–16. [PubMed] [Google Scholar]
- 8.Kirschbaum T, Pourrajabi S, Zocher I, Schwendinger J, Heim V, Roschenthaler F, Kirschbaum V, Zachau HG. The 3′ part of the immunoglobulin kappa locus of the mouse. Eur J Immunol. 1998;28:1458–66. doi: 10.1002/(SICI)1521-4141(199805)28:05<1458::AID-IMMU1458>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 9.Max EE, Seidman JG, Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci USA. 1979;76:3450–54. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seidman JG, Leder P. The arrangement and rearrangement of antibody genes. Nature. 1978;276:790–95. doi: 10.1038/276790a0. [DOI] [PubMed] [Google Scholar]
- 11.Seidman JG, Max EE, Leder P. A kappa-immunoglobulin gene is formed by site-specific recombination without further somatic mutation. Nature. 1979;280:370–75. doi: 10.1038/280370a0. [DOI] [PubMed] [Google Scholar]
- 12.Zachau HG. Immunoglobulin light chain genes of the κ type in human in man and mouse. In: Honjo T, Alt FW, Rabbitts TH, editors. Immunoglobulin Genes. 1 San Diego: Harcourt Brace; 1989. pp. 91–110. [Google Scholar]
- 13.Hieter PA, Korsmeyer SJ, Waldmann TA, Leder P. Human immunoglobulin kappa light-chain genes are deleted or rearranged in lambda-producing B cells. Nature. 1981;290:368–72. doi: 10.1038/290368a0. [DOI] [PubMed] [Google Scholar]
- 14.Zachau HG. The human immunoglobulin κ genes. 1995:174–91. See Ref. 204. [Google Scholar]
- 15.Wood DL, Coleclough C. Different joining region J elements of the murine kappa immunoglobulin light chain locus are used at markedly different frequencies. Proc Natl Acad Sci USA. 1984;81:4756–60. doi: 10.1073/pnas.81.15.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Selsing E, Voss J, Storb U. Immunoglobulin gene ‘remnant’ DNA–implications for antibody gene recombination. Nucleic Acids Res. 1984;12:4229–46. doi: 10.1093/nar/12.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis S, Rosenberg N, Alt F, Baltimore D. Continuing kappa-gene rearrangement in a cell line transformed by Abelson murine leukemia virus. Cell. 1982;30:807–16. doi: 10.1016/0092-8674(82)90285-9. [DOI] [PubMed] [Google Scholar]
- 18.Feddersen RM, Van Ness BG. Double recombination of a single immunoglobulin kappa-chain allele: implications for the mechanism of rearrangement. Proc Natl Acad Sci USA. 1985;82:4793–97. doi: 10.1073/pnas.82.14.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shapiro MA, Weigert M. How immunoglobulin V kappa genes rearrange. J Immunol. 1987;139:3834–39. [PubMed] [Google Scholar]
- 20.Feddersen RM, Martin DJ, Van Ness BG. Novel recombinations of the IG kappa-locus that result in allelic exclusion. J Immunol. 1990;145:745–50. [PubMed] [Google Scholar]
- 21.Clarke S, McCray S. A shared kappa reciprocal fragment and a high frequency of secondary Jk5 rearrangements among influenza hemagglutinin specific B cell hybridomas. J Immunol. 1991;146:343–49. [PubMed] [Google Scholar]
- 22.Huber C, Klobeck HG, Zachau HG. Ongoing V kappa-J kappa recombination after formation of a productive V kappa-J kappa coding joint. Eur J Immunol. 1992;22:1561–65. doi: 10.1002/eji.1830220632. [DOI] [PubMed] [Google Scholar]
- 23.Prak EL, Trounstine M, Huszar D, Weigert M. Light chain editing in kappa-deficient animals: a potential mechanism of B-cell tolerance. J Exp Med. 1994;180:1805–15. doi: 10.1084/jem.180.5.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harada K, Yamagishi H. Lack of feedback inhibition of V kappa gene rearrangement by productively rearranged alleles. J Exp Med. 1991;173:409–15. doi: 10.1084/jem.173.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu T, Iwasato T, Yamagishi H. Deletions of immunoglobulin C kappa region characterized by the circular excision products in mouse splenocytes. J Exp Med. 1991;173:1065–72. doi: 10.1084/jem.173.5.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koop BF, Wilson RK, Wang K, Vernooij B, Zallwer D, Kuo CL, Seto D, Toda M, Hood L. Organization, structure, and function of 95 kb of DNA spanning the murine T-cell receptor C alpha/C delta region. Genomics. 1992;13:1209–30. doi: 10.1016/0888-7543(92)90039-u. [DOI] [PubMed] [Google Scholar]
- 27.Wilson RK, Koop BF, Chen C, Halloran N, Sciammis R, Hood L. Nucleotide sequence analysis of 95 kb near the 3′ end of the murine T-cell receptor alpha/delta chain locus: strategy and methodology. Genomics. 1992;13:1198–1208. doi: 10.1016/0888-7543(92)90038-t. [DOI] [PubMed] [Google Scholar]
- 28.Concannon P, Lai E, Klein M, Siu S, Strauss E, Pickering L, Kung P, Gatti R, Hood L. Human T-cell receptor genes: organization, diversity, and polymorphism. Cold Spring Harbor Symp Quant Biol. 1986;51(Pt. 2):785–95. doi: 10.1101/sqb.1986.051.01.091. [DOI] [PubMed] [Google Scholar]
- 29.Winoto A, Mjolsness S, Hood L. Genomic organization of the genes encoding mouse T-cell receptor alpha-chain. Nature. 1985;316:832–36. doi: 10.1038/316832a0. [DOI] [PubMed] [Google Scholar]
