Abstract
Purpose of review
It is unknown whether biomarkers simply correlate with or are causal for HIV-associated outcomes. Mendelian randomization (MR), is a genetic epidemiologic approach used to disentangle causation from association. Here, we discuss the potential use of MR for differentiating whether biomarkers are correlating with or causal for HIV-associated outcomes.
Recent findings
MR refers to the random allocation of alleles at the time of gamete formation. In observational epidemiology, this refers to the use of genetic variants to estimate a causal effect between a modifiable risk factor and an outcome of interest. A formal MR study using a genetic marker as a proxy for the biomarker has not been conducted in the HIV field. However, in the post-genomic era this approach is being used increasingly. Examples are evidence for the causal role of body mass index on blood pressure and non-causal role of CRP in coronary heart disease. We discuss the conceptual framework, uses and limitations of MR in the context of HIV infection as well as specific biomarkers (IL-6, CRP) and genetic determinants (e.g., in CCR5, chemokine, and DARC genes) that associate with HIV-related outcomes.
Summary
Making the distinction between correlation and causality has particular relevance when a biomarker (e.g. IL-6) is potentially modifiable, in which case a biomarker-guided targeted treatment strategy may be feasible. Although the tenets of MR rest on strong assumptions, and conducting an MR study in HIV infection presents many challenges, it may offer the potential to identify causal biomarkers for HIV-associated outcomes.
Keywords: Mendelian randomization, gene, chemokine, HIV, biomarker
Introduction
In contrast to the ‘one size fits all’ approach of contemporary medicine for common complex diseases such as cancer and coronary heart disease (CHD), the goals of personalized medicine are to use individual biological signals such as biomarkers to predict the risk of developing disease, improve disease diagnosis, risk stratification of patients with active disease, and selection of therapies. This patient-specific approach is also the hope for HIV-1 medicine.
Many challenging barriers prohibit the utility of biomarkers in personalized medicine. Foremost, it is difficult to differentiate whether a biomarker is diagnostic, prognostic, or etiologic. For prognostic research, all factors associated with the outcome are of interest, whether they are causal or not. However, in etiologic research, causality is of fundamental importance. Although significant progress has been made in the identification of biomarkers that associate with HIV-related outcomes (e.g., see articles in this series [1–4]), it is unclear whether these biomarkers simply correlate with or are causal for HIV-associated outcomes, independent of all other risk factors (i.e. ceteris paribus). Making this distinction and adopting strategies to determine whether a biomarker may be causally linked with an HIV-associated outcome may have particular relevance when the biomarker is potentially modifiable, in which case a biomarker-guided targeted treatment strategy may be feasible.
Inferring causality from observational data is difficult as it is not always clear which of the two associated variables is the cause and which the effect, or whether both are common effects of a third unobserved variable, or confounder. The direction of causality can sometimes be determined by temporal criteria (e.g., the cause must precede the effect) or from knowledge of the underlying biology. However, confounding is always hard to address fully as it is due mostly to factors that are difficult to measure and control for. Also, it is nearly impossible to be certain that all the relevant confounders have been identified and accounted for. Also, for ethical reasons, risk factors cannot be assessed by randomized controlled trials. Studies combining genetics and biomarkers may help establish causality of a biomarker with outcome, and a genetic approach has been proposed to tackle some of these tribulations of observational studies and is termed ‘Mendelian randomization’.
The term Mendelian randomization (MR) is used in observational epidemiology to refer to the use of genetic variants as an instrument to estimate a causal effect between a specific modifiable risk factor (e.g., biomarker) and a trait/disease of interest [5–13]. The goal of this approach is to overcome some of the confounding that is encountered in observational epidemiology, such as residual confounding and reverse causation, by taking advantage of the natural random allocation of alleles during meiosis. It is important to emphasize that the while the goal of the MR approach is to help identify causal biomarkers that may provide useful diagnostic and/or prognostic information, it does not ascertain factors that are prognostic/diagnostic only, i.e., with no causal element.
In principle, MR may also hold promise as a tool for evaluating whether a biomarker is causally associated with HIV-related outcomes. However, infection with HIV imposes special constraints and confounders that may limit the full utility of this approach. That is, HIV infection through diverse mechanisms may induce or depress levels of a biomarker that may or may not be in the causal pathway for a particular HIV-associated outcome. Moreover, following infection the biomarker level may be altered to such an extent that it may inflate or obscure the association of a genetic variant with biomarker concentrations observed in HIV-negative persons. Additionally, epidemiological factors (e.g. differences in cohort characteristics, HIV outcome definitions) that are discussed below may further confound the results and interpretation of genetic variant–biomarker or genetic variant–HIV outcome association studies. Hence, the goals of this review are two-fold: First, as this approach has been used in the non-HIV field with some success in making the distinction of whether a biomarker is causal with or correlated with an outcome [10,11,14], we sought to bring the principles, assumptions, strengths, limitations and potential promise of MR to the fore of HIV investigators who evaluate biomarkers or other risk factors associated with HIV-related outcomes, and second, to illustrate this approach within the context of specific biomarkers and genetic determinants that influence HIV-associated outcomes. For these reasons we do not provide a comprehensive listing/discussion of genetic variants that associate with HIV-specific biomarkers [e.g., CD4+ T cell counts, plasma HIV RNA levels (viral load)], or HIV-associated outcomes (e.g., AIDS, death, opportunistic diseases), for which the reader is referred to other outstanding reviews [15–19].
Mendelian randomization (MR): conceptual framework
MR has been used extensively in the cardiovascular field (reviewed in [11]). Recent findings pertaining to the associations among single nucleotide polymorphisms (SNP) in the gene encoding C-reactive protein (CRP), CRP levels and coronary heart disease (CHD) serve as a blueprint for evaluation of whether biomarkers are causal for disease outcome [14]. We use this case-study from the cardiology field to illustrate the concept of MR as it also has direct relevance to the HIV field with many studies suggesting that CRP serves as a biomarker of HIV-associated outcomes (see related articles in this series [2,3]).
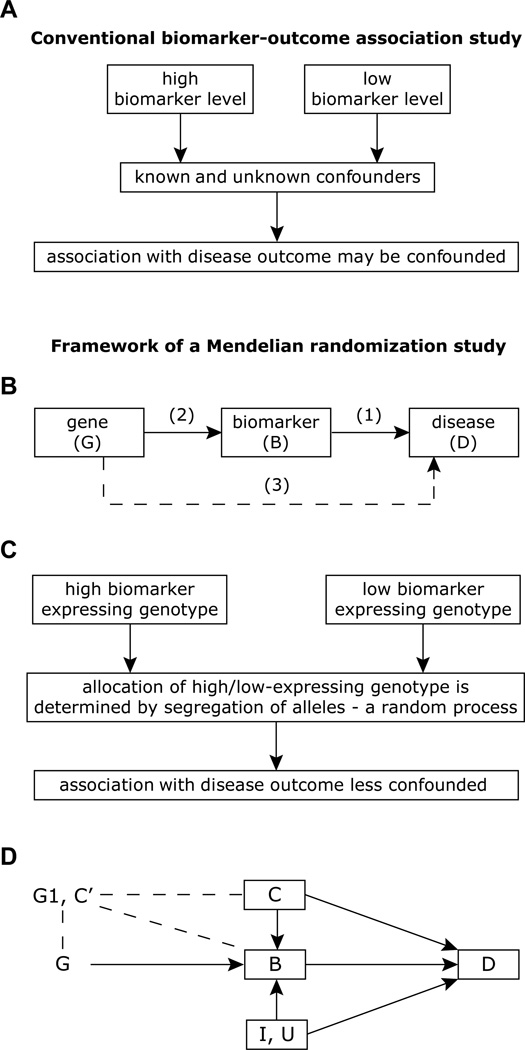
To establish the causal relationship between biomarker and disease outcome, investigators have traditionally used observational data showing an association between biomarker and outcome (Fig. 1A). However, this association may be confounded. One way to overcome this confounding is to integrate genetic strategies wherein a genetic factor is used as a proxy for the biomarker (Fig. 1B–C). Conceptually then, if a genetic variant is related to the biomarker (e.g., CRP levels) but is not otherwise associated with the outcome (e.g., CHD or HIV-related outcomes), then the relationship of the genotype with the outcome can be exploited to assess the causal relationship between biomarker and outcome (Fig. 1B–C). In the setting where the relationship between the biomarker and outcome is causal, then the genotype should associate with outcome, and the magnitude of this association should approximate the association of the genotype with the biomarker.
Fig. 1.
Approaches for evaluation of biomarkers in observational studies. (A) Evaluation of the relationship of biomarker with disease outcome using a non-randomized conventional approach. (B) Basic framework of Mendelian randomization approach. In its most basic form the approach is to study the causal relationship between a putative risk factor (biomarker levels) and the outcome (association 1) using a genetic variation that is known to influence the biomarker levels under investigation (association 2) as an instrumental variable, by estimating the association between the genetic variant and the outcome (association 3). (C) Same association study in panel A, but conducted within the principles of Mendelian randomization. (D) Detailed framework and assumptions of Mendelian randomization for evaluation of the relationship between biomarker and disease outcome. Directed acyclic graph (DAG) represents the relationships of a modifiable risk factor or biomarker B with the disease D with measured confounding variables C, unmeasured confounding variables U, unmeasured processes I impacting on both B and D, a SNP in the candidate gene designated as G that influences levels of B and which is also related to other genetic variants in another gene G1 that is possibly related to G and B, and other individual characteristics C’ possibly related to G, B, and D. In a DAG, a node represents a variable and an arrow a direct causal effect. Because a cause must precede an effect, no cycle is allowed and this is why the graph is termed acyclic (there is no loop from one node back to itself following the arrows). The methodology of Mendelian randomization uses the relationship of G with D, which presumably is unaffected by known confounding variables, C, and unknown confounders, U, to evaluate the relationship of B with D. Thus, in this situation, for causal inference of the impact of B on D, a genetic variant, G, which strongly influences the biomarker, can serve as a valuable instrumental variable. The premise of the instrumental variable approach is that integrating the estimates of the relationships of G with B and G with D can yield inferences regarding the relationship of B with D. There are many challenges to this method: the assumption that a gene influences disease solely through B is strong and unverifiable, as a single gene can influence disease risk through multiple pathways other than B (the phenomenon of pleiotropy); other alleles, G1 may be linked to G (linkage disequilibrium) and influence D through other pathways, which may result in confounding; and other characteristics of individuals at birth, C', that independently predict the development of D can be associated with G (population stratification) or influence the expression of G (e.g., epigenetics), and may void the conditions for a valid instrumental variable. Additionally, other alleles and patient characteristics can modify the effect of G on B, the effect of G on D, or both. These and other limitations are further illustrated in Figure 3. This figure is adapted from [12,13,20].
