Abstract
MicroRNAs (miRNAs) are tiny, endogenous, conserved, non-coding RNAs that negatively modulate gene expression by either promoting the degradation of mRNA or down-regulating the protein production by translational repression. They maintain optimal dose of cellular proteins and thus play a crucial role in the regulation of biological functions. Recent discovery of miRNAs in the heart and their differential expressions in pathological conditions provide glimpses of undiscovered regulatory mechanisms underlying cardiovascular diseases. Nearly 50 miRNAs are overexpressed in mouse heart. The implication of several miRNAs in cardiovascular diseases has been well documented such as miRNA-1 in arrhythmia, miRNA-29 in cardiac fibrosis, miRNA-126 in angiogenesis and miRNA-133 in cardiac hypertrophy. Aberrant expression of Dicer (an enzyme required for maturation of all miRNAs) during heart failure indicates its direct involvement in the regulation of cardiac diseases. MiRNAs and Dicer provide a particular layer of network of precise gene regulation in heart and vascular tissues in a spatiotemporal manner suggesting their implications as a powerful intervention tool for therapy. The combined strategy of manipulating miRNAs in stem cells for their target directed differentiation and optimizing the mode of delivery of miRNAs to the desired cells would determine the future potential of miRNAs to treat a disease. This review embodies the recent progress made in microRNomics of cardiovascular diseases and the future of miRNAs as a potential therapeutic target - the putative challenges and the approaches to deal with it.
Keywords: microRNA, heart failure, cardiovascular diseases, Dicer, microRNA-based therapy, stem cell
Introduction
Regulation is the key in maintaining any complex phenomenon in an arrayed manner and human physiology is one of the best examples where gene expression is precisely controlled at cellular and tissue levels in temporal and conditional manners [1, 2]. From a therapeutic perspective, complete understanding of the regulatory mechanisms is indispensible for genomic medicine [2, 3]. A newly discovered method of regulation of biological functions by microRNAs (miRNAs), a ∼22 nucleotide non-coding, endogenous and conserved RNAs has opened a new era of genomics called microRNomics [4, 5]. MiRNAs regulate protein expression primarily through base pairing at 3′ untranslated region (UTR) of target mRNAs leading to mRNA cleavage or repression of the translational machinery for protein synthesis [6]. The binding specificity of miRNA to its target is presumably dictated by 6–7 nucleotide sequence from 5′ region of miRNA, called ‘seed’ sequence [4], which nucleates binding to target mRNA and allows more 3′ region of miRNA for subsequent zippers up with the target mRNA [6]. The complimentarity of seed sequence to the target determines mRNA degradation or translational inhibition [4]. In the human genome nearly thousands of unique miRNAs are encoded, and they are predicted to regulate expressions of at least 30% of all human protein encoding genes thereby regulating almost all cellular functions [7, 8]. Here, it is germane to mention that a given miRNA is specific of a sequence and not of a single mRNA, and this is the reason that a single miRNA can modify expression of several mRNAs [9]. Some miRNAs are expressed in a tissue-specific manner [10]. For example mir-126 is endothelial specific [11] whereas miR-208 is cardiac specific [12]. There are approximately 50 miRNAs overexpressed in mouse heart [13]. Aberrant expressions of miRNAs in the heart lead to hypertrophy and other cardiac pathologies [14, 15]. The precise regulatory function of miRNAs is not restricted to the heart; rather it is involved in regulation of other organs like brain [16], gonad [17], dendritic cells [18] and many more.
The heart is the first organ formed during embryogenesis [19]. From embryogenesis to adult life, the heart is precisely regulated, and even a subtle perturbance in fine tuning of cardiac regulation causing either structural or functional remodelling leads to catastrophic consequences [20]. Despite our understanding about the biology of the heart, it is a leading cause of non-infectious infant mortality [21], and primary cause of morbidity and mortality in the industrialized world [22]. Cardiovascular diseases (the wide spectrum of diseases that affect the heart or blood vessels) is one of the major causes of death worldwide accounting for nearly 17 million deaths per annum according to the World Health Organization (http://www.who.int/cardiovascular_diseases/resources/atlas/en/). These statistics suggest a dire need for understanding the finer details of regulatory mechanisms maintaining the functions of the heart. The deep insight into the regulatory mechanisms of miRNAs which maintain the optimal dose of cellular proteins will provide a better approach than the traditional one. In this review, we summarize the therapeutic potential of miRNAs for treating cardiovascular diseases.
Biogenesis of miRNAs and their regulatory mechanisms
MiRNAs can be derived from individual miRNA genes, introns of protein coding genes or polycistronic transcripts [20]. They are transcribed by RNA polymerase II or III into primary miRNA (pri-miRNA), which are several kilobases long, capped (MGpppG) and polyadenylated [23] (Fig. 1). The pri-miRNAs are processed in nucleus by RNase III enzyme Drosha and the dsRNA binding protein Pasha (also known as DGCR8), into a hairpin-shaped structure of 70–100 nucleotides called preliminary-miRNA (pre-miRNA). Some intronic miRNA precursors, however, bypass Drosha processing to produce miRNA by Dicer, possibly representing an alternative pathway for miRNA biogenesis [24]. The pre-miRNAs are transported from the nucleus to the cytoplasm by RAN-GTP-dependant nuclear export factor, exportin 5 [25, 26] where they are processed by another RNase III enzyme, Dicer. Dicer processes pre-miRNA into a transient ∼18–24 nucleotide duplex. Out of two strands, one strand of miRNA duplex is preferentially retained in the complex that finally becomes the mature miRNA whereas the other strand, also called passenger strand or miRNA*, is degraded and eliminated from the complex [26]. The mature single stranded miRNA is loaded into the miRNA associated multi-protein RNA induced silencing complex (miRNA-RISC) that includes the Argonoute (Ago) proteins [26]. If the 3′ UTR of target sequence precisely matches the miRNA, Ago-2 will bind to the RISC complex, but if there are mismatches, Ago-1 will bind to the RISC [27].
1.

Biogenesis of miRNA and its regulatory mechanisms: miRNAs are encoded from either intergenic regions or introns and sometimes more than one encoded miRNA shares the same transcript (polycistronic). The primary miRNA (pri-miRNA), an approximately 200 kilobases long, capped and polyadenylated transcript, is transcribed by RNA polymerase III (or II) from any of the above mentioned encoding regions of miRNA. The pri-miRNA from intergenic and intronic regions is shown in the upper panel and from polycistronic region is shown in the lower panel of pri-miRNA. Drosha (an RNase III enzyme) along with Pasha (a double strand RNA binding protein) processes pri-miRNA into approximately 70 nucleotide pre-miRNA by cleaving the 5′ cap and polyadenylated tail. The pre-miRNA is transported from the nucleus to the cytoplasm by RAN-GTP exportin 5. Dicer (another RNase III endonuclease) cleaves pre-miRNA into double stranded transient duplex miRNA (22 nucleotides). The double stranded miRNA dissociates into single strands, and only one strand is retained as mature miRNA and it binds to RNA induced silencing complex (RISC). The miRNA-RISC complex regulates the dose of a particular protein by either degrading the complimentary mRNA by using RNAi mechanism or by inhibiting the translational machinery of protein.