- 30.Takeshita S, Toda M, Yamagishi H. Excision products of the T cell receptor gene support a progressive rearrangement model of the alpha/delta locus. EMBO J. 1989;8:3261–70. doi: 10.1002/j.1460-2075.1989.tb08486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrie HT, Livak F, Burtrum D, Mazel S. T cell receptor gene recombination patterns and mechanisms: cell death, rescue, and T cell production. J Exp Med. 1995;182:121–27. doi: 10.1084/jem.182.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrie HT, Livak F, Schatz DG, Strasser A, Crispe IN, Shortman K. Multiple rearrangements in T cell receptor alpha chain genes maximize the production of useful thymocytes. J Exp Med. 1993;178:615–22. doi: 10.1084/jem.178.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak F, Schatz DG. T-cell receptor alpha locus V(D)J recombination byproducts are abundant in thymocytes and mature T cells. Mol Cell Biol. 1996;16:609–18. doi: 10.1128/mcb.16.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong F, Chen CH, Cooper MD. Thymic function can be accurately monitored by the level of recent T cell emigrants in the circulation. Immunity. 1998;8:97–104. doi: 10.1016/s1074-7613(00)80462-8. [DOI] [PubMed] [Google Scholar]
- 35.Douek DC, McFarland RD, Keiser PH, Gage EA, Massey JM, Haynes BF, Polis MA, Haase AT, Feinberg MB, Sullivan JL, Jamieson BD, Zack JA, Picker LJ, Koup RA. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396:690–95. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- 36.Thompson SD, Pelkonen J, Hurwitz JL. First T cell receptor alpha gene rearrangements during T cell ontogeny skew to the 5′ region of the J alpha locus. J Immunol. 1990;145:2347–52. [PubMed] [Google Scholar]
- 37.Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med. 1998;188:2151–62. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brodeur PH, Osman GE, Mackle JJ, Lalor TM. The organization of the mouse Igh-V locus. Dispersion, interspersion, and the evolution of VH gene family clusters. J Exp Med. 1988;168:2261–78. doi: 10.1084/jem.168.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reth MG, Jackson S, Alt FW. VHDJH formation and DJH replacement during pre-B differentiation: non-random usage of gene segments. EMBO J. 1986;5:2131–38. doi: 10.1002/j.1460-2075.1986.tb04476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corbett SJ, Tomlinson IM, Sonnhammer ELL, Buck D, Winter G. Sequence of the human immunoglobulin diversity (D) segment locus: a systematic analysis provides no evidence for the use of DIR segments, inverted D segments, “minor” D segments or D-D recombination. J Mol Biol. 1997;270:587–97. doi: 10.1006/jmbi.1997.1141. [DOI] [PubMed] [Google Scholar]
- 41.Kleinfield RW, Weigert MG. Analysis of VH gene replacement events in a B cell lymphoma. J Immunol. 1989;142:4475–82. [PubMed] [Google Scholar]
- 42.Bernard O, Hozumi N, Tonegawa S. Sequences of mouse immunoglobulin light chain genes before and after somatic changes. Cell. 1978;15:1133–44. doi: 10.1016/0092-8674(78)90041-7. [DOI] [PubMed] [Google Scholar]
- 43.Brandle D, Muller C, Rulicke T, Hengartner H, Pircher H. Engagement of the T-cell receptor during positive selection in the thymus down-regulates RAG-1 expression. Proc Natl Acad Sci USA. 1992;89:9529–33. doi: 10.1073/pnas.89.20.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carson S, Wu GE. A linkage map of the mouse immunoglobulin lambda light chain locus. Immunogenetics. 1989;29:173–79. doi: 10.1007/BF00373642. [DOI] [PubMed] [Google Scholar]
- 45.Dildrop R, Gause A, Muller W, Rajewsky K. A new V gene expressed in lambda-2 light chains of the mouse. Eur J Immunol. 1987;17:731–34. doi: 10.1002/eji.1830170525. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez P, Cazenave PA. A new variable region in mouse immunoglobulin lambda light chains. J Exp Med. 1987;166:265–70. doi: 10.1084/jem.166.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selsing E, Daitch LE. Immunoglobulin λ genes. 1995:194–203. See Ref. 204. [Google Scholar]
- 48.Eisen HN, Reilly EB. Lambda chains and genes in inbred mice. Annu Rev Immunol. 1985;3:337–65. doi: 10.1146/annurev.iy.03.040185.002005. [DOI] [PubMed] [Google Scholar]
- 49.Kawasaki K, Minoshima S, Nakato E, Shibuya K, Shintani A, Schmeits JL, Wang J, Shimizu N. One-megabase sequence analysis of the human immunoglobulin lambda gene locus. Genome Res. 1997;7:250–61. doi: 10.1101/gr.7.3.250. [DOI] [PubMed] [Google Scholar]
- 50.Stiernholm NBJ, Berinstein NL. Immunoglobulin somatic variation; studies of receptor editing in a human B cell lymphoma. Leuk Lymphoma. 1994;12:333–41. doi: 10.3109/10428199409073774. [DOI] [PubMed] [Google Scholar]
- 51.Siminovitch KA, Moore MW, Durdik J, Selsing E. The human kappa deleting element and the mouse recombining segment share DNA sequence homology. Nucleic Acids Res. 1987;15:2699–705. doi: 10.1093/nar/15.6.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore MW, Durdik J, Persiani DM, Selsing E. Deletions of kappa chain constant region genes in mouse lambda chain-producing B cells involve intrachromosomal DNA recombinations similar to V-J joining. Proc Natl Acad Sci USA. 1985;82:6211–15. doi: 10.1073/pnas.82.18.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller B, Stappert H, Reth M. A physical map and analysis of the murine C kappa-RS region show the presence of a conserved element. Eur J Immunol. 1990;20:1409–11. doi: 10.1002/eji.1830200631. [DOI] [PubMed] [Google Scholar]
- 54.Klobeck HG, Zachau HG. The human CK gene segment and the kappa deleting element are closely linked. Nucleic Acids Res. 1986;14:4591–603. doi: 10.1093/nar/14.11.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durdik J, Moore MW, Selsing E. Novel kappa light-chain gene rearrangements in mouse lambda light chain-producing B lymphocytes. Nature. 1984;307:749–52. doi: 10.1038/307749a0. [DOI] [PubMed] [Google Scholar]