MR, is based on the premise of random allocation of alleles at time of gamete formation, and hence is thought to be independent of confounding factors [5–9]. In MR, a specific genotype results from two randomized transmissions, one from the paternally inherited allele and the other from the maternally inherited allele. A consequence of these randomizations is that genotypes are not expected to be associated with known (measurable or not) or unknown confounders for any outcome of interest, except for those that lie on the causal pathway between the genotype and the outcome (e.g., CRP→CHD; Fig. 1C). In theory, this should allow for the elucidation of two associations in a non-confounded manner (Fig. 1B–D): (i) genotype (G)-risk factor/biomarker (B) and (ii) the genotype (G)–disease outcome (D). By integrating appropriately the results of these two associations, one can, in theory, obtain an estimate of the risk factor/biomarker (B)-disease outcome (D) association, which is itself not confounded. Figure 1D outlines the principle and assumptions of Mendelian randomization, and it is important to emphasize that this approach does not define the association of the path from disease to biomarker (i.e., no D→B in Fig. 1B or D), a situation that might be common in the setting of HIV infection.
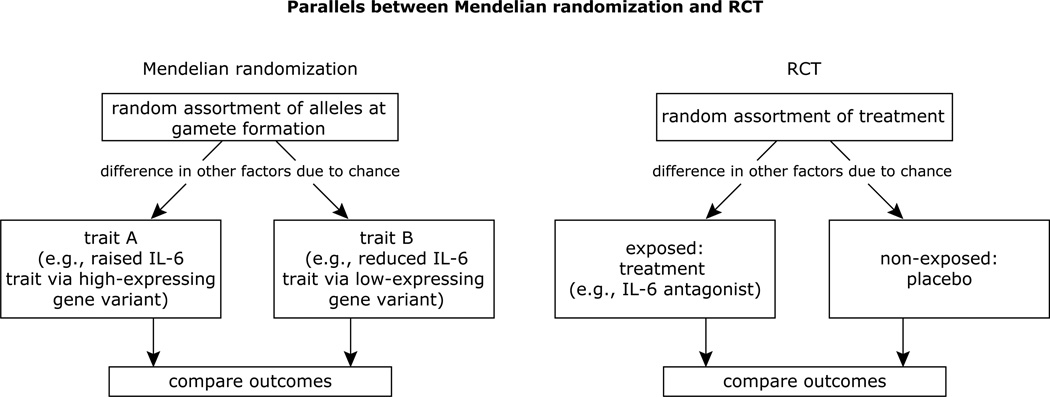
In some respects, the MR approach is analogous to randomized controlled trials (of sufficient sample size), in which the random allocation of treatment (or preventive measure) is expected to lead to an even distribution of (known or unknown) confounding factors across each group (Fig. 2). Thus, this strategy may be particularly helpful when direct intervention studies are not feasible due to the absence of a specific inhibitor of the risk factor. An additional strength of this approach is that while the levels of a biomarker may vary throughout a person’s life in response to diverse and unmeasured lifestyle and age-related biologic processes, genetic constitution is invariant. Another advantage is the accuracy of genotyping, which contrasts with biomarker measurements that may vary with time (e.g., diurnal changes, age) and there may be laboratory-to-laboratory differences in biomarker quantification. Finally, a genetic variant can be used as a proxy for a exposure that is difficult to measure (e.g., alcohol intake; [21]) and to possibly even infer biological causal pathways [22].
Fig. 2.
Similarities between Mendelian randomization and randomized clinical trials, illustrated by the example of the relationships between IL-6 gene variants, IL-6 levels and HIV outcomes. Figure adapted from [12].
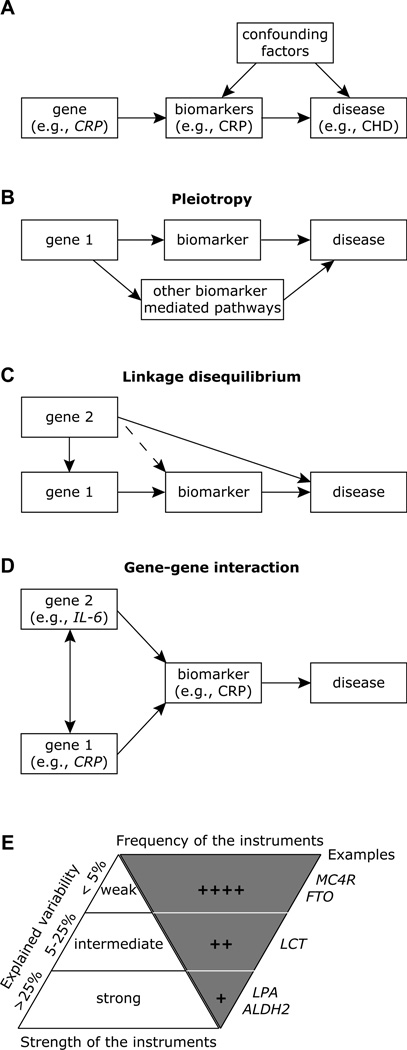
Historically, the first description of the concept of MR in observational epidemiology is attributed to Katan, who suggested the use of ApoE genotypes that associate with cholesterol levels as a way to distinguish whether low cholesterol levels were a cause of cancer or a consequence of carcinogenesis [23]. The MR concept builds on what is known as an instrumental variable method in econometrics [24]. In the case of MR, the genetic variant acts as an instrumental variable. Three essential assumptions of MR must be met to allow for accurate application of MR and interpretation: (i) genotype is independent of confounding between biomarker and outcome [i.e., the graph has no arrow (in either direction) connecting CRP gene with the confounders; Fig. 3A], (ii) genotype is associated with the biomarker (i.e, there is an arrow connecting CRP genotype to serum CRP and this relationship can be accurately quantified, with a stronger association being most favorable; Fig. 3A), and (iii) genotype is independent of outcome except as mediated through the biomarker (i.e., no single arrow between CRP gene and CHD; Fig. 3A). Although these assumptions are strong and may have some untestable aspects, as for many modeling strategies, the approach may yield useful insights.
Fig. 3.
Limitations and considerations while applying Mendelian randomization. (A) The CRP genotype as an instrumental variable in Mendelian randomization. The arrows can be considered to represent causal relationships; it is important to note that there is no arrow between CRP gene and the confounders and there is no single arrow between CRP gene and CHD, i.e., there are no other routes in the pictorial between CRP gene and CHD. (B) The problem of pleiotropy. A single gene can influence disease risk through multiple pathways other than the biomaker. This violates the second assumption of Mendelian randomization. (C) Problem of linkage disequilibrium. In this instance, the genetic variant is in linkage disequilibrium with the variant associated with, or causal for, the disease. SNP in Gene 1 is in linkage disequilibrium with another SNP in Gene 2. Here, Gene 2 directly affects the disease level or risk, and hence Gene 1 is not an adequate genetic instrument for MR. The SNP in Gene 1 could be in linkage disequilibrium with a SNP in Gene 2 that is also causal for the biomarker (dashed line). In this instance, if Gene 2 SNP only affects disease via its effects on the same biomarker, there is no violation of the assumptions, and SNP in Gene 1 can still be considered as an instrument in a Mendelian randomization analysis. (D) Problem of gene-gene interaction. Example shown is for the interaction between genetic variants in IL-6 and CRP wherein a polymorphism in IL-6 gene correlates more closely with CRP levels than IL-6 levels. (E) Low explained variability. The Mendelian randomization approach is dependent on the proportion of the variance (explained variability depicted along the left of the upright triangle) of the biomarker explained by the genetic instrument (SNP) for the genes depicted on along the right of the inverted triangle. Abbreviations for genes: MC4R, melanocortin 4 receptor gene; FTO, fast mass and obesity associated gene; CRP, C-reactive protein gene; LCT, lactase gene; LPA, lipoprotein A gene; and ALDH2, aldehyde dehydrogenase gene. Data is adapted from [11].
CRP in CHD: a case-study of Mendelian randomization
Although CRP is a well-established biomarker for inflammation and CHD, studies have been inconclusive regarding a causal role of CRP in CHD. Using a powerful multi-staged study design, Elliott and colleagues recently identified SNPs in the CRP gene that associated with CRP levels [14]. However, in this MR analysis, these CRP SNPs did not associate with CHD [14]. These results suggest that CRP may not be causal for CHD.
Considerations for Mendelian randomization
MR studies represent a special case of conventional genetic association studies and hence many of the same genetic and non-genetic parameters/features considered for interpretation of such studies also apply to MR [11,20]. Many of the assumptions, challenges, and considerations necessary to account for in a MR study are outlined in Fig. 1D. We discuss some of these considerations and limiting factors pertinent to the conduct of an MR study within the context of the results of the CRP case-study described above as well as IL-6, as both CRP and IL-6 have relevance to HIV-associated outcomes [1–4].
(i) In an MR study it is important to consider that a gene may influence disease risk through multiple pathways other than the biomarker of interest (i.e., pleiotropy, which refers to the situation where a genetic variant has more than one specific phenotypic effect; Fig. 3B). E.g., the association of CRP SNPs may be indirect, influencing other biomarkers that cause or prevent CHD. (ii) A suitable functional genetic variant to study the biomarker of interest may not be identifiable for a MR study, such that most associations will be indirect, relying on linkage disequilibrium (e.g., SNPs in the CRP gene are in close proximity to SNPs in another gene that associates with CHD, Fig. 3C). Also, linkage disequilibrium patterns (i.e., the genomic architecture of the locus of interest) may vary from one population to the other and can potentially confound analyses. Copy number variation is a distinct polymorphism that is also thought to play a major role in gene expression levels [25] and failure to account for the genomic architectural complexity of the copy number variation region may also potentially confound MR [26–30].
(iii) A MR study will need to consider the growing evidence that complex diseases are influenced by numerous gene–gene and gene–environment interactions, which again may differ from one population to the other (Fig. 3D). In this instance, a false-negative association may be due to failure to account for a second gene that modifies the levels of the biomarker of interest, e.g., a SNP in the promoter of the IL-6 gene that associates with serum IL-6 levels [31] has been shown to correlate more strongly with CRP than IL-6 levels [32], and ApoE alleles can also influence CRP concentrations [33,34]. Illustrating gene-environmental interactions, a recent study showed that environmental factors modify the effects of the functional IL-6 promoter SNP [35]. (iv) An important limitation of MR is the weakness of the instrument, i.e., most single genetic variants only explain a small proportion of the trait variance (typically <5%; Fig. 3E), and hence very large sample sizes are needed to achieve a reasonable power. (v) The assumption of linearity between biomarker and outcome may not always be true.
(vi) The success of MR heavily rests on the existence of allelic homogeneity, i.e., a common causal allele is shared by many individuals. (vii) It is difficult to exclude canalization (e.g., genetic elevations in CRP levels are counterbalanced by compensatory changes in other systems) (viii) Epigenetics may confound the MR analyses. Given that levels of cytokines such as IL-6 and CRP have a heritable component [36–38] that may be further modified by environmental influences (e.g., gender, age), MR studies need to not only assume a random distribution of alleles in the offspring, but also a random distribution of epigenetic changes (e.g., gene expression) at conception, in order for the core assumptions of the MR strategy to remain valid [39].
