Abstract
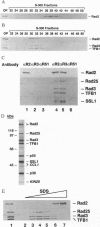
Nucleotide excision repair (NER) of ultraviolet light-damaged DNA in eukaryotes requires a large number of highly conserved protein factors. Recent studies in yeast have suggested that NER involves the action of distinct protein subassemblies at the damage site rather than the placement there of a "preformed repairosome" containing all the essential NER factors. Neither of the two endonucleases, Rad1-Rad10 and Rad2, required for dual incision, shows any affinity for ultraviolet-damaged DNA. Rad1-Rad10 forms a ternary complex with the DNA damage recognition protein Rad14, providing a means for targeting this nuclease to the damage site. It has remained unclear how the Rad2 nuclease is targeted to the DNA damage site and why mutations in the human RAD2 counterpart, XPG, result in Cockayne syndrome. Here we examine whether Rad2 is part of a higher order subassembly. Interestingly, we find copurification of Rad2 protein with TFIIH, such that TFIIH purified from a strain that overexpresses Rad2 contains a stoichiometric amount of Rad2. By several independent criteria, we establish that Rad2 is tightly associated with TFIIH, exhibiting an apparent dissociation constant < 3.3 x 10(-9) M. These results identify a novel subassembly consisting of TFIIH and Rad2, which we have designated as nucleotide excision repair factor 3. Association with TFIIH provides a means of targeting Rad2 to the damage site, where its endonuclease activity would mediate the 3' incision. Our findings are important for understanding the manner of assembly of the NER machinery and they have implications for Cockayne syndrome.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailly V., Sommers C. H., Sung P., Prakash L., Prakash S. Specific complex formation between proteins encoded by the yeast DNA repair and recombination genes RAD1 and RAD10. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8273–8277. doi: 10.1073/pnas.89.17.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankmann M., Prakash L., Prakash S. Yeast RAD14 and human xeroderma pigmentosum group A DNA-repair genes encode homologous proteins. Nature. 1992 Feb 6;355(6360):555–558. doi: 10.1038/355555a0. [DOI] [PubMed] [Google Scholar]
- Bardwell A. J., Bardwell L., Iyer N., Svejstrup J. Q., Feaver W. J., Kornberg R. D., Friedberg E. C. Yeast nucleotide excision repair proteins Rad2 and Rad4 interact with RNA polymerase II basal transcription factor b (TFIIH). Mol Cell Biol. 1994 Jun;14(6):3569–3576. doi: 10.1128/mcb.14.6.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell A. J., Bardwell L., Tomkinson A. E., Friedberg E. C. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994 Sep 30;265(5181):2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- Burns J. L., Guzder S. N., Sung P., Prakash S., Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J Biol Chem. 1996 May 17;271(20):11607–11610. doi: 10.1074/jbc.271.20.11607. [DOI] [PubMed] [Google Scholar]
- Clugston C. K., McLaughlin K., Kenny M. K., Brown R. Binding of human single-stranded DNA binding protein to DNA damaged by the anticancer drug cis-diamminedichloroplatinum (II). Cancer Res. 1992 Nov 15;52(22):6375–6379. [PubMed] [Google Scholar]
- Guzder S. N., Bailly V., Sung P., Prakash L., Prakash S. Yeast DNA repair protein RAD23 promotes complex formation between transcription factor TFIIH and DNA damage recognition factor RAD14. J Biol Chem. 1995 Apr 14;270(15):8385–8388. doi: 10.1074/jbc.270.15.8385. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Habraken Y., Sung P., Prakash L., Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995 Jun 2;270(22):12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Bailly V., Prakash L., Prakash S. RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature. 1994 Jun 16;369(6481):578–581. doi: 10.1038/369578a0. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Prakash L., Prakash S. Nucleotide excision repair in yeast is mediated by sequential assembly of repair factors and not by a pre-assembled repairosome. J Biol Chem. 1996 Apr 12;271(15):8903–8910. doi: 10.1074/jbc.271.15.8903. [DOI] [PubMed] [Google Scholar]
- Guzder S. N., Sung P., Prakash L., Prakash S. Yeast DNA-repair gene RAD14 encodes a zinc metalloprotein with affinity for ultraviolet-damaged DNA. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5433–5437. doi: 10.1073/pnas.90.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken Y., Sung P., Prakash L., Prakash S. Structure-specific nuclease activity in yeast nucleotide excision repair protein Rad2. J Biol Chem. 1995 Dec 15;270(50):30194–30198. doi: 10.1074/jbc.270.50.30194. [DOI] [PubMed] [Google Scholar]