- 56.Nadel B, Cazenave PA, Sanchez P. Murine lambda gene rearrangements: the stochastic model prevails over the ordered model. EMBO J. 1990;9:435–40. doi: 10.1002/j.1460-2075.1990.tb08128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Retter MW, Nemazee D. Receptor editing occurs frequently during normal B cell development. J Exp Med. 1998;188:1231–38. doi: 10.1084/jem.188.7.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dunda O, Corcos D. Recombining sequence recombination in normal kappa-chain-expressing B cells. J Immunol. 1997;159:4362–66. [PubMed] [Google Scholar]
- 59.Daitch LE, Moore MW, Persiani DM, Durdik JM, Selsing E. Transcription and recombination of the murine RS element. J Immunol. 1992;149:832–40. [PubMed] [Google Scholar]
- 60.Muller B, Reth M. Ordered activation of the Ig lambda locus in Abelson B cell lines. J Exp Med. 1988;168:2131–37. doi: 10.1084/jem.168.6.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Begley CG, Aplan PD, Davey MP, de Villartay JP, Cohen DI, Waldmann TA, Kirsch IR. Demonstration of delta rec-pseudo J alpha rearrangement with deletion of the delta locus in a human stem-cell leukemia. J Exp Med. 1989;170:339–42. doi: 10.1084/jem.170.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hockett RD, de Villartay JP, Pollock K, Poplack DG, Cohen DI, Korsmeyer SJ. Human T-cell antigen receptor (TCR) delta-chain locus and elements responsible for its deletion are within the TCR alpha-chain locus. Proc Natl Acad Sci USA. 1988;85:9694–98. doi: 10.1073/pnas.85.24.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verschuren MC, Wolvers-Tettero IL, Breit TM, Noordzij J, van Wering ER, van Dongen JJ. Preferential rearrangements of the T cell receptor-delta-deleting elements in human T cells. J Immunol. 1997;158:1208–16. [PubMed] [Google Scholar]
- 64.Du Pasquier L, Schwager J. Evolutions of the immune system. Prog Immunol. 1989;7:1246–55. [Google Scholar]
- 65.Ehlich A, Martin V, Muller W, Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Curr Biol. 1994;4:573–83. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 66.Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K. A critical role of lambda 5 protein in B cell development. Cell. 1992;69:823–31. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 67.Kudo A, Melchers F. A second gene, VpreB in the lambda 5 locus of the mouse, which appears to be selectively expressed in pre-B lymphocytes. EMBO J. 1987;6:2267–72. doi: 10.1002/j.1460-2075.1987.tb02500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karasuyama H, Rolink A, Melchers F. Surrogate light chain in B cell development. Adv Immunol. 1996;63:1–41. doi: 10.1016/s0065-2776(08)60853-6. [DOI] [PubMed] [Google Scholar]
- 69.Reth M, Wienands J. Initiation and processing of signals from the B cell antigen receptor. Annu Rev Immunol. 1997;15:453–79. doi: 10.1146/annurev.immunol.15.1.453. [DOI] [PubMed] [Google Scholar]
- 70.Tsubata T, Tsubata R, Reth M. Cell surface expression of the short immunoglobulin mu chain (D mu protein) in murine pre-B cells is differently regulated from that of the intact mu chain. Eur J Immunol. 1991;21:1359–63. doi: 10.1002/eji.1830210605. [DOI] [PubMed] [Google Scholar]
- 71.Tsubata T, Tsubata R, Reth M. Crosslinking of the cell surface immunoglobulin (mu-surrogate light chains complex) on pre-B cells induces activation of V gene rearrangements at the immunoglobulin kappa locus. Int Immunol. 1992;4:637–41. doi: 10.1093/intimm/4.6.637. [DOI] [PubMed] [Google Scholar]
- 72.Decker DJ, Boyle NE, Klinman NR. Predominance of nonproductive rearrangements of VH81X gene segments evidences a dependence of B cell clonal maturation on the structure of nascent H chains. J Immunol. 1991;147:1406–11. [PubMed] [Google Scholar]
- 73.Constantinescu A, Schlissel MS. Changes in locus-specific V(D)J recombinase activity induced by immunoglobulin gene products during B cell development. J Exp Med. 1997;185:609–20. doi: 10.1084/jem.185.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Born W, Rathbun G, Tucker P, Marrack P, Kappler J. Synchronized rearrangement of T-cell gamma and beta chain genes in fetal thymocyte development. Science. 1986;234:479–82. doi: 10.1126/science.3020688. [DOI] [PubMed] [Google Scholar]
- 75.Snodgrass HR, Kisielow P, Kiefer M, Steinmetz M, von Boehmer H. Ontogeny of the T-cell antigen receptor within the thymus. Nature. 1985;313:592–95. doi: 10.1038/313592a0. [DOI] [PubMed] [Google Scholar]
- 76.Davis MM. Molecular genetics of the T cell-receptor beta chain. Annu Rev Immunol. 1985;3:537–60. doi: 10.1146/annurev.iy.03.040185.002541. [DOI] [PubMed] [Google Scholar]
- 77.Honjo T, Matsuda F. Immunoglobulin heavy chain loci of mouse and human. 1995:145–71. See Ref. 204. [Google Scholar]
- 78.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 1995;375:795–98. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 79.von Boehmer H, Fehling HJ. Structure and function of the pre-T cell receptor. Annu Rev Immunol. 1997;15:433–52. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- 80.Mombaerts P, Clarke AR, Rudnicki MA, Iacomini J, Itohara S, Lafaille JJ, Wang L, Ichikawa Y, Jaenisch R, Hooper ML, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360:225–31. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 81.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–77. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 82.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–67. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 83.Uematsu Y, Ryser S, Dembic Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. In transgenic mice the introduced functional T cell receptor beta gene prevents expression of endogenous beta genes. Cell. 1988;52:831–41. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- 84.Shinkai Y, Koyasu S, Nakayama K, Murphy KM, Loh DY, Reinherz EL, Alt FW. Restoration of T cell development in RAG-2-deficient mice by functional TCR transgenes. Science. 1993;259:822–25. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 85.Marolleau JP, Fondell JD, Malissen M, Trucy J, Barbier E, Marcu KB, Cazenave PA, Primi D. The joining of germ-line V-alpha to J-alpha genes replaces the preexisting V-alpha-J-alpha complexes in a T-cell receptor-alpha, beta positive T-cell line. Cell. 1988;55:291–300. doi: 10.1016/0092-8674(88)90052-9. [DOI] [PubMed] [Google Scholar]