Mendelian randomization and HIV-AIDS: special considerations
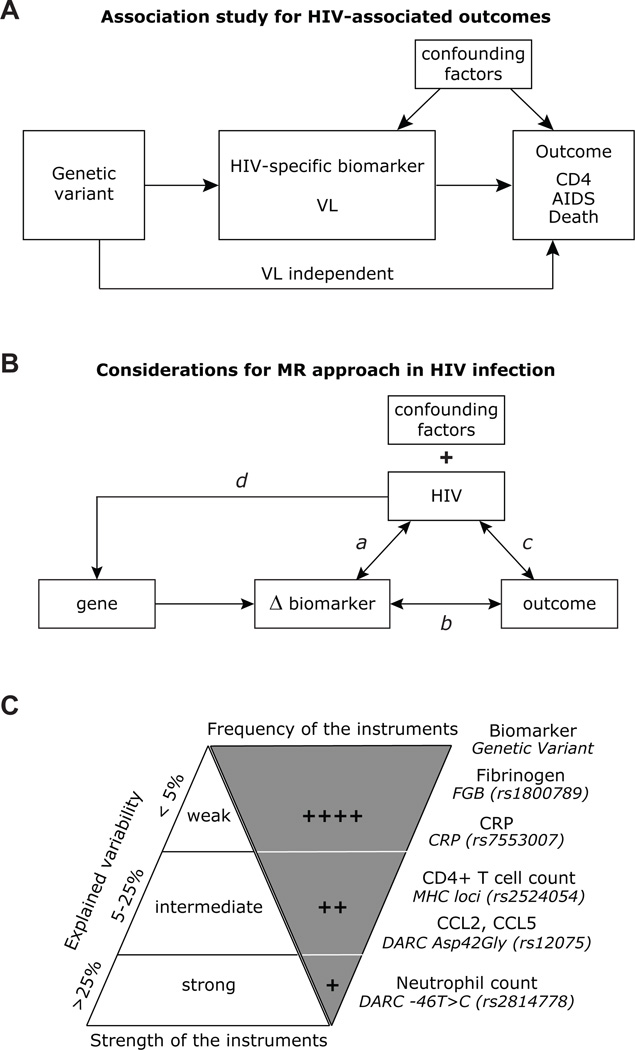
In general most association studies in the HIV field evaluate the association between a genetic variant with plasma HIV RNA levels (viral load), an intermediary phenotype, and/or with more distal outcomes such as AIDS, specific-AIDS defining illnesses or death (Fig. 4A). These studies using both candidate gene approaches and genome wide association studies have yielded many advances [15–19], including identification of genetic markers in the MHC locus that associate strongly with viral load [40], and the insight that HLA alleles such as HLA-B*57 [41] and variations in CCL3L/CCR5 genes may influence HIV pathogenesis by impacting on both viral load-dependent and –independent parameters such as cell-mediated immunity [42]. These viral load-independent associations are in accord with the notion that although highly contributory, viral load is not the sole determinant of CD4+ T cell loss and HIV-associated outcomes [43]. In some cohorts, the explained variability of viral load for rates of CD4 cell decline [44] and progression to AIDS [42] was <5% and <15%, respectively. Additionally, despite suppression of the viral load by highly active antiretroviral therapy, many parameters such as T cell activation that correlate highly with viral load persist, suggesting that mechanisms other than viral load may underlie HIV-associated outcomes [1,4,16]. These observations have prompted a search for and identification of host-centric rather than virus-centric intermediary phenotypes such as inflammatory factors (biomarkers) that associate HIV-associated outcomes (reviewed in [1]). (x) In HIV-1 infection, an MR study may need to account for the possibility of shared genetic influences. Although a significant part of the variation in inflammatory markers is due to genetic influences [36–38], almost 50% of the shared variance among biomarkers such as IL-6, CRP and fibrinogen may be due to a common genetic factor, suggesting that these biomarkers may lie within common causal pathways [37]. This has relevance to the results from the SMART and other observational studies that have found similar biomarkers to associate with HIV-associated outcomes [1].
Fig. 4.
Association study for HIV-associated outcomes, considerations for MR approach in HIV infection and candidate biomarkers for HIV-associated outcomes. (A) Schema depicting a conventional genetic association study for HIV-associated outcomes. The figure illustrates the association between a genetic variant with HIV-associated outcomes (CD4+ T cell counts, AIDS or death) via either viral load (VL)-dependent or –independent parameters such as cell-mediated immunity. (B) Considerations for MR study in HIV infection. In addition to the confounding factors that may influence any genetic association study, HIV may introduce additional variable that needs to be considered when conducting an MR study. a, HIV infection may alter biomarker levels (Δ biomarker), which in turn could impact on viral load. b, A biomarker may correlate with HIV-associated outcomes, and conversely, specific HIV-associated outcomes may influence biomarker levels. c, Specific HIV-associated outcomes (e.g., T cell activation) may influence HIV viremia, and conversely, HIV may influence outcomes through mechanisms that are independent of the biomarker of interest. d, HIV may theoretically impact on the gene locus of interest (e.g., by epigenetic reprogramming of gene locus that encodes the biomarker under investigation), and thus impact on gene expression and biomarker levels. (C) Candidate biomarkers that may influence HIV-associated outcomes and proportion of variance of the biomarker explained by the genetic instruments (SNP) depicted along the right of the inverted triangle.
In the ensuing sections of this review, we focus on the application of MR in HIV infection for investigating whether an intermediary phenotype such as a biomarker is causal with or correlating with HIV-associated outcomes. It should be noted that MR has, to date, not been discussed or conducted within the context of communicable or infectious diseases [11]. Although in theory the tenets of MR should also hold true in the context of HIV infection, both the evaluation and conduct of an MR study and meeting the assumptions of MR in HIV infection may be difficult for many reasons, some of which are summarized in Fig. 4B, and discussed below.
(i) Null or false-positive associations may arise because infection with HIV-1 may grossly alter levels of a biomarker. Fig. 4B illustrates the possible ways by which infection with HIV may distort many of the assumptions that need to be met for an MR study. We use the example of the recent associations of elevated IL-6 levels with adverse HIV-associated outcomes [1,4] to illustrate this important point. Although these associations raise the possibility that an IL-6 level-guided targeted treatment strategy could become an attractive therapeutic option, caution may need to be exercised as IL-6 levels may simply be a reflection of a more proximal and causal unmeasured biological pathway (e.g., T cell activation). Another possibility is that the impact of IL-6 on HIV-associated outcomes is via an additional biological factor. One can also envisage that the association of IL-6 or any other biomarker with disease outcome may be highly sensitive to the timing of IL-6 measurements, as associations may differ depending on whether biomarker concentrations were assessed during early or late stages of HIV disease. There is also the concern of reverse causation as CD4 cell depletion or specific forms of opportunistic infections may result in higher IL-6 levels and the pathways that mediate these raised IL-6 levels may remain activated despite suppression of viral replication.
If one were to apply the principles of MR to IL-6 in HIV infection, the same tenets described for the integration of CRP gene variant→CRP levels→CHD would have to apply, i.e., IL-6 gene variant→IL-6 levels→HIV-associated outcome. However, one will have to be very mindful of the fact that HIV infection may perturb IL-6 levels in a manner that the association of an IL-6 SNP with IL-6 levels may differ (inflated or obscured assocation) from that observed in HIV-negative persons. Consider then the following hypothetical scenario: assume for a moment that the association between IL-6 levels and HIV-associated outcomes is strong (as it appears to be the case). However, also consider that for the caveats and reasons mentioned in the preceding sections, future studies with large numbers of HIV-positive persons show that, at least based on the MR criteria, IL-6 is not causal for HIV-associated outcomes. These null genetic findings would limit our ability to distinguish whether IL-6 a risk marker or a risk factor.
Several additional types of confounding could have influenced the results of this hypothetical MR study with IL-6 in HIV (e.g., gene-gene interaction, pleiotropy). For example, the effects of IL-6 are pleiotropic, and interestingly, as noted above, a functional IL-6 SNP is known to correlate more strongly with CRP than IL-6 levels [32], and hence it is conceivable that the associations of the IL-6 SNP might not relate to IL-6 but CRP levels. Also, the interpretation of null MR studies is challenging, especially because in most instances a genetic variant explains only a small variance in the level of the biomarker (e.g., Fig. 3E and Fig. 4C). Also, the IL-6 SNP that may serve as a proxy for IL-6 is highly European-specific [45,46] and this will limit cross-comparisons of the same instrumental variable (i.e. SNP) across ethnicities. These limitations and considerations that we describe for a hypothetical MR study designed for IL-6 will also apply to any biomarker evaluated within the context of HIV infection.
(ii) Associations for gene or biomarker may vary because of differences in the criteria used to define an HIV-associated outcome. Although investigators oft-times do not differentiate between ‘soft’ non-clinical (e.g. CD4+ T cell counts or viral load) versus ‘hard’ clinical (AIDS or death, or specific AIDS-defining illnesses) endpoints, this differentiation has important consequences for any genetic or biomarker association study. Differences in the definition of the HIV-related phenotypic endpoint may greatly impact on the interpretation of association studies because genotypes have been identified (e.g. HLA alleles [47] and CCR5 haplotypes [48]) that impact on specific stages of HIV disease course (early vs late). Hence, it would not be surprising if genetic associations made for an endpoint classified as ‘progression phenotype’ and defined on the basis of the time to treatment initiation, or the time to the drop of CD4 cell counts below 350 cells/mm3 yields contrasting results when the associations for the same genotype are made with the 1987 CDC criteria of AIDS or death. Similarly, a biomarker because of its biological characteristics may causally associate with a specific AIDS-defining illnesses (e.g. cancer or HIV-associated dementia) but not with clinical AIDS (CD4 count <200 cells/mm3) and vice versa. For the same reasons, the association of a biomarker with overall AIDS (which is an amalgamation of several AIDS-defining illnesses) may be dependent on the frequency of the AIDS-defining illness present in the cohort, which may differ according to the risk characteristics of the cohort subjects (e.g. intravenous drug use versus mucosal transmission).
(iii) Associations for gene or biomarker may vary across cohorts because of differences in the epidemiologic characteristics of the cohort. Lead-time bias may confound association studies conducted in a seroprevalent cohort versus a seroconverting cohort. Other epidemiological features may also impact on the results of biomarker or genetic association studies. First, patients enrolled in distinct cohorts may have contrasting access to health care (e.g., differential antiretroviral use across cohorts in the pre-HAART era may confound associations of biomarker levels measured on samples during this time-period). Second, risk characteristics (e.g. intravenous drug use versus mucosal transmission) will impact on rates of coinfections with other viruses (e.g. Hepatitis C virus or GB virus-C) that may modify biomarker responses. Third, gender may also impact on biomarker status. For example, CCR5 levels are higher in women compared to men [49], and differential expression of biomarkers in men versus women have been observed [50]. Fourth, continent-of-origin may influence HIV outcomes. Because of environmental factors or co-infections specific to distinct geographic regions (e.g. malaria, TB), the gene-biomarker associations may differ. For example, studies have shown that some African populations have higher levels of T cell activation [51,52] and CCR5 [53], which in turn may influence biomarker levels. Also, pertinent to the evaluation of gene-biomarker associations across ethnicities, studies have suggested that compared to persons of European descent, persons of African ancestry might be over-represented and under-represented, respectively for genetic variants that associate with upregulation and downregulation of inflammation [45,54]. Finally, HIV subtypes may also associate with contrasting HIV outcomes and biomarker responses [55,56].