Mature miRNA binds to complementary sites in the target to negatively regulate gene expression in two ways depending on its complimentarity to target sequence: (1) if sequence at 3′ UTR matches perfectly to target sequence, it will be cleaved and degraded by Ago2-RISC complex [28, 29], and (2) if there is imperfect complimentarity, as in most situations in mammalian cells, it causes translational repression [30, 31]. However, few investigations revealed that even imperfect base pairing of miRNA with its target sequence can lead to a decreased abundance of the mRNA [32]. Although the prediction of target of a particular miRNA is mainly dependant on Watson-Crick seed pairing, this approach is unable to predict the functional mismatched target sites of miRNA, suggesting the need for a more advanced miRNA target prediction tool [33].
MiRNAs in cardiovascular diseases
The profiling studies revealed a large number of miRNAs which are differentially expressed in heart failure pointing to the new mode of regulation of cardiovascular diseases [13–15, 31–34]. Although there are dramatic changes (up-or down-regulation) of several miRNAs in pathological condition, it is not necessary that all miRNAs, which are aberrantly expressed, must be implicated in the disease. For example, cardiac-specific overexpression of miR-214, which is most highly up-regulated among the 24 miRNAs studied in three different types of human heart disease (ischemic cardiomyopathy, dilated cardiomyopathy [DCM] and aortic stenosis) [31], failed to induce cardiac phenotypes in transgenic mice [35]. On the other hand, knock down of miR-133 leads to cardiac hypertrophy [36]. The functional assay thus confirmed the role of several miRNAs in hypertrophy, fibrosis, arrhythmia, myocardial infarction (MI), heart failure and angiogenesis [11, 26, 37–39] (Fig. 2).
2.
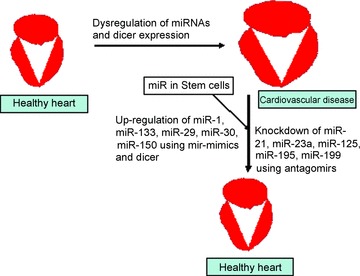
Putative therapy of cardiovascular diseases through microRNomics: in cardiovascular diseases Dicer and several miRNAs are dysregulated in heart and shows differential expressions. The pathological condition can be ameliorated by either knocking down those miRNAs, which are up-regulated or enhancing those, which are down-regulated. Another strategy can be the use of stem cells to replenish the diseased cells by new cells. The miRNAs in stem cells can be used to direct them for differentiation to produce specific type of cells required for mitigating the pathology.
MiRNAs in hypertrophy
Excessive cardiac workload leads to the enlargement of the heart (hypertrophy) in an endeavour to manage the increased haemodynamic demands, which ultimately leads to heart failure. It has two forms: (1) physiological, where the heart enlarges in healthy individuals following heavy exercise and is not associated with any cardiac damage, and (2) pathological where the size of the heart initially increases to compensate the damage to cardiac tissue but later on leads to decline in the left ventricular function representing an independent risk factor in heart failure [40]. Pathological hypertrophy is mainly caused due to hypertension, genetic polymorphisms and loss of myocytes following ischemic damage [41]. In addition to this, alterations in cardiac metabolism can also lead to hypertrophy [42]. Increase in the size of the heart develops in two ways: (1) concentric, which is caused by chronic pressure overload and leads to reduced left ventricular volume and increased wall thickness through addition of sarcomeres in parallel, and (2) eccentric that results due to volume overload and ultimately leads to dilation after thinning of the heart wall through addition of sarcomeres in series [43]. Hypertrophy is accompanied by an increase in the number of cardiac fibroblasts leading to fibrosis and myocyte loss. Myocyte loss leads to more workload [43] leading to enlargement of the heart and thus the vicious cycle of cardiac enlargement and myocyte loss continues causing heart failure. The whole process is regulated at fine level by different miRNAs such as miR-29, which is involved in fibrosis, and miR-133 and miR-195 which regulate the size of cardiomyocytes. Several miRNAs differentially expressed in cardiac hypertrophy [44] such as miR-1, miR-133, miR-29, miR-30 and miR-150 are down-regulated whereas miR-23a, miR-24, miR-125, miR-129, miR-195, miR-199, miR-208 and miR-212 are up-regulated [45]. The role of miR-21 in hypertrophy is controversial [4].
An interesting aspect of change in genetic profiling of miRNAs in failing heart is that it is reprogrammed as foetal genetic set up [46] which allows coordinated synthesis of proteins needed for increased myocyte size and adjustment to the altered energy demands of these larger cells [41].
MiRNAs in cardiac fibrosis
Ratio of collagen to elastin is crucial to maintain the extracellular matrix (ECM) in the heart. In failing heart, several structural remodelling occur, and one of them is the increase in matrix protein that enhances fibrosis. Connective tissue growth factor (CTGF), a key molecule involved in fibrosis, is shown to be regulated by two miRNAs: miR-133 and miR-30 [34]. Both miRNAs directly interact with 3′ UTR of CTGF and down-regulate its expression, which is accompanied by decreased production of collagen [34]. These exciting results suggest the involvement of miRNAs in structural heart disease. Another interesting study involving miRNA profiling at different regions of the heart after MI revealed that miR-29 is dramatically down-regulated in boarder zone flanking the infracted area [39]. Therefore, it is tempting to speculate that up-regulation of miR-29 can be a used as a good candidate for MI therapy. However, detailed analyses and characterization of individual miRNAs and their expression profiles especially associated with MI may provide a new window for concrete diagnosis and putative therapy of MI [47].