- 86.Casanova JL, Romero P, Widmann C, Kourilsky P, Maryanski JL. T cell receptor genes in a series of class I major histocompatibility complex-restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J Exp Med. 1991;174:1371–83. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jameson SC, Hogquist KA, Bevan MJ. Positive selection of thymocytes. Annu Rev Immunol. 1995;13:93–126. doi: 10.1146/annurev.iy.13.040195.000521. [DOI] [PubMed] [Google Scholar]
- 88.Borgulya P, Kishi H, Uematsu Y, von Boehmer H. Exclusion and inclusion of alpha and beta T cell receptor alleles. Cell. 1992;69:529–37. doi: 10.1016/0092-8674(92)90453-j. [DOI] [PubMed] [Google Scholar]
- 89.Kouskoff V, Vonesch JL, Benoist C, Mathis D. The influence of positive selection on RAG expression in thymocytes. Eur J Immunol. 1995;25:54–58. doi: 10.1002/eji.1830250111. [DOI] [PubMed] [Google Scholar]
- 90.Turka LA, Schatz DG, Oettinger MA, Chun JJ, Gorka C, Lee K, McCormack WT, Thompson CB. Thymocyte expression of RAG-1 and RAG-2: termination by T cell receptor cross-linking. Science. 1991;253:778–81. doi: 10.1126/science.1831564. [DOI] [PubMed] [Google Scholar]
- 91.Chien YH, Iwashima M, Kaplan KB, Elliott JF, Davis MM. A new T-cell receptor gene located within the alpha locus and expressed early in T-cell differentiation. Nature. 1987;327:677–82. doi: 10.1038/327677a0. [DOI] [PubMed] [Google Scholar]
- 92.Pardoll DM, Fowlkes BJ, Bluestone JA, Kruisbeek A, Maloy WL, Coligan JE, Schwartz RH. Differential expression of two distinct T-cell receptors during thymocyte development. Nature. 1987;326:79–81. doi: 10.1038/326079a0. [DOI] [PubMed] [Google Scholar]
- 93.Raulet DH, Garman RD, Saito H, Tonegawa S. Developmental regulation of T-cell receptor gene expression. Nature. 1985;314:103–7. doi: 10.1038/314103a0. [DOI] [PubMed] [Google Scholar]
- 94.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes BJ, Cado D, Robey E. Notch activity influences the alpha-beta versus gamma-delta T-cell lineage decision. Cell. 1997;88:833–43. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 95.Robey E, Fowlkes BJ. The alpha beta versus gamma-delta T-cell lineage choice. Curr Opin Immunol. 1998;10:181–87. doi: 10.1016/s0952-7915(98)80247-1. [DOI] [PubMed] [Google Scholar]
- 96.Li YS, Hayakawa K, Hardy RR. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J Exp Med. 1993;178:951–60. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wilson A, Held W, MacDonald HR. Two waves of recombinase gene expression in developing thymocytes. J Exp Med. 1994;179:1355–60. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cebra JJ, Colberg JE, Dray S. Rabbit lymphoid cells differentiated with respect to alpha-, gamma-, and mu-heavy polypeptide chains and to allotypic markers Aa1 and Aa2. J Exp Med. 1966;123:547–58. doi: 10.1084/jem.123.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weiler E. Differential activity of allelic gamma-globulin genes in antibody-producing cells. Proc Natl Acad Sci USA. 1965;54:1765–72. doi: 10.1073/pnas.54.6.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bernier GM, Cebra JJ. Polypeptide chains of human gamma-globulin cellular localization by fluorescent antibody. Science. 1964;144:1590–91. doi: 10.1126/science.144.3626.1590. [DOI] [PubMed] [Google Scholar]
- 101.Pernis B, Chiappino G, Kelus AS, Gell PG. Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J Exp Med. 1965;122:853–76. doi: 10.1084/jem.122.5.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsukamoto A, Weissman IL, Hunt SV. Allelic exclusion in rat kappa immunoglobulin chains: extent of Jk rearrangement in normal B lymphocytes. EMBO J. 1984;3:975–81. doi: 10.1002/j.1460-2075.1984.tb01916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kitamura D, Rajewsky K. Targeted disruption of mu chain membrane exon causes loss of heavy-chain allelic exclusion. Nature. 1992;356:154–56. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- 104.Coleclough C. Chance, necessity and antibody gene dynamics. Nature. 1983;303:23–26. doi: 10.1038/303023a0. [DOI] [PubMed] [Google Scholar]
- 105.Takeda S, Zou YR, Bluethmann H, Kitamura D, Muller U, Rajewsky K. Deletion of the immunoglobulin kappa chain intron enhancer abolishes kappa chain gene rearrangement in cis but not lambda chain gene rearrangement in trans. EMBO J. 1993;12:2329–36. doi: 10.1002/j.1460-2075.1993.tb05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Malissen M, Trucy J, Jouvin-Marche E, Cazenave PA, Scollay R, Malissen B. Regulation of TCR alpha and beta gene allelic exclusion during T-cell development. Immunol Today. 1992;13:315–22. doi: 10.1016/0167-5699(92)90044-8. [DOI] [PubMed] [Google Scholar]