These observations, in conjunction with those factors noted above that require consideration during the conduct of any genetic association study (pleiotropy, complexity of genetic architecture of the locus, gene-gene and gene-environment interactions) indicate that results of gene-biomarker, or gene-HIV disease, associations, may not necessarily be invariant across cohorts with heterogeneous epidemiological characteristics. Such a viewpoint is dogmatic and requires caution.
Mendelian randomization and HIV-associated outcomes: case studies
Many genetic variants have been identified that influence biomarker levels, and some of these may have a relationship with HIV-associated outcomes. Table 1 provides a brief listing of some potential biomarker candidates to consider for further evaluation by MR. Because of the strong in vitro, ex vivo and in vivo data linking the chemokine—chemokine receptor system to HIV-AIDS pathogenesis, we discuss CCR5 and its ligands (primarily CCL3L/CCL4L) as examples. We also discuss another gene, Duffy Antigen Receptor for Chemokines (DARC) as it is a chemokine binding receptor and variations in this gene may potentially impact on levels of chemokines and other biomarkers relevant to HIV-associated outcomes.
Table 1.
Selected biomarkers (B) and genetic instruments (G) that may have utility for evaluation in an MR study for HIV-associated outcomes.
| Category | Biomarker (B) | Genetic instruments (G) | Genetic consequence on biomarkers (G→B) | Genetic Effect on HIV disease outcome (G→D) | References |
|---|---|---|---|---|---|
| Chemokine receptor | CCR5 | CCR5 −Δ32 (rs333) CCR5 haplotypes (e.g. P1/HHE) | Altered CCR5 surface expression | Associations with HIV-AIDS susceptibility [including surrogates of disease (CD4, VL)] and immune reconstitution during HAART. | [15–19,48,57,58] |
| Chemokines | CCL3L, CCL4L | CCL3L and CCL4L various gene copy number. | Increased gene dose of CCL3L1-containing segmental duplication associates with increased protein levels (the relationship is not linear, reaching a plateau at high gene dose) | Associations of CCL3L1-containing segmental duplications with HIV-AIDS susceptibility and immune reconstitution. Specific combinatorial content of CCL3L and CCL4L genes associate with HIV-AIDS outcomes. CCL4L gene copy number associates with HIV susceptibility. | [28–30,42,59–61] |
| CCL5 | CCL5 −471G>A (rs2107538) CCL5 In1.1T>C (rs2280789) | Altered transcription and possibly protein levels | Associations with HIV-AIDS susceptibility | [62–66] | |
| CCL2 | CCL2 −2578 A>G (rs4795895) | Altered protein expression | Associations with HIV susceptibility and disease progression including HIV-associated dementia | [67,68] | |
| CCL2, CCL5 | DARC Asp42Gly (rs12075) | Altered serum level of CCL2 (MCP-1), CCL5 (RANTES) and IL-8; SNP accounts for ~20% of the variability in serum CCL2 levels | Not studied | [69] | |
| CCL4 | CCL4L2 rs4796217 C>T | Minor allele significantly associates with a low plasma level of CCL4 (MIPβ) | Not studied | [70] | |
| Peripheral blood cell counts | CD4+ T cell counts | MHC locus rs2524054 C>A | Altered CD4 levels and CD4/CD8 ratio in HIV negative individuals. Explains ~5.7% variation of CD4/CD8 ratio | The A allele of rs2524054 strongly associates with viremia when HIV controllers were compared to a group of HIV-1 progressors | [71] |
| WBC, neutrophils | DARC −46T>C (rs2814778) | Low WBC and neutrophil counts, and at the population level, explains ~27% and ~20% of the variation in neutrophil and WBC counts, respectively. | African-specific DARC −46C/C associates with an increased risk of acquiring HIV but slow disease progression. The latter association occurs mainly in those African Americans who are also leukopenic. | [72–75] | |
| Platelet counts | ATXN2 rs11065987 A>G | Minor allele associates with altered platelet counts | Not studied | [76] | |
| Cytokines/inflammatory markers | IL-6 | IL-6 −174 G>C (rs1800795) | Associates with altered IL-6 and CRP levels | Altered risk of KS development and variable recovery of CD4 cells during HAART | [77–80] |
| IL-6sR | IL-6R rs4129267 C>T | Associates with plasma levels of IL-6sR | Not studied | [70] | |
| IL-10 | IL-10 −592C>A (rs1800872) | Decreased IL-10 levels | Increased HIV-AIDS susceptibility | [81–84] | |
| IL-18 | IL-18 rs2250417 A>G | Minor allele associates with low plasma IL-18 | Not studied | [70] | |
| TNFα | ABO rs505922 T>C | Minor allele associates with low levels of TNFα | Not studied | [70] | |
| CRP | CRP rs7553007 G>A | Minor allele associates lower CRP levels | Not studied | [14] | |
| Fibrinogen | FGB rs1800789 G>A | Minor allele associates with high fibrinogen and explains less than 2% of variations in levels | Not studied | [85] |
Abbreviations: CCR5, CC chemokine receptor 5; CCL, CC ligand; DARC, Duffy Antigen Receptor for Chemokines; WBC, white blood cells; VL, viral load; IL, interleukin; KS, Kaposi sarcoma, and CRP, C-reactive protein. CCL3L denotes CCL3L-1, -2, and -3 genes; CCL4L denotes CCL4L-1 and CCL4L-2 genes.
CCR5 levels
Surface levels of CCR5, the HIV coreceptor, represent an important biomarker as it impacts on several outcomes. Variable human CCR5 expression is associated with variable HIV cell entry in vitro, HIV acquisition, AIDS progression rates, viral load and CD4 immune reconstitution during HAART [15–19,86–90] Also, CCR5 surface expression influences efficacy of CCR5 blockers and entry inhibitors [91–93], and pre-infection CCR5 levels correlate with rates of disease progression post-infection [94]. The association of low CCR5 levels with reduced AIDS susceptibility also has an evolutionary correlate: high vs. low CCR5 surface expression distinguishes pathogenic (e.g. human) vs non-pathogenic lentiviral (SIV) infection in some non-human primates, with low CCR5 levels associating with non-pathogenic infection as seen in Sooty Mangabey [95,96]. Studies have identified polymorphisms in the CCR5 coding and promoter regions that influence both CCR5 expression and HIV-AIDS susceptibility as well as CD4 recovery during HAART [15–19,42,48,57,58,61,86–90,97–105]. Thus, at many levels, the assumptions of Mendelian randomization are met in the case of CCR5. However, evaluation of CCR5 variations in MR studies requires special considerations: CCR5 SNPs display complex linkage disequilibrium patterns [97]; the associations of CCR5 haplotypes are race-specific and there is a strong interactive effect between CCR5 haplotypes with different evolutionary histories [57]; and, there is evidence for gene gene interaction. For example, a recent study demonstrated that the interaction between the CCR5 Δ32 mutation and variations in the IL-12B gene influence HIV disease progression [106]
CCR5 ligand levels
Supporting the strong in vitro results that CCR5 ligands have HIV suppressive properties, extensive data demonstrates that ex vivo expression of CCR5 ligands such as CCL3, CCL4 and CCL5 serve as a biomarker for HIV-AIDS susceptibility, with higher levels associating with beneficial effects [107–113]. However, there is a cautionary note: the biomarker potential of CCR5 ligands requires consideration of the fact that although antiviral immunity is afforded by release of HIV-suppressive CCR5 ligands from activated leukocytes, serum levels of these biomarkers may correlate poorly with HIV-AIDS outcomes [114] and high levels of chemokines by imparting pro-inflammatory effects [115] may accelerate disease progression.
Among the CCR5 ligands, CCL3L1 displays the maximal HIV-suppressive effects in vitro [116,117], suggesting that it might play a key role in HIV outcomes in vivo. Notably, CCL3L genes (CCL3L-1, -2, and -3) are subject to copy number variation (reviewed in [30,118]), with higher copies associating with increased CCL3L1 expression [59,119], and enhanced HIV-specific T cell responses [120]. These in vitro biological data along with the in vivo observations demonstrating a strong association between higher CCR5 ligand expression and reduced HIV-AIDS susceptibility, led investigators to determine the associations of the CCL3L1-containing segmental duplications with HIV-AIDS susceptibility. In several cohorts, a lower copy number of the CCL3L1-containing segmental duplication associated with lower CD4 counts, higher viral load, increased risk of acquiring HIV and faster HIV disease course (reviewed in [30]). A similar association of increased AIDS progression rates following experimental infection with SIV was observed in rhesus macaques with low CCL3L gene copies [121]. In some cohorts these associations were not observed, and some of the reasons for this discrepancy have been discussed [28,30]. Analogous to the associations of the copy number of the CCL3L1-containing segmental duplication, an association between the copy number of CCL4L genes and HIV susceptibility has also been reported [30]. Thus, multiple lines of evidence suggest that variations in CCR5 chemokine genes that influence expression levels may serve as a useful instrument to test the role for chemokines in HIV-AIDS susceptibility using the MR approach. However, a special consideration for evaluation of CCL3L/CCL4L gene dose in MR studies is the complex genomic architecture of this locus because it is likely that it is the combinatorial content of these genes that associate with HIV-AIDS outcomes[30].
DARC expression: relevance for biomarker research relevant to HIV
Duffy Antigen Receptor for Chemokines (DARC) is a CC and CXC chemokine binding protein that is highly expressed on red blood cells and endothelial cells [122]. The functions of DARC have relevance for evaluation of chemokines and other biomarkers with HIV outcome. Foremost, many chemokine-related functions have been proposed for erythroid DARC, ranging from its serving as a sink or reservoir for several chemokines (e.g., IL-8 and RANTES), as well as maintenance of chemokine plasma concentrations [reviewed in [122]]. Two polymorphisms in the DARC gene have relevance to HIV biomarker research. A close relationship between erythrocyte DARC and chemokine levels is underscored by a genome-wide association study showing that a polymorphism (Asp42Gly) in the DARC coding region is a major regulator of the DARC-mediated erythrocyte binding of CCL2, IL-8 and RANTES and in turn, the circulating serum levels of these proteins [69]. Second, is an African-specific genotype in the DARC promoter (−46C/C) that results in selective loss of expression of DARC on red blood cells (Duffy/DARC-null trait on erythroid cells) and resistance to Plasmodium vivax malaria [122]. A comparison of healthy subjects with and without DARC −46C/C revealed differences in circulating levels of chemokines, as well as coagulation responses to intravenously administered endotoxin, with Duffy-null subjects having, in general, reduced responses [123,124]. In light of these findings that DARC expression may modify the chemokine and coagulation responses to experimental endotoxemia it will be important to evaluate whether the Duffy-null state influences LPS levels during HIV disease.