MiRNAs in arrhythmia
Membrane excitability is a special feature of cardiomyocytes and it is maintained by ion channels, which are regulated by miRNA [4]. The three major intrinsic properties of cardiac excitability that are reflected through electric conductance of cardiac cells are auto-maticity (the measure of cells to generate spontaneous action potential), cardiac conduction (propagation of excitation within a cell and between cells) and membrane repolarization (property to determine the length of action potential and the effective refractory period) [48]. Arrhythmia is diagnosed with the help of electrocardiogram (ECG) which is composed of mainly PR interval and segment, QRS complex, QT interval, ST segment and the RR intervals. PR interval begins at the onset of the P wave and ends at the onset of QRS complex. PR segment represents the duration of the conduction from the atrioventricular node, down the bundle of His and through the bundle branches to the muscle. It detects atrioventricular blockade. The QRS complex begins at the Q wave and ends at the endpoint of the S wave. It represents the duration of ventricular depolarization. It diagnoses the aberrant conduction, ventricular ectopic beats, ventricular hypertrophy and electrolyte imbalance. The QT interval begins at the onset of the QRS complex and ends at the end point of the T wave. It reflects the duration from the depolarization to the repolarization of the ventricles. Although it varies with gender and age, it helps in detecting the ventricular ectopic beats and ventricular tachycardia. The ST segment begins at the end point of the S wave and ends at the onset of the T wave. It is normally isoelectric and represents the duration of relaxation of the atrial cells and contraction of ventricles. This segment diagnoses the myocardial ischemia because depression of ST segment occurs when the ventricles are starved of oxygen due to blocked of arteries. The RR interval indicates the time elapsed between the R wave of one heart beat and R wave of preceding heart. It detects the sinus node disease and supraventricular arrhythmias (http://www.cardionetics.com/cardiology/ecg-durations.php). Different categories of ion channels like Na+, Ca++ and K+ and connexin 43 (gap junction protein) are key players of membrane polarization and depolarization during contraction and relaxation of cardiomyocytes [48]. The aberrant genetic expressions of ion channel genes may cause channelopathies or dysfunction of the ion channels rendering perturbance in electrical conduction that may result in arrhythmias [49]. Intriguingly, almost all miRNAs showing differential expression during cardiac hypertrophy and ischemia, and also the muscle-specific miRNAs, are theoretically presumed to be involved in regulation of cardiac ion channel genes such as miR-23a, miR-29, miR-150, miR-193, miR-214, miR-185, miR-320, miR-351 and miR-494 [48]. During MI, miR-30 remarkably increases in expression whereas it decreases in cardiac hypertrophy [4], and it could theoretically regulate several ion channel genes including gap junction protein alpha 1 (GJA1) that encode connexin 43, calcium channel beta-2 (CACNB2) that is dihydropyridine-sensitive L-type calcium channel β subunit and potassium ion channel gene (KCNJ3), which is Kir3,1 or G protein-activated inward rectifier potassium channel (GIRK1) of acetylcholine sensitive K+ channels [48]. Similarly, miR-195 that are up-regulated during cardiac hypertrophy [35] are predicted to regulate sodium channel (SCN)5A that encodes cardiac Na+ channel α-subunit, KCNJ2 that encodes Kir2.1 (a pore-forming α-subunit of inward rectifier K+ channel) and potassium channel, voltage-dependent beta subunit (KCNAB)1, the β-subunit of Shaker-type voltage-gated K+ channels [48]. There are empirical evidences that miR-1 that is involved in regulation of GJA1 and KCNJ2 causes arrhythmias [38]. There is up-regulation of miR-1 in individuals with coronary artery disease [38]. The overexpression of miR-1 in normal rat heart widens the QRS complex and prolongs the QT interval pointing to slowing of cardiac conduction, and causes membrane depolarization through a defective inward rectifier K+ current, Ik1. Further, ablation of miR-1 with an anti-sense inhibitor was sufficient to relieve arrhythmogenesis of infarcted rat heart [38]. After MI, two targets of miR-1 (KCNJ2 and GJA1) were found to be down-regulated in mice as well as samples from coronary artery diseased patients [38]. The apoptosis during MI causing arrhythmia [50] involves miR-1 and miR-133, the two muscle-specific miRNAs that share the same transcript. Interestingly both miRNAs produce opposite effects on apoptosis induced by oxidative stress in H9c2 rat ventricular cells; miR-1 is pro-apoptotic whereas miR-133 is anti-apoptotic [48].
In another study, it has been found that miR-1–2 knockout mice that survived to adulthood shows abnormal propagation of cardiac electrical activity [37]. These mice show normal anatomy and functions but had slower heart rate, a shortened PR interval, and a broadened QRS complex, indicative of bundle branch block associated with sudden death [37]. The plausible target of miR-1 in adult mice is Irx5 (Iroquois family of homeodomain-containing transcription factor), which regulates cardiac repolarization by repressing transcription of a key K+ channel, Kcnd2 [37]. Recently, it has been shown that overexpression of miR-1 causes arrhythmogenesis by enhancing calcium release, and by selectively increasing phosphorylation of L-type and the ryanodine receptor channels by targeting PP2A regulatory subunit B56 [51].
MiRNAs in myocardial infarction
The coronary artery supplies blood to the heart muscle and its occlusion leads to MI. MI is accompanied by a pathological remodelling response that includes hypertrophy and fibrosis, and is a major cause of morbidity and mortality in human beings [52]. MI results in irreparable death of cardiomyocytes and remains as a scar at the site of infarct. It impairs contractility and causes arrhythmias [48, 53]. The ECM is a dynamic microenvironment which bridges the endothelium to myocyte and contributes to adverse ventricular remodelling after MI by enhancing collagenase activity of matrix metalloproteinases (MMPs) [54]. At the site of infarct, the myofibroblasts (phenotypically transformed fibroblast like cells) mainly contribute to fibrous tissue formation [55]. Investigations on miRNAs after MI, which are dysregulated in mouse and human hearts, revealed that there is dramatic down-regulation of miR-29 in the region of fibrotic scar after MI [39]. MiR-29 negatively regulates a plethora of mRNAs involved encoding various collagens and other ECM proteins which are involved in cardiac remodelling after MI both in vivo and in vitro[39]. It further provides the mechanistic basis of cardiac fibrosis and suggests that enhancing miR-29 expression after MI may have therapeutic value associated with fibrosis [39]. In another study, it has been found that cardioprotective heat shock enhances the expressions of miR-1, miR-21 and miR-24 in the heart, and these miRNAs reduce the infarct size after ischemia-reperfusion injury suggesting the role of endogenous miRNAs in cardioprotection [56]. Recently, it has been shown in another ischemia-reperfusion study that miR-21 regulates MMP-2 expressions in infarcted zone of MI by phosphatase and tensin homolog pathways [57].
MiRNAs in heart failure
Due to hypertrophy and other pathological conditions, the heart cannot pump an adequate amount of blood to the body. This disease is called heart failure. In extensive miRNA profile of three human heart diseases – ischemic cardiomyopathy, DCM and aortic stenosis with the normal heart – it has been found that out of 87 miRNAs detected in the heart, 7 showed the same pattern of expression and more than half were differentially expressed in at least one disease group [31]. Interestingly, the miRNAs in foetal heart and failing heart expressed in similar patterns and this repro-gramming of foetal pattern in the failing heart provides a clue to progression of heart failure [46]. The regulation of calmodulin, a critical mediator of calcium signal in cardiomyocytes, by miR-1 provides another example of the critical role of miRNAs in heart failure [14]. Although investigating the precise function of miRNAs for a particular disease is complicated because of several targets of a single miRNA and regulation of single target by several miRNAs, their role in signalling pathways particularly in electrical conductance provide a tantalizing basis to speculate that it may be an important tool for future treatment of heart failure [58]. Empirical evidences, hitherto documented, clearly show that miRNAs are a potential therapeutic candidate for heart failure, and that future studies will define the exact pros and cons of use of microRNomics in prevention and cure of cardiovascular diseases.