- 107.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Expression of two T cell receptor alpha chains: dual receptor T cells. Science. 1993;262:422–24. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 108.Sleckman BP, Khor B, Monroe R, Alt FW. Assembly of productive T cell receptor delta variable region genes exhibits allelic inclusion. J Exp Med. 1998;188:1465–71. doi: 10.1084/jem.188.8.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davodeau F, Peyrat MA, Houde I, Hallet MM, De Libero G, Vie H, Bonneville M. Surface expression of two distinct functional antigen receptors on human-gamma-delta T-cells. Science. 1993;260:1800–2. doi: 10.1126/science.8390096. [DOI] [PubMed] [Google Scholar]
- 110.Mostoslavsky R, Singh N, Kirillov A, Pelanda R, Cedar H, Chess A, Bergman Y. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–11. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alt FW, Enea V, Bothwell AL, Baltimore D. Activity of multiple light chain genes in murine myeloma cells producing a single, functional light chain. Cell. 1980;21:1–12. doi: 10.1016/0092-8674(80)90109-9. [DOI] [PubMed] [Google Scholar]
- 112.Coleclough C, Perry RP, Karjalainen K, Weigert M. Aberrant rearrangements contribute significantly to the allelic exclusion of immunoglobulin gene expression. Nature. 1981;290:372–78. doi: 10.1038/290372a0. [DOI] [PubMed] [Google Scholar]
- 113.Korsmeyer SJ, Hieter PA, Sharrow SO, Goldman CK, Leder P, Waldmann TA. Normal human B cells display ordered light chain gene rearrangements and deletions. J Exp Med. 1982;156:975–85. doi: 10.1084/jem.156.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Arakawa H, Shimizu T, Takeda S. Re-evaluation of the probabilities for productive arrangements on the kappa and lambda loci. Int Immunol. 1996;8:91–99. doi: 10.1093/intimm/8.1.91. [DOI] [PubMed] [Google Scholar]
- 115.Wabl M, Steinberg C. A theory of allelic and isotypic exclusion for immunoglobulin genes. Proc Natl Acad Sci USA. 1982;79:6976–78. doi: 10.1073/pnas.79.22.6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sonoda E, Pewzner-Jung Y, Schwers S, Taki S, Jung S, Eilat D, Rajewsky K. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–33. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 117.Nemazee D. Antigen receptor ‘capacity’ and the sensitivity of self-tolerance. Immunol Today. 1996;17:25–29. doi: 10.1016/0167-5699(96)80565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Loffert D, Ehlich A, Muller W, Rajewsky K. Surrogate light chain expression is required to establish immunoglobulin heavy chain allelic exclusion during early B cell development. Immunity. 1996;4:133–44. doi: 10.1016/s1074-7613(00)80678-0. [DOI] [PubMed] [Google Scholar]
- 119.Rusconi S, Kohler G. Transmission and expression of a specific pair of rearranged immunoglobulin mu and kappa genes in a transgenic mouse line. Nature. 1985;314:330–34. doi: 10.1038/314330a0. [DOI] [PubMed] [Google Scholar]
- 120.Sakaguchi N, Melchers F. Lambda 5, a new light-chain-related locus selectively expressed in pre-B lymphocytes. Nature. 1986;324:579–82. doi: 10.1038/324579a0. [DOI] [PubMed] [Google Scholar]
- 121.Nussenzweig MC, Shaw AC, Sinn E, Danner DB, Holmes KL, Morse HC, Leder P. Allelic exclusion in transgenic mice that express the membrane form of immunoglobulin mu. Science. 1987;236:816–19. doi: 10.1126/science.3107126. [DOI] [PubMed] [Google Scholar]
- 122.Sakaguchi N, Berger CN, Melchers F. Isolation of a cDNA copy of an RNA species expressed in murine pre-B cells. EMBO J. 1986;5:2139–47. doi: 10.1002/j.1460-2075.1986.tb04477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Manz J, Denis K, Witte O, Brinster R, Storb U. Feedback inhibition of immunoglobulin gene rearrangement by membrane mu, but not by secreted mu heavy chains. J Exp Med. 1988;168:1363–81. doi: 10.1084/jem.168.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–26. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 125.Nussenzweig MC, Shaw AC, Sinn E, Campos-Torres J, Leder P. Allelic exclusion in transgenic mice carrying mutant human IgM genes. J Exp Med. 1988;167:1969–74. doi: 10.1084/jem.167.6.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Papavasiliou F, Misulovin Z, Suh H, Nussenzweig MC. The role of Ig beta in precursor B cell transition and allelic exclusion. Science. 1995;268:408–11. doi: 10.1126/science.7716544. [DOI] [PubMed] [Google Scholar]
- 127.Papavasiliou F, Jankovic M, Suh H, Nussenzweig MC. The cytoplasmic domains of immunoglobulin (Ig) alpha and Ig beta can independently induce the precursor B cell transition and allelic exclusion. J Exp Med. 1995;182:1389–94. doi: 10.1084/jem.182.5.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weaver D, Costantini F, Imanishi-Kari T, Baltimore D. A transgenic immunoglobulin mu gene prevents rearrangement of endogenous genes. Cell. 1985;42:117–27. doi: 10.1016/s0092-8674(85)80107-0. [DOI] [PubMed] [Google Scholar]
- 129.Nussenzweig MC, Schmidt EV, Shaw AC, Sinn E, Campos-Torres J, Mathey-Prevot B, Pattengale PK, Leder P. A human immunoglobulin gene reduces the incidence of lymphomas in c-Myc-bearing transgenic mice. Nature. 1988;336:446–50. doi: 10.1038/336446a0. [DOI] [PubMed] [Google Scholar]
- 130.Keyna U, Beck-Engeser GB, Jongstra J, Applequist SE, Jack HM. Surrogate light chain-dependent selection of Ig heavy chain V regions. J Immunol. 1995;155:5536–42. [PubMed] [Google Scholar]
- 131.Keyna U, Applequist SE, Jongstra J, Beck-Engeser GB, Jack HM. Ig mu heavy chains with VH81X variable regions do not associate with lambda 5. Ann NY Acad Sci. 1995;764:39–42. doi: 10.1111/j.1749-6632.1995.tb55803.x. [DOI] [PubMed] [Google Scholar]