- Habraken Y., Sung P., Prakash L., Prakash S. Yeast excision repair gene RAD2 encodes a single-stranded DNA endonuclease. Nature. 1993 Nov 25;366(6453):365–368. doi: 10.1038/366365a0. [DOI] [PubMed] [Google Scholar]
- Hanawalt P. C. Transcription-coupled repair and human disease. Science. 1994 Dec 23;266(5193):1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- He Z., Henricksen L. A., Wold M. S., Ingles C. J. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 1995 Apr 6;374(6522):566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- Li L., Lu X., Peterson C. A., Legerski R. J. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol Cell Biol. 1995 Oct;15(10):5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madura K., Prakash S. Nucleotide sequence, transcript mapping, and regulation of the RAD2 gene of Saccharomyces cerevisiae. J Bacteriol. 1986 Jun;166(3):914–923. doi: 10.1128/jb.166.3.914-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T., Saijo M., Kuraoka I., Kobayashi T., Nakatsu Y., Nagai A., Enjoji T., Masutani C., Sugasawa K., Hanaoka F. DNA repair protein XPA binds replication protein A (RPA). J Biol Chem. 1995 Feb 24;270(8):4152–4157. doi: 10.1074/jbc.270.8.4152. [DOI] [PubMed] [Google Scholar]
- Matsunaga T., Park C. H., Bessho T., Mu D., Sancar A. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J Biol Chem. 1996 May 10;271(19):11047–11050. doi: 10.1074/jbc.271.19.11047. [DOI] [PubMed] [Google Scholar]
- Mellon I., Hanawalt P. C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989 Nov 2;342(6245):95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Mu D., Hsu D. S., Sancar A. Reaction mechanism of human DNA repair excision nuclease. J Biol Chem. 1996 Apr 5;271(14):8285–8294. doi: 10.1074/jbc.271.14.8285. [DOI] [PubMed] [Google Scholar]
- Mu D., Park C. H., Matsunaga T., Hsu D. S., Reardon J. T., Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995 Feb 10;270(6):2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- Park C. H., Mu D., Reardon J. T., Sancar A. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J Biol Chem. 1995 Mar 3;270(9):4896–4902. doi: 10.1074/jbc.270.9.4896. [DOI] [PubMed] [Google Scholar]
- Robinson G. W., Nicolet C. M., Kalainov D., Friedberg E. C. A yeast excision-repair gene is inducible by DNA damaging agents. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1842–1846. doi: 10.1073/pnas.83.6.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994 Aug 26;265(5176):1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P., Guzder S. N., Prakash L., Prakash S. Reconstitution of TFIIH and requirement of its DNA helicase subunits, Rad3 and Rad25, in the incision step of nucleotide excision repair. J Biol Chem. 1996 May 3;271(18):10821–10826. doi: 10.1074/jbc.271.18.10821. [DOI] [PubMed] [Google Scholar]
- Sung P., Prakash L., Matson S. W., Prakash S. RAD3 protein of Saccharomyces cerevisiae is a DNA helicase. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8951–8955. doi: 10.1073/pnas.84.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Prakash L., Weber S., Prakash S. The RAD3 gene of Saccharomyces cerevisiae encodes a DNA-dependent ATPase. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6045–6049. doi: 10.1073/pnas.84.17.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P., Reynolds P., Prakash L., Prakash S. Purification and characterization of the Saccharomyces cerevisiae RAD1/RAD10 endonuclease. J Biol Chem. 1993 Dec 15;268(35):26391–26399. [PubMed] [Google Scholar]
- Sung P., Watkins J. F., Prakash L., Prakash S. Negative superhelicity promotes ATP-dependent binding of yeast RAD3 protein to ultraviolet-damaged DNA. J Biol Chem. 1994 Mar 18;269(11):8303–8308. [PubMed] [Google Scholar]
- Svejstrup J. Q., Feaver W. J., LaPointe J., Kornberg R. D. RNA polymerase transcription factor IIH holoenzyme from yeast. J Biol Chem. 1994 Nov 11;269(45):28044–28048. [PubMed] [Google Scholar]
- Svejstrup J. Q., Wang Z., Feaver W. J., Wu X., Bushnell D. A., Donahue T. F., Friedberg E. C., Kornberg R. D. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell. 1995 Jan 13;80(1):21–28. doi: 10.1016/0092-8674(95)90447-6. [DOI] [PubMed] [Google Scholar]
- van Hoffen A., Natarajan A. T., Mayne L. V., van Zeeland A. A., Mullenders L. H., Venema J. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 1993 Dec 25;21(25):5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]