MiRNAs in angiogenesis
Neoangiogenesis after MI plays vital role in cardiac repair and maintaining normal myocardial vascularization. Vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) are angiogenic growth factors required for generation of new blood vessels after MI. To maintain vascular integrity after MI, endothelial-specific miR-126 acts on mitogen-activated protein kinase signalling downstream to VEGF and FGF. Up-regulation of miR-126 favours angiogenesis enhancing cardiac repair after MI whereas miR-126 knockout mice either die during embryogenesis or if alive are prone to cardiac rupture following MI due to impaired neovascu-larization [11]. The implications of miRNAs in regulating various aspects of angiogenesis may be utilized in novel therapeutic approaches for diseases where insufficient or excess vasculature are involved [59, 60].
MiRNAs in diabetic cardiomyopathy
Diabetes is a physiological condition where glucose level goes high due to defects in insulin production, secretion and its signalling [61]. Diabetes may be caused either due to self-destruction of the pancreatic β cells (type 1 diabetes) or due to defects in insulin production or action (type 2 diabetes) [61]. Interestingly, both types of diabetes are associated with secondary complications and cardiac dysfunction is a most common manifestation of type 2 diabetes [61]. The pancreatic β-cells that produce insulin are regulated by miRNAs [62, 63] and particularly miR-375 that regulates insulin secretion negatively by targeting myotrophin [62]. Myotrophin acts as a transcriptional activator of NF-κB in cardiomyocytes that is associated with cardiac hypertrophy [64]. However, the first miRNA reported to be differentially expressed in diabetic heart is miR-133, and it plays key role in a long QT syndrome (LQTS) [65] and cardiac hypertrophy [36]. Mir-133 represses expression of human ether-a-go-go related gene that is associated with LQTS and encodes a cardiac potassium channel responsible for rapid delayed rectifier K+ current (IKr) [66]. Nevertheless, it is noteworthy that overexpression of miR-133 induces LQTS whereas down-regulation of miR-133 leads to cardiac hypertrophy in diabetic heart, and the significance of this intriguing regulation remains to be determined [67]. The role of miR-29 in both hyperglycemia and hyperinsulinemia [68], and its association with cardiac fibrosis [39] tempted us to speculate that it may be another potential candidate for future studies in diabetic cardiomyopathy.
MiRNAs in antherogenesis
Antherogenesis, also called atherosclerosis, is defined as a process of forming atheromas, plaques in the inner lining of arteries, and it has been established as a chronic inflammatory disease [69]. The low-density lipoprotein (LDL) when oxidized by oxygen free radicals (reactive oxygen species) and coming into contact with arterial wall, causes atherosclerotic lesions leading to increase in endothelial permeability and adhesiveness [70]. In response to the damage to the artery wall and endothelial dysfunction, the immune system responds by recruiting white blood cells (macrophages and T-lymphocytes) to adsorb the oxidized-LDL. The oxidized LDL molecule is globular shaped with a hollow core for carrying cholesterol. It is not processed by white blood cells that ultimately grow and rupture releasing cytokines and chemokines, which in turn trigger more white blood cells and the cycle continues. The complications of advanced atherosclerosis are chronic, slowly progressive and cumulative, and result in thrombosis that decreases or stops blood flow. The coronary thrombosis causing heart attack (MI) and thrombosis in the arteries to the brain causing stroke are deadly forms of the disease. This vascular inflammation process is also regulated by miRNA [71]. The vascular cell adhesion molecule 1 (VCAM-1) that mediates leucocyte adherence to endothelial cells is regulated by miR-126 [71]. The decrease in expression of miR-126 increases TNFα-stimulated VCAM-1 expression, which in turn enhances adherence of leucocytes to the endothelium [71]. Another candidate miRNA that regulates the neointima lesion formation is miR-21, which if down-regulated causes decrease in neointima formation in rat carotid artery after angioplasty [72]. Recently, miR-155 have been implicated in down-modulating the inflammatory cytokine production in response to lipopolysaccharides in human monocyte-derived dendritic cells [18].The association of miR-155 in regulation of angiotensin II type 1 receptor +1166 A/C polymorphism [73], inflammatory response [74] and endotoxin shock [75] makes it a very promising therapeutic candidate for inflammatory diseases.
Role of Dicer in cardiomyopathy
The fact that Dicer is biologically important for maturation of all miRNAs has been established in various cell types including embryonic stem cells [76], germline cells [77] as well as specialized cell types, such as pancreatic islet cells [78], immune cells [79], neural cells [80] and endothelial cells [81]. In order to understand the global role of miRNAs in cardiovascular diseases, a new approach developed where Dicer was knocked out. It resulted in death at embryonic day 12.5 with hearts displaying pericardial oedema and an insufficiently developed ventricular myocardium [37]. There was up-regulation of those genes that were activated after cardiac stress in the adult heart [37]. In another study Dicer was depleted in 3 weeks (young) and 8 weeks (adult) mice to understand its effect on postnatal cardiomyocytes [82]. It has been found that in the cardiomyocytes of young mice there was only a mild form of ventricular remodelling, and mice died due to sudden cardiac death within 1–2 weeks, probably due to arrhythmias [82]. In contrast to that, adult mice survived for the monitored time but displayed severe heart failure with all typical characteristics (ventricular enlargement, myocyte disarray, cardiac hypertrophy, fibrosis, etc.) [82] suggesting the temporal role of Dicer in cardiomyopathy. It is also vital for normal angiogenesis [83] and endothelial cell function [81, 84].
Functional studies show that cardiac-specific knockout of Dicer leads to rapidly progressive DCM, heart failure and postnatal lethality [44]. The contractile proteins were misexpressed and there was profound sarcomeres disarray in Dicer mutant mice [44]. An important caveat is that Dicer expression decreased in end-stage human DCM and in failing heart but significantly increased in those hearts which were assisted with left ventricle assist devices to improve cardiac functions [44], suggesting that Dicer/ miRNAs play critical roles in both normal cardiac functions as well as under pathological conditions. Further, increase in Dicer expression generating abundant miRNAs during compensatory phase of heart failure and decrease in Dicer expression in decom-pensatory/end stage/failing heart indicates that cardiac functions are highly regulated by Dicer in spatiotemporal manner.
MiRNAs – a new strategy for treatment of cardiovascular diseases
MiRNA – an innovative therapeutic approach
Despite tremendous efforts in the traditional approach to reduce mortality by heart disease, there is high rate of mortality and increasing number of morbidity. Therefore, it poses a great challenge to the therapeutic strategy adopted until now. In the traditional method of drug design involving enzymes, cell surface receptors might not bring much success in treatment of cardiovascular diseases due to the high sensitive nature of the heart. In this dismaying scenario, the discovery of an entirely new method of regulation by miRNAs, and their validation as markers and modulators of cardiac remodelling during pathological conditions engendered a new hope for innovative therapy. Recent investigations on the regulation of cellular response to stress [35], cardiac and disease development [85] and the reprogramming of foetal gene expression during heart failure [46] point to the major involvement of miRNAs in heart diseases. The fact that overexpression of single miRNA, miR-195, can induce pathological cardiac hypertrophy and failure [35] indicates the power of individual miRNA in regulating the coordination of target genes involved in complex physiological and disease phenotypes. The most important difference between microRNomics and traditional approach of therapy is that traditional drugs have specific cellular targets whereas miRNAs modulate the entire functional network [86].