In addition to impacting on chemokine homeostasis, DARC expression may also influence cellular biomarkers relevant to HIV-AIDS pathogenesis. There are striking inter-racial differences in white blood cell (WBC) counts, a cellular biomarker. Compared to persons of European ancestry, individuals of African descent have lower WBC counts, attributable mainly to reduced neutrophil counts [125–128]. This hematological condition is referred to as “ethnic leukopenia or neutropenia” [125–129]. Although generally thought to be without consequence, inter-racial differences in WBC counts may influence treatment outcomes [129] and infectious disease susceptibility [130]. Conclusive data now demonstrates that the Duffy-null mutation is causal for the leukopenia/neutropenia found commonly in persons of African ancestry [72,73]. Whether the Duffy null-associated neutropenia influences HIV-AIDS pathogenesis merits further investigation for several reasons. HIV-negative healthy subjects with low neutrophil counts have elevated CRP levels [131], and whether this association is a correlate of HIV acquisition is unknown. Also, Detels et al (1) demonstrated that resistance to acquiring HIV in men followed prospectively in the Multicenter AIDS Cohort Study (MACS) was associated with high neutrophil and CD8+ T cell counts (1) and He et al showed that the Duffy-null state associated with an increased risk of acquiring HIV infection [74]. In light of the aforementioned findings, one possibility is that the association of Duffy-null state with increased HIV risk may reflect the association of this trait with low neutrophil counts, or that the DARC-null state associates with increased HIV risk specifically in those persons with this trait who also have low neutrophil counts. However, additional mechanisms may be operative as suggested by the binding of HIV-suppressive and inflammatory chemokines [74,122] and HIV-1 viral particles to DARC on RBC [74].
The recently described association of the DARC-null state with low WBC counts in persons of African descent [73] prompted investigators to evaluate further the relationships among the DARC-null state, leukopenia and HIV disease course in HIV-positive European Americans and African Americans [74,75]. They found that although leukopenia occurs frequently during HIV infection, its influence on disease outcome differs depending on both race and DARC genotype [75]. In HIV-infected European Americans, leukopenia was associated with an accelerated disease course. By contrast, the association of Duffy-null state with HIV disease course differed according to WBC but not CD4+ T-cell counts, such that leukopenic but not nonleukopenic HIV-positive African Americans with the Duffy-null trait had a survival advantage compared with all Duffy-positive subjects. This survival advantage became increasingly pronounced in those with progressively lower WBC counts. [75]. These findings suggested that in African Americans, the DARC genotype may influence HIV disease course within the context of a specific cellular milieu, as reflected by leukocyte cell counts.
It is important to note that others did not identify an association of DARC genotype with HIV disease course [132–135]. However, as posited by He et al in their riposte to these correspondences, these disparate findings may in part be explained by differences in cohort characteristics and study design, including study endpoints but not population stratification as suggested by some [19,132–135]. Further it is possible that the recently described relationships between DARC genotype, WBC counts and HIV disease progression [75] may have played a role in these null associations and may help account for the different findings reported in these cohorts [74,132–135].
Taken together, much more needs to be learnt about DARC function and biology in health and disease. Nonetheless, these observations collectively highlight the importance of being mindful that DARC expression may impact on evaluation of cellular (WBC and neutrophil counts) and inflammatory (chemokine) biomarkers relevant to HIV-associated outcomes as well as influence the chemokine and coagulation response to endotoxemia.
Conclusions
The biomarker field in HIV medicine is clearly at a very exciting point. The hope is that some of these biomarkers would be identified as risk factors for HIV-associated outcomes and could be used for both risk assessment and to individualize specific therapies. We review briefly the conceptual framework, strengths, and limitations of MR, an application that has been employed in other fields with some success to evaluate the possible causal role of biomarkers.
However, MR is not, and should not be viewed as a panacea, and the limitations of MR should be clearly acknowledged. Although MR can make a distinctive contribution to the epidemiology of biomarkers and HIV-associated outcomes, it is important to emphasize what MR is not. (a) MR is not fundamentally about discovering how genetic variation influences HIV-associated outcomes. In the context in which we have discussed MR, genotype is used strictly as a proxy for a biomarker. (b) MR is not a strategy for detecting genes that confer a higher risk of HIV-associated outcomes. (c) MR is an epidemiologic tool, not a molecular or physiologic inquiry into underlying mechanism of HIV-associated outcomes, albeit it may indirectly provide information regarding potentially critical pathways underlying HIV-associated outcomes. HIV infection may further constrain the ability to meet the assumptions of MR.
Nonetheless, with full acknowledgement of the limitations and caveats related to MR, and under the appropriate clinical settings where many confounding factors can be accounted for (e.g. SMART study), and including especially the availability of a genetic instrument that is both strongly associated with the biomarker and independent of HIV-associated outcome, MR can complement the findings from traditional observational and functional studies. We envisage that large scale genetic studies and MR as highlighted by the studies of Elliott et al on CRP and CHD [14] may be necessary for definitive evaluation of the causal role of biomarkers in HIV-associated outcomes. Together these complementary strategies offer the potential of identifying which biomarkers may be useful as specific drug targets and have a role in risk assessment and therapeutic selection. Undoubtedly, better biomarkers and innovative strategies for combining them will be needed, along with statistical and functional evaluation of causality, to fulfill the promise of biomarkers for personalized medicine.
Acknowledgements
We thank Drs. Robert Clark and Gabriel Catano for critical reading of the manuscript, and Robert Maldonado and Una Aluyen for graphical work. The opinions or ascertains contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Air Force or the Department of Defense.
This work was supported by the Veterans Administration HIV/AIDS Center of the South Texas Veterans Health Care System, NIH (R37046326), and the Doris Duke Distinguished Clinical Scientist Award to S.K.A. S.K.A. is also supported by a VA MERIT award and the Burroughs Wellcome Clinical Scientist Award in Translational Research.
Footnotes
The authors declare no conflict of interest.
Reference
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Neaton J, Emery S, Neuhaus J. Soluble biomarkers and morbidity and mortality among people infected with HIV: summary of published resports from 1997–2010. Current Opinion in HIV and AIDS. 2010;5 doi: 10.1097/COH.0b013e32833ed75d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker J, Duprez D. Biomarkers and HIV-associated cardiovascular disease. Current Opinion in HIV and AIDS. 2010;5 doi: 10.1097/COH.0b013e32833ed7ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worm S, Hsue P. Role of biomarkers in predicting CVD risk in the setting of HIV infection? Current Opinion in HIV and AIDS. 2010;5 doi: 10.1097/COH.0b013e32833f2ea6. [DOI] [PubMed] [Google Scholar]
- 4.Niton D, Landay A. Biomarkers of immune dysfunction in HIV. Current Opinion in HIV and AIDS. 2010 doi: 10.1097/COH.0b013e32833ed6f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas DC, Conti DV. Commentary: the concept of 'Mendelian Randomization'. Int J Epidemiol. 2004;33:21–25. doi: 10.1093/ije/dyh048. [DOI] [PubMed] [Google Scholar]
- 6.Davey Smith G, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 7.Ebrahim S, Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum Genet. 2008;123:15–33. doi: 10.1007/s00439-007-0448-6. [DOI] [PubMed] [Google Scholar]
- 8.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 9.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16:309–330. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 10.Sheehan NA, Didelez V, Burton PR, Tobin MD. Mendelian randomisation and causal inference in observational epidemiology. PLoS Med. 2008;5:e177. doi: 10.1371/journal.pmed.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bochud M, Rousson V. Usefulness of Mendelian randomization in observational epidemiology. Int J Environ Res Public Health. 2010;7:711–728. doi: 10.3390/ijerph7030711. ** Introduction, theoretical basis, assumptions, challenges, limitations and literature review of Mendelian randomization approach.
- 12.Verduijn M, Siegerink B, Jager KJ, Zoccali C, Dekker FW. Mendelian randomization: use of genetics to enable causal inference in observational studies. Nephrol Dial Transplant. 2010;25:1394–1398. doi: 10.1093/ndt/gfq098. [DOI] [PubMed] [Google Scholar]
- 13.Glynn RJ. Promises and limitations of mendelian randomization for evaluation of biomarkers. Clin Chem. 2010;56:388–390. doi: 10.1373/clinchem.2009.142513. [DOI] [PubMed] [Google Scholar]
- 14. Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, Erdmann J, Braund P, Engert JC, Bennett D, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. Jama. 2009;302:37–48. doi: 10.1001/jama.2009.954. ** Comprehensive study that used extensive data from genome wide-association study approaches and Mendelian randomization to evaluate whether CRP is causal for coronary heart disease. This approach is a case-study for MR.
- 15.Kaslow RA, Dorak T, Tang JJ. Influence of host genetic variation on susceptibility to HIV type 1 infection. J Infect Dis. 2005;191(Suppl 1):S68–S77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- 16.Hunt P, Carrington M. Host genetic determinants of HIV pathogenesis: an immunologic perspective. Immune correlates of protection, activation and exhaustion. Current Opinion in HIV & AIDS. 2008;3:342–348. doi: 10.1097/COH.0b013e3282fbaa92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fellay J. Host genetics influences on HIV type-1 disease. Antivir Ther. 2009;14:731–738. doi: 10.3851/IMP1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Telenti A, Carrington M. Host factors associated with outcome from primary human immunodeficiency virus-1 infection. Current Opinion in HIV & AIDS. 2008;3:28–35. doi: 10.1097/COH.0b013e3282f18ac0. [DOI] [PubMed] [Google Scholar]
- 19.An P, Winkler CA. Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet. 2010;26:119–131. doi: 10.1016/j.tig.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nitsch D, Molokhia M, Smeeth L, DeStavola BL, Whittaker JC, Leon DA. Limits to causal inference based on Mendelian randomization: a comparison with randomized controlled trials. Am J Epidemiol. 2006;163:397–403. doi: 10.1093/aje/kwj062. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Davey Smith G, Harbord RM, Lewis SJ. Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS Med. 2008;5:e52. doi: 10.1371/journal.pmed.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Wang M, Irigoyen P, Gregersen PK. Inferring causal relationships among intermediate phenotypes and biomarkers: a case study of rheumatoid arthritis. Bioinformatics. 2006;22:1503–1507. doi: 10.1093/bioinformatics/btl100. [DOI] [PubMed] [Google Scholar]
- 23.Katan MB. Apolipoprotein E isoforms, serum cholesterol, and cancer. Lancet. 1986;1:507–508. doi: 10.1016/s0140-6736(86)92972-7. [DOI] [PubMed] [Google Scholar]
- 24.Greenland S. An introduction to instrumental variables for epidemiologists. Int J Epidemiol. 2000;29:722–729. doi: 10.1093/ije/29.4.722. [DOI] [PubMed] [Google Scholar]
- 25.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry GH, Ben-Dor A, Tsalenko A, Sampas N, Rodriguez-Revenga L, Tran CW, Scheffer A, Steinfeld I, Tsang P, Yamada NA, et al. The fine-scale and complex architecture of human copy-number variation. Am J Hum Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alkan C, Kidd JM, Marques-Bonet T, Aksay G, Antonacci F, Hormozdiari F, Kitzman JO, Baker C, Malig M, Mutlu O, et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat Genet. 2009 doi: 10.1038/ng.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He W, Kulkarni H, Castiblanco J, Shimizu C, Aluyen U, Maldonado R, Carrillo A, Griffin M, Lipsitt A, Beachy L, et al. Reply to:"Experimental aspects of copy number variant assays at CCL3L1". Nat Med. 2009;15:1117–1120. doi: 10.1038/nm1009-1117. [DOI] [PubMed] [Google Scholar]
- 29. Shostakovich-Koretskaya L, Catano G, Chykarenko ZA, He W, Gornalusse G, Mummidi S, Sanchez R, Dolan MJ, Ahuja SS, Clark RA, et al. Combinatorial content of CCL3L and CCL4L gene copy numbers influence HIV-AIDS susceptibility in Ukrainian children. Aids. 2009;23:679–688. doi: 10.1097/QAD.0b013e3283270b3f. *This study shows that both CCL3L1 and CCL4L gene copy number as well as their combinatorial content at the 17q11 chemokine gene enriched copy number variation region influences HIV-AIDS susceptibility.