- 132.Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–19. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–25. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gu H, Kitamura D, Rajewsky K. B cell development regulated by gene rearrangement: arrest of maturation by membrane-bound D mu protein and selection of DH element reading frames. Cell. 1991;65:47–54. doi: 10.1016/0092-8674(91)90406-o. [DOI] [PubMed] [Google Scholar]
- 135.Tornberg UC, Bergqvist I, Haury M, Holmberg D. Regulation of B lymphocyte development by the truncated immunoglobulin heavy chain protein Dmu. J Exp Med. 1998;187:703–9. doi: 10.1084/jem.187.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.O’Shea CC, Thornell AP, Rosewell IR, Hayes B, Owen MJ. Exit of the pre-TCR from the ER/cis-Golgi is necessary for signaling differentiation, proliferation, and allelic exclusion in immature thymocytes. Immunity. 1997;7:591–99. doi: 10.1016/s1074-7613(00)80380-5. [DOI] [PubMed] [Google Scholar]
- 137.Irving BA, Alt FW, Killeen N. Thymocyte development in the absence of pre-T cell receptor extracellular immunoglobulin domains. Science. 1998;280:905–8. doi: 10.1126/science.280.5365.905. [DOI] [PubMed] [Google Scholar]
- 138.Chan VW, Meng F, Soriano P, DeFranco AL, Lowell CA. Characterization of the B lymphocyte populations in Lyn-deficient mice and the role of Lyn in signal initiation and down-regulation. Immunity. 1997;7:69–81. doi: 10.1016/s1074-7613(00)80511-7. [DOI] [PubMed] [Google Scholar]
- 139.Gollahon KA, Hagman J, Brinster RL, Storb U. Ig lambda-producing B cells do not show feedback inhibition of gene rearrangement. J Immunol. 1988;141:2771–80. [PubMed] [Google Scholar]
- 140.Pauza ME, Rehmann JA, LeBien TW. Unusual patterns of immunoglobulin gene rearrangement and expression during human B cell ontogeny: human B cells can simultaneously express cell surface kappa and lambda light chains. J Exp Med. 1993;178:139–49. doi: 10.1084/jem.178.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Giachino C, Padovan E, Lanzavecchia A. Kappa(+)lambda(+) dual receptor B cells are present in the human peripheral repertoire. J Exp Med. 1995;181:1245–50. doi: 10.1084/jem.181.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ritchie KA, Brinster RL, Storb U. Allelic exclusion and control of endogenous immunoglobulin gene rearrangement in kappa transgenic mice. Nature. 1984;312:517–20. doi: 10.1038/312517a0. [DOI] [PubMed] [Google Scholar]
- 143.Carmack CE, Camper SA, Mackle JJ, Gerhard WU, Weigert MG. Influence of a V kappa 8 L chain transgene on endogenous rearrangements and the immune response to the HA(Sb) determinant on influenza virus. J Immunol. 1991;147:2024–33. [PubMed] [Google Scholar]
- 144.Pelanda R, Schaal S, Torres RM, Rajewsky K. A prematurely expressed Ig(kappa) transgene, but not V(kappa)J(kappa) gene segment targeted into the Ig(kappa) locus, can rescue B cell development in lambda5-deficient mice. Immunity. 1996;5:229–39. doi: 10.1016/s1074-7613(00)80318-0. [DOI] [PubMed] [Google Scholar]
- 145.Ma A, Fisher P, Dildrop R, Oltz E, Rathbun G, Achacoso P, Stall A, Alt FW. Surface IgM mediated regulation of RAG gene expression in E mu-N-myc B cell lines. EMBO J. 1992;11:2727–34. doi: 10.1002/j.1460-2075.1992.tb05338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rolink A, Grawunder U, Haasner D, Strasser A, Melchers F. Immature surface Ig+ B cells can continue to rearrange kappa and lambda L chain gene loci. J Exp Med. 1993;178:1263–70. doi: 10.1084/jem.178.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Levy S, Campbell MJ, Levy R. Functional immunoglobulin light chain genes are replaced by ongoing rearrangements of germline V kappa genes to downstream J kappa segment in a murine B cell line. J Exp Med. 1989;170:1–13. doi: 10.1084/jem.170.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Berinstein N, Levy S, Levy R. Activation of an excluded immunoglobulin allele in a human B lymphoma cell line. Science. 1989;244:337–39. doi: 10.1126/science.2496466. [DOI] [PubMed] [Google Scholar]
- 149.Verkoczy LK, Stiernholm BJ, Berinstein NL. Up-regulation of recombination activating gene expression by signal transduction through the surface Ig receptor. J Immunol. 1995;154:5136–43. [PubMed] [Google Scholar]
- 150.Prak EL, Weigert M. Light chain replacement: a new model for antibody gene rearrangement. J Exp Med. 1995;182:541–48. doi: 10.1084/jem.182.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pelanda R, Schwers S, Sonoda E, Torres RM, Nemazee D, Rajewsky K. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 1997;7:765–75. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- 152.Chen C, Prak EL, Weigert M. Editing disease-associated autoantibodies. Immunity. 1997;6:97–105. doi: 10.1016/s1074-7613(00)80673-1. [DOI] [PubMed] [Google Scholar]
- 153.Nossal GJ. Negative selection of lymphocytes. Cell. 1994;76:229–39. doi: 10.1016/0092-8674(94)90331-x. [DOI] [PubMed] [Google Scholar]
- 154.Goodnow CC. Balancing immunity and tolerance: deleting and tuning lymphocyte repertoires. Proc Natl Acad Sci USA. 1996;93:2264–71. doi: 10.1073/pnas.93.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nemazee D, Buerki K. Clonal deletion of autoreactive B lymphocytes in bone marrow chimeras. Proc Natl Acad Sci USA. 1989;86:8039–43. doi: 10.1073/pnas.86.20.8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–66. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 157.Hartley SB, Crosbie J, Brink R, Kantor AB, Basten A, Goodnow CC. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature. 1991;353:765–69. doi: 10.1038/353765a0. [DOI] [PubMed] [Google Scholar]