MiRNAs in stem cell therapy
MiRNAs are also involved in regulation of stem cell division [87]. The role of miRNA in regulating mesenchymal stem cell-derived neuronal cells [88] and in other cell functions has been documented [89]. Embryonic stem cells are pluripotent (having capability to form any type of cell in the body) and four master transcription factors, namely Oct3/4, Sox2, Klf 4 and c-Myc, are reported to reprogram the differentiated somatic cells into pluripotent stem cells [90]. It is a milestone discovery for stem cell therapy because it will facilitate creating patient- and disease-specific stem cells. Additionally, understanding the role of miRNAs in regulating these transcription factors and manipulating them for stem cell differentiation into a specific type of cell will improve stem cell therapy. Human embryonic stem cells, cardiac progenitor cells, adult stem cells from bone marrow, mesenchymal stem cells and several other cells have been used in experiments for improving MI by replacing dead cardiomyocytes with healthy ones [91]. Although the success in transplantation of stem cells in the heart is promising and the delivery vectors for treatment of the heart is improving [92], stem cell therapy still needs several improvements for translation from bench to bed side. A problem with mesenchymal stem cell engraftment was formation of bone and cartilage in the heart after transplantation [93]. The microenvironments at the region of engraftment of stem cells are very important and it may cause differentiation of stem cells into unwanted cell types (teratomas). The immunogenic reactions of the host, the successful integration of engrafted stem cells to the infarction zone in case of MI, and the coupling of contractility of transplanted stem cells to the host cardiomyocytes in case of arrhythmia are indispensible issues to be addressed for successful stem cell therapy. MiRNAs may play a crucial role in resolving these issues. The combined approach involving Dicer and miRNAs in host cells, and miRNAs in engrafted stem cells can be used as better strategy for future therapy of cardiovascular diseases.
A new candidate in microRNomics
A distinct type of cell is reported in myocardium called interstitial Cajal-like cells (ICLC) [94, 95] which are speculated to be progenitor/stem cells in myocardium and are expected to be a therapeutic target of regenerative cardiovascular medicine [96]. Although the function of ICLC is yet to be established, their putative therapeutic potential will make them a promising candidate to be included in microRNomics.
Therapeutic challenges and their remedies
Since the first discovery of miRNA lin-4, which target lin-14 during temporal pattern formation in Caenorhabditis elegans[97], and their use in genetic manipulation to silence mRNA by RNA interference [98], a large number of miRNAs have been discovered that regulate cardiac functions. However, the major challenge in the functional studies of miRNAs is the fact that each miRNA has a large number of targets and each gene can also be regulated by several different miRNAs [99]. Additionally, the recent findings that miRNAs can also stimulate translation of mRNAs [100] and regulation of one miRNA by another [101] makes the understanding of functional aspects of miRNAs more complex. Nevertheless, the development of several tools for either to overexpress miRNAs in specific tissues [102] or to block the function of individual miRNAs [103, 104] will facilitate the future investigations of tissue-specific roles of individual miRNAs.
To pinpoint the role of a specific miRNA in a particular time and condition requires focused pursuit on alteration of expressions of that miRNA in different stages of the cardiovascular diseases. Investigations on Dicer expression in different age groups’ heart failure [82], and comparison of genetic profiles of foetal, adult and failing heart [46] are pioneer works in materializing this approach.
The general strategy of therapy is to reverse the pathological changes that resulted due to disease. Therefore, those miRNAs, which are up-regulated in pathological conditions should be knocked down and those down-regulated must be supplied from outside to cure the disease. In the case of miRNA therapy to cardiovascular diseases, this approach is feasible by supplying antagomiRs (synthetic reverse compliments of oligonucleotides) that bind to the up-regulated miRNA, and silence it or using miRNA mimics that act in similar way as miRNAs and enhance the expressions of down-regulated miRNAs [105]. However, there are practical limitations to this approach due to our limited understanding about the implications of these methods on biological functions at cellular level and on overall genetic set up. Nevertheless, miRNA biology can contribute to the identification of new disease targets and personalized strategies for heart failure patients, and a few miRNAs may become therapeutic targets for heart failure [106].
Mode of delivery of miRNAs
The mode of delivery of miRNAs will play significant role in therapy. For treatment of heart disease, which is entering phase II/III clinical trials, the major challenge is to improve the efficacy of gene delivery vectors to the target tissue [92] There are several approaches for delivery of intact gene to the nucleus of cells in the target tissues: (1) non-viral DNA delivery where plasmid DNA is directly injected into the myocardium. It is a safe method, and minimizes the complications raised due to insertional mutagenesis and immunogenicity of viral carrier [92]. However, it is less preferred because (i) the efficiency of uptake of plasmid DNA by target tissue is much less as in case of coronary infusion where the injected DNA is cleared by blood flow [107], and (ii) it requires invasive delivery techniques [108]. (2) Adenoviral and adeno-associated virus vectors acted better than the non-viral DNA because of their more efficient delivery [109–111]. Adenovirus vectors in particular are approved for clinical trials and have extensive data from several phase II/III clinical trials for cardiac gene therapy [112]. Nevertheless, they create high immunogenicity and need further evaluation via clinical trials. (3) For cardiac gene therapy, lentivirus particularly those that derived from HIV type 1 (HIV-1) are emerging candidates [92]. Although, the main safety concern with these vectors is their chances of homologous recombination producing wild type HIV [113], designing of new lentivirus where U3 promoter region of the long terminal repeats are deleted (self inactivating lentivirus) resolved this issue [114]. The success in current form of lentivirus vector [115] is making it a promising vector for future cardiac therapy.
For miRNA delivery, there can be two approaches: (1) Direct delivery of miRNA mimics (oligonucleotides) or antigomirs for enhancing and blocking the expression of a particular miRNA, respectively. For example, expression of miR-30c and miR-133 is inhibited by their antigomirs in cardiac myocytes [34, 36]. Interestingly this approach resembles pharmacological intervention and there is no immunogenic safety issue as happened with most delivery vectors as mentioned above. However, the major drawback is repeated delivery of doses required for therapeutic effect, which become a critical issue when the route of delivery is an invasive procedure [92]. (2) The second approach is to use gene delivery vectors expressing the miRNA mimics or antigomirs to cardiomyocytes. The use of lentivirus for the overexpression of miR-133b in isolated rat cardiac fibroblast has been recently reported [34]. This approach will further simplify the delivery of several miRNA mimics/antagomiRs at single dose due to small size of miRNA coding sequence, and would allow external drug-mediated regulation of expression [92]. Nonetheless, this approach includes the entire disadvantage mentioned above for other gene delivery vectors. The detailed understanding of the biology of miRNAs and optimization of gene delivery vectors would determine the future of miRNAs in therapy.