- 30. Colobran R, Pedrosa E, Carretero-Iglesia L, Juan M. Copy number variation in chemokine superfamily: the complex scene of CCL3L–CCL4L genes in health and disease. Clin Exp Immunol. 2010 doi: 10.1111/j.1365-2249.2010.04224.x. **This review highlights the complexity of the genetic architecture of the 17q12 chemokine gene rich region and summarizes the associations for HIV-AIDS and other diseases for CCL3L and CCL4L chemokine genes.
- 31.Vickers MA, Green FR, Terry C, Mayosi BM, Julier C, Lathrop M, Ratcliffe PJ, Watkins HC, Keavney B. Genotype at a promoter polymorphism of the interleukin-6 gene is associated with baseline levels of plasma C-reactive protein. Cardiovasc Res. 2002;53:1029–1034. doi: 10.1016/s0008-6363(01)00534-x. [DOI] [PubMed] [Google Scholar]
- 32.Walston JD, Fallin MD, Cushman M, Lange L, Psaty B, Jenny N, Browner W, Tracy R, Durda P, Reiner A. IL-6 gene variation is associated with IL-6 and C-reactive protein levels but not cardiovascular outcomes in the Cardiovascular Health Study. Hum Genet. 2007;122:485–494. doi: 10.1007/s00439-007-0428-x. [DOI] [PubMed] [Google Scholar]
- 33.Berrahmoune H, Herbeth B, Siest G, Visvikis-Siest S. Heritability of serum hs-CRP concentration and 5-year changes in the Stanislas family study: association with apolipoprotein E alleles. Genes Immun. 2007;8:352–359. doi: 10.1038/sj.gene.6364395. [DOI] [PubMed] [Google Scholar]
- 34.Lange LA, Burdon K, Langefeld CD, Liu Y, Beck SR, Rich SS, Freedman BI, Brosnihan KB, Herrington DM, Wagenknecht LE, et al. Heritability and expression of C-reactive protein in type 2 diabetes in the Diabetes Heart Study. Ann Hum Genet. 2006;70:717–725. doi: 10.1111/j.1469-1809.2006.00280.x. [DOI] [PubMed] [Google Scholar]
- 35. Cole SW, Arevalo JM, Takahashi R, Sloan EK, Lutgendorf SK, Sood AK, Sheridan JF, Seeman TE. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci U S A. 2010;107:5681–5686. doi: 10.1073/pnas.0911515107. **This study highlights gene x environment interactions for a functional SNP in the promoter of IL-6 gene that influences IL-6 levels.
- 36.Worns MA, Victor A, Galle PR, Hohler T. Genetic and environmental contributions to plasma C-reactive protein and interleukin-6 levels--a study in twins. Genes Immun. 2006;7:600–605. doi: 10.1038/sj.gene.6364330. [DOI] [PubMed] [Google Scholar]
- 37.Su S, Snieder H, Miller AH, Ritchie J, Bremner JD, Goldberg J, Dai J, Jones L, Murrah NV, Zhao J, et al. Genetic and environmental influences on systemic markers of inflammation in middle-aged male twins. Atherosclerosis. 2008;200:213–220. doi: 10.1016/j.atherosclerosis.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rahman I, Bennet AM, Pedersen NL, de Faire U, Svensson P, Magnusson PK. Genetic dominance influences blood biomarker levels in a sample of 12,000 Swedish elderly twins. Twin Res Hum Genet. 2009;12:286–294. doi: 10.1375/twin.12.3.286. [DOI] [PubMed] [Google Scholar]
- 39.Ogbuanu IU, Zhang H, Karmaus W. Can we apply the Mendelian randomization methodology without considering epigenetic effects? Emerg Themes Epidemiol. 2009;6:3. doi: 10.1186/1742-7622-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, Urban TJ, Zhang K, Gumbs CE, Smith JP, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. ** This study represents a comprehensive assessment of common human genetic variation in HIV-1 control in Caucasians.
- 41.Catano G, Kulkarni H, He W, Marconi VC, Agan BK, Landrum M, Anderson S, Delmar J, Telles V, Song L, et al. HIV-1 disease-influencing effects associated with ZNRD1, HCP5 and HLA-C alleles are attributable mainly to either HLA-A10 or HLA-B*57 alleles. PLoS ONE. 2008;3:e3636. doi: 10.1371/journal.pone.0003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dolan MJ, Kulkarni H, Camargo JF, He W, Smith A, Anaya JM, Miura T, Hecht FM, Mamtani M, Pereyra F, et al. CCL3L1 and CCR5 influence cell-mediated immunity and affect HIV-AIDS pathogenesis via viral entry-independent mechanisms. Nat Immunol. 2007;8:1324–1336. doi: 10.1038/ni1521. * Illustrates that genetic variations may influence AIDS pathogenesis by impacting on viral load-dependent and –independent parameters such as cell-mediated immunity.
- 43.Henry WK, Tebas P, Lane HC. Explaining, predicting, and treating HIV-associated CD4 cell loss: after 25 years still a puzzle. Jama. 2006;296:1523–1525. doi: 10.1001/jama.296.12.1523. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez B, Sethi AK, Cheruvu VK, Mackay W, Bosch RJ, Kitahata M, Boswell SL, Mathews WC, Bangsberg DR, Martin J, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. Jama. 2006;296:1498–1506. doi: 10.1001/jama.296.12.1498. [DOI] [PubMed] [Google Scholar]
- 45.Ness RB, Haggerty CL, Harger G, Ferrell R. Differential distribution of allelic variants in cytokine genes among African Americans and White Americans. Am J Epidemiol. 2004;160:1033–1038. doi: 10.1093/aje/kwh325. [DOI] [PubMed] [Google Scholar]
- 46.Smith AJ, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev. 2009;20:43–59. doi: 10.1016/j.cytogfr.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Gao X, Bashirova A, Iversen AK, Phair J, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Altfeld M, O'Brien SJ, et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. 2005;11:1290–1292. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- 48.Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, Lee B, Doms RW, Margolick J, Buchbinder S, et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 49.Portales P, Clot J, Corbeau P. Sex differences in HIV-1 viral load due to sex difference in CCR5 expression. Ann Intern Med. 2001;134:81–82. doi: 10.7326/0003-4819-134-1-200101020-00023. [DOI] [PubMed] [Google Scholar]
- 50.Planchard D, Loriot Y, Goubar A, Commo F, Soria JC. Differential expression of biomarkers in men and women. Semin Oncol. 2009;36:553–565. doi: 10.1053/j.seminoncol.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Koesters SA, Matu L, Kiama P, Anzala O, Embree J, Plummer FA, Kimani J, Fowke KR. Elevation of immune activation in kenyan women is associated with alterations in immune function: implications for vaccine development. J Clin Immunol. 2004;24:702–709. doi: 10.1007/s10875-004-6238-1. [DOI] [PubMed] [Google Scholar]
- 52.Eggena MP, Barugahare B, Okello M, Mutyala S, Jones N, Ma Y, Kityo C, Mugyenyi P, Cao H. T cell activation in HIV-seropositive Ugandans: differential associations with viral load, CD4+ T cell depletion, and coinfection. J Infect Dis. 2005;191:694–701. doi: 10.1086/427516. [DOI] [PubMed] [Google Scholar]
- 53.Kalinkovich A, Borkow G, Weisman Z, Tsimanis A, Stein M, Bentwich Z. Increased CCR5 and CXCR4 expression in Ethiopians living in Israel: environmental and constitutive factors. Clin Immunol. 2001;100:107–117. doi: 10.1006/clim.2001.5040. [DOI] [PubMed] [Google Scholar]
- 54.Zabaleta J, Schneider BG, Ryckman K, Hooper PF, Camargo MC, Piazuelo MB, Sierra RA, Fontham ET, Correa P, Williams SM, et al. Ethnic differences in cytokine gene polymorphisms: potential implications for cancer development. Cancer Immunol Immunother. 2008;57:107–114. doi: 10.1007/s00262-007-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaleebu P, French N, Mahe C, Yirrell D, Watera C, Lyagoba F, Nakiyingi J, Rutebemberwa A, Morgan D, Weber J, et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002;185:1244–1250. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- 56.Vasan A, Renjifo B, Hertzmark E, Chaplin B, Msamanga G, Essex M, Fawzi W, Hunter D. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis. 2006;42:843–852. doi: 10.1086/499952. [DOI] [PubMed] [Google Scholar]
- 57.Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, Cabrera S, McBride M, Cao XH, Merrill G, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci U S A. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh P, Kaur G, Sharma G, Mehra NK. Immunogenetic basis of HIV-1 infection, transmission and disease progression. Vaccine. 2008;26:2966–2980. doi: 10.1016/j.vaccine.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 60.Huik K, Sadam M, Karki T, Avi R, Krispin T, Paap P, Ruutel K, Uuskula A, Talu A, Abel-Ollo K, et al. CCL3L1 copy number is a strong genetic determinant of HIV seropositivity in Caucasian intravenous drug users. J Infect Dis. 2010;201:730–739. doi: 10.1086/650491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahuja SK, Kulkarni H, Catano G, Agan BK, Camargo JF, He W, O'Connell RJ, Marconi VC, Delmar J, Eron J, et al. CCL3L1-CCR5 genotype influences durability of immune recovery during antiretroviral therapy of HIV-1-infected individuals. Nat Med. 2008;14:413–420. doi: 10.1038/nm1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu H, Chao D, Nakayama EE, Taguchi H, Goto M, Xin X, Takamatsu JK, Saito H, Ishikawa Y, Akaza T, et al. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci U S A. 1999;96:4581–4585. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McDermott DH, Beecroft MJ, Kleeberger CA, Al-Sharif FM, Ollier WE, Zimmerman PA, Boatin BA, Leitman SF, Detels R, Hajeer AH, et al. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. Aids. 2000;14:2671–2678. doi: 10.1097/00002030-200012010-00006. [DOI] [PubMed] [Google Scholar]
- 64.Gonzalez E, Dhanda R, Bamshad M, Mummidi S, Geevarghese R, Catano G, Anderson SA, Walter EA, Stephan KT, Hammer MF, et al. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci U S A. 2001;98:5199–5204. doi: 10.1073/pnas.091056898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.An P, Nelson GW, Wang L, Donfield S, Goedert JJ, Phair J, Vlahov D, Buchbinder S, Farrar WL, Modi W, et al. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci U S A. 2002;99:10002–10007. doi: 10.1073/pnas.142313799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duggal P, Winkler CA, An P, Yu XF, Farzadegan H, O'Brien SJ, Beaty TH, Vlahov D. The effect of RANTES chemokine genetic variants on early HIV-1 plasma RNA among African American injection drug users. J Acquir Immune Defic Syndr. 2005;38:584–589. doi: 10.1097/01.qai.0000134741.49208.03. [DOI] [PubMed] [Google Scholar]
- 67.Modi WS, Goedert JJ, Strathdee S, Buchbinder S, Detels R, Donfield S, O'Brien SJ, Winkler C. MCP-1–MCP-3-Eotaxin gene cluster influences HIV-1 transmission. Aids. 2003;17:2357–2365. doi: 10.1097/00002030-200311070-00011. [DOI] [PubMed] [Google Scholar]
- 68.Gonzalez E, Rovin BH, Sen L, Cooke G, Dhanda R, Mummidi S, Kulkarni H, Bamshad MJ, Telles V, Anderson SA, et al. HIV-1 infection and AIDS dementia are influenced by a mutant MCP-1 allele linked to increased monocyte infiltration of tissues and MCP-1 levels. Proc Natl Acad Sci U S A. 2002;99:13795–13800. doi: 10.1073/pnas.202357499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schnabel RB, Baumert J, Barbalic M, Dupuis J, Ellinor PT, Durda P, Dehghan A, Bis JC, Illig T, Morrison AC, et al. Duffy antigen receptor for chemokines (Darc) polymorphism regulates circulating concentrations of monocyte chemoattractant protein-1 and other inflammatory mediators. Blood. 2009 doi: 10.1182/blood-2009-05-221382. *This study found a polymorphism (Asp42Gly) in the coding region of the DARC gene is a major regulator of CCL2 and CCL5 and IL-8 serum level, it accounted for approximately 20% of the variability in serum MCP-1 concentrations.