- 158.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. J Exp Med. 1993;177:1009–20. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. J Exp Med. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Radic MZ, Erikson J, Litwin S, Weigert M. B lymphocytes may escape tolerance by revising their antigen receptors. J Exp Med. 1993;177:1165–73. doi: 10.1084/jem.177.4.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Radic MZ, Mascelli MA, Erikson J, Shan H, Weigert M. Ig H and L chain contributions to autoimmune specificities. J Immunol. 1991;146:176–82. [PubMed] [Google Scholar]
- 162.Erikson J, Radic MZ, Camper SA, Hardy RR, Carmack C, Weigert M. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature. 1991;349:331–34. doi: 10.1038/349331a0. [DOI] [PubMed] [Google Scholar]
- 163.Pewzner-Jung Y, Friedmann D, Sonoda E, Jung S, Rajewsky K, Eilat D. B cell deletion, anergy, and receptor editing in “knock in” mice targeted with a germ-line-encoded or somatically mutated anti-DNA heavy chain. J Immunol. 1998;161:4634–45. [PubMed] [Google Scholar]
- 164.Taki S, Meiering M, Rajewsky K. Targeted insertion of a variable region gene into the immunoglobulin heavy chain locus. Science. 1993;262:1268–71. doi: 10.1126/science.8235657. [DOI] [PubMed] [Google Scholar]
- 165.Chen C, Nagy Z, Prak EL, Weigert M. Immunoglobulin heavy chain gene replacement: a mechanism of receptor editing. Immunity. 1995;3:747–55. doi: 10.1016/1074-7613(95)90064-0. [DOI] [PubMed] [Google Scholar]
- 166.Kleinfield R, Hardy RR, Tarlinton D, Dangl J, Herzenberg LA, Weigert M. Recombination between an expressed immunoglobulin heavy-chain gene and a germline variable gene segment in a Ly 1+ B-cell lymphoma. Nature. 1986;322:843–46. doi: 10.1038/322843a0. [DOI] [PubMed] [Google Scholar]
- 167.Cascalho M, Ma A, Lee S, Masat L, Wabl M. A quasi-monoclonal mouse. Science. 1996;272:1649–52. doi: 10.1126/science.272.5268.1649. [DOI] [PubMed] [Google Scholar]
- 168.Bertrand FE, Golub R, Wu GE. VH gene replacement occurs in the spleen and bone marrow of non-autoimmune quasi-monoclonal mice. Eur J Immunol. 1998;28:3362–70. doi: 10.1002/(SICI)1521-4141(199810)28:10<3362::AID-IMMU3362>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 169.Fanning L, Bertrand FE, Steinberg C, Wu GE. Molecular mechanisms involved in receptor editing at the Ig heavy chain locus. Int Immunol. 1998;10:241–46. doi: 10.1093/intimm/10.2.241. [DOI] [PubMed] [Google Scholar]
- 170.Nadel B, Tang A, Feeney AJ. V(H) replacement is unlikely to contribute significantly to receptor editing due to an ineffectual embedded recombination signal sequence. Mol Immunol. 1998;35:227–32. doi: 10.1016/s0161-5890(98)00029-7. [DOI] [PubMed] [Google Scholar]
- 171.Lawton AR, Cooper MD. Modification of B lymphocyte differentiation by anti-immunoglobulins. Contemp Top Immunobiol. 1974;3:193–225. doi: 10.1007/978-1-4684-3045-5_8. [DOI] [PubMed] [Google Scholar]
- 172.Nossal GJ. Cellular mechanisms of immunologic tolerance. Annu Rev Immunol. 1983;1:33–62. doi: 10.1146/annurev.iy.01.040183.000341. [DOI] [PubMed] [Google Scholar]
- 173.Sidman CL, Unanue ER. Receptor-mediated inactivation of early B lymphocytes. Nature. 1975;257:149–51. doi: 10.1038/257149a0. [DOI] [PubMed] [Google Scholar]
- 174.Raff MC, Owen JJ, Cooper MD, Lawton AR, Megson M, Gathings WE. Differences in susceptibility of mature and immature mouse B lymphocytes to anti-immunoglobulin-induced immunoglobulin suppression in vitro. Possible implications for B-cell tolerance to self. J Exp Med. 1975;142:1052–64. doi: 10.1084/jem.142.5.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Norvell A, Mandik L, Monroe JG. Engagement of the antigen-receptor on immature murine B-lymphocytes results in death by apoptosis. J Immunol. 1995;154:4404–13. [PubMed] [Google Scholar]
- 176.Monroe JG. Tolerance sensitivity of immature-stage B-cells: Can developmentally regulated B-cell antigen receptor (BCR) signal transduction play a role? J Immunol. 1996;156:2657–60. [PubMed] [Google Scholar]
- 177.Sandel PC, Monroe JG. Negative selection of immature B cells by receptor editing or deletion is determined by site of antigen encounter. Immunity. 1999;10:289–99. doi: 10.1016/s1074-7613(00)80029-1. [DOI] [PubMed] [Google Scholar]
- 178.Hartley SB, Cooke MP, Fulcher DA, Harris AW, Cory S, Basten A, Goodnow CC. Elimination of self-reactive B-lymphocytes proceeds in two stages: arrested development and cell death. Cell. 1993;72:325–35. doi: 10.1016/0092-8674(93)90111-3. [DOI] [PubMed] [Google Scholar]
- 179.Hertz M, Nemazee D. BCR ligation induces receptor editing in IgM+IgD– bone marrow B cells in vitro. Immunity. 1997;6:429–36. doi: 10.1016/s1074-7613(00)80286-1. [DOI] [PubMed] [Google Scholar]
- 180.Melamed D, Benschop RJ, Cambier JC, Nemazee D. Developmental regulation of B lymphocyte immune tolerance compartmentalizes clonal selection from receptor selection. Cell. 1998;92:173–82. doi: 10.1016/s0092-8674(00)80912-5. [DOI] [PubMed] [Google Scholar]
- 181.Melamed D, Nemazee D. Self-antigen does not accelerate immature B cell apoptosis, but stimulates receptor editing as a consequence of developmental arrest. Proc Natl Acad Sci USA. 1997;94:9267–72. doi: 10.1073/pnas.94.17.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lang J, Arnold B, Hammerling G, Harris AW, Korsmeyer S, Russell D, Strasser A, Nemazee D. Enforced Bcl-2 expression inhibits antigen-mediated clonal elimination of peripheral B-cells in an antigen dose-dependent manner and promotes receptor editing in autoreactive, immature B-cells. J Exp Med. 1997;186:1513–22. doi: 10.1084/jem.186.9.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fang W, Weintraub BC, Dunlap B, Garside P, Pape KA, Jenkins MK, Goodnow CC, Mueller DL, Behrens TW. Self-reactive B lymphocytes overexpressing Bcl-xL escape negative selection and are tolerized by clonal anergy and receptor editing. Immunity. 1998;9:35–45. doi: 10.1016/s1074-7613(00)80586-5. [DOI] [PubMed] [Google Scholar]