Role of microenvironment
The microenvironment of target tissue has a great impact on desired therapeutic outcome after miRNA delivery. The result from in vitro experiments where the microenvironments are controlled by the researcher may not perfectly match with the in vivo results where transplanted gene/miRNA are exposed to a dynamic microenvironment of the host [116]. From therapeutic point of view, creating the in vitro models that mimic the local milieu of the target tissue and pursuing experiments in that microenvironment to understand the ill effects of the gene/miRNA at in vivo condition will be of tremendous help to assess the tentative causes that impede the desired therapeutic outcome [116]. Additionally, this approach will increase the chances of success of the gene/drug/miRNA during clinical trial. Microenvironment has tremendous effect on the expression of apoptotic genes. Several miRNAs are involved in regulation of cell differentiation, proliferation and death [117–121], and depending on the microenvironment some of them cause cell proliferation (cancer) even though they are involved in regulation of cell death (apoptosis) [122].
MicroRNomics approach to cardiovascular therapy
There can be two approaches for investigating miRNAs in cardiovascular therapy: (i) at individual miRNA level and (ii) at global level where Dicer expression plays a key role. In the first approach, the major challenge is to understand the detailed mechanism involved in regulating expression of a specific miRNA in spatial and temporal manner specifically in pathological conditions. In addition to that it is essential to know how many targets are regulated by that specific miRNA, and how other miRNAs are influencing that target. Nonetheless, the detailed dissection of transcriptional and translational modifications of individual miRNA in different stages of pathology can provide many opportunities to manipulate individual miRNA for therapy. The second approach seems easier as it is broad and does not require the finer dissection of mechanistic regulation of individual miRNAs. Increasing the dose of Dicer can increase processing of all miRNAs required to reverse heart failure conditions. The large number of miRNAs will maintain the regulatory network layer and regain the optimal dose of proteins required for the normal heart functions. Till now, the only known function of Dicer is in the maturation of miRNAs. However, taking into consideration the infancy of microRNomics, we cannot rule out its other functions. Therefore, manipulating the Dicer expression for therapy of cardiovascular diseases should be investigated with caution. Other aspects of therapy include better methods of delivery of antagomiRs and miR mimics for knocking down and overexpression of miRNAs, respectively. The delivery method should be non-toxic, specific and should not influence other normal biological processes [92].
Although recent investigations are focusing on miRNAs in cardiomyocytes for cardiovascular diseases, the surrounding structures like smooth muscle layer, endothelium and ECM might be contributing significantly to the pathology. For example, ECM plays a pivotal role in cardiac remodelling [123] that leads to heart failure. Therefore, for complete understanding of cardiac pathology and for its better treatment, we must explore the involvement and mechanisms of action of miRNAs regulating ECM, endothelium and smooth muscle layers.
In conclusion, the discovery of differential expression of miRNAs causing gain and loss of function leading to cardiovascular diseases has provided an exciting new hope for its use in therapy. However, understanding the detailed molecular mechanisms underlying spatiotemporal miRNA expression, analysing the different targets of a single miRNA and regulatory mechanisms of different miRNAs on the same target, and optimizing the successful delivery of miRNAs to the target site will be the major challenges ahead which would determine the therapeutic potential of miRNAs for cardiovascular diseases.
Acknowledgments
This work was supported in part by the NIH grants HL-71010, HL-74185, HL-88012 and NS-51568.
References
- 1.Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–5. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 2.Little PF. Structure and function of the human genome. Genome Res. 2005;15:1759–66. doi: 10.1101/gr.4560905. [DOI] [PubMed] [Google Scholar]
- 3.Eckhardt F, Beck S, Gut IG, Berlin K. Future potential of the human epigenome project. Expert Rev Mol Diagn. 2004;4:609–18. doi: 10.1586/14737159.4.5.609. [DOI] [PubMed] [Google Scholar]
- 4.Latronlco MV, Cataluccl D, Condorelll G. Emerging role of microRNAs in cardiovascular biology. Circ Res. 2007;101:1225–36. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C. MicroRNomics: a newly emerging approach for disease biology. Physiol Genomics. 2008;33:139–47. doi: 10.1152/physiolgenomics.00034.2008. [DOI] [PubMed] [Google Scholar]
- 6.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Berezikov E, Guryev V, Van De BJ, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 10.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van RE, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–9. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 13.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–40. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR, Pu WT. microRNA-1 negatively regulates expression of the hypertrophy-associated genes calmodulin and Mef2a. Mol Cell Biol. 2009 doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–41. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, Roussel MF. The miR-17∼92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci USA. 2009;106:2812–7. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sirotkin AV, Ovcharenko D, Grossmann R, Laukova M, Mlyncek M. Identification of MicroRNAs controlling human ovarian cell steroidogenesis via a genome-scale screen. J Cell Physiol. 2009;219:415–20. doi: 10.1002/jcp.21689. [DOI] [PubMed] [Google Scholar]
- 18.Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. 2009;106:2735–40. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–7. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van RE, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369–76. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman JI. Incidence of congenital heart disease: II. Prenatal incidence. Pediatr Cardiol. 1995;16:155–65. doi: 10.1007/BF00794186. [DOI] [PubMed] [Google Scholar]
- 22.Rosamond W, Flegal K, Furie K, Go A, Greenlund K, Haase N, Hailpern SM, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O’Donnell C, Roger V, Sorlie P, Steinberger J, Thom T, Wilson M, Hong Y. Heart disease and stroke statistics–2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117:e25–146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- 23.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 24.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin Sci. 2008;114:699–706. doi: 10.1042/CS20070211. [DOI] [PubMed] [Google Scholar]