- 70.Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, Rafferty I, Lauretani F, Murray A, Gibbs JR, Paolisso G, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ferreira MA, Mangino M, Brumme CJ, Zhao ZZ, Medland SE, Wright MJ, Nyholt DR, Gordon S, Campbell M, McEvoy BP, et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am J Hum Genet. 2010;86:88–92. doi: 10.1016/j.ajhg.2009.12.008. **This study found a polymorphism in the MHC locus that associated with CD4+ T cell counts and CD4/CD8 T cell ratio in HIV-negative subjects. This polymorphism also associated with viral load in HIV-positive subjects.
- 72. Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, Mullikin J, Hsueh WC, Cheng CY, Coresh J, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. **This study using an exhaustive admixture mapping approach and demonstrated that a polymorphism in the promoter region of DARC is highly predictive of WBC and neutrophil counts in persons of African descent.
- 73.Nalls MA, Wilson JG, Patterson NJ, Tandon A, Zmuda JM, Huntsman S, Garcia M, Hu D, Li R, Beamer BA, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am J Hum Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He W, Neil S, Kulkarni H, Wright E, Agan BK, Marconi VC, Dolan MJ, Weiss RA, Ahuja SK. Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility. Cell Host Microbe. 2008;4:52–62. doi: 10.1016/j.chom.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kulkarni H, Marconi VC, He W, Landrum ML, Okulicz JF, Delmar J, Kazandjian D, Castiblanco J, Ahuja SS, Wright EJ, et al. The Duffy-null state is associated with a survival advantage in leukopenic HIV-infected persons of African ancestry. Blood. 2009;114:2783–2792. doi: 10.1182/blood-2009-04-215186. *This study showed that the interaction between DARC genotype and the cellular milieu defined by white blood cell counts may influence HIV disease course.
- 76.Soranzo N, Spector TD, Mangino M, Kuhnel B, Rendon A, Teumer A, Willenborg C, Wright B, Chen L, Li M, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Foster CB, Lehrnbecher T, Samuels S, Stein S, Mol F, Metcalf JA, Wyvill K, Steinberg SM, Kovacs J, Blauvelt A, et al. An IL6 promoter polymorphism is associated with a lifetime risk of development of Kaposi sarcoma in men infected with human immunodeficiency virus. Blood. 2000;96:2562–2567. [PubMed] [Google Scholar]
- 78.Fernandez S, Rosenow AA, James IR, Roberts SG, Nolan RC, French MA, Price P. Recovery of CD4+ T Cells in HIV patients with a stable virologic response to antiretroviral therapy is associated with polymorphisms of interleukin-6 and central major histocompatibility complex genes. J Acquir Immune Defic Syndr. 2006;41:1–5. doi: 10.1097/01.qai.0000188990.57760.e3. [DOI] [PubMed] [Google Scholar]
- 79.Nattermann J, Vogel M, Berg T, Danta M, Axel B, Mayr C, Bruno R, Tural C, Klausen G, Clotet B, et al. Effect of the interleukin-6 C174G gene polymorphism on treatment of acute and chronic hepatitis C in human immunodeficiency virus coinfected patients. Hepatology. 2007;46:1016–1025. doi: 10.1002/hep.21778. [DOI] [PubMed] [Google Scholar]
- 80.Saumoy M, Lopez-Dupla M, Veloso S, Alonso-Villaverde C, Domingo P, Broch M, Miranda M, Coll B, Sauri A, Vendrell J, et al. The IL-6 system in HIV-1-infection and in HAART-related fat redistribution syndromes. Aids. 2008;22:893–896. doi: 10.1097/QAD.0b013e3282f4dde7. [DOI] [PubMed] [Google Scholar]
- 81.Mahajan SD, Agosto-Mojica A, Aalinkeel R, Reynolds JL, Nair BB, Sykes DE, Martinez J, Adams J, Singh N, Bernstein Z, et al. Role of chemokine and cytokine polymorphisms in the progression of HIV-1 disease. Biochem Biophys Res Commun. 396:348–352. doi: 10.1016/j.bbrc.2010.04.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Naicker DD, Werner L, Kormuth E, Passmore JA, Mlisana K, Karim SA, Ndung'u T. Interleukin-10 Promoter Polymorphisms Influence HIV-1 Susceptibility and Primary HIV-1 Pathogenesis. J Infect Dis. 2009 doi: 10.1086/600072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shin HD, Winkler C, Stephens JC, Bream J, Young H, Goedert JJ, O'Brien TR, Vlahov D, Buchbinder S, Giorgi J, et al. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci U S A. 2000;97:14467–14472. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oleksyk TK, Shrestha S, Truelove AL, Goedert JJ, Donfield SM, Phair J, Mehta S, O'Brien SJ, Smith MW. Extended IL10 haplotypes and their association with HIV progression to AIDS. Genes Immun. 2009;10:309–322. doi: 10.1038/gene.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dehghan A, Yang Q, Peters A, Basu S, Bis JC, Rudnicka AR, Kavousi M, Chen MH, Baumert J, Lowe GD, et al. Association of novel genetic Loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ Cardiovasc Genet. 2009;2:125–133. doi: 10.1161/CIRCGENETICS.108.825224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 coreceptors--central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res Hum Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 87.Arenzana-Seisdedos F, Parmentier M. Genetics of resistance to HIV infection: Role of co-receptors and co-receptor ligands. Semin Immunol. 2006;18:387–403. doi: 10.1016/j.smim.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 88.Lederman MM, Penn-Nicholson A, Cho M, Mosier D. Biology of CCR5 and its role in HIV infection and treatment. Jama. 2006;296:815–826. doi: 10.1001/jama.296.7.815. [DOI] [PubMed] [Google Scholar]
- 89.Lederman MM, Sieg SF. CCR5 and its ligands: a new axis of evil? Nat Immunol. 2007;8:1283–1285. doi: 10.1038/ni1207-1283. [DOI] [PubMed] [Google Scholar]
- 90.Corbeau P, Reynes J. CCR5 antagonism in HIV infection: ways, effects, and side effects. Aids. 2009;23:1931–1943. doi: 10.1097/QAD.0b013e32832e71cd. [DOI] [PubMed] [Google Scholar]
- 91.Heredia A, Amoroso A, Davis C, Le N, Reardon E, Dominique JK, Klingebiel E, Gallo RC, Redfield RR. Rapamycin causes down-regulation of CCR5 and accumulation of anti-HIV beta-chemokines: an approach to suppress R5 strains of HIV-1. Proc Natl Acad Sci U S A. 2003;100:10411–10416. doi: 10.1073/pnas.1834278100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heredia A, Gilliam B, DeVico A, Le N, Bamba D, Flinko R, Lewis G, Gallo RC, Redfield RR. CCR5 density levels on primary CD4 T cells impact the replication and Enfuvirtide susceptibility of R5 HIV-1. Aids. 2007;21:1317–1322. doi: 10.1097/QAD.0b013e32815278ea. [DOI] [PubMed] [Google Scholar]
- 93.Heredia A, Latinovic O, Gallo RC, Melikyan G, Reitz M, Le N, Redfield RR. Reduction of CCR5 with low-dose rapamycin enhances the antiviral activity of vicriviroc against both sensitive and drug-resistant HIV-1. Proc Natl Acad Sci U S A. 2008;105:20476–20481. doi: 10.1073/pnas.0810843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Roda Husman AM, Blaak H, Brouwer M, Schuitemaker H. CC chemokine receptor 5 cell-surface expression in relation to CC chemokine receptor 5 genotype and the clinical course of HIV-1 infection. J Immunol. 1999;163:4597–4603. [PubMed] [Google Scholar]
- 95.Pandrea I, Apetrei C, Gordon S, Barbercheck J, Dufour J, Bohm R, Sumpter B, Roques P, Marx PA, Hirsch VM, et al. Paucity of CD4+CCR5+ T cells is a typical feature of natural SIV hosts. Blood. 2007;109:1069–1076. doi: 10.1182/blood-2006-05-024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pandrea I, Onanga R, Souquiere S, Mouinga-Ondeme A, Bourry O, Makuwa M, Rouquet P, Silvestri G, Simon F, Roques P, et al. Paucity of CD4+ CCR5+ T cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J Virol. 2008;82:5501–5509. doi: 10.1128/JVI.02555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mummidi S, Bamshad M, Ahuja SS, Gonzalez E, Feuillet PM, Begum K, Galvis MC, Kostecki V, Valente AJ, Murthy KK, et al. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA. Potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem. 2000;275:18946–18961. doi: 10.1074/jbc.M000169200. [DOI] [PubMed] [Google Scholar]
- 98.Shieh B, Liau YE, Hsieh PS, Yan YP, Wang ST, Li C. Influence of nucleotide polymorphisms in the CCR2 gene and the CCR5 promoter on the expression of cell surface CCR5 and CXCR4. Int Immunol. 2000;12:1311–1318. doi: 10.1093/intimm/12.9.1311. [DOI] [PubMed] [Google Scholar]
- 99.Kawamura T, Gulden FO, Sugaya M, McNamara DT, Borris DL, Lederman MM, Orenstein JM, Zimmerman PA, Blauvelt A. R5 HIV productively infects Langerhans cells, and infection levels are regulated by compound CCR5 polymorphisms. Proc Natl Acad Sci U S A. 2003;100:8401–8406. doi: 10.1073/pnas.1432450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Salkowitz JR, Bruse SE, Meyerson H, Valdez H, Mosier DE, Harding CV, Zimmerman PA, Lederman MM. CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-1 propagation in vitro. Clin Immunol. 2003;108:234–240. doi: 10.1016/s1521-6616(03)00147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hladik F, Liu H, Speelmon E, Livingston-Rosanoff D, Wilson S, Sakchalathorn P, Hwangbo Y, Greene B, Zhu T, McElrath MJ. Combined effect of CCR5-Delta32 heterozygosity and the CCR5 promoter polymorphism-2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J Virol. 2005;79:11677–11684. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thomas SM, Tse DB, Ketner DS, Rochford G, Meyer DA, Zade DD, Halkitis PN, Nadas A, Borkowsky W, Marmor M. CCR5 expression and duration of high risk sexual activity among HIV-seronegative men who have sex with men. Aids. 2006;20:1879–1883. doi: 10.1097/01.aids.0000244207.49123.ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O'Brien TR, McDermott DH, Ioannidis JP, Carrington M, Murphy PM, Havlir DV, Richman DD. Effect of chemokine receptor gene polymorphisms on the response to potent antiretroviral therapy. Aids. 2000;14:821–826. doi: 10.1097/00002030-200005050-00008. [DOI] [PubMed] [Google Scholar]