- 184.Spanopoulou E, Roman CA, Corcoran LM, Schlissel MS, Silver DP, Nemazee D, Nussenzweig MC, Shinton SA, Hardy RR, Baltimore D. Functional immunoglobulin transgenes guide ordered B-cell differentiation in Rag-1-deficient mice. Genes Dev. 1994;8:1030–32. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 185.Xu H, Li H, Suri-Payer E, Hardy RR, Weigert M. Regulation of anti-DNA B cells in recombination-activating gene-deficient mice. J Exp Med. 1998;188:1247–54. doi: 10.1084/jem.188.7.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Carsetti R, Kohler G, Lamers MC. Transitional B cells are the target of negative selection in the B cell compartment. J Exp Med. 1995;181:2129–40. doi: 10.1084/jem.181.6.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Allman DM, Ferguson SE, Cancro MP. Peripheral B cell maturation. I Immature peripheral B cells in adults are heat-stable antigen hi and exhibit unique signaling characteristics. J Immunol. 1992;149:2533–40. [PubMed] [Google Scholar]
- 188.Melamed D, Kench JA, Grabstein K, Rolink A, Nemazee D. A functional B cell receptor transgene allows efficient IL-7-independent maturation of B cell precursors. J Immunol. 1997;159:1233–39. [PubMed] [Google Scholar]
- 189.Lin WC, Desiderio S. V(D)J recombination and the cell cycle. Immunol Today. 1995;16:279–89. doi: 10.1016/0167-5699(95)80182-0. [DOI] [PubMed] [Google Scholar]
- 190.Hikida M, Mori M, Takai T, Tomochika K, Hamatani K, Ohmori H. Reexpression of RAG-1 and RAG-2 genes in activated mature mouse B cells. Science. 1996;274:2092–94. doi: 10.1126/science.274.5295.2092. [DOI] [PubMed] [Google Scholar]
- 191.Han S, Dillon SR, Zheng B, Shimoda M, Schlissel MS, Kelsoe G. V(D)J recombinase activity in a subset of germinal center B lymphocytes. Science. 1997;278:301–5. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- 192.Papavasiliou F, Casellas R, Suh H, Qin XF, Besmer E, Pelanda R, Nemazee D, Rajewsky K, Nussenzweig MC. V(D)J recombination in mature B cells: a mechanism for altering antibody responses. Science. 1997;278:298–301. doi: 10.1126/science.278.5336.298. [DOI] [PubMed] [Google Scholar]
- 193.Hikida M, Ohmori H. Rearrangement of lambda light chain genes in mature B cells in vitro and in vivo. Function of reexpressed recombination-activating gene (RAG) products. J Exp Med. 1998;187:795–99. doi: 10.1084/jem.187.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Han S, Zheng B, Schatz DG, Spanopoulou E, Kelsoe G. Neoteny in lymphocytes: Rag1 and Rag2 expression in germinal center B cells. Science. 1996;274:2094–97. doi: 10.1126/science.274.5295.2094. [DOI] [PubMed] [Google Scholar]
- 195.Hikida M, Mori M, Kawabata T, Takai T, Ohmori H. Characterization of B cells expressing recombination activating genes in germinal centers of immunized mouse lymph nodes. J Immunol. 1997;158:2509–12. [PubMed] [Google Scholar]
- 196.Han S, Zheng B, Takahashi Y, Kelsoe G. Distinctive characteristics of germinal center B cells. Semin Immunol. 1997;9:255–60. doi: 10.1006/smim.1997.0081. [DOI] [PubMed] [Google Scholar]
- 197.Hikida M, Nakayama Y, Yamashita Y, Kumazawa Y, Nishikawa SI, Ohmori H. Expression of recombination activating genes in germinal center B cells: involvement of interleukin 7 (IL-7) and the IL-7 receptor. J Exp Med. 1998;188:365–72. doi: 10.1084/jem.188.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hertz M, Kouskoff V, Nakamura T, Nemazee D. V(D)J recombinase induction in splenic B lymphocytes is inhibited by antigen-receptor signalling. Nature. 1998;394:292–95. doi: 10.1038/28419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Meffre E, Papavasiliou F, Cohen P, de Bouteiller O, Bell D, Karasuyama, Schiff C, Banchereau J, Liu YJ, Nussenzweig MC. Antigen receptor engagement turns off the V(D)J recombination machinery in human tonsil B cells. J Exp Med. 1998;188:765–72. doi: 10.1084/jem.188.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.de Wildt R, Hoet RMA, van Venrooij WJ, Tomlinson IM, Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol. 1999;285:895–901. doi: 10.1006/jmbi.1998.2396. [DOI] [PubMed] [Google Scholar]
- 201.Qin XF, Schwers S, Yu W, Papavasiliou F, Suh HY, Nussenzweig A, Rajewsky K, Nussenzweig MC. Secondary V(D)J recombination in B-1 cells. Nature. 1999;397:355–59. doi: 10.1038/16933. [DOI] [PubMed] [Google Scholar]
- 202.Wang F, Huang CY, Kanagawa O. Rapid deletion of rearranged T cell antigen receptor (TCR) Valpha-Jalpha segment by secondary rearrangement in the thymus: role of continuous rearrangement of TCR alpha chain gene and positive selection in the T cell repertoire formation. Proc Natl Acad Sci USA. 1998;95:11834–39. doi: 10.1073/pnas.95.20.11834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.McMahan CJ, Fink PJ. RAG reexpression and DNA recombination at T cell receptor loci in peripheral CD4+ T cells. Immunity. 1998;9:637–47. doi: 10.1016/s1074-7613(00)80661-5. [DOI] [PubMed] [Google Scholar]
- 204.Honjo T, Alt FW, editors. Immunoglobulin Genes. 2 San Diego: Harcourt Brace; 1995. [Google Scholar]