- 27.Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–97. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–23. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 30.Davison TS, Johnson CD, Andruss BF. Analyzing micro-RNA expression using microarrays. Methods Enzymol. 2006;411:14–34. doi: 10.1016/S0076-6879(06)11002-2. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31:367–73. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 32.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, Van Der Made I, Herias V, Van Leeuwen RE, Schellings MW, Barenbrug P, Maessen JG, Heymans S, Pinto YM, Creemers EE. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–8. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 35.Van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Ransom JF, Li A, Vedantham V, Von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 38.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–91. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 39.Van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy D, Garrison RJ, Savage DD, Kannel WB, Castelli WP. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N Engl J Med. 1990;322:1561–6. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 41.Barry SP, Davidson SM, Townsend PA. Molecular regulation of cardiac hypertrophy. Int J Biochem Cell Biol. 2008;40:2023–39. doi: 10.1016/j.biocel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 42.Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007;12:331–43. doi: 10.1007/s10741-007-9034-1. [DOI] [PubMed] [Google Scholar]
- 43.Wakatsuki T, Schlessinger J, Elson EL. The biochemical response of the heart to hypertension and exercise. Trends Biochem Sci. 2004;29:609–17. doi: 10.1016/j.tibs.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang DZ. Targeted deletion of dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105:2111–6. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latronico MV, Elia L, Condorelli G, Catalucci D. Heart failure: targeting transcriptional and post-transcriptional control mechanisms of hypertrophy for treatment. Int J Biochem Cell Biol. 2008;40:1643–8. doi: 10.1016/j.biocel.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, Van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–67. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 48.Yang B, Lu Y, Wang Z. Control of cardiac excitability by microRNAs. Cardiovasc Res. 2008;79:571–80. doi: 10.1093/cvr/cvn181. [DOI] [PubMed] [Google Scholar]
- 47.Diez J. Do microRNAs regulate myocardial fibrosis? Nat Clin Pract Cardiovasc Med. 2009;6:88–9. doi: 10.1038/ncpcardio1415. [DOI] [PubMed] [Google Scholar]
- 49.Marban E. Cardiac channelopathies. Nature. 2002;415:213–8. doi: 10.1038/415213a. [DOI] [PubMed] [Google Scholar]
- 50.Nerheim P, Krishnan SC, Olshansky B, Shivkumar K. Apoptosis in the genesis of cardiac rhythm disorders. Cardiol Clin. 2001;19:155–63. doi: 10.1016/s0733-8651(05)70201-0. [DOI] [PubMed] [Google Scholar]
- 51.Terentyev D, Belevych AE, Terentyeva R, Martin MM, Malana GE, Kuhn DE, Abdellatif M, Feldman DS, Elton TS, Gyorke S. miR-1 overexpression enhances Ca2+ release and promotes cardiac arrhythmogenesis by targeting PP2A regulatory subunit B56{alpha} and causing CaMKII-dependent hyperphosphorylation of RyR2. Circ Res. 2009;104:514–21. doi: 10.1161/CIRCRESAHA.108.181651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fox CS, Coady S, Sorlie PD, D’Agostino RB, Sr, Pencina MJ, Vasan RS, Meigs JB, Levy D, Savage PJ. Increasing cardiovascular disease burden due to diabetes mellitus: the Framingham Heart Study. Circulation. 2007;115:1544–50. doi: 10.1161/CIRCULATIONAHA.106.658948. [DOI] [PubMed] [Google Scholar]
- 53.Swynghedauw B. Molecular mechanisms of myocardial remodeling. Physiol Rev. 1999;79:215–62. doi: 10.1152/physrev.1999.79.1.215. [DOI] [PubMed] [Google Scholar]
- 54.Bouzeghrane F, Reinhardt DP, Reudelhuber TL, Thibault G. Enhanced expression of fibrillin-1, a constituent of the myocardial extracellular matrix in fibrosis. Am J Physiol Heart Circ Physiol. 2005;289:H982–91. doi: 10.1152/ajpheart.00151.2005. [DOI] [PubMed] [Google Scholar]
- 55.Powell DW, Mifflin RC, Valentlch JD, Crowe SE, Saada Jl, West AB. Myofibroblasts. I. Paracrine cells important in health and disease. Am J Physiol. 1999;277:C1–9. doi: 10.1152/ajpcell.1999.277.1.C1. [DOI] [PubMed] [Google Scholar]
- 56.Yin C, Wang X, Kukreja RC. Endogenous microRNAs induced by heat-shock reduce myocardial infarction following ischemia-reperfusion in mice. FEBS Lett. 2008;582:4137–42. doi: 10.1016/j.febslet.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ, Sen CK. microRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast MMP2 via PTEN. Cardiovasc Res. 2009;82:21–9. doi: 10.1093/cvr/cvp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zorio E, Medina P, Rueda J, Millan JM, Arnau MA, Beneyto M, Marin F, Gimeno JR, Osca J, Salvador A, Espana F, Estelles A. Insights into the role of microRNAs in cardiac diseases: from biological signalling to therapeutic targets. Cardiovasc Hematol Agents Med Chem. 2009;7:82–90. doi: 10.2174/187152509787047676. [DOI] [PubMed] [Google Scholar]
- 59.Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2009;2:pe1. doi: 10.1126/scisignal.252pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377–82. doi: 10.4161/cc.8.3.7526. [DOI] [PubMed] [Google Scholar]
- 61.Cohen A, Horton ES. Progress in the treatment of type 2 diabetes: new pharmacologic approaches to improve glycemic control. Curr Med Res Opin. 2007;23:905–17. doi: 10.1185/030079907x182068. [DOI] [PubMed] [Google Scholar]
- 62.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–30. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 63.Walker MD. Role of MicroRNA in pancreatic beta-cells: where more is less. Diabetes. 2008;57:2567–8. doi: 10.2337/db08-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gupta S, Sen S. Myotrophin-kappaB DNA interaction in the initiation process of cardiac hypertrophy. Biochim Biophys Acta. 2002;1589:247–60. doi: 10.1016/s0167-4889(02)00178-7. [DOI] [PubMed] [Google Scholar]
- 65.Xiao J, Luo X, Lin H, Zhang Y, Lu Y, Wang N, Zhang Y, Yang B, Wang Z. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2007;282:12363–7. doi: 10.1074/jbc.C700015200. [DOI] [PubMed] [Google Scholar]
- 66.Paulussen A, Yang P, Pangalos M, Verhasselt P, Marrannes R, Verfaille C, Vandenberk I, Crabbe R, Konings F, Luyten W, Armstrong M. Analysis of the human KCNH2(HERG) gene: identification and characterization of a novel mutation Y667X associated with long QT syndrome and a non-pathological 9 bp insertion. Hum Mutat. 2000;15:483. doi: 10.1002/(SICI)1098-1004(200005)15:5<483::AID-HUMU18>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 67.Tang X, Tang G, Ozcan S. Role of microRNAs in diabetes. Biochim Biophys Acta. 2008;1779:697–701. doi: 10.1016/j.bbagrm.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol. 2007;21:2785–94. doi: 10.1210/me.2007-0167. [DOI] [PubMed] [Google Scholar]