- 104.Hendrickson SL, Jacobson LP, Nelson GW, Phair JP, Lautenberger J, Johnson RC, Kingsley L, Margolick JB, Detels R, Goedert JJ, et al. Host genetic influences on highly active antiretroviral therapy efficacy and AIDS-free survival. J Acquir Immune Defic Syndr. 2008;48:263–271. doi: 10.1097/QAI.0b013e31816fdc5f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 106.Gabutero E, Moore C, Mallal S, Stewart G, Williamson P. Interaction between allelic variation in IL12B and CCR5 affects the development of AIDS: IL12B/CCR5 interaction and HIV/AIDS. Aids. 2007;21:65–69. doi: 10.1097/QAD.0b013e3280117f49. [DOI] [PubMed] [Google Scholar]
- 107.Paxton WA, Martin SR, Tse D, O'Brien TR, Skurnick J, VanDevanter NL, Padian N, Braun JF, Kotler DP, Wolinsky SM, et al. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 108.Zagury D, Lachgar A, Chams V, Fall LS, Bernard J, Zagury JF, Bizzini B, Gringeri A, Santagostino E, Rappaport J, et al. C-C chemokines, pivotal in protection against HIV type 1 infection. Proc Natl Acad Sci U S A. 1998;95:3857–3861. doi: 10.1073/pnas.95.7.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Furci L, Scarlatti G, Burastero S, Tambussi G, Colognesi C, Quillent C, Longhi R, Loverro P, Borgonovo B, Gaffi D, et al. Antigen-driven C-C chemokine-mediated HIV-1 suppression by CD4(+) T cells from exposed uninfected individuals expressing the wild-type CCR-5 allele. J Exp Med. 1997;186:455–460. doi: 10.1084/jem.186.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zagury D, Lachgar A, Chams V, Fall LS, Bernard J, Zagury JF, Bizzini B, Gringeri A, Santagostino E, Rappaport J, et al. Interferon alpha and Tat involvement in the immunosuppression of uninfected T cells and C-C chemokine decline in AIDS. Proc Natl Acad Sci U S A. 1998;95:3851–3856. doi: 10.1073/pnas.95.7.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ullum H, Cozzi Lepri A, Victor J, Aladdin H, Phillips AN, Gerstoft J, Skinhoj P, Pedersen BK. Production of beta-chemokines in human immunodeficiency virus (HIV) infection: evidence that high levels of macrophage inflammatory protein-1beta are associated with a decreased risk of HIV disease progression. J Infect Dis. 1998;177:331–336. doi: 10.1086/514192. [DOI] [PubMed] [Google Scholar]
- 112.Garzino-Demo A, Moss RB, Margolick JB, Cleghorn F, Sill A, Blattner WA, Cocchi F, Carlo DJ, DeVico AL, Gallo RC. Spontaneous and antigen-induced production of HIV-inhibitory beta-chemokines are associated with AIDS-free status. Proc Natl Acad Sci U S A. 1999;96:11986–11991. doi: 10.1073/pnas.96.21.11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cocchi F, DeVico AL, Yarchoan R, Redfield R, Cleghorn F, Blattner WA, Garzino-Demo A, Colombini-Hatch S, Margolis D, Gallo RC. Higher macrophage inflammatory protein (MIP)-1alpha and MIP-1beta levels from CD8+ T cells are associated with asymptomatic HIV-1 infection. Proc Natl Acad Sci U S A. 2000;97:13812–13817. doi: 10.1073/pnas.240469997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ye P, Kazanjian P, Kunkel SL, Kirschner DE. Lack of good correlation of serum CC-chemokine levels with human immunodeficiency virus-1 disease stage and response to treatment. J Lab Clin Med. 2004;143:310–319. doi: 10.1016/j.lab.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 115.Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- 116.Nibbs RJ, Yang J, Landau NR, Mao JH, Graham GJ. LD78beta, a non-allelic variant of human MIP-1alpha (LD78alpha), has enhanced receptor interactions and potent HIV suppressive activity. J Biol Chem. 1999;274:17478–17483. doi: 10.1074/jbc.274.25.17478. [DOI] [PubMed] [Google Scholar]
- 117.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–481. doi: 10.1016/s1359-6101(02)00045-x. [DOI] [PubMed] [Google Scholar]
- 118.Gornalusse G, Mummidi S, He W, Silvestri G, Bamshad M, Ahuja SK. CCL3L Copy number variation and the co-evolution of primate and viral genomes. PLoS Genet. 2009;5:e1000359. doi: 10.1371/journal.pgen.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Townson JR, Barcellos LF, Nibbs RJ. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur J Immunol. 2002;32:3016–3026. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 120.Shalekoff S, Meddows-Taylor S, Schramm DB, Donninger SL, Gray GE, Sherman GG, Coovadia AH, Kuhn L, Tiemessen CT. Host CCL3L1 gene copy number in relation to HIV-1-specific CD4+ and CD8+ T-cell responses and viral load in South African women. J Acquir Immune Defic Syndr. 2008;48:245–254. doi: 10.1097/QAI.0b013e31816fdc77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Degenhardt JD, de Candia P, Chabot A, Schwartz S, Henderson L, Ling B, Hunter M, Jiang Z, Palermo RE, Katze M, et al. Copy number variation of CCL3-like genes affects rate of progression to simian-AIDS in Rhesus Macaques (Macaca mulatta) PLoS Genet. 2009;5:e1000346. doi: 10.1371/journal.pgen.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rot A, Horuk R. Chapter 9 the duffy antigen receptor for chemokines. Methods Enzymol. 2009;461:191–206. doi: 10.1016/S0076-6879(09)05409-3. [DOI] [PubMed] [Google Scholar]
- 123.Mayr FB, Spiel AO, Leitner JM, Firbas C, Jilma-Stohlawetz P, Chang JY, Key NS, Jilma B. Racial differences in endotoxin-induced tissue factor-triggered coagulation. J Thromb Haemost. 2009;7:634–640. doi: 10.1111/j.1538-7836.2009.03307.x. [DOI] [PubMed] [Google Scholar]
- 124. Mayr FB, Spiel AO, Leitner JM, Firbas C, Kliegel T, Jilma-Stohlawetz P, Derendorf H, Jilma B. Duffy antigen modifies the chemokine response in human endotoxemia. Crit Care Med. 2008;36:159–165. doi: 10.1097/01.CCM.0000297875.55969.DB. **This study showed that DARC expression may modify the chemokine response in human endotoxemia. In a subsequent paper by the same group they showed that DARC expression may modify the coagulation response in human endotoxemia.
- 125.Haddy TB, Rana SR, Castro O. Benign ethnic neutropenia: what is a normal absolute neutrophil count? J Lab Clin Med. 1999;133:15–22. doi: 10.1053/lc.1999.v133.a94931. [DOI] [PubMed] [Google Scholar]
- 126.Reed WW, Diehl LF. Leukopenia, neutropenia, and reduced hemoglobin levels in healthy American blacks. Arch Intern Med. 1991;151:501–505. [PubMed] [Google Scholar]
- 127.Freedman DS, Gates L, Flanders WD, Van Assendelft OW, Barboriak JJ, Joesoef MR, Byers T. Black/white differences in leukocyte subpopulations in men. Int J Epidemiol. 1997;26:757–764. doi: 10.1093/ije/26.4.757. [DOI] [PubMed] [Google Scholar]
- 128.Grann VR, Bowman N, Joseph C, Wei Y, Horwitz MS, Jacobson JS, Santella RP, Hershman DL. Neutropenia in 6 ethnic groups from the Caribbean and the U.S. Cancer. 2008;113:854–860. doi: 10.1002/cncr.23614. [DOI] [PubMed] [Google Scholar]
- 129.Hershman D, Weinberg M, Rosner Z, Alexis K, Tiersten A, Grann VR, Troxel A, Neugut AI. Ethnic neutropenia and treatment delay in African American women undergoing chemotherapy for early-stage breast cancer. J Natl Cancer Inst. 2003;95:1545–1548. doi: 10.1093/jnci/djg073. [DOI] [PubMed] [Google Scholar]
- 130.Martineau AR, Newton SM, Wilkinson KA, Kampmann B, Hall BM, Nawroly N, Packe GE, Davidson RN, Griffiths CJ, Wilkinson RJ. Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Invest. 2007;117:1988–1994. doi: 10.1172/JCI31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Grann VR, Ziv E, Joseph CK, Neugut AI, Wei Y, Jacobson JS, Horwitz MS, Bowman N, Beckmann K, Hershman DL. Duffy (Fy), DARC, and neutropenia among women from the United States, Europe and the Caribbean. Br J Haematol. 2008;143:288–293. doi: 10.1111/j.1365-2141.2008.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Julg B, Reddy S, van der Stok M, Kulkarni S, Qi Y, Bass S, Gold B, Nalls MA, Nelson GW, Walker BD, et al. Lack of Duffy antigen receptor for chemokines: no influence on HIV disease progression in an African treatment-naive population. Cell Host Microbe. 2009;5:413–415. doi: 10.1016/j.chom.2009.04.009. author reply 418–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Walley NM, Julg B, Dickson SP, Fellay J, Ge D, Walker BD, Carrington M, Cohen MS, de Bakker PI, Goldstein DB, et al. The Duffy antigen receptor for chemokines null promoter variant does not influence HIV-1 acquisition or disease progression. Cell Host Microbe. 2009;5:408–410. doi: 10.1016/j.chom.2009.04.011. author reply 418-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Horne KC, Li X, Jacobson LP, Palella F, Jamieson BD, Margolick JB, Martinson J, Turkozu V, Visvanathan K, Woolley IJ. Duffy antigen polymorphisms do not alter progression of HIV in African Americans in the MACS cohort. Cell Host Microbe. 2009;5:415–417. doi: 10.1016/j.chom.2009.04.013. author reply 418–419. [DOI] [PubMed] [Google Scholar]
- 135.Winkler CA, An P, Johnson R, Nelson GW, Kirk G. Expression of Duffy antigen receptor for chemokines (DARC) has no effect on HIV-1 acquisition or progression to AIDS in African Americans. Cell Host Microbe. 2009;5:411–413. doi: 10.1016/j.chom.2009.04.010. author reply 418–419. [DOI] [PMC free article] [PubMed] [Google Scholar]