- 69.Ross R. Atherosclerosis -- an inflammatory disease. N Engl J Med. 1999;340:115–26. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 70.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–8. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 71.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–21. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–88. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 73.Martin MM, Buckenberger JA, Jiang J, MaIana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem. 2007;282:24262–9. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–9. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol. 2007;179:5082–9. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 76.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 77.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for dicer in the female germline. Genes Dev. 2007;21:682–93. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–45. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 79.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–74. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 80.Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28:4322–30. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of dicer and drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 82.Da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, Van Oort RJ, Pinto YM, Molkentin JD, De Windt LJ. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–76. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 83.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–5. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 84.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–73. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 85.Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–70. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 86.Van RE, Olson EN. microRNAs put their signatures on the heart. Physiol Genomics. 2007;31:365–6. doi: 10.1152/physiolgenomics.00206.2007. [DOI] [PubMed] [Google Scholar]
- 87.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–8. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 88.Greco SJ, Rameshwar P. MicroRNAs regulate synthesis of the neurotransmitter substance P in human mesenchymal stem cell-derived neuronal cells. Proc Natl Acad Sci USA. 2007;104:15484–9. doi: 10.1073/pnas.0703037104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331:57–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 91.Passier R, Van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature. 2008;453:322–9. doi: 10.1038/nature07040. [DOI] [PubMed] [Google Scholar]
- 92.Gray SJ, Samulski RJ. Optimizing gene delivery vectors for the treatment of heart disease. Expert Opin Biol Ther. 2008;8:911–22. doi: 10.1517/14712598.8.7.911. [DOI] [PubMed] [Google Scholar]
- 93.Breitbach M, Bostani T, Roell W, Xia Y, Dewald O, Nygren JM, Fries JW, Tiemann K, Bohlen H, Hescheler J, Welz A, Bloch W, Jacobsen SE, Fleischmann BK. Potential risks of bone marrow cell transplantation into infarcted hearts. Blood. 2007;110:1362–9. doi: 10.1182/blood-2006-12-063412. [DOI] [PubMed] [Google Scholar]
- 94.Gherghiceanu M, Hinescu ME, Popescu LM. Myocardial interstitial Cajal-like cells (ICLC) in caveolin-1 KO mice. J Cell Mol Med. 2009;13:202–6. doi: 10.1111/j.1582-4934.2008.00615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hinescu ME, Popescu LM. Interstitial Cajal-like cells (ICLC) in human atrial myocardium. J Cell Mol Med. 2005;9:972–5. doi: 10.1111/j.1582-4934.2005.tb00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kostin S, Popescu LM. A distinct type of cell in myocardium: interstitial Cajal-like cells (ICLC) J Cell Mol Med. 2009;13:295–308. doi: 10.1111/j.1582-4934.2008.00668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 98.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 99.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 100.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 101.Zhao Y, He S, Liu C, Ru S, Zhao H, Yang Z, Yang P, Yuan X, Sun S, Bu D, Huang J, Skogerbo G, Chen R. MicroRNA regulation of messenger-like noncoding RNAs: a network of mutual microRNA control. Trends Genet. 2008;24:323–7. doi: 10.1016/j.tig.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 102.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA. 2007;104:20844–9. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–7. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 105.Van RE, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–28. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res. 2008;103:1072–83. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ly H, Kawase Y, Yoneyama R, Hajjar RJ. Gene therapy in the treatment of heart failure. Physiology. 2007;22:81–96. doi: 10.1152/physiol.00037.2006. [DOI] [PubMed] [Google Scholar]
- 108.Lin H, Parmacek MS, Morle G, Bolling S, Leiden JM. Expression of recombinant genes in myocardium in vivo after direct injection of DNA. Circulation. 1990;82:2217–21. doi: 10.1161/01.cir.82.6.2217. [DOI] [PubMed] [Google Scholar]
- 109.Thirion C, Lochmuller H, Ruzsics Z, Boelhauve M, Konig C, Thedieck C, Kutik S, Geiger C, Kochanek S, Volpers C, Burgert HG. Adenovirus vectors based on human adenovirus type 19a have high potential for human muscle-directed gene therapy. Hum Gene Ther. 2006;17:193–205. doi: 10.1089/hum.2006.17.193. [DOI] [PubMed] [Google Scholar]
- 110.Volpers C, Kochanek S. Adenoviral vectors for gene transfer and therapy. J Gene Med. 2004;6:S164–71. doi: 10.1002/jgm.496. [DOI] [PubMed] [Google Scholar]
- 111.Vandendriessche T, Thorrez L, Costa-Sanchez A, Petrus I, Wang L, Ma L, DE Waele L, Iwasaki Y, Gillijns V, Wilson JM, Collen D, Chuah MK. Efficacy and safety of adeno-associated viral vectors based on serotype 8 and 9 vs. lentiviral vectors for hemophilia B gene therapy. J Thromb Haemost. 2007;5:16–24. doi: 10.1111/j.1538-7836.2006.02220.x. [DOI] [PubMed] [Google Scholar]
- 112.Rissanen TT, Yla-Herttuala S. Current status of cardiovascular gene therapy. Mol Ther. 2007;15:1233–47. doi: 10.1038/sj.mt.6300175. [DOI] [PubMed] [Google Scholar]
- 113.Cockrell AS, Kafri T. Gene delivery by lentivirus vectors. Mol Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- 114.Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–7. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yoshimitsu M, Higuchi K, Dawood F, Rasaiah VI, Ayach B, Chen M, Liu P, Medin JA. Correction of cardiac abnormalities in fabry mice by direct intraventricular injection of a recombinant lentiviral vector that engineers expression of alpha-galactosidase A. Circ J. 2006;70:1503–8. doi: 10.1253/circj.70.1503. [DOI] [PubMed] [Google Scholar]
- 116.Heinrich AC, Patel SA, Reddy BY, Milton R, Rameshwar P. Multi- and interdisciplinary science in personalized delivery of stem cells for tissue repair. Curr Stem Cell Res Ther. 2009;4:16–22. doi: 10.2174/157488809787169075. [DOI] [PubMed] [Google Scholar]
- 117.Nagel S, Venturini L, Przybylski GK, Grabarczyk P, Schmidt CA, Meyer C, Drexler HG, Macleod RA, Scherr M. Activation of miR-17–92 by NK-like home-odomain proteins suppresses apoptosis via reduction of E2F1 in T-cell acute lymphoblastic leukemia. Leuk Lymphoma. 2009;50:101–8. doi: 10.1080/10428190802626632. [DOI] [PubMed] [Google Scholar]
- 118.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–42. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 119.Lin Y, Liu X, Cheng Y, Yang J, Huo Y, Zhang C. Involvements of microRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem. 2009;284:7903–13. doi: 10.1074/jbc.M806920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu X, Nelson A, Wang X, Kanaji N, Kim M, Sato T, Nakanishi M, Li Y, Sun J, Michalski J, Patil A, Basma H, Rennard SI. MicroRNA-146a modulates human bronchial epithelial cell survival in response to the cytokine-induced apoptosis. Biochem Biophys Res Commun. 2009;380:177–82. doi: 10.1016/j.bbrc.2009.01.066. [DOI] [PubMed] [Google Scholar]
- 121.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104:476–87. doi: 10.1161/CIRCRESAHA.108.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–87. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 123.Tyagi SC, Hoit BD. Metalloproteinase in myocardial adaptation and maladaptation. J Cardiovasc Pharmacol Ther. 2002;7:241–6. doi: 10.1177/107424840200700407. [DOI] [PubMed] [Google Scholar]


